Abstract
Recognition of viral double-stranded RNA (dsRNA) activates interferon production and immune signaling in host cells. Crystal structures of ebolavirus VP35 show that it caps dsRNA ends to prevent sensing by pattern recognition receptors such as RIG-I. In contrast, structures of marburgvirus VP35 show that it primarily coats the dsRNA backbone. Here, we demonstrate that ebolavirus VP35 also coats the dsRNA backbone in solution, although binding to the dsRNA ends probably constitutes the initial binding event.
TEXT
Upon the recognition of double-stranded RNA (dsRNA), host pattern recognition receptors (PRRs) such as RIG-I, MDA-5, LGP2, and TLR-3 activate a signaling cascade that results in the generation of type I interferons (IFN) in infected and neighboring cells. In response, certain viruses, including the filoviruses, have evolved to encode specific proteins that counter innate immune detection by binding to and shielding their dsRNA from the PRRs (1–7). Indeed, the survivability of a filovirus infection is linked to the ability of the host to respond to pathogen-associated molecular patterns like dsRNA in order to mount a strong immune response in the first few days after exposure (8–10).
The two major genera within the family Filoviridae are Ebolavirus and Marburgvirus. The genus Ebolavirus contains five viral species (Ebola virus [EBOV], Reston virus [RESTV], Sudan virus, Bundibugyo virus, and Taï Forest virus). The genus Marburgvirus contains only one viral species, termed Marburg virus (MARV) and its variant Ravn (11).
In all filoviruses, the viral protein that inhibits the IFN response by binding dsRNA is termed VP35 (12), and this function is encoded in its C-terminal ∼130-amino-acid dsRNA-binding domain (RBD). The VP35 protein otherwise is multifunctional and plays essential roles in viral transcription, replication, and nucleocapsid assembly (13–16).
Crystal structures of the VP35 RBDs from EBOV, RESTV, and MARV have been determined alone and in complex with dsRNA. These structures have illustrated that the RBD contains two subdomains, of which one is α-helical and the other is primarily β-sheet (1, 4, 5, 17–19). The β-sheet subdomain contains a conserved basic patch that includes K271, R294, K298, R301, R311, and K328 (RESTV numbering). In structures of the complexes, we observe that VP35s from ebolaviruses bind the dsRNA ends, while VP35s from marburgvirus binds along dsRNA, primarily in the middle.
In VP35 structures from ebolaviruses (EBOV and RESTV), two copies of the VP35 RBDs form an asymmetric dimer to bind to each terminus of the dsRNA. One VP35 RBD, termed the “end-capping” molecule, stacks against the terminal bases of the dsRNA, while the paired VP35 RBD, termed the “backbone-binding” molecule, dimerizes with the end-capping molecule and contacts the neighboring sugar-phosphate backbone (4). In the EBOV structure, an 8-bp dsRNA was crystallized. The four bound VP35 RBDs cover the 8 bp completely (5). In the RESTV structure, an 18-bp dsRNA was crystallized. The same two VP35s bind at each end, but the longer expanse of dsRNA backbone in the center remains exposed; no VP35s are bound to it (4).
In contrast, two crystal structures of MARV VP35 RBD-dsRNA complexes demonstrate that the central dsRNA backbone is bound while the ends remain free (1, 20). Masking of the dsRNA backbone may be essential for immune suppression, as it has been demonstrated that PRRs, like RIG-I and MDA5, oligomerize and recognize the dsRNA backbone in a length-dependent manner (21–23). This apparent requirement for complete backbone coating, coupled with absence of coating in the RESTV structure, made us wonder if the ebolaviruses also coat the dsRNA backbone in solution or if ebolaviruses, indeed, only cap the dsRNA ends (4, 5).
To determine if ebolavirus VP35s also coat the backbone of dsRNA, we performed isothermal titration calorimetry (ITC) analysis of RESTV VP35 RBD binding to various dsRNA molecules. The VP35 proteins were expressed and purified in Escherichia coli, and RNA was obtained (IDT Technologies) and duplexed as previously described (1). VP35 RBD and dsRNA were dialyzed overnight against 4 liters of a mixture containing 10 mM Tris (pH 8.0), 200 mM NaCl, and 2 mM tris(2-carboxyethyl)phosphine (TCEP) buffer. ITC experiments were carried out in duplicate with a MicroCal iTC200 instrument (GE). In these experiments, RESTV VP35, at concentrations ranging from 1 to 2 mM, was loaded into the syringe, while dsRNA, at concentrations ranging from 15 to 40 μM, was loaded into the cell. Each experiment consisted of either 16 injections of 2.5 μl each or 32 injections of 1.25 μl each at intervals of 180 s at a reference power of 5 μcal. In each experiment, these injections were preceded by an injection of 0.5 μl, which was removed from the data analysis (standard procedure, as the dispensed volume of the first injection is often inaccurate). The integrated titration peaks were analyzed and fitted with Origin 7.0 software by using either a “one-set-of-sites” or a “two-sets-of-sites” binding model, as appropriate.
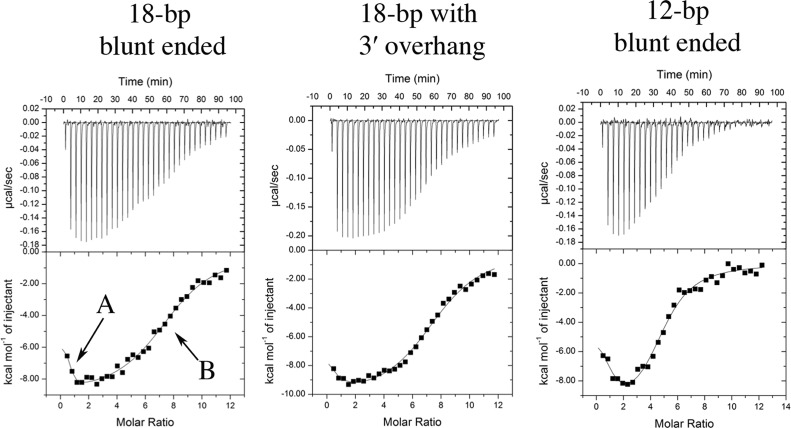
RESTV VP35 RBD binds blunt-ended dsRNA with a complex thermodynamic profile involving two separate events, A and B (Fig. 1). For RESTV, addition of a 3′ overhang to the dsRNA results in significant loss-of-binding event A (Fig. 1 and Table 1) and likely represents end capping. Indeed, crystal structures reveal that the 3′ end of the dsRNA butts into the dimer interface of VP35 from both RESTV and EBOV and that the addition of an overhang at the 3′ end would sterically prevent end capping. Binding event B likely represents backbone binding, as it is unaffected by a 3′ overhang and because more copies of VP35 contribute to binding event B when the dsRNA is longer. RESTV VP35 thus appears to be able to both end cap and backbone coat in solution, although in previous crystal structures, only end capping was observed with a stoichiometry of 4:1 (protein-to-dsRNA ratio) in the complex.
Fig 1.
Isotherms for binding of RESTV VP35 RBD molecules to 18-bp dsRNA with blunt ends, 18-bp dsRNA with a 3′ overhang, and 12-bp dsRNA with blunt ends. The top of each panel shows enthalpic heat released during titration, and the bottom panel shows integrated binding isotherms with the proposed fitted model of binding. Binding events A and B are labeled in the leftmost isotherm.
Table 1.
Thermodynamic parameters of binding determined by ITC for binding of RESTV VP35 to dsRNA molecules that vary in length and the presence of a 3′ overhang at each enda
| RESTV VP35 dsRNA | ΔGAb (kcal/mol) | nA | KDA (μM) | ΔGB (kcal/mol) | nB | KDB (μM) | nAB |
|---|---|---|---|---|---|---|---|
| 18 bp | −8.9 | 0.9 | 0.3 | −7.6 | 6.9 | 2.8 | 7–8 |
| 18 bp + 3′ overhang | Unmeasurablec | 0.2 | N/A | −7.8 | 7.8 | 2.0 | 7–8 |
| 12 bp | −9.6 | 1.2 | 0.1 | −7.5 | 3.6 | 3.0 | 4–5 |
All values represent averages of two independent measurements. Superscripts A and B refer to events A and B wherever applicable. n represents the number of molecules binding in that event.
The change in Gibbs free energy (ΔG) was determined as follows: ΔGbinding = RTlnKD (29).
There are too few measured points to confidently determine the ΔGbinding or the KD of the first binding event (stoichiometry of <0.2).
We crystallized RESTV VP35 RBD (residues 205 to 329) with a 12-bp palindromic dsRNA at a protein-to-dsRNA ratio of 1.2:1. RESTV VP35 RBD-dsRNA complexes crystallize over a period of 2 to 3 days in a mixture of 0.2 M Li2SO4, 0.1 M phosphate-citrate (pH 4.2), and 20% (wt/vol) polyethylene glycol 1000. Data were collected at beam line 5.0.3 of the Advanced Light Source (Berkeley, CA) and integrated with the program D*TREK, and the data collection statistics are summarized in Table 2 (24). The structure of the complex was determined by molecular replacement with MOLREP with the RESTV VP35 RBD (PDB 3KS4) as the starting model (25, 26). Model building was performed with COOT, and structure refinement was performed with PHENIX (27, 28). The final refinement statistics are given in Table 2.
Table 2.
Data collection and refinement statistics
| Parameter | Valuea |
|---|---|
| Wavelength (Å) | 0.9765 |
| Space group | P212121 |
| a (Å) | 55.09 |
| b (Å) | 72.87 |
| c (Å) | 175.55 |
| Resolution (Å) | 39.3–2.7 (2.84–2.70) |
| α = β = γ (°) | 90.0 |
| Total/unique reflections | 87,616/20,077 |
| Redundancy | 4.4 (4.5) |
| % Completion | 99.7 (100.0) |
| I/σ | 9.0 (1.9) |
| Rsym (%)b | 9.0 (57.4) |
| Rcrystc/Rfreed | 0.209/0.262 |
| Root mean square deviations | |
| Bonds (Å) | 0.007 |
| Angles | 1.06 |
| Dihedrals | 15.09 |
| Average B values (Å2) | |
| Main chain | 41.5 |
| Side chains and water | 48.9 |
| All atoms | 45.8 |
| Ramachandran plot | |
| Most favored region (%) | 94.6 |
| Additional favored region (%) | 5.4 |
Values in parentheses are for the highest-resolution shell.
Rsym = ΣΣi| Ii − <I> |/Σ<I>, where <I> is the mean intensity of the n reflections with intensities Ii and common indices h, k, and l.
R factor = Σhkl||Fobs|−k|Fcal|/Σhkl|Fobs|, where Fobs and Fcal are observed and calculated structure factors, respectively.
For Rfree, the sum is extended over a subset of reflections (∼5.0%) excluded from all stages of refinement. Of a total of 20,077 reflections, 1,016 were used for Rfree calculations.
The diffraction data were indexed in orthorhombic space group P212121 with two protein-RNA complexes in the asymmetric unit, each of which contains two RBDs bound to one molecule of dsRNA. Residues 208 to 329 in each copy of the RBD and base pairs 1 to 12 of one dsRNA and 1 to 9 of the other are visible in electron density maps. The N-terminal three residues of each RBD (205 to 207) and base pairs 10 to 12 of the second dsRNA are disordered.
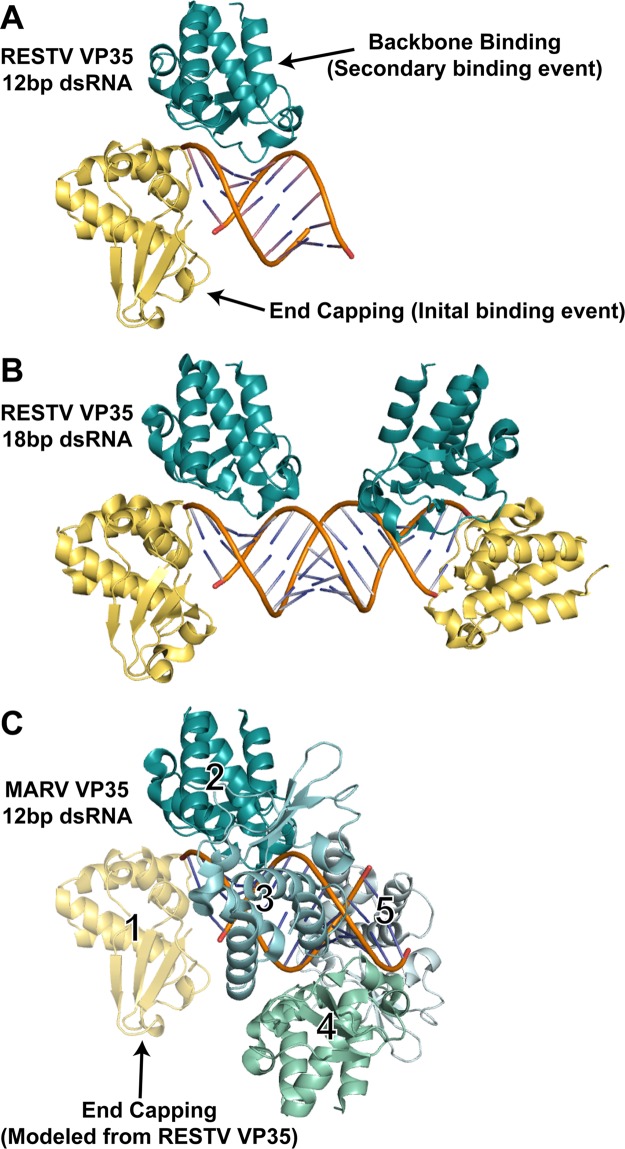
Interestingly, this crystal structure shows that the two RBDs together form one end-capping dimer while the other end of the dsRNA and the central backbone remain unbound (Fig. 2A). This crystal structure suggests that the initial event in dsRNA recognition by ebolavirus VP35 may be the formation of the dimeric end-capping/backbone binding pair on one end rather than, for example, the binding of one end-capping molecule to each end before the attachment of any backbone-binding copies adjacent to them. The previous RESTV complex may thus represent the second event in dsRNA recognition, in which a second end-capping/backbone binding pair caps the other end (4) (Fig. 2B).
Fig 2.
dsRNA recognition events observed in different filovirus VP35-dsRNA complexes. (A) Crystal structure of the RESTV VP35 RBD in complex with a 12-bp dsRNA. The end-capping monomer (number 1 in panel C) is shown in yellow, and the backbone-binding member of the dimer (number 2 in panel C) is shown in teal. (B) Previous crystal structure of the RESTV VP35 RBD in complex with an 18-bp dsRNA (PDB accession no. 3KS8). (C) Continuous-coating crystal structure of the MARV VP35 RBD determined in complex with a 12-bp dsRNA (PDB accession no. 4GHA). Only backbone binding interactions are observed in the crystal structure (numbered 2 to 5, different shades of blue and green). An end-capping interaction (yellow), modeled from RESTV structures by the superimposition of the dsRNA and backbone-binding copy number 2 indicates that no steric clash would occur if MARV also bound an RBD on the dsRNA end (1).
ITC performed on RESTV VP35 suggests that in solution, both end capping and backbone coating occur when sufficient protein copies are available (ITC events A and B, respectively) and that multiple copies of the VP35 RBD can exhibit the backbone-binding function. These results, together with the crystal structures, suggest that for RESTV, and perhaps for ebolaviruses in general, continued backbone coating occurs only after the capping of each available end. Continued coating by ebolavirus VP35s probably resembles the arrangement observed in the MARV structures. Indeed, modeling of the RESTV or EBOV end-capping dimers onto the MARV VP35 continuous-coating structure reveals no steric clashing of the end-capping pair with any continued-coating RBDs (Fig. 2C). The ITC-suggested ability of ebolavirus VP35s to also mask the backbone and the absence of likely structural clashes between the crystallized end-capping VP35s and potential backbone-coating VP35s suggest that ebolavirus VP35, like marburgvirus VP35, is also capable of preventing length-dependent dsRNA recognition by PRRs like RIG-I and MDA-5. The crystal structures, however, appear to reflect the type of interaction that is most stable or most preferred. Higher protein-to-dsRNA ratios, designed to illustrate complete coating and capping, have not yet yielded crystals.
VP35 from the ebolaviruses, but not marburgvirus, thus appears to prefer to end cap as an initial or preferred binding event. Initial end capping may afford ebolavirus VP35 stronger immune suppression than marburgvirus VP35. Indeed, the ebolavirus VP35s have an overall higher affinity for dsRNA and more efficient antagonism of the IFN response than does marburgvirus VP35 (1).
Protein structure accession number.
Coordinates and structure factors for the complex of RESTV VP35 with the 12-bp palindromic dsRNA have been deposited in the Protein Data Bank (PDB) under accession number 4LG2.
ACKNOWLEDGMENTS
We thank Michelle Zandonatti, Marnie L. Fusco, and Dafna M. Abelson for technical assistance and Ian MacRae and Jane Dyson for helpful discussions. We thank the staff of beam line 5.0.3 of the Advanced Light Source for assistance in data collection.
We acknowledge support from The Skaggs Institute for Chemical Biology (E.O.S. and I.A.W.), an Investigators in the Pathogenesis of Infectious Diseases award from the Burroughs Wellcome Fund (E.O.S.), NIAID grant R43 AI1088843 (E.O.S.), training grant 2T32AI007244 to the TSRI Department of Immunology and Microbial Science (Z.A.B.), a Canadian Institutes of Health Research fellowship (J.-P.J.), and NIH grant AI184817 (I.A.W.).
Footnotes
Published ahead of print 3 July 2013
This is report 22083 from The Scripps Research Institute.
REFERENCES
- 1. Bale S, Julien JP, Bornholdt ZA, Kimberlin CR, Halfmann P, Zandonatti MA, Kunert J, Kroon GJ, Kawaoka Y, Macrae IJ, Wilson IA, Saphire EO. 2012. Marburg virus VP35 can both fully coat the backbone and cap the ends of dsRNA for interferon antagonism. PLoS Pathog. 8:e1002916. 10.1371/journal.ppat.1002916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chao JA, Lee JH, Chapados BR, Debler EW, Schneemann A, Williamson JR. 2005. Dual modes of RNA-silencing suppression by Flock House virus protein B2. Nat. Struct. Mol. Biol. 12:952–957 [DOI] [PubMed] [Google Scholar]
- 3. Cheng A, Wong SM, Yuan YA. 2009. Structural basis for dsRNA recognition by NS1 protein of influenza A virus. Cell Res. 19:187–195 [DOI] [PubMed] [Google Scholar]
- 4. Kimberlin CR, Bornholdt ZA, Li S, Woods VL, Jr, MacRae IJ, Saphire EO. 2010. Ebolavirus VP35 uses a bimodal strategy to bind dsRNA for innate immune suppression. Proc. Natl. Acad. Sci. U. S. A. 107:314–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Leung DW, Prins KC, Borek DM, Farahbakhsh M, Tufariello JM, Ramanan P, Nix JC, Helgeson LA, Otwinowski Z, Honzatko RB, Basler CF, Amarasinghe GK. 2010. Structural basis for dsRNA recognition and interferon antagonism by Ebola VP35. Nat. Struct. Mol. Biol. 17:165–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Masliah G, Barraud P, Allain FH. 2013. RNA recognition by double-stranded RNA binding domains: a matter of shape and sequence. Cell. Mol. Life Sci. 70:1875–1895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vargason JM, Szittya G, Burgyan J, Hall TM. 2003. Size selective recognition of siRNA by an RNA silencing suppressor. Cell 115:799–811 [DOI] [PubMed] [Google Scholar]
- 8. Baize S, Leroy EM, Georges-Courbot MC, Capron M, Lansoud-Soukate J, Debre P, Fisher-Hoch SP, McCormick JB, Georges AJ. 1999. Defective humoral responses and extensive intravascular apoptosis are associated with fatal outcome in Ebola virus-infected patients. Nat. Med. 5:423–426 [DOI] [PubMed] [Google Scholar]
- 9. Leroy EM, Baize S, Volchkov VE, Fisher-Hoch SP, Georges-Courbot MC, Lansoud-Soukate J, Capron M, Debre P, McCormick JB, Georges AJ. 2000. Human asymptomatic Ebola infection and strong inflammatory response. Lancet 355:2210–2215 [DOI] [PubMed] [Google Scholar]
- 10. Mahanty S, Gupta M, Paragas J, Bray M, Ahmed R, Rollin PE. 2003. Protection from lethal infection is determined by innate immune responses in a mouse model of Ebola virus infection. Virology 312:415–424 [DOI] [PubMed] [Google Scholar]
- 11. Kuhn JH, Bao Y, Bavari S, Becker S, Bradfute S, Brister JR, Bukreyev AA, Chandran K, Davey RA, Dolnik O, Dye JM, Enterlein S, Hensley LE, Honko AN, Jahrling PB, Johnson KM, Kobinger G, Leroy EM, Lever MS, Muhlberger E, Netesov SV, Olinger GG, Palacios G, Patterson JL, Paweska JT, Pitt L, Radoshitzky SR, Saphire EO, Smither SJ, Swanepoel R, Towner JS, van der Groen G, Volchkov VE, Wahl-Jensen V, Warren TK, Weidmann M, Nichol ST. 2013. Virus nomenclature below the species level: a standardized nomenclature for natural variants of viruses assigned to the family Filoviridae. Arch. Virol. 158:301–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cárdenas WB, Loo YM, Gale M, Jr, Hartman AL, Kimberlin CR, Martinez-Sobrido L, Saphire EO, Basler CF. 2006. Ebola virus VP35 protein binds double-stranded RNA and inhibits alpha/beta interferon production induced by RIG-I signaling. J. Virol. 80:5168–5178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Basler CF, Mikulasova A, Martinez-Sobrido L, Paragas J, Muhlberger E, Bray M, Klenk HD, Palese P, Garcia-Sastre A. 2003. The Ebola virus VP35 protein inhibits activation of interferon regulatory factor 3. J. Virol. 77:7945–7956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Haasnoot J, de Vries W, Geutjes EJ, Prins M, de Haan P, Berkhout B. 2007. The Ebola virus VP35 protein is a suppressor of RNA silencing. PLoS Pathog. 3:e86. 10.1371/journal.ppat.0030086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Johnson RF, McCarthy SE, Godlewski PJ, Harty RN. 2006. Ebola virus VP35-VP40 interaction is sufficient for packaging 3E-5E minigenome RNA into virus-like particles. J. Virol. 80:5135–5144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Leung DW, Prins KC, Basler CF, Amarasinghe GK. 2010. Ebolavirus VP35 is a multifunctional virulence factor. Virulence 1:526–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bale S, Dias JM, Fusco ML, Hashiguchi T, Wong AC, Liu T, Keuhne AI, Li S, Woods VL, Jr, Chandran K, Dye JM, Saphire EO. 2012. Structural basis for differential neutralization of ebolaviruses. Viruses 4:447–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Leung DW, Ginder ND, Fulton DB, Nix J, Basler CF, Honzatko RB, Amarasinghe GK. 2009. Structure of the Ebola VP35 interferon inhibitory domain. Proc. Natl. Acad. Sci. U. S. A. 106:411–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Leung DW, Shabman RS, Farahbakhsh M, Prins KC, Borek DM, Wang T, Muhlberger E, Basler CF, Amarasinghe GK. 2010. Structural and functional characterization of Reston Ebola virus VP35 interferon inhibitory domain. J. Mol. Biol. 399:347–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ramanan P, Edwards MR, Shabman RS, Leung DW, Endlich-Frazier AC, Borek DM, Otwinowski Z, Liu G, Huh J, Basler CF, Amarasinghe GK. 2012. Structural basis for Marburg virus VP35-mediated immune evasion mechanisms. Proc. Natl. Acad. Sci. U. S. A. 109:20661–20666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Berke IC, Li Y, Modis Y. 2013. Structural basis of innate immune recognition of viral RNA. Cell. Microbiol. 15:386–394 [DOI] [PubMed] [Google Scholar]
- 22. Binder M, Eberle F, Seitz S, Mucke N, Huber CM, Kiani N, Kaderali L, Lohmann V, Dalpke A, Bartenschlager R. 2011. Molecular mechanism of signal perception and integration by the innate immune sensor retinoic acid-inducible gene-I (RIG-I). J. Biol. Chem. 286:27278–27287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Peisley A, Lin C, Wu B, Orme-Johnson M, Liu M, Walz T, Hur S. 2011. Cooperative assembly and dynamic disassembly of MDA5 filaments for viral dsRNA recognition. Proc. Natl. Acad. Sci. U. S. A. 108:21010–21015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pflugrath JW. 1999. The finer things in X-ray diffraction data collection. Acta Crystallogr. D Biol. Crystallogr. 55:1718–1725 [DOI] [PubMed] [Google Scholar]
- 25.Collaborative Computational Project, Number 4 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. 50(Pt 5):760–763 [DOI] [PubMed] [Google Scholar]
- 26. Vagin A, Teplyakov A. 1997. MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 30:1022–1025 [Google Scholar]
- 27. Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66:213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Emsley P, Cowtan K. 2004. COOT: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132 [DOI] [PubMed] [Google Scholar]
- 29. de Azevedo WF, Jr, Dias R. 2008. Experimental approaches to evaluate the thermodynamics of protein-drug interactions. Curr. Drug Targets 9:1071–1076 [DOI] [PubMed] [Google Scholar]