Abstract
Experimental vaccine antigens based upon the HIV-1 envelope glycoproteins (Env) have failed to induce neutralizing antibodies (NAbs) against the majority of circulating viral strains as a result of antibody evasion mechanisms, including amino acid variability and conformational instability. A potential vaccine design strategy is to stabilize Env, thereby focusing antibody responses on constitutively exposed, conserved surfaces, such as the CD4 binding site (CD4bs). Here, we show that a largely trimeric form of soluble Env can be stably cross-linked with glutaraldehyde (GLA) without global modification of antigenicity. Cross-linking largely conserved binding of all potent broadly neutralizing antibodies (bNAbs) tested, including CD4bs-specific VRC01 and HJ16, but reduced binding of several non- or weakly neutralizing antibodies and soluble CD4 (sCD4). Adjuvanted administration of cross-linked or unmodified gp140 to rabbits generated indistinguishable total gp140-specific serum IgG binding titers. However, sera from animals receiving cross-linked gp140 showed significantly increased CD4bs-specific antibody binding compared to animals receiving unmodified gp140. Moreover, peptide mapping of sera from animals receiving cross-linked gp140 revealed increased binding to gp120 C1 and V1V2 regions. Finally, neutralization titers were significantly elevated in sera from animals receiving cross-linked gp140 rather than unmodified gp140. We conclude that cross-linking favors antigen stability, imparts antigenic modifications that selectively refocus antibody specificity and improves induction of NAbs, and might be a useful strategy for future vaccine design.
INTRODUCTION
A prophylactic vaccine against HIV-1 infection is urgently required to reduce the magnitude of the global pandemic. However, the manifold mechanisms HIV-1 has evolved for evasion of the adaptive immune response, in particular, antigenic variation, make vaccine design a major challenge (1). Transfer of broadly neutralizing antibodies (bNAbs) into macaques completely protects them from infection by immunodeficiency viruses, providing a clear correlate of protection that informs antibody-based vaccine design (2). Moreover, results from the recent RV144 efficacy trial imply that antibodies, not cytotoxic T cells, were responsible for the modest protection (3–5). However, a major hurdle to overcome is the design of HIV-1 vaccine antigens able to elicit neutralizing antibodies by active immunization (6). Most studies to date have used soluble components of HIV-1 Env, either the receptor-binding monomeric gp120 subunit or trimeric gp140 complexes containing gp120 coupled to the membrane-external domain of the transmembrane subunit gp41 (7, 8). These studies have consistently revealed that although NAb responses can be elicited in various animal models and trimeric Env elicits improved NAb responses compared to those of monomeric gp120 (9, 10), virus neutralization remains strain specific and relatively weak. Strain specificity is a result of the bulk of NAb responses being made against variable surfaces on Env that the virus can modify without major fitness cost, a dominant HIV-1 antibody evasion strategy (2). An approach to overcome the dramatic variability of Env is to target conserved regions as defined by the isolation of bNAbs (1, 11–15). One such region is the CD4 binding site (CD4bs) on gp120 that contains a highly conserved site of vulnerability (16). bNAbs to the CD4bs isolated from HIV-1-infected individuals can neutralize from ∼30 to 90% of circulating viral strains (17–21), indicating that under the appropriate circumstances, conserved surfaces within the CD4bs are immunogenic in vivo.
A second antibody evasion mechanism relates to Env conformational instability. The Env trimer is held together by relatively labile noncovalent interactions that lead to shedding of gp120 producing nonfunctional gp41 “stumps” that will elicit a mostly nonneutralizing antibody response (18, 22–25). Moreover, probing of gp120, gp140, and cell surface-anchored Env with monoclonal antibodies (MAbs) (26, 27) and cryoelectron tomography of both in situ viral Env spikes and soluble gp140 trimers (28, 29) has revealed that Env can fluctuate between different conformational states, potentially modulating B cell recognition of particular conformational NAb epitopes, such as the CD4bs (30).
Therefore, we hypothesized that stabilization of Env tertiary and quaternary structure prevented trimer dissociation and allowed for a more predictable and robust B cell recognition of otherwise conformationally unstable neutralization epitopes. Aldehyde cross-linking of Env in situ on HIV-1-infected (31) or Env-expressing transfected cells (26, 27), or of gp120 and gp140 in solution (26, 27), has revealed selective preservation of MAb epitopes. Of particular interest, binding of CD4bs-specific bNAbs was maintained after mild aldehyde cross-linking (26, 27, 31), implying that components of the CD4bs are stabilized without deleterious modification of associated bNAb epitopes. To further investigate this, we treated predominantly trimeric HIV-1 gp140 derived from a clade C (CN54) HIV-1 isolate with glutaraldehyde (GLA) and characterized resulting effects upon antigenicity in vitro and immunogenicity in vivo. GLA reacts rapidly with amine groups, particularly lysines, at around neutral pH, and it is more efficient than other aldehydes in generating thermally and chemically stable cross-links (32). We confirm that GLA cross-linking preserves multiple bNAb epitopes, including those within the CD4bs, and demonstrate that immunization with GLA-cross-linked gp140 resulted in higher titers of CD4bs-specific antibodies, increased recognition of the C1 and V1V2 regions of gp120, and more potent neutralization. Thus, gp140 stabilization favorably modifies the responding B cell repertoire and may be a useful strategy in antigen design for eliciting NAbs to conserved surfaces on HIV-1.
MATERIALS AND METHODS
Proteins and antibodies.
Nontagged soluble CN54 gp140 was derived from HIV-197CN54 (GenBank accession number AF286226) (33, 34). The gp140 terminating at ALDSWKN and lacking the 2F5 and 4E10 epitopes was produced in CHO cells, purified on an affinity chromatography column using gp41-specific MAb 5F3 (35), and manufactured to good manufacturing practice specifications by Polymun Scientific, Vienna, Austria (36, 37). sCD4, CD4-IgG2, 17b, F105, 15e, b12, 2G12, PG9, and PGT antibodies were obtained from the IAVI Neutralizing Antibody Consortium. 5F3 was from Polymun Inc. (35), and F105 was from the National Institutes of Health (NIH) AIDS Research and Reference Reagent Program. HGP68, HR10, and HJ16 were a kind gift from D. Corti and A. Lanzavecchia. WT and D368R mutant gp120 were produced by transient transfection of HEK293T cells and purified as described previously (38). VRC01, RSC3, and ΔRSC3 were a kind gift from J. R. Mascola. The eOD construct employed here was a variant of the “eOD Base” reported in reference 39 that contained two additional mutations (Huang et al., unpublished data) and was produced in HEK293S cells as described previously (39, 40).
GLA cross-linking of gp140.
Minor contamination (<2%) of the CN54 gp140 by human 5F3 MAb carryover from the purification procedure was removed using protein A agarose (Pierce), and the buffer was exchanged to phosphate-buffered saline (PBS). Cross-linking was performed by addition of glutaraldehyde (GLA; Agar Scientific) to a final concentration of 7.5 mM for 5 min at room temperature (RT), after which the reaction was stopped by addition of 75 mM Tris buffer (pH 7.4) and then buffer exchanged into PBS. Successful cross-linking was tested by denaturing SDS-PAGE or native blue PAGE analysis on 3 to 8% Tris-glycine or 3 to 12% native page gels (Invitrogen), respectively, according to the manufacturer's instructions. Gels were stained using the SilverQuest staining kit (Invitrogen), and analysis was performed using the AIDA image analyzer v. 3.27.
ELISAs.
Enzyme-linked immunosorbent assays (ELISAs) were performed essentially as described previously (37). Briefly, antigens were coated onto ELISA plates (Greiner Bio-One) at 0.5 or 2.0 μg/ml for detection of polyclonal sera or MAbs, respectively. Wells were blocked with 2% bovine serum albumin (BSA) in PBS plus 0.05% Tween, followed by incubation with a dilution series of MAbs or sera for 2 h. Bound antibodies were labeled with horseradish peroxidase-coupled goat anti-human IgG (Sigma-Aldrich) or goat anti-rabbit IgG (Cayman Chemical Company-Aldrich) and detected using TMB-ultra substrate (Thermo Fisher Scientific), and absorption was read at 450 nm. Curves were fitted to the data using Prism V6 (GraphPad Software), and endpoint titers were defined as the concentration that corresponded to an optical density at 450 nm (OD450) of 0.01, which was always greater than 3 standard deviations (SD) above background levels. RSC3 ELISAs were run similarly, but serum dilutions were preincubated with 10 μg/ml ΔRSC3 before addition to RSC3-coated plates. GLA binding indices were calculated by dividing the log10-transformed titers against GLA antigen by the respective log10-transformed titers against unmodified antigen. Similarly, D368R sensitivity values were calculated using the following formula: binding sensitivity = 100% − log10 IC50(D368R)/log10 IC50(wt) × 100%. Cross-competition ELISAs were performed similarly to direct ELISAs, with the difference that competing MAb was added to the wells at the respective 50% binding concentration directly after the addition of 30-fold-diluted sera. Background-subtracted data were normalized to positive-control wells containing MAb without competing serum using the following formula: percent inhibition = 100% − ODsample/ODpositive control × 100%. Preimmune sera or serum pools were tested in parallel in all ELISAs and showed responses that were undetectable or insignificantly above plate background levels compared to postimmunization samples.
SPR.
Surface plasmon resonance (SPR) data were collected at 37°C on a Biacore 3000 instrument (GE Healthcare) at a flow rate of 50 μl/min in HBS-EP buffer (GE Healthcare). Rabbit anti-human IgG antibody (Jackson ImmunoResearch) was immobilized on a CM5 sensor chip (GE Healthcare) using standard amine coupling at pH 4.5. In each cycle, approximately 700 RUs of anti-HIV-1 MAbs or irrelevant human control MAbs were captured onto the flow cells, and 2-fold concentration series of WT or GLA-treated gp140, starting at 256 nM, were passed over the captured MAbs for 5 min, followed by a 5-min dissociation time. For kinetic analysis of MAbs with very slow dissociation times, an extended dissociation time of 30 min was used for the highest concentration. Between cycles, the sensor surface was regenerated by two 30-s injections of 10 mM glycine, pH 2.0. Data analysis was performed with the BIA evaluation software (version 4.0.1) by subtracting the data from the control antibody flow cell followed by subtraction of a blank injection cycle (double referencing). The binding index was calculated using the area under the curve (AUC) as a read-out, since this is sensitive to changes in on-rate, off-rate, and maximum binding and circumvents problems with fitting data from a 3:2 binding event. The formula used was AUC(GLA)/AUC(WT) × CL(WT)/CL(GLA), where AUC is the area under the curve and CL is the capture level of the MAb. Kinetic data were obtained by fitting data to a 1:1 Langmuir model.
CELISA.
A modified version of the cell-based ELISA (CELISA) developed by Haim et al. (65) was used for this study. Briefly, HEK293A cells were grown and transfected with UG37 gp160ΔCT or control DNA by calcium-phosphate precipitation in 15-cm dishes. Twenty-four h after transfection, they were lifted and reseeded into tissue culture-treated, clear-bottom, black-well 96-well plates. The next day, cells were left untreated or were cross-linked with 7.5 mM GLA in wash buffer (25 mM HEPES, 140 mM NaCl, 1.8 mM CaCl2, 1 mM MgCl2, pH 7.4) for 5 min, and the reaction was stopped by addition of an excess of Tris buffer. Cells were blocked and primary antibody was added in blocking buffer (10% fetal calf serum [FCS] in Dulbecco's modified Eagle medium [DMEM]). Bound MAb was labeled with horseradish peroxidase (HRP)-coupled goat anti-human IgG (Sigma) secondary antibody and detected using 35 μl Western Lightning reagents supplemented with 150 mM NaCl on a Spectromax X5. Data analysis was performed as described above for the standard ELISA.
Flow cytometry.
HEK293T cells were grown and transfected with ZM53 gp160ΔCT or control DNA by calcium-phosphate precipitation in 15-cm dishes. Forty-eight h after transfection, cells were lifted with 10 mM EDTA in PBS and incubated with serial dilutions of sera. Bound antibodies were detected using phycoerythrin (PE)-conjugated goat anti-rabbit IgG (Jackson ImmunoResearch) on a FACSCalibur flow cytometer. Data were analyzed as described for ELISAs using the mean fluorescence intensity (MFI) of cell populations gated in FlowJo (VX.0.6).
Animals and immunizations.
New Zealand White rabbits (n = 8 per group) were immunized intramuscularly with 20 μg unmodified (WT group) or GLA cross-linked (GLA group) CN54 gp140 per dose by Covance Inc. Immunizations were performed at weeks 0, 6, 12, and 22 using Carbopol 971p (1% wt/vol; 1 ml) for the prime and the first boost and polyethyleneimine (PEI; 25 kDa; branched; 1 mg/ml; 1 ml) for both successive boosts. Serum samples were collected at week 0 and 2 weeks after each immunization.
Peptide array serum specificity mapping.
Reactivity of sera (n = 4 per group, including 3 randomly chosen animals plus the highest responder from each group) to immobilized 15-mer peptides covering homologous CN54 gp140 were measured by Pepscan peptide binding analysis in an ELISA-based format in which the colored substrate was quantified with a charge-coupled device (CCD) camera and an image processing system. Statistical analysis was performed by averaging the responses to 8 regions for each animal and performing a one-way analysis of variance (ANOVA) with Bonferroni's posttest. Further epitope mapping of heterologous strains was performed essentially as previously described (42). Briefly, a peptide library of overlapping peptides (15-mers overlapping by 12), covering 7 full-length HIV-1 gp160 Env consensus sequences (clades A, B, C, and D, group M, CRF1, and CRF2) and 6 vaccine and laboratory strain gp120 sequences (A244_1, TH023_1, MN_B, 1086_C, TV1_C, and ZM651_C), was printed onto epoxy glass slides (provided by JPT Peptide Technologies GmbH [Germany]). All serum samples were diluted 1:250 and hybridized to the slides using a Tecan HS4000 hybridization workstation, followed by incubation with DyLight 649-conjugated goat anti-rabbit IgG (Jackson ImmunoResearch). Fluorescence intensity was measured using a GenePix 4300 scanner (Molecular Devices) and analyzed with GenePix software. Binding intensity of the postimmunization serum to each peptide was corrected with its own background value, which was defined as the median signal intensity of the prebleed serum for that peptide plus 3 times the standard errors among the 3 subarray replicates present on each slide.
Neutralization assays.
Neutralizing titers were measured in TZM-bl or A3R5 cells as described previously (43, 44). Briefly, pseudovirus (TZM-bl assay) or replication-competent luciferase reporter viruses (A3R5 assay) were incubated with serial dilutions of sera and added to their respective target cells. Luciferase expression was measured after 2 days (TZM-bl) or 4 days (A3R5), and IC50s were determined as the serum concentration that reduced the background-subtracted relative light units by 50% compared to virus-only control wells. For V3 inhibition assays, sera were preincubated with an excess of V3 peptide (TRPNNNTRKSIRIGPGQTFYATGDIIGNIRQAH), scrambled control, or PBS only. RSC3 inhibition assays were performed in TZM-bl cells as described above with HIV-1 clade C clone NL-LucR.T2A-MW965.ecto under single-cycle conditions. Serum dilutions were preincubated with 10 μg/ml circularized CN54 V3 loop (CTRPGNNTRKSIRIGPGQTFYATGDIIGDIRQAHCGC) plus either 20 μg/ml RSC or ΔRSC3. Statistical analysis of neutralizing titers was performed using a one-way ANOVA of log-transformed data with Bonferroni's posttest. Statistical analysis of overall differences was performed using a Mantel-Cox log-rank test on magnitude-breadth curves. Comparison of RSC3 neutralization curves was performed by an F-test of pooled curves from all animals of each group. Fold inhibition was calculated as IC50 (no V3 loop)/IC50 (V3 loop) for the V3 loop and IC50 (V3 + ΔRSC3)/IC50 (V3 + RSC3) for the CD4bs.
Computational modeling of modification sites.
Crystal structures of gp120 or fragments of gp120 in complex with CD4 (Protein Data Bank [PBD] entry 2NXY), 17b (2NXY), F105 (3HI1), b12 (2NY7), VRC01 (3NGB), or PG9 (3U4E) were analyzed and rendered using UCSF Chimera 1.6.2 (45). Lysine residues were identified based on alignments of the sequence of CN54 (GenBank accession number AF286226) or UG37 (AY494974) with the respective crystal structure gp120 sequence using the EMBL-EBI EMBOSS Needle online service (46).
RESULTS
Stability and antigenicity of GLA cross-linked gp140.
Soluble HIV-1 trimeric gp140 from the clade C CN54 isolate (gp140CN54) was cross-linked with 7.5 mM GLA for 5 min, free aldehydes were neutralized, and the resulting glycoprotein was desalted. Control gp140CN54 received the same treatment without addition of GLA. Unmodified control protein migrated predominantly as a mixture of trimeric ∼450-kDa complexes and higher-molecular-mass complexes by blue native PAGE (Fig. 1). GLA treatment yielded a band that ran slightly faster than the untreated material, potentially as a result of modest trimer compaction by cross-linking. Reducing SDS-PAGE disassembled the untreated gp140 into monomeric species, whereas GLA cross-linked material preserved trimers and smaller amounts of dimer and higher order oligomers.
Fig 1.
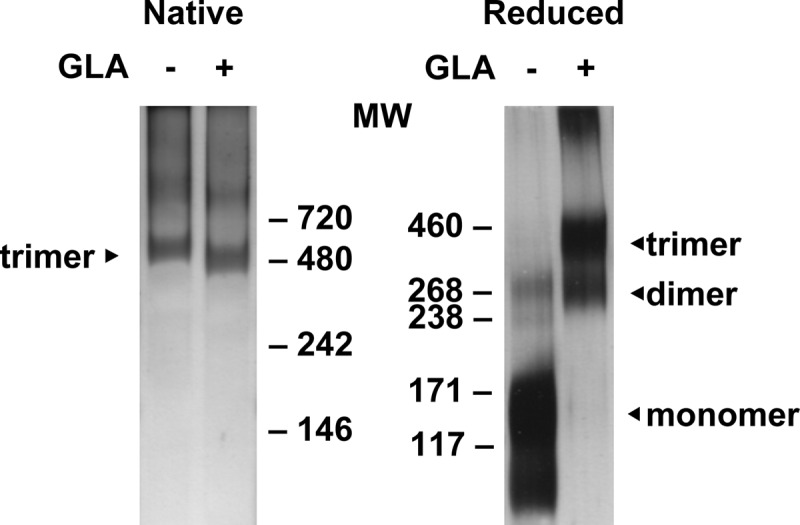
PAGE analysis of unmodified and GLA cross-linked CN54 gp140. HIV-1 gp140, untreated or treated with GLA, was migrated by native polyacrylamide gel electrophoresis (PAGE; left) or denaturing SDS-PAGE (right). Monomeric, dimeric, and trimeric bands are indicated according to molecular weight (MW; in thousands) analysis.
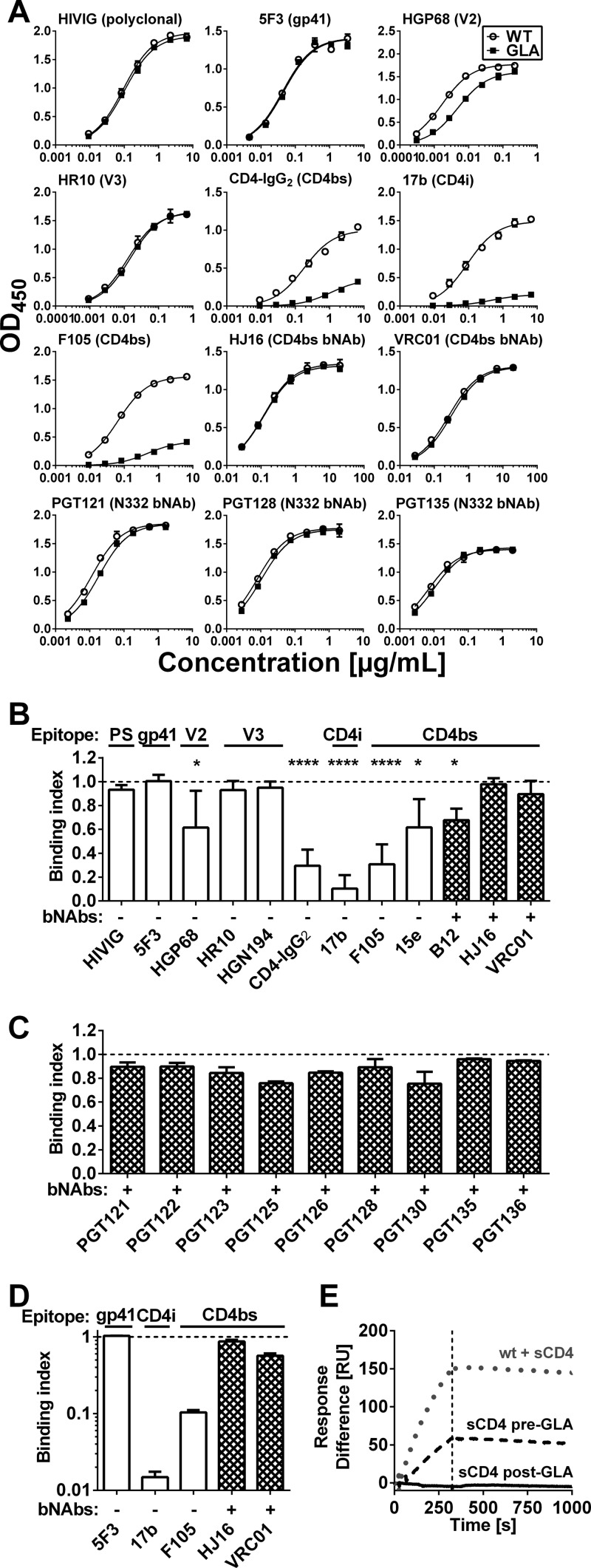
We analyzed the effect of GLA cross-linking on antibody recognition of gp140CN54 by ELISA. Polyclonal HIVIG binding was not significantly affected by cross-linking, indicating no major overall modification in antigenicity (Fig. 2). Binding of the nonneutralizing gp41-specific MAb 5F3 (35) was likewise unaltered. Binding of MAbs HGP68 (gp120 V2 loop) and HR10 (gp120 V3 loop) (18) was weakly affected (P = 0.02) or unaffected by GLA treatment, respectively. Binding of a second V3 loop-specific MAb (HGN194) was similarly unaffected by GLA treatment (Fig. 2B). In contrast, binding of a soluble sCD4-IgG fusion protein (CD4-IgG2) to gp140CN54 was highly significantly (P < 0.0001) reduced after cross-linking, consistent with the inhibition of structural rearrangements in gp120 required for high-affinity CD4 binding (30, 47). In line with this result, the 17b MAb that binds a CD4-induced (CD4i) epitope (48) showed dramatically reduced binding to GLA cross-linked gp140 (P < 0.0001). The binding of non- or weakly neutralizing CD4bs MAbs 15e and F105 was substantially reduced (P = 0.02 and P < 0.0001, respectively). This was expected, because, similar to CD4, these MAbs require conformational rearrangement of gp120 for tight binding (30) and might also be subject to steric inhibition in a rigid trimer due to a predicted restricted angle of approach (49). This may also explain the partial reduction in IgG1b12 binding after GLA treatment (Fig. 2B). In contrast, binding of the more broadly neutralizing CD4bs MAbs VRC01 and HJ16 was unaffected by gp140 cross-linking, confirming a predicted lack of requirement for gp120 conformational change during binding. Several bNAbs (PGT121, PGT122, PGT123, PGT128, PGT135, and 2G12) recognize an epitope centered on an N-linked glycan at position N332. None of these MAbs showed any significant difference in binding to GLA-treated and untreated gp140 (Fig. 2C). We further investigated the effects of cross-linking by SPR. Using this more sensitive technique, a modest reduction in binding of CD4bs bNAb VRC01 was detected, although the effect was far less pronounced than that with the nonneutralizing CD4bs F105 (Fig. 2D; also see Fig. S1 in the supplemental material). Furthermore, binding of the CD4i antibody 17b was undetectable when sCD4 and 17b were added post-GLA treatment (Fig. 2D and E; also see Fig. S1). However, if an excess of sCD4 was added to gp140 prior to cross-linking, detectable 17b binding was observed, indicating that cross-linking interferes with structural rearrangements required for the exposure of the CD4i epitope (Fig. 2E; also see Fig. S1).
Fig 2.
Antigenicity of GLA cross-linked gp140. Antibody binding to unmodified (WT) or cross-linked (GLA) gp140 from strain CN54 (clade C) was analyzed by ELISA (A to C) and SPR (D and E). (A) Representative titration series against CN54 gp140 using MAbs covering tested epitopes (indicated in parentheses) show selective decreases of binding of nonneutralizing CD4bs antibody and CD4i antibodies. Curves indicate results of the nonlinear fit. (B and C) The binding index (cross-linked/unmodified) for CN54 gp140 is shown for all antibodies tested, with glycan-dependent antibodies presented in panel C. Values lower than 1.0 indicate loss of binding. Error bars indicate SD from at least five (B) or two (C) independent experiments. (D) The binding index from SPR measurements of CN54 gp140 is shown, with error bars indicating SD of technical repeats. (E) SPR traces are shown for unmodified or cross-linked CN54 gp140 binding to the CD4i MAb 17b. sCD4 was added either before or after cross-linking as indicated. The dotted vertical line indicates the end of the injection. PS, polyclonal serum; *, P < 0.05; ****, P < 0.0001 according to a Kruskal-Wallis test.
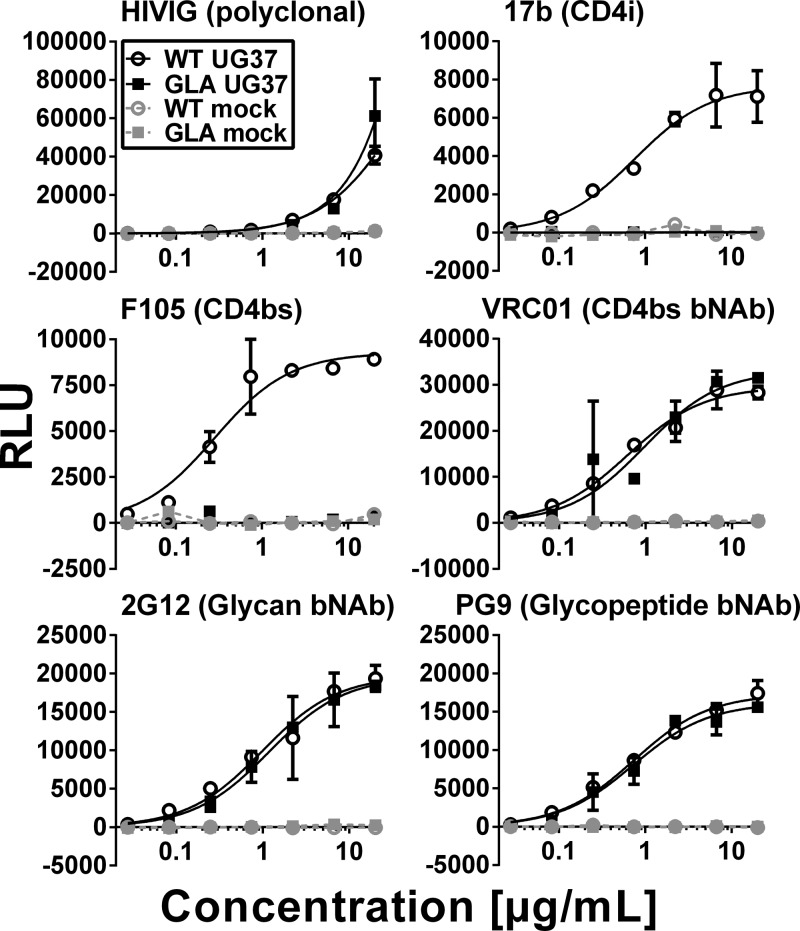
To determine whether GLA cross-linking modified antigenicity of membrane-associated gp160 in a manner similar to that of soluble gp140, we tested a panel of MAbs for binding using a cell-based ELISA (CELISA) using gp160-expressing HEK293 cells. The same trends were observed, in that HIVIG and VRC01 bound equivalently to the cross-linked and unmodified gp160, whereas the binding of the weak CD4bs NAb F105 and the CD4i antibody 17b were abolished (Fig. 3). Finally, binding of the bNAbs 2G12, which has a glycan epitope, and PG9, whose glycopeptide epitope is dependent upon conservation of the quaternary Env structure, was equivalent in GLA-treated and untreated cells, showing complete preservation of these epitopes (Fig. 3). Lysine residues, which are potential GLA modification sites, were present in several of the analyzed MAb epitopes (see Fig. S2 in the supplemental material). Interestingly, the bNAbs VRC01 and PG9 both directly interacted with two lysine residues on Env, yet neither showed substantially reduced binding to cross-linked protein, suggesting minor or negligible steric effects of the modifications. The epitopes of several other ligands for which crystal structures are available (CD4, 17b, F105, and b12) also contained lysines, the modification of which by GLA may have influenced binding. Thus, in summary, GLA cross-linking selectively modifies gp140 antigenicity, largely preserving all bNAb epitopes tested but reducing binding of several weakly or nonneutralizing MAbs by conformational stabilization, lysine modification within the epitope, or a combination of both.
Fig 3.
Antigenicity of GLA cross-linked gp160. HEK293-A cells were transiently transfected with UG37 gp160ΔCT (clade A) or mock transfected as indicated. Antibody binding to unmodified (WT) or cross-linked (GLA) gp160 was analyzed by CELISA. Representative titration series of MAbs (epitopes are indicated in parentheses) show maintenance (HIVIG, VRC01, 2G12, and PG9) or selective decreases (17b and F105) of binding. Lines indicate results of the nonlinear fit where possible.
Immunogenicity of GLA cross-linked gp140.
To investigate how changes in antigenicity translate into immunogenicity in vivo, New Zealand White rabbits were immunized with GLA cross-linked or unmodified gp140CN54. Immunizations were carried out at weeks 0, 6, 12, and 22 in carbopol (50) and PEI (51) adjuvants, a combination of adjuvants that induces increased antibody titer and avidity compared to either adjuvant alone (unpublished results). Serum samples were collected before the start of the immunization regimen and 2 weeks after each immunization. Indistinguishable binding antibody responses were induced by both antigens at all time points regardless of whether sera were titrated onto untreated (Fig. 4A) or GLA-treated gp140 (data not shown). In contrast, significantly higher (P < 0.01) titers were obtained from sera from the GLA group against cell surface-expressed Env (Fig. 4B) than for the WT group, implying better recognition of native Env species. To determine whether antibodies were elicited against the GLA modifications themselves, binding of sera to GLA cross-linked and unmodified irrelevant antigen (hen egg lysozyme [HEL]) was tested. The GLA binding index was calculated as the ratio of the log10 endpoint titer against GLA cross-linked HEL divided by the log10 endpoint titer against unmodified HEL (Fig. 4C). There was no difference between animals immunized with unmodified or GLA cross-linked gp140 (P = 0.91). Thus, the immunization regimen induced strong binding antibody responses to gp140 and membrane-bound gp160 but did not induce GLA cross-reactive antibodies.
Fig 4.

Immunogenicity of GLA cross-linked CN54 gp140. (A) CN54 gp140-specific total IgG endpoint titers of sera from New Zealand White rabbits (n = 8 per group) immunized with unmodified (WT) or GLA cross-linked (GLA) CN54 gp140 showed no significant differences at any time point (one-way ANOVA of log-transformed data with Bonferroni's posttest). Inject represents time points of immunizations. (B) Binding of week 24 sera to cell surface-expressed heterologous ZM53 Env was analyzed by fluorescence-activated cell sorting (FACS). **, P < 0.01 according to a t test of log-transformed data. (C) Binding of sera to a GLA cross-linked or unmodified irrelevant protein (HEL) was analyzed with week 24 sera, and results are expressed as an index of binding to GLA-treated/untreated HEL. Student's t test was performed for statistical analysis.
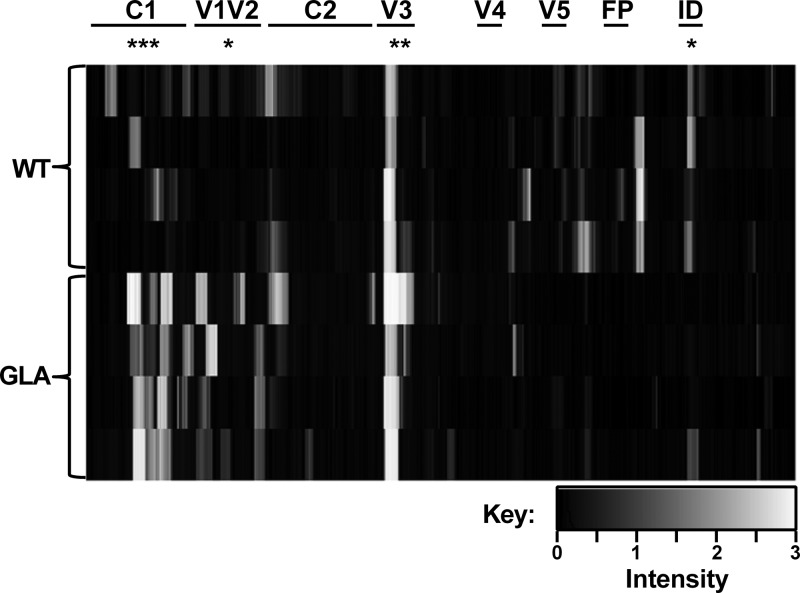
To determine whether GLA cross-linking modified B cell gp140 epitope recognition and antibody production in vivo, we first carried out an analysis of linear sequences recognized by sera from animals immunized with WT or GLA-treated antigen. For this, we used a printed overlapping 15-mer peptide Pepscan array spanning the gp140CN54 sequence. This assay revealed striking differences in the epitopes recognized by sera from GLA or WT gp140-immunized rabbits (Fig. 5; also see Fig. S3 in the supplemental material). Sera from the GLA-gp140 group bound significantly more strongly to peptides from the C1 and V1V2 regions than sera from untreated (WT) gp140-immunized animals (P = 0.0005 and P = 0.0293, respectively). All animals showed strong responses to the V3 loop, although the responses were higher in the GLA group (P = 0.0045). Animals from the WT gp140-immunized group showed strong binding to epitopes in the C terminus of gp120 and gp41, with responses to the immunodominant region being significantly higher than those in the GLA group (P = 0.0169). Similar results were obtained when sera were mapped using peptide libraries derived from a panel of gp120s from diverse clades and origins (see Fig. S4). Thus, immunization with cross-linked gp140 refocused antibody responses toward N-terminal regions of gp120, including the V1V2 loop, and reduced responses to more C-terminal regions and elements of gp41.
Fig 5.
Peptide scanning of week 24 serum responses. Binding of sera to 15-mer peptides (n = 4 per group) was tested by peptide array. Binding intensity is plotted in arbitrary units. Regions of gp140 are marked at the top. Statistical analysis for individual regions was performed by one-way ANOVA with Bonferroni's posttest. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
GLA cross-linked gp140 induces increased CD4bs antibody responses.
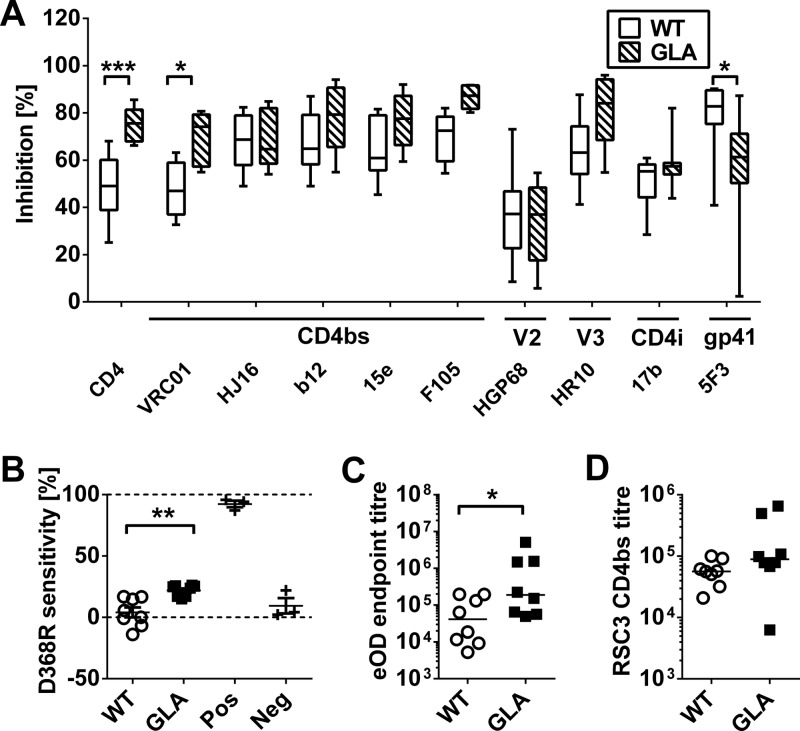
Since most bNAb epitopes are conformational in nature and not represented by linear peptide epitopes, we performed epitope mapping by cross-competition using a panel of well-characterized MAbs. Strong competitive responses were induced by WT gp140 against CD4bs, CD4i epitope, V3 loop, and gp41-specific MAbs and weaker responses against a V2 MAb (Fig. 6A). Significant differences in cross-competition were detected with sera from animals immunized with GLA-treated gp140: competition was increased for sCD4 (P = 0.0006) and the broadly neutralizing CD4bs MAb VRC01 (P = 0.012). Interestingly, cross-competition for the CD4bs MAbs IgG1b12 and HJ16 did not differ significantly between groups, demonstrating selectivity within the CD4bs epitope cluster. Of note, cross-competition was significantly reduced for the 5F3 epitope, suggesting a reduced immunogenicity of this epitope. The increased competition for CD4bs MAbs in sera from GLA-gp140-immunized animals suggested that the modified antigenicity of GLA-treated gp140 was influencing the immunogenicity of this region. To confirm that increased levels of CD4bs antibodies were elicited in animals receiving GLA-treated gp140, we performed three further assays. First, we measured the sensitivity of binding to the D368R mutation introduced into gp120, which abolishes binding of most CD4bs-specific MAbs. Loss of binding to D368R gp120 compared to WT protein was significantly (P = 0.0011) stronger in animals receiving GLA cross-linked gp140 than in those receiving unmodified material (Fig. 6B). Second, binding to an engineered gp120 outer domain construct (39, 40), which lacks variable loops and almost exclusively displays elements of the CD4bs, was significantly (P = 0.021) increased in the GLA group (Fig. 6C). Three animals in the GLA group showed extremely high titers of binding (>106) compared to the mean of the non-GLA group (which was ∼4 × 104). Finally, binding to the RSC3 probe (20) was analyzed, which displays the HIV-1 CD4bs in the context of a background engineered to contain non-HIV-1 residues in order to selectively assess HIV-1 CD4bs antibody responses. Binding to this probe in the presence of an excess of RSC3 Δ371I/P363N protein (20), which has two mutations in the CD4bs that abolish CD4bs MAb binding and is used to compete out any non-CD4bs binding to the probe, was increased 3-fold in the GLA group, although this was not statistically significant (Fig. 6D). Taken together, these findings strongly support the increased elicitation of CD4bs-specific antibodies in animals immunized with GLA-treated gp140.
Fig 6.
Conformational epitope analysis of week 24 serum responses. (A) Cross-competition ELISA of sera against a panel of gp140-specific human MAbs. (B) D368R sensitivity ELISA shows amount of CD4bs antibodies. Pos, panel of CD4bs MAbs; Neg, panel of non-CD4bs MAbs. (C) Endpoint titers against an engineered outer domain of ENV. (D) Binding of sera to the CD4 binding site probe RSC3 in the presence of an excess of competing RSC3 Δ371I/P363N CD4bs knockout protein. A one-way ANOVA with Bonferroni's posttest (A), Student's t test (B), and Student's t test of log-transformed data (C and D) were used for statistical analysis. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
GLA cross-linking induces more potent neutralizing responses than unmodified protein.
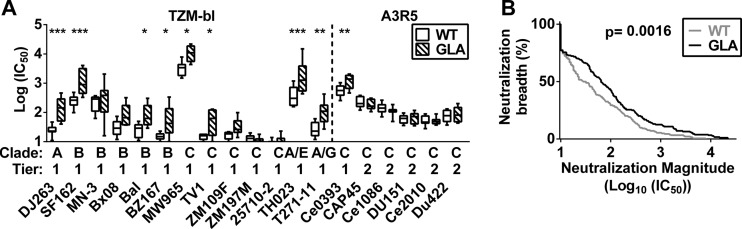
To assess whether these changes in epitope recognition modified serum neutralization activity, we performed TZM-bl-based neutralization assays (43) using week 24 sera (2 weeks after final boost). All animals showed neutralization of all tier 1 pseudoviruses expressing Envs from 5 different clades (Fig. 7A; also see Table S1 in the supplemental material). Furthermore, sera from the GLA-gp140-immunized group exhibited significantly increased (P = 0.0002) neutralization potency against the tier 1 viruses. This increased neutralizing potency could not be detected in the sera at the earlier time point at week 14 (see Table S1). No neutralization of tier 2 viruses could be detected in the TZM-bl assay for either time point (see Table S1); thus, we used the more sensitive A3R5 assay (44) to test a panel of clade C viruses, including 4 tier 2 viruses (Fig. 6; also see Table S2). All animals showed neutralization against all tier 2 viruses tested (Fig. 7A; also see Table S2). Compared to the TZM-bl assay, differences were smaller in this assay, and only one virus (Ce0393, tier 1, clade C) showed significantly (multiple-comparison adjusted P of 0.0042) enhanced neutralization by the GLA-gp140 group sera. Pooling all data together to carry out a magnitude-breadth plot (53), we observed a significant (P = 0.0016) (Fig. 7B) increase in neutralization in sera from the GLA group compared to the WT group. Thus, immunization with GLA cross-linked gp140 led to the induction of higher titers of neutralizing antibodies than unmodified protein.
Fig 7.
Neutralization assays. (A) Neutralization of week 24 sera from animals immunized with unmodified (WT) or GLA cross-linked (GLA) gp140 was tested in the TZM-bl assay against tier 1 viruses (left) and in the more sensitive A3R5 assay against tier 2 viruses (right). (B) Magnitude-breadth curves are shown for pooled data from all animal sera from both assays. A one-way ANOVA of log-transformed data with Bonferroni's posttest (A) and a Mantel-Cox log-rank test (B) were used for statistical analysis. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Since tier 1 virus serum neutralizing activity in immunized animals is frequently associated with V3 loop recognition, we analyzed the ability of saturating concentrations of a V3 loop peptide to reduce neutralization against a tier 1 clade C virus (MW965). Neutralization titers were reduced by about 10-fold for sera from both WT and GLA-treated gp140-immunized animals, showing a strong component of V3-mediated neutralization (Fig. 8A). However, strong neutralization remained in most sera, suggesting the presence of other species of NAb. Moreover, the significantly greater neutralization activity of the sera from GLA-treated gp140-immunized animals was maintained, implying that the increased neutralizing activity in these sera was mostly non-V3 specific. To test whether the observed increase in CD4bs-specific antibodies was responsible for the increased neutralization in the GLA group, we performed further neutralization assays in which sera were preabsorbed with either RSC3 or ΔRSC3. To increase the sensitivity, we performed this assay in the presence of competing V3-peptide in all tests. Curve comparison showed that the GLA group displayed significantly (P = 0.022) higher neutralization activity when absorbed with the negative control ΔRSC3 (Fig. 8B, left). However, the difference in neutralization disappeared when the CD4bs probe RSC3 was used (Fig. 8B, right). Comparison of the loss of binding between ΔRSC3 and RSC3 showed a significant (P = 0.0076) difference between the WT and GLA groups (Fig. 8C). The total effect of the CD4bs neutralization was much less pronounced than the effect of the V3 loop (Fig. 8C), which might be partially due to the tier 1 viruses used for this assay, which are known to be very sensitive to V3-mediated neutralization. Taken together, these data show that immunization with GLA cross-linked gp140 leads to the induction of higher titers of neutralizing activity than unmodified protein, which is at least partially due to CD4bs-mediated neutralization.
Fig 8.
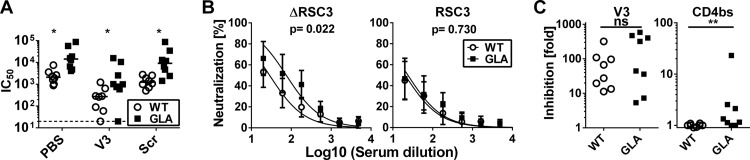
Characterization of neutralizing responses. (A) Neutralization of HIV-1 strain MW965 by week 24 sera in the presence of V3 peptide (V3), scrambled control (Scr), or vehicle alone (PBS) in TZM-bl cells. (B) Neutralization curves of MW965 by week 24 in the presence of V3 peptide plus either the CD4bs probe RSC3 or the corresponding negative control, ΔRSC3. Curves show averages of all animals from the two groups. (C) Loss of neutralization upon addition of inhibitor is shown as the ratio IC50 (no V3 loop)/IC50 (V3 loop) (left) or IC50 (V3 + ΔRSC3)/IC50 (V3 + RSC3) (right). A one-way ANOVA of log-transformed data with Bonferroni's posttest (A), a curve-comparison F test (B), and a Mann-Whitney test (C) were used for statistical analysis. *, P < 0.05; **, P < 0.01.
DISCUSSION
GLA treatment results in covalent cross-linking of proteins, allowing their conformational stabilization. GLA-modified gp140 largely conserved all bNAb epitopes tested but selectively reduced binding of weakly neutralizing CD4bs MAbs and some other nonneutralizing MAbs. These findings are in line with previous studies that suggested that structural flexibility within CD4bs epitopes is a mechanism by which HIV-1 circumvents the induction and high-affinity binding of CD4bs-directed antibodies, termed “conformational masking” (30). It has further been suggested that weakly neutralizing CD4bs MAbs, such as F105, only recognize one specific gp140 conformation, whereas bNAbs recognize many or all conformations (26, 27). Our data are consistent with this proposal and extend it to include representatives of known bNAb epitope clusters within gp120, including the quaternary epitope-specific bNAbs PG9 and PG16. On the basis of these data, we hypothesize that cross-linking lowers the effective concentration of conformations recognized by nonneutralizing MAbs and redirects immune responses toward features recognized by bNAbs that are conserved between different gp140 conformations. The loss of binding of specific MAbs was unlikely to be due to direct modification of their epitopes by GLA adduction to lysines, since bNAbs VRC01 and PG9 directly interact with two lysine residues each and still retained strong to complete binding. However, we cannot rule out a substantial effect of lysine modification on some of the weakly or nonneutralizing MAbs that lost binding after GLA treatment. A further useful effect of cross-linking the antigen may be that of increasing in vivo lifetime after administration, particularly of conformational epitopes such as those contained within the CD4bs that might otherwise be rapidly lost upon protease cleavage or other degradation of the antigen.
Consistent with the antigenicity results, immunization with cross-linked gp140 resulted in substantially different specificities of serum antibody production compared to unmodified gp140, as determined by peptide mapping and binding of sera to the CD4bs. GLA treatment reduced the immunogenicity of C-terminal regions, including the gp41 immunodominant domain, which are considered to elicit nonneutralizing antibodies and are of reduced interest to vaccine design, but the treatment elicited increased antibody responses to the C1 region and the V1V2 loop. Binding of IgG to the V1V2 loop has recently been shown to inversely correlate with the rate of infection in the RV144 trial (3, 5, 54, 55). Although the causality of the relationship between binding V1V2 antibodies and protection from HIV-1 infection is currently unclear, induction of this type of antibody response might be a desirable property of an HIV-1 vaccine.
Higher titers of CD4bs-specific antibodies were detected in the GLA-gp140-immunized group. Interestingly, the increased immunogenicity of the CD4bs in vivo was achieved despite the reduction in binding of weakly neutralizing CD4bs MAbs in vitro. Since elements of the CD4bs, particularly those associated with the gp120 outer domain (16, 56–58), are well conserved and the target of broadly and potently NAb, such as VRC01 (20, 21), rendering this site more immunogenic is considered to be a beneficial property of an HIV-1 vaccine (6, 38). The selectively increased cross-competition of sCD4 and VRC01 but not HJ16, IgG1b12, 15e, and F105 by sera from the GLA group suggest that a very specific subset of CD4bs antibodies have been induced that react predominantly with the outer domain of CD4. This hypothesis is supported by the outer domain binding observed in the GLA group, particularly in some animals that showed 30- to 100-fold increases over the non-GLA group (Fig. 6C). The increase in immunogenicity of the CD4bs in the cross-linked material could be further increased by additional boosting with the homologous material or may be focused using boosting with outer domain constructs, such as that used here as a probe. The reduction in binding of sCD4 induced by GLA treatment might have additional benefits for immunization in humans, since Env binding to cell surface CD4 leads to high titers of CD4i-specific antibodies (59) that are weakly or nonneutralizing (60–62). The neutralizing responses induced by the cross-linked material, although significantly increased in breadth and potency compared to that of the WT material, nevertheless were weaker than desired for vaccine design purposes. Moreover, differences were not observed between sera from animals immunized with GLA-treated and WT gp140 in pseudoviruses carrying tier 2 Envs, suggesting that these Envs have more stringent requirements for CD4bs Nab binding than tier 1 Envs. There are several possible reasons for this limited CD4bs-directed neutralizing response, including insufficient affinity maturation to achieve adequate occupancy at the maximum serum concentration used and an angle of approach to the trimer on the clones of pseudovirus used for neutralization that is suboptimal for tight binding (49). We propose that these limitations can be overcome by modified immunization protocols that allow greater affinity maturation and boosting with other Env-based constructs to focus antibody responses that have the appropriate angle of approach to the trimer.
We have used a recombinant gp140 that, although predominantly trimeric, contained some dimeric and higher order oligomeric species that were stabilized by cross-linking. Although the higher order species could be detected in both unmodified and cross-linked material in a native page, we cannot rule out that covalent stabilization of the multimers influences immunogenicity. Our GLA-treated material will contain a mixture of Env conformers (27), some of which may have antigenicity and immunogenicity profiles more suitable than others for induction of NAb. Finally, the gp140 used here shows a lack of binding to bNAbs with epitopes dependent upon Env quaternary structure, such as PG9 and PG16. This is typical for soluble Env constructs (63) and suggests a degree of misfolding. Thus, several optimization steps are immediately obvious that might improve the antigenicity of cross-linked gp140, particularly purifying out the trimeric species, potentially leading to better presentation of bNAb epitopes and enhanced immunogenicity and induction of NAbs.
Sera from animals that were immunized with GLA-treated gp140 elicited higher tier 1 virus neutralization titers in the TZM-bl assay than the unmodified gp120-immunized animals. The specificity of this increased activity does not appear to be V3 specific but seems to be at least partially due to CD4bs-mediated neutralization. Robust neutralization of tier 1 viruses after rabbit immunization with Env-based antigens is not in itself novel. However, the fact that higher titers of CD4bs antibodies and neutralization were elicited using an antigen that had undergone a simple modification with a chemical species (aldehyde) used to treat other vaccine antigens with extensive and safe use in humans (64) suggests that this approach deserves further investigation.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the Bill and Melinda Gates Foundation to the Weiss Neutralizing Antibody Vaccine Design Consortium, the International AIDS Vaccine Initiative (IAVI) Neutralizing Antibody Consortium, and Dormeur Investment Services Ltd. Q.J.S. is a Jenner Vaccine Institute Investigator and a James Martin Senior Fellow.
Footnotes
Published ahead of print 10 July 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.01161-13.
REFERENCES
- 1. Burton DR, Poignard P, Stanfield RL, Wilson IA. 2012. Broadly neutralizing antibodies present new prospects to counter highly antigenically diverse viruses. Science 337:183–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mascola JR, Montefiori DC. 2010. The role of antibodies in HIV vaccines. Annu. Rev. Immunol. 28:413–444 [DOI] [PubMed] [Google Scholar]
- 3. Haynes BF, Gilbert PB, McElrath MJ, Zolla-Pazner S, Tomaras GD, Alam SM, Evans DT, Montefiori DC, Karnasuta C, Sutthent R, Liao HX, DeVico AL, Lewis GK, Williams C, Pinter A, Fong Y, Janes H, DeCamp A, Huang Y, Rao M, Billings E, Karasavvas N, Robb ML, Ngauy V, de Souza MS, Paris R, Ferrari G, Bailer RT, Soderberg KA, Andrews C, Berman PW, Frahm N, De Rosa SC, Alpert MD, Yates NL, Shen X, Koup RA, Pitisuttithum P, Kaewkungwal J, Nitayaphan S, Rerks-Ngarm S, Michael NL, Kim JH. 2012. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N. Engl. J. Med. 366:1275–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, Premsri N, Namwat C, de Souza M, Adams E, Benenson M, Gurunathan S, Tartaglia J, McNeil JG, Francis DP, Stablein D, Birx DL, Chunsuttiwat S, Khamboonruang C, Thongcharoen P, Robb ML, Michael NL, Kunasol P, Kim JH. 2009. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N. Engl. J. Med. 361:2209–2220 [DOI] [PubMed] [Google Scholar]
- 5. Rolland M, Edlefsen PT, Larsen BB, Tovanabutra S, Sanders-Buell E, Hertz T, deCamp AC, Carrico C, Menis S, Magaret CA, Ahmed H, Juraska M, Chen L, Konopa P, Nariya S, Stoddard JN, Wong K, Zhao H, Deng W, Maust BS, Bose M, Howell S, Bates A, Lazzaro M, O'Sullivan A, Lei E, Bradfield A, Ibitamuno G, Assawadarachai V, O'Connell RJ, de Souza MS, Nitayaphan S, Rerks-Ngarm S, Robb ML, McLellan JS, Georgiev I, Kwong PD, Carlson JM, Michael NL, Schief WR, Gilbert PB, Mullins JI, Kim JH. 2012. Increased HIV-1 vaccine efficacy against viruses with genetic signatures in Env V2. Nature 490:417–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kong L, Sattentau QJ. 2011. Antigenicity and immunogenicity in HIV-1 antibody-based vaccine design. J. Acquir. Immune Defic. Syndr. Clin. Res. S. 8:003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Karlsson Hedestam GB, Fouchier RA, Phogat S, Burton DR, Sodroski J, Wyatt RT. 2008. The challenges of eliciting neutralizing antibodies to HIV-1 and to influenza virus. Nature Reviews. Microbiology 6:143–155 [DOI] [PubMed] [Google Scholar]
- 8. Schief WR, Ban YE, Stamatatos L. 2009. Challenges for structure-based HIV vaccine design. Curr. Opin. HIV AIDS 4:431–440 [DOI] [PubMed] [Google Scholar]
- 9. Keele BF, Giorgi EE, Salazar-Gonzalez JF, Decker JM, Pham KT, Salazar MG, Sun C, Grayson T, Wang S, Li H, Wei X, Jiang C, Kirchherr JL, Gao F, Anderson JA, Ping LH, Swanstrom R, Tomaras GD, Blattner WA, Goepfert PA, Kilby JM, Saag MS, Delwart EL, Busch MP, Cohen MS, Montefiori DC, Haynes BF, Gaschen B, Athreya GS, Lee HY, Wood N, Seoighe C, Perelson AS, Bhattacharya T, Korber BT, Hahn BH, Shaw GM. 2008. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc. Natl. Acad. Sci. U. S. A. 105:7552–7557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yang X, Wyatt R, Sodroski J. 2001. Improved elicitation of neutralizing antibodies against primary human immunodeficiency viruses by soluble stabilized envelope glycoprotein trimers. J. Virol. 75:1165–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Burton DR, Weiss RA. 2010. AIDS/HIV. A boost for HIV vaccine design. Science 329:770–773 [DOI] [PubMed] [Google Scholar]
- 12. Kwong PD, Mascola JR, Nabel GJ. 2011. Rational design of vaccines to elicit broadly neutralizing antibodies to HIV-1. Cold Spring Harbor Perspect. Med. 1:a007278. 10.1101/cshperspect.a007278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nabel GJ. 2013. Designing tomorrow's vaccines. N. Engl. J. Med. 368:551–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nabel GJ, Kwong PD, Mascola JR. 2011. Progress in the rational design of an AIDS vaccine. Philos. Trans. R. S. London Ser. B Biol. Sci. 366:2759–2765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sattentau QJ, McMichael AJ. 2010. New templates for HIV-1 antibody-based vaccine design. F1000 Biol. Rep. 2:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhou T, Xu L, Dey B, Hessell AJ, Van Ryk D, Xiang SH, Yang X, Zhang MY, Zwick MB, Arthos J, Burton DR, Dimitrov DS, Sodroski J, Wyatt R, Nabel GJ, Kwong PD. 2007. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature 445:732–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Burton DR, Pyati J, Koduri R, Sharp SJ, Thornton GB, Parren PW, Sawyer LS, Hendry RM, Dunlop N, Nara PL, Lamacchia M, Garratty E, Stiehm ER, Bryson YJ, Cao Y, Moore JP, Ho DD, Barbas CF., III 1994. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 266:1024–1027 [DOI] [PubMed] [Google Scholar]
- 18. Corti D, Langedijk JP, Hinz A, Seaman MS, Vanzetta F, Fernandez-Rodriguez BM, Silacci C, Pinna D, Jarrossay D, Balla-Jhagjhoorsingh S, Willems B, Zekveld MJ, Dreja H, O'Sullivan E, Pade C, Orkin C, Jeffs SA, Montefiori DC, Davis D, Weissenhorn W, McKnight A, Heeney JL, Sallusto F, Sattentau QJ, Weiss RA, Lanzavecchia A. 2010. Analysis of memory B cell responses and isolation of novel monoclonal antibodies with neutralizing breadth from HIV-1-infected individuals. PLoS One 5:e8805. 10.1371/journal.pone.0008805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Scheid JF, Mouquet H, Ueberheide B, Diskin R, Klein F, Oliveira TY, Pietzsch J, Fenyo D, Abadir A, Velinzon K, Hurley A, Myung S, Boulad F, Poignard P, Burton DR, Pereyra F, Ho DD, Walker BD, Seaman MS, Bjorkman PJ, Chait BT, Nussenzweig MC. 2011. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science 333:1633–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, Longo NS, McKee K, O'Dell S, Louder MK, Wycuff DL, Feng Y, Nason M, Doria-Rose N, Connors M, Kwong PD, Roederer M, Wyatt RT, Nabel GJ, Mascola JR. 2010. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329:856–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhou T, Georgiev I, Wu X, Yang ZY, Dai K, Finzi A, Kwon YD, Scheid JF, Shi W, Xu L, Yang Y, Zhu J, Nussenzweig MC, Sodroski J, Shapiro L, Nabel GJ, Mascola JR, Kwong PD. 2010. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science 329:811–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Herrera C, Spenlehauer C, Fung MS, Burton DR, Beddows S, Moore JP. 2003. Nonneutralizing antibodies to the CD4-binding site on the gp120 subunit of human immunodeficiency virus type 1 do not interfere with the activity of a neutralizing antibody against the same site. J. Virol. 77:1084–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Moore PL, Crooks ET, Porter L, Zhu P, Cayanan CS, Grise H, Corcoran P, Zwick MB, Franti M, Morris L, Roux KH, Burton DR, Binley JM. 2006. Nature of nonfunctional envelope proteins on the surface of human immunodeficiency virus type 1. J. Virol. 80:2515–2528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Parren PW, Burton DR, Sattentau QJ. 1997. HIV-1 antibody–debris or virion? Nat. Med. 3:366–367 [DOI] [PubMed] [Google Scholar]
- 25. Poignard P, Moulard M, Golez E, Vivona V, Franti M, Venturini S, Wang M, Parren PW, Burton DR. 2003. Heterogeneity of envelope molecules expressed on primary human immunodeficiency virus type 1 particles as probed by the binding of neutralizing and nonneutralizing antibodies. J. Virol. 77:353–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Haim H, Salas I, Sodroski J. 2013. Proteolytic processing of the human immunodeficiency virus envelope glycoprotein precursor decreases conformational flexibility. J. Virol. 87:1884–1889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yuan W, Bazick J, Sodroski J. 2006. Characterization of the multiple conformational States of free monomeric and trimeric human immunodeficiency virus envelope glycoproteins after fixation by cross-linker. J. Virol. 80:6725–6737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Harris A, Borgnia MJ, Shi D, Bartesaghi A, He H, Pejchal R, Kang YK, Depetris R, Marozsan AJ, Sanders RW, Klasse PJ, Milne JL, Wilson IA, Olson WC, Moore JP, Subramaniam S. 2011. Trimeric HIV-1 glycoprotein gp140 immunogens and native HIV-1 envelope glycoproteins display the same closed and open quaternary molecular architectures. Proc. Natl. Acad. Sci. U. S. A. 108:11440–11445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. 2008. Molecular architecture of native HIV-1 gp120 trimers. Nature 455:109–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kwong PD, Doyle ML, Casper DJ, Cicala C, Leavitt SA, Majeed S, Steenbeke TD, Venturi M, Chaiken I, Fung M, Katinger H, Parren PW, Robinson J, Van Ryk D, Wang L, Burton DR, Freire E, Wyatt R, Sodroski J, Hendrickson WA, Arthos J. 2002. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature 420:678–682 [DOI] [PubMed] [Google Scholar]
- 31. Sattentau QJ. 1995. Conservation of HIV-1 gp120 neutralizing epitopes after formalin inactivation. AIDS 9:1383–1385 [DOI] [PubMed] [Google Scholar]
- 32. Migneault I, Dartiguenave C, Bertrand MJ, Waldron KC. 2004. Glutaraldehyde: behavior in aqueous solution, reaction with proteins, and application to enzyme crosslinking. Biotechniques 37:790–796, 798–802 [DOI] [PubMed] [Google Scholar]
- 33. Rodenburg CM, Li Y, Trask SA, Chen Y, Decker J, Robertson DL, Kalish ML, Shaw GM, Allen S, Hahn BH, Gao F. 2001. Near full-length clones and reference sequences for subtype C isolates of HIV type 1 from three different continents. AIDS Res. Hum. Retrovir. 17:161–168 [DOI] [PubMed] [Google Scholar]
- 34. Su L, Graf M, Zhang Y, von Briesen H, Xing H, Kostler J, Melzl H, Wolf H, Shao Y, Wagner R. 2000. Characterization of a virtually full-length human immunodeficiency virus type 1 genome of a prevalent intersubtype (C/B′) recombinant strain in China. J. Virol. 74:11367–11376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Buchacher A, Predl R, Strutzenberger K, Steinfellner W, Trkola A, Purtscher M, Gruber G, Tauer C, Steindl F, Jungbauer A, Katinger H. 1994. Generation of human monoclonal antibodies against HIV-1 proteins; electrofusion and Epstein-Barr virus transformation for peripheral blood lymphocyte immortalization. AIDS Res. Hum. Retrovir. 10:359–369 [DOI] [PubMed] [Google Scholar]
- 36. Lewis DJ, Fraser CA, Mahmoud AN, Wiggins RC, Woodrow M, Cope A, Cai C, Giemza R, Jeffs SA, Manoussaka M, Cole T, Cranage MP, Shattock RJ, Lacey CJ. 2011. Phase I randomised clinical trial of an HIV-1(CN54), clade C, trimeric envelope vaccine candidate delivered vaginally. PLoS One 6:e25165. 10.1371/journal.pone.0025165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wegmann F, Krashias G, Luhn K, Laamanen K, Vieira S, Jeffs SA, Shattock RJ, Sattentau QJ. 2011. A novel strategy for inducing enhanced mucosal HIV-1 antibody responses in an anti-inflammatory environment. PLoS One 6:e15861. 10.1371/journal.pone.0015861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kong L, Sheppard NC, Stewart-Jones GB, Robson CL, Chen H, Xu X, Krashias G, Bonomelli C, Scanlan CN, Kwong PD, Jeffs SA, Jones IM, Sattentau QJ. 2010. Expression-system-dependent modulation of HIV-1 envelope glycoprotein antigenicity and immunogenicity. J. Mol. Biol. 403:131–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jardine J, Julien JP, Menis S, Ota T, Kalyuzhniy O, McGuire A, Sok D, Huang PS, Macpherson S, Jones M, Nieusma T, Mathison J, Baker D, Ward AB, Burton DR, Stamatatos L, Nemazee D, Wilson IA, Schief WR. 2013. Rational HIV immunogen design to target specific germline B cell receptors. Science 340:711–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pejchal R, Doores KJ, Walker LM, Khayat R, Huang PS, Wang SK, Stanfield RL, Julien JP, Ramos A, Crispin M, Depetris R, Katpally U, Marozsan A, Cupo A, Maloveste S, Liu Y, McBride R, Ito Y, Sanders RW, Ogohara C, Paulson JC, Feizi T, Scanlan CN, Wong CH, Moore JP, Olson WC, Ward AB, Poignard P, Schief WR, Burton DR, Wilson IA. 2011. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science 334:1097–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Reference deleted.
- 42. Tomaras GD, Binley JM, Gray ES, Crooks ET, Osawa K, Moore PL, Tumba N, Tong T, Shen X, Yates NL, Decker J, Wibmer CK, Gao F, Alam SM, Easterbrook P, Abdool Karim S, Kamanga G, Crump JA, Cohen M, Shaw GM, Mascola JR, Haynes BF, Montefiori DC, Morris L. 2011. Polyclonal B cell responses to conserved neutralization epitopes in a subset of HIV-1-infected individuals. J. Virol. 85:11502–11519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li M, Gao F, Mascola JR, Stamatatos L, Polonis VR, Koutsoukos M, Voss G, Goepfert P, Gilbert P, Greene KM, Bilska M, Kothe DL, Salazar-Gonzalez JF, Wei X, Decker JM, Hahn BH, Montefiori DC. 2005. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J. Virol. 79:10108–10125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Montefiori DC, Karnasuta C, Huang Y, Ahmed H, Gilbert P, de Souza MS, McLinden R, Tovanabutra S, Laurence-Chenine A, Sanders-Buell E, Moody MA, Bonsignori M, Ochsenbauer C, Kappes J, Tang H, Greene K, Gao H, LaBranche CC, Andrews C, Polonis VR, Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Self SG, Berman PW, Francis D, Sinangil F, Lee C, Tartaglia J, Robb ML, Haynes BF, Michael NL, Kim JH. 2012. Magnitude and breadth of the neutralizing antibody response in the RV144 and Vax003 HIV-1 vaccine efficacy trials. J. Infect. Dis. 206:431–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605–1612 [DOI] [PubMed] [Google Scholar]
- 46. Rice P, Longden I, Bleasby A. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16:276–277 [DOI] [PubMed] [Google Scholar]
- 47. Myszka DG, Sweet RW, Hensley P, Brigham-Burke M, Kwong PD, Hendrickson WA, Wyatt R, Sodroski J, Doyle ML. 2000. Energetics of the HIV gp120-CD4 binding reaction. Proc. Natl. Acad. Sci. U. S. A. 97:9026–9031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Thali M, Moore JP, Furman C, Charles M, Ho DD, Robinson J, Sodroski J. 1993. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J. Virol. 67:3978–3988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chen L, Kwon YD, Zhou T, Wu X, O'Dell S, Cavacini L, Hessell AJ, Pancera M, Tang M, Xu L, Yang ZY, Zhang MY, Arthos J, Burton DR, Dimitrov DS, Nabel GJ, Posner MR, Sodroski J, Wyatt R, Mascola JR, Kwong PD. 2009. Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120. Science 326:1123–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Krashias G, Simon AK, Wegmann F, Kok WL, Ho LP, Stevens D, Skehel J, Heeney JL, Moghaddam AE, Sattentau QJ. 2010. Potent adaptive immune responses induced against HIV-1 gp140 and influenza virus HA by a polyanionic carbomer. Vaccine 28:2482–2489 [DOI] [PubMed] [Google Scholar]
- 51. Wegmann F, Gartlan KH, Harandi AM, Brinckmann SA, Coccia M, Hillson WR, Kok WL, Cole S, Ho LP, Lambe T, Puthia M, Svanborg C, Scherer EM, Krashias G, Williams A, Blattman JN, Greenberg PD, Flavell RA, Moghaddam AE, Sheppard NC, Sattentau QJ. 2012. Polyethyleneimine is a potent mucosal adjuvant for viral glycoprotein antigens. Nat. Biotechnol. 30:883–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Reference deleted.
- 53. Huang Y, Gilbert PB, Montefiori DC, Self SG. 2009. Simultaneous evaluation of the magnitude and breadth of a left and right censored multivariate response, with application to HIV vaccine development. Stat. Biopharm. Res. 1:81–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Karasavvas N, Billings E, Rao M, Williams C, Zolla-Pazner S, Bailer RT, Koup RA, Madnote S, Arworn D, Shen X, Tomaras GD, Currier JR, Jiang M, Magaret C, Andrews C, Gottardo R, Gilbert P, Cardozo TJ, Rerks-Ngarm S, Nitayaphan S, Pitisuttithum P, Kaewkungwal J, Paris R, Greene K, Gao H, Gurunathan S, Tartaglia J, Sinangil F, Korber BT, Montefiori DC, Mascola JR, Robb ML, Haynes BF, Ngauy V, Michael NL, Kim JH, de Souza MS. 2012. The Thai phase III HIV type 1 vaccine trial (RV144) regimen induces antibodies that target conserved regions within the V2 loop of gp120. AIDS Res. Hum. Retrovir. 28:1444–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zolla-Pazner S, Decamp AC, Cardozo T, Karasavvas N, Gottardo R, Williams C, Morris DE, Tomaras G, Rao M, Billings E, Berman P, Shen X, Andrews C, O'Connell RJ, Ngauy V, Nitayaphan S, de Souza M, Korber B, Koup R, Bailer RT, Mascola JR, Pinter A, Montefiori D, Haynes BF, Robb ML, Rerks-Ngarm S, Michael NL, Gilbert PB, Kim JH. 2013. Analysis of V2 antibody responses induced in vaccinees in the ALVAC/AIDSVAX HIV-1 vaccine efficacy trial. PLoS One 8:e53629. 10.1371/journal.pone.0053629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bhattacharyya S, Rajan RE, Swarupa Y, Rathore U, Verma A, Udaykumar R, Varadarajan R. 2010. Design of a non-glycosylated outer domain-derived HIV-1 gp120 immunogen that binds to CD4 and induces neutralizing antibodies. J. Biol. Chem. 285:27100–27110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Chen H, Xu X, Jones IM. 2007. Immunogenicity of the outer domain of a HIV-1 clade C gp120. Retrovirology 4:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yang X, Tomov V, Kurteva S, Wang L, Ren X, Gorny MK, Zolla-Pazner S, Sodroski J. 2004. Characterization of the outer domain of the gp120 glycoprotein from human immunodeficiency virus type 1. J. Virol. 78:12975–12986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Forsell MN, Dey B, Morner A, Svehla K, O'Dell S, Hogerkorp CM, Voss G, Thorstensson R, Shaw GM, Mascola JR, Karlsson Hedestam GB, Wyatt RT. 2008. B cell recognition of the conserved HIV-1 co-receptor binding site is altered by endogenous primate CD4. PLoS Pathog. 4:e1000171. 10.1371/journal.ppat.1000171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Decker JM, Bibollet-Ruche F, Wei X, Wang S, Levy DN, Wang W, Delaporte E, Peeters M, Derdeyn CA, Allen S, Hunter E, Saag MS, Hoxie JA, Hahn BH, Kwong PD, Robinson JE, Shaw GM. 2005. Antigenic conservation and immunogenicity of the HIV coreceptor binding site. J. Exp. Med. 201:1407–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Labrijn AF, Poignard P, Raja A, Zwick MB, Delgado K, Franti M, Binley J, Vivona V, Grundner C, Huang CC, Venturi M, Petropoulos CJ, Wrin T, Dimitrov DS, Robinson J, Kwong PD, Wyatt RT, Sodroski J, Burton DR. 2003. Access of antibody molecules to the conserved coreceptor binding site on glycoprotein gp120 is sterically restricted on primary human immunodeficiency virus type 1. J. Virol. 77:10557–10565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Xiang SH, Wang L, Abreu M, Huang CC, Kwong PD, Rosenberg E, Robinson JE, Sodroski J. 2003. Epitope mapping and characterization of a novel CD4-induced human monoclonal antibody capable of neutralizing primary HIV-1 strains. Virology 315:124–134 [DOI] [PubMed] [Google Scholar]
- 63. Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL, Lehrman JK, Priddy FH, Olsen OA, Frey SM, Hammond PW, Kaminsky S, Zamb T, Moyle M, Koff WC, Poignard P, Burton DR. 2009. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326:285–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Rappuoli R. 1994. Toxin inactivation and antigen stabilization: two different uses of formaldehyde. Vaccine 12:579–581 [DOI] [PubMed] [Google Scholar]
- 65. Haim H, Si Z, Madani N, Wang L, Courter JR, Princiotto A, Kassa A, DeGrace M, McGee-Estrada K, Mefford M, Gabuzda D, Smith AB, III, Sodroski J. 2009. Soluble CD4 and CD4-mimetic compounds inhibit HIV-1 infection by induction of a short-lived activated state. PLoS Pathog. 5:e1000360. 10.1371/journal.ppat.1000360 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.