Abstract
Although prior studies have characterized the neutralizing activities of monoclonal antibodies (MAbs) against dengue virus (DENV) serotypes 1, 2, and 3 (DENV-1, DENV-2, and DENV-3), few reports have assessed the activity of MAbs against DENV-4. Here, we evaluated the inhibitory activity of 81 new mouse anti-DENV-4 MAbs. We observed strain- and genotype-dependent differences in neutralization of DENV-4 by MAbs mapping to epitopes on domain II (DII) and DIII of the envelope (E) protein. Several anti-DENV-4 MAbs inefficiently inhibited at least one strain and/or genotype, suggesting that the exposure or sequence of neutralizing epitopes varies within isolates of this serotype. Remarkably, flavivirus cross-reactive MAbs, which bound to the highly conserved fusion loop in DII and inhibited infection of DENV-1, DENV-2, and DENV-3, more weakly neutralized five different DENV-4 strains encompassing the genetic diversity of the serotype after preincubation at 37°C. However, increasing the time of preincubation at 37°C or raising the temperature to 40°C enhanced the potency of DII fusion loop-specific MAbs and some DIII-specific MAbs against DENV-4 strains. Prophylaxis studies in two new DENV-4 mouse models showed that neutralization titers of MAbs after preincubation at 37°C correlated with activity in vivo. Our studies establish the complexity of MAb recognition against DENV-4 and suggest that differences in epitope exposure relative to other DENV serotypes affect antibody neutralization and protective activity.
INTRODUCTION
Dengue virus (DENV) is a member of the Flaviviridae family of RNA viruses and is genetically related to other human pathogens of global concern, including yellow fever, West Nile (WNV), and Japanese encephalitis viruses. In nature, DENV disease occurs exclusively in humans after Aedes aegypti or Aedes albopictus mosquito inoculation, with clinical disease ranging from a debilitating febrile illness (dengue fever [DF]) to a life-threatening hemorrhagic and capillary leak syndrome (dengue hemorrhagic fever [DHF]/dengue shock syndrome [DSS]). No approved antiviral treatment is currently available, although candidate tetravalent vaccines are in advanced clinical trials (reviewed in references 1 and 2). Because of the increased geographic range of its mosquito vectors, urbanization, and international travel, DENV continues to spread worldwide and now causes an estimated 390 million infections and 500,000 cases of DHF/DSS per year, with 3.6 billion people at risk (3, 4). Given that the most advanced live-attenuated DENV vaccine candidate showed a poor 30% overall efficacy rate in a recently published phase 2b clinical trial (5), there is an urgent need for understanding the correlates of protection, especially those of neutralizing antibodies.
DENV is an enveloped virus with a single-stranded, positive-polarity RNA genome. Based on experiments with DENV-1 and DENV-2 isolates, the mature DENV virion is ∼500 Å in diameter with a highly organized outer protein shell, a 50-Å lipid membrane bilayer, and a nucleocapsid core (6–8). At 28°C, mature DENV virions are covered by 90 antiparallel envelope (E) protein homodimers, arranged flat along the surface with quasi-icosahedral symmetry. At higher temperatures (e.g., greater than 35°C), structural changes occur and DENV virions acquire a bumpy appearance with a diameter of ∼550 Å and some exposed membrane (9, 10). The immature virion, which lacks cleavage of the premembrane (prM) protein, has a rough surface with 60 spikes, each composed of three prM-E heterodimers (11, 12). Exposure to mildly acidic conditions in the trans-Golgi network promotes virus maturation through a structural rearrangement of the flavivirus E proteins and cleavage of prM to membrane (M) protein by a furin-like protease (13–16). The ectodomain of DENV E protein is comprised of three discrete domains (17–20). Domain I (DI) is a central, eight-stranded β-barrel. Domain II (DII) is a long, finger-like protrusion from DI with the highly conserved fusion peptide at its distal end and an N-linked glycan that recognizes DC-SIGN (21–24). Domain III (DIII), which adopts an immunoglobulin-like fold, has been suggested to contain cell surface receptor recognition sites (25–27).
Significant diversity exists among DENV strains, including four unique serotypes (DENV-1, -2, -3, and -4) that differ at the amino acid level by 25 to 40%. Additional complexity occurs within each serotype, as genotypes vary by up to 3% at the amino acid level (28, 29). DENV-4 has been suggested to have four distinct genotypes (I, II, III, and sylvatic), whose circulation patterns differ in time and space: genotype I includes strains from the Philippines, Thailand, Vietnam, Malaysia, Sri Lanka, and India; genotype II is composed of two lineages with strains from Thailand, Malaysia, Taiwan, and the Americas; genotype III includes Thai strains from 1997 to 2001; sylvatic strains were isolated from monkeys or mosquitoes in the 1970s in Malaysia (30). Although sylvatic DENV strains show the greatest sequence differences among the genotypes within a serotype, these viruses still can replicate and cause disease in humans (30–32).
Infection with one DENV serotype is believed to confer long-term durable immunity against strains of the homologous but not heterologous DENV serotypes due to the specificity of neutralizing antibodies and protective CD8+ T cells (33). Recently, in the context of DENV-1, DENV-2, and DENV-3, the question of how intergenotypic or even strain variation within a serotype affects the protective efficacy of neutralizing antibodies has begun to be addressed. Neutralizing antibodies or polyclonal serum with inhibitory activity against one genotype within a serotype may show significant loss of potency against heterologous genotypes (34–38). This concept may be important, because the development of tetravalent DENV vaccines with prototype strains assumes that neutralizing antibody responses, which are lower during vaccination than natural infection, will protect completely against all genotypes within a given serotype (5, 39).
While our laboratory and others have generated panels of inhibitory monoclonal antibodies (MAbs) that react specifically with DENV-1, DENV-2, or DENV-3, to date, remarkably few strongly neutralizing type-specific anti-DENV-4 MAbs have been reported: these include three unmapped mouse MAbs (UH-6C3, D4-II-12B2, and D4-I-11D11) (40, 41), two chimpanzee MAbs (5D9 and 5H2), one of which is localized to an epitope in domain I of the E protein (42, 43), and one human MAb DV22.3, which recognizes an epitope in DI-DII (44). In the current study, we developed a panel of 81 new DENV-4 mouse MAbs and examined their neutralization potentials initially against a genotype II strain (DENV-4 1036; Indonesia 1976) that was used for immunization. Somewhat surprisingly, in our initial functional screens, we identified only six type-specific DENV-4 MAbs that efficiently blocked infection of DENV-4 1036, with the majority of these mapping to distinct epitopes on DIII of the E protein. Because so few of our type-specific MAbs inhibited DENV-4 efficiently, we tested the inhibitory activities of four previously generated cross-reactive fusion loop-specific MAbs (E60, E86, E106, and E119); although these MAbs were originally generated against WNV (45), they have strongly neutralizing activities against other DENV serotypes, with some demonstrating therapeutic activity against DENV-2-induced vascular leakage syndrome in mice (46). These fusion loop MAbs also exhibited weakly neutralizing activity against DENV-4 1036 and several additional strains, including those belonging to the other DENV-4 genotypes. However, when we increased the time and/or temperature of preincubation, all fusion loop-specific cross-reactive MAbs showed enhanced inhibitory activity against the DENV-4 strains. These time and temperature of incubation experimental results suggest that at steady state, DENV-4 strains may exist as a unique ensemble of conformations compared to other DENV serotypes, which would obscure epitope exposure and limit neutralizing activities of fusion loop MAbs.
To begin to assess the significance of these findings, we developed two new lethal peripheral infection models in AG129 IFN-αβR−/− × IFN-γR−/− mice with DENV-4 strains (H-241 and TVP-376) corresponding to distinct genotypes and used them to assess MAb protection by passive transfer. Remarkably, the highly cross-reactive fusion loop MAbs, which potently inhibited infection of DENV-1, DENV-2, and DENV-3, showed marginal protection against genotype I DENV-4 H-241 and enhanced protection against genotype II DENV-4 TVP-376. These in vivo results correlated with focus reduction neutralization titers of MAbs after preincubation at 37°C. Our studies establish the complexity of MAb recognition against the strains and genotypes of the DENV-4 serotype and suggest that differences in DENV-4 epitope exposure relative to other DENV serotypes modulate the protective capacity of antibodies.
MATERIALS AND METHODS
Cells and viruses.
BHK21-15 and Vero cells were cultured in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS; Omega Scientific) and antibiotics (penicillin G and streptomycin). Raji-DC-SIGN cells were cultured in RPMI 1640 medium supplemented with 10% FBS and antibiotics. DENV-4 strains used in this study included 1036 (genotype II; Indonesia, 1976), H-241 (genotype I; Philippines, 1956), TVP-376 (genotype II; Puerto Rico, 1982), TVP-986 (genotype I; Brazil, 1982) (47, 48), and P75-514 (sylvatic; Malaysia, 1975) (30). DENV strains from other serotypes (DENV-1, 16007; DENV-2, 16681; DENV-3, 16652) were obtained from colleagues (A. de Silva, University of North Carolina, and R. Tesh, University of Texas Medical Branch). All stock viruses were propagated in C6/36 Aedes albopictus cells according to established protocols (49).
DENV-4 MAbs.
To generate anti-DENV-4 MAbs, C57BL/6 mice deficient in IFN-αβ receptors (Ifnar−/−) were infected with 105 PFU of a mixture (1:1) of DENV-4 strains 1036 and H-241 via the intraperitoneal route and rechallenged 3 weeks later with the same strains. Subsets of mice with serum showing the highest binding titer to permeabilized Raji-DC-SIGN cells infected with DENV-4 strain 1036 (assayed at a dilution of ∼1:10,000) were immunized with purified DENV-4 DIII (50 μg; strain 1036) in phosphate-buffered saline (PBS) as a final intravenous boost. Three days later, splenocytes were fused to P3X63Ag8.6.5.3 myeloma cells by using polyethylene glycol 1500 (50). Hybridoma cells producing anti-DENV-4 MAbs (see Table S1 in the supplemental material) were subcloned by limiting dilution and isotyped using an enzyme-linked immunosorbent assay (ELISA) kit (Southern Biotech). The fusion loop-specific MAbs (E60, E86, E106, and E119) cross-react with all DENV serotypes and have been described previously (45). Unless otherwise indicated, all MAbs were purified from hybridoma supernatants by protein A or protein G immunoaffinity chromatography.
In vitro neutralization assays.
Focus reduction neutralization titer (FRNT) assays were performed with the different DENV-4 strains and MAbs on Vero cells in a manner analogous to that described previously for WNV (51). Serial dilutions of purified MAbs or hybridoma supernatants were mixed with 102 focus-forming units (FFU) of different DENV-1, DENV-2, DENV-3, or DENV-4 viruses for specified times (1 or 3 h) and temperatures (37 or 40°C). Subsequently, virus-MAb mixtures were added to Vero cell monolayers for 1 h, and then a 1% carboxymethylcellulose overlay was added. Two days later, the overlays were removed and monolayers were fixed with 1% paraformaldehyde (1 hour at room temperature), permeabilized with 0.1% saponin in PBS, and incubated with human-mouse chimeric WNV E18 MAb (200 ng/ml) (45), which recognizes the conserved fusion loop epitope. Following several washes, wells were incubated with horseradish peroxidase-conjugated anti-human IgG antibody (Sigma; 250 ng/ml in saponin buffer) for 1 h at room temperature. Wells were washed, and infectious foci were visualized with TrueBlue substrate (KPL) after a 10-minute incubation at room temperature. Wells were rinsed with water and analyzed with a Biospot counter (Cellular Technology) and Immunocapture software. Neutralization curves (the percent reduction in spot numbers in virus samples preincubated with MAbs compared to medium alone) were graphed using Prism software, and the 50% and 90% effective concentration (EC50 and EC90) values were calculated by nonlinear regression using a variable slope.
E gene sequencing.
DENV-4 E genes were amplified by reverse transcription-PCR (RT-PCR) using primers (see Table S2 in the supplemental material), a high-fidelity Taq polymerase, and RNA extracted directly from infected C6/36 cells. Each amplicon was sequenced on both strands by conventional capillary sequencing on an Applied Biosystems 3730 genetic analyzer using previously reported primers (30, 34). The resulting sequence reads were assembled, and the E gene sequences were aligned using the Lasergene suite (DNAStar).
Domain mapping by yeast surface display.
The DNA fragments encoding amino acid residues 1 to 292 (DI-DII) and 293 to 409 (DIII) of the DENV-4 E protein were amplified from DENV-4 strain 1036 by RT-PCR with KpnI and XhoI or BamHI and XhoI sites added at the 5′ and 3′ ends, respectively. The PCR product was cloned as a downstream fusion to Aga2 and Xpress epitope tag genes in the yeast surface display vector pYD1 (Invitrogen) and transformed into Saccharomyces cerevisiae strain EBY100 (Invitrogen) to generate yeast that expressed DENV-4 DI-DII or DIII or the E ectodomain, as described previously (49, 52).
Amino acid mapping of epitopes by shotgun mutagenesis and expression in mammalian cells.
A DENV-4 prM-E protein expression construct (strain TVP-376) was subjected to high-throughput alanine-scanning mutagenesis to generate a comprehensive mutation library. Primers were designed to mutate each residue within prM-E to alanine, with alanine codons mutated to serine. In total, 660 DENV-4 mutant proteins with sequence confirmation (>97% coverage) were generated and arrayed into 384-well plates (one clone per well). Anti-DENV-4 MAbs were tested for reactivity against either the entire library of mutations or a selected subset of the library containing the most relevant clones. Each prM-E mutant was transfected into HEK-293T cells and allowed to express for 22 h. Cells then were fixed in 4% (vol/vol) paraformaldehyde (Electron Microscopy Sciences) and permeabilized with 0.1% (wt/vol) saponin (Sigma) in PBS plus calcium and magnesium (PBS++). Cells were stained with purified MAb DV4-E75 (0.2 μg/ml) or DV4-E88 (0.2 μg/ml) or hybridoma supernatant DV4-E4 (1:30), DV4-E29 (1:90), DV4-E33 (1:180), DV4-E40 (1:270), DV4-E76 (1:60), DV4-E78 (1:30), DV4-E87 (1:60), or DV4-E121 (1:60) diluted in 10% normal goat serum (NGS; Sigma) and 0.1% saponin, pH 9.0. Primary antibody concentrations were determined using an independent immunofluorescence titration curve against wild-type DENV-4 prM-E to ensure that signals were within the linear range of detection. Antibodies were detected using 3.75 μg/ml of AlexaFluor 488-conjugated secondary antibody (Jackson ImmunoResearch Laboratories) in 10% NGS (Sigma) and 0.1% saponin. Cells were washed three times with PBS supplemented with 0.1% saponin, 1 mM MgCl2, and CaCl2 followed by two washes in PBS. The mean cellular fluorescence was detected using the a high-throughput flow cytometer (HTFC; Intellicyt). Antibody reactivity against each mutant E protein clone was calculated relative to wild-type E protein reactivity by subtracting the signal from mock-transfected controls and normalizing to the signal from wild-type prM-E-transfected controls. Mutations were identified as critical to the MAb epitope if they did not support reactivity of the test MAb but did support reactivity of other DENV-4 MAbs. This counterscreen strategy facilitates the exclusion of E mutants that are locally misfolded or have expression defects (53).
Immunostaining of DENV-infected cells.
To assess binding of DENV-4 MAbs to different DENV strains, Raji DC-SIGN-R or C6/36 cells were infected at a multiplicity of infection (MOI) of 0.5 or 1. Depending on the strain, cells were harvested 48, 72, or 96 h after infection. Cells were washed, fixed in PBS with 1% paraformaldehyde, permeabilized, incubated with MAbs, and processed by flow cytometry as described previously (35).
Generation of MAb-resistant virus escape mutants.
DENV-4 (1036 or TVP-376; 5 × 105 PFU) was incubated with 25 μg/ml of MAb DV4-75 or DV4-E88 for 1 h at 37°C in DMEM. The mixture was added to 5 × 105 Vero cells in a 6-well plate. After infection for 2 h at 37°C, wells were washed thrice with DMEM, and fresh medium containing 5 μg/ml of MAb was added. Virus growth under antibody selection proceeded for 72 h at 37°C. At each passage, half of the supernatant was mixed 1:1 with 10 μg/ml of MAb for 1 h. The remaining half was stored at −80°C. After three to six passages under MAb selection, virus-containing supernatants were tested by plaque reduction assay for escape from neutralization of DV4-75 or DV4-88. After confirming the escape phenotype, an aliquot of the supernatant was used for a Vero cell plaque assay under MAb selection. Plaque-purified virus was amplified further under MAb selection (25 μg/ml) overnight at 37°C. Vero cells were scraped from wells, and total cellular RNA was isolated using an RNeasy kit (Qiagen). cDNA was amplified with random hexamers as well as a DENV-4 primer by using a SuperScript III first-strand synthesis system for RT-PCR (Invitrogen) and served as a template for PCR amplification using forward and reverse primers (see Table S2 in the supplemental material). Amplicons were sequenced, and the neutralization escape mutant sequence was compared to the parent virus stock that was passaged in parallel in the absence of MAb selection.
Mouse experiments.
All mouse studies were approved and performed according to the guidelines of the Washington University School of Medicine Animal Safety Committee. Ifnar−/− mice on the C57BL/6 background were a gift of Jonathan Sprent (The Scripps Research Institute). The IFN-αβR−/− × IFN-γR−/− mice on a 129 Sv background (AG129) have been described previously (54). All mice were housed in pathogen-free barrier facilities. In prophylaxis experiments, mice were administered a single dose of individual MAbs via the intraperitoneal route 1 day before infection. AG129 mice were challenged with DENV-4 H-241 (genotype I) or DENV-4 TVP-376 (genotype II) intravenously, and mortality was monitored.
Generation of mouse-adapted DENV-4 strains.
To develop a mouse model of DENV-4 infection with the H-241 genotype I strain, virus was passaged once in AG129 newborn pups after intraperitoneal infection; moribund mice were sacrificed and brains were harvested, homogenized, and virus was clarified by centrifugation (1,600 × g for 5 min). The supernatant was used to infect 6-week-old AG129 mice via an intravenous route. Brains from mice that succumbed to infection were harvested and serially passaged in 6-week-old AG129 for two additional rounds using the same protocol. Virus isolated from the brains of AG129 mice infected at passage five was used to infect C6/36 cells and generate a virus stock used for experiments.
The TVP-376 genotype II mouse model was developed using a different approach. TVP-376 was amplified in C6/36 cells, and virus was concentrated from supernatant by ultracentrifugation (67,000 × g for 3 h over a 25% glycerol cushion in 10 mM Tris, 150 mM NaCl, and 1 mM EDTA in an SW32 rotor). Six-week-old AG129 mice were infected with 1 × 104 PFU of TVP-376 virus via the intravenous route. Four days later, mice were bled and serum was added to C6/36 cells to generate a seed stock that was purified by ultracentrifugation and used for infection of AG129 mice.
Statistical analysis.
All data were analyzed using Prism software (GraphPad, San Diego, CA). Kaplan-Meier survival curves were analyzed by the log rank test. Log EC50 values were compared using Student's t test when comparing two samples or a one-way analysis of variance when comparing more than two samples.
Nucleotide sequence accession numbers.
Two new E gene sequences, TVP-376 and TVP-986, were deposited in GenBank with accession numbers of KC963424 and KC963425, respectively.
RESULTS
Functional properties of newly generated MAbs against DENV-4.
In prior studies, we generated type-specific and cross-reactive neutralizing MAbs against DENV-1, DENV-2, and DENV-3 after immunization of Ifnar−/− C57BL/6 mice with live DENV and recombinant E proteins (35, 37, 38). As a first step toward creating an analogous panel of MAbs that neutralized DENV-4 infection, we infected Ifnar−/− C57BL/6 mice with a mixture of the DENV-4 strains 1036 (genotype II) and H-241 (genotype I) (Table 1) and boosted mice with homologous viruses and recombinant DIII. Although DENV-4 failed to cause a lethal infection in Ifnar−/− mice, it replicated to higher titers than in wild-type (WT) mice, which resulted in the induction of a stronger neutralizing antibody response (data not shown). After screening more than 6,000 hybridoma clones as part of 10 independent fusions, we isolated 81 new MAbs that recognized permeabilized cells infected with DENV-4 (see Table S1 in the supplemental material). Somewhat surprisingly (compared to studies with newly generated MAbs against other DENV serotypes [35, 37, 38]), after a 1-h preincubation at 37°C, only six (DV4-E3, DV4-E33, DV4-E75, DV4-E76, DV4-E88, and DV4-E121) MAbs inhibited infection of DENV-4 1036 as neat hybridoma supernatant by greater than 90% in focus reduction assays, with only a subset showing an ability to completely neutralize DENV-4 infection (Table 2, Fig. 1, and data not shown). The remainder of the panel had less inhibitory activity against DENV-4 1036 when used under similar conditions (Table 2 and data not shown).
Table 1.
Characteristics of DENV-4 strains used in the studya
| Virus | Origin, yr | Genotype | Reference(s) | GenBank accession no(s). |
|---|---|---|---|---|
| 1036 | Indonesia, 1976 | II | 34 | U18429 |
| H-241 | Philippines, 1956 | I | 30, 34 | AY947539, U18433 |
| TVP-376 | Colombia, 1982 | II | Unpublished | KC963424 |
| TVP-986 | Brazil, 1982 | II | 47 | KC963425 |
| P75-514 | Malaysia, 1975 | Sylvatic | 30 | JF262780 |
The genotypes were assigned based on alignment of E gene nucleotide sequences and creation of a dendrogram. The results were confirmed using the Genotype Determination and Recombination Detection online software from the Virus Pathogen Resource (http://www.viprbrc.org/). The TVP-376 sequence has not been published and was obtained from the World Arbovirus Collection.
Table 2.
Neutralizing MAbs against DENV-4
| MAb | % neutralization (undiluted supernatant)a | Isotypeb | Domain localizationc | Cross-reactivityd |
|---|---|---|---|---|
| DV4-E3 | 94 | IgM | DI-II | DENV-1, -2, and -3 |
| DV4-E4 | 84 | IgG1 | DI-II | DENV-1, -2, and -3 |
| DV4-E27 | 68 | IgG2a | DI-II | DENV-1, -2, and -3 |
| DV4-E29 | 69 | IgG1 | DIII | None |
| DV4-E33 | 90 | IgG2a | DIII | None |
| DV4-E40 | 79 | IgG2a | DIII | None |
| DV4-E68 | 63 | IgG2a | DIII | None |
| DV4-E75 | 100 | IgG3 | DIII | None |
| DV4-E76 | 91 | IgG2c | DIII | DENV-2 |
| DV4-E78 | 79 | IgG2c | DIII | DENV-2 |
| DV4-E87 | 68 | IgG2c | DIII | None |
| DV4-E88 | 100 | IgG2c | DIII | None |
| DV4-E121 | 100 | IgG2c | DIII | None |
Undiluted hybridoma supernatant (∼1 to 10 μg/ml) was incubated with 102 FFU of DENV-4 (strain 1036) for 1 h at 37°C. Virus-MAb mixtures were added to Vero cell monolayers for 1 h at 37°C prior to addition of a methylcellulose overlay (described in Materials and Methods). DENV-infected foci were stained with a human-mouse chimeric E18 MAb. The percent neutralization was determined compared to medium alone. Results are representative of three independent experiments.
The MAb isotype was determined using a commercial ELISA kit.
Domain localization was determined by yeast surface display or mapping with mutant structural proteins (see Fig. 4). DV4-E75 and DV4-E88 did not bind to DIII when expressed on the surface of yeast.
Cross-reactivity to other DENV serotypes was established after intracellular staining of C6/36 cells infected with DENV-1 (strain 16007), DENV-2 (strain 16681), or DENV-3 (strain 16652).
Fig 1.
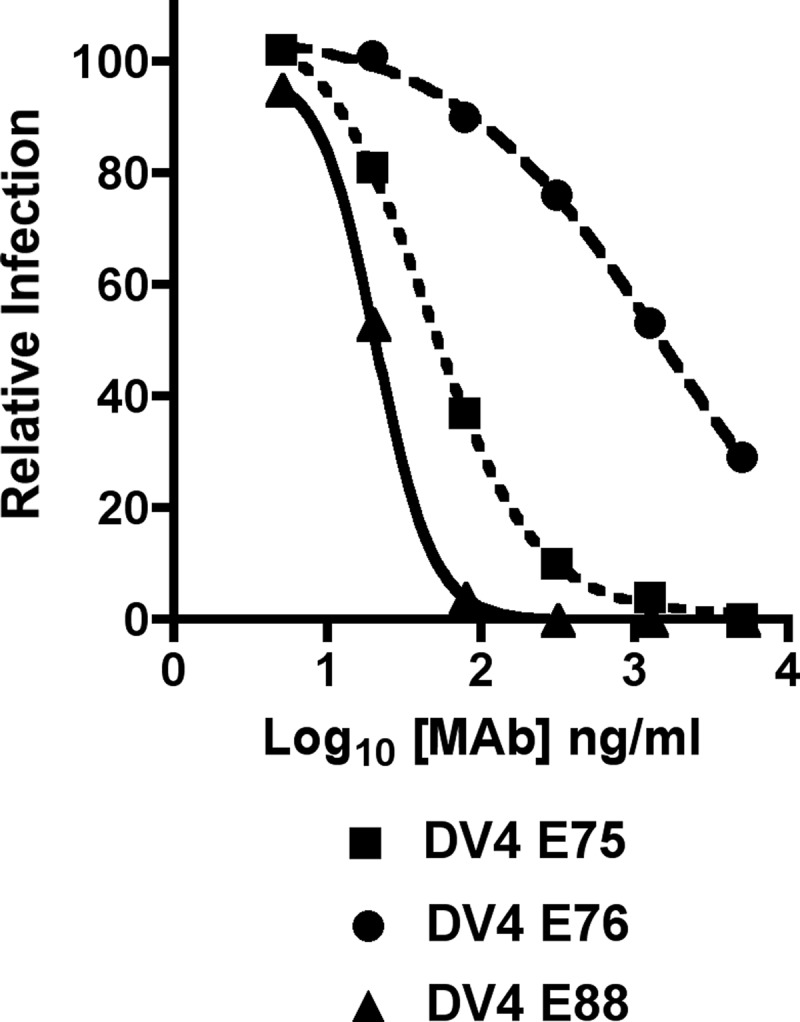
Neutralization of DENV-4 by type-specific MAbs. Increasing concentrations of purified MAbs (DV4-E75, DV4-E76, and DV4-E88) were mixed with 102 PFU of DENV-4 strain 1036 for 1 h at 37°C, and inhibition was assessed in a FRNT assay in Vero cells and compared to virus that was incubated with no antibody. The graph was generated after regression analysis using Prism statistical software. The data are representative of at least three independent experiments performed in duplicate.
Because these results were unexpected, we questioned whether it was intrinsically more difficult for MAbs to inhibit DENV-4 than other serotypes. We reasoned this was possible, as we previously had difficulty isolating potently neutralizing antibodies against hepatitis C virus (HCV), a distantly related member of the Flaviviridae family (55). To begin to assess this, we tested whether MAbs recognizing the highly conserved fusion loop on domain II neutralized infection of DENV-4. We utilized a panel of cross-reactive fusion loop-specific MAbs that we had generated previously after sequential infection of C57BL/6 mice with WNV and DENV-2 (45). Four of these MAbs (E60, E86, E106, and E119) were selected for testing with DENV-4, because these efficiently neutralized (EC50 values from 17 to 612 ng/ml) infection of DENV-1, DENV-2, and DENV-3 (Fig. 2A and Table 3), with one MAb (E60) having postexposure therapeutic activity against DENV-2 in a mouse model of DSS (46, 56). Remarkably, despite binding efficiently to DENV-4-infected cells (Fig. 2B), these four MAbs showed relatively weak inhibitory activities (EC50 of 822 to 2,786 ng/ml and EC90 of >5,000 ng/ml) against DENV-4 1036 after a 1-h preincubation at 37°C (Fig. 2C and Table 4).
Fig 2.
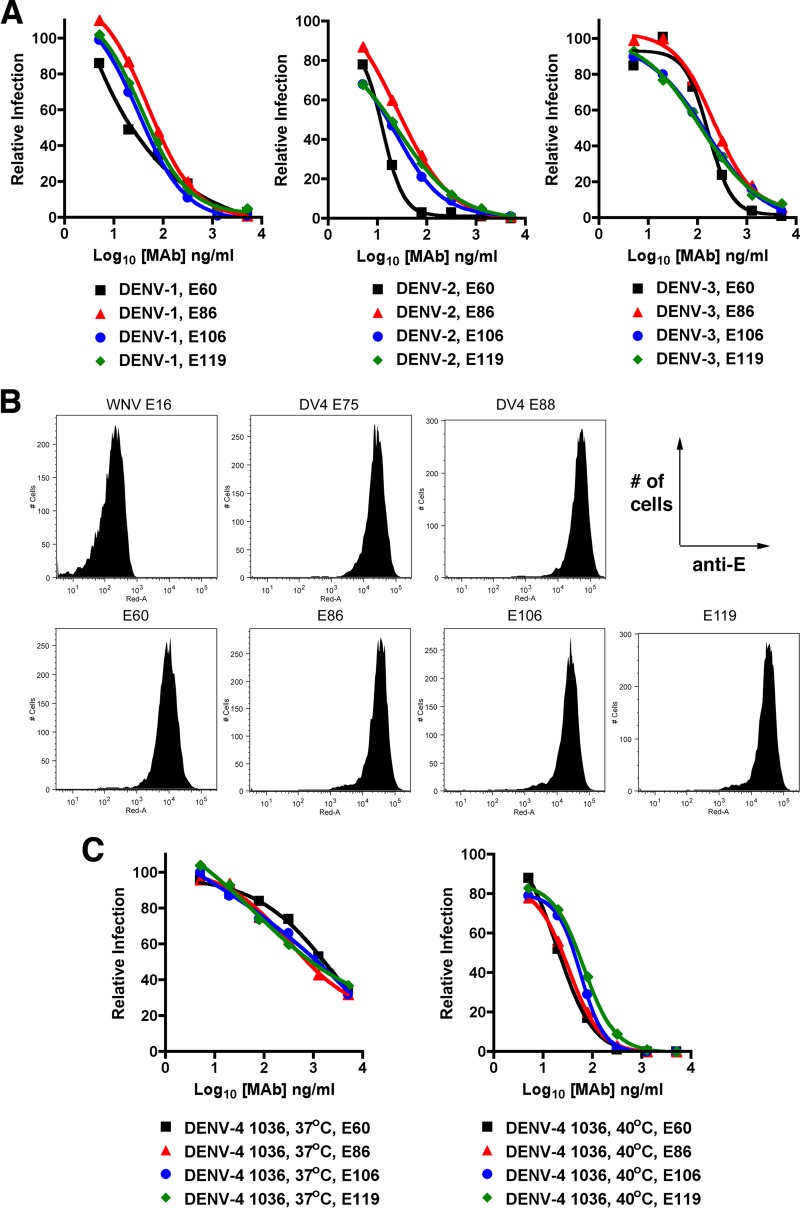
Neutralization of DENV serotypes by cross-reactive MAbs that map to fusion loop epitope in DII. (A) Increasing concentrations of purified fusion loop-specific MAbs (E60, E86, E106, and E119) were mixed with 102 PFU of DENV-1 (strain 16007), DENV-2 (strain 16681), or DENV-3 (strain 16652) (left, middle, and right, respectively) for 1 h at 37°C. Virus-MAb complexes were added to Vero cells, and neutralization was assessed in a FRNT assay after comparison to virus that was incubated with no antibody. The data are representative of at least three independent experiments performed in duplicate. (B) Flow cytometry histograms of Vero cells infected with DENV-4 (strain 1036) and stained with WNV E16 (negative control), DV4-E75, DV4-E88, E60, E86, E106, or E119. Results are representative of several independent experiments. All histograms show the number of cells on the y axis and staining with individual anti-E MAbs on the x axis. (C) Increasing concentrations of E60, E86, E106, and E119 were mixed with 102 PFU of the DENV-4 (strain 1036) for 1 h at 37°C (left) or 40°C (right), and neutralization was assessed in a FRNT assay. The data are representative of at least three independent experiments performed in duplicate.
Table 3.
Neutralization of DENV-1, DENV-2, and DENV-3 by cross-reactive fusion loop-specific MAbsa
| MAb | Isotype | DENV-1 |
DENV-2 |
DENV-3 |
|||
|---|---|---|---|---|---|---|---|
| EC50 (ng/ml) | EC90 (ng/ml) | EC50 (ng/ml) | EC90 (ng/ml) | EC50 (ng/ml) | EC90 (ng/ml) | ||
| E60 | IgG2a | 21 | 661 | 119 | 537 | 612 | >5,000 |
| E86 | IgG2a | 83 | 450 | 34 | 343 | 264 | 2,089 |
| E106 | IgG2a | 47 | 309 | 15 | 275 | 84 | >5,000 |
| E119 | IgG2a | 59 | 406 | 17 | 491 | 130 | 2,500 |
Increasing concentrations (5 to 5,000 ng/ml) of the indicated MAbs were premixed with 102 FFU of DENV-1 (strain 16007), DENV-2 (strain 16681), or DENV-3 (strain 16652) for 1 h at 37°C. Virus-MAb mixtures were added to Vero cell monolayers for 1 h at 37°C prior to addition of a methylcellulose overlay. DENV-infected foci were stained with a human-mouse chimeric E18 MAb. EC50 and EC90 values were determined by using nonlinear regression models (GraphPad Prism). The results are the averages of three independent experiments performed in triplicate.
Table 4.
Neutralization of DENV-4 strain 1036 by cross-reactive fusion loop- and type-specific MAbsa
| MAb | Temp (°C) | Time (h) | EC50 (ng/ml) | EC90 (ng/ml) |
|---|---|---|---|---|
| E60 | 37 | 1 | 2,786 | >5,000 |
| E60 | 37 | 3 | 833 | >5,000 |
| E60 | 40 | 1 | 34 | 195 |
| E86 | 37 | 1 | 822 | >5,000 |
| E86 | 37 | 3 | 232 | >5,000 |
| E86 | 40 | 1 | 23 | 125 |
| E106 | 37 | 1 | 1,141 | >5,000 |
| E106 | 37 | 3 | 271 | >5,000 |
| E106 | 40 | 1 | 35 | 178 |
| E119 | 37 | 1 | 1,730 | >5,000 |
| E119 | 37 | 3 | 598 | >5,000 |
| E119 | 40 | 1 | 73 | 442 |
| DV4-E75 | 37 | 1 | 63 | 258 |
| DV4-E75 | 37 | 3 | 36 | 181 |
| DV4-E75 | 40 | 1 | 36 | 103 |
| DV4-E88 | 37 | 1 | 18 | 49 |
| DV4-E88 | 37 | 3 | 9 | 34 |
| DV4-E88 | 40 | 1 | 17 | 51 |
Increasing concentrations (5 to 5,000 ng/ml) of the indicated MAbs were premixed with 102 FFU of DENV-4 (strain 1036) for 1 h at 37°C, 3 h at 37°C, or 1 h at 40°C. Virus-MAb mixtures were added to Vero cell monolayers for 1 h at 37°C prior to addition of a methylcellulose overlay. DENV-infected foci were stained with a human-mouse chimeric E18 MAb. EC50 and EC90 values were determined by using nonlinear regression models (GraphPad Prism). The results are the averages of three independent experiments performed in triplicate.
Neutralization of DENV-4 by fusion loop MAbs is enhanced by increasing the time and temperature of preincubation.
One possible explanation as to why the fusion loop MAbs neutralized DENV-1, DENV-2, and DENV-3 but not DENV-4, despite binding to infected cells, was that the fusion loop epitope was not equivalently accessible on the surface of the DENV-4 virion as on the other DENV serotypes. Recent studies with WNV, DENV-1, DENV-2, and HCV have suggested that increasing the time and/or temperature of preincubation facilitates exposure of buried epitopes on different virion conformational ensembles, which results in enhanced neutralizing activity of some MAbs (57–60). To test this hypothesis, we repeated the experiments with DENV-4 1036 and E60, E86, E106, or E119 for a longer time (37°C, 3 h) or at a higher temperature (40°C, 1 h) for preincubation. Notably, preincubation of the fusion loop MAbs for 3 h at 37°C slightly improved neutralization of DENV-4 1036 (EC50 values 3- to 4-fold lower than with the 1-h incubation), although three (E60, E86, and E119) of the four MAbs still failed to achieve a measurable EC90 value (Table 4). Preincubation of DENV-4 1036 with the fusion loop MAbs at 40°C for 1 h more strongly enhanced their inhibitory activities (Fig. 2C, EC50 values 24- to 82-fold lower than at 37°C incubation; P < 0.05). Remarkably, the 40°C incubation improved the capacity to neutralize most of the DENV-4 virus within the population, as the EC90 values now ranged from 125 to 442 ng/ml (Table 4). In comparison, changing the time or temperature of incubation had small effects on the activities of selected type-specific neutralizing MAbs DV4-E75 (2- to 2.5-fold decrease in EC50 and EC90 values; P > 0.05) and DV4-E88 (2- to 1.5-fold change in EC50 and EC90 values; P > 0.05).
Effects of time and temperature of preincubation on MAb neutralization of other DENV-4 strains.
Neutralization of DENV-1, DENV-2, and DENV-3 infection by MAbs can vary in a genotype-dependent manner (35–38, 60). Given this observation, we assessed whether the preincubation conditions differentially affected neutralization by cross-reactive fusion loop MAbs of several genetically distinct DENV-4 strains (Tables 4 and 5). We tested four additional strains (H-241, TVP-376, TVP-986, and P75-514 [Table 1]), corresponding to three DENV-4 genotypes (I, II, and sylvatic), which varied from strain 1036 by 0.4% (TVP-376 and TVP-396), 3% (H-241), and 4% (P75-514) at the amino acid level for the E protein. When these DENV-4 strains were preincubated with fusion loop MAbs (E60, E86, E106, and E119) for 1 h at 37°C, we observed weak neutralizing activity for H-241 (EC50 of ≥4,500 ng/ml; EC90 of >5,000 ng/ml), TVP-376 (EC50 of 509 to 1,165 ng/ml; EC90 of >5,000 ng/ml), and TVP-986 (EC50 of 458 to 900 ng/ml; EC90 of >5,000 ng/ml). Under the same preincubation conditions, higher levels of neutralization were observed against the sylvatic strain P75-514 (EC50 of 98 to 267 ng/ml; EC90 of 1,312 to 2,317 ng/ml).
Table 5.
Neutralization of other DENV-4 strains by cross-reactive fusion loop MAbs
| Virusa | Genotypeb | MAb | Temp (°C) | Time (h) | EC50 (ng/ml) | EC90 (ng/ml) |
|---|---|---|---|---|---|---|
| H241 | I | E60 | 37 | 1 | >5,000 | >5,000 |
| H241 | I | E60 | 37 | 3 | 1,171 | >5,000 |
| H241 | I | E60 | 40 | 1 | 65 | 711 |
| H241 | I | E86 | 37 | 1 | >5,000 | >5,000 |
| H241 | I | E86 | 37 | 3 | 2,534 | >5,000 |
| H241 | I | E86 | 40 | 1 | 95 | 700 |
| H241 | I | E106 | 37 | 1 | 4,500 | >5,000 |
| H241 | I | E106 | 37 | 3 | 1,472 | >5,000 |
| H241 | I | E106 | 40 | 1 | 183 | 1,750 |
| H241 | I | E119 | 37 | 1 | >5,000 | >5,000 |
| H241 | I | E119 | 37 | 3 | >5,000 | >5,000 |
| H241 | I | E119 | 40 | 1 | 267 | >5,000 |
| TVP-376 | II | E60 | 37 | 1 | 1,054 | >5,000 |
| TVP-376 | II | E60 | 37 | 3 | 153 | 3,524 |
| TVP-376 | II | E60 | 40 | 1 | 10 | 113 |
| TVP-376 | II | E86 | 37 | 1 | 594 | >5,000 |
| TVP-376 | II | E86 | 37 | 3 | 95 | 2,104 |
| TVP-376 | II | E86 | 40 | 1 | 9 | 285 |
| TVP-376 | II | E106 | 37 | 1 | 510 | >5,000 |
| TVP-376 | II | E106 | 37 | 3 | 121 | 2,864 |
| TVP-376 | II | E106 | 40 | 1 | 9 | 166 |
| TVP-376 | II | E119 | 37 | 1 | 1,165 | >5,000 |
| TVP-376 | II | E119 | 37 | 3 | 213 | >5,000 |
| TVP-376 | II | E119 | 40 | 1 | 105 | 640 |
| TVP-986 | II | E60 | 37 | 1 | 900 | >5,000 |
| TVP-986 | II | E60 | 37 | 3 | 56 | 548 |
| TVP-986 | II | E60 | 40 | 1 | 11 | 282 |
| TVP-986 | II | E86 | 37 | 1 | 756 | >5,000 |
| TVP-986 | II | E86 | 37 | 3 | 22 | 832 |
| TVP-986 | II | E86 | 40 | 1 | 31 | 644 |
| TVP-986 | II | E106 | 37 | 1 | 458 | >5,000 |
| TVP-986 | II | E106 | 37 | 3 | 51 | 809 |
| TVP-986 | II | E106 | 40 | 1 | 22 | 411 |
| TVP-986 | II | E119 | 37 | 1 | 546 | >5,000 |
| TVP-986 | II | E119 | 37 | 3 | 60 | 2,259 |
| TVP-986 | II | E119 | 40 | 1 | 37 | 621 |
| P75-514 | S | E60 | 37 | 1 | 268 | 2,317 |
| P75-514 | S | E60 | 37 | 3 | 45 | 313 |
| P75-514 | S | E60 | 40 | 1 | 38 | 454 |
| P75-514 | S | E86 | 37 | 1 | 98 | 1,758 |
| P75-514 | S | E86 | 37 | 3 | 25 | 214 |
| P75-514 | S | E86 | 40 | 1 | 58 | 306 |
| P75-514 | S | E106 | 37 | 1 | 136 | 1,312 |
| P75-514 | S | E106 | 37 | 3 | 30 | 285 |
| P75-514 | S | E106 | 40 | 1 | 33 | 210 |
| P75-514 | S | E119 | 37 | 1 | 99 | 1,507 |
| P75-514 | S | E119 | 37 | 3 | 23 | 380 |
| P75-514 | S | E119 | 40 | 1 | 7 | 115 |
Increasing concentrations (5 to 5,000 ng/ml) of the indicated MAbs were premixed with 102 FFU of DENV-4 (strain H241, TVP-376, TVP-986, or P75-514) for 1 h at 37°C, 3 h at 37°C, or 1 h at 40°C. Virus-MAb mixtures were added to Vero cell monolayers for 1 h at 37°C prior to addition of a methylcellulose overlay. DENV-4-infected foci were stained with a human-mouse chimeric E18 MAb. EC50 and EC90 values were determined by using nonlinear regression models (GraphPad Prism). The results are the averages of three independent experiments performed in triplicate.
Summary of genotypes: H-241 is genotype I; 1036, TVP-376, and TVP-986 are genotype II; P75-514 is the sylvatic genotype (S).
When experiments were repeated with longer preincubation times, analogous to the findings with DENV-4 strain 1036, we observed modestly improved inhibitory activity by fusion loop MAbs against TVP-376, TVP-986, H-241, and P75-514. Preincubation for 3 h at 37°C improved neutralization of DENV-4 TVP-376 (EC50 values 4- to 7-fold lower than with the 1-h incubation), and three of the four (E60, E86, and E106) achieved measurable EC90 values (2,103 to 3,523 ng/ml) (Table 5). Similar results were observed with DENV-4 P75-514 (EC50 values 4- to 6-fold lower and EC90 values 4- to 8-fold lower than with the 1-h incubation at 37°C; P < 0.05), although smaller effects were observed with H-241, which still remained relatively resistant to neutralization. In comparison, a longer incubation at 37°C improved the inhibitory activity of the fusion loop MAbs against DENV-4 TVP-986 to an even greater level (EC50 values 9- to 35-fold lower than with the 1-h incubation; P < 0.05).
Enhanced neutralization with fusion loop-specific MAbs also was observed after preincubation of different DENV-4 strains at 40°C. Preincubation for 1 h at 40°C improved neutralization of TVP-376 (EC50 values 57- to 105-fold lower; P < 0.02), TVP-986 (EC50 values 15- to 80-fold lower; P < 0.05), and H-241 (EC50 values 5- to 14-fold lower) compared to experiments at 37°C. This pattern also was seen for EC90 values after 40°C preincubation with fusion loop MAbs and TVP-376 (EC90 of 112 to 640 ng/ml) and TVP-986 (EC90 of 281 to 644 ng/ml). A more modest improvement in neutralization by fusion loop-specific MAbs was observed with P75-514 (EC50 values 2- to 15-fold lower), but the change did not attain statistical significance, likely secondary to the better baseline inhibition achieved after incubation at 37°C. Even under conditions of a higher-temperature incubation, H-241 remained relatively resistant to neutralization by several of the fusion loop-specific MAbs.
As our data with DENV-4 1036 suggested that modifying the incubation conditions had less impact on the inhibitory activity of the type-specific MAbs DV4-E75 and DV4-E88, we evaluated this with the strains H-241, TVP-376, TVP-986, and P75-514 (Table 6). Under standard 1-h preincubation conditions at 37°C, DV4-E75 neutralized the homologous genotype II strains TVP-376 and TVP-986 efficiently, with EC50 and EC90 values of 91 to 140 ng/ml and 769 to 1,127 ng/ml, respectively. However, DV4-E75 had little or no inhibitory activity against the heterologous sylvatic and genotype I strains P75-514 and H-241 (EC50, >5,000 ng/ml), which is consistent with the genotype-dependent effects on neutralization that have been observed with MAbs against DENV-2 and DENV-3 (36, 37, 61). Under similar conditions, DV4-E88 efficiently neutralized TVP-376 and TVP-986 (EC50 and EC90 of 60 to 80 ng/ml and 317 to 407 ng/ml, respectively), showed an ∼10-fold decrease in activity against P75-514 (EC50 of 530 ng/ml and EC90 of 3,420 ng/ml), and poorly inhibited infection by H-241 (EC50 of 5,000 ng/ml and EC90 of >5,000 ng/ml).
Table 6.
Neutralization of other DENV-4 strains by type-specific MAbsa
| Virus | Genotype | MAb | Temp (°C) | Time (h) | EC50 (ng/ml) | EC90 (ng/ml) |
|---|---|---|---|---|---|---|
| H241 | I | DV4-E75 | 37 | 1 | >5,000 | >5,000 |
| H241 | I | DV4-E75 | 37 | 3 | >5,000 | >5,000 |
| H241 | I | DV4-E75 | 40 | 1 | >5,000 | >5,000 |
| H241 | I | DV4-E88 | 37 | 1 | >5,000 | >5,000 |
| H241 | I | DV4-E88 | 37 | 3 | 4,593 | >5,000 |
| H241 | I | DV4-E88 | 40 | 1 | >5,000 | >5,000 |
| TVP-376 | II | DV4-E75 | 37 | 1 | 140 | 1,127 |
| TVP-376 | II | DV4-E75 | 37 | 3 | 73 | 656 |
| TVP-376 | II | DV4-E75 | 40 | 1 | 57 | 471 |
| TVP-376 | II | DV4-E88 | 37 | 1 | 80 | 407 |
| TVP-376 | II | DV4-E88 | 37 | 3 | 58 | 337 |
| TVP-376 | II | DV4-E88 | 40 | 1 | 47 | 331 |
| TVP-986 | II | DV4-E75 | 37 | 1 | 91 | 769 |
| TVP-986 | II | DV4-E75 | 37 | 3 | 29 | 311 |
| TVP-986 | II | DV4-E75 | 40 | 1 | 47 | 417 |
| TVP-986 | II | DV4-E88 | 37 | 1 | 60 | 317 |
| TVP-986 | II | DV4-E88 | 37 | 3 | 25 | 171 |
| TVP-986 | II | DV4-E88 | 40 | 1 | 46 | 294 |
| P75-514 | S | DV4-E75 | 37 | 1 | >5,000 | >5,000 |
| P75-514 | S | DV4-E75 | 37 | 3 | 2,522 | >5,000 |
| P75-514 | S | DV4-E75 | 40 | 1 | 2,873 | >5,000 |
| P75-514 | S | DV4-E88 | 37 | 1 | 531 | 3,420 |
| P75-514 | S | DV4-E88 | 37 | 3 | 248 | 2,535 |
| P75-514 | S | DV4-E88 | 40 | 1 | 352 | 2,133 |
Increasing concentrations (5 to 5,000 mg/ml) of the indicated DV4 type-specific MAbs were premixed with 102 FFU of DENV-4 (strain H241, TVP-376, TVP-986, or P75-514) for 1 h at 37°C, 3 h at 37°C, or 1 h at 40°C. Virus-MAb mixtures were added to Vero cell monolayers for 1 h at 37°C prior to addition of a methylcellulose overlay. DENV-4-infected foci were stained with a human-mouse chimeric E18 MAb. EC50 and EC90 values were determined by using nonlinear regression models (GraphPad Prism). The results are the averages of three independent experiments performed in triplicate. Summary of genotypes: H-241 is genotype I; 1036, TVP-376, and TVP-986 are genotype II; P75-514 is the sylvatic genotype (S).
Increasing the duration of incubation at 37°C or raising the temperature to 40°C had a smaller impact on neutralization of the other DENV-4 strains by the DV4-E75 and DV4-E88 MAbs (Table 6). For example, against TVP-376 and TVP-986, the EC50 and EC90 values improved marginally from 1.5- to 3-fold after a 3-h incubation at 37°C or 1-h incubation at 40°C. We also observed only slightly improved inhibition of P75-514 by DV4-E75 under analogous conditions, with EC50 values of 2,522 and 2,873 ng/ml. For DV4-E88, less-than-2-fold changes in neutralization of P75-514 were observed under conditions of a longer incubation or higher temperature. Moreover, little improvement in neutralization of H-241 was observed under these conditions. As neutralization activities of several genetically diverse DENV-4 strains by DV4-E75 and DV4-E88 were not altered by preincubation conditions, their epitopes likely are exposed similarly or not at all on the different temperature-dependent conformational ensembles of the virus.
Effect of time and temperature on other DENV-4 MAbs.
As the neutralizing activities of the fusion loop MAbs against DENV-4 were considerably stronger after incubation at 40°C, we speculated that some of our other newly generated DENV-4 MAbs with modest neutralizing activities against DENV-4 1036 (Table 2) might show greater inhibitory activity under similar conditions, or against the relatively divergent DENV-4 P75-514 strain. To test this, we compared the inhibitory activities of neat and diluted (1/5 and 1/25) hybridoma supernatants after preincubation of DENV-4 for 1 h at 37°C or 40°C (Table 7). Many of the MAbs tested (DV4-E3, DV4-E4, DV4-E27, DV4-E29, DV4-E33, DV4-E40, DV4-E68, DV4-E72, DV4-E76, DV4-E78, and DV4-E87) showed improved inhibitory activity against one or both of the DENV-4 1036 or P75-514 strains, and this was most apparent at higher dilutions of supernatant. In comparison, some MAbs (DV4-E121 and DV4-E138) showed marginal improvement when higher temperatures of incubation were used.
Table 7.
Neutralization of DENV-4 strains 1036 (genotype I) and P75-514 (sylvatic genotype) based on a panel of DENV-4 MAbsa
| Virus | MAb | Temp (°C) | % neutralization of supernatant at dilution of: |
||
|---|---|---|---|---|---|
| Neat | 1:5 | 1:25 | |||
| 1036 | DV4-E3 | 37 | 94 | 58 | 29 |
| 1036 | DV4-E3 | 40 | 100 | 98 | 72 |
| P75-514 | DV4-E3 | 37 | 99 | 91 | 63 |
| P75-514 | DV4-E3 | 40 | 99 | 99 | 86 |
| 1036 | DV4-E4 | 37 | 84 | 23 | 1 |
| 1036 | DV4-E4 | 40 | 99 | 83 | 33 |
| P75-514 | DV4-E4 | 37 | 94 | 69 | 35 |
| P75-514 | DV4-E4 | 40 | 97 | 92 | 78 |
| 1036 | DV4-E27 | 37 | 68 | 24 | 6 |
| 1036 | DV4-E27 | 40 | 82 | 38 | 12 |
| P75-514 | DV4-E27 | 37 | 89 | 53 | 29 |
| P75-514 | DV4-E27 | 40 | 98 | 92 | 70 |
| 1036 | DV4-E29 | 37 | 69 | 36 | 15 |
| 1036 | DV4-E29 | 40 | 95 | 78 | 50 |
| P75-514 | DV4-E29 | 37 | 60 | 31 | 20 |
| P75-514 | DV4-E29 | 40 | 78 | 59 | 51 |
| 1036 | DV4-E33 | 37 | 90 | 70 | 32 |
| 1036 | DV4-E33 | 40 | 99 | 92 | 65 |
| P75-514 | DV4-E33 | 37 | 99 | 90 | 80 |
| P75-514 | DV4-E33 | 40 | 100 | 100 | 97 |
| 1036 | DV4-E40 | 37 | 79 | 40 | 23 |
| 1036 | DV4-E40 | 40 | 98 | 78 | 50 |
| P75-514 | DV4-E40 | 37 | 95 | 79 | 58 |
| P75-514 | DV4-E40 | 40 | 99 | 98 | 89 |
| 1036 | DV4-E68 | 37 | 63 | 32 | 16 |
| 1036 | DV4-E68 | 40 | 88 | 70 | 29 |
| P75-514 | DV4-E68 | 37 | 79 | 39 | 11 |
| P75-514 | DV4-E68 | 40 | 95 | 82 | 47 |
| 1036 | DV4-E72 | 37 | 16 | 0 | 0 |
| 1036 | DV4-E72 | 40 | 21 | 4 | 0 |
| P75-514 | DV4-E72 | 37 | 43 | 7 | 11 |
| P75-514 | DV4-E72 | 40 | 84 | 74 | 70 |
| 1036 | DV4-E76 | 37 | 91 | 49 | 23 |
| 1036 | DV4-E76 | 40 | 100 | 96 | 61 |
| P75-514 | DV4-E76 | 37 | 95 | 64 | 30 |
| P75-514 | DV4-E76 | 40 | 97 | 84 | 68 |
| 1036 | DV4-E78 | 37 | 79 | 32 | 6 |
| 1036 | DV4-E78 | 40 | 97 | 76 | 34 |
| P75-514 | DV4-E78 | 37 | 85 | 33 | 5 |
| P75-514 | DV4-E78 | 40 | 91 | 60 | 6 |
| 1036 | DV4-E87 | 37 | 68 | 27 | 9 |
| 1036 | DV4-E87 | 40 | 90 | 56 | 24 |
| P75-514 | DV4-E87 | 37 | 87 | 50 | 17 |
| P75-514 | DV4-E87 | 40 | 90 | 73 | 32 |
| 1036 | DV4-E121 | 37 | 100 | 93 | 57 |
| 1036 | DV4-E121 | 40 | 100 | 96 | 64 |
| P75-514 | DV4-E121 | 37 | 45 | 4 | 16 |
| P75-514 | DV4-E121 | 40 | 24 | 14 | 31 |
| 1036 | DV4-E138 | 37 | 28 | 7 | 3 |
| 1036 | DV4-E138 | 40 | 31 | 8 | 9 |
| P75-514 | DV4-E138 | 37 | 52 | 18 | 5 |
| P75-514 | DV4-E138 | 40 | 53 | 28 | 25 |
Hybridoma supernatant (neat or diluted) was incubated with 102 FFU of DENV-4 (strain 1036 or P75-514) for 1 h at 37°C or 40°C. Virus-MAb mixtures were added to Vero cell monolayers for 1 h at 37°C prior to addition of a methylcellulose overlay. DENV-infected foci were stained with a human-mouse chimeric E18 MAb. The percent neutralization was determined compared to medium alone. Results are representative of three independent experiments.
Epitope mapping of neutralizing DENV-4 MAbs.
To understand in greater detail the underlying basis of variation in inhibitory potential of the neutralizing MAbs, we mapped the domains and amino acid residues required for MAb binding by using multiple approaches.
(i) Domain recognition.
MAbs were screened initially for E protein domain recognition by using yeast that expressed DENV-4 DI-II, DIII, or the E ectodomain (DI-DII-DIII) (Table 2 and data not shown; see also Table S1 in the supplemental material). Seventeen MAbs in the panel bound to yeast expressing DI-II, 20 MAbs recognized DIII on yeast, and 10 MAbs failed to bind to either DI-DII or DIII yet still bound to the E ectodomain on yeast, suggesting a recognition site that possibly spans multiple domains. Thirty-one MAbs failed to bind any of the three yeast constructs, and three (nonneutralizing) MAbs were not tested. Of the 13 MAbs in our panel with relatively moderate to strong neutralizing activity against DENV-4 strains (including under higher-temperature or longer-incubation conditions), 3 bound epitopes in DI-II and 10 recognized sites in DIII.
(ii) Neutralization escape.
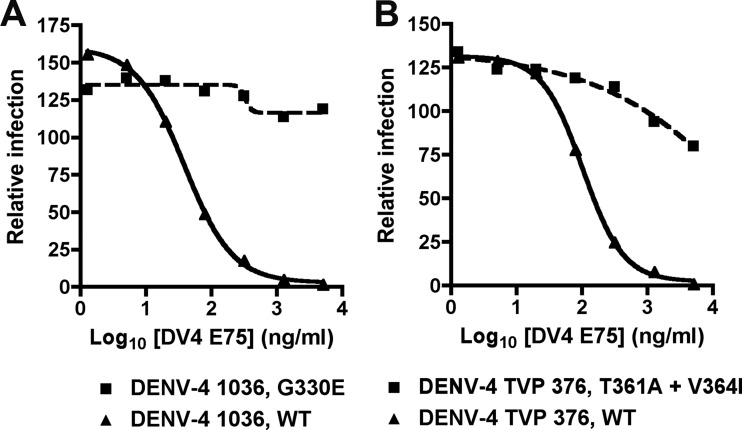
As two of our strongly inhibitory MAbs (DV4-E75 and DV4-E88) recognized a determinant on DENV-4 that was not present on yeast-displayed DI-DII or DIII, we selected neutralization escape mutants against strain DENV-4 1036 and DENV-4 TVP-376 to begin to define their epitopes. After sequential passage of DENV-4 1036 on Vero cells under DV4-E75 selection, we identified an escape variant (Fig. 3A) that was no longer neutralized efficiently. Viral sequence from plaque-purified escape variants was compared to virus passaged in parallel in the absence of MAb selection. All (6 of 6) escape variants uniquely had a G330E mutation, which corresponds to a site within the BC loop of DIII. When DENV-4 TVP-376 was passaged under DV4-E75 selection, we identified a different escape variant (Fig. 3B). Sequencing (6 of 6) purified plaques revealed two mutations, at T361A and V364I, which correspond to residues in the CC′ loop of DIII. For DV4-E88, we were unable to select a neutralization escape variant against either DENV-4 1036 or TVP-376.
Fig 3.
Selection of neutralization escape mutants of DV4-E75. The graphs show results of the focus reduction neutralization assays with bulk virus (DENV-4 1036 (A) or DENV-4 TVP-376 (B) obtained after six passages under selection with medium (WT) or DV4-E75 on Vero cells. Results are representative of at least three independent experiments performed in triplicate and were normalized to the no-MAb control.
(iii) Binding to mutant prM-E proteins in mammalian cells.
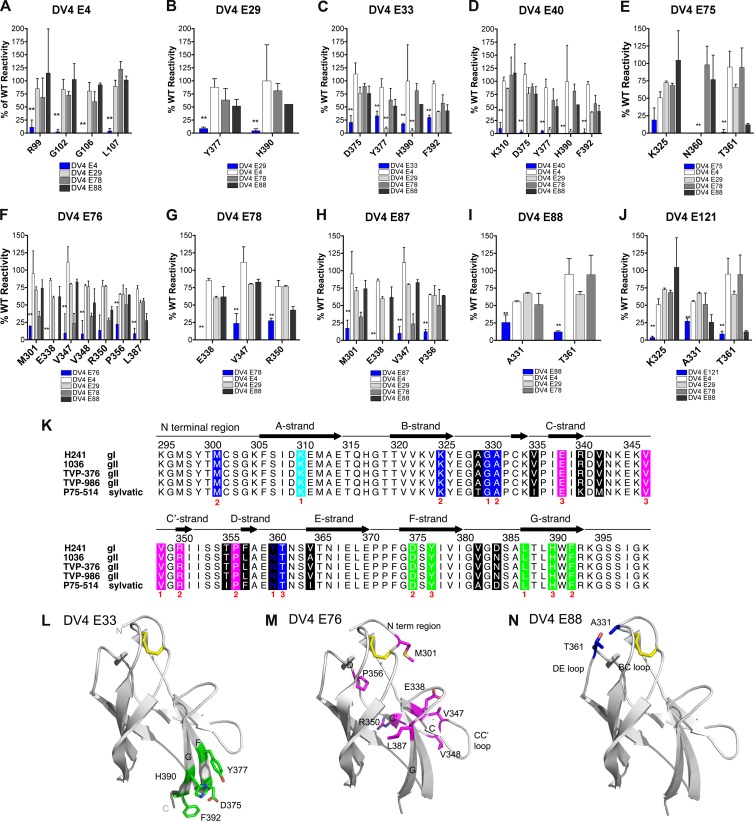
To gain additional insight as to where our strongly neutralizing anti-DENV-4 MAbs bound, we screened DV4-E4, DV4-E29, DV4-E33, DV4-E40, DV4-E75, DV4-E76, DV4-E78, DV4-E87, DV4-E88, and DV4-E121 against a comprehensive DENV-4 mutation library in which nearly every residue within prM and E (see Materials and Methods) was mutated individually to an alanine (alanine residues were mutated to serine). Each mutant protein clone was expressed in HEK-293T cells and assessed for intracellular MAb antibody binding by using high-throughput flow cytometry on permeabilized cells. The mean fluorescence was determined, and MAb reactivity to each mutant was calculated relative to reactivity to the WT DENV-4 prM-E protein (Fig. 4 and Table 8). Clones were identified as crucial for binding if they had low reactivity to individual MAbs but high reactivity to other DENV-4-specific control MAbs. This counterscreen strategy facilitated the exclusion of prM-E mutants that are globally or locally misfolded or that have an expression defect (53).
Fig 4.
Structural basis of neutralization of DENV-4 by MAbs. (A to J) Mapping of MAbs based on prM-E protein epitope mutants that were expressed intracellularly in HEK-293T cells (see Materials and Methods). For each panel, the blue bar reflects binding of the interrogated MAb. Although binding to a limited set of mutants is shown for each MAb, no differences were observed with other prM-E protein variants corresponding to changes in virtually any amino acids of the E protein (data not shown). The results are the averages of at least two independent experiments for each MAb with each mutant protein. Error bars indicate ranges, and asterisks denote differences that were statistically significant compared to binding to the WT prM-E protein (P < 0.05). (K) Sequence alignment of different DENV-4 genotypes and mapping of neutralizing MAbs. The sequences from five strains of DENV-4 DIII (strain H241, genotype 1; strain 1036, genotype II; strain TVP-376, genotype II; strain TVP-986, genotype II; strain P75-514, sylvatic) were aligned and assigned the secondary structure described for the DENV-4 DIII structure (PDB ID 3UAJ) for residues 295 to 400. Black blocks highlight the sites of genotypic variation. Results of the epitope mapping are denoted underneath in red according to the number of neutralizing MAbs in our panel that lost binding when a specific amino acid was mutated. The colored boxes correspond to specific neutralizing antibody and structural recognition determinates, as follows: lateral ridge, blue; A-strand, cyan; C-strand, CC′ loop, and C′-strand, magenta; F- and G-strands, green. (L to N) Localization of neutralizing epitopes on DENV-4 DIII, as determined by neutralization escape and staining of permeabilized cells expressing mutant prM-E proteins. The ribbon diagram shows the DENV-4 DIII structure (PDB ID 3UAJ) with amino acid residues that affect binding of neutralizing MAbs colored according to the major epitope. (L) DV4-E33 (F- and G-strand epitopes; green); (M) DV4-E76 (CC′ loop epitope; magenta); (N) DV4-E88 (lateral ridge epitope; blue).
Table 8.
Loss-of-binding residues identified by prM-E protein display on HEK 293T cells
| MAb | Domain | Amino acid residue(s)a | Epitopeb |
|---|---|---|---|
| DV4-E4 | DII | R99, G102, G106, L107 | Fusion loop |
| DV4-E29 | DIII | Y377 | F-strand |
| H390 | G-strand | ||
| DV4-E33 | DIII | D375, Y377 | F-strand |
| H390, F392 | G-strand | ||
| DV4-E40 | DIII | K310 | A-strand |
| D375, Y377 | F-strand | ||
| H390, F392 | G-strand | ||
| DV4-E75 | DIII | K325 | B-strand |
| N360, T361 | DE loop | ||
| DV4-E76 | DIII | M301 | N-terminal linker |
| E338, V347, V348 | B-strand | ||
| R350 | CC′ loop | ||
| P356 | D-strand | ||
| L387 | G-strand | ||
| DV4-E78 | DIII | E338, V347, R350 | CC′ loop |
| DV4-E87 | DIII | M301 | N-terminal linker |
| E338, V347 | CC′ loop | ||
| P356 | D-strand | ||
| DV4-E88 | DIII | A331 | BC loop |
| T361 | DE loop | ||
| DV4-E121 | DIII | K325 | B-strand |
| A331 | BC loop | ||
| T361 | DE loop |
Amino acids were determined based on loss of binding to the indicated MAb by using prM-E display of a shotgun alanine-scanning mutagenesis library (see Materials and Methods for details).
For MAb DV4-E4, four amino acids within the fusion loop of DII were identified as critical for recognition (Fig. 4A). When residue R99, G102 G106, or L107 was mutated to alanine, DV4-E4 binding activity was significantly lower than binding to the WT protein, whereas other MAbs showed strong reactivities against these fusion loop mutants. The remaining neutralizing MAbs in our panel that were subjected to mapping by this method showed loss of binding when residues in DIII were mutated that corresponded to previously described epitopes for mouse and human MAbs against other DENV serotypes (35–38, 44). These included residues in the N-terminal linker region (M301; DV4-E75 and DV4-E87), A-strand (K310; DV4-E40), B-strand (K325; DV4-E76 and DV4-E121), BC loop (A331; DV4-E88 and DV4-E121), CC′ loop (E338 [DV4-E76, DV4-E78, and DV4-E87]; V347 DV4-[E76 and DV4-E87]; V348 [DV4-E76]), D-strand (P356; DV4-E76 and DV4-E87), DE loop (T361; DV4-E75, DV4-E88, and DV4-E121), F-strand (D375 [DV4-E33 and DV4-E40]; Y377 [DV4-E29, DV4-E33, and DV4-E40]), and G-strand (L387 [DV4-E88]; H390 [DV4-E29, DV4-E33, and DV4-E40]; F392 [DV4-E33 and DV4-E40]) (Fig. 4B to J and L to N).
Three of the type-specific neutralizing MAbs (DV4-E75, DV4-E88, and DV4-E121) that mapped to epitopes within DIII showed variable inhibitory activities against strains corresponding to heterologous genotypes. Given this, we sequenced the prM and E genes from all five viral strains (H-241 [genotype I], 1036 [genotype II], TVP-376 [genotype II], TVP-986 [genotype II], and P75-514 ([sylvatic genotype]) and performed an alignment to assess strain variation in amino acid residues that were identified as important for MAb binding. Notably, we observed variation in the D-strand and the DE loop among the different strains (Fig. 4K). H-241 uniquely had a bulky tyrosine at position 360 instead of an asparagine, whereas H-241 and P75-514 both had a large phenylalanine at residue 357 compared to the genotype II strains which encoded a leucine. These sequence changes in combination with the mapping data likely explain the loss of neutralizing activities of DV4-E75, DV4-E88, and DV4-E121 against the genotype II and/or the sylvatic strain.
Protective activities in mice of type-specific and cross-reactive anti-DENV-4 MAbs.
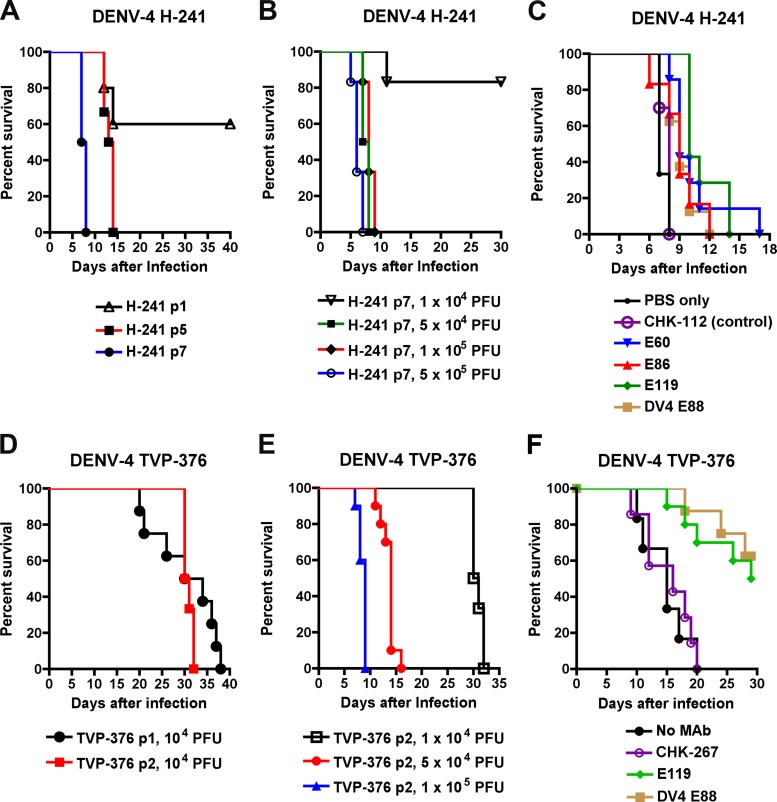
To define the relationship between neutralization in cell culture and protection in vivo, we developed two new mouse models of DENV-4 infection, corresponding to infection with strains of two different genotypes. While prior studies have published mouse models of DENV-1, DENV-2, and DENV-3 infection, no lethal peripheral inoculation model of adult mice with DENV-4 has been published. Based on experiments with other DENV serotypes (54, 62), we infected IFN-αβR−/− × IFN-γR−/− immunodeficient AG129 mice with 104 to 106 PFU of different DENV-4 strains (1036, H-241, TVP-986, and TVP-376). Although mice were followed for 40 days, no morbidity or mortality was observed after infection with 1036, H-241 or TVP-986, and only some mice became ill after infection with TVP-376 infection, with this occurring late in the time course. We passaged the H-241 genotype I strain between mice and C6/36 insect cells, as was done previously to generate a mouse-adapted DENV-2 strain (62). After several passages, we identified an isolate that exhibited virulence in AG129 mice, resulting in 100% lethality at day 8 after infection with 5 × 104 PFU of virus (Fig. 5A and B). We used this model to test the efficacy of several of our MAbs in vivo, even though in general, H-241 was relatively resistant to neutralization in vitro compared to the other DENV-4 strains (Tables 5 and 6). A single dose (100 μg) of DV4-E88 or three different cross-reactive fusion loop MAbs (E60, E86, and E119) prior to infection provided only modest protection, as the mean survival time was extended by 2 to 3 days (P < 0.004) compared to animals who received PBS or the isotype control (CHK-112) MAb (Fig. 5C). However, these MAbs failed to decrease the mortality rate after infection with DENV-4 H-241, even when higher doses (500 μg) were used (data not shown).
Fig 5.
Prophylactic efficacies of neutralizing antibodies in mice after DENV-4 infection. (A) Development of the AG129 mouse model with DENV-4 H-241. Alternating passage of DENV-4 H-241 between mice and C6/36 cells was performed (see Materials and Methods). Virus (1 × 105 PFU) generated from the indicated passage (p) cycle was used for intravenous injection of AG129 mice. Animals were monitored for survival over 40 days. Data were generated from two independent experiments, with 5 to 6 mice per experimental group. (B) Dose response of H-241 p7 virus in AG129 mice. Virus was administered by intravenous injection, and survival was followed for 30 days. Data were generated from two independent experiments with 6 mice per experimental group. (C) AG129 mice were passively transferred via the intraperitoneal route either PBS or 100 μg of CHK-112 (isotype control MAb), E60, E86, E119, or DV4-E88 1 day before infection with 5 × 104 PFU of H-241 (genotype I) by an intravenous route. Mice were monitored for survival for 17 days after infection. The survival curves were constructed from data from three independent experiments. The number of animals ranged from 6 to 14 per group. (D) Development of the AG129 mouse model with DENV-4 TVP-376. Successive passage of DENV-4 TVP-376 between mice was performed (see Materials and Methods). Virus (5 × 104 PFU) generated from the indicated passage (p) cycle was used for intravenous injection of AG129 mice. Animals were monitored for survival over 40 days. Data were generated from two independent experiments with 8 to 10 mice per experimental group. (E) Dose response of TVP-376 p2 virus in AG129 mice. Virus was administered by intravenous injection, and survival was followed for 30 days. Data were generated from two independent experiments with 6 to 10 mice per experimental group. (F) AG129 mice were passively transferred via the intraperitoneal route either no MAb or 250 μg of CHK-267 (isotype control MAb), E119, or DV4-E88 1 day before infection with 1 × 105 PFU of TVP-376 (genotype II) by the intravenous route. Mice were monitored for survival for 30 days after infection. The survival curves were constructed from data from three independent experiments. The number of animals ranged from 6 to 9 per group.
Because the DENV-4 H-241 was resistant to neutralization and protection by several MAbs, we passaged the TVP-376 (genotype I) strain for further analysis. After one passage in mice, an isolate was obtained that caused 100% lethality in AG129 mice (Fig. 5D). After two successive passages, inoculation with a higher dose (p2; 105 PFU) resulted in more rapid death (Fig. 5E). A single dose (250 μg) of DV4-E88 or of the cross-reactive fusion loop MAb E119 prior to infection provided significant protection (50 to 63%; P < 0.001) against DENV-4 TVP-376 compared to PBS or the isotype control (CHK-267) MAb (Fig. 5F). Thus, for DENV-4 strains H-241 and TVP-376, the in vivo protection results correlated with focus reduction neutralization titers of type-specific or cross-reactive MAbs after preincubation at 37°C.
DISCUSSION
Despite the induction of neutralizing antibodies against all four DENV serotypes, the most advanced tetravalent live-attenuated DENV vaccine candidate for humans showed a poor 30% overall efficacy rate in a recently published phase 2b clinical trial (5). Thus, there is a pressing need for an improved understanding of the correlates of protection, especially those of neutralizing antibodies. To define in greater detail the structural and molecular correlates of antibody protection against DENV, we and others have generated and evaluated panels of mouse and human MAbs for their neutralizing activities, epitope binding patterns, and in vivo protective capacities (35–38, 44, 49, 63–69). In this study, we developed a new panel of anti-DENV-4 MAbs by using an immunization strategy with infectious DENV-4 that had successfully generated neutralizing antibodies against DENV-1 (35), DENV-2 (38), DENV-3 (37), and WNV (52). Despite screening 6,000 hybridomas as part of 10 independent fusions, unexpectedly, we identified a relatively small number of MAbs that inhibited DENV-4, and none that reached the potency that was observed with MAbs against other flaviviruses. Remarkably, we observed strain- and temperature-dependent neutralization by MAbs that mapped to distinct epitopes on DII and DIII of the E protein. This was most apparent with DII fusion loop MAbs, which strongly neutralized DENV-1, DENV-2, and DENV-3 yet only weakly neutralized five different DENV-4 strains despite complete epitope sequence conservation. Thus, DENV-4 may have a unique virion conformation relative to other DENV serotypes that results in poor fusion loop accessibility.
Neutralization of DENV-4 by MAbs.
While many neutralizing MAbs against DENV-1, DENV-2, and DENV-3 have been reported, curiously, fewer inhibitory MAbs against DENV-4 have been described. Of the few type-specific strongly neutralizing anti-DENV-4 MAbs that have been characterized, fewer data exist regarding strain dependence, epitope specificity, or functional activity in vivo. A study by Morens et al. (40) produced 19 mouse MAbs against DENV-4 (strain 4328-S): only one (UH-6C3) was serotype specific and strongly neutralizing (PRNT50 of ∼50 ng/ml), and no additional functional testing was performed with different DENV-4 strains. Forty-three MAbs against DENV-4 (strain H-241) were generated by Yamanaka et al. (41): only two were serotype specific (D4-II-12B2 and D4-I-11D11) and neutralizing, and no strain dependence or mapping was performed. Two type-specific anti-DENV-4 chimpanzee MAbs (5D9 and 5H2) were generated after immunization with DENV-4 (strain 814669; Caribbean); these MAbs inhibited infection of the homologous strain (PRNT50 of 580 and 240 ng/ml, respectively) (42). The 5H2 MAb, which localized to an epitope in domain I (amino acid residues K174 and P176) of the E protein (43), neutralized two other DENV-4 isolates (H-241 [Philippines] and 341750 [Caribbean]) from similar and distinct geographic regions (42) and also protected suckling mice from lethal DENV-4 H-241 infection when administered as prophylaxis. A recombinant variant of 5H2 lacking the ability to bind Fcγ receptors protected monkeys from viremia after challenge with DENV-4 814669. Only two human type-specific neutralizing MAbs (DV16.8 and DV22.3) against DENV-4 have been reported (44). These MAbs were generated from a subject 200 days after primary DENV-4 infection, recognized an epitope in DI-DII, and neutralized efficiently (EC50 of 6 to 39 ng/ml by flow cytometric assay) infection by a DENV-4 vaccine strain. Why have so relatively few type-specific DENV-4-neutralizing MAbs been described? The answer remains uncertain, although it could reflect a bias of study in the DENV field toward detailed investigation with other serotypes, or a difficulty in identifying strongly inhibitory anti-DENV-4 MAbs when using conventional neutralization assays.
An intriguing finding of our study was that cross-reactive DII-specific fusion loop MAbs, which efficiently inhibited infection of other DENV serotypes, poorly neutralized all five DENV-4 strains after a standard 37°C 1-h preincubation. Thus, under these experimental conditions, fusion loop MAbs neutralized DENV-4 inefficiently, in a manner analogous to that seen with mature forms of WNV (45, 70, 71). However, lengthening the time or increasing the temperature of incubation resulted in improved neutralization of DENV-4 strains by all fusion loop MAbs tested. This result suggests that under standard 37°C preincubation conditions, the fusion-loop epitope on DENV-4, in contrast to other DENV serotypes, is not sufficiently accessible to reach a stoichiometry of binding required for neutralization (72). However, increasing the time and/or temperature of preincubation enhances exposure of the fusion loop epitope, which allows MAb binding and DENV-4 neutralization. What is unique about DENV-4 compared to the other DENV serotypes? We hypothesize that two interrelated physical processes govern this: (i) DENV-4 “breathing” creates unique structural ensembles (57, 58, 60) compared to other DENV serotypes under standard incubation conditions, such that the fusion loop epitope remains inaccessible; (ii) DENV-4 is more mature than other DENV serotypes. The maturation state of the virion, which is modified by cleavage of prM by furin-like proteases in the trans-Golgi network, modulates the sensitivity to neutralization by many classes of MAbs, including those binding fusion loop-specific epitopes. Given that the neutralization phenotype of fusion loop MAbs of DENV-4 resembles that seen with WNV (45, 58, 71), we speculate that the structure and/or behavior of the DENV-4 virion may be more similar to WNV than other DENV serotypes.
DENV-4 MAb epitope exposure.
Many of our anti-DENV-4-neutralizing MAbs that mapped to DIII also showed time- and temperature-dependent improvement in neutralization of DENV-4 1036, with the exception of three, DV4-E75, DV4-E88, and DV4-E121. These three MAbs shared a loss-of-binding phenotype at residues T361 in the DE loop in contrast to the others, which recognized sites in BC loop, CC′ loop, FG loop, or G-strand. These data suggest that for DENV-4 1036, the DE loop epitope is highly accessible on DIII. Of note, several of the DIII-specific anti-DENV-4 MAbs showed less temperature-dependent neutralization with the sylvatic strain P75-514, which suggests there may be strain- or genotype-dependent differences in virion structure, maturation, or ensembles of conformations.
Variation in neutralization of different genotypes within a serotype by type-specific antibodies against DENV-1, DENV-2, and DENV-3 suggests that the limited amino acid dissimilarity that occurs within a DENV serotype can impact the broad-spectrum inhibitory activity of some antibodies (35–37, 60). Indeed, this property may apply to polyclonal responses and has been suggested as an explanation for the poor protective efficacy of a live tetravalent DENV vaccine against a circulating DENV-2 strain (5, 73). Our results confirmed this observation for DENV-4. DV4-E75 and DV4-E88 strongly neutralized genotype II strains (1036, TVP-376, and TVP-986) but either failed to or only weakly inhibited infection with genotype I or sylvatic strains. Analogously, DV4-E121 showed a markedly reduced capacity to neutralize infection of the sylvatic strain. Sequence alignment of all five strains revealed genotypic variation in amino acids of the D-strand and DE loop, which corresponds to residues that contribute to binding of these MAbs. The DE loop is a solvent-accessible region on DIII, which has been implicated in the binding of strongly neutralizing antibodies against other flaviviruses, including WNV (74, 75). Based on these data and published results with other MAbs, we suggest that it may be important to assess whether polyclonal antibody responses in the context of a vaccine generated against an individual strain will neutralize infection of heterologous genotypes effectively. This may be especially true for DENV-4, given the variation of both type-specific and cross-reactive MAbs in neutralizing infections with strains corresponding to different genotypes. Indeed, sera from humans immunized with a DENV-4 live-attenuated vaccine candidate (DENV4Δ30) showed weak to modest neutralization (PRNT60 of 20 to 40 at 180 days) of heterologous DENV-4 strains, with some individuals failing to neutralize infection at all of the strains corresponding to heterologous DENV-4 genotypes (32).
Some of our DENV-4 DIII-specific MAbs (e.g., DV4-E75 and DV4-E88) mapped to DIII by neutralization escape or shotgun mutagenesis yet failed to bind DIII when displayed on the surface of yeast. Although structural studies are warranted to definitively resolve this apparent paradox, we suggest three possible explanations. (i) DIII displayed on yeast fails to adopt a native structure and, thus, does not present all MAb epitopes. While possible, this seems less likely, given that the vast majority of our DIII-specific MAbs against DENV-1, DENV-2, DENV-3, and WNV have recognized a similarly designed yeast-displayed DIII (35, 37, 38, 49, 52). Moreover, even in our current study, most of the DENV-4 DIII-specific neutralizing and nonneutralizing MAbs recognized DIII when displayed on yeast. (ii) DV4-E75 and DV4-E88 MAbs have recognition sites in DIII (identified by neutralization escape or shotgun mutagenesis) and other regions of the E protein that would not be presented on isolated DIII proteins. Indeed, prior studies with neutralizing anti-WNV or anti-DENV human MAbs identified composite structural epitopes comprised of sites in multiple domains on adjacent E protein dimers (69, 76, 77). (iii) These MAbs require bivalent interactions to bind DIII, such that monovalent binding on the surfaces of yeast cells is too weak in nature to detect. Indeed, we recently identified bivalent neutralizing MAbs against DENV-1 that map to adjacent epitopes on DIII (M. Barrow, M. Diamond, and D. Fremont, unpublished observations).
Mouse models of DENV-4 infection.
In this study, we developed two new models of lethal DENV-4 infection via a peripheral route in AG129 mice with virus strains corresponding to genotypes I (H-241) and II (TVP-376) and used these to assess the protective efficacy of selected cross-reactive and type-specific anti-DENV-4 MAbs. Prior to this, only one strain of DENV-1 (West Pac-74) (35) and two strains of DENV-2 (D2S10 and D2Y98P) (62, 78, 79) had been reported to cause lethal infection in AG129 mice after peripheral inoculation, with the DENV-2 strains promoting a vascular leakage phenotype that to some extent resembled the DSS seen in humans. In preliminary studies using high infectious doses of the two DENV-4 strains, we saw a similar rapid death phenotype consistent with the shock syndrome that is observed with mouse-adapted DENV-2 strains (46, 62, 80). In comparison, at lower DENV-4 doses, the mice survived longer and developed signs of central nervous system disease, which rarely occurs in human disease (81). Two mouse-adapted strains (DENV-2 and DENV-3) have been reported to cause lethal infection in wild-type mice (82–84), although mechanistically how this occurs remains unknown, as DENV isolates uniformly fail to antagonize key innate immune restriction signaling molecules in mouse cells and in mice, including MITA (also known as STING) (85, 86) and STAT2 (87, 88). Passive transfer studies in our mouse models of DENV-4 infection correlated with in vitro neutralization data: poor inhibitory activity in vitro against DENV-4 H-241 was associated with only modest protection in vivo, whereas greater neutralizing activity against TVP-376 corresponded to more complete protection against lethal infection. Remarkably, even the highly cross-reactive fusion loop-specific MAbs that neutralized all other DENV serotypes and had postexposure therapeutic activity against DENV-2 infection in vivo (46, 56) showed a differential ability to protect against the two DENV-4 strains in mice. Thus, the two DENV-4 models in AG129 mice likely will be useful for future therapeutic testing, as currently most preclinical studies rely exclusively on protection data from the DENV-2 D2S10 strain. Future studies will be directed at defining the genetic determinants that enabled these DENV-4 strains to cause more severe disease in mice.
In summary, we developed a panel of 81 new DENV-4 MAbs and examined their cross-reactivities, epitope specificities, neutralization potential levels in cell culture, and the protective capacities of a subset of these MAbs in vivo. Our studies established the complexity of MAb recognition against the genetically divergent DENV-4 and suggest that differences in epitope exposure and possibly virion structure relative to other DENV serotypes impact antibody neutralization. These results may be directly relevant for understanding serotype-specific protective efficacy in ongoing human clinical trials with several live-attenuated tetravalent DENV vaccine candidates.
Supplementary Material
ACKNOWLEDGMENTS
We thank A. de Silva, R. Tesh, and the World Arbovirus Collection (University of Texas Medical Branch, Galveston, TX) for providing the DENV strains and C. Nelson for cloning the DIII of DENV-4. We thank A. Barrett for discussion of unpublished studies on DENV-4.
This work was supported by the Burroughs Wellcome Fund, NIH grants R01-AI077955 and U01-AI061373, and NIAID contract HHSN272200900055C.
Footnotes
Published ahead of print 19 June 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.01314-13.
REFERENCES
- 1. Durbin AP, Whitehead SS. 2010. Dengue vaccine candidates in development. Curr. Top. Microbiol. Immunol. 338:129–143 [DOI] [PubMed] [Google Scholar]
- 2. Guy B, Barrere B, Malinowski C, Saville M, Teyssou R, Lang J. 2011. From research to phase III: preclinical, industrial and clinical development of the Sanofi Pasteur tetravalent dengue vaccine. Vaccine 29:7229–7241 [DOI] [PubMed] [Google Scholar]
- 3. Gubler DJ. 2012. The economic burden of dengue. Am. J. Trop. Med. Hyg. 86:743–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O, Myers MF, George DB, Jaenisch T, Wint GR, Simmons CP, Scott TW, Farrar JJ, Hay SI. 2013. The global distribution and burden of dengue. Nature 496:504–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sabchareon A, Wallace D, Sirivichayakul C, Limkittikul K, Chanthavanich P, Suvannadabba S, Jiwariyavej V, Dulyachai W, Pengasa K, Wartel TA, Moureau A, Saville M, Bouckenooghe A, Viviani S, Tornieporth NG, Lang J. 2012. Protective efficacy of the recombinant, live-attenuated, CYD tetravalent dengue vaccine in Thai schoolchildren: a randomised, controlled phase 2b trial. Lancet 380:1559–1567 [DOI] [PubMed] [Google Scholar]
- 6. Kuhn RJ, Zhang W, Rossmann MG, Pletnev SV, Corver J, Lenches E, Jones CT, Mukhopadhyay S, Chipman PR, Strauss EG, Baker TS, Strauss JH. 2002. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell 108:717–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhang X, Ge P, Yu X, Brannan JM, Bi G, Zhang Q, Schein S, Zhou ZH. 2013. Cryo-EM structure of the mature dengue virus at 3.5-Å resolution. Nat. Struct. Mol. Biol. 20:105–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kostyuchenko VA, Zhang Q, Tan JL, Ng TS, Lok SM. 2013. Immature and mature dengue serotype 1 virus structures provide insight into the maturation process. J. Virol. 87:7700–7707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang X, Sheng J, Plevka P, Kuhn RJ, Diamond MS, Rossmann MG. 2013. Dengue structure differs at the temperatures of its human and mosquito hosts. Proc. Natl. Acad. Sci. U. S. A. 110:6795–6799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fibriansah G, Ng TS, Kostyuchenko VA, Lee J, Lee S, Wang J, Lok SM. 2013. Structural changes of dengue virus when exposed to a temperature of 37°C. J. Virol. 87:7585–7592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang Y, Corver J, Chipman PR, Zhang W, Pletnev SV, Sedlak D, Baker TS, Strauss JH, Kuhn RJ, Rossmann MG. 2003. Structures of immature flavivirus particles. EMBO J. 22:2604–2613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cherrier MV, Kaufmann B, Nybakken GE, Lok SM, Warren JT, Chen BR, Nelson CA, Kostyuchenko VA, Holdaway HA, Chipman PR, Kuhn RJ, Diamond MS, Rossmann MG, Fremont DH. 2009. Structural basis for the preferential binding of immature flaviviruses by a fusion-loop specific antibody. EMBO 28:3269–3276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wengler G. 1989. Cell-associated West Nile flavivirus is covered with E+pre-M protein heterodimers which are destroyed and reorganized by proteolytic cleavage during virus release. J. Virol. 63:2521–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li L, Lok SM, Yu IM, Zhang Y, Kuhn RJ, Chen J, Rossmann MG. 2008. The flavivirus precursor membrane-envelope protein complex: structure and maturation. Science 319:1830–1834 [DOI] [PubMed] [Google Scholar]
- 15. Yu IM, Zhang W, Holdaway HA, Li L, Kostyuchenko VA, Chipman PR, Kuhn RJ, Rossmann MG, Chen J. 2008. Structure of the immature dengue virus at low pH primes proteolytic maturation. Science 319:1834–1837 [DOI] [PubMed] [Google Scholar]
- 16. Yu IM, Holdaway HA, Chipman PR, Kuhn RJ, Rossmann MG, Chen J. 2009. Association of the pr peptides with dengue virus at acidic pH blocks membrane fusion. J. Virol. 83:12101–12107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Modis Y, Ogata S, Clements D, Harrison SC. 2003. A ligand-binding pocket in the dengue virus envelope glycoprotein. Proc. Natl. Acad. Sci. U. S. A. 100:6986–6991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Modis Y, Ogata S, Clements D, Harrison SC. 2004. Structure of the dengue virus envelope protein after membrane fusion. Nature 427:313–319 [DOI] [PubMed] [Google Scholar]
- 19. Modis Y, Ogata S, Clements D, Harrison SC. 2005. Variable surface epitopes in the crystal structure of dengue virus type 3 envelope glycoprotein. J. Virol. 79:1223–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nayak V, Dessau M, Kucera K, Anthony K, Ledizet M, Modis Y. 2009. Crystal structure of dengue virus type 1 envelope protein in the postfusion conformation and its implications for membrane fusion. J. Virol. 83:4338–4344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mondotte JA, Lozach PY, Amara A, Gamarnik AV. 2007. Essential role of dengue virus envelope protein N glycosylation at asparagine-67 during viral propagation. J. Virol. 81:7136–7148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pokidysheva E, Zhang Y, Battisti AJ, Bator-Kelly CM, Chipman PR, Xiao C, Gregorio GG, Hendrickson WA, Kuhn RJ, Rossmann MG. 2006. Cryo-EM reconstruction of dengue virus in complex with the carbohydrate recognition domain of DC-SIGN. Cell 124:485–493 [DOI] [PubMed] [Google Scholar]
- 23. Tassaneetrithep B, Burgess T, Granelli-Piperno A, Trumpfheller C, Finke J, Sun W, Eller M, Pattanapanyasat K, Sarasombath S, Birx D, Steinman RM, Schlesinger S, Marovich M. 2003. DC-SIGN (CD209) mediates dengue virus infection of human dendritic cells. J. Exp. Med. 197:823–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Navarro-Sanchez E, Altmeyer R, Amara A, Schwartz O, Fieschi F, Virelizier JL, Arenzana-Seisdedos F, Despres P. 2003. Dendritic-cell-specific ICAM3-grabbing non-integrin is essential for the productive infection of human dendritic cells by mosquito-cell-derived dengue viruses. EMBO Rep. 4:723–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bhardwaj S, Holbrook M, Shope RE, Barrett AD, Watowich SJ. 2001. Biophysical characterization and vector-specific antagonist activity of domain III of the tick-borne flavivirus envelope protein. J. Virol. 75:4002–4007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Volk DE, Beasley DW, Kallick DA, Holbrook MR, Barrett AD, Gorenstein DG. 2004. Solution structure and antibody binding studies of the envelope protein domain III from the New York strain of West Nile virus. J. Biol. Chem. 279:38755–38761 [DOI] [PubMed] [Google Scholar]
- 27. Yu S, Wuu A, Basu R, Holbrook MR, Barrett AD, Lee JC. 2004. Solution structure and structural dynamics of envelope protein domain III of mosquito- and tick-borne flaviviruses. Biochemistry 43:9168–9176 [DOI] [PubMed] [Google Scholar]
- 28. Rico-Hesse R. 1990. Molecular evolution and distribution of dengue viruses type 1 and 2 in nature. Virology 174:479–493 [DOI] [PubMed] [Google Scholar]
- 29. Holmes EC, Twiddy SS. 2003. The origin, emergence and evolutionary genetics of dengue virus. Infect. Genet. Evol. 3:19–28 [DOI] [PubMed] [Google Scholar]
- 30. Rossi SL, Nasar F, Cardosa J, Mayer SV, Tesh RB, Hanley KA, Weaver SC, Vasilakis N. 2012. Genetic and phenotypic characterization of sylvatic dengue virus type 4 strains. Virology 423:58–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vasilakis N, Fokam EB, Hanson CT, Weinberg E, Sall AA, Whitehead SS, Hanley KA, Weaver SC. 2008. Genetic and phenotypic characterization of sylvatic dengue virus type 2 strains. Virology 377:296–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Durbin AP, Mayer SV, Rossi SL, Amaya-Larios IY, Ramos-Castaneda J, Eong Ooi E, Jane Cardosa M, Munoz-Jordan JL, Tesh RB, Messer WB, Weaver SC, Vasilakis N. 2013. Emergence potential of sylvatic dengue virus type 4 in the urban transmission cycle is restrained by vaccination and homotypic immunity. Virology 439:34–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rothman AL. 2004. Dengue: defining protective versus pathologic immunity. J. Clin. Invest. 113:946–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lanciotti RS, Gubler DJ, Trent DW. 1997. Molecular evolution and phylogeny of dengue-4 viruses. J. Gen. Virol. 78:2279–2284 [DOI] [PubMed] [Google Scholar]
- 35. Shrestha B, Brien JD, Sukupolvi-Petty S, Austin SK, Edeling MA, Kim T, O'Brien KM, Nelson CA, Johnson S, Fremont DH, Diamond MS. 2010. The development of therapeutic antibodies that neutralize homologous and heterologous genotypes of dengue virus type 1. PLoS Pathog. 6(4):e1000823. 10.1371/journal.ppat.1000823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wahala WM, Donaldson EF, de Alwis R, Accavitti-Loper MA, Baric RS, de Silva AM. 2010. Natural strain variation and antibody neutralization of Dengue serotype 3 viruses. PLoS Pathog. 6(3):e1000821. 10.1371/journal.ppat.1000821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Brien JD, Austin SK, Sukupolvi-Petty S, O'Brien KM, Johnson S, Fremont DH, Diamond MS. 2010. Genotype specific neutralization and protection by antibodies against dengue virus type 3. J. Virol. 84:10630–10643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sukupolvi-Petty S, Austin SK, Engle M, Brien JD, Dowd KA, Williams KL, Johnson S, Rico-Hesse R, Harris E, Pierson TC, Fremont DH, Diamond MS. 2010. Structure and function analysis of therapeutic monoclonal antibodies against dengue virus type 2. J. Virol. 84:9227–9239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Whitehead SS, Blaney JE, Durbin AP, Murphy BR. 2007. Prospects for a dengue virus vaccine. Nat. Rev. Microbiol. 5:518–528 [DOI] [PubMed] [Google Scholar]
- 40. Morens DM, Venkateshan CN, Halstead SB. 1987. Dengue 4 virus monoclonal antibodies identify epitopes that mediate immune infection enhancement of dengue 2 viruses. J. Gen. Virol. 68:91–98 [DOI] [PubMed] [Google Scholar]
- 41. Yamanaka A, Kosugi S, Konishi E. 2008. Infection-enhancing and -neutralizing activities of mouse monoclonal antibodies against dengue type 2 and 4 viruses are controlled by complement levels. J. Virol. 82:927–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Men R, Yamashiro T, Goncalvez AP, Wernly C, Schofield DJ, Emerson SU, Purcell RH, Lai CJ. 2004. Identification of chimpanzee Fab fragments by repertoire cloning and production of a full-length humanized immunoglobulin G1 antibody that is highly efficient for neutralization of dengue type 4 virus. J. Virol. 78:4665–4674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lai CJ, Goncalvez AP, Men R, Wernly C, Donau O, Engle RE, Purcell RH. 2007. Epitope determinants of a chimpanzee dengue virus type 4 (DENV-4)-neutralizing antibody and protection against DENV-4 challenge in mice and rhesus monkeys by passively transferred humanized antibody. J. Virol. 81:12766–12774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Beltramello M, Williams KL, Simmons CP, Macagno A, Simonelli L, Quyen NT, Sukupolvi-Petty S, Navarro-Sanchez E, Young PR, de Silva AM, Rey FA, Varani L, Whitehead SS, Diamond MS, Harris E, Lanzavecchia A, Sallusto F. 2010. The human immune response to Dengue virus is dominated by highly cross-reactive antibodies endowed with neutralizing and enhancing activity. Cell Host Microbe 8:271–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Oliphant T, Nybakken GE, Engle M, Xu Q, Nelson CA, Sukupolvi-Petty S, Marri A, Lachmi BE, Olshevsky U, Fremont DH, Pierson TC, Diamond MS. 2006. Antibody recognition and neutralization determinants on domains I and II of West Nile virus envelope protein. J. Virol. 80:12149–12159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Balsitis SJ, Williams KL, Lachica R, Flores D, Kyle JL, Mehlhop E, Johnson S, Diamond MS, Beatty PR, Harris E. 2010. Lethal antibody enhancement of dengue disease in mice is prevented by Fc modification. PLoS Pathog. 6(2):e1000790. 10.1371/journal.ppat.1000790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Temporao JG, Penna GO, Carmo EH, Coelho GE, do Socorro Silva Azevedo R, Teixeira Nunes MR, da Costa Vasconcelos PF. 2011. Dengue virus serotype 4, Roraima State, Brazil. Emerg. Infect. Dis. 17:938–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Vasilakis N, Weaver SC. 2008. The history and evolution of human dengue emergence. Adv. Virus Res. 72:1–76 [DOI] [PubMed] [Google Scholar]
- 49. Sukupolvi-Petty S, Austin SK, Purtha WE, Oliphant T, Nybakken G, Schlesinger JJ, Roehrig JT, Gromowski GD, Barrett AD, Fremont DH, Diamond MS. 2007. Type- and subcomplex-specific neutralizing antibodies against domain III of dengue virus type 2 envelope protein recognize adjacent epitopes. J. Virol. 81:12816–12826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Harlow E, Lane D. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 51. Fuchs A, Pinto AK, Schwaeble WJ, Diamond MS. 2011. The lectin pathway of complement activation contributes to protection from West Nile virus infection. Virology 412:101–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Oliphant T, Engle M, Nybakken G, Doane C, Johnson S, Huang L, Gorlatov S, Mehlhop E, Marri A, Chung KM, Ebel GD, Kramer LD, Fremont DH, Diamond MS. 2005. Development of a humanized monoclonal antibody with therapeutic potential against West Nile virus. Nat. Med. 11:522–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Paes C, Ingalls J, Kampani K, Sulli C, Kakkar E, Murray M, Kotelnikov V, Greene TA, Rucker JB, Doranz BJ. 2009. Atomic-level mapping of antibody epitopes on a GPCR. J. Am. Chem. Soc. 131:6952–6954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Johnson AJ, Roehrig JT. 1999. New mouse model for dengue virus vaccine testing. J. Virol. 73:783–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sabo MC, Luca VC, Prentoe J, Hopcraft SE, Blight KJ, Yi M, Lemon SM, Ball JK, Bukh J, Evans MJ, Fremont DH, Diamond MS. 2011. Neutralizing monoclonal antibodies against hepatitis C virus E2 protein bind discontinuous epitopes and inhibit infection at a postattachment step. J. Virol. 85:7005–7019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Williams KL, Sukupolvi-Petty S, Beltramello M, Johnson S, Sallusto F, Lanzavecchia A, Diamond MS, Harris E. 2013. Therapeutic efficacy of antibodies lacking FcγR against lethal dengue virus infection is due to neutralizing potency and blocking of enhancing antibodies. PLoS Pathog. 9(2):e1003157. 10.1371/journal.ppat.1003157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lok SM, Kostyuchenko V, Nybakken GE, Holdaway HA, Battisti AJ, Sukupolvi-Petty S, Sedlak D, Fremont DH, Chipman PR, Roehrig JT, Diamond MS, Kuhn RJ, Rossmann MG. 2008. Binding of a neutralizing antibody to dengue virus alters the arrangement of surface glycoproteins. Nat. Struct. Mol. Biol. 15:312–317 [DOI] [PubMed] [Google Scholar]
- 58. Dowd KA, Jost CA, Durbin AP, Whitehead SS, Pierson TC. 2011. A dynamic landscape for antibody binding modulates antibody-mediated neutralization of West Nile virus. PLoS Pathog. 7(6):e1002111. 10.1371/journal.ppat.1002111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sabo MC, Luca VC, Ray SC, Bukh J, Fremont DH, Diamond MS. 2012. Hepatitis C virus epitope exposure and neutralization by antibodies is affected by time and temperature. Virology 422:174–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Austin SK, Dowd KA, Shrestha B, Nelson CA, Edeling MA, Johnson S, Pierson TC, Diamond MS, Fremont DH. 2012. Structural basis of differential neutralization of DENV-1 genotypes by an antibody that recognizes a cryptic epitope. PLoS Pathog. 8(10):e1002930. 10.1371/journal.ppat.1002930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Halstead SB, Venkateshan CN, Gentry MK, Larsen LK. 1984. Heterogeneity of infection enhancement of dengue 2 strains by monoclonal antibodies. J. Immunol. 132:1529–1532 [PubMed] [Google Scholar]
- 62. Shresta S, Sharar KL, Prigozhin DM, Beatty PR, Harris E. 2006. Murine model for dengue virus-induced lethal disease with increased vascular permeability. J. Virol. 80:10208–10217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Gromowski GD, Barrett AD. 2007. Characterization of an antigenic site that contains a dominant, type-specific neutralization determinant on the envelope protein domain III (ED3) of dengue 2 virus. Virology 366:349–360 [DOI] [PubMed] [Google Scholar]
- 64. Gromowski GD, Barrett ND, Barrett AD. 2008. Characterization of dengue virus complex-specific neutralizing epitopes on envelope protein domain III of dengue 2 virus. J. Virol. 82:8828–8837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Matsui K, Gromowski G, Li L, Barrett AD. 2010. Characterization of a dengue type-specific epitope on dengue 3 virus envelope protein domain III. J. Gen. Virol. 91:2249–2253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Matsui K, Gromowski GD, Li L, Schuh AJ, Lee JC, Barrett AD. 2009. Characterization of dengue complex-reactive epitopes on dengue 3 virus envelope protein domain III. Virology 384:16–20 [DOI] [PubMed] [Google Scholar]
- 67. Costin JM, Zaitseva E, Kahle KM, Nicholson CO, Rowe DK, Graham AS, Bazzone LE, Hogancamp G, Figueroa Sierra M, Fong RH, Yang ST, Lin L, Robinson JE, Doranz BJ, Chernomordik LV, Michael SF, Schieffelin JS, Isern S. 2013. Mechanistic study of broadly neutralizing human monoclonal antibodies against dengue virus that target the fusion loop. J. Virol. 87:52–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. de Alwis R, Beltramello M, Messer WB, Sukupolvi-Petty S, Wahala WM, Kraus A, Olivarez NP, Pham Q, Brien JD, Tsai WY, Wang WK, Halstead S, Kliks S, Diamond MS, Baric R, Lanzavecchia A, Sallusto F, de Silva AM. 2011. In-depth analysis of the antibody response of individuals exposed to primary dengue virus infection. PLoS Negl. Trop. Dis. 5(6):e1188. 10.1371/journal.pntd.0001188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. de Alwis R, Smith SA, Olivarez NP, Messer WB, Huynh JP, Wahala WM, White LJ, Diamond MS, Baric RS, Crowe JE, Jr, de Silva AM. 2012. Identification of human neutralizing antibodies that bind to complex epitopes on dengue virions. Proc. Natl. Acad. Sci. U. S. A. 109:7439–7444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Vogt MR, Dowd KA, Engle M, Tesh RB, Johnson S, Pierson TC, Diamond MS. 2011. Poorly neutralizing cross-reactive antibodies against the fusion loop of West Nile virus envelope protein protect in vivo via Fc-γ receptor and complement-dependent effector mechanisms. J. Virol. 22:11567–11580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Nelson S, Jost CA, Xu Q, Ess J, Martin JE, Oliphant T, Whitehead SS, Durbin AP, Graham BS, Diamond MS, Pierson TC. 2008. Maturation of West Nile virus modulates sensitivity to antibody-mediated neutralization. PLoS Pathog. 4(5):e1000060. 10.1371/journal.ppat.1000060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Pierson TC, Xu Q, Nelson S, Oliphant T, Nybakken GE, Fremont DH, Diamond MS. 2007. The stoichiometry of antibody-mediated neutralization and enhancement of West Nile virus infection. Cell Host Microbe 1:135–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Halstead SB. 2012. Dengue vaccine development: a 75% solution? Lancet 380:1535–1536 [DOI] [PubMed] [Google Scholar]
- 74. Nybakken G, Oliphant T, Johnson S, Burke S, Diamond MS, Fremont DH. 2005. Structural basis for neutralization of a therapeutic antibody against West Nile virus. Nature 437:764–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Choi KS, Nah JJ, Ko YJ, Kim YJ, Joo YS. 2007. The DE loop of the domain III of the envelope protein appears to be associated with West Nile virus neutralization. Virus Res. 123:216–218 [DOI] [PubMed] [Google Scholar]
- 76. Kaufmann B, Vogt MR, Goudsmit J, Holdaway HA, Aksyuk AA, Chipman PR, Kuhn RJ, Diamond MS, Rossmann MG. 2010. Neutralization of West Nile virus by cross-linking of its surface proteins with Fab fragments of the human monoclonal antibody CR4354. Proc. Natl. Acad. Sci. U. S. A. 107:18950–18955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Teoh EP, Kukkaro P, Teo EW, Lim AP, Tan TT, Yip A, Schul W, Aung M, Kostyuchenko VA, Leo YS, Chan SH, Smith KG, Chan AH, Zou G, Ooi EE, Kemeny DM, Tan GK, Ng JK, Ng ML, Alonso S, Fisher D, Shi PY, Hanson BJ, Lok SM, MacAry PA. 2012. The structural basis for serotype-specific neutralization of dengue virus by a human antibody. Sci. Transl. Med. 4:139ra183. [DOI] [PubMed] [Google Scholar]
- 78. Tan GK, Ng JK, Trasti SL, Schul W, Yip G, Alonso S. 2010. A non mouse-adapted dengue virus strain as a new model of severe dengue infection in AG129 mice. PLoS Negl. Trop. Dis. 4(4):e672. 10.1371/journal.pntd.000672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Grant D, Tan GK, Qing M, Ng JK, Yip A, Zou G, Xie X, Yuan Z, Schreiber MJ, Schul W, Shi PY, Alonso S. 2011. A single amino acid in nonstructural protein NS4B confers virulence to dengue virus in AG129 mice through enhancement of viral RNA synthesis. J. Virol. 85:7775–7787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Zellweger RM, Prestwood TR, Shresta S. 2010. Enhanced infection of liver sinusoidal endothelial cells in a mouse model of antibody-induced severe dengue disease. Cell Host Microbe 7:128–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Agarwal JP, Bhattacharyya PC, Das SK, Sharma M, Gupta M. 2009. Dengue encephalitis. Southeast Asian J. Trop. Med. Public Health 40:54–55 [PubMed] [Google Scholar]
- 82. Atrasheuskaya A, Petzelbauer P, Fredeking TM, Ignatyev G. 2003. Anti-TNF antibody treatment reduces mortality in experimental dengue virus infection. FEMS Immunol. Med. Microbiol. 35:33–42 [DOI] [PubMed] [Google Scholar]
- 83. Costa VV, Fagundes CT, Valadao DF, Cisalpino D, Dias AC, Silveira KD, Kangussu LM, Avila TV, Bonfim MR, Bonaventura D, Silva TA, Sousa LP, Rachid MA, Vieira LQ, Menezes GB, de Paula AM, Atrasheuskaya A, Ignatyev G, Teixeira MM, Souza DG. 2012. A model of DENV-3 infection that recapitulates severe disease and highlights the importance of IFN-gamma in host resistance to infection. PLoS Negl. Trop. Dis. 6(5):e1663. 10.1371/journal.pntd.0001663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Souza DG, Fagundes CT, Sousa LP, Amaral FA, Souza RS, Souza AL, Kroon EG, Sachs D, Cunha FQ, Bukin E, Atrasheuskaya A, Ignatyev G, Teixeira MM. 2009. Essential role of platelet-activating factor receptor in the pathogenesis of dengue virus infection. Proc. Natl. Acad. Sci. U. S. A. 106:14138–14143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Yu CY, Chang TH, Liang JJ, Chiang RL, Lee YL, Liao CL, Lin YL. 2012. Dengue virus targets the adaptor protein MITA to subvert host innate immunity. PLoS Pathog. 8(6):e1002780. 10.1371/journal.ppat.1002780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Aguirre S, Maestre AM, Pagni S, Patel JR, Savage T, Gutman D, Maringer K, Bernal-Rubio D, Shabman RS, Simon V, Rodriguez-Madoz JR, Mulder LC, Barber GN, Fernandez-Sesma A. 2012. DENV inhibits type I IFN production in infected cells by cleaving human STING. PLoS Pathog. 8(10):e1002934. 10.1371/journal.ppat.1002934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Ashour J, Laurent-Rolle M, Shi PY, Garcia-Sastre A. 2009. NS5 of dengue virus mediates STAT2 binding and degradation. J. Virol. 83:5408–5418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Ashour J, Morrison J, Laurent-Rolle M, Belicha-Villanueva A, Plumlee CR, Bernal-Rubio D, Williams KL, Harris E, Fernandez-Sesma A, Schindler C, Garcia-Sastre A. 2010. Mouse STAT2 restricts early dengue virus replication. Cell Host Microbe 8:410–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Cockburn JJ, Navarro Sanchez ME, Fretes N, Urvoas A, Staropoli I, Kikuti CM, Coffey LL, Arenzana Seisdedos F, Bedouelle H, Rey FA. 2012. Mechanism of dengue virus broad cross-neutralization by a monoclonal antibody. Structure 20:303–314 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.