Abstract
Cellular cytidine deaminases from the APOBEC3 family are potent restriction factors that are able to block the replication of retroviruses. Consequently, retroviruses have evolved a variety of different mechanisms to counteract inhibition by APOBEC3 proteins. Lentiviruses such as human immunodeficiency virus (HIV) express Vif, which interferes with APOBEC3 proteins by targeting these restriction factors for proteasomal degradation, hence blocking their ability to access the reverse transcriptase complex in the virions. Other retroviruses use less-well-characterized mechanisms to escape the APOBEC3-mediated cellular defense. Here we show that the prototype foamy virus Bet protein can protect foamy viruses and an unrelated simian immunodeficiency virus against human APOBEC3G (A3G). In our system, Bet binds to A3G and prevents its encapsidation without inducing its degradation. Bet failed to coimmunoprecipitate with A3G mutants unable to form homodimers and dramatically reduced the recovery of A3G proteins from soluble cytoplasmic cell fractions. The Bet-A3G interaction is probably a direct binding interaction and seems to be independent of RNA. Together, these data suggest a novel model whereby Bet uses two possibly complementary mechanisms to counteract A3G: (i) Bet prevents encapsidation of A3G by blocking A3G dimerization, and (ii) Bet sequesters A3G in immobile complexes, impairing its ability to interact with nascent virions.
INTRODUCTION
APOBEC3G (apolipoprotein B mRNA-editing, enzyme-catalytic, polypeptide-like 3G; also called A3G) is a cytidine deaminase of the APOBEC family. There are seven A3 genes (A3A to -D and A3F to -H) found in humans and most primates, one gene in rodents, and four genes in cats, showing that the A3 genes evolved in lineage-specific compositions in placental mammals (1, 2).
Human immunodeficiency virus type 1 with a deleted vif gene (HIV-1 Δvif) is efficiently inhibited by A3G. Viral particle-encapsidated A3G interferes with viral replication by multiple mechanisms: it deaminates cytosines to uracils in the viral cDNA generated during reverse transcription, reduces the efficiency of reverse transcription, and impairs integration (for recent reviews, see references 3 and 4). The HIV-1 Vif protein counteracts A3G by triggering its polyubiquitination and degradation (5). Vif also employs other mechanisms to inhibit A3G. In HeLa cells, production of infectious HIV-1 was shown to be independent of A3G depletion (6–8). Vif bound to A3G might inhibit the deamination activity of A3G (9), especially when both proteins are encapsidated in virions as a complex (10). Some of the A3G molecules escape the action of Vif and can even edit wild-type HIV-1, causing mutations which may contribute to viral diversity (see reference 3 for a review).
HIV-1 is not the only virus that faces antiviral A3 proteins: retroviruses, retroelements, hepadnaviruses, and certain RNA and DNA viruses show some restriction (11). Most other lentiviruses also use their Vif proteins to destroy cellular A3s: human T cell lymphotropic virus type 1 escapes the virion encapsidation of A3G, but not that of A3A, A3B, and A3H, through its unique nucleocapsid protein (12, 13); murine leukemia virus mitigates murine A3 activity by its glycosylated Gag protein (glyco-Gag) (14, 15); and foamy retroviruses express the accessory protein Bet to counteract A3 proteins (16–20).
Species-specific foamy viruses (FVs) are found in many mammals, and simian foamy viruses (SFVs) infect Old World monkeys, New World monkeys, and apes. In chimpanzees, the chimpanzee SFV (SFVcpz) is widely distributed, and prevalence rates from 44% to 100% have been reported (21). The prototype foamy virus (PFV) was isolated from human cell cultures but identified as a chimpanzee virus (22). Humans are occasionally infected by diverse simian foamy viruses (23–25), but neither in their natural host species nor after cross-species transmission to humans do foamy retroviruses induce disease, and infected humans are dead-end hosts (26–28). The reason that humans do not transmit FVs is not clear.
Bet is an accessory protein expressed in all foamy viruses. PFV Bet has been suggested to be involved in resistance to viral superinfection (29) and as a negative regulator of the basal activity of the internal PFV promoter (30). The feline foamy virus (FFV) Bet protein inhibits antiviral feline A3 proteins without inducing their degradation (16–18). A mechanism for FFV Bet activity has not been identified, but an interaction of FFV Bet and feline A3s was demonstrated to be important (16). For the PFV system, controversial data regarding the function of Bet as an A3G antagonist have been reported (20, 31). Russell et al. described that PFV Bet was able to rescue the infectivity of PFV Δbet and Vif-deficient HIV-1 in the presence of A3G (20). In contrast, Delebecque et al. found that PFV is sensitive to A3G, independent of Bet (31). Like the Bet activity of FFV, the mechanism of PFV Bet's inhibition of the antiviral activity of A3G has not been resolved.
We aimed here to address whether PFV Bet can protect PFV and lentiviruses against the antiviral activity of human A3G (huA3G) and to understand the fate of A3G bound to Bet.
MATERIALS AND METHODS
Plasmids.
Reporter viruses for the simian immunodeficiency virus SIVAGMTAN-1 (pSIVAGM-luc-R−E−Δvif) have been described previously (32). The PFV vector pczDWP002 is a variant of the PFV Gag/Pol expression vector pczDWP001 (33) in which the NLS-LacZ sequence replaces the enhanced green fluorescent protein (EGFP) open reading frame (ORF). The PFV Env expression construct pczHFVenvEM002 has been described previously (34). Human (hu), African green monkey (AGM), and chimpanzee (cpz) A3G-HA expression constructs were kindly provided by N. R. Landau (32). Myc-tagged huA3G (pcDNA3.1 expressing human APOBEC3G-Myc-6His) has been described previously (35). Plasmids encoding huA3G Y124A, Y125A, F126L, and W127A mutants were described previously (36). pBetPFV (19) was generated by insertion of the PFV Bet sequence, which was obtained from HEL299 cells (ATCC CCL-137) infected with the PFV isolated by Achong et al. (37), into the HindIII and SmaI sites of pBC12-CMV (38). pVifHIV-1 (39) (referred to here as pcDNA3.1Vif) and the analogous pVifAGM.TAN-1 plasmid were kindly provided by N. R. Landau. To generate pBet-V5-His, the forward primer KpnI 5′ Bet (5′-TAGGGTACCGTATCATGGATTCCTACGAA-3′) and the reverse primer ApaI 3′ Bet (5′-CTAGGGCCCGAAGGGTCCATCTGAGTCAC-3′) were used. The amplicons were cloned into the KpnI and ApaI sites of pcDNA4/V5-HisA (Life Technologies, Darmstadt, Germany). Subsequent amplification of the 3′ part of Bet containing V5 and His sequences by use of primers HFV-Bet824-844f (5′-GTTTCAAGGTAATTTATGAAG-3′) and G-6xH-stop-XbaI_rc (5′-GCTCTAGAGCTCAATGGTGATGGTGATGATG-3′) generated a fragment which was cloned into pBetPFV via BglII and XbaI sites. pA3G-α (40), expressing A3G fused to the α-fragment of β-galactosidase, was a gift of N. R. Landau. The ω-fragment of β-galactosidase (pCMV-ω [41]; formerly known as pSCTZ-omega [42]) was a gift of S. Rusconi.
Cell culture.
Human HOS and 293T cells were maintained in Dulbecco's modified Eagle's complete medium (PAN-Biotech, Aidenbach, Germany) supplemented with 10% fetal bovine serum (FBS), 0.29 mg/ml l-glutamine, and 100 U/ml penicillin-streptomycin at 37°C in a humidified atmosphere of 5% CO2. Plasmid transfection into 293T cells was done with Lipofectamine LTX (Life Technologies), except that PFV plasmid transfection was accomplished with Polyfect (Qiagen, Hilden, Germany), with a DNA/Polyfect ratio of 1:2.5. Reporter lentiviruses were generated by transfection into 6-well plates with 1.5 μg pSIVAGM-luc-E−R−Δvif, 0.5 μg vesicular stomatitis virus G glycoprotein (VSV-G) expression plasmid pMD.G, 1 μg A3 expression plasmid, and 2 μg pVifHIV, pVifAGM.TAN-1, or pBetPFV. PFV reporter vectors were generated in 12-well plates with 1 μg pczDWP002, 1 μg pczHFVenvEM02, 0.33 μg A3 expression plasmid, and 0.66 μg pBetPFV. Total plasmid DNA (4.5 or 3 μg) was maintained by the addition of pcDNA3.1 as needed. Reverse transcriptase (RT) activity of SIV-containing supernatants was determined using a Cavidi HS Lenti RT kit or a C-type RT activity kit (Cavidi Tech, Uppsala, Sweden) for PFV vectors. For infectivity assays, HOS cells were transduced in 96-well plates in triplicate with a virus amount equivalent to 10 pg of RT for HIV or SIV vectors and 20 mU for PFV. At 3 days postinfection (dpi), luciferase activity was measured by using a Steadylite HTS kit (PerkinElmer, Rodgau, Germany), or β-galactosidase activity was determined by using a Tropix Galacto-Star system (Life Technologies). Data are presented as counts/s normalized to that for virus obtained without A3 and Bet.
α-Complementation assay.
293T cells (seeded in 96-well plates) were transfected in triplicate with an expression vector for the ω-subunit of β-galactosidase and an APOBEC3G α-subunit expression vector (huA3G-α) in combination with BetPFV, VifSIVagm, or VifHIV-1. The activity of β-galactosidase was measured 3 days later by using a Tropix Galacto-Star system (Life Technologies).
Immunoblot analysis.
To generate protein lysates, cells were washed with phosphate-buffered saline (PBS) and lysed in radioimmunoprecipitation assay (RIPA) buffer for 5 min on ice. Lysates were clarified by centrifugation. For mild cell lysis, cells were treated with mild lysis buffer (50 mM HEPES, 125 mM NaCl, 0.1 mM phenylmethylsulfonyl fluoride [PMSF], 0.2% NP-40, pH 7.5, and 1× complete protease inhibitor cocktail [Merck Calbiochem]) for 30 min on ice and then centrifuged (10 min, 300 × g, 4°C), and the supernatant was separated from the pellet. The pellet was treated with RIPA buffer for 5 min on ice and then centrifuged (10 min, 300 × g, 4°C), and the supernatant was taken. For the differential detergent fractionation of proteins from cells, a ProteoExtract subcellular proteome extraction kit (Merck, Darmstadt, Germany) was used according to the manufacturer's specifications. To generate virus lysates, supernatant samples were layered on a 2-ml 20% (wt/vol) sucrose cushion (in PBS), and virions were pelleted at 35,000 rpm (SW-41 rotor; Beckman Coulter, Krefeld, Germany) for 2 h at 4°C. Pellets were resuspended with RIPA buffer and normalized to RT values for 500 pg determined from original supernatants. The levels of p27 and hemagglutinin (HA)-tagged A3 proteins were analyzed by immunoblotting. For coimmunoprecipitation (co-IP) of Bet and A3G, cells were transfected with 2 μg of pBetPFV and 1 μg of A3G expression plasmid. After 2 days, cells were lysed, lysates were clarified by centrifugation, and supernatants were incubated with anti-HA beads (Roche Diagnostics, Berlin, Germany) for 60 min at 4°C and washed 5 times with RIPA buffer. For immunoblot analysis, samples were boiled in NuPAGE SDS sample buffer and NuPAGE sample reducing agent (Life Technologies) and subjected to SDS-PAGE followed by transfer to a polyvinylidene difluoride (PVDF) membrane. Proteins were detected using an anti-HA antibody (Ab) (1:104 dilution) (MMS-101P; Covance, Munich, Germany) or an anti-c-Myc monoclonal Ab (MAb) (1:200 dilution) (9E10; Sigma-Aldrich, Munich, Germany). For the detection of endogenous A3G, rabbit antiserum (anti-Apo C17; 1:104 dilution) was used (7). For the detection of BetPFV, a Bel2/Bet-specific hyperimmune serum (1:50,000 dilution) was used (43). Equal loading of cell lysates was confirmed for immunoblots probed with anti-tubulin Ab (1:104 dilution) (B5-1-2; Sigma-Aldrich), and equal loading of virions was confirmed by use of anti-p24/p27 MAb AG3.0 (44). Anti-mouse– and anti-rabbit–horseradish peroxidase (GE Healthcare, Freiburg, Germany) were used as secondary Abs. Signals were visualized using ECL reagent (GE Healthcare).
Purification of Bet-A3G complexes.
To purify A3G-Bet complexes, expression plasmids for A3G-HA and Bet-V5-His were cotransfected into 293T cells. Cells were harvested at 48 h posttransfection, washed with PBS, and lysed with lysis buffer (50 mM Tris, pH 8.0, 10% glycerol, 0.8% [vol/vol] NP-40, 1 mM PMSF, and 1× complete protease inhibitor cocktail [Merck Calbiochem]). Cell suspensions were placed on ice for 15 min, and the debris was pelleted by centrifugation at 13,000 rpm for 20 min. Prior to RNase A (Thermo Scientific, Schwerte, Germany) treatment (50 mg/ml), the soluble fraction was adjusted to 0.8 M NaCl and incubated for 15 min at 37°C and for 1 h at 25°C. After treatment, the lysate was mixed with 20 μl of equilibrated Ni-nitrilotriacetic acid (Ni-NTA) agarose (Life Technologies) and kept at 4°C for 2 h, turning in an end-over-end fashion. Beads were washed extensively with wash buffer (50 mM Tris, pH 8.0, 0.3 M NaCl, 10% [vol/vol] glycerol, 20 mM imidazole), and Bet-A3G complexes were eluted with elution buffer (50 mM Tris, pH 8.0, 0.3 M NaCl, 10% [vol/vol] glycerol, 250 mM imidazole).
Sequencing of viral reverse transcripts.
SIVAGM-lucΔvif (VSV-G) reporter viruses generated in the presence of huA3G with and without Bet and treated with 20 U/ml DNase I (Thermo Scientific) for 1 h at 37°C were used for infection of 293T cells (5 × 105). After 10 h, the cells were washed and total DNA was isolated using a DNeasy DNA blood and tissue kit (Qiagen). A 600-bp luc fragment was amplified with DreamTaq DNA polymerase (Thermo Scientific) (denaturation at 95°C for 5 min followed by 30 cycles of annealing at 61°C for 30 s and denaturation at 94°C for 30 s) and primers Luc-Fw (5′-GATATGTGGATTTCGAGTCGTC-3′) and Luc-Rev (5′-GTCATCGTCTTTCCGTGCTC-3′) and then cloned into the pJet blunt cloning vector (Thermo Scientific). The nucleotide sequences of 10 independent clones were analyzed and the G-to-A conversion presented using the Hypermut online tool (http://www.hiv.lanl.gov/content/sequence/HYPERMUT/hypermut.html).
In vitro DNA cytidine deamination assay of A3G-Bet complexes.
A3G deamination reactions were performed as described previously (45), using a 10-μl reaction volume containing 25 mM Tris, pH 7.0, and 10 fmol single-stranded DNA (ssDNA) substrate (5′-GGATTGGTTGGTTATTTGTTTAAGGAAGGTGGATTAAAGGCCCAAGAAGGTGATGGAAGTTATGTTTGGTAGATTGATGG-3′). Reaction mixtures were incubated for 3 h at 37°C, and reactions were terminated by incubation at 95°C for 5 min. One femtomole of the reaction mixture was used for PCR amplification with DreamTaq polymerase (Thermo Scientific) (denaturation at 95°C for 3 min followed by 19 cycles of annealing at 61°C for 30 s and denaturation at 94°C for 30 s) and the following primers: forward, 5′-GGATTGGTTGGTTATTTGTTTAAGGA-3′; and reverse, 5′-CCATCAATCTACCAAACATAACTTCCA-3′. PCR products were digested with the restriction enzyme Eco147I (StuI) (Thermo Scientific) for 1 h at 37°C, resolved by 15% PAGE, and stained with SYBR gold (Life Technologies). A positive-control substrate oligonucleotide with CCU instead of CCA was used to control the Eco147I digestion. To determine the deamination activity of purified A3G in the presence of purified Bet, A3G-Myc-His and Bet-V5-His were purified separately as described above, except that the RNase A treatment step was omitted in the Bet purification. Purified His-tagged protein concentrations were determined spectrophotometrically by measuring the A280, using their (theoretical) extinction coefficients and molecular masses. The A3G deamination reaction was conducted as described above. For the titration experiments, 10 fmol single-stranded DNA substrate was mixed with 20 fmol A3G-His protein and increasing amounts of Bet protein (10 to 1,000 nM).
Sucrose density gradient centrifugation.
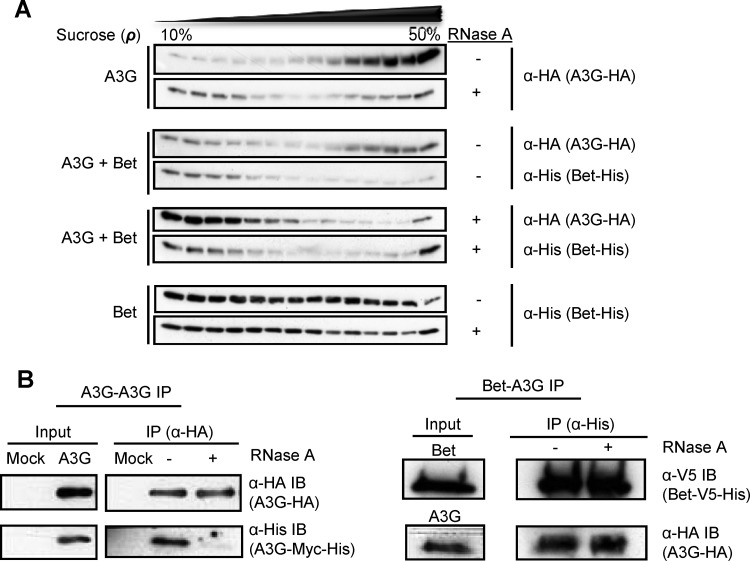
293T cells were transfected with expression plasmids for 2 μg A3G-HA, 2 μg Bet-His, or 2 μg A3G-HA plus 2 μg Bet-His. After 24 h, cells were lysed with lysis buffer (0.626% NP-40, 100 mM NaCl, 50 mM potassium acetate, 10 mM EDTA, 10 mM Tris, pH 7.4, and complete protease inhibitor cocktail [Merck Calbiochem]) and then clarified by centrifugation for 10 min at 162 × g followed by a short spin at 18,000 × g for 30 s. A half-portion of the sample was aliquoted to a new tube, to which RNase A (Thermo Scientific) (70 μg/ml) was added and incubated for 30 min at 37°C. Samples were then overlaid on top of a 10%-15%-20%-30%-50% sucrose step gradient in lysis buffer and centrifuged for 45 min at 163,000 × g at 4°C in an MLS-50 rotor (Beckman Coulter, Fullerton, CA). After centrifugation, the samples were sequentially removed from the top of the gradient, resolved by SDS-PAGE, and analyzed by immunoblotting with anti-HA and anti-Bet antibodies to detect A3G and Bet, respectively.
Coimmunoprecipitation assays. (i) Interaction of purified A3G-Myc-His and Bet-V5-His proteins.
A total of 1.5 μg of mouse monoclonal anti-Myc antibody (AbD Serotec, Düsseldorf, Germany) was mixed with purified A3G-Myc-His and Bet-V5-His proteins (1:2 ratio) in a buffer (50 mM Tris, pH 7.4, and 150 mM NaCl) and incubated overnight at 4°C with end-over-end rotation. The next day, 20 μl of protein A/G Plus agarose (Santa Cruz, Heidelberg, Germany) was added and incubated for 3 h at 4°C. After binding, the beads were washed 4 times with the same buffer, and the Bet-A3G complexes were eluted by boiling the beads at 95°C for 5 min in native loading buffer. The supernatant was further heated at 95°C for 5 min with 2-mercaptoethanol (2-ME) before being subjected to SDS-PAGE. Immunoblots were developed using PentaHis antibody (Qiagen) analysis. For better visualization of the A3G-Bet interaction, the gel was stained with Coomassie brilliant blue staining solution after the immunoblot transfer and was destained with destaining solution.
(ii) RNA-dependent interaction.
The protocol to assay RNA-dependent interactions was adapted from a previously described protocol (46). 293T cells were transfected with 1 μg pcDNA3.1 (Invitrogen) empty vector (mock) and expression plasmids for 1 μg A3G-HA plus 1 μg A3G-Myc-His or 1 μg A3G-HA plus 1 μg Bet-V5-His in a six-well culture plate. At 24 h posttransfection, cells were harvested and lysed with lysis buffer (0.5% Triton X-100, 287 mM NaCl, 3 mM KCl, 50 mM Tris, pH 7.5, and Complete protease inhibitor mixture [Roche]). The cleared lysates were incubated with 30 μl anti-HA affinity matrix beads (Roche) for A3G-A3G co-IP and with 30 μl Ni-NTA agarose (Life Technologies) for 2 h at 4°C, with end-over-end rotation. After binding, the beads were washed twice, with lysis buffer and lysis buffer containing 20 mM imidazole for A3G-A3G and A3G-Bet, respectively, and half-portions of the samples were aliquoted to new tubes. RNase A (70 μg/ml) was added to one aliquot of the sample and incubated at 37°C for 10 min and at 22°C for 40 min. Samples were further washed thrice with lysis buffer and lysis buffer containing 20 mM imidazole for A3G-A3G and A3G-Bet, respectively. A3G-A3G co-IP products were eluted by boiling beads in SDS gel loading buffer at 95°C for 5 min, and A3G-Bet co-IP products were eluted with an elution buffer containing 300 mM imidazole; the supernatant containing the eluate was heated at 95°C for 5 min with 2-ME before being subjected to SDS-PAGE and detected by immunoblotting with respective antibodies. Results are representative of three independent experiments.
Immunofluorescence microscopy.
Immunofluorescence studies were performed in HeLa cells 2 days after transfection by applying FuGENE technology (Roche Applied Science). Cells were fixed in 4% paraformaldehyde in PBS for 20 min, washed twice in PBS, permeabilized in 0.1% Triton X-100 in PBS for 15 min, washed twice in PBS, and blocked with blocking solution (1% milk powder in PBS) for 45 min. For A3G-HA staining, cells were incubated for 1 h with an anti-HA antibody (MMS-101P; Covance) in a 1:1,000 dilution in blocking solution. Bet was stained with Bet antibody at a 1:1,000 dilution in blocking solution. Anti-mouse–Alexa Fluor 488 (Life Technologies) and anti-rabbit–Alexa Fluor 594 were used as secondary antibodies at a 1:300 dilution in blocking solution for 1 h. Subsequently, 4′,6-diamidino-2-phenylindole staining (1:1,000; Millipore, Darmstadt, Germany) was performed for 10 min. Finally, cells were washed twice in PBS and analyzed by laser scanning microscopy (LSM 510 Meta; Zeiss, Göttingen, Germany). Images were acquired using a 40× objective.
Model structure.
For structure-guided mutation analysis, the previously published homology model of the A3G N terminus-N terminus dimer interface (36) was used. This model was generated using the MODELLER program (47); the crystal structure of the human A3G C-terminal domain (48) served as the template (for detailed methods, see reference 36). The graphical visualization presented in Fig. 5C was constructed using PyMOL (PyMOL Molecular Graphics System, version 1.5.0.4; Schrödinger, Portland, OR).
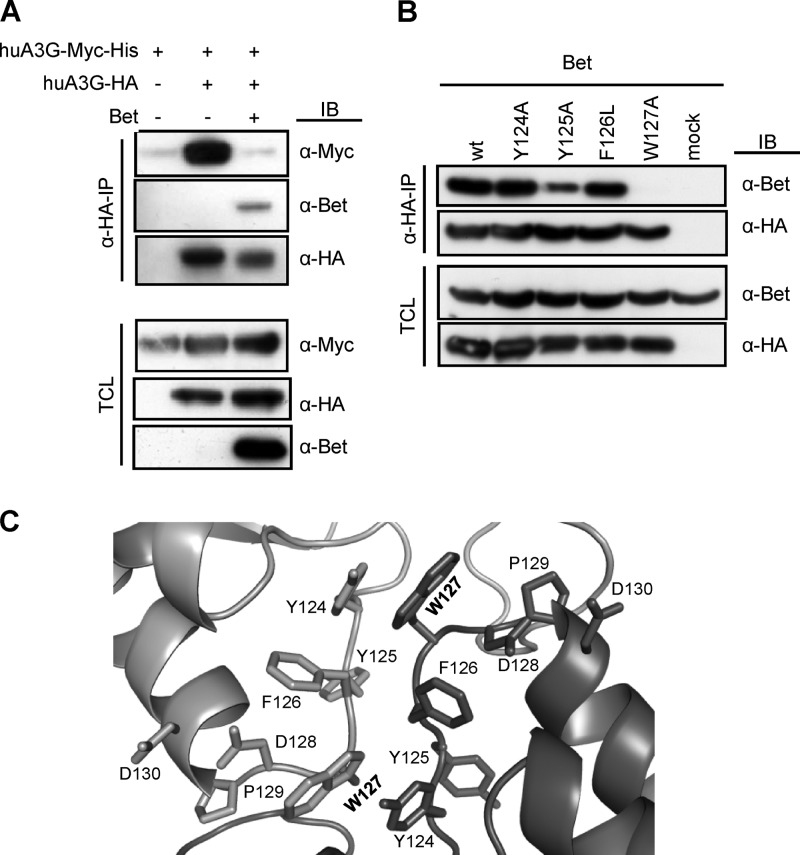
Fig 5.
Bet prevents A3G dimerization. (A) Cells were transfected with Myc-tagged huA3G alone or also with HA-tagged huA3G in the presence and absence of Bet. Cell lysates were prepared, and anti-HA immunoprecipitation (α-HA-IP) was performed. huA3G-Myc was coprecipitated with HA-tagged A3G, Bet was detected by immunoblot (IB) analysis using Bet antibody, and A3G-Myc was detected using an anti-Myc MAb. The HA-tagged A3G protein was detected with an anti-HA MAb. TCL, total cell lysate. (B) Cells were transfected with Bet alone or with expression plasmids for HA-tagged wild-type (wt) huA3G or the Y124A, Y125A, F126L, or W127A mutant. Cell lysates were prepared, and anti-HA immunoprecipitation (α-HA-IP) was performed. A3G-associated Bet was detected by immunoblot (IB) analysis using Bet antiserum. A3G proteins were detected in anti-HA IP samples and in cell lysates by use of anti-HA MAbs, as well as Bet and tubulin antibodies in cell lysates (TCL). (C) Model of A3G N-terminal dimeric interface. The cartoon representation of the A3G dimer interface depicts the spatial arrangements of amino acids 124 to 127, which are involved in dimerization, and HIV-1 Vif binding residues 128 to 130. Dark gray and light gray represent separate N-terminal parts of two A3G proteins, and the Trp127 residue is highlighted in bold.
Statistical analysis.
Data are presented as means with standard deviations (SD) in all bar diagrams. Statistically significant differences between two groups were analyzed using unpaired Student's t test in GraphPad Prism, version 5 (GraphPad Software, San Diego, CA). A minimum P value of 0.05 was considered statistically significant.
RESULTS
PFV Bet counteracts A3G in a dose-dependent manner.
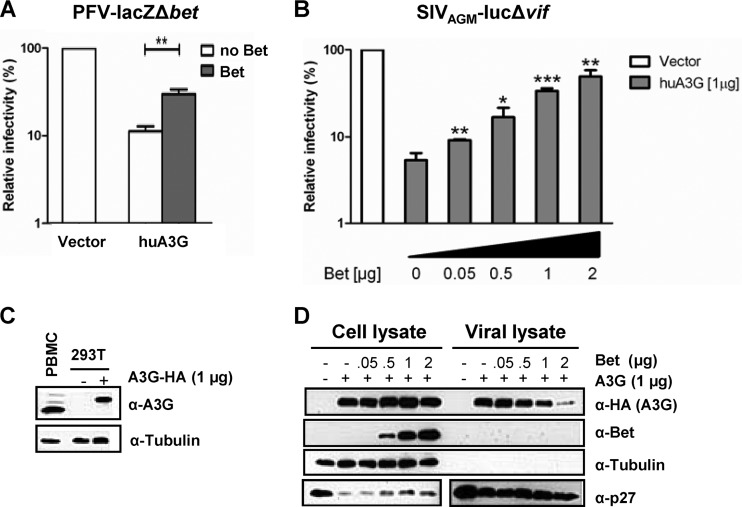
Conflicting data on the functional effect of PFV Bet on the activity of A3G have been reported (20, 31). These data prompted us to further investigate possible Bet activity toward A3G. First, we tested A3G by using a Bet-deficient single-round reporter vector based on PFV and expressing the lacZ gene for β-galactosidase in 293T cells transfected with an expression plasmid for A3G alone and together with Bet. Normalized amounts of particles were used to transduce human osteosarcoma cells, and the β-galactosidase levels were quantified at 3 days postransduction (Fig. 1A). A3G reduced the infectivity of PFV vectors 10-fold, and the expression of Bet rescued the PFV infectivity to half the levels attained by PFV vectors generated without A3G. Previously, it was shown that Bet can also protect heterologous lentiviruses against A3 proteins (19, 20). Thus, for the next experiments, we used a luciferase reporter virus based on the SIV derived from AGMs (SIVAGM), with an inactivated vif gene (referred to here as SIVAGM-lucΔvif) (32). SIVAGM-lucΔvif was generated in the presence of 1 μg A3G plasmid and increasing amounts of Bet plasmid (0 to 2 μg). Viral particles were normalized for reverse transcription activity and used for transduction. Figure 1B shows that the SIV reporter virus could be inhibited >10-fold by A3G under these experimental conditions. The A3G expression plasmid yielded physiological levels of A3G protein compared to the level of A3G detectable in activated human peripheral blood mononuclear cells (PBMCs) (Fig. 1C). The coexpression of Bet counteracted the antiviral activity of A3G in a dose-dependent way, and virus made with A3G and 2 μg of Bet plasmid had an infectivity similar to that of SIV without A3G (Fig. 1B). Note that neither Bet expression in the PFV system nor that in the SIV system completely restored infectivity, showing that human A3G can be counteracted only partially by PFV Bet. Bet expression also caused a dose-dependent inhibition of A3G encapsidation, as demonstrated by immunoblotting of viral particles produced in the presence of 1 μg A3G plasmid and increasing amounts of Bet expression plasmid (Fig. 1D). Under these conditions, the viral particles did not package the Bet protein.
Fig 1.
Bet counteracts A3G in a dose-dependent manner and independent of the virus background. (A) Bet-deficient lacZ reporter viruses derived from PFV were produced in the presence and absence of human APOBEC3G (huA3G) and Bet. Infectivities of viruses were determined by quantification of galactosidase expression at 3 dpi. Infectivities relative to that of the virus generated in the absence of huA3G and Bet are given. (B) SIVAGM-lucΔvif viruses were produced in the presence and absence of huA3G and increasing amounts of Bet. Infectivities of equal amounts of viruses, relative to that of the virus generated in the absence of huA3G and Bet, were determined by quantification of luciferase activity at 3 dpi. (C) Endogenous level of A3G in activated human PBMCs compared to plasmid-derived A3G level in transfected 293T cells (1 μg A3G-HA plasmid), detected by using anti-A3G antiserum. Equal loading of cell lysate samples was confirmed with an anti-tubulin antibody. (D) Packaging of A3G in SIVAGM-lucΔvif virions. Virions were produced in the presence of an empty expression plasmid (−; pcDNA3.1) or 1 μg A3G expression plasmid and increasing amounts of Bet expression plasmid (0 μg, 0.05 μg, 0.5 μg, 1 μg, and 2 μg). Supernatants were filtered and concentrated through a 20% sucrose cushion by centrifugation. The cell and viral lysates were analyzed by immunoblotting and were probed for A3G (α-HA, anti-HA antibody), Bet (α-Bet, anti-Bet antibody), capsid (α-p27, anti-p27 antibody), and tubulin (α-Tubulin, anti-tubulin antibody). Unpaired t tests were computed to determine whether differences between samples in the presence of A3G protein with and without Bet reached the level of statistical significance (*, P < 0.05; **, P < 0.001; and ***, P < 0.0001), using GraphPad Prism 5 software.
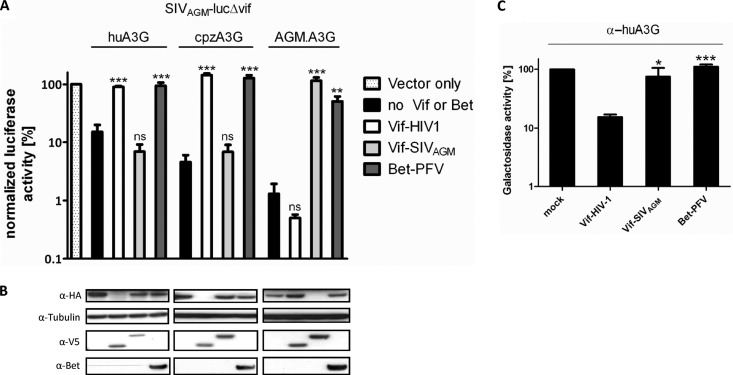
We also used the SIVAGM-lucΔvif reporter virus to ask whether PFV Bet inhibits the antiviral activity of A3G proteins derived from chimpanzees (cpzA3G) and AGMs (AGM.A3G). To control for specificity, we directly compared Bet to HIV-1 Vif and SIVAGM Vif. All three A3Gs inhibited the SIV reporter virus 10- to 100-fold, and Bet completely counteracted cpzA3G and largely counteracted AGM.A3G and human A3G (Fig. 2A). HIV-1 Vif inhibited and degraded huA3G and cpzA3G but was not active on AGM.A3G. Only SIVAGM Vif was active upon and degraded AGM.A3G, as seen previously (32). In contrast to the sensitivity of A3Gs to Vif, immunoblots of the viral producer cells showed no degradation of A3Gs in samples where Bet was produced (Fig. 2B). To further support the absence of A3G degradation by Bet, we applied the α-complementation assay to A3G (40). This assay is based on α-complementation, the ability of β-galactosidase fragments to complement in trans. Figure 2C shows that A3G fused to the α-peptide complemented a coexpressed ω-fragment of β-galactosidase. HIV-1 Vif synthesis reduced the galactosidase activity 10-fold, likely due to A3G degradation, as described previously (40). In contrast, neither SIVAGM Vif nor PFV Bet reduced the β-galactosidase activity, further indicating the lack of a Bet-triggered degradation of A3G.
Fig 2.
Bet exhibits a broader spectrum of anti-A3G activity than Vif, without leading to A3G degradation. (A) SIVAGM-lucΔvif viruses were produced in the presence and absence (vector only) of the indicated HA-tagged A3G, V5-tagged HIV-1 Vif and SIVAGM Vif, and PFV Bet expression plasmids. Relative infectivities (relative to the virus generated in the absence of A3G and Bet) of equal amounts of the reporter viruses are shown. (B) Immunoblot analysis performed with lysates of virus-producing cells. A3G was detected with an anti-HA MAb, tubulin with an anti-tubulin MAb, Vif with an anti-V5 MAb, and Bet with Bet antiserum. (C) α-Complementation assay of β-galactosidase. 293T cells were transfected in triplicate with the ω-subunit expression plasmid of β-galactosidase and an α-subunit–A3G expression plasmid in combination with PFV Bet, SIVAGM Vif, or HIV-1 Vif. The activity of β-galactosidase was measured 3 days later. Unpaired t tests were computed to determine whether differences between samples in the presence of A3G, with and without Vif or Bet, reached the level of statistical significance (*, P < 0.05; **, P < 0.001; ***, P < 0.0001; and ns, not significant), using GraphPad Prism 5 software.
In summary, PFV Bet has a broad A3G tropism, does not deplete cellular A3G proteins of human, chimpanzee, or AGM origin, and can rescue the infectivity of PFV and of a heterologous lentivirus produced in the presence of A3G.
Bet prevents antiviral editing but does not inhibit A3G enzymatic activity.
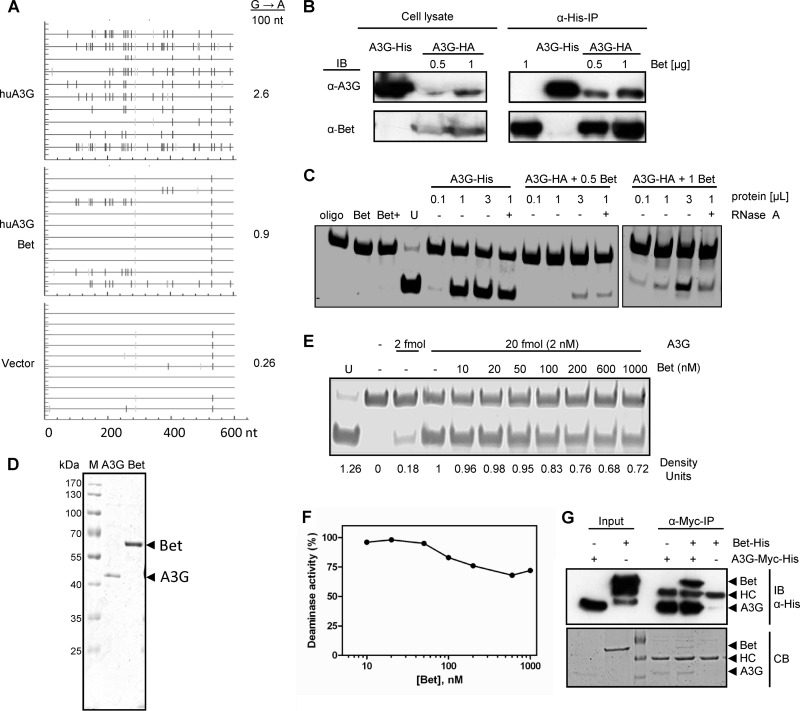
Next, we were interested in studying the functional consequences of the Bet interaction with A3G. Published data support Bet binding to A3G, so one could postulate that Bet might impair the enzymatic activity of A3G. To test A3G-induced genome editing, SIVAGM-lucΔvif reporter particles were generated with A3G alone and with A3G and Bet. We used these virions to infect 293T cells, isolated the viral genomic DNA by PCR at 10 h postinfection, and analyzed the sequence for signs of G-to-A mutations on the positive (+) strand, which are indicative of cytidine deaminations in DNA of the viral negative (−) strand. A3G induced G-to-A changes at a mutation rate of 2.6%, in contrast to virions made without A3G, which had a very low G-to-A mutation rate of 0.26% (Fig. 3A). The viral cDNA prepared from virions made in the presence of A3G and Bet showed a reduced but important G-to-A mutation rate of 0.9%, supporting the conclusion that Bet partially counteracts the antiviral activity of A3G (Fig. 3A).
Fig 3.
Bet reduces antiviral editing of A3G but does not affect enzymatic activity of A3G. (A) A fragment of the luciferase gene was amplified from reverse transcripts of SIVAGM-lucΔvif viruses, generated in the presence and absence of huA3G in combination with Bet, at 12 h postinfection. Ten independent nucleotide sequences were determined. The G-to-A conversions (vertical lines) are shown for each clone (horizontal line). (B) The A3G cytidine deamination activity of Bet-A3G complexes was analyzed in Bet-precipitated complexes. For immunoprecipitation of Bet-A3G complexes, 0.5 μg or 1 μg of Bet-V5-His expression plasmid was transfected alone or together with a constant amount (0.5 μg) of A3G-HA into 293T cells. Blots were probed separately with anti-Bet antibody and anti-A3G antibody. (C) Immunoprecipitated complexes and A3G-His and Bet alone were tested in the in vitro deamination assay. Protein complexes (0.1 μl, 1 μl, and 3 μl) were used with each Bet concentration (for cells transfected with either 0.5 μg or 1 μg Bet expression plasmid). In addition, 1 μl was purified after RNase A treatment (+) and compared with the untreated sample. An oligonucleotide with CCU in place of CCA served as a marker/digestion control (U). A3G-His (0.5 μg) served as a positive control for the cytidine deamination reaction, whereas Bet alone was the negative control. (D) Coomassie blue-stained SDS-PAGE gel indicating the purity of huA3G-Myc-His and Bet-V5-His proteins after purification from transfected 293T cells by use of Ni-NTA agarose. (E) In vitro deamination assay using 293T cell-purified huA3G and Bet proteins. The A3G-Myc-His activity of 20 fmol (2 nM) of protein was tested in the presence of increasing amounts of Bet-His (0 to 1,000 μM). (F) Deaminase activities determined in panel E, in terms of density units plotted against the concentration of Bet. The plot represents relative deaminase activities, with 100% activity adjusted to deamination in the absence of Bet. (G) Anti-Myc immunoprecipitation (IP) shows the interaction of purified A3G-Myc-His and Bet-His in vitro. HC, anti-Myc antibody heavy chain. (Top) Immunoblot (IB) of IP and input samples, using anti-His antibody. (Bottom) The same SDS-PAGE gel stained with Coomassie blue (CB).
To test whether Bet can inhibit the cytidine deaminase activity of A3G, we purified Bet by immunoprecipitation from 293T cells producing either Bet alone or Bet together with A3G. These experiments were performed using His-tagged Bet that showed full anti-APOBEC3 activity (data not shown). The immunoprecipitated samples were subjected to Western blotting (Fig. 3B) and an in vitro deamination assay (45) (Fig. 3C). We tested two different conditions and transfected either 0.5 μg or 1 μg Bet expression plasmid together with an expression plasmid for A3G (A3G-HA). Immunoprecipitated Bet protein showed binding to A3G as demonstrated with an anti-A3G antibody (Fig. 3B, α-His-IP panel). We used different volumes (0.1, 1, and 3 μl) of the immunoprecipitated protein samples and asked whether A3G bound to Bet is able to deaminate single-stranded DNA in vitro (Fig. 3C). In addition, we treated the 1-μl samples with RNase A to remove potential inhibitory RNA (49). As a positive control for in vitro cytidine deamination, we directly immunoprecipitated A3G having a His tag (A3G-Myc-6XHis). This sample contained severalfold more A3G protein than the amount of A3G immunoprecipitated by Bet. The PCR-based deamination assay depends on the sequence change caused by A3G converting a cytidine to uridine in an 80-nucleotide (nt) ssDNA substrate. Deamination of C to U by A3G is then followed by a PCR that replaces the uridine with thymidine, generating a StuI restriction site. The efficiency of StuI digestion is monitored by using a similar 80-nt ssDNA substrate containing a uridine instead of a cytidine in the StuI recognition site (Fig. 3C, lane U). Polyacrylamide gel electrophoresis separates deaminated from nondeaminated DNA substrates after StuI cleavage of the PCR product. While we found a faint band after StuI cleavage in the 3-μl input of the sample generated with 0.5 μg Bet plasmid, much more cleavage product was detectable using the 3-μl input of cells transfected with 1 μg Bet plasmid (Fig. 3C). Smaller volumes (0.1 and 1 μl) of this sample showed a reduced, dose-dependent deamination of cytidine. Immunoprecipitated Bet-A3G complexes treated with RNase A displayed little change in deamination activity compared to the corresponding sample prepared without RNase A treatment (Fig. 3C). Very importantly, immunoprecipitated Bet protein from cells that did not produce A3G did not show cytidine deamination activity, irrespective of RNase A treatment. Similar results were obtained using chimpanzee A3G instead of human A3G (data not shown). In addition, recombinant A3G and Bet proteins were purified from 293T cells by use of Ni-NTA agarose (Fig. 3D), and the in vitro deamination assay was conducted with a constant level of A3G (2 nM) and a gradient of Bet (Fig. 3E). A deamination activity chart (Fig. 3F) was plotted according to the corresponding density units (Fig. 3E) and illustrates the activity of A3G in the presence of Bet. The slightly reduced activity (to 83%) at the 100 nM Bet concentration may have been due to precipitation of Bet proteins. Using even higher Bet concentrations, a further minor drop in deamination activity, to about 70%, was seen. The interaction between the separately purified A3G and Bet proteins was confirmed by coimmunoprecipitation using an anti-Myc antibody. The Myc antibody bound to protein A/G Plus agarose precipitated A3G-Myc-His and Bet-V5-His complexes but not Bet-V5-His alone, as shown in the anti-His immunoblot and Coomassie blue-stained gel (Fig. 3G). Together, these observations indicate that A3G bound to Bet is not inhibited in its cytidine deamination activity.
Bet interaction is RNA independent.
Since A3G-A3G interaction is known to be bridged by RNAs (46), we asked whether A3G-Bet interaction follows a similar phenomenon. To test this, we first performed velocity sedimentation of cell lysates containing A3G alone, A3G with Bet, or Bet alone, with and without RNase A treatment. 293T cells were transfected with expression vectors for A3G alone or together with Bet. Part of the lysates was subjected to RNase A treatment for an hour, and all samples were layered on 10 to 50% sucrose gradients. A3G without Bet showed the expected high-molecular-mass (HMM) complexes that were sensitive to RNase A (Fig. 4A) (50). In contrast, Bet was distributed throughout the fractions, irrespective of RNase A treatment, indicating that Bet forms complexes of various molecular masses that are RNA independent. Cell lysates of A3G with Bet showed that A3G was found partially in HMM complexes but also in lower-molecular-mass (LMM) complexes. In the presence of A3G, the Bet protein delocalized from HMM to LMM complexes, consistent with a Bet-A3G interaction. RNase A did not change the detection of Bet in the coexpressing cells but removed the HMM complexes of A3G that did not show copurification with Bet (Fig. 4A). While Bet oligomerization appears to be RNA independent, Bet could require RNA to bind to A3G. To directly check the influence of RNA on A3G-Bet interaction, we performed a co-IP analysis of A3G-HA and Bet-V5-His produced in 293T cells with and without RNase treatment. The cell lysates were allowed to bind to Ni-NTA agarose first and then split into two parts for RNase A treatment. The immunoblot indicates no difference between RNase-treated and untreated Bet-A3G samples (Fig. 4B, right panel), demonstrating that Bet does not bind to A3G via an RNA molecule. In contrast, the parallel A3G-A3G co-IP with anti-HA beads showed a dramatic reduction of A3G-A3G interaction when lysates were treated with RNase A (Fig. 4B, left panel).
Fig 4.
A3G-Bet interaction is RNA independent. (A) Velocity sedimentation of RNase A-treated and untreated cell lysates of 293T cells that were transfected with expression plasmids for A3G alone, A3G and Bet, or Bet alone through sucrose gradients (10% to 50%). Samples were examined by use of specific antibodies: A3G was detected with an anti-HA MAb, and Bet was detected with an anti-His antibody. (B) 293T cells were cotransfected with expression plasmids for A3G-HA and A3G-Myc-His for A3G-A3G immunoprecipitation (IP) and with Bet-V5-His and A3G-HA expression plasmids for Bet-A3G IP. Immunoblots (IB) were incubated with specific antibodies: A3G was detected with an anti-HA MAb, and Bet was detected with an anti-His antibody. IP samples were treated with RNase A or left untreated while binding to the beads.
Bet prevents A3G-A3G interaction.
We recently reported that PFV Bet binds to A3C and reduces A3C dimer formation (19). To determine if Bet also inhibits A3G-A3G complex formation, we studied the oligomerization of A3G in the presence of Bet. Coprecipitation experiments were done with HA-tagged A3G and Myc-tagged A3G. As expected, A3G-HA coprecipitated with A3G-Myc (Fig. 5A), but interestingly, this A3G-A3G interaction was not detectable when Bet was coexpressed. The observation that Bet prevented A3G dimerization allowed us to identify relevant residues in A3G regulating the Bet interaction. Amino acids in the putative A3G dimer interface were recently described (36, 46), so we tested the Y124A, Y125A, F126L, and W127A A3G mutants (36) that partake in stabilizing the A3G dimer for interaction with Bet. Of these mutants, the W127A mutant is unable to homodimerize and to inhibit HIV-1 Δvif due to a lack of encapsidation (36, 46).
The Y124A and F126L A3G mutants coimmunoprecipitated with Bet, like wild-type A3G, whereas the F126A and W127A A3G mutants showed partial and complete loss of coimmunoprecipitation, respectively (Fig. 5B). The predicted localization of the analyzed amino acids is shown in Fig. 5C, using a head-to-head structural model of the A3G homodimer (36, 48). Together, these data suggest that Bet interacts with A3G and disrupts its ability to dimerize.
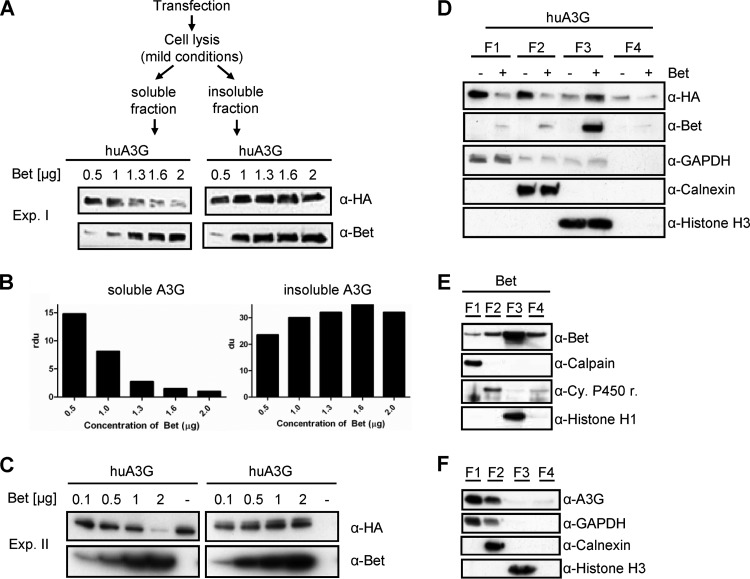
Bet impairs the cytosolic solubility of A3G.
Pilot experiments indicated that the choice of cell lysis buffer strongly affected immunoblot detection of A3G in Bet-cotransfected cells (data not shown). To describe any Bet-dependent recovery of A3G under mild lysis conditions, we performed a Western blot titration experiment. After cotransfection of A3G and different amounts of Bet plasmid (0.5 to 2 μg in experiment I and 0.1 to 2 μg in experiment II), we used a mild lysis buffer, separated the soluble and insoluble fractions by centrifugation, and solubilized the insoluble fraction with RIPA buffer. Normalized protein samples were analyzed by SDS electrophoresis and immunoblotting (Fig. 6A). Increasing amounts of Bet protein reduced the detectable amount of A3G in the soluble fraction in a dose-dependent way, and the sample generated with 2 μg Bet plasmid lost most of the A3G signal (Fig. 6A to C). In contrast, the insoluble cell fractions showed no A3G depletion, but rather an increased amount of A3G with increased expression of Bet. The Bet protein itself was detectable in the soluble as well as insoluble cell fraction.
Fig 6.
Bet impairs the cytosolic solubility and modifies the buffer-dependent detection of huA3G. (A) 293T cells transfected with huA3G and increasing amounts of Bet expression plasmid (0.5, 1, 1.3, 1.6, and 2 μg; Exp. I) were lysed under mild conditions. Soluble and insoluble fractions were separated by centrifugation and analyzed by SDS electrophoresis and subsequent immunoblotting. HA-tagged A3G was detected with an anti-HA MAb, and Bet was detected with Bet antiserum. (B) Relative density units (rdu) of A3G signals shown in panel A. (C) Independent experiment II (Exp. II) was performed as described for panel A, except that 0.1, 0.5, 1, and 2 μg of Bet expression plasmid were used. (D) Differential detergent fractionation of proteins of 293T cells transfected with expression plasmids for A3G-HA, with and without Bet, according to their solubilities in buffers specific for the following subcellular fractions: cytoplasm (F1; digitonin extraction), membranes and organelles (F2; Triton X-100 extraction), nucleus (F3; Tween20/deoxycholate extraction), and cytoskeleton (F4; SDS extraction). HA-tagged A3G on immunoblots was detected with an anti-HA MAb, and Bet was detected with Bet antiserum. Equal loading of samples and proper fractionations were confirmed with an anti-GAPDH MAb for cytosolic fractions, anti-calnexin MAb for membrane/organelle fractions, and anti-histone H3 MAb for nuclear fractions. (E) Immunoblot analysis of Bet without huA3G in different fractions of 293T cells. The F1 fraction was detected using an anti-calpain MAb, the F2 fraction using an anti-cytochrome 450 reductase MAb, and the F3 fraction using an anti-histone H1 MAb. (F) Immunoblot analysis of endogenous levels of huA3G in different fractions of activated human peripheral blood mononuclear cells, using huA3G antiserum.
To determine whether the presence of Bet affects the subcellular recovery of A3G, proteins were fractionated according to their subcellular locations by applying a ProteoExtract subcellular proteome extraction kit. For this approach, proteomes of 293T cells containing A3G in combination with Bet were separated by taking advantage of the different solubilities of certain subcellular compartments in four selected reagents. This system, also known as differential detergent fractionation (DDF), uses different detergents to sequentially extract cellular proteins prior to homogenization while keeping the cell architecture intact (51). Interestingly, using DDF, the resulting fractions reflect not only the characteristics of the protein's solubility but also its localization within the cell (52). The cellular fractions of proteins in their native state, representing enriched samples of cytosolic proteins (F1; digitonin extraction), proteins of membranes and organelles (F2; Triton X-100 extraction), soluble and DNA-associated nuclear proteins (F3; Tween 20/deoxycholate extraction), and cytoskeleton proteins (F4; SDS extraction), were analyzed by immunoblotting for the presence of A3G and Bet. Proper fractionation was controlled by the use of MAbs specific for the cytosolic proteins glyceraldehyde-3-phosphate dehydrogenase (GAPDH), calpain, and calnexin, the mitochondrial protein cytochrome P450 reductase, and the nuclear histone proteins. The results show that Bet reduced the amount of A3G in the cytosolic (F1), membrane/organelle (F2), and cytoskeleton (F4) fractions while increasing the amount of A3G protein in the nuclear (F3) fraction (Fig. 6D). Bet was found in all fractions, independent of the presence or absence of A3G (Fig. 6E), with the largest amounts in the nuclear fraction (F3). The control proteins GAPDH, calpain, calnexin, cytochrome P450 reductase, and histones were detected only in the expected fractions (Fig. 6D to F), and their recovery was not influenced by Bet or A3G (data not shown). To rule out the possibility that an artificially elevated level of A3G was modulating its detection, we performed immunoblot analysis of subcellular fractions derived from human PBMCs, which possess endogenous A3G. The results revealed that endogenous A3G was localized predominately in fractions F1 and F2 (Fig. 6F), similar to the localization of A3G in transfected 293T cells (Fig. 6D).
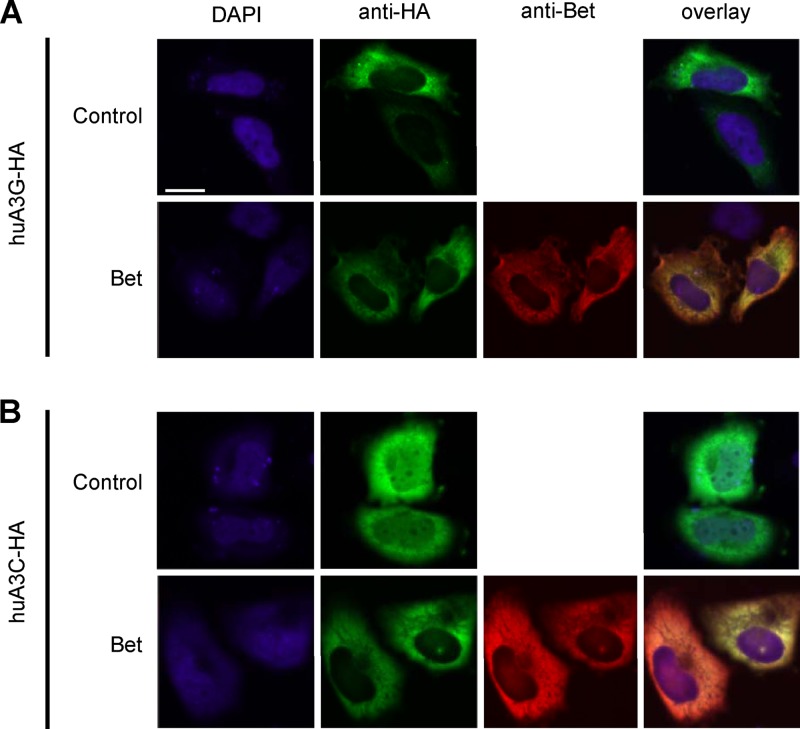
To further characterize whether Bet modifies the subcellular localization of A3G, we analyzed cells expressing huA3G together with Bet by confocal microscopy. Using anti-HA antibodies directed against HA-tagged A3G, immunofluorescence studies with transiently transfected HeLa cells showed that huA3G was distributed in the cytoplasm (Fig. 7A). In the presence of Bet, the cytoplasmic localization of A3G did not change. The Bet protein was detectable only in the cytoplasm, and expression of A3G did not affect its localization (Fig. 7A and data not shown). Because both A3G and Bet showed a cytoplasmic localization, we also tested A3C in the presence of Bet (Fig. 7B). A3C has a cytoplasmic and nuclear localization and is sensitive to Bet (19). Interestingly, in the presence of Bet, the nuclear A3C was removed, indicating that complexes of Bet and A3C proteins change their subcellular localization. Taking the data together, we concluded that Bet either modulates the subcellular localization of A3G via sequestration to a not yet identified cytoplasmic region or impairs its solubility, thus reducing its capacity for encapsidation.
Fig 7.
Influence of Bet on subcellular localization of APOBEC3G. HeLa cells were transfected with expression plasmids for HA-tagged huA3G and huA3G with Bet (A) or HA-tagged human APOBEC3C (huA3C-HA) and huA3C-HA with Bet (B). To detect APOBEC3 (green) and Bet (red), immunofluorescence staining was performed with anti-HA antibodies and Bet antiserum, respectively. Nuclei (blue) were visualized by 4′,6-diamidino-2-phenylindole (DAPI) staining. Bar = 20 μm.
DISCUSSION
In this study, we extend previous reports that PFV Bet can counteract human APOBEC3 proteins (19, 20). Our data are in full agreement with those of Russell et al., who reported that PFV Bet antagonizes A3G and can protect foamy viral and unrelated lentiviral particles (20). In our study, we showed that SIVAGMΔvif can be defended by Bet from A3Gs of human, chimpanzee, and AGM origin (Fig. 2). We also confirmed a molecular interaction between Bet and A3G that does not induce degradation of A3G. We found that Bet showed anti-A3G activity in a dose-dependent manner but that even the largest transfectable amounts of Bet expression plasmid could not fully restore viral infectivity (Fig. 1B). Thus, it is likely that the expression levels of Bet, like the case for A3G, are critical for the replication of wild-type PFV. The amount of A3G protein in the poorly described in vivo target cells of FVs is unknown.
Our aim was to obtain information about the functional consequences of Bet binding to A3G. We have demonstrated that Bet inhibits A3G-A3G complex formation in an RNA-independent manner. Very similarly, we recently showed that PFV Bet changes the capacity of A3C to form homodimers (19). The amino acid W127 in A3G was found to be critical for forming a complex with Bet. Changing this tryptophan to alanine abolished the binding to Bet. W127 is in the dimerization interface of A3G and was found to be important for A3G to form A3G-A3G dimers (36, 46). However, the importance of A3G dimerization is controversial, and Khan et al. did not see inhibition of A3G dimerization of the W127A mutant (53).
Our data allowed us to conclude that Bet prevents the formation of A3G dimers by its interaction, likely with the dimer interface. The interaction of Bet and A3G is either direct via W127 or indirectly regulated by W127. Interestingly, despite the positive selection in genes for A3G, the residues in the A3G dimer interface, including W127, are highly conserved between primates (36). Thus, targeting these residues essential for antiviral activity might explain the ability of PFV Bet to counteract diverse A3Gs of AGM, macaque, chimpanzee, and human origin (Fig. 2) (19). Amino acids in the dimer interface are necessary for A3G to gain access to budding viral particles. The packaging-defective phenotype of A3G W127A can be rescued by forcing its incorporation into HIV by fusion with Vpr (36). These data and the DNA editing shown in an A3G W127A bacterial expression system (54) demonstrate that this protein can deaminate cytidine. Our results show that A3G bound to Bet is not inhibited in cytidine deamination (Fig. 3C and E), providing further evidence that monomeric A3G is able to deaminate cytidines. In a first model, we propose that Bet counteracts A3s by preventing the A3 homodimerization that is required for viral encapsidation.
While W127 is important for A3G's interaction with Bet, very interestingly, the HIV-1 Vif interaction with A3G is regulated by amino acids 128 to 130, which are not involved in encapsidation (54). Thus, the presumed interaction sites of these two viral inhibitors are in remarkably close proximity on A3G (Fig. 5C). The molecular consequences of Vif interaction on A3G encapsidation in experimental situations where Vif-induced degradation of A3G is prevented are controversially described. Yu et al. showed that A3G is detectable in HIV particles when the Vif-induced degradation of A3G is blocked by the proteasome inhibitor MG132 (55). Very similarly, blocking the Vif-mediated degradation of A3G by inhibition of NEDD8 conjugation of the Cullin-5 compound of the ubiquitin ligase restored the restriction of HIV-1 by A3G and the viral A3G encapsidation (56). In a different system, Vif inhibited the packaging of a degradation-resistant mutant of A3G (A3G.C97A), suggesting that Vif, like Bet, might be able to interfere with the encapsidation of A3G without inducing A3G degradation (8).
Because A3G most likely interacts with nascent virions in the cytoplasm to become encapsidated, we were interested in the subcellular fate of A3G in the presence of Bet. We found that A3G disappeared from cytoplasmic extracts in a Bet-dependent manner (Fig. 6). Immunofluorescence microscopy did not display Bet-dependent redistribution of A3G within the cell as was seen for A3C (Fig. 7). It remains unknown whether Bet drags A3G to a subcellular compartment that resists simple detection in the microscope and sequesters it away from progeny virions or forms insoluble protein aggregates that inhibit the interaction with assembling viral proteins. Our data revealed the intriguing possibility that Bet uses a second mechanism to counteract A3G, sequestering A3G in insoluble complexes.
Because the A3 genes have undergone a turbulent evolution and are found only in placental mammals (2), the recent discoveries of endogenous foamy viruses in the aye-aye (Daubentonia madagascariensis, a strepsirrhine primate) and the two-toed sloth (Choloepus hoffmanni) and of foamy virus-like insertions within the genome of a “living fossil,” the coelacanth (Latimeria chalumnae), provide a unique opportunity to study the evolution of the foamy virus bet gene in species with and without A3 genes (57–59).
ACKNOWLEDGMENTS
We thank Nathaniel R. Landau, Dirk Lindemann, Martin Löchelt, and Sandro Rusconi for reagents and Wioletta Hörschken and Marion Battenberg for excellent technical assistance. The following reagents were obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: a monoclonal antibody to HIV-1 p24 (AG3.0), from Jonathan Allan; anti-ApoC17, from Klaus Strebel; and pcDNA3.1 human APOBEC3G-Myc-6XHis, from David Kabat.
This project was funded by DFG grant MU 1608/3-1. M.P. was also supported by the Forschungskommission of the Medical Faculty, Heinrich Heine University Düsseldorf. A.A.J.V. was supported by the International NRW Research School BioStruct, under a grant from the Ministry of Innovation, Science and Research of the North Rhine-Westphalia State, by the Heinrich Heine University Düsseldorf, and by the Entrepreneur Foundation at the Heinrich Heine University Düsseldorf. C.M. was supported by the Heinz-Ansmann Foundation for AIDS Research.
Footnotes
Published ahead of print 12 June 2013
REFERENCES
- 1. LaRue RS, Jonsson SR, Silverstein KA, Lajoie M, Bertrand D, El-Mabrouk N, Hotzel I, Andresdottir V, Smith TP, Harris RS. 2008. The artiodactyl APOBEC3 innate immune repertoire shows evidence for a multi-functional domain organization that existed in the ancestor of placental mammals. BMC Mol. Biol. 9:104. 10.1186/1471-2199-9-104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Münk C, Willemsen A, Bravo IG. 2012. An ancient history of gene duplications, fusions and losses in the evolution of APOBEC3 mutators in mammals. BMC Evol. Biol. 12:71. 10.1186/1471-2148-12-71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Münk C, Jensen BE, Zielonka J, Häussinger D, Kamp C. 2012. Running loose or getting lost: how HIV-1 counters and capitalizes on APOBEC3-induced mutagenesis through its Vif protein. Viruses 4:3132–3161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sheehy AM, Erthal J. 2012. APOBEC3 versus retroviruses, immunity versus invasion: clash of the titans. Mol. Biol. Int. 2012:974924. 10.1155/2012/974924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chiu YL, Greene WC. 2009. APOBEC3G: an intracellular centurion. Philos. Trans. R. Soc. Lond. B Biol. Sci. 364:689–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kao S, Goila-Gaur R, Miyagi E, Khan MA, Opi S, Takeuchi H, Strebel K. 2007. Production of infectious virus and degradation of APOBEC3G are separable functional properties of human immunodeficiency virus type 1 Vif. Virology 369:329–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kao S, Miyagi E, Khan MA, Takeuchi H, Opi S, Goila-Gaur R, Strebel K. 2004. Production of infectious human immunodeficiency virus type 1 does not require depletion of APOBEC3G from virus-producing cells. Retrovirology 1:27. 10.1186/1742-4690-1-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Opi S, Kao S, Goila-Gaur R, Khan MA, Miyagi E, Takeuchi H, Strebel K. 2007. Human immunodeficiency virus type 1 Vif inhibits packaging and antiviral activity of a degradation-resistant APOBEC3G variant. J. Virol. 81:8236–8246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Santa-Marta M, da Silva FA, Fonseca AM, Goncalves J. 2005. HIV-1 Vif can directly inhibit apolipoprotein B mRNA-editing enzyme catalytic polypeptide-like 3G-mediated cytidine deamination by using a single amino acid interaction and without protein degradation. J. Biol. Chem. 280:8765–8775 [DOI] [PubMed] [Google Scholar]
- 10. Britan-Rosich E, Nowarski R, Kotler M. 2011. Multifaceted counter-APOBEC3G mechanisms employed by HIV-1 Vif. J. Mol. Biol. 410:1065–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Arias JF, Koyama T, Kinomoto M, Tokunaga K. 2012. Retroelements versus APOBEC3 family members: no great escape from the magnificent seven. Front. Microbiol. 3:275. 10.3389/fmicb.2012.00275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Derse D, Hill SA, Princler G, Lloyd P, Heidecker G. 2007. Resistance of human T cell leukemia virus type 1 to APOBEC3G restriction is mediated by elements in nucleocapsid. Proc. Natl. Acad. Sci. U. S. A. 104:2915–2920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ooms M, Krikoni A, Kress AK, Simon V, Münk C. 2012. APOBEC3A, APOBEC3B, and APOBEC3H haplotype 2 restrict human T-lymphotropic virus type 1. J. Virol. 86:6097–6108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kolokithas A, Rosenke K, Malik F, Hendrick D, Swanson L, Santiago ML, Portis JL, Hasenkrug KJ, Evans LH. 2010. The glycosylated Gag protein of a murine leukemia virus inhibits the antiretroviral function of APOBEC3. J. Virol. 84:10933–10936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Stavrou S, Nitta T, Kotla S, Ha D, Nagashima K, Rein AR, Fan H, Ross SR. 2013. Murine leukemia virus glycosylated Gag blocks apolipoprotein B editing complex 3 and cytosolic sensor access to the reverse transcription complex. Proc. Natl. Acad. Sci. U. S. A. 110:9078–9083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chareza S, Slavkovic LD, Liu Y, Rathe AM, Münk C, Zabogli E, Pistello M, Löchelt M. 2012. Molecular and functional interactions of cat APOBEC3 and feline foamy and immunodeficiency virus proteins: different ways to counteract host-encoded restriction. Virology 424:138–146 [DOI] [PubMed] [Google Scholar]
- 17. Löchelt M, Romen F, Bastone P, Muckenfuss H, Kirchner N, Kim YB, Truyen U, Rosler U, Battenberg M, Saib A, Flory E, Cichutek K, Münk C. 2005. The antiretroviral activity of APOBEC3 is inhibited by the foamy virus accessory Bet protein. Proc. Natl. Acad. Sci. U. S. A. 102:7982–7987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Münk C, Beck T, Zielonka J, Hotz-Wagenblatt A, Chareza S, Battenberg M, Thielebein J, Cichutek K, Bravo IG, O'Brien SJ, Löchelt M, Yuhki N. 2008. Functions, structure, and read-through alternative splicing of feline APOBEC3 genes. Genome Biol. 9:R48. 10.1186/gb-2008-9-3-r48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Perkovic M, Schmidt S, Marino D, Russell RA, Stauch B, Hofmann H, Kopietz F, Kloke BP, Zielonka J, Strover H, Hermle J, Lindemann D, Pathak VK, Schneider G, Löchelt M, Cichutek K, Münk C. 2009. Species-specific inhibition of APOBEC3C by the prototype foamy virus protein Bet. J. Biol. Chem. 284:5819–5826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Russell RA, Wiegand HL, Moore MD, Schafer A, McClure MO, Cullen BR. 2005. Foamy virus Bet proteins function as novel inhibitors of the APOBEC3 family of innate antiretroviral defense factors. J. Virol. 79:8724–8731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu W, Worobey M, Li Y, Keele BF, Bibollet-Ruche F, Guo Y, Goepfert PA, Santiago ML, Ndjango JB, Neel C, Clifford SL, Sanz C, Kamenya S, Wilson ML, Pusey AE, Gross-Camp N, Boesch C, Smith V, Zamma K, Huffman MA, Mitani JC, Watts DP, Peeters M, Shaw GM, Switzer WM, Sharp PM, Hahn BH. 2008. Molecular ecology and natural history of simian foamy virus infection in wild-living chimpanzees. PLoS Pathog. 4:e1000097. 10.1371/journal.ppat.1000097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Herchenroder O, Renne R, Loncar D, Cobb EK, Murthy KK, Schneider J, Mergia A, Luciw PA. 1994. Isolation, cloning, and sequencing of simian foamy viruses from chimpanzees (SFVcpz): high homology to human foamy virus (HFV). Virology 201:187–199 [DOI] [PubMed] [Google Scholar]
- 23. Locatelli S, Peeters M. 2012. Cross-species transmission of simian retroviruses: how and why they could lead to the emergence of new diseases in the human population. AIDS 26:659–673 [DOI] [PubMed] [Google Scholar]
- 24. Sandstrom PA, Phan KO, Switzer WM, Fredeking T, Chapman L, Heneine W, Folks TM. 2000. Simian foamy virus infection among zoo keepers. Lancet 355:551–552 [DOI] [PubMed] [Google Scholar]
- 25. Switzer WM, Bhullar V, Shanmugam V, Cong ME, Parekh B, Lerche NW, Yee JL, Ely JJ, Boneva R, Chapman LE, Folks TM, Heneine W. 2004. Frequent simian foamy virus infection in persons occupationally exposed to nonhuman primates. J. Virol. 78:2780–2789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Heneine W, Schweizer M, Sandstrom P, Folks T. 2003. Human infection with foamy viruses. Curr. Top. Microbiol. Immunol. 277:181–196 [DOI] [PubMed] [Google Scholar]
- 27. Khan AS. 2009. Simian foamy virus infection in humans: prevalence and management. Expert Rev. Anti Infect. Ther. 7:569–580 [DOI] [PubMed] [Google Scholar]
- 28. Linial M. 2000. Why aren't foamy viruses pathogenic? Trends Microbiol. 8:284–289 [DOI] [PubMed] [Google Scholar]
- 29. Bock M, Heinkelein M, Lindemann D, Rethwilm A. 1998. Cells expressing the human foamy virus (HFV) accessory Bet protein are resistant to productive HFV superinfection. Virology 250:194–204 [DOI] [PubMed] [Google Scholar]
- 30. Meiering CD, Linial ML. 2002. Reactivation of a complex retrovirus is controlled by a molecular switch and is inhibited by a viral protein. Proc. Natl. Acad. Sci. U. S. A. 99:15130–15135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Delebecque F, Suspene R, Calattini S, Casartelli N, Saib A, Froment A, Wain-Hobson S, Gessain A, Vartanian JP, Schwartz O. 2006. Restriction of foamy viruses by APOBEC cytidine deaminases. J. Virol. 80:605–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mariani R, Chen D, Schröfelbauer B, Navarro F, König R, Bollman B, Münk C, Nymark-McMahon H, Landau NR. 2003. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell 114:21–31 [DOI] [PubMed] [Google Scholar]
- 33. Duda A, Stange A, Luftenegger D, Stanke N, Westphal D, Pietschmann T, Eastman SW, Linial ML, Rethwilm A, Lindemann D. 2004. Prototype foamy virus envelope glycoprotein leader peptide processing is mediated by a furin-like cellular protease, but cleavage is not essential for viral infectivity. J. Virol. 78:13865–13870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pietschmann T, Heinkelein M, Heldmann M, Zentgraf H, Rethwilm A, Lindemann D. 1999. Foamy virus capsids require the cognate envelope protein for particle export. J. Virol. 73:2613–2621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Marin M, Rose KM, Kozak SL, Kabat D. 2003. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat. Med. 9:1398–1403 [DOI] [PubMed] [Google Scholar]
- 36. Bulliard Y, Turelli P, Rohrig UF, Zoete V, Mangeat B, Michielin O, Trono D. 2009. Functional analysis and structural modeling of human APOBEC3G reveal the role of evolutionarily conserved elements in the inhibition of human immunodeficiency virus type 1 infection and Alu transposition. J. Virol. 83:12611–12621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Achong BG, Mansell PW, Epstein MA, Clifford P. 1971. An unusual virus in cultures from a human nasopharyngeal carcinoma. J. Natl. Cancer Inst. 46:299–307 [PubMed] [Google Scholar]
- 38. Cullen BR. 1986. Trans-activation of human immunodeficiency virus occurs via a bimodal mechanism. Cell 46:973–982 [DOI] [PubMed] [Google Scholar]
- 39. Muckenfuss H, Kaiser JK, Krebil E, Battenberg M, Schwer C, Cichutek K, Münk C, Flory E. 2007. Sp1 and Sp3 regulate basal transcription of the human APOBEC3G gene. Nucleic Acids Res. 35:3784–3796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fang L, Landau NR. 2007. Analysis of Vif-induced APOBEC3G degradation using an alpha-complementation assay. Virology 359:162–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Holland AU, Munk C, Lucero GR, Nguyen LD, Landau NR. 2004. Alpha-complementation assay for HIV envelope glycoprotein-mediated fusion. Virology 319:343–352 [DOI] [PubMed] [Google Scholar]
- 42. Moosmann P, Rusconi S. 1996. Alpha complementation of LacZ in mammalian cells. Nucleic Acids Res. 24:1171–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Löchelt M, Zentgraf H, Flugel RM. 1991. Construction of an infectious DNA clone of the full-length human spumaretrovirus genome and mutagenesis of the bel1 gene. Virology 184:43–54 [DOI] [PubMed] [Google Scholar]
- 44. Simm M, Shahabuddin M, Chao W, Allan JS, Volsky DJ. 1995. Aberrant Gag protein composition of a human immunodeficiency virus type 1 vif mutant produced in primary lymphocytes. J. Virol. 69:4582–4586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nowarski R, Britan-Rosich E, Shiloach T, Kotler M. 2008. Hypermutation by intersegmental transfer of APOBEC3G cytidine deaminase. Nat. Struct. Mol. Biol. 15:1059–1066 [DOI] [PubMed] [Google Scholar]
- 46. Huthoff H, Autore F, Gallois-Montbrun S, Fraternali F, Malim MH. 2009. RNA-dependent oligomerization of APOBEC3G is required for restriction of HIV-1. PLoS Pathog. 5:e1000330. 10.1371/journal.ppat.1000330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sali A, Blundell TL. 1993. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234:779–815 [DOI] [PubMed] [Google Scholar]
- 48. Holden LG, Prochnow C, Chang YP, Bransteitter R, Chelico L, Sen U, Stevens RC, Goodman MF, Chen XS. 2008. Crystal structure of the anti-viral APOBEC3G catalytic domain and functional implications. Nature 456:121–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Soros VB, Yonemoto W, Greene WC. 2007. Newly synthesized APOBEC3G is incorporated into HIV virions, inhibited by HIV RNA, and subsequently activated by RNase H. PLoS Pathog. 3:e15. 10.1371/journal.ppat.0030015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kreisberg JF, Yonemoto W, Greene WC. 2006. Endogenous factors enhance HIV infection of tissue naive CD4 T cells by stimulating high molecular mass APOBEC3G complex formation. J. Exp. Med. 203:865–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ramsby M, Makowski G. 2011. Differential detergent fractionation of eukaryotic cells. Cold Spring Harb. Protoc. 2011:prot5592. 10.1101/pdb.prot5592 [DOI] [PubMed] [Google Scholar]
- 52. Ramsby ML, Makowski GS. 1999. Differential detergent fractionation of eukaryotic cells. Analysis by two-dimensional gel electrophoresis. Methods Mol. Biol. 112:53–66 [DOI] [PubMed] [Google Scholar]
- 53. Khan MA, Goila-Gaur R, Kao S, Miyagi E, Walker RC, Jr, Strebel K. 2009. Encapsidation of APOBEC3G into HIV-1 virions involves lipid raft association and does not correlate with APOBEC3G oligomerization. Retrovirology 6:99. 10.1186/1742-4690-6-99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Huthoff H, Malim MH. 2007. Identification of amino acid residues in APOBEC3G required for regulation by human immunodeficiency virus type 1 Vif and virion encapsidation. J. Virol. 81:3807–3815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yu X, Yu Y, Liu B, Luo K, Kong W, Mao P, Yu XF. 2003. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 302:1056–1060 [DOI] [PubMed] [Google Scholar]
- 56. Stanley DJ, Bartholomeeusen K, Crosby DC, Kim DY, Kwon E, Yen L, Cartozo NC, Li M, Jager S, Mason-Herr J, Hayashi F, Yokoyama S, Krogan NJ, Harris RS, Peterlin BM, Gross JD. 2012. Inhibition of a NEDD8 cascade restores restriction of HIV by APOBEC3G. PLoS Pathog. 8:e1003085. 10.1371/journal.ppat.1003085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Han GZ, Worobey M. 2012. An endogenous foamy virus in the aye-aye (Daubentonia madagascariensis). J. Virol. 86:7696–7698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Han GZ, Worobey M. 2012. An endogenous foamy-like viral element in the coelacanth genome. PLoS Pathog. 8:e1002790. 10.1371/journal.ppat.1002790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Katzourakis A, Gifford RJ, Tristem M, Gilbert MT, Pybus OG. 2009. Macroevolution of complex retroviruses. Science 325:1512. 10.1126/science.1174149 [DOI] [PubMed] [Google Scholar]