Abstract
RNA interference (RNAi) is an important antiviral defense response in plants and invertebrates; however, evidences for its contribution to mammalian antiviral defense are few. In the present study, we demonstrate the anti-dengue virus role of RNAi in mammalian cells. Dengue virus infection of Huh 7 cells decreased the mRNA levels of host RNAi factors, namely, Dicer, Drosha, Ago1, and Ago2, and in corollary, silencing of these genes in virus-infected cells enhanced dengue virus replication. In addition, we observed downregulation of many known human microRNAs (miRNAs) in response to viral infection. Using reversion-of-silencing assays, we further showed that NS4B of all four dengue virus serotypes is a potent RNAi suppressor. We generated a series of deletion mutants and demonstrated that NS4B mediates RNAi suppression via its middle and C-terminal domains, namely, transmembrane domain 3 (TMD3) and TMD5. Importantly, the NS4B N-terminal region, including the signal sequence 2K, which has been implicated in interferon (IFN)-antagonistic properties, was not involved in mediating RNAi suppressor activity. Site-directed mutagenesis of conserved residues revealed that a Phe-to-Ala (F112A) mutation in the TMD3 region resulted in a significant reduction of the RNAi suppression activity. The green fluorescent protein (GFP)-small interfering RNA (siRNA) biogenesis of the GFP-silenced line was considerably reduced by wild-type NS4B, while the F112A mutant abrogated this reduction. These results were further confirmed by in vitro dicer assays. Together, our results suggest the involvement of miRNA/RNAi pathways in dengue virus establishment and that dengue virus NS4B protein plays an important role in the modulation of the host RNAi/miRNA pathway to favor dengue virus replication.
INTRODUCTION
RNA interference (RNAi) is a conserved, sequence-specific, gene-regulatory mechanism that plays an important role(s) in host cell defense against viral pathogens and transposons in plants, insects, and nematodes (1, 2). A number of reports have implied a direct role of RNAi in regulating viral infections in mammalian cells (3). Furthermore, it was found that mutations in components of RNAi machinery enhanced the replication of vesicular stomatitis virus (VSV), influenza virus, and human immunodeficiency virus (HIV) in mammalian cells (3). Viruses in turn have evolved mechanisms that counteract this host cell defense by encoding factors known as RNAi suppressors (4). The suppressor proteins and noncoding viral RNAs can inhibit the RNAi (microRNA [miRNA]/small interfering RNA [siRNA]) pathway through different mechanisms (5–7). Suppression of RNAi by viral proteins was first discovered in plants. In 1998, the Baulcombe group showed convincingly the suppression of trans-gene silencing by HC-Pro of potato virus Y (PVY) and the 2b protein of cucumber mosaic virus (CMV2b) (8). Subsequently, several viral suppressors of RNA silencing (VSRs) have been identified in plant and animal viruses based on their ability to interfere with antiviral responses and endogenous silencing pathways (5, 7). A majority of plant VSRs that have been identified so far have functions other than suppression of RNAi. These proteins also function as coat proteins, replicases, movement proteins, and helper components for viral transmission, replication, or transcriptional regulation, etc. (6).
Although small RNA-based mechanisms such as RNAi or miRNA levels have been studied mainly in plant and invertebrate systems, a recent study by Parameswaran et al. (9) highlighted the importance of these mechanisms in vertebrate systems. Human viruses, like plant viruses, encode suppressor proteins or RNAs that block or modulate the RNAi pathway. The best-studied mammalian VSRs are Ebola virus VP35, VP30, and VP40 proteins; severe acute respiratory syndrome (SARS) virus 7a accessory protein; NS1 of influenza A virus; HIV-1 TAT; capsid protein of hepatitis C virus (HCV); and primate foamy virus Tas (5, 10, 11). Viral suppressors are diverse within and across kingdoms, exhibiting little or no sequence similarities, and they act via different mechanisms (7). However, a few functional motifs, such as double-stranded-RNA (dsRNA)-binding properties and/or GW repeat motifs, have been demonstrated for some VSRs (5, 12, 13). As mentioned above, VSRs interfere with the endogenous silencing pathways at different steps. P14 of Pothos latent aureus virus and p38 of turnip crinkle virus (TCV) inhibit the processing of dsRNA to siRNAs (14, 15). We have recently shown that flock house virus B2 protein (FHVB2) binds Dicer, thus preventing dsRNA processing (16). p19 of tombusviruses, tomato aspermy cucumovirus 2b protein, and B2 of flock house virus bind siRNAs and sequester them, thus preventing siRNA/miRNA RISC assembly (17–19). The prevention of RNA-induced silencing complex (RISC) assembly also occurs through direct or indirect interactions between VSRs and the protein components of RISC. Fny-CMV2b protein physically interacts with the PAZ domain and part of the PIWI domain of Ago1 (20). A few VSRs use the GW/WG AGO hook to inhibit siRNA/miRNA-loaded RISC activity and also target the amplification of antiviral silencing. The vast majority of mammalian viral proteins have not been assessed for potential RNA silencing suppressor (RSS) activities.
Dengue virus (DV) is the causative agent of dengue fever, the most prevalent arthropod-borne viral illness in humans, with 50 to 100 million individuals infected annually worldwide. The role of the host RNAi pathway in mosquitoes upon arboviral infection has been extensively studied (21). The four serotypes of DV identified so far (DV1 to DV4) have a single-stranded genomic RNA of positive polarity that serves as the mRNA for the translation of a large polyprotein that is co- and posttranslationally processed. Most, if not all, nonstructural (NS) proteins are involved in the replication of dengue virus RNA. NS5 is the RNA-dependent RNA polymerase, NS3 acts as the viral serine protease as well as the RNA helicase, and the glycoprotein NS1 probably plays a role at an early step of viral RNA replication. Little is known about the functions of the small hydrophobic proteins NS2A, NS4A, and NS4B. NS4B is the largest of the small hydrophobic NS proteins of dengue virus, consisting of 248 amino acids (aa). It has been shown to be part of the viral replication complex and is also implicated in DV pathogenicity (22, 23).
Relatively little is known about the role of host RNAi/miRNA pathways, including the role of viral suppressor proteins, in flavivirus replication in mammalian cells (24). In the present study, we showed that DV infection downregulates components of host RNAi/miRNA machinery affecting the biogenesis of many known human miRNAs. Depletion of some of the RNAi components by siRNA-mediated silencing leads to increases in DV replication. This indicates that the RNAi machinery is involved in controlling DV replication in mammalian cells, and one or more of the DV proteins may counteract this host defense. We identified NS4B as one of the DV proteins with RNAi suppression activity and showed that this activity is independent of the interferon (IFN) inhibition functions of NS4B. Furthermore, the NS4B protein failed to bind dsRNA but interfered with dicing activity. Together, our results provide evidence for the antiviral function of the host RNAi/miRNA machinery in dengue virus replication in mammalian cells and implicate NS4B as a VSR that might promote virus replication.
MATERIALS AND METHODS
Cell lines and viruses.
Huh 7 and HEK293T cells were propagated in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 2 mM l-glutamine, 100 units/ml penicillin G sodium, and 100 μg/ml streptomycin sulfate and nonessential amino acids at 37°C and 5% CO2.
DV2 (P23085 INDI-60) was obtained from the National Institute of Virology, Pune, India, and propagated in C6/36 mosquito cells. Infected culture supernatant was used throughout the study.
Gene synthesis and generation of plasmid constructs.
To express NS4B in the Sf21 cell line, the ns4b gene was amplified by using primers ns4b forward (5′-GGA ATG GGA AAC GAG ATG GGT TTC CTA-3′) and ns4b reverse (5′-GTC GAC TTA CCT TCT TGT GTT GGT TGT GTT-3′). Plasmid pTM1.4-DV-2K(1–249) GFP (a kind gift of Ralf Bartenschlager, University of Heidelberg, Heidelberg, Germany) served as a template for PCR amplification of 2K-NS4B using primers 2k-ns4b forward (5′-GAA ATG GAA TTA ACA CCC CAA GAT AAC CAA TTG-3′) and 2k-ns4b reverse (5′-GTC GAC TTA CCT TCT TGT GTT GGT TGT GTT-3′). Additionally, NS4B sequences from the remaining serotypes of dengue virus were amplified by using the primers listed in Table 1. The amplified products were ligated into the pIB-V5/His-TOPO vector (Invitrogen, Grand Island, NY, USA) under the control of a baculovirus early promoter, OpIE2, for better expression levels.
Table 1.
List of primers for different serotypes of dengue virus ns4b
| Dengue virus serotype | Primer sequence of NS4B (5′–3′)a |
|---|---|
| DV1 | GGA ATG GGA AAT GAG ATG GGA TTA CTG GAA (For) |
| TTA TCT CCT ACC TCC TCC TAG AGA TTT (Rev) | |
| DV3 | GGA ATG GGA AAT GAA ATG GGA CTG CTG GAA (For) |
| TTA TCT TTT TCC TGT TCC AAC TGA TTT (Rev) | |
| DV4 | GGA ATG GGA AAC GAG ATG GGG CTG ATT GAA (For) |
| TTA CCT CCT AGG GGT TTG TGC ATT CTT (Rev) |
For, forward; Rev, reverse.
To express NS4B in the leaves of Nicotiana xanthi, the ns4b gene was amplified by using primers ns4b forward (5′-GGA TCC GGA ATG GGA AAC GAG ATG GGT-3′) and ns4b reverse (5′-GAG CTC TTA CCT TCT TGT GTT GGT TGT-3′) and cloned into the plant binary vector pBI121 under the control of the cauliflower mosaic virus (CaMV) 35S promoter.
To express NS4B proteins of all serotypes and 2K-NS4B in a mammalian cell line (HEK293T), DNAs corresponding to these genes were amplified and cloned into pIB-V5/His-TOPO. The cloned fragments were mobilized into the mammalian expression vector pCDNA 3.1+ (Invitrogen, Grand Island, NY, USA) under the control of the human cytomegalovirus (CMV) promoter.
siRNA transfection and virus infection.
Silencer Select siRNAs for Dicer, Drosha, Ago1, and Ago2 were purchased from Ambion. The knockdown studies were performed by transfecting 10 nM siRNA duplex Huh 7 cells by using a reverse transfection protocol with Lipofectamine 2000 (Invitrogen, Grand Island, NY, USA). At 48 h posttransfection, cells were infected with virus at a multiplicity of infection (MOI) of 10. The virus was diluted in DMEM containing 2% FBS, and cells were incubated for 1 h at 37°C on a rocker. After 1 h, cells were washed twice with PBS and grown in complete growth medium. Cells were collected at 48 h postinfection by trypsinization, and the cell pellets were washed once with phosphate-buffered saline (PBS) and lysed in TRIzol (Invitrogen, Grand Island, NY, USA) for RNA isolation. RNA was prepared according to the manufacturer's protocol. DNase I-treated total RNA was used for quantitative real-time PCR (qRT-PCR).
Quantitative real-time PCR.
The comparative threshold cycle (CT) method with SYBR green was conducted for mRNA quantification of host RNAi factors using primers described previously (25), and viral genomic RNA was quantitated by using TaqMan primer-probe mix as described previously (26). Genome copy numbers were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA levels. Relative expression was calculated by using the comparative threshold cycle method. Similarly, miRNA quantifications were performed by using the TaqMan microRNA assay system according to the manufacturer's instructions.
Small RNA library preparation and sequencing.
Huh 7 cells were infected with DV2 at an MOI of 10. At 48 h postinfection, the cells were harvested, and total RNA was extracted by using TRIzol according to the manufacturer's protocol. The total RNA of the uninfected and infected cells was used to prepare a small RNA library using Ture Seq small RNA library preparation by Illumina Inc. The PCR products of the library corresponding to a length of 145 to 160 bp were considered and sequenced by using Solexa, a massively parallel sequencing technology.
Analysis of sequence reads for known miRNAs.
Small 36-nucleotide (nt) RNA reads were produced by using the Illumina genome analyzer. Low-quality reads were trimmed with our own perl script. Adaptor sequences were accurately clipped with the aid of a dynamic programming algorithm. After elimination of redundancy, sequences of ≥18 nt and <26 nt were used to create a set of unique reads. The unique reads were matched to miRBase v19 and mapped to known human miRNAs. Sequences that perfectly matched the reported human miRNA sequences were considered for subsequent analysis.
Generation and overexpression of NS4B mutants.
To generate NS4B deletion mutants, ns4b gene segments with transmembrane domain (TMD) deletions were amplified by using specific primers. The primers used for the generation of the deletion mutants are listed in Table 2. The amplified products were ligated into the pIB-V5/His-TOPO vector, and the orientation of the genes was confirmed by PCR and restriction analysis.
Table 2.
List of primer sequences used for generation of ns4b deletion mutants
| Deletion mutant | Primer sequence (5′–3′) |
|---|---|
| AB | GGA ATG GGA AAC GAG ATG GGT TTC CTA (For) |
| CAG AGC CCA TGT AGT CCT CAT CAT CAA (Rev) | |
| BC | GGA ATG GGA CTC GCC ATT GGA TGC TAC TCA (For) |
| CCT TCT TGT GTT GGT TGT GTT CTT CAT (Rev) | |
| A | GGA ATG GGA AAC GAG ATG GGT TTC CTA (For) |
| GGA TCC TTA AAG GGG AAC TCC GAT GTC CAT (Rev) | |
| B | GGA ATG GGA CTC GCC ATT GGA TGC TAC TCA (For) |
| CAG AGC CCA TGT AGT CCT CAT CAT CAA (Rev) | |
| C | GGA ATG GGA TGT GAG GCT TTA ACC TTA GCT (For) |
| CCT TCT TGT GTT GGT TGT GTT CTT CAT (Rev) | |
| TMD123 | GGA ATG GGA AAC GAG ATG GGT TTC CTA (For) |
| GTC GAC TTA AGT TGG GTT TTT CAT GAT GCC (Rev) | |
| TMD45 | GAA ATG GTC GAT GGA ATA ACA GTG ATT (For) |
| GTC GAC TTA CCT TCT TGT GTT GGT TGT GTT (Rev) |
Conserved and alanine scan mutants in the TMD3 region of the ns4b gene were constructed by using the QuikChange site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA, USA) according to the manufacturer's protocol. Briefly, PCR was performed on the wild-type (wt) ns4b gene cloned into a pIB-V5/His-TOPO vector using primer pairs with the corresponding mutation. Pfu Turbo DNA polymerase (Agilent Technologies, Santa Clara, CA, USA) was used for PCR. The mismatched oligonucleotides used for site-directed mutagenesis are shown in Table 3. After amplification, the PCR mixture was subsequently treated with DpnI enzyme to remove methylated parental DNA prior to transformation into Escherichia coli XL1B cells. Three clones were selected for each mutant, and the plasmid from each colony was isolated and sequenced to confirm the incorporation of the desired mutation. All of the deletion and substitution mutants were further cloned into pCDNA3.1+ as described above for transfection into the HEK293T cell line.
Table 3.
List of primer sequences used for various substitution mutants of ns4b
| Substitution mutation | Primer sequence (5′–3′) |
|---|---|
| P104L | TCA CAA GTC AAC CTC ATA ACT CTC ACA (For) |
| TGT GAG AGT TAT GAG GTT GAC TTG TGA (Rev) | |
| P104A | TCA CAA GTC AAC GCC ATA ACT CTC ACA (For) |
| TGT GAG AGT TAT GGC GTT GAC TTG TGA (Rev) | |
| T108I | CCC ATA ACT CTC ATA GCA GCT CTT TTC (For) |
| GAA AAG AGC TGC TAT GAG AGT TAT GGG (Rev) | |
| T108A | CCC ATA ACT CTC GCA GCA GCT CTT TTC (For) |
| GAA AAG AGC TGC TGC GAG AGT TAT GGG (Rev) | |
| F112L | ACA GCA GCT CTT CTC TTA TTG GTA GCA (For) |
| TGC TAC CAA TAA GAG AAG AGC TGC TGT (Rev) | |
| F112A | ACA GCA GCT CTT GCC TTA TTG GTA GCA (For) |
| TGC TAC CAA TAA GGC AAG AGC TGC TGT (Rev) |
Suppression-of-RNAi assay in the Sf21 cell line.
To analyze dengue virus NS4B suppressor activity in the Sf21 cell line, a previously developed Sf21 RNAi sensor line (experimental line 1 [green fluorescent protein {GFP}-silenced cells in which GFP expression levels are reduced to the minimum by constitutively expressing GFP and short hairpin RNA {shRNA} of gfp under the control of the same promoter, OpIE2]) was employed (16). Briefly, Sf21 cells were seeded into 6-well plates in BD Baculogold TNM-FH insect medium containing 10% fetal bovine serum with gentamicin and 300 μg/ml zeocin. The line was transfected with 1 μg of plasmid pIB-V5/His-TOPO containing the ns4B gene by using Cellfectin II reagent (Invitrogen, Grand Island, NY, USA) in BD Baculogold max-XP serum-free medium. After 4 h, the serum-free medium was replaced with serum-containing medium, and the cells were allowed to transiently express NS4B. The vector pIB-V5/His-TOPO was transfected independently to serve as a control. GFP expression in the transfected lines was quantified by fluorescence-activated cell sorter (FACS) analysis.
Suppression-of-RNAi assay in plant leaves.
The suppression assay in plant leaves was performed as described previously (27). Briefly, NS4B was transiently expressed by agroinfiltrating a recombinant pBI121 vector harboring the ns4B gene into plant tissues of N. xanthi expressing a GFP reporter gene and GFP shRNA, and the leaves were observed at 8 days postinfiltration (dpi). As a control, the vector pBI121 was agroinfiltrated into leaf tissues.
Suppression-of-RNAi assay in a mammalian (HEK293T) cell line.
The suppression assay in a mammalian (HEK293T) cell line was performed as described previously (27). Briefly, HEK293T cells were seeded into 12-well plates in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum. The cells were transfected with 1 μg each of the pSIREN-RetroQ, pSIREN-RetroQ-shRNA gfp, and pSIREN-RetroQ-shRNA gfp vectors with NS4B/pCDNA3.1+ or NS4B deletion/substitution mutants by using JetPRIME reagent (Polyplus Transfection; BP, Illkirch, France) in jetPRIME buffer. After 4 h of transfection, the serum-free medium was replaced by serum-supplemented medium, and the cells were allowed to transiently express NS4B and/or its deletion as well as substitution mutants. The GFP expression in the transfected lines was quantified by FACS analysis.
Flow cytometric (FACS) analysis of various cell lines.
The GFP expression levels in two different cell lines were quantified by FACS analysis. For the FACS analysis, Sf21 cells or HEK293T cells were washed with FACS-grade PBS (BD Biosciences) and resuspended in 400 μl of PBS. The GFP fluorescence of the cells was determined by FACSCalibur flow cytometry (Becton, Dickinson, Franklin Lakes, NJ, USA). The fluorescence analysis was performed by using Cellquest software (Becton, Dickinson, Franklin Lakes, NJ, USA). All the experiments involving quantification of GFP-fluorescing cells by FACS analysis were carried out three times, and statistical significance of the results was determined by the use of Student's t test.
IFN assay.
For the IFN assay, HeLa cells were considered instead of HEK293T cells. Unlike HEK293T cells, HeLa cells express TLR3 (Toll-like receptor 3) on their surface, which can recognize poly(I·C) to stimulate an IFN response inside the cell. The cells were cotransfected with 100 ng of pISRE-Luc (a firefly luciferase expression plasmid under the control of the interferon-stimulated response element [ISRE] promoter) (Agilent Technologies, Santa Clara, CA, USA) and 500 ng of plasmids encoding wt NS4B, 2K-NS4B, and domain BC/NS4B. Subsequently, at 24 h posttransfection, cells were transfected with poly(I·C) (20 μg/ml) by using Lipofectamine 2000 CD reagent (Invitrogen, Grand Island, NY, USA). The cells were lysed 7 h after the poly(I·C) treatment, and the luciferase expression level was determined by using the dual-luciferase reporter assay (Promega, Madison, WI, USA) according to the manufacturer's protocol.
Protein purification.
NS4B is predicted to be a membrane protein. The protein was purified in a recombinant manner according to a protocol described previously (28). Briefly, protein, expressed in BL21 cells, was localized in inclusion bodies, which were isolated as described previously (28). Purified inclusion bodies were solubilized in 50 mM 3-(cyclohexylamino)-1-propanesulfonic acid (CAPS) buffer (pH 11.0) containing 1.5% N-lauryl sarcosine and 0.3 M NaCl for 30 min, and the solubilized protein was separated by centrifugation (10,000 × g for 30 min). Protein was purified by affinity chromatography on a His-Trap column, a high-performance nickel affinity column (Qiagen), using an imidazole gradient in 50 mM CAPS buffer (pH 11.0) containing 0.3% N-lauryl sarcosine and 0.3 M NaCl. Protein-containing fractions were pooled, purified to homogeneity, and concentrated by ultrafiltration at 3,000 × g (Centricon-30 with a cutoff of 10 kDa).
Preparation of gfp dsRNA.
For both in vitro RNA-binding and dicing assays, the substrate, gfp dsRNA, was prepared by using the Riboprobe Combination System-SP6/T7 RNA polymerase (Promega, Madison, WI, USA), but for the RNA-binding assay, the substrate was incorporated with [α-32P]UTP. At the end of the reaction, the substrate was subjected to RNase-free DNase I digestion and purified on an Illustra MicroSpin G-25 column (GE Healthcare, Piscataway, NJ, USA).
Electrophoretic mobility shift assay.
Electrophoretic mobility shift assays (EMSAs) were performed at 37°C for 20 min with 32P-labeled probes (∼150 bp) (30,000 cpm per reaction) and 3 μg of poly(dI·dC) (Sigma-Aldrich, St. Louis, MO, USA) in 1× binding buffer. The binding buffer contained 20 mM HEPES (pH 7.5), 1 mM dithiothreitol, 1 mM EDTA (pH 8.0), 1.5 mM MgCl2, and 4% glycerol in a final volume of 30 μl. The RNA-protein complex was resolved on a 6% acrylamide nondenaturing gel (prerun for 1 h at 4°C) in 1× Tris-borate-EDTA (TBE) buffer at 200 V at 4°C. Gels were dried and exposed for autoradiography. Scanning was performed by using the Typhoon 9210 variable-mode imager from Amersham Biosciences.
Northern blotting.
Total low-molecular-weight RNA was isolated from the GFP-expressing, GFP-silenced, and GFP-reverted cells by using a mirVANA miRNA isolation kit (Ambion, Austin, TX, USA). Ten micrograms of each low-molecular-weight RNA was resolved by electrophoresis on a 20% polyacrylamide gel containing 8 M urea. RNA was electroblotted for 90 min at 15 V onto a Hybond-N+ membrane (GE Healthcare, Piscataway, NJ, USA) and immobilized by UV cross-linking at 1,200 × 100 μJ. The complementary gfp probe was labeled with [γ-32P]ATP by 5′-end labeling using T4 kinase and was allowed to hybridize at room temperature (RT) overnight. The membrane was washed three times in 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) with 0.2% SDS and once in 6× SSC with 0.2% SDS at 42°C and then exposed to a phosphorimager screen (Amersham Biosciences, USA) overnight and scanned at 200 μm with a Typhoon-9210 instrument (Amersham Biosciences, USA).
In vitro Dicer assay.
dsRNA of ∼900 bp of the GFP gene was transcribed in vitro by using the Riboprobe Combination System-SP6/T7 RNA polymerase (Promega, Madison, WI, USA). dsRNA was digested into siRNA (∼22 bp) by recombinant human Dicer (Genlantis) either in the absence or in the presence of decreasing amounts of recombinant protein NS4B, F112A (29). Reaction products were loaded onto an RNase-free 3% agarose gel and stained with ethidium bromide.
RESULTS
RNAi factors modulate dengue virus replication in a human cell line (Huh 7).
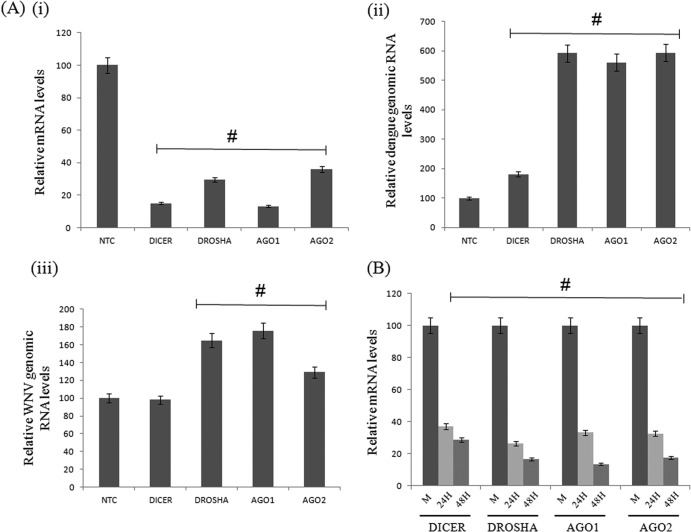
Although host RNAi machinery has been shown to restrict dengue virus replication in insect cell lines (30, 31), limited data are available on the anti-dengue virus response of RNAi machinery in mammalian cells (24). To examine the role of host RNAi in dengue virus replication, the main catalytic components of host RNAi, namely, Dicer, Drosha, Ago1, and Ago2, were knocked down by the corresponding siRNAs in Huh 7 cells, and the levels of knockdown were analyzed by qRT-PCR of the corresponding mRNAs in transfected cells (Fig. 1Ai). We then carried out infection of the knocked-down cells with DV and consequently measured DV RNA from cells at 48 h postinfection. A 6-fold increase in DV genomic RNA levels was observed in the cases of Drosha, Ago1, and Ago2 knockdowns, and a nearly 2-fold increase was seen in the case of Dicer at 48 h postinfection (Fig. 1Aii), compared to cells transfected with siRNA corresponding to a nontarget control (NTC). We further assessed the effect of DV infection on the host RNAi pathway by measuring the mRNA levels of these components in DV-infected cells. As shown in Fig. 1B, we observed a >65% reduction in the levels of these mRNAs, suggesting a negative regulation of RNAi machinery by DV infection. These data support the notion that downregulation of RNAi factors helps dengue virus multiplication.
Fig 1.
Host RNAi regulates dengue virus replication. (Ai) Reduction in dicer, drosha, ago1, and ago2 mRNA levels at 48 h posttransfection in Huh 7 cells transfected with the respective siRNA duplexes (#, P < 0.001). (ii) TaqMan analysis of dengue virus genomic RNA in knocked-down cells at 48 h postinfection (#, P < 0.03). (iii) TaqMan analysis of WNV genomic RNA in knocked-down cells at 48 h postinfection (#, P < 0.02). (B) Downregulation of mRNA levels of dicer, drosha, ago1, and ago2 genes at 24 and 48 h postinfection in dengue virus-infected Huh 7 cells (#, P < 0.05). Data shown are means ± standard deviations from three independent experiments. The # symbols indicate a statistically significant difference in terms of the P value.
Similar results were observed in the case of West Nile virus (WNV) replication upon siRNA knockdown of RNAi components. As shown by our findings, there was an enhancement of WNV replication in the cases of Drosha and Ago1 knockdowns (Fig. 1B), although the levels were not high compared to levels of dengue virus replication. These results indicated that RNAi machinery modulates dengue virus replication in mammalian cells, and the pattern may be conserved among flaviviruses.
Host response to dengue virus infection.
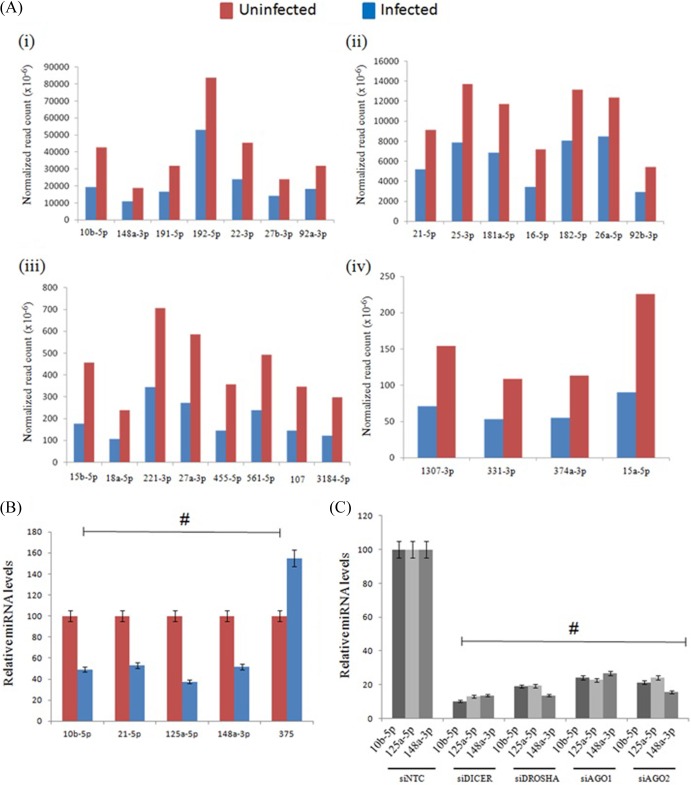
The fact that Dicer, Drosha, Ago1, and Ago2 knockdowns impacted viral replication in mammalian cells (Fig. 1) suggested that both the cellular RNAi and miRNA pathways might be influenced by viral infection in mammalian cells. Since miRNAs have been shown to play an important role(s) in cellular development, metabolism, and proliferation, it is likely that the profile of host miRNAs might alter to cope with viral pathogenesis (11). Therefore, we examined the human miRNA responses to dengue virus infection by examining the changes in the host miRNA profile in cells infected with DV at 48 h postinfection. The population of small RNAs extracted from both the uninfected and infected cells was sequenced separately by using the Illumina next generation sequencing (NGS) platform. The total number of reads was normalized to 1 million for each sample for analysis purposes. The NGS data analysis revealed that the abundance of a majority of known human microRNAs had changed following DV infection. Of the total (151) known microRNAs randomly examined, 143 (94.7%) were downregulated, 3 (1.9%) (hsa-miR-149-5p, hsa-mir-204-5p, and hsa-miR-375) were upregulated, and 5 did not change. These results are not unexpected in view of the general downregulation of drosha and dicer levels following DV infection. A few of the downregulated miRNAs are displayed in Fig. 2Ai to iv, and these results were once again confirmed by stem-loop qRT-PCR analysis for a few of these miRNAs, namely, hsa-miR-10b-5p, hsa-miR-21-5p, hsa-miR-125a-5p, hsa-miR-148a-3p, and hsa-miR-375 (Fig. 2B). In addition, we performed stem-loop qRT analysis of some human miRNAs upon siRNA knockdown of RNAi components in Huh 7 cells. As seen from our results (Fig. 2C), similar reductions in miRNA levels were observed compared to changes in the miRNA profile upon dengue virus infection (Fig. 1B and 2C). These results clearly indicate that the host miRNA/siRNA pathway plays an important role in controlling viral replication in mammalian cell lines.
Fig 2.
Host miRNA response to dengue virus infection. (A) Changes in the host miRNA profile were observed upon dengue virus infection at 48 h in Huh 7 cells. The abundance of downregulated miRNAs (>1.5-fold) are presented in terms of the normalized read counts (i to iv) in both uninfected and infected samples. (B) Validation by stem-loop qRT analysis of some of the miRNAs down- and upregulated upon dengue virus infection at 48 h (#, P < 0.006). (C) Stem-loop qRT analysis of host miRNAs that are affected upon siRNA knockdowns in Huh 7 cells (#, P < 0.005). Data shown in panels B and C are means ± standard deviations from three independent experiments. The # symbols indicate a statistically significant difference in terms of the P value.
Dengue virus NS4B is a viral RNAi suppressor protein.
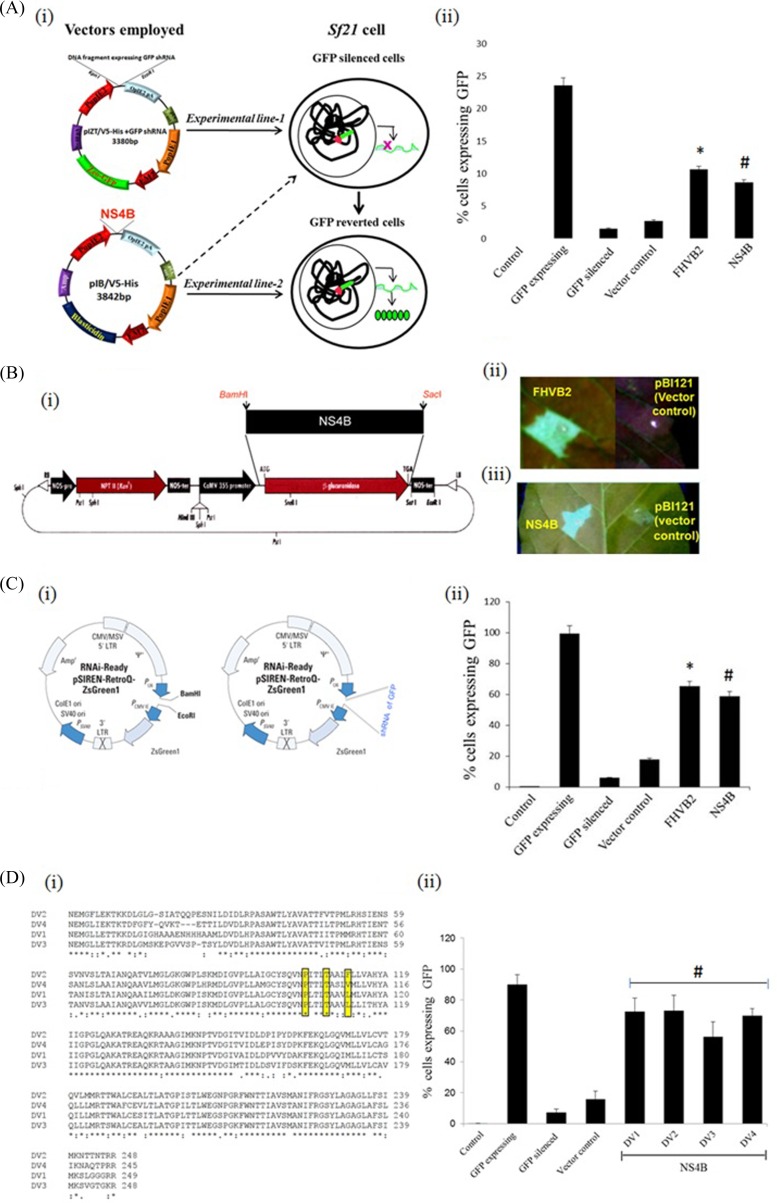
Plant viruses have been shown to suppress the RNAi/miRNA pathway by producing virus-encoded RNAi suppressor proteins (32). A number of approaches that involve transient/stable expression of virus-encoded proteins in an RNAi sensor line expressing the reporter genes have been used to analyze viral suppressor proteins (33, 34). We have used similar approaches to test the RNAi suppressor activity of a number of viral proteins (16, 27). In the present study, we analyzed the NS4B protein for its RNAi suppressor activity. NS4B was selected based on its previously demonstrated role in alpha/beta interferon signaling inhibition (22, 23, 35). To assess the RNAi suppressor function of NS4B, we used a previously established Sf21 RNAi sensor line (experimental line 1) that was used to screen RNAi suppression properties of FHVB2 (16). This cell line is particularly useful in discriminating between the interferon response and the RNAi pathway, because Sf21 cells lack the machinery to elicit an interferon response to viral infection or the presence of dsRNA/dsDNA-like structures in their cytoplasm. We transfected the Sf21 RNAi sensor line with NS4B/pIB at 48 h postinfection and analyzed the cells for GFP expression. As shown in Fig. 3Aii, NS4B showed a significant reversion in the expression levels of GFP, similarly to FHVB2, a known RNAi suppressor (experimental line 2 [GFP-reverted cells]). To further confirm the RNAi suppressor properties of NS4B, we cloned the ns4b gene into the plant binary vector pBI121 (Fig. 3Bi) and carried out reversal of silencing by agroinfiltration-mediated transient ectopic expression of NS4B in tobacco leaf tissues. The infiltrated leaves showed a good amount of reversal of GFP activity at 8 days postinfiltration, while the empty control vector failed to show such a reversal (Fig. 3Bii). The RNAi suppressor properties of NS4B were further confirmed in a GFP-expressing mammalian sensor cell line, HEK293T. The bicistronic vector Retro-Q, which encodes GFP and carries a short hairpin gfp RNA, was cotransfected with either a plasmid containing NS4B (pCDNA-NS4B) or the empty vector alone (Fig. 3Ci). Cotransfection of the NS4B expression plasmid dramatically restored the number of GFP-expressing cells, as was the case with pCDNA-FHVB2, a known RNAi suppressor-expressing plasmid. However, no such reversal was observed when only the empty control plasmid without any suppressor gene was used (Fig. 3Cii). Expression of NS4B in transfected HEK293T cells was confirmed by Western blot analysis (see Fig. 7v). Taken together, these results clearly demonstrated that the dengue virus NS4B protein acts as a suppressor of RNAi.
Fig 3.
Reversion-of-silencing assays show that NS4B is an RNAi suppressor. (Ai) Schematic representation of plasmid constructs used for the generation of the RNAi sensor line (experimental line 1) and the GFP-reverted cells (the RNAi sensor line that overexpressed NS4B and FHVB2) (experimental line 2). (ii) Bar graph representation of the FACS results for FHVB2 and NS4B in the Sf21 sensor cell line, with the percentage of cells expressing GFP represented on the y axis (*, P < 0.008; #, P < 0.05). (Bi) Schematic representation of plasmid construct used for overexpressing NS4B in GFP-silenced leaves of N. xanthi through agroinfiltration. (ii) GFP fluorescence pictures of agroinfiltrated leaves at 8 dpi taken under a UV lamp. The empty vector was used as a mock control, while FHVB2 was used as a positive control in the assay. The tested NS4B protein showed positive reversal of silencing. (Ci) Schematic representation of plasmid constructs used for studying the suppressor activity of NS4B in a mammalian cell line (HEK293T). LTR, long terminal repeat; MSV, mouse sarcoma virus. (ii) Bar graph representation of the FACS results for FHVB2 and NS4B in HEK293T cells, with the percentage of cells expressing GFP represented on the y axis (*, P < 0.001; #, P < 0.003). (Di) Sequence alignment of NS4B proteins from all the four serotypes of dengue virus. (ii) Bar graph representation of the FACS results for NS4B in HEK293T cells, with the percentage of cells expressing GFP represented on the y axis and the dengue virus serotype represented on the x axis. Data shown in panels A, C, and D are means ± standard deviations from three independent experiments. The * and # symbols indicate statistically significant differences at a P value of <0.00004.
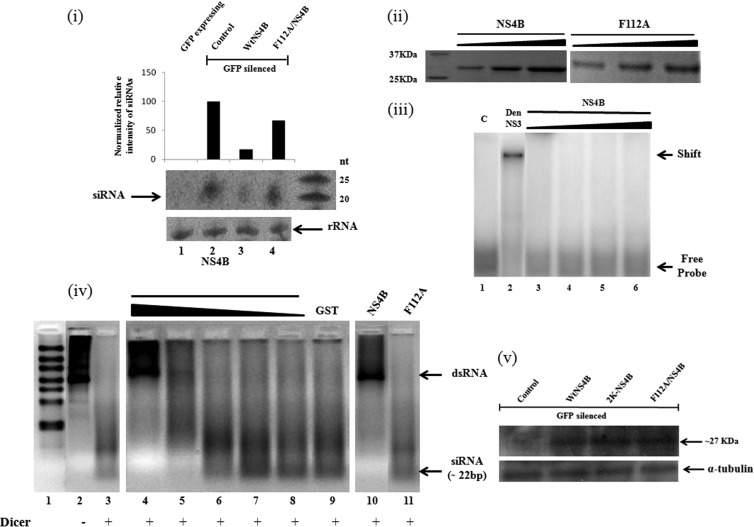
Fig 7.
NS4B inhibits the processing of shRNA/dsRNA into siRNAs both in vivo and in vitro. (i) Histogram showing levels of gfp-specific siRNA in HEK293T cells transfected for a GFP reversion assay and Northern blot analysis of GFP siRNAs in transfected cell lines (HEK293T). Lane 1, GFP-expressing cells; lane 2, GFP-silenced cells; lane 3, GFP-reverted cells expressing wt NS4B; lane 4, GFP-reverted cells expressing the NS4B F112A protein with rRNA loading controls. (ii) Purification of recombinant NS4B F112A proteins run on 12% SDS-PAGE gels in increasing concentrations. (iii) EMSAs using labeled dsRNA probes with purified NS4B protein. Lane 1, free probe; lane 2, dengue virus NS3 (1.0 μg); lanes 3 to 6, NS4B protein (0.1, 0.5, 1.0, and 2.0 μg, respectively). (iv) In vitro dicer assay in the presence of recombinant NS4B F112A proteins in various concentrations. The purified dsRNA could be a mixture of dsRNAs of various sizes, but the majority of the dsRNA molecules were ∼900 bp. Lane 1, ladder; lane 2, dsRNA substrate; lane 3, control dicing reaction; lanes 4 to 11, dicing in the presence of recombinant proteins at various concentrations (lanes 4 to 8, NS4B protein [1.0, 0.5, 0.2, 0.1, and 0.01 μg, respectively]; lane 9, glutathione S-transferase [GST] [1 μg]; lane 10, NS4B [1 μg]; lane 11, F112A mutant [3 μg]). The average size of the diced siRNAs was 22 bp. (v) Western analysis for expression check of NS4B and its F112A mutant in reversal-of-silencing assays in HEK293T cells.
Four serotypes of DV (DV1 to DV4) have been identified so far, and we compared the sequences of NS4B proteins of these serotypes. As shown in Fig. 3Di, differences were observed among the different serotypes with respect to the NS4B protein, especially at the N and C termini. Therefore, we tested the suppressor activities of all four NS4B proteins in a reversal-of-silencing assay using the HEK293T cell line. Interestingly, NS4B proteins of all serotypes showed similar RNAi suppressor activities (Fig. 3Dii), thus advocating a conserved role of the protein as a suppressor in the viral life cycle.
NS4B deletion mutants reveal that transmembrane domains (TMD3 and TMD5) are responsible for RNAi suppression.
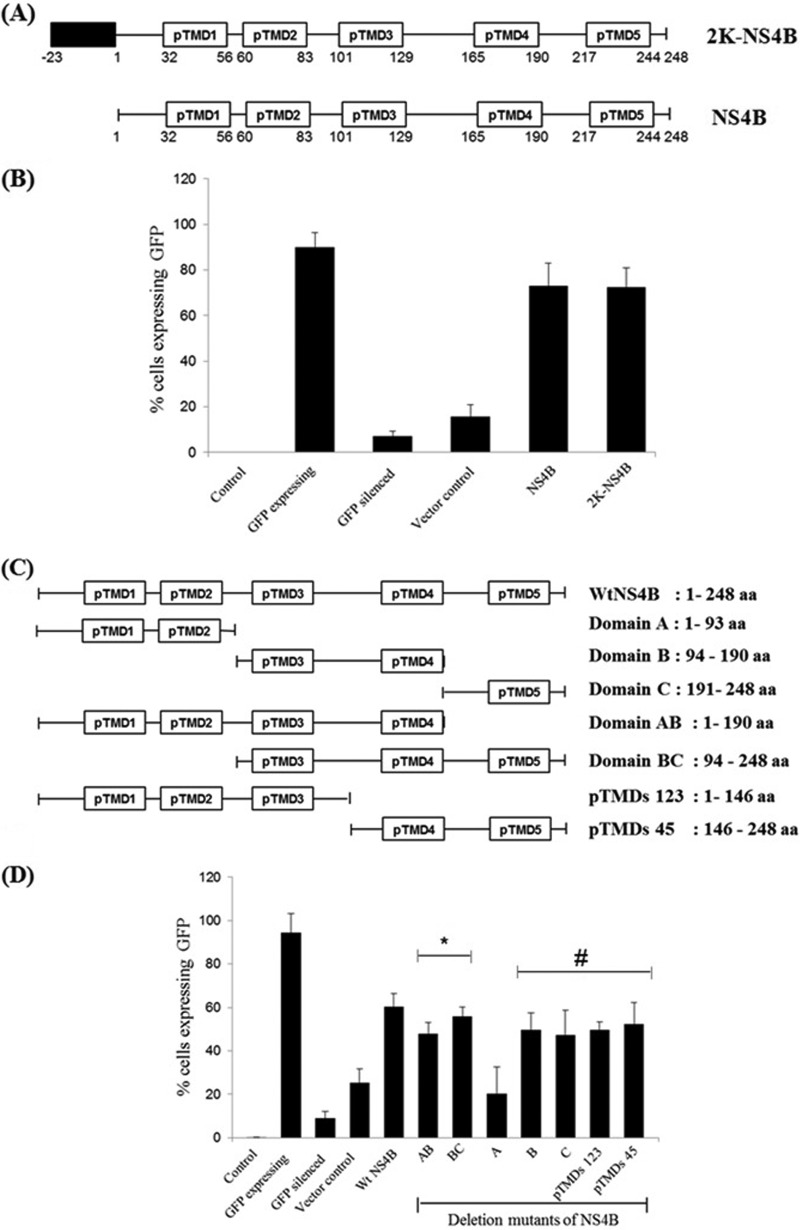
NS4B consists of 248 amino acids and contains a 23-residue signal sequence, 2K, at its N terminus that translocates NS4B into the lumen of the ER (2K-NS4B). The domains of NS4B and 2K-NS4B are schematically shown in Fig. 4A. We compared the RNAi suppressor activities of NS4B and 2K-NS4B in the HEK293T sensor line. As shown in Fig. 4B, both these proteins were able to reverse the effect of RNAi on the sensor lines of HEK293T cells effectively, thereby suggesting that the 2K signal sequence does not significantly influence the RNAi suppressor activity of NS4B.
Fig 4.
Analysis of deletion mutants for RNAi suppressor activity of NS4B. (A) Schematic representation of predicted transmembrane domains of NS4B with and without its signal peptide 2K. (B) Bar graph representation of the FACS results for NS4B and 2K-NS4B in HEK293T cells, represented on the x axis, with the percentage of cells expressing GFP represented on the y axis (#, P < 0.0004). (C) Schematic representation of deletion mutants of NS4B. (D) Bar graph representation of the FACS results for different deletion mutants of NS4B in HEK293T cells, represented on the x axis, with the percentage of cells expressing GFP represented on the y axis (*, P < 0.0004; #, P < 0.005). Data shown in panels B and D are means ± standard deviations from three independent experiments. The * and # symbols indicate statistically significant differences in terms of the P value.
DV NS4B is an integral membrane protein, and several membrane topology models of NS4B predicted the presence of five TMDs (36). To delineate the NS4B regions involved in RNAi suppression, we generated several NS4B deletion mutants encompassing the entire length of the ns4b gene and analyzed the RNAi suppressor activity of each mutant in a reversal-of-RNAi assay using the HEK293T cell line. Figure 4C shows the schematics of the deletion mutants employed in the present study. As evident from Fig. 4D, a deletion mutant that retains only the TMD1 and TMD2 regions (domain A) had negligible RNAi suppressor activity, while deletion mutant B, consisting of the TMD3 and TMD4 regions, and deletion mutant C, consisting of the TMD5 region, showed RNAi suppressor activities similar to those of wild-type NS4B. Thus, the TMD1 and TMD2 regions play no role in RNAi suppression. This observation is noteworthy, as the N-terminal region (domain A) of NS4B was shown previously to inhibit alpha/beta interferon signaling (37). Deletion of the extreme C-terminal region (aa 191 to 248) of NS4B slightly affected its RNAi suppressor activity. Together, the results of the deletion mutant analysis showed that the TMD3 and TMD5 regions of NS4B are required for the RNAi suppressor activity, either as an individual unit or as a combinatorial group. Thus, the domain for the interferon response is distinct from that for the RNAi suppression activity of NS4B.
The TMD3 region of NS4B is important for its RNAi suppressor activity.
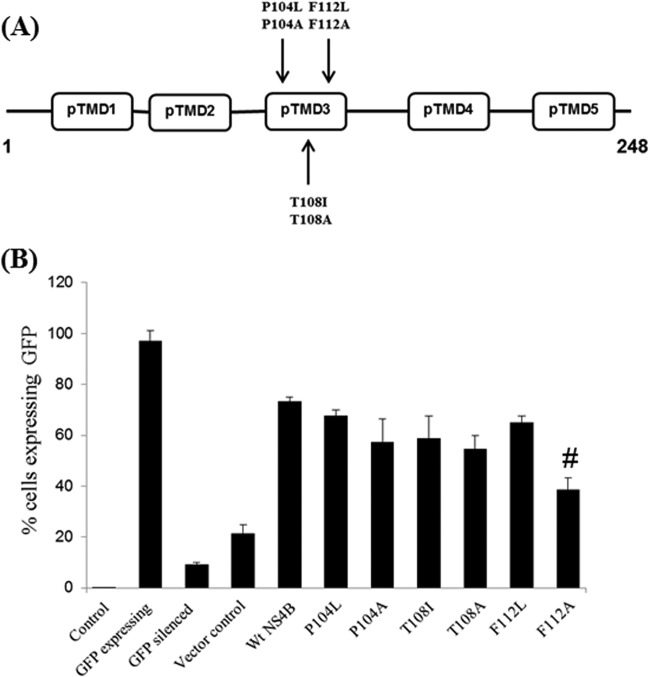
A series of substitution mutations in the ns4B gene had previously revealed that the TMD3 region is required for virus replication (38). To investigate the role of TMD3 in RNAi suppressor activity, we further generated six substitution mutants in the NS4B TMD3 region, namely, P104L/A, T108I/A, and F112L/A, and confirmed their sequences with automated gene sequencing. Figure 5A shows the location of the substitution mutants generated in the present study. RNAi suppression studies using the reversion-of-RNAi assay in a mammalian cell line were performed with the six mutant plasmids. As shown in Fig. 5B, a double-nucleotide modification (T340G and T341C) resulting in the amino acid change of F112A caused a significant reduction (∼40%) of RNAi suppressor activity of NS4B, while a change of F112L did not (Fig. 5B). Expression of the F112A NS4B mutant was also confirmed by Western blot analysis (see Fig. 7v). The other four substitution mutations, P104L/A and T108I/A, did not change the RNAi suppression activities of NS4B significantly (Fig. 5B). The results from a previous study (38) and the present study together suggest that the F112A substitution in the TMD3 region affects RNAi suppressor activity as well as viral replication.
Fig 5.
Localization of amino acid residues responsible for RNAi suppression activity of NS4B. (A) Positions of the DV2 NS4B mutations generated in this study. Mutation sites are described as the amino acid positions in DV2 NS4B. pTMD refers to the predicted transmembrane domain. (B) Bar graph representation of the FACS results for different mutants studied in the pTMD3 region of NS4B in HEK293T cells, represented on the x axis, with the percentage of cells expressing GFP represented on the y axis. Data shown in panel B are means ± standard deviations from three independent experiments. The # symbols indicate a statistically significant difference at a P value of <0.02.
NS4B harbors distinct domains for IFN-antagonistic and RNAi suppressor activities.
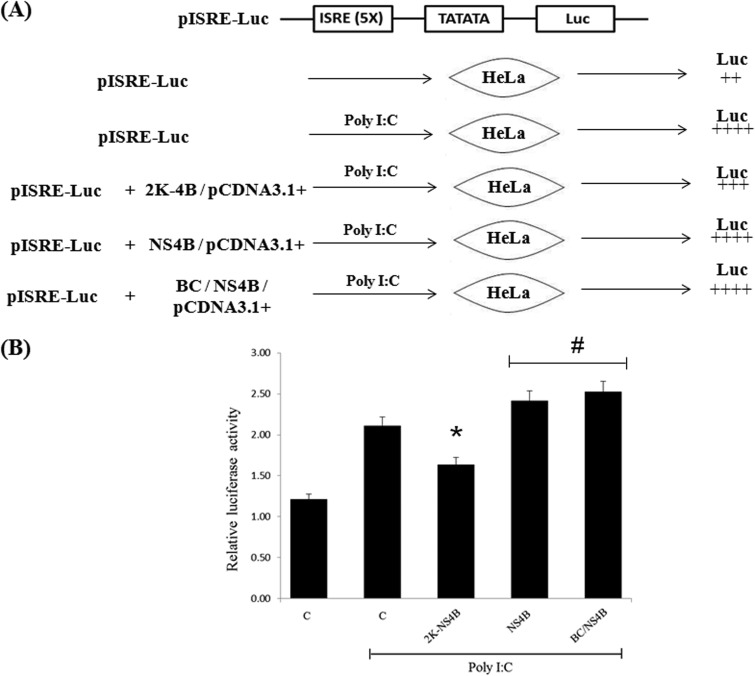
DV NS4B has been shown to inhibit the IFN signaling cascade (23), and intriguingly, most identified VSRs of mammalian viruses also have IFN- or protein kinase R (PKR)-antagonistic properties that may be linked to their RNAi suppressor function (39–41). To delineate the NS4B domains required for IFN-antagonistic activity, a HeLa cell line was transfected with a firefly luciferase expression plasmid under the control of the ISRE (interferon-stimulated response element) promoter pISRE-Luc (Agilent Technologies, Santa Clara, CA, USA), and Renilla luciferase was used as an internal control to normalize the transfection efficiency among all the samples (Fig. 6A). As a positive control, firefly luciferase expression was induced by poly(I·C), which produces a stimulatory IFN response. Cotransfection of a plasmid harboring ns4B along with pISRE-Luc did not result in IFN-antagonistic activity. However, transfection with 2K-NS4B resulted in a small but consistent reduction in the luciferase expression level (Fig. 6B), thereby suggesting that the N-terminal region of NS4B (2K region) is required for its IFN-antagonistic activity. As the “BC” domain of NS4B did not show a reduction of the luciferase expression level, the RNAi suppression domain of NS4B is thus clearly distinct from the IFN-antagonistic activity.
Fig 6.
Delineation of domains responsible for IFN antagonism and RNAi suppressor activity of NS4B. (A) Schematic representation of structural features of pISRE-Luc. Upon transfection into mammalian cells (HeLa), the luciferase gene was expressed from a spliced transcript. The change in luciferase expression was observed upon poly(I·C) treatment in the 2K-NS4B-, wt NS4B-, and BC/NS4B-transfected lines. (B) HeLa cells were cotransfected with expression vectors encoding firefly luciferase under the control of an IFN-β-inducible promoter (pISRE-Luc) (100 ng), Renilla luciferase, and 2K-NS4B, wt NS4B, and BC/NS4B at 500 ng each in either the presence or absence of poly(I·C). Luciferase expression was measured at 31 h posttransfection. The relative luciferase expression level corrected for the internal Renilla control (firefly/Renilla) is shown. Data shown in panel B are statistically significant at P values of <0.02 (*) and <0.05 (#).
NS4B does not bind dsRNA.
Many VSRs have been shown to cause RNAi suppression by binding to dsRNA/siRNA (14–16, 18, 40, 42). Although NS4B has been shown to dissociate NS3 from single-stranded RNA (ssRNA), there has been no report of its interaction with dsRNAs/siRNAs (43). To test the NS4B activity for dsRNA/siRNA binding, we expressed the protein in a heterologous E. coli expression system. The recombinant protein was purified to apparent homogeneity and was tested for dsRNA/siRNA binding by an electrophoretic mobility shift assay (EMSA). However, we did not observe any significant binding of NS4B with labeled dsRNA (Fig. 7iii), but the recombinant dengue virus NS3 protein shifted the labeled dsRNA very well in the same assay, signifying the affinity of NS3 for dsRNA. These results suggest that NS4B suppressor activity might not involve dsRNA binding.
NS4B inhibits Dicer processing of dsRNA into siRNA.
In order to explore other mechanism of RNAi suppression mediated by NS4B, we looked at the possibility of blockage of siRNA biogenesis by NS4B. Hence, we performed Northern analysis for gfp siRNA in GFP reversion assays with expression of either NS4B or its F112A mutant. As observed from our results, NS4B reduced the biogenesis of GFP-siRNA (Fig. 7i, lane 3) whereas the F112A mutant failed to significantly inhibit siRNA generation (Fig. 7i, lane 4). The level of expression of the mutant NS4B protein was similar to that of the wt NS4B protein (not shown). Hence, the in vivo reduction of siRNA biogenesis by NS4B seems clear.
Also, we investigated the in vitro processing of long dsRNA substrates by dicer in the presence of NS4B and its F112A mutant. An in vitro dicer assay was performed according to the manufacturer's instructions (Genlantis, USA), in the presence of recombinant NS4B F112A proteins with increasing concentrations. Figure 7iv (lane 3) reveals that 1 unit of human dicer was able to process the dsRNA substrates into siRNAs (∼22 bp). However, this processing was inhibited by NS4B in a dose-dependent manner (lanes 5 to 8). While 1 μg of NS4B protein blocked dicer processing almost completely (lane 4), about 3 μg of the F112A protein blocked processing by not more than 10%. These results clearly indicate that NS4B suppresses the RNAi pathway by inhibiting siRNA biogenesis.
DISCUSSION
Increasing evidence has emerged to suggest that RNAi pathways are the evolutionarily conserved defense mechanism against pathogenic viral infections (3). In early reports, plant RNAi was shown to act as a systemic antiviral defense against cytoplasmic RNA and nuclear DNA viruses (1). As in plants, the defense function of the RNAi pathway was also shown in Caenorhabditis elegans and Drosophila melanogaster, where mutations in the RNAi factor(s) showed increased susceptibility of these organisms to infection by viruses (44). The existence of analogous systems in vertebrates has been questioned simply because of the prevalence of interferon-based defense mechanisms (3). Recently, a number of studies of mammalian cells have advocated the possible role of RNAi pathways against animal viruses (3, 9, 11). Mutations in RNAi factors or perturbations of RNAi pathways have been shown to increase vesicular stomatitis virus (VSV), influenza A virus, and human immunodeficiency virus type 1 (HIV-1) titers (3). In addition, HIV-1 and human T-cell leukemia virus type 1 (HTLV-1) infections in mammalian cells cause host cell miRNA profile changes (45), thus suggesting the role of RNAi/miRNA pathways in viral infections. Recently, a detailed analysis of small RNA repertoires in vertebrate and invertebrate systems using six mammalian RNA viruses unequivocally advocated an interplay between viral small RNAs (vsRNAs) and virus infection in vertebrate systems (9).
Given the importance of siRNAs/miRNAs in establishing viral infections, in the present study, we looked for the role(s) of host miRNA/RNAi factors, especially Dicer, Drosha, Ago1, and Ago2, in establishing dengue virus infections in mammalian cells. siRNA-mediated knockdown of these genes resulted in significant increases in replication of the dengue virus genome. We also observed reduced mRNA levels of these genes in dengue virus type 2-infected Huh 7 cells, and similar effects were observed in the case of WNV replication upon siRNA knockdowns. Together, these results suggested a role of the RNAi machinery in limiting dengue virus replication. Similar results were previously observed in the cases of equine encephalitis virus, respiratory syncytial virus, and influenza virus (46–48). Although this is the first report of a link between host miRNA/RNAi machineries and dengue virus infection in a mammalian system, a number of studies have reported the role of RNAi machinery in the replication of flaviviruses, especially dengue virus, in insect cells (30, 31, 37). Since the proteins Dicer, Drosha, Ago1, and Ago2 seem to be involved in modulating dengue virus replication in human cell lines, as evident from the present study, it suggested that both siRNA- as well as miRNA-mediated pathways may play an important role(s) in dengue virus replication in mammalian cells.
We next analyzed the changes in miRNA profiles in dengue virus serotype 2-infected Huh 7 cells. As observed in the cases of human retroviruses, hepatitis B virus (HBV), HCV, and herpesvirus (11), we observed a considerable reduction in the human miRNA profile in infected cells. As the drosha and dicer levels were downregulated following DV infection, the reduction in the levels of human miRNAs of the virus-infected cells was not very surprising. Together, these results suggest that both RNAi and miRNA pathways probably regulate dengue virus replication and pathogenesis in mammalian cells.
To overcome antiviral RNAi/miRNA responses, many plant, insect, and human viruses have been shown to encode suppressor of RNA silencing, also referred to as VSR (3, 5–7, 11). In this study, we analyzed dengue virus NS4B for its RNAi suppressor properties. NS4B was selected because it has been shown to inhibit alpha/beta interferon signaling, and a number of recent reports suggest a link between the IFN response and RNAi machinery in regulating viral replication in mammalian cells (3). Along with NS4B, we also examined NS4B with signal peptide 2K (2K-NS4B) for suppressor activity using reversal-of-silencing assays in three independent systems: the Sf21 line, the HEK293T line, and plant leaves that were silenced for the expression of the reporter protein GFP. Such transient assays have been applied successfully to identify many plant and animal RNAi suppressors (16, 27, 33, 34). Both the NS4B and 2K-NS4B proteins were effectively able to revert back GFP expression in GFP-silenced lines, thereby highlighting the role of NS4B as an RNAi suppressor in three different sensor lines. Most importantly, NS4B proteins from all four serotypes of dengue virus, serotypes 1 to 4, were able to reverse RNA silencing to a similar extent. It thus appears that NS4B-mediated characteristics are probably conserved across the eukaryotic kingdom.
A consensus in silico model for the topology of NS4B suggested that it contains five trans-membrane domains, designated TMD1, -2, -3, -4, and -5. Furthermore, expression studies using plasmids containing different NS4B predicted TMDs (pTMDs) fused to enhanced GFP (eGFP) at their COOH termini showed that the sequences containing pTMD3, -4, and -5, but not those containing pTMD1 and -2, mediate membrane targeting of eGFP (36). Based on these analyses, a number of NS4B deletion mutants were generated, and their suppression activities were analyzed. The NS4B deletion mutants retaining pTMD3 and -5 reversed RNA silencing to a level similar to that shown by wild-type NS4B. This observation is significant because the NS4B anti-IFN function is mediated by the pTMD1 and -2 regions of NS4B (35). Hence, NS4B showed the unique characteristic of possessing distinct domains for IFN antagonism and RNAi suppression.
Dengue virus NS4B has been shown to interact with the NS3 protein, and this interaction dissociates NS3 from single-stranded RNA. Furthermore, it has been shown that a single-amino-acid mutation in pTMD3 of NS4B (P104L), which had a pleiotropic effect on dengue virus replication in mosquito, disrupted this interaction (49). An independent study recently showed that two mutations in the NS4B pTMD3 region (C7152U and G7165U) attenuated the growth of wild-type DV1 and restored the replication of replication-defective dengue virus type 1 bearing a mutation in the C-terminal cytoplasmic portion of NS4A (38). These findings thus suggested that NS4B, particularly TMD3, contributes to the adaptation of DV for efficient replication. Viral replication is also linked with RNAi and its suppression. Thus, to investigate the details of the TMD3 involvement in RNAi suppression, we generated six different substitution mutants, P104L/A, T108I/A, and F112L/A, in the NS4B pTMD3 region and tested the suppressor activity of these mutant proteins. Three of these substitutions were the same as those described previously (38), while the other three were alanine substitutions for the same amino acids. Surprisingly, two substitution mutations, namely, P104L and F112L, which attenuated the growth of wild-type dengue virus serotype 1 in Vero cells in a previous study, did not have a substantial effect on the RNAi suppressor activity of NS4B. However, a substitution mutation, F112A, resulted in a considerable loss of RNAi suppression activity. These results are consistent with previous findings that suggested that NS4B, particularly its TMD3 region, contributes to the adaptation of DV for efficient replication in mammalian cells.
Experiments elaborating the mechanism of NS4B-mediated RNAi suppression revealed interesting insights into the mechanism of suppression. EMSA analysis with labeled dsRNA showed no affinity binding of NS4B. These results suggested that NS4B could be targeting one or a few components of the RNAi machinery for inactivation. The observed reduction in biogenesis of gfp siRNA from GFP reversion assays in HEK293T cells suggested the possible involvement of NS4B in dicing activity. This possibility was substantiated by the observed inhibition of dicer activity by NS4B in in vitro assays in a dose-dependent manner.
Furthermore, in vivo experiments revealed reduced dicer mRNA levels upon DV infection. In addition, our data suggest that dicer activity could also be reduced following viral infection. Thus, DVs have evolved many counterstrategies to defeat the RNAi-mediated host defense. It would be interesting to discover if any other RNAi components of the host are also affected by NS4B.
In summary, three major conclusions can be drawn from this work. RNAi/miRNA factors regulate dengue virus replication. Dengue virus NS4B acts as a viral RNAi suppressor, and the TMD3 region of NS4B, which has been shown to be involved in viral replication, is also important for its suppressor activity. We furthermore show that the NS4B protein suppresses the RNAi response by directly inhibiting the dicing process. As the profiles of human miRNAs change following virus infection, the abundance of RNAi factors might also consequently change, impacting virus replication and propagation indirectly. Thus, the modulation of DV replication within the human host could be due to the direct interference of NS4B in Dicer activity as well as the indirect effects mediated by the changes in the host microRNA profiles. Although the present study does not elaborate on the mechanism of NS4B suppression in detail, it is clear that NS4B suppresses RNAi by interfering with dicer activity. It is possible that NS4B might interact with another host RNAi factor(s) and inhibit the biochemical activities of the factor, resulting in an overall downregulation of the host RNAi response. Nevertheless, the present study provides a lead to elucidate such mechanisms and also advocates the development of inhibitors against NS4B so that viral replication can be blocked by chemical means.
ACKNOWLEDGMENTS
We thank Ralf Bartenschlager (University of Heidelberg, Heidelberg, Germany) for providing 2K-NS4B plasmid DNA and David J. Blackbourn (The Science Park, Cambridge, United Kingdom) for providing pISRE-Luc plasmid DNA. We also thank Ravinder Kumar (International Centre for Genetic Engineering and Biotechnology) for helping to make the Sf21 permanent cell line.
This work was supported by a financial grant from the Department of Biotechnology, Government of India.
Footnotes
Published ahead of print 5 June 2013
REFERENCES
- 1. Ding SW, Voinnet O. 2007. Antiviral immunity directed by small RNAs. Cell 130:413–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ding SW. 2010. RNA-based antiviral immunity. Nat. Rev. Immunol. 10:632–644 [DOI] [PubMed] [Google Scholar]
- 3. Jeang KT. 2012. RNAi in the regulation of mammalian viral infections. BMC Biol. 10:58. 10.1186/1741-7007-10-58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Voinnet O, Pinto YM, Baulcombe DC. 1999. Suppression of gene silencing: a general strategy used by diverse DNA and RNA viruses of plants. Proc. Natl. Acad. Sci. U. S. A. 96:14147–14152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bivalkar-Mehla S, Vakharia J, Mehla R, Abreha M, Kanwar JR, Tikoo A, Chauhan A. 2011. Viral RNA silencing suppressors (RSS): novel strategy of viruses to ablate the host RNA interference (RNAi) defense system. Virus Res. 155:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Burgyán J, Havelda Z. 2011. Viral suppressors of RNA silencing. Trends Plant Sci. 16:265–272 [DOI] [PubMed] [Google Scholar]
- 7. Song L, Gao S, Jiang W, Chen S, Liu Y, Zhou L, Huang W. 2011. Silencing suppressors: viral weapons for countering host cell defenses. Protein Cell 2:273–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brigneti G, Voinnet O, Li WX, Ji LH, Ding SW, Baulcombe DC. 1998. Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J. 17:6739–6746 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9. Parameswaran P, Sklan E, Wilkins C, Burgon T, Samuel MA, Lu R, Ansel KM, Heissmeyer V, Einav S, Jackson W, Doukas T, Paranjape S, Polacek C, dos Santos FB, Jalili R, Babrzadeh F, Gharizadeh B, Grimm D, Kay M, Koike S, Sarnow P, Ronaghi M, Ding SW, Harris E, Chow M, Diamond MS, Kirkegaard K, Glenn JS, Fire AZ. 2010. Six RNA viruses and forty-one hosts: viral small RNAs and modulation of small RNA repertoires in vertebrate and invertebrate systems. PLoS Pathog. 6:e1000764. 10.1371/journal.ppat.1000764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. de Vries W, Berkhout B. 2008. RNAi suppressors encoded by pathogenic human viruses. Int. J. Biochem. Cell Biol. 40:2007–2012 [DOI] [PubMed] [Google Scholar]
- 11. Houzet L, Jeang KT. 2011. MicroRNAs and human retroviruses. Biochim. Biophys. Acta 1809:686–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Karlowski WM, Zielezinski A, Carrère J, Pontier D, Lagrange T, Cooke R. 2010. Genome-wide computational identification of WG/GW Argonaute-binding proteins in Arabidopsis. Nucleic Acids Res. 38:4231–4245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Till S, Ladurner AG. 2007. RNA Pol IV plays catch with Argonaute 4. Cell 131:643–645 [DOI] [PubMed] [Google Scholar]
- 14. Mérai Z, Kerényi Z, Molnár A, Barta E, Válóczi A, Bisztray G, Havelda Z, Burgyán J, Silhavy D. 2005. Aureus virus P14 is an efficient RNA silencing suppressor that binds double-stranded RNAs without size specificity. J. Virol. 79:7217–7226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mérai Z, Kerényi Z, Kertész S, Magna M, Lakatos L, Silhavy D. 2006. Double-stranded RNA binding may be a general plant RNA viral strategy to suppress RNA silencing. J. Virol. 80:5747–5756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Singh G, Popli S, Hari Y, Malhotra P, Mukherjee S, Bhatnagar RK. 2009. Suppression of RNA silencing by flock house virus B2 protein is mediated through its interaction with the PAZ domain of Dicer. FASEB J. 23:1845–1857 [DOI] [PubMed] [Google Scholar]
- 17. Chen HY, Yang J, Lin C, Yuan YA. 2008. Structural basis for RNA-silencing suppression by tomato aspermy virus protein 2b. EMBO Rep. 9:754–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chao JA, Lee JH, Chapados BR, Debler EW, Schneemann A, Williamson JR. 2005. Dual modes of RNA-silencing suppression by flock house virus protein B2. Nat. Struct. Mol. Biol. 12:952–957 [DOI] [PubMed] [Google Scholar]
- 19. Silhavy D, Molnár A, Lucioli A, Szittya G, Hornyik C, Tavazza M, Burgyán J. 2002. A viral protein suppresses RNA silencing and binds silencing-generated, 21- to 25-nucleotide double-stranded RNAs. EMBO J. 21:3070–3080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang X, Yuan YR, Pei Y, Lin SS, Tuschl T, Patel DJ, Chua NH. 2006. Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes Dev. 20:3255–3268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Blair CD. 2011. Mosquito RNAi is the major innate immune pathway controlling arbovirus infection and transmission. Future Microbiol. 6:265–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bartenschlager R, Miller S. 2008. Molecular aspects of dengue virus replication. Future Microbiol. 3:155–165 [DOI] [PubMed] [Google Scholar]
- 23. Muñoz-Jordan JL, Sánchez-Burgos GG, Laurent-Rolle M, García-Sastre A. 2003. Inhibition of interferon signaling by dengue virus. Proc. Natl. Acad. Sci. U. S. A. 100:14333–14338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen S, Chahar HS, Abraham S, Wu H, Pierson TC, Wang XA, Manjunath N. 2011. Ago-2-mediated slicer activity is essential for anti-flaviviral efficacy of RNAi. PLoS One 6:e27551. 10.1371/journal.pone.0027551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. 2004. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell 15:185–197 [DOI] [PubMed] [Google Scholar]
- 26. Gurukumar KR, Priyadarshini D, Patil JA, Bhagat A, Singh A, Shah PS, Cecilia D. 2009. Development of real time PCR for detection and quantitation of dengue viruses. Virol. J. 6:10. 10.1186/1743-422X-6-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Karjee S, Minhas A, Sood V, Ponia SS, Banerjea AC, Chow VT, Mukherjee SK, Lal SK. 2010. The 7a accessory protein of severe acute respiratory syndrome coronavirus acts as an RNA silencing suppressor. J. Virol. 84:10395–10401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jani D, Nagarkatti R, Beatty W, Angel R, Slebodnick C, Andersen J, Kumar S, Rathore D. 2008. HDP—a novel heme detoxification protein from the malaria parasite. PLoS Pathog. 4:e1000053. 10.1371/journal.ppat.1000053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chen W, Zhang Z, Chen J, Zhang J, Zhang J, Wu Y, Huang Y, Cai X, Huang A. 2008. HCV core protein interacts with Dicer to antagonize RNA silencing. Virus Res. 133:250–258 [DOI] [PubMed] [Google Scholar]
- 30. Sánchez-Vargas I, Scott JC, Poole-Smith BK, Franz AW, Barbosa-Solomieu V, Wilusz J, Olson KE, Blair CD. 2009. Dengue virus type 2 infections of Aedes aegypti are modulated by the mosquito's RNA interference pathway. PLoS Pathog. 5:e1000299. 10.1371/journal.ppat.1000299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hess AM, Prasad AN, Ptitsyn A, Ebel GD, Olson KE, Barbacioru C, Monighetti C, Campbell CL. 2011. Small RNA profiling of dengue virus-mosquito interactions implicates the PIWI RNA pathway in anti-viral defense. BMC Microbiol. 11:45. 10.1186/1471-2180-11-45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dunoyer P, Lecellier CH, Parizotto EA, Himber C, Voinnet O. 2004. Probing the microRNA and small interfering RNA pathways with virus-encoded suppressors of RNA silencing. Plant Cell 16:1235–1250 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33. McClure LV, Seo GJ, Sullivan CS. 2011. Reporter-based assays for analyzing RNA interference in mammalian cells. Methods Mol. Biol. 725:173–189 [DOI] [PubMed] [Google Scholar]
- 34. Van Cleef KW, van Mierlo JT, van den Beek M, van Rij RP. 2011. Identification of viral suppressors of RNAi by a reporter assay in Drosophila S2 cell culture. Methods Mol. Biol. 721:201–213 [DOI] [PubMed] [Google Scholar]
- 35. Muñoz-Jordán JL, Laurent-Rolle M, Ashour J, Martínez-Sobrido L, Ashok M, Lipkin WI, García-Sastre A. 2005. Inhibition of alpha/beta interferon signaling by the NS4B protein of flaviviruses. J. Virol. 79:8004–8013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Miller S, Sparacio S, Bartenschlager R. 2006. Subcellular localization and membrane topology of the dengue virus type 2 non-structural protein 4B. J. Biol. Chem. 281:8854–8863 [DOI] [PubMed] [Google Scholar]
- 37. Mukherjee S, Hanley KA. 2010. RNA interference modulates replication of dengue virus in Drosophila melanogaster cells. BMC Microbiol. 10:127. 10.1186/1471-2180-10-127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tajima S, Takasaki T, Kurane I. 2011. Restoration of replication-defective dengue type 1 virus bearing mutations in the N-terminal cytoplasmic portion of NS4A by additional mutations in NS4B. Arch. Virol. 156:63–69 [DOI] [PubMed] [Google Scholar]
- 39. de Vries W, Haasnoot J, Fouchier R, de Haan P, Berkhout B. 2009. Differential RNA silencing suppression activity of NS1 proteins from different influenza A virus strains. J. Gen. Virol. 90:1916–1922 [DOI] [PubMed] [Google Scholar]
- 40. Haasnoot J, de Vries W, Geutjes EJ, Prins M, de Haan P, Berkhout B. 2007. The Ebola virus VP35 protein is a suppressor of RNA silencing. PLoS Pathog. 3:e86. 10.1371/journal.ppat.0030086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fabozzi G, Nabel CS, Dolan MA, Sullivan NJ. 2011. Ebolavirus proteins suppress the effects of small interfering RNA by direct interaction with the mammalian RNA interference pathway. J. Virol. 85:2512–2523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schnettler E, de Vries W, Hemmes H, Haasnoot J, Kormelink R, Goldbach R, Berkhout B. 2009. The NS3 protein of rice hoja blanca virus complements the RNAi suppressor function of HIV-1 Tat. EMBO Rep. 10:258–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Umareddy I, Chao A, Sampath A, Gu F, Vasudevan SG. 2006. Dengue virus NS4B interacts with NS3 and dissociates it from single-stranded RNA. J. Gen. Virol. 87:2605–2614 [DOI] [PubMed] [Google Scholar]
- 44. Flynt A, Liu N, Martin R, Lai EC. 2009. Dicing of viral replication intermediates during silencing of latent Drosophila viruses. Proc. Natl. Acad. Sci. U. S. A. 106:5270–5275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Van Duyne R, Guendel I, Klase Z, Narayanan A, Coley W, Jaworski E, Roman J, Popratiloff A, Mahieux R, Kehn-Hall K, Kashanchi F. 2012. Localization and sub-cellular shuttling of HTLV-1 tax with the miRNA machinery. PLoS One 7:e40662. 10.1371/journal.pone.0040662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kehn-Hall K. 2012. Venezuelan equine encephalitis virus interacts with host cellular micro-RNA processing machinery to facilitate viral replication. Abstr. 2nd World Congr. Virol, Las Vegas, NV [Google Scholar]
- 47. Inchley CS, Sonerud T, Fjærli HO, Nakstad B. 2011. Reduced Dicer expression in the cord blood of infants admitted with severe respiratory syncytial virus disease. BMC Infect. Dis. 11:59. 10.1186/1471-2334-11-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Matskevich AA, Moelling K. 2007. Dicer is involved in protection against influenza A virus infection. J. Gen. Virol. 88:2627–2635 [DOI] [PubMed] [Google Scholar]
- 49. Hanley KA, Manlucu LR, Gilmore LE, Blaney JE, Hanson CT, Jr, Murphy BR, Whitehead SS. 2003. A trade-off in replication in mosquito versus mammalian systems conferred by a point mutation in the NS4B protein of dengue virus type 4. Virology 312:222–232 [DOI] [PubMed] [Google Scholar]