Abstract
The replication complexes of positive-strand RNA viruses are always associated with cellular membranes. The morphology of the replication-associated membranes is altered in different ways in different viral systems, but many viruses induce small membrane invaginations known as spherules as their replication sites. We show here that for Semliki Forest virus (SFV), an alphavirus, the size of the spherules is tightly connected with the length of the replicating RNA template. Cells with different model templates, expressed in trans and copied by the viral replicase, were analyzed with correlative light and electron microscopy. It was demonstrated that the viral-genome-sized template of 11.5 kb induced spherules that were ∼58 nm in diameter, whereas a template of 6 kb yielded ∼39-nm spherules. Different sizes of viral templates were replicated efficiently in trans, as assessed by radioactive labeling and Northern blotting. The replication of two different templates, in cis and trans, yielded two size classes of spherules in the same cell. These results indicate that RNA plays a crucial determining role in spherule assembly for SFV, in direct contrast with results from other positive-strand RNA viruses, in which either the presence of viral RNA or the RNA size do not contribute to spherule formation.
INTRODUCTION
Most plant viruses and many important human and animal pathogens are positive-strand RNA viruses. Upon infection, the positive-strand RNA acts both as an mRNA for the viral replication proteins, and subsequently as a template for the synthesis of complementary minus-strand RNA. The minus strands are produced only in small quantities, and are confined to replication sites, where they act as templates for multiple rounds of positive-strand RNA synthesis (1). The RNA replication of positive-strand RNA viruses always takes place in association with cytoplasmic membranes, which undergo extensive morphological and functional modification during infection (2). Different virus groups utilize different membranes for replication. The membranous environment is essential for replication, and has been proposed to act as a concentrating device, a structural scaffold, a protective environment, as well as an activator of replication proteins (3–5). The mechanisms leading to the formation of virus-modified membranes remain poorly understood.
Alphaviruses, including Semliki Forest virus (SFV) and Sindbis virus, are positive-strand RNA viruses that are usually transmitted by mosquitoes between mammalian and avian hosts. In humans, several alphaviruses cause febrile disease connected with encephalitis or arthritis (6). The alphavirus genome is ∼11.5 kb in length, contains a 5′ cap structure, a 3′ poly(A) tail, and two open reading frames. The first open reading frame covers almost two-thirds of the genome and encodes the replication proteins of the virus. They are expressed as a polyprotein precursor P1234 that is cleaved to four final components, nonstructural proteins nsP1-nsP4. The second open reading frame, encoding the structural proteins, is expressed via a subgenomic mRNA, which is produced through internal initiation on the minus-strand template (7). SFV RNA replication takes place in small membrane invaginations termed spherules (8, 9). The alphavirus spherule has a diameter of ∼60 nm, and its interior is always connected to the cytoplasm by a narrow neck structure. Thousands of spherules first arise at the plasma membrane and are later found also on the surfaces of endo/lysosomal vacuoles (10, 11). Recent electron tomography analyses have indicated that the replication sites of flaviviruses, such as Dengue virus and West Nile virus, are also membrane invaginations that resemble the classical spherule invaginations, although they are much larger in size (average diameter almost 90 nm) (12, 13).
Many plant viruses produce spherule structures that are strikingly similar to those of SFV in size and appearance. The plant virus spherules are most commonly found on the endoplasmic reticulum membranes, and have been extensively characterized for brome mosaic virus (BMV) (1, 14). A third well-studied virus generating spherules is Flock house virus (FHV), a nodavirus unrelated to BMV or SFV. FHV infects insects and induces spherules on mitochondrial membranes (15). As in the case of alphaviruses, for BMV and FHV there is also good evidence that RNA synthesis takes place at the spherules (14, 15). BMV encodes only two replication proteins, 1a and 2apol. It is notable that BMV 1a, a distant homolog of alphavirus nsP1 and nsP2, gives rise to spherule structures when expressed alone in the absence of viral RNA (14). In striking contrast, alphavirus or FHV replicase proteins are incapable of producing spherules on their own. FHV and SFV additionally require a viral RNA template and an active viral polymerase for spherule formation. This implies that there are at least two distinct pathways by which spherules are generated (16, 17). For FHV, it has been shown that the size of the spherules remains the same, even if the length of the viral RNA template varies over a 10-fold range (16).
We demonstrate here that for SFV, the length of the replicating RNA template determines the size of the replication complex spherules. Thus, the assembly mechanism of SFV replication spherules must differ from those of BMV and FHV, and factors other than proteins need to be explicitly considered in the models describing the formation of membranous replication structures.
MATERIALS AND METHODS
Cell culture, viruses, and infection.
Baby hamster kidney cells (BHK-21) were cultured in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum, 2 mM l-glutamine, 100 U of penicillin/ml, and 100 μg of streptomycin/ml. BSR T7/5 cells, a derivative of BHK cells stably expressing T7 RNA polymerase (18), were cultured in the same medium further supplemented with 2% Bacto tryptose phosphate broth, 1% nonessential amino acids, and 1 mg of G418/ml for selection of T7 polymerase expression. Cells for correlative light and electron microscopy (CLEM) experiments were grown on no. 2 glass-bottom P35G-2-14-C-Grid dishes for transfections or on P35G-1.5-14-C dishes for infections (MatTek).
Wild-type SFV (wtSFV) was propagated as described previously (4). The replicon construct pSFV1-ZsG and the luciferase-expressing virus SFV-Rluc have been described (10, 19). For EM experiments, BSR T7/5 cells were infected with wtSFV at 50 PFU/cell and fixed at 3 h postinfection.
Plasmid constructs.
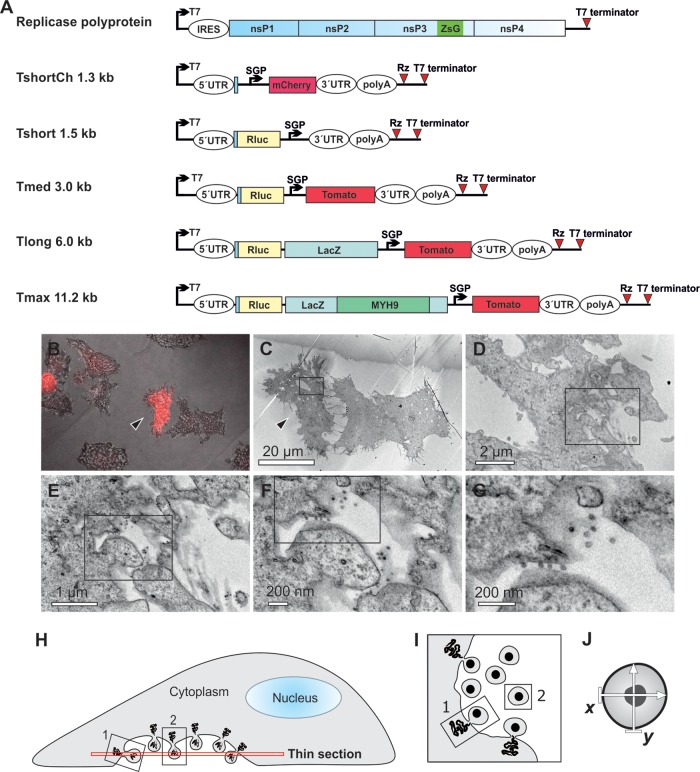
Plasmids for P123Z4 and templates Tshort, Tmed, and Tlong have been described (17). To construct a short template suitable for CLEM experiments, the marker gene mCherry was PCR amplified with primers creating BglII and a SalI restriction sites and cloned under the control of the subgenomic promoter of the pUC18 templ+ construct (17). This template was designated as TshortCh. A Tmax template construct was created by cloning human nonmuscle myosin 9 heavy chain (MYH9; human ORFeome library, clone ID 100000287; Open Biosystems) into the Tlong template. MYH9 was PCR amplified with primers creating Bsu36I and BspDI restriction sites and cloned inside the β-galactosidase gene.
DNA and RNA transfections.
For CLEM experiments, BSR T7/5 cells were transfected with plasmids expressing polyproteins and the templates Tshort, Tmed, or Tlong with jetPRIME transfection reagent (Polyplus transfection) according to the manufacturer's instructions. One microgram each of polyprotein and template plasmid was cotransfected per 35-mm dish in 2 ml of regular BSR medium. To maximize Tmax template replication, more template plasmid (3.75 μg) was used in cotransfection with polyprotein plasmid (1 μg) in 2 ml of minimal essential medium (MEM) supplemented with 0.2% bovine serum albumin, 20 mM HEPES, and 2 mM l-glutamine using Lipofectamine LTX (Invitrogen) reagent. The cells were fixed at 24 h (Tmax) or 16 h (other templates) posttransfection.
For RNA labeling and luciferase measurement experiments BSR T7/5 cells were transfected with Lipofectamine LTX transfection reagent according to manufacturer's instructions. For six-well plate, 1 μg of polyprotein plasmid and 1.25 μg of template plasmid were cotransfected per well in 2 ml of MEM supplemented as described above. For the luciferase assay with the Tmax template, a 3-fold-higher amount of template DNA was used. Renilla luciferase activity was measured as previously described (17).
To transfect linearized DNA, the Tshort and Tmed template plasmids were cleaved to completion with SacI and Tlong with RsrII, followed by purification with a GeneJET gel extraction kit (Thermo Scientific). Then, 1 μg of polyprotein plasmid and 1.25 μg of linearized template were cotransfected with Lipofectamine LTX for all experiments, and analysis was carried out at 16 h posttransfection.
For CLEM experiments with the SFV replicon, the plasmids pSFV1-ZsG and Tmed were first linearized with SpeI and SacI, respectively. In vitro transcription was performed with SP6 or T7 RNA polymerases for pSFV1-ZsG and Tmed, respectively, in a mixture containing 1 mM (each) ATP, CTP, and UTP, 0.5 mM GTP, 1 mM cap analog m7G(5′)ppp(5′)G (New England BioLabs), 5 mM dithiothreitol, and 50 U of RNasin (Promega). Finally, BHK cells were transfected with 1 μg of either replicon RNA alone or together with 1 μg of Tmed RNA by using Lipofectamine LTX and fixed at 12 h posttransfection.
RNA synthesis in trans-replication system.
BSR T7/5 cells were transfected with plasmids expressing polyproteins and templates or infected with wtSFV at 1 PFU/cell. Cells transfected with Tmed plasmid only, polyprotein plasmid only or mock-transfected cells served as controls. Duplicate dishes of each set were treated with 2 μg of actinomycin D/ml, starting 1 h prior to labeling. Cells were labeled at 3, 5, and 7 h posttransfection with 30 μCi of [3H]uridine/dish for 1 h in the presence of actinomycin D (2 μg/ml) in 1 ml of medium. Samples were washed five times with cold phosphate-buffered saline (PBS) and were lysed with 2% sodium dodecyl sulfate (SDS) in PBS at 72°C. Labeled RNAs were precipitated with 10% trichloroacetic acid (TCA) on ice for 30 min. RNA was collected on glass fiber filters (GF/C; Whatman) and washed once with 10% TCA, four times with 5% TCA, and once with 96% ethanol. The amount of incorporated label was determined by liquid scintillation. After subtraction of mock background, the results were divided by the number of uridine residues in each RNA to obtain the relative numbers of labeled RNAs. In control experiments it was verified that T7-driven transcription of template plasmid did not give rise to additional background compared to mock-transfected samples, since the DNA-driven transcription of T7 was also inhibited by actinomycin D.
RNA isolation and Northern blotting.
BSR T7/5 cells were transfected with plasmids expressing templates alone or cotransfected with plasmids expressing replicase polyproteins by using Lipofectamine LTX reagent and incubated for 16 h. BHK cells were infected with SFV-Rluc at 10 PFU/cell for 4 h. Cells were lysed and collected with TRIsure reagent (Bioline), followed by RNA isolation according to the manufacturer's instructions except that an additional phenol (pH 5.0)-chloroform extraction was performed prior to precipitation. A 2-μg portion of total RNA was fractionated on a denaturing 1% agarose gel and transferred to a positively charged Amersham Hybond-N+ nylon filter (GE Healthcare) by capillary blotting overnight. RNA was cross-linked to the membrane with Stratalinker (Stratagene). 32P-labeled antisense probes were made by in vitro transcription with T7 RNA polymerase from PCR-amplified DNA fragments containing sequences of the Rluc gene and the T7 promoter. The probe for positive-strand RNA detection corresponded to nucleotides 32 to 676, and the probe for negative-strand RNA detection corresponded to nucleotides 38 to 623 of the Rluc gene as present on template constructs. Prehybridization and hybridization were performed as described previously (20), except that 106 cpm of the RNA probe was included. Hybridization was performed at 60°C overnight. The membrane was washed as described previously (21).
CLEM.
All CLEM samples were fixed with 2% glutaraldehyde in 0.1 M sodium-cacodylate buffer for 30 min at room temperature and washed with the buffer. Cells were immediately imaged with a Leica SP2 confocal microscope using HC PL APO 20×/0.7 CS (air) and HCX PL APO 63×/1.2 W Corr/0.17 CS (water) objective lenses or a Leica TCS SP5II HCS A confocal microscope using an HC PL APO 20×/0.7 CS (air) objective lens. Fluorescence mode was used to obtain images from RNA-replicating cells and reflection or differential interference contrast mode to image the grid of the dish. Samples were then prepared for transmission electron microscopy. Briefly, samples were stained with reduced buffered osmium tetroxide and uranyl acetate and processed for flat embedding and ultrathin sectioning as previously described (22). Positive cells were relocated on electron microscopy based on previously taken fluorescence and reflection images and imaged with JEOL 1200 EX II transmission electron microscope operated at 80 kV. Images were acquired with Gatan Erlangshen ES5000 W, model 782 (Gatan, Inc.). Alternatively, a newer microscope model, JEOL JEM-1400 (80 kV), and bottom-mounted camera, Gatan Orius SC 1000B, were used for EM imaging.
Spherule measurements.
The spherules were analyzed from electron micrographs with Image Pro Plus software (Media Cybernetics). Reference calibrations were made for every image. The average diameter for each individual spherule was calculated from two values measured orthogonally. The average was rounded to the nearest full nm, and the values were plotted as histograms to visualize the distribution of diameters.
Western blotting.
BSR T7/5 cells were transfected with plasmids as described above by using Lipofectamine LTX reagent and incubated for 16 h. BHK cells were infected with SFV-Rluc at 10 PFU/cell for 4 h. Total cell lysates were fractionated on 10% SDS-polyacrylamide gels, followed by transfer to Hybond-ECL (Amersham Biosciences). Filters were blocked against nonspecific binding using 5% nonfat dry milk powder and probed with specific antibodies against SFV nsP3 and nsP4 (9). Equal loading was confirmed by probing the same filter with an antibody for β-actin (Sigma-Aldrich). Signals were obtained by incubating the filters with secondary antibodies IRDye800CW donkey anti-rabbit IgG (Li-Cor Biosciences) and Alexa Fluor 680 anti-mouse IgG (Invitrogen) and scanning the filters with Odyssey system (Li-Cor).
RESULTS
The length of replicating RNA affects the size of spherules.
We have recently established an efficient plasmid-based replication system for SFV (17). In this system, replication proteins expressed from a separate plasmid can replicate RNA templates expressed in trans. Membranous replication structures arising during replication were studied by CLEM, showing that SFV replication proteins alone cannot induce the formation of spherules, but an RNA template and functional polymerase are both required (17). Since the trans-replication system permits the use of different kinds of template RNAs, we wanted to assess the role of a template in the assembly of replication complexes. First, a short RNA template suitable for CLEM experiments was constructed. TshortCh expresses the red fluorescent protein mCherry from the subgenomic promoter of SFV and, to minimize template size, it lacks the Renilla luciferase (Rluc) marker present in the other templates (Fig. 1A).
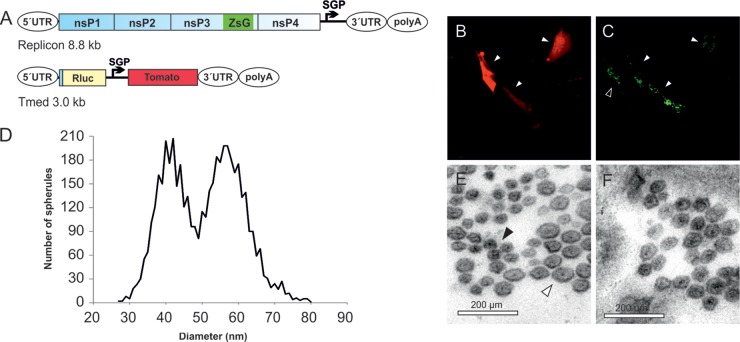
Fig 1.
Visualization of the replication of RNA templates by CLEM and analysis of spherule size. (A) Schematic of the SFV replicase polyprotein and template constructs used in the present study. The size of the templates, excluding poly(A), is indicated. Both the polyprotein mRNA (containing an internal ribosome entry site [IRES]) and the template RNAs (containing viral 5′ and 3′ untranslated regions [UTRs]) are expressed from T7 promoters. ZsG indicates the fluorescent protein ZsGreen fused with replicase protein nsP3, and SGP indicates SFV subgenomic promoter, which directs the expression of red fluorescent markers mCherry or Tomato from the template RNA. Rluc, Renilla luciferase; LacZ, β-galactosidase; MYH9, myosin 9 heavy chain; Rz, hepatitis delta virus antigenomic ribozyme. (B to G) After cotransfection of polyprotein and TshortCh template constructs, CLEM technique was used to visualize the identified replication-positive cells (red in panel B, marked with an arrowhead in panels B and C) at the EM level (C to G). Boxes indicate the area shown in the subsequent higher-magnification image. (H to J) Schematic of the sectioning of cells and EM analysis of spherules. Thin sections were made horizontally starting from the bottom (H). Plentiful spherules were commonly detected on the bottom plasma membrane that was facing the cell culture dish. In these sections, most spherules are cut as indicated for spherule number 2 and therefore appear to be dissociated from the plasma membrane (I), although they are always connected to it by a neck structure above the plane of the section. In the bottom sections spherules that are cut “sideways,” like spherule number 1, and thus showing the neck, are more rare. We have not observed differences in size when spherule sizes have been measured in the two different cutting orientations. The diameter of each spherule was defined as the average of two orthogonally measured distances (J).
When the TshortCh template-producing plasmid was transfected to cells alone, no fluorescence was detected. As expected, when TshortCh was cotransfected with replicase polyprotein expressing plasmid, red fluorescence was detected in some of the cells, indicating active RNA replication (Fig. 1B). After the CLEM procedure, EM analysis revealed abundant spherule-like structures at the plasma membrane in all of the replication-positive cells (Fig. 1C to G) but not in negative cells. It was evident that the spherules produced during TshortCh replication were much smaller than those observed during SFV infection. Therefore, a comprehensive analysis of spherule size was undertaken for different sizes of RNA templates using thin-section EM images derived from CLEM samples, as shown schematically in Fig. 1H to J.
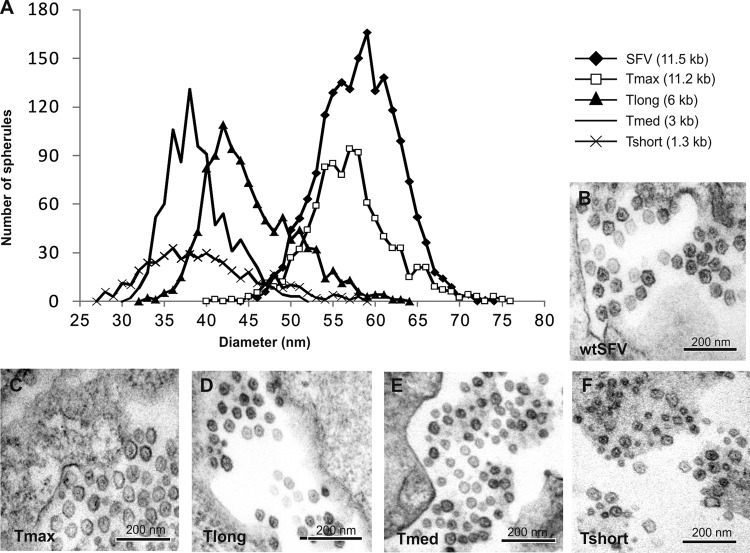
Cells replicating short (∼1.3-kb), medium-sized (∼3.0-kb), and long (∼6.0-kb) templates in trans were analyzed with CLEM and compared to SFV-infected cells. The diameters of altogether 4,229 spherules derived from multiple cells in three to four independent experiments for each construct/virus were measured. The size distribution was plotted in a histogram as explained in Materials and Methods (Fig. 2A). In SFV infected-cells (genome, ∼11.5 kb), the average spherule diameter was 58 nm with a standard deviation of ± 4.5 nm (n = 1807) (Fig. 2B), whereas the long template yielded an average of 45 ± 5.2 nm (n = 1067) (Fig. 2D), the medium-sized template yielded an average of 39 ± 3.6 nm (n = 898) (Fig. 2E), and short template yielded an average of 40 ± 6.4 nm (n = 457) (Fig. 2F). The spherule size distribution for the virus-infected sample and the medium-sized template was essentially nonoverlapping (Fig. 2A). The spherule-like structures produced during short template replication were more difficult to identify and analyze. The size distribution was wider, and it mostly overlapped with that of the medium-sized template (Fig. 2A). However, there were also smaller structures, which may be underrepresented, since they were difficult to find and definitely identify as “spherules” (Fig. 2F).
Fig 2.
Spherule sizes induced by different lengths of replicating RNA. (A) Distribution of spherule diameters for different lengths of RNA templates. The number of spherules detected having a particular diameter is plotted as a histogram for each of the indicated templates. (B to E) Representative images of spherules during SFV infection (B), and during Tmax (C), Tlong (D), Tmed (E), and Tshort (F) replication. All of the images are in the same scale; the template is indicated for each panel.
After the detection of spherules remarkably different in size between the trans-replication system and wtSFV, it was necessary to study whether the difference was caused by the transfection-based system. The template construct Tmax (11.2 kb) was created to address this question. Similarly to the other template constructs, Tmax had a fluorescent marker to identify the replicating cells and Rluc marker at the 5′ end of the genome (Fig. 1A). Although the replication efficiency of Tmax was relatively low (see below), cells with efficient replication were found for CLEM analysis, and spherules were measured in two independent experiments. The average size was found to be 57 ± 4.9 nm (n = 950), which is very close to that observed for wtSFV (Fig. 2A and C). Therefore, the smaller spherules induced by the trans-replication system cannot be due to a specific property of the system itself.
Efficiency of replication for different sizes of RNA templates.
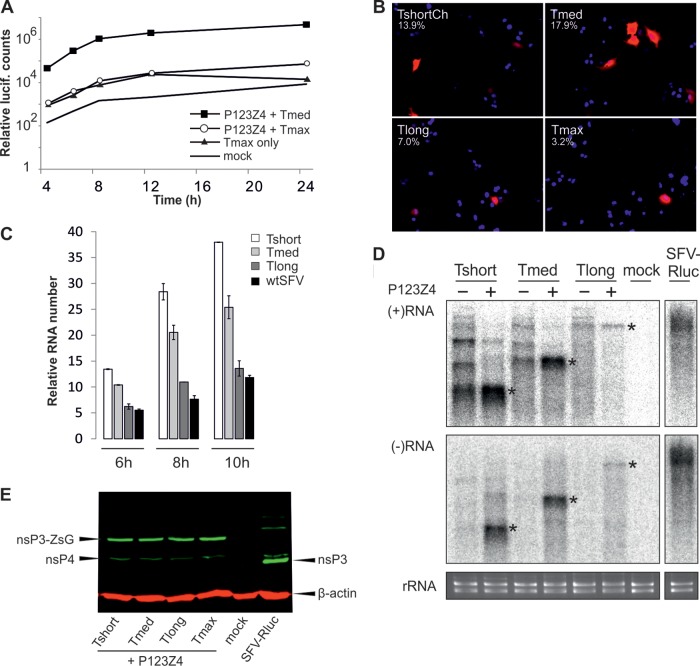
We have previously characterized luciferase activity after Tshort, Tmed, and Tlong transfection and shown that the shorter templates produced the highest activities, which appeared relatively early after transfection (17). The replication efficiency of Tmax was also first established with luciferase assays. The luciferase activity after Tmax plus replicase polyprotein transfection was much lower than that observed for the efficient Tmed template (Fig. 3A). Furthermore, for Tmax the difference between the template alone versus template plus replicase was only observed at late time points of >12 h after transfection (Fig. 3A). In accordance with these findings, the red fluorescent signal indicating replication also appeared very late and in only a few cells for Tmax, compared to the other templates, giving only ∼3% of replication-positive cells (Fig. 3B). This may be due to the low transfection efficiency of the large plasmid and/or to the lower stability of the large RNA template. However, these few cells represent true replication, since transfection of a polymerase mutant with the templates did not yield any positive cells. Due to the low efficiency at the cell population level, Tmax was omitted from the following experiments.
Fig 3.
Replication of different RNA templates. (A) Replication efficiency of genome-size construct (Tmax) was studied with Rluc assay. Triplicate samples were analyzed at the indicated time points. Mock transfection, Tmax alone and Tmed replication served as controls. (B) Efficiency of replication as analyzed by fluorescence. BSR T7/5 cells transfected with the indicated template together with the polyprotein were analyzed for replication, as indicated by Cherry/Tomato signal at 16 h (other templates) or 24 h (Tmax) posttransfection. The percentage of positive cells is indicated. The data are from one of multiple, reproducible experiments. Nuclei have been stained blue with DAPI (4′,6′-diamidino-2-phenylindole). (C) RNA synthesis levels were analyzed by labeling infected (wtSFV), control (mock), or transfected cells for 1 h, ending at the time points indicated with 3[H]uridine in the presence of actinomycin D. Tshort, Tmed, or Tlong templates were transfected with the viral polyprotein. After subtraction of mock background, the radioactive counts in precipitated RNA were divided by the number of uridine residues in each RNA to visualize the relative numbers of labeled RNAs. (D) Northern blot of positive-sense and negative-sense RNAs in the presence or absence of replicase polyprotein as indicated. RNA from SFV-Rluc-infected cells (in the same blot) is shown as a control. The negative-strand blot was exposed eight times longer than the positive-strand blot. The bands corresponding to the sizes of the three templates are marked with asterisks, and ethidium bromide staining of rRNAs is shown below as a loading control. (E) Western blot of the replicase proteins when the different templates were replicated. SFV-Rluc infection is shown as a control. The arrows indicate nsP4, nsP3 (during virus infection), nsP3-ZsG fusion (produced during trans-replication), and β-actin loading control.
To measure the RNA replication activity of the other templates directly, cells replicating different RNAs were labeled with 1-h pulses of [3H]uridine in the presence of actinomycin D, and incorporation into virus-specific template RNA was measured. When compared to wtSFV, the numbers of RNA molecules synthesized in trans-replication complexes were high with the Tshort, Tmed, and Tlong templates (Fig. 3C). The RNA synthesis increased slightly from 6-h to 10-h time points with all templates and with wtSFV, indicating that new replication complexes were being formed at these time points. The high efficiency of RNA replication demonstrated that the different sizes of spherules were fully functional.
Northern blotting showed that both the positive-strand and negative-strand RNAs generated from Tshort, Tmed, and Tlong templates were of the expected sizes (Fig. 3D). It appears that the template constructs alone also produce some larger RNA species, which may be due to the well-known poor efficiency of T7 polymerase termination signals (23). However, these longer positive-sense RNAs did not give rise to equivalent quantities of negative-strand products during replication. In the case of Tshort, the longer negative strands were most prominent but amounted to <10% of the total negative strands. Overall, in this Northern blot analysis of RNAs within the cell population, the accumulated levels of Tshort and Tmed RNAs in the presence of the polyprotein were higher than that of Tlong, a finding which is in agreement with the result that fewer cells were replication positive after Tlong transfection (Fig. 3B). Tlong was reproducibly seen in lower quantities in Northern blotting than in RNA labeling experiments. The reasons for this are not understood but could be related to the lower stability of longer RNAs, since labeling was used to measure RNAs synthesized during 1 h at earlier time points, whereas Northern blotting detected the total accumulated RNAs at 16 h. Finally, it was also verified that the expression level of the replicase remained constant when different templates were used (Fig. 3E), indicating that the amount of the replicase proteins present did not influence the results of RNA replication levels or spherule size.
Transfection of linearized DNA.
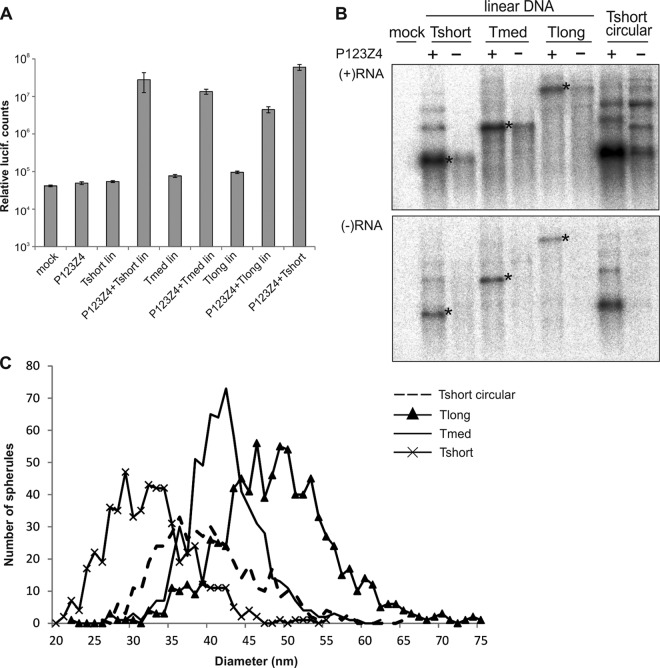
To avoid the synthesis of extended positive-strand templates by the T7 polymerase (Fig. 3D), the template plasmids were linearized after the virus-specific sequences, as described in Materials and Methods. Although linear DNA is not commonly used for transfection, in this particular case it worked well. Replication initiated by linearized template together with circular replicase expression plasmid yielded 100- to 1,000-fold more luciferase activity than the template alone (Fig. 4A). This result is comparable to that obtained with circular plasmids (17) and also the overall level of activity for the most efficient template, Tshort, was only slightly lower for linear DNA than for circular DNA (Fig. 4A).
Fig 4.
trans-Replication initiated by transfection of linear DNA. (A) The indicated templates, with or without replicase expression plasmids, were transfected to BSR cells, and triplicate samples were analyzed at 16 h posttransfection for luciferase activity. Lin, linearized plasmid. (B) Northern blot of positive-sense and negative-sense RNAs in the presence or absence of replicase polyprotein as indicated. The negative-strand blot was exposed four times longer than the positive-strand blot. The bands corresponding to the sizes of the three templates are marked with asterisks. Tshort circular is included as a control. (C) Distribution of spherule diameters for different lengths of RNA templates. The number of spherules detected having a particular diameter is plotted as a histogram for each of the indicated templates. Tshort circular data from Fig. 2A is included for comparison and marked with a dotted line.
In Northern blotting, transfection of linear DNA only yielded a single correct-size band of T7 transcript in the absence of viral replicase for each of the templates (Fig. 4B). However, in the presence of viral replicase, larger negative-strand bands were still visible, essentially as before (∼10% of for Tshort) (Fig. 4B). This suggests that the viral replicase may synthesize small amounts of concatemeric RNAs or covalently linked negative and positive strands, since similar larger RNAs were present for the positive-strand RNAs in the presence of the replicase (Fig. 4B). Thus, the larger RNAs appear in the system independent of the original T7 transcripts and could not be completely eliminated.
In CLEM experiments with this transfection setup, Tshort yielded spherules of 32 ± 5.4 nm (n = 545), Tmed 42 ± 4.6 nm (n = 654), and Tlong 48 ± 7.3 nm (n = 839). Therefore, the, spherules of Tmed and Tlong were unchanged compared to the previous results (Fig. 2), whereas the spherules of Tshort appeared slightly smaller than before, and smaller than those produced by Tmed (Fig. 4C). Thus, these results confirmed that template RNA length is a crucial determinant of spherule size. They also suggested further experimental approaches to examine the types of RNAs incorporated into a forming spherule (see Discussion).
Generation of two sizes of spherules in the same cell.
The influence of RNA size was further corroborated by analyzing cells transfected with SFV replicons. These are RNAs replicating in cis that lack the structural open reading frame and are thus intermediate in size between SFV genome and the long trans-replication template (Fig. 5A). For the replicon alone, 2,890 spherules were analyzed, and their average size was found to be 56 ± 8 nm. This value was larger than expected and relatively close to that produced by the full-length genome of SFV. However, the replicon experiment was carried out in BHK cells in contrast to all of the previous experiments in BSR cells. We therefore measured the replicon spherules again in BSR cells and obtained a value of 49.5 ± 2.9 nm (n = 749), which was intermediate between Tlong and Tmax, as expected. Similarly, SFV itself produced slightly larger spherules in BHK cells than in BSR cells (data not shown). This difference between the cell lines appeared consistently but, due to the small magnitude of the effect, it was not pursued further. It could be due to biological differences (e.g., lipid composition) causing a difference in actual spherule size or to different behaviors of the structures under EM fixation and staining conditions in the two cell lines.
Fig 5.
Two sizes of spherules detected in the same cell. (A) Schematic of the SFV replicon and Tmed RNAs prepared by in vitro transcription. (B and C) The two RNAs were cotransfected, and cells expressing both red and green fluorescence (filled arrowheads), indicating the replication of both Tmed and the replicon, were chosen for spherule measurements after CLEM. The cell population also contains cells expressing green fluorescence only (open arrowhead) derived from the replicon but lacking Tmed replication. (D) The distribution of spherule diameters in cells replicating both RNAs was plotted as a histogram. (E) Larger (white arrowhead) and smaller (black arrowhead) spherules visualized in the same EM field from a cell replicating the two RNAs. (F) Spherules produced by the transfection of replicon RNA alone were uniform in size and corresponded to the larger spherules.
In three independent cotransfection experiments of SFV-based replicon RNA, together with Tmed template RNA in BHK cells, 4,746 spherules were studied. In this system, both cis-replication of replicon and trans-replication of medium size template take place in many of the transfected cells (Fig. 5B and C). The coreplication gave rise to two clearly distinct populations of spherules with different diameters, indicating that two sizes of spherules could be produced in the cells at the same time (Fig. 5D and E). The diameters of the two populations closely corresponded to those observed for each of the templates independently (Fig. 2E and 5F). This experiment also supported the high efficiency of trans-replication for the Tmed template, since it produced approximately as many spherules as the cis-replication of the replicon RNA in cells replicating both RNAs. A further interesting observation was that the two sizes of spherules were intermixed with each other (Fig. 5E) and not concentrated to separate regions of the cell.
DISCUSSION
We have shown here that the size of the membranous replication complex, usually called a spherule, induced by SFV strongly correlates with the length of the template RNA. The viral RNA of 11.5 kb replicating normally in cis and a trans-replication template of similar size both yielded spherules of ∼58 nm in diameter (Fig. 2A). It is currently unknown whether an even longer RNA would yield larger spherules, since the trans-replication of Tmax was only observed in few cells, probably due to limitations of transfection efficiency and/or RNA stability, and thus experiments with longer RNAs would require further technical advances. At the other extreme, we consistently observed spherules of ∼39 nm in diameter for Tmed template of 3 kb (Fig. 2A). The smallest spherules thus far were observed with the linearized Tshort template, giving a diameter of ∼32 nm (Fig. 4C). The definite recognition of very small spherules is challenging, which may limit our observations at the smaller end of scale.
The current results are in direct contrast to what has been observed for two other positive-strand RNA viruses, FHV and BMV (14, 16), indicating that the formation of SFV spherules is a flexible process that utilizes the replicating RNA in a different manner than the two other viruses. FHV and SFV are similar in that for both of them spherule formation requires the replicase proteins, RNA, and an active polymerase (16, 17). However, for FHV it has been demonstrated that the spherule size is independent of the length of the RNA template varying over a 10-fold range (16), and thus far only one size class of spherules has been observed.
In BMV the replicase protein 1a alone can form spherules (14), and therefore it is reasonable that the properties of this protein determine the main features of the process, and the viral RNA appears to have no effect. Interestingly, smaller spherules have been observed in the BMV system with some 1a protein mutants, most having alterations in the membrane binding alpha helix of 1a (24, 25). Deletion of the genes for host reticulon proteins that are components of the BMV spherules, and reduction in the amounts of cellular unsaturated fatty acids also gave rise to smaller spherules (26, 27). In several cases, the small spherule phenotype also dramatically reduced viral RNA synthesis, suggesting that altered spherule assembly could lead to functional defects. In the case of the SFV spherules observed here, the replication of small RNAs in smaller spherules proceeded very efficiently (Fig. 3), indicating that they were fully competent for RNA replication.
It should be noted that both BMV and FHV have segmented genomes, with the segment sizes varying between 2.1 to 3.2 kb and 1.4 to 3.1 kb, respectively. During virus multiplication, each of the segments has to be replicated efficiently and packed to progeny particles. Based on volume calculations of the spherules, it has been speculated that several small segments might be incorporated into a single spherule simultaneously (15). In contrast, the SFV genome is ca. 11.5 kb in length and, based on similar considerations, we hypothesize that SFV is likely to have only one copy of the negative strand within a spherule. Since SFV spherules arise during active RNA synthesis (17) (see Fig. 6 for models), the forming spherule will contain the initial positive-strand template and the nascent/completed negative strand. Therefore, it could be the length or the positive strand or the combined length of the positive and negative strands that determines the spherule size. Interestingly, our results tentatively suggest a different size distribution for Tshort spherules when the initial mix of positive strands contained also 3′-extended RNA species produced by T7 polymerase (slightly larger spherule sizes overall) versus when only proper Tshort-sized RNAs were initially present (linearized Tshort templates produced slightly smaller spherules) (Fig. 4C). To confirm and extend this result regarding the size of the initial template, we are currently studying the replication competence, negative strands and spherules produced by “short” positive-strand templates that do not contain proper Tshort-sized RNAs but consist only of 3′-extended RNA species.
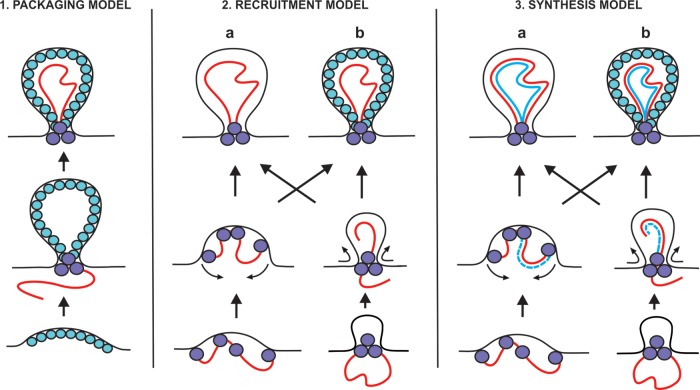
Fig 6.
Three classes of models for spherule formation: packaging, recruitment, and synthesis models. The mature spherules, depicted at the top, could either contain an inner shell (variant b) or be devoid of the shell (variant a). Intermediates in spherule formation could potentially include “open” dome-like structures (variant a, middle row) or “closed” structures already containing a neck-like constriction (variant b). Positive-strand RNA is shown in red, and negative-strand RNA is shown in blue. Spheres represent proteins that could be located at the neck and/or form an inner shell. See the text for further discussion.
At least three different models can be proposed for the formation of spherules in relation to the template RNA (Fig. 6): (i) in the packaging model, RNA template is packaged to spherules, which have been preformed by viral protein components; (ii) in the recruitment model, spherules are formed by RNA and protein together at the stage of RNA recruitment preceding replication; and (iii) in the synthesis model, the formation of spherules takes place concomitantly with RNA replication. Further variation in these models can be introduced by the presence or absence of an inner protein shell and by the nature of the intermediate stages during formation (Fig. 6). For BMV, the packaging model has been proposed as a possibility (16), whereas both FHV and SFV need an active polymerase and appear to represent variants of the synthesis model. In the recruitment and synthesis models, RNA could theoretically determine the size of the spherule, if the mechanism for spherule formation is flexible in this regard. For FHV, the size of the spherules remains fixed (16) and might be determined, for instance, by the packing of the replication proteins within the spherules.
It has been proposed, with some good evidence from EM labeling and from calculations based on purified membrane preparations, that BMV and FHV spherules contain >100 copies of the major virus-encoded replicase component, which could form an inner shell that stabilizes the spherule (14, 15). This shell could also include host proteins, such as reticulons for BMV or amphiphysins for SFV (26, 28). In the case of SFV, there is no information regarding the presence of a protein shell. Interestingly, the formation of membrane invaginations without an inner coating is catalyzed by the endosomal sorting complex required for transport (ESCRT) involved in the formation of multivesicular bodies (MVBs) (29). The intermediates in the formation of the inner vesicles in MVBs have a strong resemblance to spherules, and their sizes in mammalian cells vary between 40 and 100 nm, with an average of 56 nm (30). There is evidence from tomato bushy stunt virus, another spherule-generating plant virus (31), that ESCRT proteins are crucial for the formation of viral replication complexes (32). The ESCRT proteins are also involved in retrovirus budding during particle production (33), which therefore might share some common features with spherule formation, with the crucial difference that the spherules remain attached to membranes via the neck structure and do not pinch off.
The intermediate stages of spherule formation (Fig. 6) have thus far not been detected (1), which might be due to their transient nature. Currently, we are addressing the question of intermediates by analyzing early time points from a very high-multiplicity SFV infection (10) using electron tomography. An interesting hypothesis for further study is that the SFV RNA could act as a structural component within the spherule in such a way that the inclusion of the entire RNA within the forming spherule would contribute to the determination of its size. The size determination could be effected through the binding of multiple copies of proteins to the RNA or, given the requirement for an active polymerase, it could be connected to the completed replication of the entire RNA.
ACKNOWLEDGMENTS
We thank Mervi Lindman and Arja Strandell for excellent technical assistance in EM.
This study was supported by the Academy of Finland (grant 127214) and the Sigrid Jusélius Foundation. K.K. was supported in part by a fellowship from the Helsinki Graduate Program in Biotechnology and Molecular Biology, and K.H. was supported by an Academy of Finland postdoctoral fellowship.
Footnotes
Published ahead of print 12 June 2013
REFERENCES
- 1. den Boon JA, Ahlquist P. 2010. Organelle-like membrane compartmentalization of positive-strand RNA virus replication factories. Annu. Rev. Microbiol. 64:241–256 [DOI] [PubMed] [Google Scholar]
- 2. Belov G, van Kuppeveld FJM. 2012. (+)RNA viruses rewire cellular pathways to build replication organelles. Curr. Opin. Virol. 2:740–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Salonen A, Ahola T, Kääriäinen L. 2005. Viral RNA replication in association with cellular membranes. Curr. Top. Microbiol. Immunol. 285:139–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Spuul P, Salonen A, Merits A, Jokitalo E, Kääriäinen L, Ahola T. 2007. Role of the amphipathic peptide of Semliki Forest virus replicase protein nsP1 in membrane association and virus replication. J. Virol. 81:872–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Miller S, Krijnse-Locker J. 2008. Modification of intracellular membrane structures for virus replication. Nat. Rev. Microbiol. 6:363–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Griffin DE. 2007. Alphaviruses, p 1023–1067 In Knipe DM, Howley PM. (ed), Fields virology, 5th ed, vol 1 Lippincott/The Williams & Wilkins Co, Philadelphia, PA [Google Scholar]
- 7. Kääriäinen L, Ahola T. 2002. Functions of alphavirus nonstructural proteins in RNA replication. Prog. Nucleic Acids Res. Mol. Biol. 71:187–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Grimley PM, Berezesky IK, Friedman RM. 1968. Cytoplasmic structures associated with an arbovirus infection: loci of viral ribonucleic acid synthesis. J. Virol. 2:1326–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kujala P, Ikäheimonen A, Ehsani N, Vihinen H, Auvinen P, Kääriäinen L. 2001. Biogenesis of the Semliki Forest virus RNA replication complex. J. Virol. 75:3873–3884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Spuul P, Balistreri G, Kääriäinen L, Ahola T. 2010. Phosphatidylinositol 3-kinase-, actin-, and microtubule-dependent transport of Semliki Forest virus replication complexes from the plasma membrane to modified lysosomes. J. Virol. 84:7543–7557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Frolova EI, Gorchakov R, Pereboeva L, Atasheva S, Frolov I. 2010. Functional Sindbis virus replicative complexes are formed at the plasma membrane. J. Virol. 84:11679–11695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Welsch S, Miller S, Romero-Brey I, Merz A, Bleck CK, Walther P, Fuller SD, Antony C, Krijnse-Locker J, Bartenschlager R. 2009. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe 5:365–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gillespie LK, Hoenen A, Morgan G, Mackenzie JM. 2010. The endoplasmic reticulum provides the membrane platform for biogenesis of the flavivirus replication complex. J. Virol. 84:10438–10447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schwartz M, Chen J, Janda M, Sullivan M, den Boon J, Ahlquist P. 2002. A positive-strand RNA virus replication complex parallels form and function of retrovirus capsids. Mol. Cell 9:505–514 [DOI] [PubMed] [Google Scholar]
- 15. Kopek BG, Perkins G, Miller DJ, Ellisman MH, Ahlquist P. 2007. Three-dimensional analysis of a viral RNA replication complex reveals a virus-induced mini-organelle. PLoS Biol. 5:e220. 10.1371/journal.pbio.0050220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kopek BG, Settles EW, Friesen PD, Ahlquist P. 2010. Nodavirus-induced membrane rearrangement in replication complex assembly requires replicase protein A, RNA templates, and polymerase activity. J. Virol. 84:12492–12503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Spuul P, Balistreri G, Hellström K, Golubtsov AV, Jokitalo E, Ahola T. 2011. Assembly of alphavirus replication complexes from RNA and protein components in a novel trans-replication system in mammalian cells. J. Virol. 85:4739–4751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Buchholz UJ, Finke S, Conzelmann KK. 1999. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J. Virol. 73:251–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pohjala L, Barai V, Azhayev A, Lapinjoki S, Ahola T. 2008. A luciferase-based screening method for inhibitors of alphavirus replication applied to nucleoside analogues. Antivir. Res. 78:215–222 [DOI] [PubMed] [Google Scholar]
- 20. Tarn WY, Yario TA, Steitz JA. 1995. U12 snRNA in vertebrates: evolutionary conservation of 5′ sequences implicated in splicing of pre-mRNAs containing a minor class of introns. RNA 1:644–656 [PMC free article] [PubMed] [Google Scholar]
- 21. Tarn WY, Steitz JA. 1996. A novel spliceosome containing U11, U12, and U5 snRNPs excises a minor class (AT-AC) intron in vitro. Cell 84:801–811 [DOI] [PubMed] [Google Scholar]
- 22. Puhka M, Vihinen H, Joensuu M, Jokitalo E. 2007. Endoplasmic reticulum remains continuous and undergoes sheet-to-tubule transformation during cell division in mammalian cells. J. Cell Biol. 179:895–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Macdonald LE, Zhou Y, McAllister WT. 1993. Termination and slippage by bacteriophage T7 RNA polymerase. J. Mol. Biol. 232:1030–1047 [DOI] [PubMed] [Google Scholar]
- 24. Wang X, Lee WM, Watanabe T, Schwartz M, Janda M, Ahlquist P. 2005. Brome mosaic virus 1a nucleoside triphosphatase/helicase domain plays crucial roles in recruiting RNA replication templates. J. Virol. 79:13747–13758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu L, Westler WM, den Boon JA, Wang X, Diaz A, Steinberg HA, Ahlquist P. 2009. An amphipathic alpha-helix controls multiple roles of brome mosaic virus protein 1a in RNA replication complex assembly and function. PLoS Pathog. 5:e1000351. 10.1371/journal.ppat.1000351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Diaz A, Wang X, Ahlquist P. 2010. Membrane-shaping host reticulon proteins play crucial roles in viral RNA replication compartment formation and function. Proc. Natl. Acad. Sci. U. S. A. 107:16291–16296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang J, Diaz A, Mao L, Ahlquist P, Wang X. 2012. Host acyl coenzyme A binding protein regulates replication complex assembly and activity of a positive-strand RNA virus. J. Virol. 86:5110–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Neuvonen M, Kazlauskas A, Martikainen M, Hinkkanen A, Ahola T, Saksela K. 2011. SH3 domain-mediated recruitment of host cell amphiphysins by alphavirus nsP3 promotes viral RNA replication. PLoS Pathog. 7:e1002383. 10.1371/journal.ppat.1002383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wollert T, Hurley JH. 2010. Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature 464:864–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Murk JLAN, Humbel BM, Ziese U, Griffith JM, Posthuma G, Slot JW, Koster AJ, Verkleij AJ, Geuze HJ, Kleijmeer MJ. 2003. Endosomal compartmentalization in three dimensions: implications for membrane fusion. Proc. Natl. Acad. Sci. U. S. A. 100:13332–13337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. McCartney AW, Greenwood JS, Fabian MR, White KA, Mullen RT. 2005. Localization of the tomato bushy stunt virus replication protein p33 reveals a peroxisome-to-endoplasmic reticulum sorting pathway. Plant Cell 17:3513–3531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Barajas D, Jiang Y, Nagy PD. 2009. A unique role for the host ESCRT proteins in replication of tomato bushy stunt virus. PLoS Pathog. 5:e1000705. 10.1371/journal.ppat.1000705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Carlton JG, Martin-Serrano J. 2009. The ESCRT machinery: new functions in viral and cellular biology. Biochem. Soc. Trans. 37:195–199 [DOI] [PubMed] [Google Scholar]