Abstract
Human and animal hemorrhagic viruses initially target dendritic cells (DCs). It has been proposed, but not documented, that both plasmacytoid DCs (pDCs) and conventional DCs (cDCs) may participate in the cytokine storm encountered in these infections. In order to evaluate the contribution of DCs in hemorrhagic virus pathogenesis, we performed a genome-wide expression analysis during infection by Bluetongue virus (BTV), a double-stranded RNA virus that induces hemorrhagic fever in sheep and initially infects cDCs. Both pDCs and cDCs accumulated in regional lymph nodes and spleen during BTV infection. The gene response profiles were performed at the onset of the disease and markedly differed with the DC subtypes and their lymphoid organ location. An integrative knowledge-based analysis revealed that blood pDCs displayed a gene signature related to activation of systemic inflammation and permeability of vasculature. In contrast, the gene profile of pDCs and cDCs in lymph nodes was oriented to inhibition of inflammation, whereas spleen cDCs did not show a clear functional orientation. These analyses indicate that tissue location and DC subtype affect the functional gene expression program induced by BTV and suggest the involvement of blood pDCs in the inflammation and plasma leakage/hemorrhage during BTV infection in the real natural host of the virus. These findings open the avenue to target DCs for therapeutic interventions in viral hemorrhagic diseases.
INTRODUCTION
Bluetongue virus (BTV) is a double-stranded RNA virus of the Reoviridae family that is transmitted by insect vectors to ruminants. Whereas BTV infection has been endemic for several centuries in Southern Africa, it recently caused major outbreaks in several northern European countries with severe economical consequences in sheep and cattle (1, 2). Bluetongue disease (BT) is characterized by fever, vascular injury with hemorrhages, tissue infarctions, and widespread edema, lesions that are consistent with those of the so-called viral hemorrhagic fevers (3–6). After inoculation in the skin, the virus migrates, associated to conventional dendritic cells (cDCs), to the regional lymph node, where initial replication occurs (7, 8). BTV is then disseminated to a variety of tissues, and it replicates in monocytes (9), endothelial cells (10), and γ/δ T lymphocytes (11).
In several hemorrhagic viral infections, dendritic cells (DCs) have been proposed to be implicated in the virus dispersion around the body as well as in the fever, the inflammatory status, and the altered vascular functions, although in vivo evidence supporting the importance of their role is lacking (12). DCs include two major subsets, the cDCs, which are specialized in antigen processing and presentation to T lymphocytes, and the plasmacytoid DCs (pDCs), which produce large amounts of type I interferon (IFN) and other inflammatory cytokines upon virus infection. The cDCs are the early cell targets of BTV (8) and of other hemorrhagic viruses (13–16). BTV replicates both in cDCs and in pDCs and activates them in vitro (8, 17). Thus, BTV infection in sheep appears as a pertinent infection model to study the contributions of DCs in hemorrhagic fever virus pathogenesis in a natural host. Furthermore, this relatively convenient animal model allows extraction of both pDCs and cDCs from different lymphoid organs in order to perform analyses of gene expression during infection. Indeed, lymphoid compartments are differentially involved during BTV infection: BTV first replicates in the lymph node that drains the inoculation site and then disseminates to other secondary lymphoid organs via blood (18, 19). Using genome-wide expression profiling, we present evidence that BTV infection modifies the gene expression programs in pDCs and cDCs depending on the lymphoid organs and suggest their role in the expression of clinical symptoms and in the control of the disease.
MATERIALS AND METHODS
Sheep infection and sample collections.
BTV-negative male lambs of Prealpes breed (4 to 5 months old) were used in this study. Nineteen sheep were infected subcutaneously in the shoulder area and intravenously with 1 and 5 ml, respectively, of blood from a viremic BTV8-infected animal (8.1 × 106 BTV8 RNA copies/ml). This procedure was previously shown by us to induce high viremia and clinical disease (20). Eleven sheep were used as negative controls. Animal experimentation was performed as prescribed by the guidelines of the European Community Council on Animal Care (86/609/CEE) in the biosafety level 3 (BSL3) facilities of the INRA-UE 1277 “Plateforme d'Infectiologie Expérimentale” in Nouzilly, France, under the “Direction départementale de la protection des populations,” authorization number B-37-175-3. The animal experiments were carried out under licenses issued by the Direction of the Veterinary Services of Versailles (accreditation numbers B78-93) and of Toulouse (311-055-524). Clinical signs and rectal temperature were recorded every day as described in a previous study (20). BTV RNA was measured by quantitative reverse transcription-PCR (qRT-PCR) assays of blood collected in EDTA, sampled every 2 days beginning 3 days before virus inoculation until the end of the experiment (day 21). A standard plasmid curve for quantification was obtained by dilutions of plasmid pBTVM containing the L1 gene replicon (kindly provided by E. Sellal, Laboratoire Service International). Three control sheep were sacrificed at day 0 (D0) before BTV8 infection. At days 2, 6, and 10 postinoculation, 3 infected sheep were euthanized for collection of tissues. Just before these slaughters, 500 ml of blood per sheep was collected on sodium citrate. The prescapular lymph node draining the subcutaneous inoculation site, and a slice of the spleen were harvested and weighed. The complexity of the experiment in a large-animal BSL3 facility, the logistics, and the high cost dictated the relatively low number of sheep used in this study.
Cell isolation from lymph node, spleen, and blood.
Lymph nodes were cut in small pieces and treated with collagenase (2 mg/ml; Roche), DNase (2 mg/ml; Roche), and dispase (0.5 mg/ml; Gibco-BRL) in RPMI 1640 supplemented with 10% heat-inactivated fetal calf serum (FCS) for 1 h at 37°C. The lymph node cell suspension was filtered through a 100-μm polypropylene cell strainer (Becton, Dickinson). The spleen tissue was cut in small pieces, and the suspension was sequentially filtered through a 500-μm steel strainer and a 100-μm polypropylene cell strainer. The splenic cells were further purified on a 1.076 density Percoll gradient (GE Healthcare), following the same protocol as for sheep peripheral blood mononuclear cells (PBMCs) (21). The total number of spleen and lymph node cells per gram of tissue was calculated after cell counting with a hemocytometer. Low-density (LD) lymph node, spleen, and PBMCs were obtained by centrifugation on a 1.065 density iodixanol gradient (Optiprep; Nycomed Pharma) as previously described (22). LD cells were counted after each preparation. LD lymph node and spleen cells and LD PBMCs were frozen in FCS containing 10% dimethyl sulfoxide (Sigma) and stored in liquid nitrogen.
DC staining and isolation.
cDCs and pDCs were stained and sorted from LD frozen spleen, blood cells, and nodes. For the staining of spleen and blood cDCs and pDCs, cells were reacted with anti-CD11c monoclonal antibody (MAb) (2 μg/ml, OM1 clone, IgG1) followed by a saturating concentration of Alexa Fluor 488-conjugated anti-mouse IgG donkey Fab (50 μg/ml). After extensive washing, cells were further incubated with 2 μg/ml of anti-CD45RB (CC76 clone, IgG1), anti-B cells (DU2-104 clone, IgM), anti-CD8 (7C2 clone, IgG2a), anti-TCRγ/δ (CC15 clone, IgG2a), and anti-CD11b (ILA-130 clone, IgG2a), followed by phycoerythrin- and Alexa Fluor 647-conjugated goat anti-mouse isotype-specific antibodies (Caltag). For the staining of lymph node cDCs and pDCs, cells were processed as described above, except that cDCs were labeled with anti-CD1b MAb (Th97A, IgG2a). After the final wash, cells were resuspended in Hanks balanced salt solution (HBSS) + 1% heat-inactivated FCS for cytometry sorting or in phosphate-buffered saline (PBS) for fluorescence-activated cell sorter (FACS) analysis. The pDCs and cDCs were sorted by flow cytometry on the Imagif Cytometry platform using the analyzer-sorter MoFlo XDP cytometer and the Summit 5.2 software (purity, >98%; Beckman Coulter). The FACS analyses of the pDC and cDC representation in lymphoid organs were done with a FACSCalibur using the CELLQuest software (Becton, Dickinson). The percent representation of each DC subset in LD cells was used to calculate their number from gram of tissue (lymph node/spleen) or from ml of blood, based on the numbers of collected LD cells and of total cells.
RNA extraction and hybridization on microarrays.
Total RNA from pDCs and cDCs was extracted using the Arcturus PicoPure RNA Isolation kit (Arcturus Life Technologies) and checked for quality with an Agilent 2100 Bioanalyzer using RNA 6000 Nano or Pico Kits (Agilent Technologies). All RNA samples had an RNA integrity number (RIN) above 8. RNA amplification and labeling were performed using the one-color Low Input Quick Amp Labeling kit (Agilent Technologies) according to the manufacturer's recommendations. Each RNA sample (25 to 50 ng) was amplified and cyanin 3 (Cy3) labeled, and subsequently the cRNA was checked for quality on Nanodrop and Agilent 2100 Bioanalyzer. Subsequently the cRNA (600 ng) was fragmented and used for hybridization on custom-designed Agilent ovine arrays (ID 29915). Our custom-designed ovine array is based on the commercial ovine Agilent array (15,208 60mer probes; AMADID, 1992; Agilent Technologies). About 5,000 probes of the commercial ovine array were replaced, based on quality analyses made by the Sigreannot program from the Sigenae group (23). New probes were designed with the e-array software from Agilent Technologies (custom AMADID, 29915). The new probes (4,921 in total) included ovine gene sequences derived from (i) ovine Sigenae contigs (http://www.sigenae.org/) and (ii) ovine or bovine expressed sequence tags corresponding to genes known to be selectively expressed in human and mouse DC subsets (24). The ovine microarray has been annotated by the Sigreannot program (23) and by blasting the ovine expressed sequence tags against bovine sequences (RefSeq Bos taurus) in order to identify the putative bovine orthologous gene (>200 nucleotides [nt] with >92% identity). After hybridization of the cRNA on the custom-designed ovine array, the chips were washed according to the manufacturer's protocol. The chips were then scanned by using a G2505C scanner (Agilent Technologies) at a resolution of 5 μm. Raw data were extracted using the Feature Extraction v7.10.3.1 software (Agilent Technologies). All the protocols used can be obtained by contacting the CRB GADIE facility (http://crb-gadie.inra.fr/).
Microarray analyses.
The median normalized microarray data have been used to compute the hierarchical clustering (Pearson correlation distance function and average linkage method) and unsupervised principal component analysis (PCA), using the R software and the FactoMineR package. The robustness of the hierarchical clustering was tested with the support tree function of the MeV software (using the Pearson correlation coefficient, average linkage, and 100 iterations): subgroups of gene expression data were permutated between samples (bootstrap), and the classification was repeated 100 times, generating a percentage of stability for each branch. For the PCA, the eigenvalues correspond to the percentage of the total variance of normalized signal data, and they are represented by a given component (axis).
The lists of genes modulated by BTV infection in pDCs and cDCs from the different lymphoid tissues (differentially expressed genes [DEG]) were determined with methods that depended on the number of RNA samples available from different sheep (biological replicates). For lymph node cDCs and pDCs, hybridization was conducted with samples from 3 control and 3 BTV-infected sheep (6 days postinfection). In these cases, the probe signals corresponding to genes significantly modulated during BTV infection over control were analyzed by a linear model in the Limma R package (25). The P values obtained have been corrected for multiple testing using the Benjamini and Hochberg procedure (26). By convention, genes were considered differentially expressed in BTV infection (up- or downmodulated) when their corresponding mean probe signal showed a >2- or <0.5-fold change compared to the control condition, with an adjusted P value of <0.05. For blood pDCs and for splenic cDCs, sufficient quantities of RNA for hybridization on microarrays were only obtained from 2 controls and 2 infected sheep and from 3 control and 2 infected sheep, respectively, which did not allow reliable P values calculation. Genes were considered differentially expressed by BTV infection in blood pDCs and splenic cDCs when their corresponding Agilent probe signal showed a >2- or <0.5-fold change in the DCs from the 2 infected sheep compared to all control sheep DC samples. The final DEG list was prepared by keeping the genes from reliably annotated probes, removing duplicates, and assigning the human official gene symbol to the probes for subsequent meta-analysis.
For functional analysis, the DEG lists were submitted to Ingenuity Pathway Analysis (IPA) software (Ingenuity Systems). Canonical pathway analyses, Ingenuity downstream effects analyses (DEA), and Upstream Regulator Prediction were conducted by the IPA software. The DEA examines genes in the DEG lists that are known to affect a given biological function and compares their direction of change (up- or downmodulation) to what is expected from the literature. When the direction of change is consistent with the literature across the majority of genes, then the function is predicted to be increased in the biological sample (z-score > 2), whereas if the direction of change is mostly anticorrelated with the literature, then the function is predicted to be decreased in the biological sample (z-score < −2). If there is no clear pattern, then there is no prediction either way. The Upstream Regulator Prediction analysis examines how many known targets of each transcription regulator are present in the DEG data set and also compares their direction of change to what is expected from the literature in order to predict likely relevant transcriptional regulators. If the observed direction of change is mostly consistent with a particular activation state of the transcriptional regulator (“activated” or “inhibited”), then a prediction is made about that activation state (z-score > 2 for activation, z-score < −2 for inhibition). The white paper regarding the algorithm used for DEA and Upstream Regulator is available on the IPA website.
Real-time qPCR.
For quantitation of gene expression in DCs, RNA (200 ng) was reverse transcribed using random primers and the Multiscribe reverse transcriptase (Applied Biosystem). Real-time qPCR was carried out using 5 ng cDNA with 300 nM primers in a final reaction volume of 25 μl of 1× SYBR green PCR Master Mix (Applied Biosystem). The primers used to amplify ovine cDNA were designed with the Primer Express software (v2.0) using publicly available GenBank sequences. The primers used in this study are as follows: GAPDH (glyceraldehyde-3-phosphate dehydrogenase) (forward, CACCATCTTCCAGGAGCGAG, reverse, CCAGCATCACCCCACTTGAT); cyclophilin (forward, TGACTTCACACGCCATAATGG T, reverse, CATCATCAAATTTCTCGCCATAGA); XCL1 (forward, TGAGCCAGAGCAAGCCTACA, reverse, TCACTACCCAGTCAGGGTCACA-AAG GAA CGC AAG AAC AGA ATG AA); XCR1 (forward, TGCCATCTTCCACAAGGTGTT, reverse, ACGGAGGCGAGGAACCA); TCF4 (forward, TGGTCTGGCCTCAGGGTATG, reverse, GGCCCCAACCATGAGTGA); FLT3 (forward, TGTTCACGCTGAATATAAGAAGGAA, reverse, GGAGCAGGAAGCCTGACTTG); ID2 (forward, CAAGAAGGTGAGCAAGATGGAA, reverse, CGCGATCTGCAAGTCCAA); SKT17 (forward, GAAACACCCATGGCTGACTCA, reverse, GGCCCCTCTGACCTTGAAA); CCR7 (forward, CCAGATGGTGGTAGGCTTCCT, reverse, GCGGATGATGACAAGGTAGCA); EIF2AK2 (forward, AGGTTGGTCAAGGATTTCACAGA, reverse, TCTGTGTTCGGCTTGAAAACTC); C-C motif ligand 5 (CCL5) (forward, GTGGGTGCGAGAGTACATCAAC, reverse, GGCGCAAGTTCAGGTTCAAG); tryptophan-degrading enzyme indoleamine 2,3 dioxygenase (IDO1) (forward, GCCTCCGAGGCCACAAG, reverse, CATACGCCATGGTGATGTATCC); C-X-C motif ligand 10 (CXCL10) (forward, GGTCCTTAGAAAAACTTGAACTGATTC, reverse, TCCTTTTCATTGTGGCAATAATCTC); IFN regulatory factor 8 (IRF8) (forward, GGAGTGTGGGCGCTCTGA, reverse, TCACCATCCCCATGTAGTCATC); interleukin-1B (IL-1B) (forward, CGAACATGTCTTCCGTGATG, reverse, TCTCTGTCCTGGAGTTTGCAT); IL-8 (forward, TTCCAAGCTGGCTGTTGCTCTCTT, reverse, GCATTGGCATCGAAGTTCTGTACTC); tumor necrosis factor (TNF) (forward, CAAGGGCCAGGGTTCTTACC, reverse, GCCCACCCATGTCAAGTTCT); IFNG (forward, TGATTCAAATTCCGGTGGATG, reverse, TTCATTGATGGCTTTGCGC). PCR cycling conditions were 95°C for 10 min, linked to 40 cycles of 95°C for 15 s and 60°C for 1 min. Real-time qPCR data were collected by the Mastercycler ep realplex-Eppendorf system, and 2−ΔCT calculations for the relative expression of the different genes (arbitrary units) were performed with the Realplex software using GAPDH for normalization in the pDC and cDC canonical gene expression experiment and cyclophilin for normalization of the qPCR control experiment of the microarray data. All qPCRs showed >95% efficacy. Viral RNA detection was performed using a commercial pan-BTV real time-RT-PCR (Adiavet BTV Real-time A352; Adiagene, France) targeting segment 10 of BTV (NS3), which is highly conserved among BTV serotypes, and the GAPDH as an internal control.
Statistics for cell count data.
Cell count data are presented as mean values ± standard errors of the means (SEM). The statistical significance between sets of cell count data was assessed with the two-tailed unpaired Student t test.
Microarray data accession number.
The data discussed in this article have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO series accession number GSE48450.
RESULTS
BTV infection modifies the distribution of cDCs and pDCs in lymphoid compartments.
All the BTV8-infected sheep became febrile 4 days after virus inoculation (D4) and developed moderate disease by D5 to D17 (data not shown). Clinical signs were characterized by fever, prostration, nasal discharge, conjunctivitis, facial edema and congestion, hemorrhages, and ulceration of the oral cavity. Hyperthermia (>40.5°C) was detected in all infected animals from D5 to D9 with a mean peak of 41.2°C at D6. Neither clinical signs nor hyperthermia was observed in the noninfected control group. BTV8 RNA copies were detected in all infected sheep from D1 to D19 and reached between 4.2 and 6.6 log10 RNA copies per ml of blood with peaks of RNA viral detection between D5 and D9. In parallel, virus isolation was performed and confirmed the infectivity of the blood samples. These viral and clinical observations were similar to those described in other published studies (5, 6, 20, 27, 28).
In lymph nodes, sheep cDCs had been previously identified as CD1b+ cells (29). Sheep pDCs were previously extensively identified as CD45RB+CD1b−CD11c−CD8−TCRγ/δ−B−CD11b− cells in afferent skin lymph and blood (17, 30). We used a combination of markers to simultaneously label cDCs and pDCs among lymph node LD cells (see Fig. S1A in the supplemental material). The cDCs (CD1b+ cells) and pDCs (CD45RB+ cells) were isolated from the FSChiCD8−TCRγ/δ−B−CD11b− cells. CD14+ and B cells were isolated as controls. In order to demonstrate the validity of our cell isolation strategies, the expression of cDC and pDC canonical genes was analyzed by qPCR in the sorted subsets. The expression of the cDC canonical FLT3 (31) and XCR1 (32) genes was selectively found in the CD1b+ cells, whereas the expression of the pDC canonical TCF4 gene (33) was selectively found in the CD45RB+ cells (see Fig. S1A in the supplemental material). Because the CD1b marker does not label cDCs in blood and spleen, we identified cDCs as CD11c+ cells in these compartments (see Fig. S1B in the supplemental material). In both blood and spleen, the TCF4 gene was expressed mainly in the CD45RB+ population, whereas the FLT3 and XCR1 genes were selectively expressed in the CD11c+ cells and not in the CD11c−CD45RB− cells (see Fig. S1B in the supplemental material). These double-negative cells strongly expressed XCL1 mRNA, suggesting that this population included NK and/or activated CD8+ T cells (32).
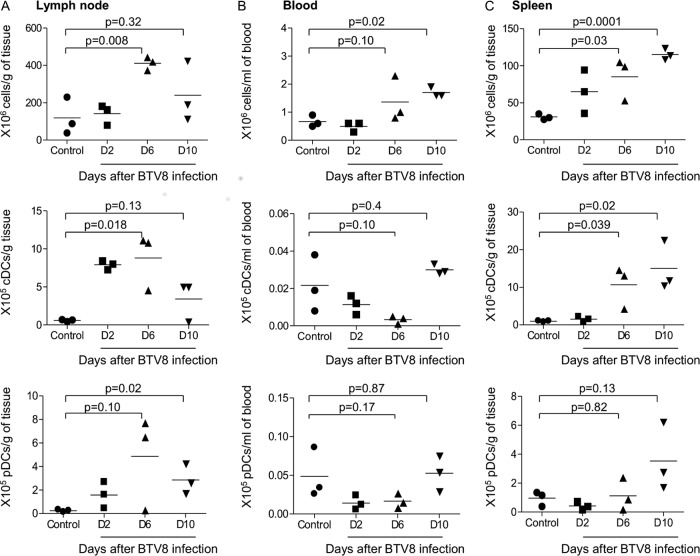
An analysis of the representation of pDCs and cDCs was conducted in the draining lymph nodes, blood, and spleen during BTV infection. Three time points were chosen: at D2 postinfection, when type I IFN was induced in the serum (17); at D6 postinfection, when the viremia peaked and symptoms appeared (this study and others [5]); and at D10, when symptoms and virus counts started to decrease (this study and others [5, 27]). As shown in Fig. 1, the global leukocyte cellularity (top panel) was higher in lymph nodes and spleen at D6 postinfection than in controls. The cDC numbers per gram of local lymph node and spleen were elevated in infected sheep at D6 compared to controls (Fig. 1, middle panels; P = 0.018 and P = 0.039, respectively), whereas they tended to be lower than in controls in blood at D6 postinfection. The reduction of cDC numbers in blood was transient and reached control values by D10. The effect of BTV infection on pDCs was less evident (Fig. 1, lower panels), and only trends in pDC number modifications could be inferred. This was in part due to the low number of sheep that we could handle under the BSL3 conditions. The pDCs accumulated in the draining lymph node at D10 (P = 0.02), and a small increase was observed in spleen at D10 (P = 0.13). Conversely, pDC numbers tended to be reduced in blood at D2 and D6 postinfection (P = 0.17). Thus, altogether, these analyses indicate that cDCs and pDCs accumulate in lymphoid organs and their numbers tend to transiently decrease in blood during the first 10 days post-BTV infection.
Fig 1.
Representation of cDCs and pDCs in the draining lymph nodes (A), blood (B), and spleen (C) during BTV infection. cDCs and pDCs were labeled from LD cell preparations of lymph nodes, blood, and spleen as in Fig. S1 in the supplemental material. The percentage of DCs among other cells was used to calculate their total number representation within the initial tissue and blood sample. Total numbers of leukocytes (top panels), cDCs (middle panels), and pDCs (bottom panels) were reported per gram of tissue or per ml of blood. Three different sheep were used per time point (control and D2, D6, and D10 postinfection).
BTV infection induces gene expression profiles in DCs that depend on the pDC/cDC subtype and organ localization.
In order to gain insight into the role of cDCs and pDCs in the physiopathology of BTV infection, we chose to isolate cDCs and pDCs from different lymphoid compartments at D6 following viral inoculation, i.e., at the onset of symptoms. Blood, spleen, and regional draining lymph nodes are hypothesized to be important sites in BT pathogenesis: cDCs were shown to strongly migrate from skin to draining lymph node upon intradermal inoculation of BTV and to be cell targets that disseminate the virus (8); furthermore, the BTV load is important in the blood compartment where cDCs and pDCs circulate (1, 34). From the lymph nodes of our control and BTV-infected sheep, we isolated sufficient numbers of cDCs and pDCs (>50,000 cells) to enable three independent RNA hybridization replicates. We isolated sufficient RNA quantities from 2 independent replicates of blood pDCs and spleen cDCs. However, spleen pDCs and blood cDCs could not be collected in sufficient numbers from infected sheep.
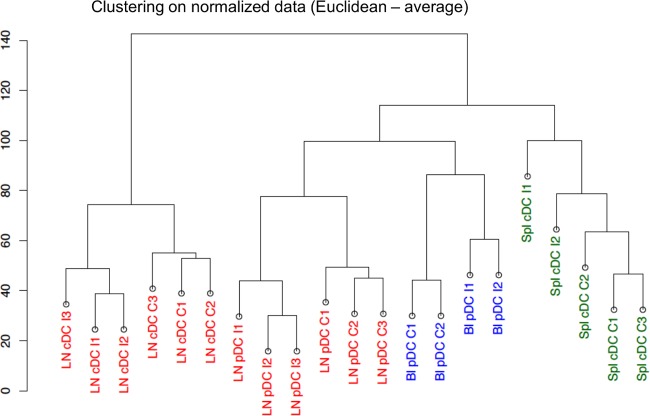
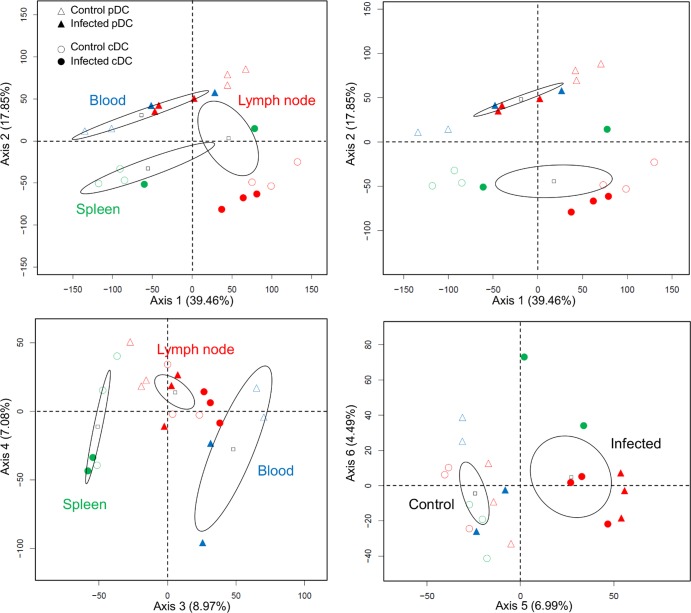
As a first exploratory analysis, the unsupervised hierarchical clustering of normalized probe signals showed that the biological duplicates and/or triplicates from different sheep all clustered together, indicating similarity of gene expression profiles between replicates (Fig. 2). Furthermore, a bootstrap analysis using gene data permutations with the MeV software (see Materials and Methods) revealed a strong stability within the arms of the hierarchical clustering (over 90%; data not shown), confirming the similarity between experimental replicates. The hierarchical clustering also reveals that the data group primarily according to the tissue of origin and to the cDC and pDC types and secondly according to the infected versus noninfected status (Fig. 2). A principal component analysis (PCA) (Fig. 3) showed that the axes 1 and 3, both corresponding to the “tissue effect,” accounted for 39.46% and 8.97% of the data variance; the axis 2, corresponding to the “DC subtype effect,” accounted for 17.85% of the data variance; and finally the axis 5, corresponding to the “infection effect,” accounted for only 6.99% of the data variance. Thus, both the hierarchical clustering and PCA suggest that the pDC/cDC types and the lymphoid tissue of origin have more impact on the global gene expression than the DC response to infection.
Fig 2.
Hierarchical clustering of the microarray data from node, spleen, and blood cDCs and pDCs in control and BTV-infected sheep (D6). Hierarchical clustering (Pearson-average method) was done on the normalized gene chips data. The data from three control sheep (C1, C2, and C3) and from 3 infected sheep at day 6 (I1, I2, and I3) are reported. The samples grouped first according to their spleen (Spl), lymph node (LN), and blood (Bl) origin and to the type of DCs (pDCs or cDCs) and secondly according to the BTV-infected or control status.
Fig 3.
PCA on the microarray data from node, spleen, and blood cDCs and pDCs in control and BTV-infected sheep (D6). A PCA of the normalized gene chips data from the spleen, lymph node, and blood pDCs (triangles) and cDCs (circles) of control (empty symbols) and infected sheep (filled symbols) generated 6 axes. Axis 1 separated samples according to lymphoid organs, axis 2 according to DC types, axis 3 according to organs, and axis 5 according to BTV infection status.
In order to analyze the effects of BTV infection on gene expression in DCs, the lists of the differentially expressed genes (DEG) modulated upon BTV infection were established for the blood pDCs (238 genes; see Table S1 in the supplemental material), the lymph node pDCs (654 genes; see Table S2 in the supplemental material), the lymph node cDCs (489 genes; see Table S3 in the supplemental material), and the spleen cDCs (161 genes; see Table S4 in the supplemental material), as described in Materials and Methods. The modulation of gene expression determined in microarray analyses was confirmed using qPCR for several selected genes in all DC subsets of different lymphoid compartments (see Table S5 in the supplemental material), supporting the reliability of the microarray data. BTV viral RNA was detected in cDCs and pDCs from infected lymph nodes, suggesting that both cell types were infected in vivo by BTV (see Fig. S2 in the supplemental material). A comparison of these DEG lists was made in a Venn diagram (Fig. 4) to identify genes modulated in common for the different DC subsets. Only 3 downmodulated and 6 upregulated genes were common to all the DEG lists. The majority of the upregulated (>63%) and repressed (>58%) genes were exclusive to each DC subset of a given lymphoid compartment. Thus, our global gene expression data analysis indicates that the cDC/pDC type and the lymphoid organ location have an impact on the gene expression program induced by BTV infection.
Fig 4.
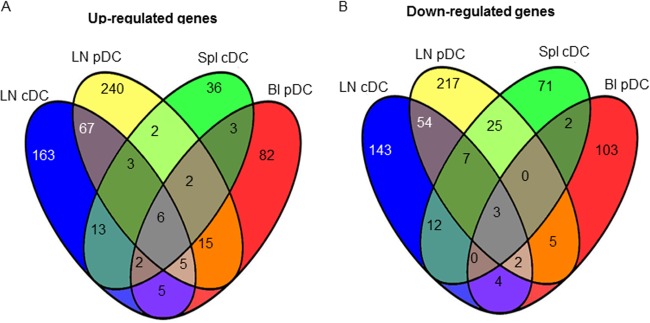
Venn diagram comparison of the genes upregulated (A) and downmodulated (B) by BTV in lymph node pDCs, in lymph node cDCs, in blood pDCs, and in spleen cDCs. Each of the four circles represents the list of the DG in the different DC subsets from the mentioned lymphoid organ (Spl, spleen; Bl, blood; LN, lymph node). Numbers in the intersections between circles represent the numbers of common genes that are modulated in two, three, or four subset types.
The functional gene expression profiles of DCs modulated during BTV infection depend on the lymphoid compartments and on the cDC/pDC types and suggest their distinct roles in BT physiopathology.
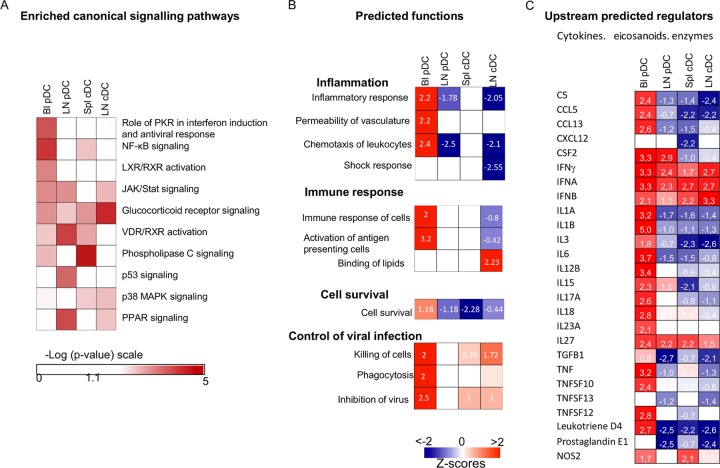
In order to get insight into the possible functions of DCs in BT physiopathology, the DEG lists were submitted to the integrative systems level analysis of the IPA software. The predominant signaling pathways triggered in the DC subsets from different compartments upon BTV infection were clearly distinct: the dominant signaling pathways were the phospholipase C signaling in spleen cDCs, the glucocorticoid receptor signaling in lymph node cDCs, the NF-κB and protein kinase R (PKR) signaling in blood pDCs, and the vitamin D receptor/retinoid X receptor (VDR/RXR) and peroxisome proliferator-activated receptor (PPAR) signaling in lymph node pDCs (Fig. 5A; see Table S6 in the supplemental material).
Fig 5.
Enrichment for canonical pathways and prediction of functions and of upstream regulators in blood and lymph node pDCs and in lymph node and spleen cDCs upon BTV infection. The DEG lists in blood and node pDCs and in node and spleen cDCs were processed by the IPA software and the DEA tool. (A) Heat map of the enriched canonical signaling pathways [−Log (P values) in a red scale]. (B and C) Heat map of the generated statistically significant prediction values for activated and inhibited functions (B) and upstream regulators (C), with z-scores of >2 for activation and <−2 for inhibition. z-scores are reported in each square.
The DEA feature of IPA (see Materials and Methods) was used to identify the biological functions that were expected to be increased or decreased given the observed gene expression changes upon BTV infection. Strikingly, many functions related to inflammation and immunity were predicted to be activated in blood pDCs upon BTV infection (Fig. 5B; see Table S7 in the supplemental material), and conversely these functions were not activated, or even inhibited, in the lymph node pDCs and cDCs. Indeed, in blood pDCs, chemotaxis (z-score = 2.4), the inflammatory response (z-score = 2.2), and the permeability of vasculature (z-score = 2.2) were all predicted activated functions, in agreement with the activation of the NF-κB pathways (Fig. 5A). The increased expression of inflammatory chemokine and cytokine genes (CCL2, CCL4, CXCL10, IL-8, IFNG, TNF; see Table S7 in the supplemental material) and of other genes involved in increased permeability of vasculature such as PTX3 (pentraxin 3) and THSB1 (thrombospondin-1) suggests that blood pDCs could be involved in the plasma leakages and the general inflammatory state encountered in BTV infection. The upregulation of the CFB (complement factor B) and F13A1 (coagulation factor XIII) genes (see Table S1 in the supplemental material) suggests that pDCs may also participate in blood coagulation disorders. The observed upregulation of the gene expression of homing receptor CCR7 (see Table S1 in the supplemental material) can be related to the accumulation of pDCs in lymph nodes during BTV infection (35). Finally, blood pDCs showed activation of functions related to the control of viral infection with inhibition of virus (z-score = 2.5) and killing of cells (z-score = 2) that include upregulation of the GZMB (granzyme B) gene (see Table S7 in the supplemental material). Conversely, pDCs from regional node did not display at all the proinflammatory gene expression profile expressed by blood pDCs. In agreement with the dominant involvement of the anti-inflammatory PPAR pathway (36) (Fig. 5A), node pDCs rather expressed a trend to inhibition of inflammation (z-score = −1.778) and a significant prediction for reduction of chemotaxis (z-score = −2.597) with downregulated expression of CCL4 and upregulated expression of SOCS3 (suppressor of cytokine signaling 3). The reduction of CCR7, CXCR4, and CXCR7 expression in node pDCs is in accordance with the accumulation of pDCs in the regional lymph node during BTV infection (see Table S2 in the supplemental material). While expressing distinct genes upon BTV infection (<30% in common with pDCs [Fig. 4]), cDCs from lymph nodes also displayed gene expression profiles predictive of inhibition of inflammation and shock response (z-score = −2 and −2.5, respectively [Fig. 5B]) as well as of cell death (z-score = 2.17) and lipid binding (z-score = 2.4). The suppression of inflammation unraveled by the DEA can be related to the involvement of the glucocorticoid signaling pathway in node cDCs (Fig. 5A), which has been associated in several instances to inhibition of T cell activation by DCs (37). In the same trend toward immune regulation or suppression, the expression of the IDO (tryptophan-degrading enzyme indoleamine 2,3 dioxygenase) gene was strongly increased (3.8-fold; see Table S3 in the supplemental material), and the ADA (adenosine deaminase expression) gene was also strongly suppressed in node cDCs (0.26-fold; see Table S3 in the supplemental material). The activation of IDO has been shown to be strongly involved in the induction of regulatory T cells by DCs (38), and the reduction of ADA was described to lead to inhibition of T cell activation and to immunodeficiency (39, 40). The reduction of the S1PR3 (sphingosine-1-phosphate receptor 3) gene expression (0.43-fold) can be related to the accumulation of cDCs within lymph nodes, as reduced expression of this receptor has been previously demonstrated to play an important role in cDC sequestration in lymph nodes (41).
An analysis of the upstream regulators provided by the IPA software (Fig. 5C) indicates that many cytokines, eicosanoids, and enzymes are predicted to be activators responsible for the gene expression modulated by BTV in blood pDCs, whereas in many instances their signaling is predicted to be inhibited in lymph node pDCs, spleen cDCs, and lymph node cDCs. Interestingly, however, IFN is considered by IPA to be a positive regulator in all the DCs irrespective of the DC types or of the lymphoid compartment (Fig. 5C), although the IFN-induced genes were different between the DC subsets (see Table S8 in the supplemental material).
Overall, our gene expression meta-analysis shows that the pDC/cDC type and the lymphoid organ location have a strong impact on the dominant signaling pathways and on the functional gene expression profiles expressed by DCs upon BTV infection. Notably, the transcriptomic profile of blood pDCs at the onset of symptoms was associated with promotion of inflammation and plasma leakages, whereas pDCs in the regional lymph node displayed an anti-inflammatory profile. In addition, cDCs in the regional lymph node exhibited an anti-inflammatory and immunoregulatory gene expression profile. Thus, the transcriptomic analysis suggests a role for DCs in BT pathogenesis that depends on the DC subtypes and the organs in which these DCs reside at the onset of symptoms.
DISCUSSION
We demonstrate that cDC and pDC subsets from different lymphoid compartments selectively activated or repressed different sets of genes (from 36 to 240) upon BTV infections and possibly contributed to viral pathogenesis. Our in vivo investigation in a pertinent natural host allowed extraction of DC subsets from different lymphoid organs in sufficient quantities for microarray studies, which would not have been technically possible using nonhuman primate models. Even in our large-animal model, extraction of enough DCs for microarray studies was not possible in all instances, such as in the case of blood cDCs, due the paucity of DC representation in our preserved samples.
As reported for several human viral infections such as human immunodeficiency virus (HIV), hepatitis B and C viruses (HBV and HCV), and dengue virus (42–45), the pDC and cDC numbers appeared to decrease in blood during BTV infection. Given the low numbers of sheep that we could technically handle during this experimentation under BSL3 conditions, statistical significance could not always be achieved; however, we were able to show tendencies for decrease with cellular data that we obtained from 3 sheep per group. A strong decrease in pDC and cDC numbers in blood has been described in HIV patients, and the restoration of their numbers after primary HIV infection correlated with higher CD4+ T cell numbers and lower viral load (45). Similarly, in dengue virus infection, low numbers of circulating cDCs and pDCs in humans and monkeys correlated with disease severity (42, 46). In mice, a strong reduction of circulating pDCs was encountered in herpes simplex virus, vesicular stomatitis virus, and murine cytomegalovirus (MCMV) infections and was related to pDC apoptosis induced by type I IFN (47), thus presumably avoiding excessive systemic responses. In the case of BTV infection, blood pDCs did not show gene signatures related to apoptosis but rather a trend to increased cell survival (Fig. 5B; see Table S7 in the supplemental material). In BTV infection, the accumulation of pDCs and cDCs in lymph nodes may explain their reduction in blood. It should be noted that their numbers were back to normal in blood at day 10. As suggested for HIV (45), the recovery of a steady-state DC representation in blood might be important in the resolution of BT disease.
Recently, pDCs were shown to play a pivotal role in the systemic inflammation induced by toll-like receptor 9 (TLR9) triggering or by bacterial infections in vivo in mice (48). We show here that a strong inflammatory gene profile was induced by BTV in blood pDCs in vivo at day 6 postinfection, at the onset of symptoms, with expression of genes encoding cytokines such as CCL2, CCL4, TNF, IL-8, and CXCL10 (see Table S7 in the supplemental material). Notably, the in vitro stimulation of sheep pDCs with replicative and UV-irradiated BTV8 triggered the upregulation of TNF transcripts, but not that of IL-8 or CCL4 (S.R. and I.S.-C., data not shown), indicating that in vitro stimulation by BTV does not faithfully mimic the viral stimulation of pDCs in vivo. Finally, the permeability of vasculature was predicted to be an increased function of pDCs during BTV infection, indicating that this cell type could be involved in the hemodynamic disorders encountered in BTV infection. This finding is of importance, because it allows us to hypothesize that blood pDCs are important cells involved in the general symptoms of fever, inflammation, and bleeding disorders in BT and possibly in other viral hemorrhagic diseases. It should be pointed out that our experimental infection in the Prealpes sheep breed led to a moderate severity of disease expression, with facial edema and limited hemorrhagic lesions in the oral cavity. A similar experiment using more sensitive breeds such as Dorset Poll sheep would possibly unravel more-extensive gene expression alterations in pDCs. Unfortunately, we could not extract sufficient numbers of cDCs from blood. It can thus not be known whether the proinflammatory profile found for blood pDCs extends to the cDC type in blood.
We found that cDCs from spleen and lymph nodes and pDCs from blood and lymph nodes expressed different functional gene expression profiles, suggesting that the lymphoid tissue microenvironment shapes the DC transcriptomic program upon viral infection. Under basal conditions, resident cDC (CD4+ and CD8+ types), but not pDCs, exhibit different genomic programs across lymphoid organs (49). However, no studies to our knowledge previously evaluated the influence of the local milieu on the global DC response during viral infection, although some suggested that lymphoid organ location affects pDC response: during systemic MCMV infection, pDCs produced type I IFN in a lymphoid organ-specific manner (50); during lymphocytic choriomeningitis virus (LCMV) infection, pDCs were activated for type I IFN production in the pancreas (51), whereas they adopted a tolerogenic profile in the pancreatic draining lymph node, under the control of local NKT cells (52). However, it remains possible that the difference of gene expression profile in lymph node and blood pDCs is independent of the lymphoid organ environment but reflects a difference of time lapse after stimulation by BTV: in that scenario, pDCs stimulated in blood by BTV would undergo a proinflammatory transcriptomic program that would be followed by an anti-inflammatory transcriptomic program concomitant with entry into lymph nodes. The use of CCR7-deficient pDCs or of anti-CCR7 MAb treatments to impede homing in lymph nodes may allow determining whether the change of transcriptomic profile in BTV-exposed pDCs is cell intrinsic or modified by the lymphoid environment; however, such investigations are not feasible in sheep for the moment due to the lack of appropriate tools. The lymph node environment may also partly explain the difference of responses to BTV between isolated lymph cDCs and cDCs in the node: indeed, we previously reported that BTV induced upregulation of IL-12p40, IL-1β, and IL-6 transcripts on isolated lymph cDCs in vitro (8), whereas these transcripts were not upregulated in vivo in lymph nodes in the present study. Finally, regardless of the DC types and lymphoid tissue location, the only common predicted activator among all the analyzed DCs appeared to be IFN, which was previously shown by us to be detected in blood from day 2 to day 6 in the same sheep study (published in reference 17). This finding suggests that the IFN response to BTV infection is a major convergent response in DCs.
The transcriptomic analysis was conducted at day 6 postinfection, a time that corresponds to the peak of viremia and the beginning of symptoms. However, it is possible that the major impact of BTV infection on the gene expression reprogramming in DCs would occur at an earlier time, as observed in the case of MCMV infection in mice, where the maximum of DC activation occurs at day 2 postinfection (53). Thus, the relatively low impact of BTV infection on DC gene expression reprogramming observed in our study may have been higher, or different, at an earlier time postinfection.
The majority of genes activated and repressed in pDCs and cDCs from lymph nodes during BTV infection were distinct and corresponded to the enrichments of different canonical pathways (Fig. 5A). A recent study of the gene expression program modulated by MCMV infection also reported that hundreds of genes were differentially modulated in splenic pDCs and cDCs and specific functional gene sets were induced by the infection in pDCs, while they were unchanged or even repressed in cDC. This included genes involved in mitosis/cell cycle and coexpressed with Ccnb2 or induced by IL-15 (53). Together with these results, our data indicate that viral infection modulates the gene expression program in DCs depending on the DC type. The reason for the distinct responses between the two DC types may be related to differences in the basal genetic programs, in viral receptors, and in dominant signaling pathways and could also be related to differences in viral infections. However, we found viral RNA both in cDCs and in pDCs from lymph nodes at day 6, suggesting that both cell types are infected by the virus, in agreement with our previous in vivo and in vitro published results (see Fig. S2 in the supplemental material and references 8 and 17). As our present study relies on global RNA expression in the DCs, we cannot evaluate whether the gene expression program in DCs is directly related to viral infection of the same cells or is induced by sensing without concomitant DC infection. Despite the modulation of different sets of genes by BTV, both cDCs and pDCs from lymph nodes displayed anti-inflammatory and antichemotactic signatures that may contribute to avoiding the general cytokine storm and possibly to controlling an excess of immune responses that could lead to immunopathology. However, the extent and the duration of the expression of regulatory or suppressive genes may interfere with the establishment of protective immunity and suggest that immunosuppression may be generated in some individuals (54).
From these data, a BT physiopathology scenario involving DCs can be speculated upon: together with the expression of their potent viral defense, circulating pDCs participate in the triggering of strong systemic inflammation, increased vasculature permeability, and possibly bleeding disorders. Once in lymph nodes at day 6, pDCs and cDCs express an anti-inflammatory profile, avoiding excessive inflammation and immunopathology. It can be inferred from this scenario that defects or excesses in the response of pDCs and/or cDCs to BTV could impact the host response to this virus, thus explaining the interindividual clinical sensitivity to BTV8 infection, which generally leads to a <30% case fatality rate under natural conditions (55). It suggests the hypothesis that genetic resistance of ruminants to the disease might implicate DCs, although the responses of other hematopoietic cells and endothelial cells may also be involved. Extension of our findings to other hemorrhagic viruses could lead the way to the developments of treatments that modulate the responses of cDCs and pDCs, especially at the acute phase of the viral hemorrhagic disease manifestation.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Plateforme d'Infectiologie Expérimentale (PFIE) INRA (Nouzilly, France) for BTV8 infections and serum collections, especially Olivier Boulesteix and David Gauthier for the animal experiment, Céline Barc for her assistance with the work in the PFIE BSL3 facility, and Bertrand Schwartz for his support. We are grateful to Bernard Charley, Sabine Riffault, Aude Rémot, and Frédérick Arnaud for critical review of the manuscript.
This work was supported by the Agence Nationale pour la Recherche (VacGenDC ANR 06 GANI 015-03, PhylogenDC ANR-09-BLAN-0073) and by the EMIDA Era-Net (OrbiNet 2009).
Footnotes
Published ahead of print 19 June 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.00631-13.
REFERENCES
- 1. Schwartz-Cornil I, Mertens PP, Contreras V, Hemati B, Pascale F, Breard E, Mellor PS, Maclachlan NJ, Zientara S. 2008. Bluetongue virus: virology, pathogenesis and immunity. Vet. Res. 39:46. 10.1051/vetres:2008023 [DOI] [PubMed] [Google Scholar]
- 2. Wilson AJ, Mellor PS. 2009. Bluetongue in Europe: past, present and future. Philos. Trans. R. Soc. Lond. B Biol. Sci. 364:2669–2681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Drew CP, Gardner IA, Mayo CE, Matsuo E, Roy P, MacLachlan NJ. 2010. Bluetongue virus infection alters the impedance of monolayers of bovine endothelial cells as a result of cell death. Vet. Immunol. Immunopathol. 136:108–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Elbers AR, Backx A, Ekker HM, van der Spek AN, van Rijn PA. 2008. Performance of clinical signs to detect bluetongue virus serotype 8 outbreaks in cattle and sheep during the 2006-epidemic in The Netherlands. Vet. Microbiol. 129:156–162 [DOI] [PubMed] [Google Scholar]
- 5. Moulin V, Noordegraaf CV, Makoschey B, van der Sluijs M, Veronesi E, Darpel K, Mertens PP, de Smit H. 2012. Clinical disease in sheep caused by bluetongue virus serotype 8, and prevention by an inactivated vaccine. Vaccine 30:2228–2235 [DOI] [PubMed] [Google Scholar]
- 6. Veronesi E, Darpel KE, Hamblin C, Carpenter S, Takamatsu HH, Anthony SJ, Elliott H, Mertens PP, Mellor PS. 2010. Viraemia and clinical disease in Dorset Poll sheep following vaccination with live attenuated bluetongue virus vaccines serotypes 16 and 4. Vaccine 28:1397–1403 [DOI] [PubMed] [Google Scholar]
- 7. Barratt-Boyes SM, MacLachlan NJ. 1995. Pathogenesis of bluetongue virus infection of cattle. J. Am. Vet. Med. Assoc. 206:1322–1329 [PubMed] [Google Scholar]
- 8. Hemati B, Contreras V, Urien C, Bonneau M, Takamatsu HH, Mertens PP, Breard E, Sailleau C, Zientara S, Schwartz-Cornil I. 2009. Bluetongue virus targets conventional dendritic cells in skin lymph. J. Virol. 83:8789–8799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brodie SJ, Wilson WC, O'Hearn PM, Muthui D, Diem K, Pearson LD. 1998. The effects of pharmacological and lentivirus-induced immune suppression on orbivirus pathogenesis: assessment of virus burden in blood monocytes and tissues by reverse transcription-in situ PCR. J. Virol. 72:5599–5609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. DeMaula CD, Jutila MA, Wilson DW, MacLachlan NJ. 2001. Infection kinetics, prostacyclin release and cytokine-mediated modulation of the mechanism of cell death during bluetongue virus infection of cultured ovine and bovine pulmonary artery and lung microvascular endothelial cells. J. Gen. Virol. 82:787–794 [DOI] [PubMed] [Google Scholar]
- 11. Takamatsu H, Mellor PS, Mertens PP, Kirkham PA, Burroughs JN, Parkhouse RM. 2003. A possible overwintering mechanism for bluetongue virus in the absence of the insect vector. J. Gen. Virol. 84:227–235 [DOI] [PubMed] [Google Scholar]
- 12. Gowen BB, Holbrook MR. 2008. Animal models of highly pathogenic RNA viral infections: hemorrhagic fever viruses. Antiviral Res. 78:79–90 [DOI] [PubMed] [Google Scholar]
- 13. Geisbert TW, Hensley LE, Larsen T, Young HA, Reed DS, Geisbert JB, Scott DP, Kagan E, Jahrling PB, Davis KJ. 2003. Pathogenesis of Ebola hemorrhagic fever in cynomolgus macaques: evidence that dendritic cells are early and sustained targets of infection. Am. J. Pathol. 163:2347–2370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hensley LE, Alves DA, Geisbert JB, Fritz EA, Reed C, Larsen T, Geisbert TW. 2011. Pathogenesis of Marburg hemorrhagic fever in cynomolgus macaques. J. Infect. Dis. 204(Suppl 3):S1021–S1031 [DOI] [PubMed] [Google Scholar]
- 15. Hensley LE, Smith MA, Geisbert JB, Fritz EA, Daddario-DiCaprio KM, Larsen T, Geisbert TW. 2011. Pathogenesis of Lassa fever in cynomolgus macaques. Virol. J. 8:205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kyle JL, Beatty PR, Harris E. 2007. Dengue virus infects macrophages and dendritic cells in a mouse model of infection. J. Infect. Dis. 195:1808–1817 [DOI] [PubMed] [Google Scholar]
- 17. Ruscanu S, Pascale F, Bourge M, Hemati B, Elhmouzi-Younes J, Urien C, Bonneau M, Takamatsu H, Hope J, Mertens P, Meyer G, Stewart M, Roy P, Meurs EF, Dabo S, Zientara S, Breard E, Sailleau C, Chauveau E, Vitour D, Charley B, Schwartz-Cornil I. 2012. The double-stranded RNA bluetongue virus induces type I interferon in plasmacytoid dendritic cells via a MYD88-dependent TLR7/8-independent signaling pathway. J. Virol. 86:5817–5828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Barratt-Boyes SM, MacLachlan NJ. 1994. Dynamics of viral spread in bluetongue virus infected calves. Vet. Microbiol. 40:361–371 [DOI] [PubMed] [Google Scholar]
- 19. MacLachlan NJ. 1994. The pathogenesis and immunology of bluetongue virus infection of ruminants. Comp. Immunol. Microbiol. Infect. Dis. 17:197–206 [DOI] [PubMed] [Google Scholar]
- 20. Top S, Foucras G, Deplanche M, Rives G, Calvalido J, Comtet L, Bertagnoli S, Meyer G. 2012. Myxomavirus as a vector for the immunisation of sheep: protection study against challenge with bluetongue virus. Vaccine 30:1609–1616 [DOI] [PubMed] [Google Scholar]
- 21. Chevallier N, Berthelemy M, Le Rhun D, Laine V, Levy D, Schwartz-Cornil I. 1998. Bovine leukemia virus-induced lymphocytosis and increased cell survival mainly involve the CD11b+ B-lymphocyte subset in sheep. J. Virol. 72:4413–4420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schwartz-Cornil I, Epardaud M, Bonneau M. 2006. Cervical duct cannulation in sheep for collection of afferent lymph dendritic cells from head tissues. Nat. Protoc. 1:874. [DOI] [PubMed] [Google Scholar]
- 23. Casel P, Moreews F, Lagarrigue S, Klopp C. 2009. sigReannot: an oligo-set re-annotation pipeline based on similarities with the Ensembl transcripts and Unigene clusters. BMC Proc. 3(Suppl 4):S3. 10.1186/1753-6561-3-S4-S3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Robbins SH, Walzer T, Dembele D, Thibault C, Defays A, Bessou G, Xu H, Vivier E, Sellars M, Pierre P, Sharp FR, Chan S, Kastner P, Dalod M. 2008. Novel insights into the relationships between dendritic cell subsets in human and mouse revealed by genome-wide expression profiling. Genome Biol. 9:R17. 10.1186/gb-2008-9-1-r17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Smyth G. 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3:Article 3. http://www.statsci.org/smyth/pubs/ebayes.pdf [DOI] [PubMed] [Google Scholar]
- 26. Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Statist. Soc. B 57:289–300 [Google Scholar]
- 27. Matsuo E, Celma CC, Boyce M, Viarouge C, Sailleau C, Dubois E, Breard E, Thiery R, Zientara S, Roy P. 2011. Generation of replication-defective virus-based vaccines that confer full protection in sheep against virulent BTV challenge. J. Virol. 85:10213–10221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Perrin A, Albina E, Breard E, Sailleau C, Prome S, Grillet C, Kwiatek O, Russo P, Thiery R, Zientara S, Cetre-Sossah C. 2007. Recombinant capripoxviruses expressing proteins of bluetongue virus: evaluation of immune responses and protection in small ruminants. Vaccine 25:6774–6783 [DOI] [PubMed] [Google Scholar]
- 29. Contreras V, Urien C, Guiton R, Alexandre Y, Vu Manh TP, Andrieu T, Crozat K, Jouneau L, Bertho N, Epardaud M, Hope J, Savina A, Amigorena S, Bonneau M, Dalod M, Schwartz-Cornil I. 2010. Existence of CD8{alpha}-like dendritic cells with a conserved functional specialization and a common molecular signature in distant mammalian species. J. Immunol. 185:3313–3325 [DOI] [PubMed] [Google Scholar]
- 30. Pascale F, Contreras V, Bonneau M, Courbet A, Chilmonczyk S, Bevilacqua C, Eparaud M, Niborski V, Riffault S, Balazuc AM, Foulon E, Guzylack-Piriou L, Riteau B, Hope J, Bertho N, Charley B, Schwartz-Cornil I. 2008. Plasmacytoid dendritic cells migrate in afferent skin lymph. J. Immunol. 180:5963–5972 [DOI] [PubMed] [Google Scholar]
- 31. Karsunky H, Merad M, Cozzio A, Weissman IL, Manz MG. 2003. Flt3 ligand regulates dendritic cell development from Flt3+ lymphoid and myeloid-committed progenitors to Flt3+ dendritic cells in vivo. J. Exp. Med. 198:305–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Crozat K, Guiton R, Contreras V, Feuillet V, Dutertre CA, Ventre E, Vu Manh TP, Baranek T, Storset AK, Marvel J, Boudinot P, Hosmalin A, Schwartz-Cornil I, Dalod M. 2010. The XC chemokine receptor 1 is a conserved selective marker of mammalian cells homologous to mouse CD8{alpha}+ dendritic cells. J. Exp. Med. 207:1283–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cisse B, Caton ML, Lehner M, Maeda T, Scheu S, Locksley R, Holmberg D, Zweier C, den Hollander NS, Kant SG, Holter W, Rauch A, Zhuang Y, Reizis B. 2008. Transcription factor E2-2 is an essential and specific regulator of plasmacytoid dendritic cell development. Cell 135:37–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. MacLachlan NJ, Nunamaker RA, Katz JB, Sawyer MM, Akita GY, Osburn BI, Tabachnick WJ. 1994. Detection of bluetongue virus in the blood of inoculated calves: comparison of virus isolation, PCR assay, and in vitro feeding of Culicoides variipennis. Arch. Virol. 136:1–8 [DOI] [PubMed] [Google Scholar]
- 35. Umemoto E, Otani K, Ikeno T, Verjan Garcia N, Hayasaka H, Bai Z, Jang MH, Tanaka T, Nagasawa T, Ueda K, Miyasaka M. 2012. Constitutive plasmacytoid dendritic cell migration to the splenic white pulp is cooperatively regulated by CCR7- and CXCR4-mediated signaling. J. Immunol. 189:191–199 [DOI] [PubMed] [Google Scholar]
- 36. Wahli W, Michalik L. 2012. PPARs at the crossroads of lipid signaling and inflammation. Trends Endocrinol. Metab. 23:351–363 [DOI] [PubMed] [Google Scholar]
- 37. Szatmari I, Nagy L. 2008. Nuclear receptor signalling in dendritic cells connects lipids, the genome and immune function. EMBO J. 27:2353–2362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mellor AL, Baban B, Chandler P, Marshall B, Jhaver K, Hansen A, Koni PA, Iwashima M, Munn DH. 2003. Cutting edge: induced indoleamine 2,3 dioxygenase expression in dendritic cell subsets suppresses T cell clonal expansion. J. Immunol. 171:1652–1655 [DOI] [PubMed] [Google Scholar]
- 39. Pacheco R, Martinez-Navio JM, Lejeune M, Climent N, Oliva H, Gatell JM, Gallart T, Mallol J, Lluis C, Franco R. 2005. CD26, adenosine deaminase, and adenosine receptors mediate costimulatory signals in the immunological synapse. Proc. Natl. Acad. Sci. U. S. A. 102:9583–9588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sauer AV, Aiuti A. 2009. New insights into the pathogenesis of adenosine deaminase-severe combined immunodeficiency and progress in gene therapy. Curr. Opin. Allergy Clin. Immunol. 9:496–502 [DOI] [PubMed] [Google Scholar]
- 41. Niessen F, Schaffner F, Furlan-Freguia C, Pawlinski R, Bhattacharjee G, Chun J, Derian CK, Andrade-Gordon P, Rosen H, Ruf W. 2008. Dendritic cell PAR1-S1P3 signalling couples coagulation and inflammation. Nature 452:654–658 [DOI] [PubMed] [Google Scholar]
- 42. De Carvalho Bittencourt M, Martial J, Cabie A, Thomas L, Cesaire R. 2012. Decreased peripheral dendritic cell numbers in dengue virus infection. J. Clin. Immunol. 32:161–172 [DOI] [PubMed] [Google Scholar]
- 43. Duan XZ, Wang M, Li HW, Zhuang H, Xu D, Wang FS. 2004. Decreased frequency and function of circulating plasmocytoid dendritic cells (pDC) in hepatitis B virus infected humans. J. Clin. Immunol. 24:637–646 [DOI] [PubMed] [Google Scholar]
- 44. Kanto T, Inoue M, Miyatake H, Sato A, Sakakibara M, Yakushijin T, Oki C, Itose I, Hiramatsu N, Takehara T, Kasahara A, Hayashi N. 2004. Reduced numbers and impaired ability of myeloid and plasmacytoid dendritic cells to polarize T helper cells in chronic hepatitis C virus infection. J. Infect. Dis. 190:1919–1926 [DOI] [PubMed] [Google Scholar]
- 45. Pacanowski J, Kahi S, Baillet M, Lebon P, Deveau C, Goujard C, Meyer L, Oksenhendler E, Sinet M, Hosmalin A. 2001. Reduced blood CD123+ (lymphoid) and CD11c+ (myeloid) dendritic cell numbers in primary HIV-1 infection. Blood 98:3016–3021 [DOI] [PubMed] [Google Scholar]
- 46. Pichyangkul S, Endy TP, Kalayanarooj S, Nisalak A, Yongvanitchit K, Green S, Rothman AL, Ennis FA, Libraty DH. 2003. A blunted blood plasmacytoid dendritic cell response to an acute systemic viral infection is associated with increased disease severity. J. Immunol. 171:5571–5578 [DOI] [PubMed] [Google Scholar]
- 47. Swiecki M, Wang Y, Vermi W, Gilfillan S, Schreiber RD, Colonna M. 2011. Type I interferon negatively controls plasmacytoid dendritic cell numbers in vivo. J. Exp. Med. 208:2367–2374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Guillerey C, Mouries J, Polo G, Doyen N, Law HK, Chan S, Kastner P, Leclerc C, Dadaglio G. 2012. Pivotal role of plasmacytoid dendritic cells in inflammation and NK-cell responses after TLR9 triggering in mice. Blood 120:90–99 [DOI] [PubMed] [Google Scholar]
- 49. Elpek KG, Bellemare-Pelletier A, Malhotra D, Reynoso ED, Lukacs-Kornek V, DeKruyff RH, Turley SJ. 2011. Lymphoid organ-resident dendritic cells exhibit unique transcriptional fingerprints based on subset and site. PLoS One 6:e23921. 10.1371/journal.pone.0023921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zucchini N, Bessou G, Robbins SH, Chasson L, Raper A, Crocker PR, Dalod M. 2008. Individual plasmacytoid dendritic cells are major contributors to the production of multiple innate cytokines in an organ-specific manner during viral infection. Int. Immunol. 20:45–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Diana J, Griseri T, Lagaye S, Beaudoin L, Autrusseau E, Gautron AS, Tomkiewicz C, Herbelin A, Barouki R, von Herrath M, Dalod M, Lehuen A. 2009. NKT cell-plasmacytoid dendritic cell cooperation via OX40 controls viral infection in a tissue-specific manner. Immunity 30:289–299 [DOI] [PubMed] [Google Scholar]
- 52. Diana J, Brezar V, Beaudoin L, Dalod M, Mellor A, Tafuri A, von Herrath M, Boitard C, Mallone R, Lehuen A. 2011. Viral infection prevents diabetes by inducing regulatory T cells through NKT cell-plasmacytoid dendritic cell interplay. J. Exp. Med. 208:729–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Baranek T, Manh TP, Alexandre Y, Maqbool MA, Cabeza JZ, Tomasello E, Crozat K, Bessou G, Zucchini N, Robbins SH, Vivier E, Kalinke U, Ferrier P, Dalod M. 2012. Differential responses of immune cells to type I interferon contribute to host resistance to viral infection. Cell Host Microbe 12:571–584 [DOI] [PubMed] [Google Scholar]
- 54. Umeshappa CS, Singh KP, Nanjundappa RH, Pandey AB. 2010. Apoptosis and immuno-suppression in sheep infected with bluetongue virus serotype-23. Vet. Microbiol. 144:310–318 [DOI] [PubMed] [Google Scholar]
- 55. Conraths FJ, Gethmann JM, Staubach C, Mettenleiter TC, Beer M, Hoffmann B. 2009. Epidemiology of bluetongue virus serotype 8, Germany. Emerg. Infect. Dis. 15:433–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.