Abstract
Human T lymphotropic virus type 1 (HTLV-1) mainly causes adult T cell leukemia and predominantly immortalizes/transforms CD4+ T cells in culture. HTLV-2 is aleukemic and predominantly immortalizes/transforms CD8+ T cells in culture. We have shown previously that the viral envelope is the genetic determinant of the differential T cell tropism in culture. The surface component (SU) of the HTLV-1 envelope is responsible for binding to the cellular receptors for entry. Here, we dissect the HTLV-1 SU further to identify key domains that are involved in determining the immortalization tropism. We generated HTLV-1 envelope recombinant virus containing the HTLV-2 SU domain. HTLV-1/SU2 was capable of infecting and immortalizing freshly isolated peripheral blood mononuclear cells in culture. HTLV-1/SU2 shifted the CD4+ T cell immortalization tropism of wild-type HTLV-1 (wtHTLV-1) to a CD8+ T cell preference. Furthermore, a single amino acid substitution, N195D, in HTLV-1 SU (Ach.195) resulted in a shift to a CD8+ T cell immortalization tropism preference. Longitudinal phenotyping analyses of the in vitro transformation process revealed that CD4+ T cells emerged as the predominant population by week 5 in wtHTLV-1 cultures, while CD8+ T cells emerged as the predominant population by weeks 4 and 7 in wtHTLV-2 and Ach.195 cultures, respectively. Our results indicate that SU domain independently influences the preferential T cell immortalization tropism irrespective of the envelope counterpart transmembrane (TM) domain. We further showed that asparagine at position 195 in HTLV-1 SU is involved in determining this CD4+ T cell immortalization tropism. The slower emergence of the CD8+ T cell predominance in Ach.195-infected cultures suggests that other residues/domains contribute to this tropism preference.
INTRODUCTION
Human T lymphotropic virus type 1 (HTLV-1) and type 2 (HTLV-2) are complex retroviruses that share a genome structure (1). In addition to the structural proteins (Gag, Pol, Pro, and Env), they encode regulatory proteins (Tax and Rex) and accessory proteins, including an antisense protein, HBZ (HTLV-1) or APH-2 (HTLV-2) (2–5). Despite their closely related genomic structures, HTLV-1 and HTLV-2 display distinct pathogenic properties. HTLV-1 causes adult T cell leukemia (ATL), HTLV-1-associated myelopathy/tropical spastic paraparesis (HAM/TSP), and some noninflammatory disorders (6–9). HTLV-2 does not cause leukemia and has been associated with a HAM/TSP-like neurological disease only infrequently (10–12). Another feature that differentiates HTLV-1 and HTLV-2 is the ability to predominantly immortalize (interleukin-2 [IL-2]-dependent growth) or transform (IL-2-independent growth) CD4+ and CD8+ T cells, respectively, in culture (13–15). The in vitro immortalization/transformation preference for CD4+ T cells by HTLV-1 is recapitulated in vivo, as ATL is a CD4+ T cell leukemia. However, due to its nonleukemogenic nature, HTLV-2-induced CD8+ T cell immortalization/transformation is primarily an in vitro phenomenon.
We have previously shown that, although the viral Tax protein is indispensable for viral replication and cellular transformation, the preferential immortalization or transformation tropism of HTLV-1 and HTLV-2 is determined by the viral envelope (14, 15). Since the primary function of the viral envelope is to facilitate entry into new target cells, it was hypothesized that the cellular receptor complex requirements for HTLV-1 and HTLV-2 could be different. Subsequently, a number of studies reported that HTLV-1 and HTLV-2 slightly differ in their requirement of host cellular receptors. HTLV-1 requires heparan sulfate proteoglycans (HSPGs) and neuropilin-1 (NRP-1) for initial binding and glucose transporter-1 (GLUT-1) for subsequent membrane fusion and entry. Although HTLV-2 shares NRP-1 and GLUT-1 with HTLV-1 for both binding and entry, HSPGs interfere with HTLV-2 binding (16–19). Therefore, together these findings suggested a potential role for the viral envelope in mediating preferential T cell transformation, probably at the stage of virus binding to the host cell receptor.
The viral envelope is generated as a polyprecursor protein (gp61) comprised of 488 amino acids which is cleaved into the surface domain (SU-gp46) and transmembrane domain (TM-gp21) (20, 21). SU binds to the cellular receptor(s), and then SU and TM undergo significant conformational remodeling, thereby exposing TM to facilitate membrane fusion and subsequent entry into the cell. Functional mapping analysis of the HTLV-1 SU using soluble SU fusion proteins and in vitro binding assays revealed that the C terminus of the HTLV-1 SU (SU1) binds to the CD4+ T cells with a higher efficiency than the HTLV-2 SU (SU2) (18). SU is comprised of a receptor binding domain (RBD) at the N terminus, a proline-rich region (PRR) which carries an immunodominant epitope (SU1175–199 in HTLV-1 and SU2182–199 in HTLV-2), and a C terminus. A number of groups have studied the importance of the various amino acid residues of SU for their contribution to or effect on several biological properties of the virus. Delamarre et al. (22) showed that the SU domain tolerates only conservative amino acid substitutions in the positions conserved between HTLV-1, HTLV-2, and STLV-1. Previous studies from three different research groups have evaluated a N-to-D substitution at position 195 of the SU1 domain (the corresponding amino acid at position 191 in HTLV-2 SU is a D). The N195D displayed normal intracellular maturation and syncytium formation of the envelope (22); it resulted in active infection and immortalization of freshly isolated peripheral blood mononuclear cells (PBMCs) in vitro (23); and the virus efficiently infected and persisted in rabbits (24). However, rabbits infected with the N195D mutant virus exhibited a weaker humoral response to SU1 antigen than the rabbits infected with wild-type HTLV-1 (wtHTLV-1). Additionally, one rabbit infected with the N195D mutant virus generated a strong antibody response to the SU2 antigen. Taken together, these results suggest that the N195D mutation in SU1 could exhibit certain biological properties in vivo that are similar to those of the HTLV-2 envelope.
In this study, we further dissected the role of HTLV-1 SU in the distinct in vitro immortalization/transformation tropism characteristics. We first generated and characterized recombinant HTLV-1 provirus containing the SU region of wtHTLV-2 envelope (HTLV-1/SU2). HTLV-1/SU2 actively replicated in freshly isolated PBMCs, and additionally, HTLV-1/SU2 predominantly immortalized CD8+ T cells similarly to wtHTLV-2. This suggests that the viral envelope domain SU contributes to the preferential immortalization tropism. We further showed that the N195D substitution in the immunodominant epitope of HTLV-1 SU (Ach.195) also shifted the immortalization tropism toward a CD8+ T cell preference. This finding indicates that HTLV-1 mediates the preferential CD4+ T cell immortalization tropism through the envelope SU protein and that residue 195 plays a critical role in this preference.
MATERIALS AND METHODS
Cells.
729Achneo and 729pH6neo are stable wtHTLV-1 and wtHTLV-2 producer cell lines that were generated and characterized previously and designated 729.HTLV-1 and 729.HTLV-2 (14). The parental 729 B cell line was used as the negative control. All 729 parental and derivative cell lines were maintained in Iscove's Dulbecco's minimum essential medium (Cellgro; Mediatech, Manassas, VA). Ach.95 and Ach.195 (a generous gift from Lee Ratner) are human T cell lines that stably express HTLV-1 containing an asparagine-to-aspartic acid mutation at residues 95 and 195 of the Env coding sequence, respectively. Ach.95-, Ach.195-, wtHTLV-1-, and wtHTLV-2-producing T cell lines were maintained in RPMI 1640 (Gibco-Invitrogen, Grand Island, NY) and 10 U/ml recombinant human interleukin-2 (rhIL-2; Roche Applied Biosciences, Indianapolis, IN). All media were supplemented to contain 10% fetal bovine serum (Gemini Bio-Products, Sacramento, CA), 2 mM glutamine, penicillin (100 U/ml), and streptomycin (100 μg/ml; Gibco-Invitrogen, Grand Island, NY). Human peripheral blood mononuclear cells (PBMCs) were isolated from the blood of normal donors by centrifugation over Ficoll-Paque (GE Life Sciences, Piscataway, NY) and were cocultured with irradiated virus producer cell lines in RPMI 1640 medium supplemented with 20% fetal bovine serum (FBS), 2 mM glutamine, antibiotics, and 10 U/ml rhIL-2.
Plasmids.
The wild-type (wt) HTLV-1 proviral clone ACH (25) and wtHTLV-2 proviral clone pH6neo (26) were used to generate recombinant proviral clones for this study. To assist in the generation of the recombinant HTLV proviral clone, a restriction enzyme site was introduced using a QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). Specifically, a KpnI site was generated in the envelope region of the wtHTLV-2 proviral clone (6106CGTTCC6111 to GGTACC); a KpnI site is already present at the corresponding location in the HTLV-1 provirus that marks the end of the SU polypeptide (Fig. 1A). The HTLV-1/SU2 recombinant proviral clone was generated by replacing the HTLV-1 SU fragment with an HTLV-2 SU fragment (NcoI-KpnI). The recombined sequences in the HTLV-1/SU2 proviral clone were confirmed by DNA sequencing and diagnostic restriction enzyme digestions.
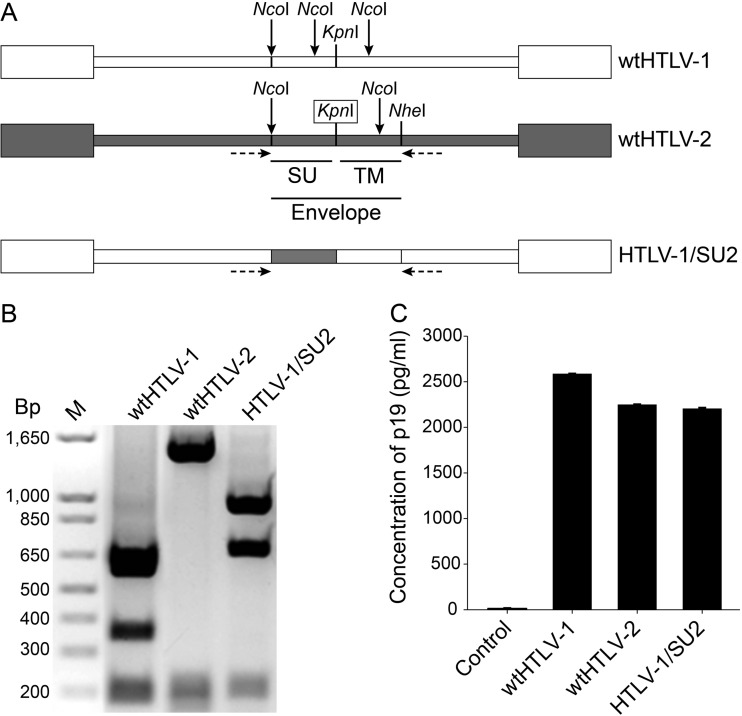
Fig 1.
Generation and characterization of the recombinant HTLV-1/SU2 provirus. (A) Genomic organization of the wild-type and recombinant proviral clones used in this study. White and gray boxes indicate wtHTLV-1 and wtHTLV-2 origin, respectively. The envelope region, along with the individual SU domain as well as the specific restriction enzymes used to construct the recombinants, is shown. The missing restriction enzyme site that was generated by site-directed mutagenesis is boxed. The multiple NcoI sites throughout the envelope region are shown by arrows. The horizontal arrows below the proviruses denote PCR primer positions. (B) Recombinant HTLV-1/SU2 provirus contained the exchanged envelope sequences. Approximately 1.8 kb of the viral genome encompassing the envelope region was amplified using a PCR oligonucleotide primer pair (shown in panel A) and the genomic DNA extracted from 729 cell lines producing wtHTLV-1, wtHTLV-2, or HTLV-1/SU2 viruses. The amplified products were digested with NcoI, and the fragments were resolved by 1% agarose gel electrophoresis and visualized with ethidium bromide. The expected fragments were observed as follows: for wtHTLV-1, 693 bp, 614 bp, 361 bp, and 205 bp; for wtHTLV-2, 1,491 bp, 205 bp, and 157 bp; and for HTLV-1/SU2, 963 bp, 693 bp, and 204 bp. (C) Viral protein production from the wild-type and recombinant virus producer cell lines. One million cells per virus producer cell line or the negative 729B cell line were cultured in 24-well plates for 72 h in triplicate. The culture supernatant from each cell line was collected and tested for p19 Gag output using a commercial ELISA kit. The bars depict the average values of the results from three independent experiments, and the error bars depict the standard deviations.
Stable transfection.
To generate stable transfectants, the proviral plasmid clone (HTLV-1/SU2) containing the Neor gene was introduced into 729 B cells using a nucleofection V kit (Amaxa; Lonza, AG, Cologne, Germany) according to the manufacturer's instructions. Stable transfectants containing the desired proviral clones were isolated following incubation in 24-well culture plates in medium containing 1 mg/ml of Geneticin. After 4 to 5 weeks of selection, viable cells were expanded and maintained in culture for further analysis. The clones were screened for p19 Gag expression in the cell supernatants by an enzyme-linked immunosorbent assay (ELISA) (Zeptometrix; Buffalo, NY) per the manufacturer's instructions.
PCR-based restriction enzyme digestion.
Genomic DNA was extracted from the stable virus producer cell lines using a DNeasy kit (Qiagen, Valencia, CA) per the manufacturer's instructions. The primers designed to amplify a 1.8-kb product containing the envelope region are Achneo4974(S) (49745′-CATTGGTATTATTTCAAGCTTC4995-3′) and Achneo6827(AS) (68275′-AGGAAAGAAAAAATGCAGGAGT6848-3′) for wtHTLV-1 and pH6neo4974(S) (49745′-CAAATGGTTCTATTATAAACTC4995-3′) and pH6neo6827(AS) (68275′-TTCAGGGTTATGTGGATTTC6846-3′) for wtHTLV-2. DNA was subjected to PCR performed with a 50-μl mixture containing a 160 nM concentration of each of the corresponding primer pairs. The cycling profile was a single cycle of 94°C for 10 min followed by 40 cycles of 94°C for 1 min, annealing at 50°C for Achneo primers or 47°C for pH6neo primers for 1 min and 72°C for 1 min, and a single-cycle final extension at 72°C for 5 min. The amplified product was digested with NcoI, and the fragments were resolved on a 1% agarose gel containing ethidium bromide.
Immortalization assay.
Approximately 1 million gamma-irradiated (10,000 rads) 729 (negative control), 729.HTLV-1, 729.HTLV-2, 729.HTLV-1/SU2, Ach.95, or Ach.195 cells (the number of virus producer cells was normalized based on p19 Gag output as measured by ELISA) were cocultured with 2 × 106 freshly isolated normal human PBMCs in 24-well plates for 8 weeks with weekly changes of media. The cultures were monitored by measuring viability using trypan blue exclusion and p19 Gag production using ELISA on a weekly basis. After 8 weeks, the cultures were harvested and stained using fluorescein isothiocyanate (FITC)-conjugated anti-human CD3 (clone UCHT1), allophycocyanin (APC)-conjugated anti-human CD4 (clone RPA-T4), and phycoerythrin (PE)-conjugated anti-human CD8 (clone HIT8a) antibodies (BD Biosciences, San Jose, CA) and analyzed by flow cytometry (14).
Internalization assay.
Studies to examine the internalization of HTLV virions were performed essentially as described previously (27) with the following modifications. Following incubation with virus-containing supernatants from HTLV-1-producing cells or uninfected controls for 2 h at 37°C, CD4+ T cells were fixed and permeabilized using commercially available reagents (BD Cytofix/Cytoperm fixation/permeabilization kit; BD Biosciences). The amount of internalized virions was determined using an antibody directed against the HTLV-1 p19 protein (TP-7; Zeptometrix) as previously described (27).
Statistical analysis.
The means of the normalized percentages of CD4+ T cells and CD8+ T cells from the wtHTLV-1- versus the HTLV-1/SU2-immortalized cultures, as well as wtHTLV-1- versus Ach.195-immortalized cultures, were compared for significant differences using analysis of variance (ANOVA). The normalized percentages of the proliferating T cells at each week in the wtHTLV-1-, Ach.95-, and Ach.195-immortalized cultures were compared using a generalized linear model. This model included the following factors: virus group (wtHTLV-1 or Ach.95/Ach.195), time, and the interaction of the two main effects. The P values of the pairwise comparisons versus the control (wtHTLV-1) at each week were adjusted by Dunnett's method.
RESULTS
Generation and characterization of the recombinant HTLV-1/SU2 provirus.
Previously, using recombinant viruses in which the env sequences were exchanged between HTLV-1 and HTLV-2, we observed that the viral envelope confers the distinct in vitro immortalization/transformation tropism (14). Subsequently, Jones et al. (18) reported that HTLV-1 and HTLV-2 differ slightly in their host cell receptor requirement for initial binding by SU, although they share a receptor to facilitate fusion and entry. Furthermore, the same group also showed that the C terminus of the HTLV-1 SU (SU1) is essential for efficient binding to CD4+ T cells (18). The amino acid homology between HTLV-1 Env and HTLV-2 Env is approximately 63% for the SU glycoprotein. To further dissect the role of the SU region in the distinct in vitro T cell immortalization tropism, we constructed an HTLV-1 recombinant that contains SU2 from wtHTLV-2 (HTLV-1/SU2; Fig. 1A). In HTLV-1, since the open reading frames of env and Hbz do not overlap, the recombinations in the envelope region do not affect HBZ. Additionally, the poly(A) signal and site are in similar locations for Hbz and Aph-2, at the 5′ end of SU1 and SU2, respectively. To determine the ability of recombinant proviral clones to replicate, synthesize viral proteins, and induce cellular immortalization, permanent 729 B cell transfectants expressing wt and recombinant proviral clones were generated and further characterized. Each of the stable transfectants contained complete copies of the provirus (data not shown), and the presence of the expected env sequences was confirmed by DNA PCR followed by restriction digestion (Fig. 1B). To monitor the production of viral protein in these stable transfectants, the concentration of p19 Gag in the culture supernatants was quantified by ELISA. As shown in Fig. 1C, representative stable cell clones selected for this study had p19 Gag expression levels similar to those of the wt virus producer cell lines.
HTLV-1 recombinants containing HTLV-2 SU exhibited a shift in the immortalization tropism to CD8+ T cells.
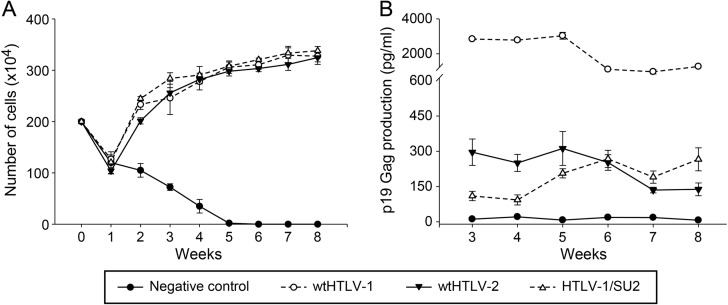
We next determined whether the recombinant viruses had the ability to immortalize human PBMCs using a well-established cocultivation immortalization assay (14). Cell number and viability were examined at weekly intervals to monitor the cellular transformation process and the characteristic expansion of cells from the mixed PBMC population. A growth curve from a representative experiment is depicted in Fig. 2A. When cocultured with irradiated uninfected 729 cells, PBMCs showed a progressive loss of viable cells over time and eventually died off approximately 5 to 6 weeks postplating. This observation is consistent with previously published reports (14, 15). In contrast, the immortalization of PBMCs was apparent in coculture wells containing producer cells of 729.HTLV-1 (wtHTLV-1), 729.HTLV-2 (wtHTLV-2), or 729.HTLV-1/SU2. This initial drop in cell numbers at week 1 in these cocultures could be attributed to HTLV-1 infection-induced apoptosis/senescence and normal cell death due to lack of antigenic stimuli. Cell numbers and viabilities were similar for all virus-producing cells throughout the experiment. Viral replication was assessed by quantitation of p19 Gag production in the culture supernatant starting at 3 weeks postcocultivation. At that time point, HTLV-infected PBMCs typically produce viral particles and the virion production from the residual irradiated viral producer cells becomes negligible (Fig. 2B). All cultures containing wt or recombinant HTLV showed p19 Gag production. However, note that this assay is principally used as a surrogate marker to ensure that the increasing proliferation of T cells (Fig. 2A) is induced by virus infection (Fig. 2B) rather than as a quantitative measure of differences in virus replication between the recombinant and wt viruses.
Fig 2.
Recombinant HTLV-1/SU2 provirus can infect and immortalize fresh PBMCs in culture. Approximately 106 virus producer cells (the number of cells was normalized to the equivalent p19 Gag output) or 729 negative-control cells were irradiated (10,000 rads) and cocultured with 2 × 106 freshly isolated human PBMCs in 24-well plates with 10 U/ml of rhIL-2. These cultures were fed with fresh media on a weekly basis. Data represent the results from a representative of two independent experiments. (A) Cell viability was determined by trypan blue exclusion using three random wells from each of the cocultures on a weekly basis. The symbols denote the averages and the error bars denote the standard deviations at each time point for each of the corresponding cocultures. (B) The active replication of HTLV in each of these cocultures was determined by measuring p19 Gag output in the supernatant using ELISA. The symbols denote the averages and the error bars denote the standard deviations in the p19 Gag levels from three random wells of the corresponding cocultures.
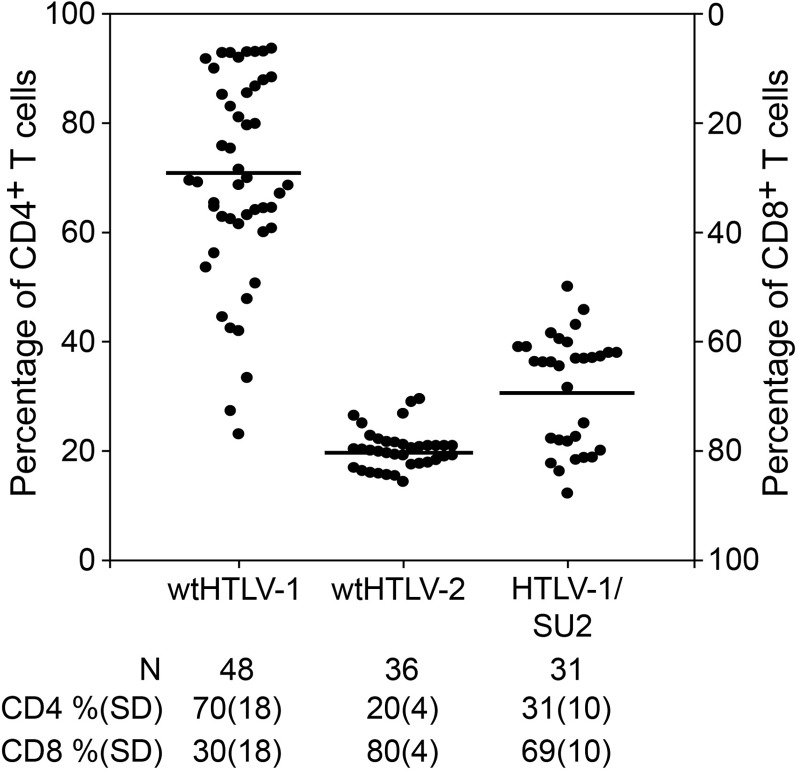
To determine if the exchange of SU sequence altered the immortalization tropism, we evaluated phenotypes of cells immortalized by wtHTLV-1, wtHTLV-2, or HTLV-1/SU2. Individual wells of cells at 8 weeks postcoculture were stained with anti-CD3, anti-CD4, and anti-CD8 antibodies and analyzed by flow cytometry. The data from multiple wells in two independent experiments are depicted in Fig. 3. Since HTLV-1 and HTLV-2 mainly immortalize/transform CD4+ and CD8+ T cells, respectively, the actively proliferating cells at the end of 8 weeks in culture most likely represent a combination of these two T cell types. Therefore, we normalized the actual percentages of CD3+ CD4+ and CD3+ CD8+ T cells to 100 in each of the wells analyzed. Our results revealed that wtHTLV-1 immortalized 70% CD4+ T cells and 30% CD8+ T cells, while wtHTLV-2 immortalized 20% CD4+ T cells and 80% CD8+ T cells. This finding is consistent with our previously published results (14, 15). HTLV-1/SU2 immortalized 31% CD4+ T cells and 69% CD8+ T cells. This indicates that the SU2 component shifted the immortalization tropism of HTLV-1 from predominantly CD4+ T cells toward a predominantly CD8+ T cell population, which is similar to the wtHTLV-2 data. The differential tropism results seen in comparisons of wtHTLV-1 and HTLV-1/SU2 were statistically significant (P < 0.0001; Holm-Sidak method).
Fig 3.
Recombinant HTLV-1/SU2 shifted the CD4+ T cell immortalization preference. The immortalization cocultures set up as described in the Fig. 2 legend were harvested after 8 weeks, stained with anti-CD3, anti-CD4, and anti-CD8 antibodies, and analyzed by flow cytometry. The average actual percentages of CD4+ and CD8+ T cells in the two freshly isolated PBMCs were 58% and 30%, respectively. The percentages of CD3+ CD4+ and CD3+ CD8+ T cells in each of the outgrowing wells were determined. The percentages in each well were normalized to 100, and the percentages of CD4+ T cells are plotted in the graph. The cultures from two independent experiments were combined and plotted. The graph shows the percentages of CD4+ T cells in ascending order in the left y axis and the percentages of CD8+ T cells in descending order in the right y axis for each well denoted by a symbol. The horizontal bars denote the average percentage of CD4+ T cells immortalized by each of the viruses. The average percentages along with standard deviations (SD) and the total number of wells analyzed for each of the virus-immortalized cultures are listed below the graph. The average CD4+ T cell percentages of HTLV-1/SU2-immortalized cultures were significantly different from that of the wtHTLV-1-immortalized cultures (P < 0.0001; Holm-Sidak method).
Thus, taken together, the data indicate that recombinant HTLV-1/SU2, like the parental viruses, was replication competent and was capable of inducing sustained proliferation and immortalization of newly isolated PBMCs. However, the recombinant HTLV-1/SU2 virus shifted the immortalization tropism preference from CD4+ to CD8+ T cells. Therefore, SU of the viral envelope contributes to the preferential immortalization/transformation tropism of HTLV-1. Moreover, this shift in tropism also helps explain the dramatic difference in p19 Gag production levels between HTLV-1- and HTLV-1/SU2- or HTLV-2-infected PBMCs shown in Fig. 2B. HTLV expression in CD8+ T cells is generally lower than in CD4+ T cells. Thus, the levels of expression of p19 are similar for the viruses present in the expanding CD8+ cells, and those levels are much lower than that seen with wtHTLV-1, which favors immortalization of CD4+ T cells.
N195D substitution in the HTLV-1 SU immunodominant domain alters T cell immortalization tropism.
At position 195 of the HTLV-1 SU domain, a conservative amino acid substitution (N195D) did not alter the function of the envelope, but a nonconservative amino acid substitution was not tolerated. In previous studies, an HTLV-1 provirus carrying the N195D substitution (Ach.195) was able to efficiently infect and immortalize fresh PBMCs in vitro (23) and replicate in vivo in newly infected rabbits (24). However, 2 of 4 Ach.195-infected rabbits mounted an altered humoral response; one rabbit had no antibody response, and the other rabbit had a stronger antibody response to SU2 than to SU1 antigen. Although the altered immune response was demonstrated in a very small number of animals, it is interesting that a single amino acid change can trigger such an altered response, suggesting an important role of this particular amino acid and position in the conformation of SU1. A similar N-to-D substitution at position 95 in HTLV-1 (Ach.95) elicited a humoral response similar to that seen with the wtHTLV-1, in addition to other similarities in the biologic properties. Interestingly, the SU2 domain carries a Q at position 91 and a D at position 191, corresponding to positions 95 and 195 in the SU1 domain, respectively, suggesting that the HTLV-1 envelope with N195D might be more like the HTLV-2 envelope functionally.
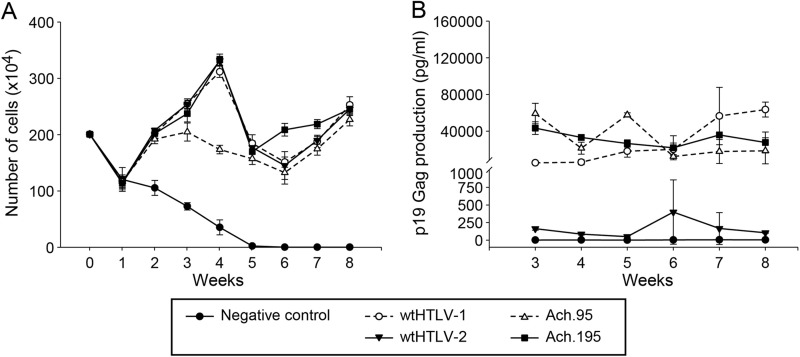
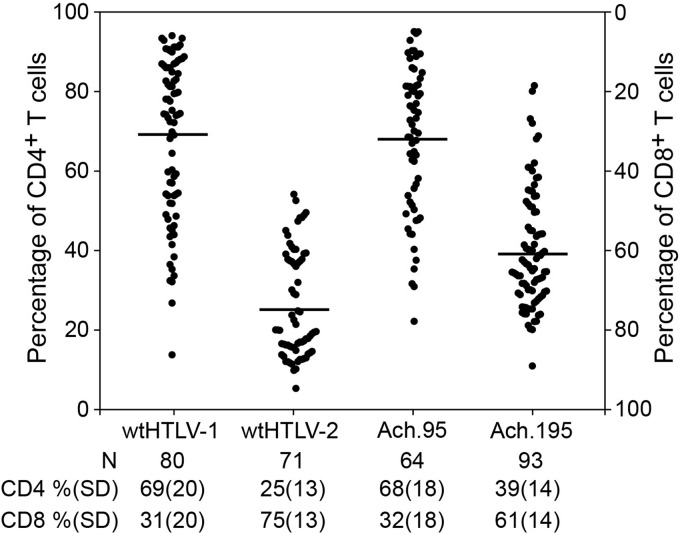
Therefore, we assessed the in vitro T cell immortalization ability of Ach.95 and Ach.195. The weekly growth curves indicated that Ach.95 and Ach.195, in similarity to the parental wtHTLV-1 and the wtHTLV-2 strains, were capable of immortalizing freshly isolated PBMCs in culture (Fig. 4A). The virus-immortalized cultures also revealed constant p19 Gag production, confirming active viral replication in these proliferating cells (Fig. 4B). PBMCs cultured with uninfected 729 cells died by week 4 to 5 (Fig. 4A), and the supernatants did not have any detectable p19 Gag levels throughout the 8-week time course (Fig. 4B). We note in this assay that the levels of p19 Gag production in cultures infected with the wtHTLV-1 (Ach) and the Ach.95 and Ach.195 were similar, whereas HTLV-2-infected cultures expressed p19 Gag at levels that were much lower but above the level seen with the negative control. Differences in protein production levels between HTLV-1 and HTLV-2 have been reported by us and others previously and are primarily attributed to Tax-1 and Tax-2 differential transactivation functions (5, 28, 29). The cultures were analyzed by flow cytometry to determine the CD4/CD8 phenotype of the predominantly proliferating CD3+ T cell type (Fig. 5). Our results showed that wtHTLV-1-immortalized population was composed of 69% CD4+ T cells and 31% CD8+ T cells, while the wtHTLV-2-immortalized population was composed of 25% CD4+ T cells and 75% CD8+ T cells. Again, these results were consistent with the results presented in Fig. 3 and our previously published reports (14, 15). The cell population immortalized by Ach.95, like that immortalized by wtHTLV-1, was composed of 68% CD4+ T cells and 32% CD8+ T cells. In contrast, the cells immortalized by Ach.195 consisted of 39% CD4+ T cells and 61% CD8+ T cells; thus, there was a shift from a CD4+ T cell to a CD8+ T cell immortalization preference. The difference corresponding to this shift between wtHTLV-1 and Ach.195 was statistically significant (P < 0.0001; Dunn's method). Therefore, asparagine at position 195 in the SU domain of the HTLV-1 envelope is involved in dictating this CD4+ T cell immortalization tropism in culture.
Fig 4.
HTLV-1 envelope SU domain mutant viruses Ach.95 and Ach.195 were capable of infecting and immortalizing fresh PBMCs in culture. Approximately 106 virus producer cells (the number of cells was normalized to the equivalent p19 Gag output) or 729B negative-control cells were irradiated (10,000 rads) and cocultured with 2 × 106 freshly isolated human PBMCs in 24-well plates for 8 weeks with 10 U/ml of rhIL-2. These cultures were fed with fresh media on a weekly basis. Data are representative of three independent experiments. (A) Cell viability was determined by trypan blue exclusion using three random wells from each of the cocultures on a weekly basis. The symbols denote the averages and the error bars denote the standard deviations at each time point for each of the corresponding cocultures. (B) The active replication of HTLV in each of these cocultures was determined by measuring p19 Gag output in the supernatant using ELISA. The symbols denote the averages and the error bars denote the standard deviations in the p19 Gag levels from three random wells of the corresponding cocultures.
Fig 5.
Ach.195 shifted the CD4+ T cell immortalization preference. The immortalization cocultures were set up as described in the legend to Fig. 2. The average actual percentages of CD4+ and CD8+ T cells in the freshly isolated PBMCs were 56% and 30%, respectively. The graph shows the percentages of the CD4+ T cells in ascending order in the left y axis and the percentages of CD8+ T cells in descending order in the right y axis for each well denoted by a symbol. The bar denotes the average percentage of CD4+ T cells transformed by each of the viruses. The average percentages along with standard deviations and the total number of wells analyzed for each of the virus-immortalized cultures are denoted below the graph. The average CD4+ T cell percentage in Ach.195-immortalized cultures was significantly different from that of the wtHTLV-1-immortalized cultures (P < 0.0001; Dunn's method).
N195D substitution in the SU domain of the HTLV-1 envelope dictates the shift in immortalization tropism during the selective clonal expansion process.
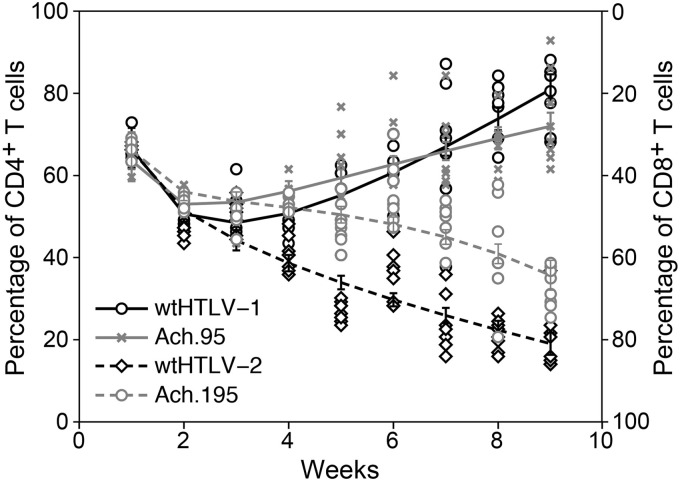
We next evaluated the T cell clonal expansion process using a 9-week in vitro immortalization assay for the preferential T cell tropism exhibited by the N-to-D substitution at residues 95 and 195 of the SU domain. For this, we analyzed the percentages of proliferating CD4+ and CD8+ T cells from eight wells of the wtHTLV-1-, wtHTLV-2-, Ach.95-, or Ach.195-immortalized cultures on a weekly basis for 9 weeks. The actual percentages of CD4+ and CD8+ T cells in the freshly isolated PBMCs were 57% and 31%, respectively. Cell viability counts and a p19 Gag ELISA performed at each week indicated efficient virus-induced immortalization by all four viruses (data not shown). As expected, the cultures with the negative-control 729 cells died within 4 to 5 weeks (data not shown). Due to the large dead cell population from week 1, the negative cultures were not phenotyped. The weekly phenotype analysis revealed that Ach.95-immortalized cultures, in similarity to the wtHTLV-1-immortalized cultures, had selectively expanded CD4+ T cells as the predominant population from week 5 onward (Fig. 6). The predominant CD8+ T cell population emerged by week 4 in wtHTLV-2-immortalized cultures. However, predominantly CD8+ T cell clonal selection in the Ach.195-immortalized cultures, which represented a shift from the predominant CD4+ T cell population in the wtHTLV-1-immortalized cultures, emerged at week 7. The differences at weeks 7 to 9 between the wtHTLV-1- and Ach.195-immortalized predominant T cell phenotypes were statistically significant (P < 0.0001).
Fig 6.
Longitudinal phenotype analysis of the T cells immortalized by wtHTLV-1, wtHTLV-2, Ach.95, or Ach.195. The immortalization cocultures were set up as described in the legend to Fig. 2. Eight wells from the cocultures with each of the four viruses were analyzed for the percentage of CD3+ CD4+ and CD3+ CD8+ T cells at each time point. The black circle symbols represent wtHTLV-1-, the black diamond symbols represent wtHTLV-2-, the gray x symbols represent Ach.95-, and the gray circle symbols represent Ach.195-immortalized cultures. The corresponding vertical bars represent the 95% confidence interval for each of the viruses at the given time point. The trend lines or fitted lines represent the overall or average trends across time points. These fitted lines were obtained by fitting a generalized linear model. The model adjusted mean values were compared at each time point. The differences in the mean values between wtHTLV-1- and Ach.195-immortalized cultures were statistically significant from week 7 onward (P < 0.0001; generalized linear model with Dunnet's method for multiplicity adjustment).
Taken together, these results indicate that the viral envelope dictates the preferential immortalization/transformation of the predominant T cell type and that residue 195 in SU is within a critical domain that contributes to the protein interactions that potentially mediate this preferential immortalization tropism in HTLV-1-infected cells. Our results suggest that preferential immortalization tropism occurs during the clonal expansion process, which likely corresponds to the clinical latency period in infected individuals, rather than during the initial virus entry stage.
DISCUSSION
Although HTLV-1 and HTLV-2 share a genomic structure, they exhibit distinct pathogeneses. HTLV-1 predominantly immortalizes/transforms CD4+ T cells in culture and mainly causes ATL, which is a CD4+ T cell leukemia. HTLV-2 predominantly immortalizes/transforms CD8+ T cells in culture (13–15) but is not associated with leukemia. Although the Tax protein is critical for transformation by HTLV-1 and HTLV-2, we have previously shown that the envelope dictates which T cell type (CD4+ or CD8+) is preferentially immortalized or transformed in culture (14). Other retroviral envelopes, including the envelopes of Jaagsiekte sheep retrovirus and Friend spleen focus-forming virus, have the capacity to transform cells or play a key role in oncogenesis (30, 31). Nevertheless, the observation was paradigm shifting for HTLV, since at that time it was generally believed that HTLV-1 and HTLV-2, along with their simian counterparts, shared common host cellular receptors for binding and entry. Subsequent studies on host cell receptor requirements provided evidence that HTLV-1 and HTLV-2 displayed slightly different receptor requirements for initial binding, which is mediated by the SU domain of the viral envelope. The main difference is the requirement of HSPGs for initial binding—HSPGs are essential for efficient HTLV-1 binding, while their presence is detrimental to HTLV-2 binding. However, it was shown that the two viruses utilized the same GLUT-1 receptor followed by subsequent fusion of the membranes and entry into the cell (16–19, 32). In this study, we characterized the viral envelope SU component to determine its independent contribution to the preferential CD4+ T cell immortalization tropism. In the context of the entire HTLV-1 provirus, the exchange of SU2 domain from wtHTLV-2 shifted the CD4+ T cell immortalization preference to a CD8+ T cell preference. This indicates that the SU domain independently influences the preferential immortalization/transformation of T cells, irrespective of the TM counterpart. This result is consistent with our recent in vivo study, which clearly confirms that HTLV-1 and HTLV-2 do not exhibit any CD4+ or CD8+ T cell preference at the infection stage. Those studies also showed that the T cell predominance emerged only after 4 to 5 weeks of coculture, providing strong evidence that the observed preferential tropism is not determined by the initial infection of the cell but by later interactions of the envelope with the infected cell (33).
The oncogenicity of the feline leukemia virus (FeLV) has been shown to be determined by the SU of that virus, along with the long terminal repeat (34). It has been shown that the exchange of the SU domain from an FeLV variant (FeLV-945) into FeLV-A altered the disease outcome entirely from a thymic lymphoma of T cell origin to a multicentric (nonthymic) lymphoma of B cell origin (34). This result indicated that the SU domain determined the tumorigenic outcome. However, in HTLV, although Tax is the key oncoprotein, the envelope mainly mediates the T cell immortalization/transformation tropism. Our previous longitudinal phenotype analysis of HTLV-immortalized in vitro cultures revealed that HTLV-1 and HTLV-2 induced the proliferation of both CD4+ and CD8+ T cells in the initial weeks postcoculture (33). However, after 4 to 5 weeks, the preferred T cell population emerged via selection and clonal expansion (33). These results, taken together with results in this study, led us to hypothesize that the SU1 protein expressed in HTLV-infected cells might interact with host cellular factors to mediate the preferential clonal expansion of CD4+ T cells. Future studies will focus on the identification of these potential factors.
Delamarre et al. (22) examined the effect of single conservative and nonconservative amino acid substitutions in the SU1 protein on intracellular maturation and function through syncytium formation. They showed that 19/23 nonconservative single amino acid substitutions impaired envelope function but 5/7 conservative single amino acid changes resulted in normal envelope function, indicating that the SU protein does not tolerate amino acid changes that are not conserved between HTLV-1, HTLV-2, and STLV-1. Two of these five amino acid residues (SU195 and SU1195) are within the immunodominant epitopes of SU1 (SU188–98 and SU1175–199) that trigger the neutralizing antibody response in HTLV-1-infected patients and asymptomatic carriers (35–38). Interestingly, both these residues, SU195 and SU1195, carry an asparagine and their species-conservative amino acid substitution was aspartic acid (N95D and N195D). Note that in HTLV-2 SU, the corresponding residue at position 91 is a glutamic acid and not aspartic acid, unlike the corresponding residue at position 191. Proviral clones of HTLV-1 carrying N95D and N195D substitutions (Ach.95 and Ach.195) were capable of infecting and immortalizing freshly isolated PBMCs (23). In vivo studies showed that rabbits inoculated with Ach.95 or Ach.195 virus-producing cells had proviral loads similar to those of rabbits infected with wtHTLV-1, indicating similar levels of infection by the wild-type and the mutant viruses. However, 2/4 Ach.195-inoculated rabbits exhibited an altered humoral response to the envelope protein compared to wtHTLV-1- or Ach.95-inoculated rabbits (differences included no humoral response or a stronger humoral response to SU2 antigen). Additionally, all four Ach.195-inoculated rabbits had an antibody response to SU1 antigen that was lower than that seen with wtHTLV-1- or Ach.95-inoculated rabbits (24). In this study, we showed that N195D shifted the CD4+ T cell immortalization tropism of wtHTLV-1 using an in vitro immortalization assay.
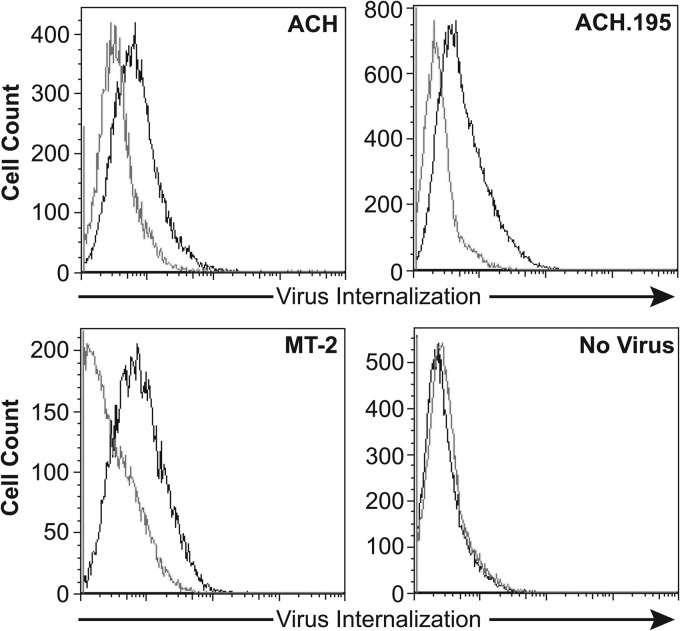
Experiments to directly compare the abilities of wtHTLV-1 virus and virus carrying the N195D mutation (Ach.195) to bind and enter cells revealed that these two viruses bound to and entered CD4+ T cells at similar levels (Fig. 7). Our longitudinal phenotyping results showed that Ach.95 and Ach.195, like wtHTLV-1 and wtHTLV-2, initially induced the proliferation of both CD4+ and CD8+ T cells. However, the selection of the corresponding predominant CD4+ or CD8+ T cell population emerged at week 5 and week 7 in Ach.95- and Ach.195-immortalized cultures, respectively. These results indicate that the asparagine at residue 195 of the SU1 domain is actively involved in potential cellular interactions that result in the predominant selection of a particular T cell clone postinfection. It is important that the percentage of CD4+ and CD8+ T cells is determined by the total number of live cells (infected and uninfected). It is possible that in the initial weeks, a small number of uninfected cells could be proliferating along with the infected cells. However, after week 5, the majority of the proliferating cells would be infected, as the uninfected cells cannot perpetuate any longer due to the lack of virus infection and immortalization as shown by the negative-control cocultures in Fig. 2A and 4A. Due to the extensive nature of these types of assays, we present here only the initial phenotyping analysis to understand the emergence of the predominantly immortalized/transformed T cell type. Further systematic analysis is required to understand the mechanism of selection in these HTLV-transformed T cells.
Fig 7.
ACH.195 binds and enters primary CD4+ T cells at levels similar to wtHTLV-1 (ACH) levels. Primary CD4+ T cells were incubated with virus supernatant from T cell producer cell lines (ACH, ACH.195, and MT-2) or uninfected control as indicated. Three hours later, the cells were washed, permeabilized, and stained with antibody to the core protein MA and the levels of internalized virions determined by flow cytometry analyses as described in Materials and Methods. Dark lines, staining with anti-HTLV p19 antibody; light lines, staining with mouse IgG1 (isotype control).
The emergence of CD8+ T cells as the predominant population in the Ach.195-immortalized cultures took 2 weeks longer than in the wtHTLV-1-immortalized cultures, suggesting that there might be other residues or domains involved in this interaction. Delamarre et al. (22) have shown that other residues, including 181 and 197 in the immunodominant epitope, are important for envelope function. This opens new avenues for exploration of the protein interactions with the immunodominant epitope of the HTLV-1 envelope that might result in the preferential transformation of CD4+ T cells. In rabbits, the first cytotoxic T cell (CTL) response is to the viral envelope, followed closely by Tax (39). Robek and Ratner (28) have shown that the transcriptional function of Tax in the CREB/ATF pathway is important for the preferential transformation of CD4+ T cells by HTLV-1. Moreover, Sibon et al. (29) showed that clonal expansion of HTLV-1-infected cells is the result of proliferation and acquisition of genetic rearrangements in CD4+ T cells versus protected accumulation of CD8+ T cells. Taking these results together, we hypothesize that the immunodominant epitope SU175–199 could play a dual role in the pathogenesis of HTLV-1. This epitope is initially expressed in large quantities and induces humoral and cytotoxic immune responses to the cells that express this protein. However, once Tax is downregulated and CTLs begin to actively remove HTLV-1 protein-expressing cells, the virus downregulates overall protein expression. The low levels of SU1175–199 probably trigger a slow progressive selection of the transformed T cell type for clonal expansion, in concert with Tax and potentially other viral proteins. Thus, our results provide novel insights into HTLV-1 pathogenesis and open avenues to explore the potential of low-level periodic booster vaccines against this SU175–199 epitope in HTLV-1-infected carriers to suppress disease progression. In addition, it might be possible to use this epitope as bait for the elucidation of the underlying mechanism for the preferential transformation tropism in HTLV-1 and HTLV-2.
ACKNOWLEDGMENTS
We thank Kate Hayes-Ozello for editorial comments on the manuscript and Tim Vogt for figure preparation.
This work was supported by grants from the National Institutes of Health (CA100730 and CA077556) to P.L.G. and in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract HHSN26120080001E (K.S.J.). This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Published ahead of print 19 June 2013
REFERENCES
- 1. Lairmore M, Franchini G. 2007. Human T-cell leukemia virus types 1 and 2, p 2071–2106 In Fields B Knipe D Howley P Chanock R Monath T Melnick J Roizman B Straus S (ed), Fields virology, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 2. Halin M, Douceron E, Clerc I, Journo C, Ko NL, Landry S, Murphy EL, Gessain A, Lemasson I, Mesnard JM, Barbeau B, Mahieux R. 2009. Human T-cell leukemia virus type 2 produces a spliced antisense transcript encoding a protein that lacks a classic bZIP domain but still inhibits Tax2-mediated transcription. Blood 114:2427–2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kannian P, Green PL. 2010. Human T Lymphotropic Virus Type 1 (HTLV-1): Molecular Biology and Oncogenesis. Viruses 2:2037–2077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Matsuoka M, Green PL. 2009. The HBZ gene, a key player in HTLV-1 pathogenesis. Retrovirology. 6:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yin H, Kannian P, Dissinger N, Haines R, Niewiesk S, Green PL. 2012. HTLV-2 APH-2 is dispensable for in vitro immortalization, but functions to repress early viral replication in vivo. J. Virol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gessain A, Barin F, Vernant JC, Gout O, Maurs L, Calender A, de Thé G. 1985. Antibodies to human T-lymphotropic virus type-I in patients with tropical spastic paraparesis. Lancet ii:407–410 [DOI] [PubMed] [Google Scholar]
- 7. Hinuma Y, Nagata K, Hanaoka M, Nakai M, Matsumoto T, Kinoshita K-I, Shirakawa S, Miyoshi I. 1981. Adult T-cell leukemia: Antigen in an ATL cell line and detection of antibodies to the antigen in human sera. Proc. Natl. Acad. Sci. U. S. A. 78:6476–6480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Osame M, Usuku K, Izumo S, Ijichi N, Amitani H, Igata A, Matsumoto M, Tara M. 1986. HTLV-I associated myelopathy, a new clinical entity. Lancet i:1031–1032 [DOI] [PubMed] [Google Scholar]
- 9. Yoshida M, Miyoshi I, Hinuma Y. 1982. Isolation and characterization of retrovirus from cell lines of human adult T-cell leukemia and its implication in the disease. Proc. Natl. Acad. Sci. U. S. A. 79:2031–2035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Araujo A, Hall WW. 2004. Human T-lymphotropic virus type II and neurological disease. Ann. Neurol. 56:10–19 [DOI] [PubMed] [Google Scholar]
- 11. Bartman MT, Kaidarova Z, Hirschkorn D, Sacher RA, Fridey J, Garratty G, Gibble J, Smith JW, Newman B, Yeo AE, Murphy EL. 2008. Long-term increases in lymphocytes and platelets in human T-lymphotropic virus type II infection. Blood 112:3995–4002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Orland JR, Engstrom J, Fridey J, Sacher RA, Smith JW, Nass C, Garratty G, Newman B, Smith D, Wang B, Loughlin K, Murphy EL. 2003. Prevalence and clinical features of HTLV neurologic disease in the HTLV Outcomes Study. Neurology 61:1588–1594 [DOI] [PubMed] [Google Scholar]
- 13. Wang T-G, Ye J, Lairmore M, Green PL. 2000. In vitro cellular tropism of human T-cell leukemia virus type 2. AIDS Res. Hum. Retroviruses 16:1661–1668 [DOI] [PubMed] [Google Scholar]
- 14. Xie L, Green PL. 2005. Envelope is a major viral determinant of the distinct in vitro cellular transformation tropism of human T-cell leukemia virus type 1 (HTLV-1) and HTLV-2. J. Virol. 79:14536–14545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ye J, Xie L, Green PL. 2003. Tax and overlapping Rex sequences do not confer the distinct transformation tropisms of HTLV-1 and HTLV-2. J. Virol. 77:7728–7735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ghez D, Lepelletier Y, Jones KS, Pique C, Hermine O. 2010. Current concepts regarding the HTLV-1 receptor complex. Retrovirology. 7:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ghez D, Lepelletier Y, Lambert S, Fourneau JM, Blot V, Janvier S, Arnulf B, van Endert PM, Heveker N, Pique C, Hermine O. 2006. Neuropilin-1 is involved in human T-cell lymphotropic virus type 1 entry. J. Virol. 80:6844–6854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jones KS, Fugo K, Petrow-Sadowski C, Huang Y, Bertolette DC, Lisinski I, Cushman SW, Jacobson S, Ruscetti FW. 2006. Human T-cell leukemia virus type 1 (HTLV-1) and HTLV-2 use different receptor complexes to enter T cells. J. Virol. 80:8291–8302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Manel N, Kim FJ, Kinet S, Taylor N, Sitbon M, Battini JL. 2003. The ubiquitous glucose transporter GLUT-1 is a receptor for HTLV. Cell 115:449–459 [DOI] [PubMed] [Google Scholar]
- 20. Hattori S, Kiyokawa T, Imagawa K-I, Shimizu F, Hashimura E, Seiki M, Yoshida M. 1984. Identification of gag and env gene products of human T-cell leukemia virus (HTLV). Virology 136:338–347 [DOI] [PubMed] [Google Scholar]
- 21. Lee TH, Coligan JE, McLane MF, Sodroski JG, Popovic M, Wong-Staal F, Gallo RC, Haseltine W, Essex M. 1984. Serological cross-reactivity between envelope gene products of type I and type II human T-cell leukemia virus. Proc. Natl. Acad. Sci. U. S. A. 81:7579–7583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Delamarre L, Pique C, Pham D, Tursz T, Dokhelar MC. 1994. Identification of functional regions in the human T-cell leukemia virus type I SU glycoprotein. J. Virol. 68:3544–3549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tsukahara T, Wielgosz MM, Ratner L. 2001. Characterization of envelope glycoprotein mutants for human T-cell leukemia virus type 1 infectivity and immortalization. J. Virol. 75:9553–9559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Silverman LR, Phipps AJ, Montgomery A, Fernandez S, Tsukahara T, Ratner L, Lairmore MD. 2005. In vivo analysis of replication and immunogenicity of proviral clones of human T-lymphotropic virus type 1 with selective envelope surface-unit mutations. Blood 106:3602–3608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kimata JT, Wong FH, Wang JJ, Ratner L. 1994. Construction and characterization of infectious human T-cell leukemia virus type I molecular clones. Virology 204:656–664 [DOI] [PubMed] [Google Scholar]
- 26. Chen IY, McLaughlin J, Gasson JC, Clark SC, Golde DW. 1983. Molecular characterization of genome of a novel human T-cell leukaemia virus. Nature 305:502–505 [DOI] [PubMed] [Google Scholar]
- 27. Jones KS, Petrow-Sadowski C, Bertolette DC, Huang Y, Ruscetti FW. 2005. Heparan sulfate proteoglycans mediate attachment and entry of human T-cell leukemia virus type 1 virions into CD4+ T cells. J. Virol. 79:12692–12702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Robek MD, Ratner L. 1999. Immortalization of CD4+ and CD8+ T-lymphocytes by human T-cell leukemia virus type 1 Tax mutants expressed in a functional molecular clone. J. Virol. 73:4856–4865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sibon D, Gabet AS, Zandecki M, Pinatel C, Thete J, Delfau-Larue MH, Rabaaoui S, Gessain A, Gout O, Jacobson S, Mortreux F, Wattel E. 2006. HTLV-1 propels untransformed CD4 lymphocytes into the cell cycle while protecting CD8 cells from death. J. Clin. Invest. 116:974–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Maeda N, Palmarini M, Murgia C, Fan H. 2001. Direct transformation of rodent fibroblasts by jaagsiekte sheep retrovirus DNA. Proc. Natl. Acad. Sci. U. S. A. 98:4449–4454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Silver JE, Fredrickson TN. 1983. Susceptibility to Friend helper virus leukemias in CXB recombinant inbred mice. J. Exp. Med. 158:1693–1702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pinon JD, Klasse PJ, Jassal SR, Welson S, Weber J, Brighty DW, Sattentau QJ. 2003. Human T-cell leukemia virus type 1 envelope glycoprotein gp46 interacts with cell surface heparan sulfate proteoglycans. J. Virol. 77:9922–9930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kannian P, Yin H, Doueiri R, Lairmore MD, Fernandez S, Green PL. 2012. Distinct transformation tropism exhibited by human T lymphotropic virus type 1 (HTLV-1) and HTLV-2 is the result of postinfection T cell clonal expansion. J. Virol. 86:3757–3766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chandhasin C, Coan PN, Levy LS. 2005. Subtle mutational changes in the SU protein of a natural feline leukemia virus subgroup A isolate alter disease spectrum. J. Virol. 79:1351–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Baba E, Nakamura M, Tanaka Y, Kuroki M, Itoyama Y, Nakano S, Niho Y. 1993. Multiple neutralizing B-cell epitopes of human T-cell leukemia virus type 1 (HTLV-1) identified by human monoclonal antibodies. A basis for the design of an HTLV-1 peptide vaccine. J. Immunol. 151:1013–1024 [PubMed] [Google Scholar]
- 36. Kuroki M, Nakamura M, Itoyama Y, Tanaka Y, Shiraki H, Baba E, Esaki T, Tatsumoto T, Nagafuchi S, Nakano S, et al. 1992. Identification of new epitopes recognized by human monoclonal antibodies with neutralizing and antibody-dependent cellular cytotoxicity activities specific for human T cell leukemia virus type 1. J. Immunol. 149:940–948 [PubMed] [Google Scholar]
- 37. Palker TJ, Riggs ER, Spragion DE, Muir AJ, Scearce RM, Randall RR, McAdams MW, McKnight A, Clapham PR, Weiss RA, et al. 1992. Mapping of homologous, amino-terminal neutralizing regions of human T-cell lymphotropic virus type I and II gp46 envelope glycoproteins. J. Virol. 66:5879–5889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tanaka Y, Tanaka R, Hoshino H. 1994. Identification of a novel neutralization epitope on envelope gp46 antigen of human T-cell-leukemia virus-type-II (HTLV-II). Int. J. Cancer 59:655–660 [DOI] [PubMed] [Google Scholar]
- 39. Haynes RA, 2nd, Phipps AJ, Yamamoto B, Green P, Lairmore MD. 2009. Development of a cytotoxic T-cell assay in rabbits to evaluate early immune response to human T-lymphotropic virus type 1 infection. Viral Immunol. 22:397–405 [DOI] [PMC free article] [PubMed] [Google Scholar]