Abstract
NS1′ is a C-terminally extended form of the NS1 protein produced only by encephalitic flaviviruses from the Japanese encephalitis virus serogroup. Here we show that West Nile virus (WNV) NS1′ and NS1 localize to the same cellular compartments when expressed from plasmid DNAs and also colocalize to viral RNA replication sites in infected cells. Using complementation analysis with NS1-deleted WNV cDNA, we demonstrated that NS1′ is able to substitute for the crucial function of NS1 in virus replication.
TEXT
West Nile virus (WNV) is a mosquito-borne flavivirus within the Japanese encephalitis virus (JEV) serogroup. This serogroup also includes other encephalitic flaviviruses, such as JEV, Murray Valley encephalitis virus, and St. Louis encephalitis virus (1). The natural transmission cycle of WNV is between birds and mosquitoes, primarily the Culex species; however, it can cause incidental infections in humans. Since the outbreak of the more pathogenic WNVNY99 strain in New York in 1999 (2), WNV has emerged as a major cause of arboviral encephalitis in the United States (3). WNV strains can be divided into two distinct lineages, lineage 1 and lineage 2. Lineage 1 includes both WNVNY99 and Kunjin (WNVKUN) (4), the prevalent strain within Australia (5). Despite high sequence similarity to the WNVNY99 strain (∼98% on the amino acid level) (6), most WNVKUN strains are highly attenuated, with only a small number of human infections and no fatalities reported (5, 7). Since its isolation in early 1960s, WNVKUN has been used extensively as a model for WNV infection (8, 9).
The WNVKUN genome is a single-stranded positive-sense RNA of 11,022 nucleotides (10–12). After translation as a single polyprotein, it is cleaved by host and viral proteases to produce 3 structural (C, prM, and E) and 7 nonstructural (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) proteins (11, 12). NS1 is a multifunctional glycoprotein that is involved in viral replication (13–16) and modulation of the immune response (17–22). The key role NS1 plays in RNA replication has been shown previously, with mutations or deletions in the NS1 gene resulting in a lack of detectable RNA replication. This function can be complemented in trans by the expression of NS1 (14, 15).
An additional nonstructural protein, NS1′, is produced exclusively by the members of the JEV serogroup due to the presence of a −1 programmed ribosomal frameshift at the beginning of the adjacent NS2A gene (23–25). This frameshift, occurring in 30 to 50% of translation events (23, 26), results in the formation of a 52-amino-acid C-terminally extended form of NS1. Although a role in neurovirulence has previously been demonstrated (23), no specific functions for NS1′ in viral replication or virus-host interactions have been identified. In the present study, we show that the NS1′ protein colocalizes with NS1 in viral RNA replication sites in the endoplasmic reticulum (ER) of infected cells and can substitute for the function of NS1 in viral replication.
Plasmid DNA-derived expression of NS1 and NS1′.
NS1′ is produced in viral infection due to the presence of a slippery heptanucleotide and 3′ pseudoknot in the viral RNA resulting in a ribosomal shift to the −1 reading frame (23, 25). This results in the addition of 52 amino acids to the C terminus of NS1 protein, including the N-terminal 9 amino acids of NS2A and 43 amino acids after the frameshift, terminating NS1′ synthesis at a stop codon (25). To enable studies on the NS1′ and NS1 proteins, CMV promoter-based plasmids expressing NS1′ or NS1 (pcDNA-NS1′ and pcDNA-NS1, respectively) were designed as shown in Fig. 1A. Both constructs contained the WNVKUN envelope protein (E) signal sequence at the N terminus followed by cDNA for the protein of interest and C-terminal Myc and Flag tags to assist in protein detection. pcDNA-NS1′ was generated by inserting an additional nucleotide (Fig. 1B, boxed nucleotide) at the frameshift site to induce the change in reading frame that leads to NS1′ synthesis. To prevent further frameshifting, we also introduced two single-nucleotide mutations into the slippery heptanucleotide of the frameshift motif, as published previously (23) (Fig. 1B, circled nucleotides). To confirm protein production from the designed plasmids, HEK293T cells transfected with either pcDNA-NS1′ or -NS1 were harvested in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris HCl, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 1% SDS) at 2 days posttransfection. Cell lysates were denatured for 10 min at 70°C, separated by electrophoresis in 10% polyacrylamide gels, transferred to nitrocellulose membranes, and analyzed by Western blotting for the presence of NS1 and NS1′. 4G4, a monoclonal antibody that recognizes both NS1 and NS1′ proteins (27), detected NS1 (monomer) in lysates from pcDNA-NS1-transfected cells and NS1′ proteins (both monomer and dimers) in lysates from pcDNA-NS1′-transfected cells (Fig. 1C). Notably, only NS1′ and not NS1 expression resulted in generation of heat-stable dimers. The results confirmed that pcDNA-NS1 and pcDNA-NS1′ plasmids express NS1 and NS1′, respectively.
Fig 1.
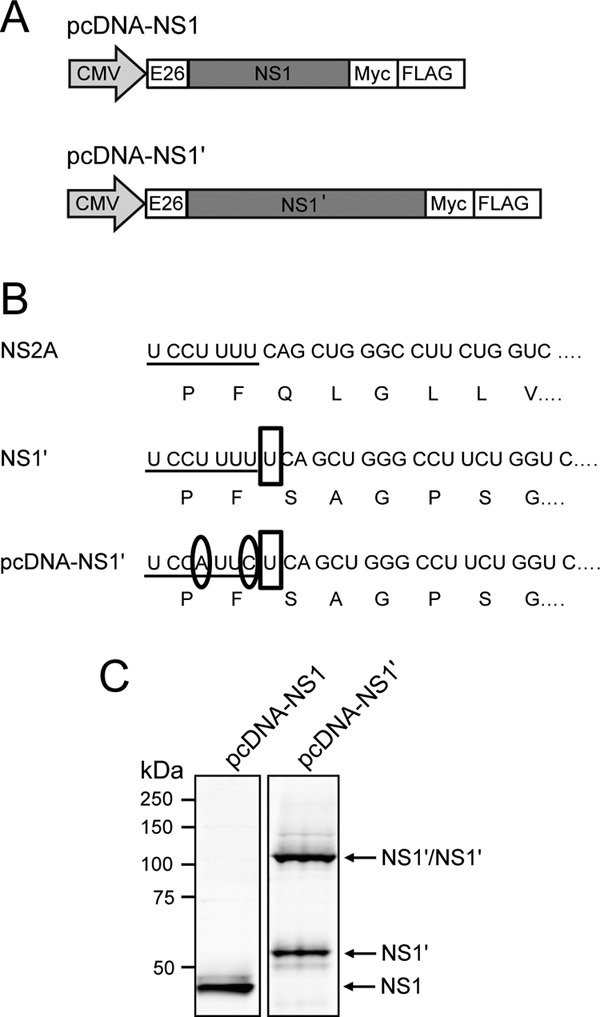
Design and characterization of plasmid DNAs expressing NS1 and NS1′ genes. (A) Plasmid constructs pcDNA-NS1 and pcDNA-NS1′, for expression of NS1 and NS1′, respectively, contain an N-terminal signal sequence consisting of the last 26 codons of the WNV E protein and Myc and Flag tags at the C terminus. (B) Alignment of the nucleic and amino acid sequences of NS2A, NS1′ and pcDNA-NS1′ showing the −1 frameshift occurring at the beginning of the NS2A gene that leads to the generation of NS1′. Underlining shows the slippery heptanucleotide of the frameshift motif, open boxes show inserted nucleotides, and circles show mutated bases. (C) Western blot showing expression of NS1 and NS1′ from pcDNA-NS1 and pcDNA-NS1′, respectively. Lysates were heat denatured and analyzed by Western blotting with anti-NS1 (4G4).
The cellular localization of NS1′ is similar to that of NS1 in plasmid DNA-transfected and virus-infected cells.
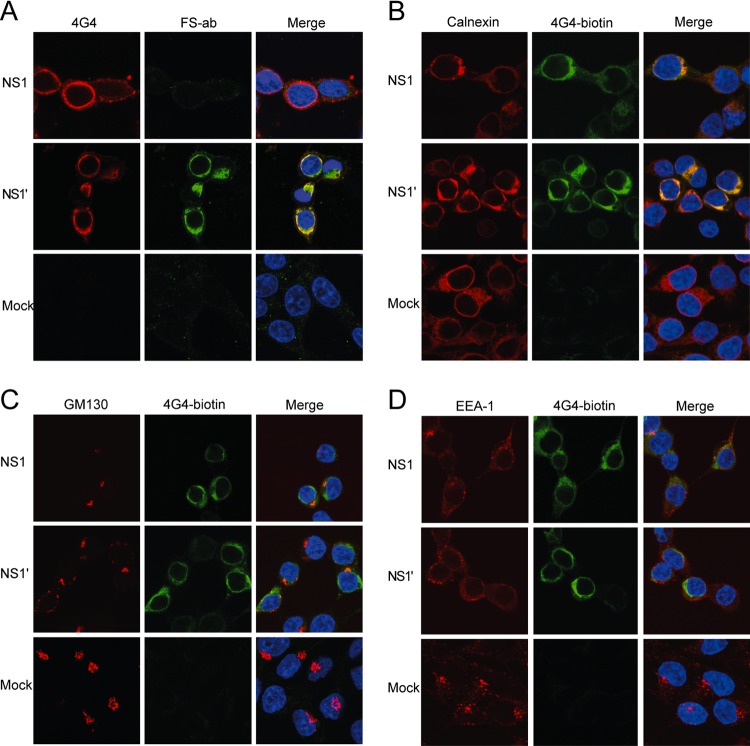
NS1 is known to localize to the ER of cells during flavivirus infection (16, 28, 29). To examine whether NS1′ also colocalizes to the ER with NS1, we carried out immunofluorescence assays on cells expressing NS1′ or NS1 proteins. HEK293T cells transfected with pcDNA-NS1 or pcDNA-NS1′ plasmids were fixed in 80% acetone–20% phosphate-buffered saline (PBS) at 48 h posttransfection and stained with 4G4, to detect both NS1 and NS1′, and with NS1′-specific antibodies (FS-Ab) (23). As expected, FS-Ab stained only pcDNA-NS1′-transfected cells, while 4G4 stained both pcDNA-NS1- and pcDNA-NS1′-transfected cells (Fig. 2A). Using 4G4 in addition to antibodies recognizing markers for various cellular compartments, we were able to further examine the localization of NS1′ and NS1 in transfected cells. HEK293T cells transfected with pcDNA-NS1 or pcDNA-NS1′ plasmids were fixed at 48 h posttransfection in 4% paraformaldehyde (PFA) with 0.1% Triton X-100 and stained with biotinylated 4G4 and antibodies recognizing markers of the ER (rabbit polyclonal antibody against calnexin [Sigma-Aldrich]), Golgi apparatus (mouse monoclonal antibody against GM130 [Becton, Dickinson]), or endosomes (mouse monoclonal antibody against EEA-1 [BD Transduction Laboratories]). Both NS1 and NS1′ localized predominantly to the ER (Fig. 2B), with a small degree of colocalization with the Golgi apparatus (Fig. 2C) and no distinct localization to the endosomes (Fig. 2D). Moreover, there did not appear to be any differences in the cellular distribution of plasmid-expressed NS1′ compared to NS1, leading us to conclude that NS1′ resides in the same cellular compartments as NS1.
Fig 2.
Cellular localization of plasmid-expressed NS1′ and NS1. (A) Immunofluorescence analysis showing production of NS1 and NS1′ using 4G4 (which stains both NS1 and NS1′) and FS-Ab (an NS1′-specific antibody) in transfected HEK293T cells. (B to D) Immunofluorescence analysis showing colocalization of plasmid-expressed NS1 and NS1′ with the ER (B), the Golgi apparatus (C), and endosomes (D). Transfected HEK293T cells were stained with antibodies to appropriate cell markers (calnexin, GM130, and EEA-1, respectively) and biotinylated anti-NS1 (4G4).
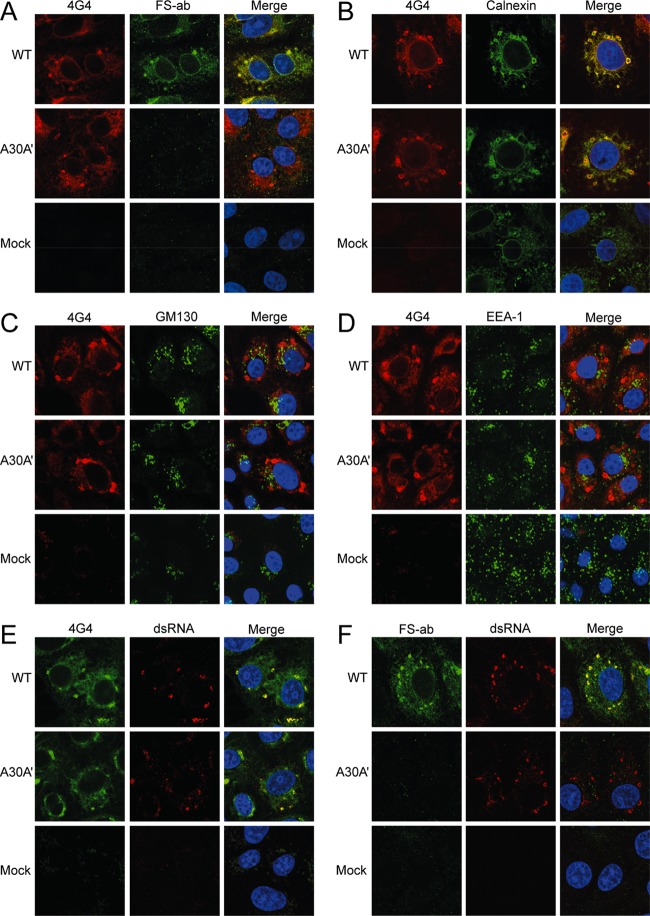
As 4G4 is able to detect both NS1 and NS1′, it is not possible to separate NS1 and NS1′ localization in cells infected with wild-type virus (WNVKUN), as it produces both proteins. FS-Ab, on the other hand, recognizes only NS1′ protein. We previously generated a mutant WNVKUN virus, A30A′, that does not produce NS1′ protein (23). Thus, we used comparative costaining with 4G4 and FS-Ab of cells infected with wild-type or A30A′ mutant WNVKUN viruses in order to assess whether NS1 and NS1′ proteins localize to the same or different cellular compartments. To examine any specific NS1 localization, Vero76 cells infected with either WNVKUN or A30A′ mutant virus at a multiplicity of infection (MOI) of 10 and fixed at 24 h postinfection (hpi) in 80% acetone were costained with 4G4 and FS-Ab. In WNVKUN-infected cells, 4G4-labeled proteins colocalized with FS-Ab-labeled proteins, indicating that NS1 and NS1′ are found in the same cellular compartments (Fig. 3A). To confirm that the cellular distribution of NS1 and NS1′ detected in transfected cells is the same as in infected cells, we carried out immunofluorescence analysis with antibodies detecting marker proteins for ER, Golgi apparatus, and endosomes. Vero76 cells infected at an MOI of 10 with WNVKUN or A30A′ viruses were fixed at 24 hpi (4% PFA in PBS with 0.1% Triton X-100) and stained with biotinylated 4G4 and either anti-calnexin, anti-GM130, or anti-EEA-1 antibodies. As in transfected cells, NS1 and NS1′ localized predominantly to the ER (Fig. 3B), with a small degree of colocalization with the Golgi apparatus (Fig. 3C) and no localization to the endosomes (Fig. 3D). A comparison between WNVKUN- and A30A′-infected cells should also indicate unique NS1′ staining. No differences in anti-NS1 staining between WNVKUN- and A30A′-infected cells indicate that NS1′ and NS1 localize to the same cellular compartments. These results, in combination with the results from plasmid-transfected cells, indicate that NS1′ protein has a cellular distribution similar to that of NS1 during viral infection or when the proteins are expressed as individual proteins.
Fig 3.
Localization of NS1′, NS1, and dsRNA in WNVKUN-infected cells. (A) Immunofluorescence analysis showing colocalization of NS1 and NS1′ in infected Vero76 cells using 4G4 (which stains both NS1 and NS1′) and FS-Ab (an NS1′-specific antibody). (B to D) Immunofluorescence analysis showing colocalization of virally expressed NS1 and NS1′ with the ER (B), the Golgi apparatus (C), and endosomes (D). Infected cells were stained with antibodies to appropriate cell markers (calnexin, GM130, and EEA-1, respectively) and biotinylated anti-NS1 (4G4). (E and F) Immunofluorescence analysis showing colocalization of NS1 and NS1′ with dsRNA in infected cells stained with anti-dsRNA and either biotinylated 4G4 (E) or FS-Ab (F).
NS1 has been shown previously to colocalize with double-stranded RNA (dsRNA) at the sites of flavivirus RNA replication in WNVKUN-infected cells (16). To determine whether NS1′ also colocalizes with dsRNA, Vero76 cells infected with WNVKUN or A30A′ (MOI of 10) were fixed at 24 hpi (80% acetone) and costained with anti-dsRNA antibodies and either biotinylated 4G4 or FS-Ab. Proteins stained with both 4G4 and FS-Ab colocalized with dsRNA (Fig. 3E and F, respectively), demonstrating that NS1′ is also associated with dsRNA and therefore with the sites of viral RNA replication.
NS1′ complements replication of NS1-deleted viral RNA.
Due to the colocalization of NS1′ with NS1 in the ER and with dsRNA, we hypothesized that NS1′, like NS1, could also play a role in virus replication. To test whether NS1′ can substitute for the function of NS1 in viral replication, we constructed a CMV promoter-driven WNVKUN genomic cDNA with a large (∼80%) internal deletion of the NS1 gene (pKUNdNS1) (Fig. 4A). This deletion (dNS1.1) has been used previously by our group in an RNA-based system to demonstrate trans-complementation of replication of NS1-deleted viral RNA by the NS1 expressed from WNVKUN replicon RNA (14). We cotransfected HEK293T cells with pKUNdNS1 and either pcDNA-NS1 or pcDNA-NS1′ plasmids to determine whether expression in trans of NS1 or NS1′ could rescue replication of replication-deficient pKUNdNS1. Transfection of repBHK cells expressing all of the WNVKUN nonstructural proteins, including NS1 and NS1′, with pKUNdNS1 was also performed as a positive control (14), and cotransfection of pKUNdNS1 with a green fluorescent protein (GFP)-expressing plasmid was included as a negative control. To examine rescue of RNA replication, total RNA was harvested from cotransfected cells at 2 or 4 days posttransfection with TriReagent (Sigma-Aldrich, St. Louis, MO), according to the manufacturer's instructions. Isolated RNA (5 μg) was subjected to denaturing 1.5% agarose gel electrophoresis followed by transfer to Hybond-N membranes (GE Healthcare Limited, Buckinghamshire, United Kingdom). RNA was cross-linked to the membrane by UV irradiation, and Northern hybridization with a 32P-labeled (PerkinElmer, Waltham, MA) WNVKUN-specific 3′ untranslated-region (UTR) probe was carried out to detect accumulation of viral RNA. No viral RNA was detected in mock-transfected cells (Fig. 4B, lanes 9 and 10), and transfection of repBHK cells with pKUNdNS1 resulted in increasing accumulation of viral genomic RNA (Fig. 4B, lanes 11 and 12). A small level of RNA accumulation in the pKUNdNS1-only-transfected cells was detected as expected (Fig. 4B, lanes 1 and 2), due to the transcription of NS1-deficient RNA driven by the CMV promoter. However, the levels of accumulated RNA were notably higher in cells cotransfected with either pcDNA-NS1 (a 5.5-fold increase at day 2 and a 10-fold increase at day 4) (Fig. 4B, lanes 3 and 4) or pcDNA-NS1′ plasmids (a 3-fold increase at day 2 and a 5-fold increase at day 4) (Fig. 4B, lanes 5 and 6) compared to those in pKUNdNS1-only-transfected cells. Notably, some variations in the Northern blot detection of complemented genomic RNA between three different complementation experiments (data not shown) were observed, producing a range of increases for pcDNA-NS1 complementation between 1.5- and 14-fold and for pcDNA-NS1′ complementation between 1- and 5-fold. No distinct band for genomic RNA was detected in cells cotransfected with GFP-expressing plasmid (Fig. 4B, lanes 7 and 8), possibly due to either an inhibitory effect of GFP expression on transcription of KUNdNS1 RNA or enhanced degradation of KUNdNS1 RNA in the presence of GFP expression. trans-complementation of viral RNA replication by NS1′ was further supported by immunofluorescence analysis of cotransfected HEK293T cells (fixed in 4% PFA with 0.1% Triton X-100 at 2 days posttransfection) using staining with mouse monoclonal anti-c-Myc antibodies (9E10 hybridoma; ATCC) (NS1 or NS1′ proteins) and rabbit polyclonal anti-NS3 antibodies (16) (pKUNdNS1-expressed NS3 protein). Increased production of NS3 protein in pcDNA-NS1- and pcDNA-NS1′-cotransfected cells, compared to pKUNdNS1-only-transfected cells (Fig. 4C), further demonstrated that trans-complementation of replication of the NS1-deleted viral RNA was successful. Therefore, we conclude that NS1′ protein can rescue replication deficiency of NS1-deleted viral RNA.
Fig 4.
NS1′ complements replication of NS1-deleted viral RNA. (A) Schematic diagram of pKUNdNS1 plasmid DNA containing a large deletion in the NS1 gene (amino acid 4 to amino acid 298) of WNVKUN genomic cDNA. CMV, cytomegalovirus promoter; HDVr, hepatitis delta virus ribozyme; pA, poly(A) signal; UTR, untranslated region. (B) Northern blot with a radiolabeled 3′-UTR probe showing replicating genomic RNA. RNA was harvested from cotransfected cells at 2 or 4 days posttransfection. (C) Immunofluorescence of HEK293T cells cotransfected with pKUNdNS1 and either NS1 or NS1′. Cotransfected cells were stained for c-Myc and NS3. (D) Immunofluorescence analysis showing production of infectious particles from cells cotransfected with pKUNdNS1 and either NS1 or NS1′. Culture fluid (CF) was harvested from cotransfected cells and used to infect repBHK cells. Infected repBHK cells were stained for E. (E) Titers were determined by infection of repBHK cells with serial dilutions of CF from cotransfected cells at day 2 (white bars) or 4 (gray bars) posttransfection and counting E-positive foci at day 2 postinfection. The graph is representative of two independent experiments. The difference between titers of complemented viruses expressing NS1 and NS1′ was not significant (P > 0.05), as determined by a standard one-way analysis of variance (ANOVA).
To detect virus production from cotransfected cells, and thus further confirm successful complementation, culture fluid from transfected HEK293T cells was harvested at 2 days posttransfection and used to infect repBHK cells. If complementation was successful, viral particles containing the NS1-deleted RNA would be able to infect the repBHK cells, replicate, and form viral particles due to the continuing expression of NS1 and NS1′ in the repBHK cells. Prior to infection, the culture fluid was treated with 10 units RQ1 DNase (Promega, Madison, WI) and 10 μg RNase A for 2 h at room temperature to digest any remaining plasmid DNA or uncoated RNA. Two days after infection, cells were fixed (4% PFA with 0.1% Triton X-100) and stained with anti-E antibodies to detect infected cells. Immunofluorescence images of repBHK cells infected with undiluted culture fluids and stained with 3.67G, a monoclonal anti-E antibody (30), showed that only cells that were cotransfected with either pcDNA-NS1 or pcDNA-NS1′ plasmids produced secreted viral particles (Fig. 4D). No secreted viral particles were produced in pKUNdNS1-only-transfected cells or in cells cotransfected with pKUNdNS1 and GFP-expressing plasmid (Fig. 4D). The lack of E-positive repBHK cells infected with DNase- and RNase-treated undiluted culture fluids from pKUNdNS1-only-transfected and pKUNdNS1-plus-GFP-expressing-plasmid-cotransfected cells also demonstrates that no transfected DNA or uncoated viral RNA was carried through to infection. To determine the titers of secreted viral particles, repBHK cells infected with serial 10-fold dilutions of collected culture fluids were stained with anti-E antibodies, and foci of E-positive cells were counted. Titration of viral particles was carried out for two independent complementation experiments, and the average viral titers were determined. The viral titers were similar from NS1- and NS1′-complemented cells, and both were similar to the viral titers obtained in repBHK cells at 2 days after transfection (Fig. 4E). Notably, the accumulation of viral RNA and viral titers in NS1 and NS1′ complementation experiments decreased from day 2 to day 4 after transfections, while transfection of pKUNdNS1 into repBHK cells led to a corresponding increase in RNA level and no decrease in viral titers (Fig. 4B and E). This is likely due to the ability of complemented virus to spread in repBHK cells, where 100% of cells express complementing NS1 and NS1′ proteins, while the spread of complemented virus is not possible in cotransfection experiments, as untransfected cells do not support replication and thus spread of complemented virus. From these results, we concluded that NS1′ could successfully complement deletion of NS1 in virus replication and that there was no significant difference in the efficiency of complementation between NS1 and NS1′.
We have shown in this study that NS1′ has a cellular distribution similar to that of NS1 with respect to the ER, Golgi apparatus, and endosomes, that both NS1 and NS1′ are colocalized in viral RNA replication sites in infected cells, and that NS1′ is able to rescue replication of NS1-deleted viral RNA. From our data it is reasonable to assume that NS1′ may not have a unique function in replication that is different from that of NS1 and thus may simply serve as an additional NS1 protein. Given that NS1′ does contain the entire NS1 protein, it was not entirely unexpected that NS1′ has a similar localization and can perform the same function(s) as NS1 in the virus life cycle. Although here we tested the function of NS1′ only in viral replication, NS1′ may also be involved in other reported functions of NS1, such as interactions with the complement system (17–20, 22) and inhibition of Toll-like receptor 3 (TLR3) signaling (21). Further studies are required to clarify this. However, the presence of the additional C-terminal 52 amino acids in the NS1′ protein compared to NS1 may also result in NS1′ having a unique function(s) in viral infection. Studies focused on identifying potential differences between the NS1 and NS1′ proteins are under way in an effort to explain the in vivo differences observed between NS1′-producing and NS1′-lacking WNVKUN viruses (23).
ACKNOWLEDGMENTS
We thank Jennifer Stow for providing antibodies to cellular markers, Roy Hall for providing various monoclonal antibodies, Paul Young for his helpful discussions, and Susann Liebscher for technical assistance with immunofluorescence.
This work was supported by National Health and Medical Research Council of Australia grant APP1009874.
Footnotes
Published ahead of print 12 June 2013
REFERENCES
- 1. Lindenbach BD, Rice CM. 2003. Molecular biology of flaviviruses. Adv. Virus Res. 59:23–61 [DOI] [PubMed] [Google Scholar]
- 2. Garmendia AE, Van Kruiningen HJ, French RA. 2001. The West Nile virus: its recent emergence in North America. Microbes Infect. 3:223–229 [DOI] [PubMed] [Google Scholar]
- 3. Rossi SL, Ross TM, Evans JD. 2010. West Nile virus. Clin. Lab. Med. 30:47–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lanciotti RS, Ebel GD, Deubel V, Kerst AJ, Murri S, Meyer R, Bowen M, McKinney N, Morrill WE, Crabtree MB, Kramer LD, Roehrig JT. 2002. Complete genome sequences and phylogenetic analysis of West Nile virus strains isolated from the United States, Europe, and the Middle East. Virology 298:96–105 [DOI] [PubMed] [Google Scholar]
- 5. Hall RA, Broom AK, Smith DW, Mackenzie JS. 2002. The ecology and epidemiology of Kunjin virus. Curr. Top. Microbiol. Immunol. 267:253–269 [DOI] [PubMed] [Google Scholar]
- 6. Audsley M, Edmonds J, Liu W, Mokhonov V, Mokhonova E, Melian EB, Prow N, Hall RA, Khromykh AA. 2011. Virulence determinants between New York 99 and Kunjin strains of West Nile virus. Virology 414:63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hall RA, Scherret JH, Mackenzie JS. 2001. Kunjin virus: an Australian variant of West Nile? Ann. N. Y. Acad. Sci. 951:153–160 [PubMed] [Google Scholar]
- 8. Westaway EG, Mackenzie JM, Khromykh AA. 2003. Kunjin RNA replication and applications of Kunjin replicons. Adv. Virus Res. 59:99–140 [DOI] [PubMed] [Google Scholar]
- 9. Westaway EG, Mackenzie JM, Khromykh AA. 2002. Replication and gene function in Kunjin virus. Curr. Top. Microbiol. Immunol. 267:323–351 [DOI] [PubMed] [Google Scholar]
- 10. Khromykh AA, Westaway EG. 1994. Completion of Kunjin virus RNA sequence and recovery of an infectious RNA transcribed from stably cloned full-length cDNA. J. Virol. 68:4580–4588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Coia G, Parker MD, Speight G, Byrne ME, Westaway EG. 1988. Nucleotide and complete amino acid sequences of Kunjin virus: definitive gene order and characteristics of the virus-specified proteins. J. Gen. Virol. 69:1–21 [DOI] [PubMed] [Google Scholar]
- 12. Speight G, Coia G, Parker MD, Westaway EG. 1988. Gene mapping and positive identification of the non-structural proteins NS2A, NS2B, NS3, NS4B and NS5 of the flavivirus Kunjin and their cleavage sites. J. Gen. Virol. 69:23–34 [DOI] [PubMed] [Google Scholar]
- 13. Chu PW, Westaway EG. 1992. Molecular and ultrastructural analysis of heavy membrane fractions associated with the replication of Kunjin virus RNA. Arch. Virol. 125:177–191 [DOI] [PubMed] [Google Scholar]
- 14. Khromykh AA, Sedlak PL, Westaway EG. 2000. cis- and trans-acting elements in flavivirus RNA replication. J. Virol. 74:3253–3263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lindenbach BD, Rice CM. 1997. trans-Complementation of yellow fever virus NS1 reveals a role in early RNA replication. J. Virol. 71:9608–9617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Westaway EG, Mackenzie JM, Kenney MT, Jones MK, Khromykh AA. 1997. Ultrastructure of Kunjin virus-infected cells: colocalization of NS1 and NS3 with double-stranded RNA, and of NS2B with NS3, in virus-induced membrane structures. J. Virol. 71:6650–6661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Avirutnan P, Punyadee N, Noisakran S, Komoltri C, Thiemmeca S, Auethavornanan K, Jairungsri A, Kanlaya R, Tangthawornchaikul N, Puttikhunt C, Pattanakitsakul SN, Yenchitsomanus PT, Mongkolsapaya J, Kasinrerk W, Sittisombut N, Husmann M, Blettner M, Vasanawathana S, Bhakdi S, Malasit P. 2006. Vascular leakage in severe dengue virus infections: a potential role for the nonstructural viral protein NS1 and complement. J. Infect. Dis. 193:1078–1088 [DOI] [PubMed] [Google Scholar]
- 18. Chung KM, Liszewski MK, Nybakken G, Davis AE, Townsend RR, Fremont DH, Atkinson JP, Diamond MS. 2006. West Nile virus nonstructural protein NS1 inhibits complement activation by binding the regulatory protein factor H. Proc. Natl. Acad. Sci. U. S. A. 103:19111–19116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kurosu T, Chaichana P, Yamate M, Anantapreecha S, Ikuta K. 2007. Secreted complement regulatory protein clusterin interacts with dengue virus nonstructural protein 1. Biochem. Biophys. Res. Commun. 362:1051–1056 [DOI] [PubMed] [Google Scholar]
- 20. Schlesinger JJ. 2006. Flavivirus nonstructural protein NS1: complementary surprises. Proc. Natl. Acad. Sci. U. S. A. 103:18879–18880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wilson JR, de Sessions PF, Leon MA, Scholle F. 2008. West Nile Virus nonstructural protein 1 inhibits TLR3 signal transduction. J. Virol. 82:8262–8271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Avirutnan P, Fuchs A, Hauhart RE, Somnuke P, Youn S, Diamond MS, Atkinson JP. 2010. Antagonism of the complement component C4 by flavivirus nonstructural protein NS1. J. Exp. Med. 207:793–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Melian EB, Hinzman E, Nagasaki T, Firth AE, Wills NM, Nouwens AS, Blitvich BJ, Leung J, Funk A, Atkins JF, Hall R, Khromykh AA. 2010. NS1′ of flaviviruses in the Japanese encephalitis virus serogroup is a product of ribosomal frameshifting and plays a role in viral neuroinvasiveness. J. Virol. 84:1641–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mason PW, McAda PC, Dalrymple JM, Fournier MJ, Mason TL. 1987. Expression of Japanese encephalitis virus antigens in Escherichia coli. Virology 158:361–372 [DOI] [PubMed] [Google Scholar]
- 25. Firth AE, Atkins JF. 2009. A conserved predicted pseudoknot in the NS2A-encoding sequence of West Nile and Japanese encephalitis flaviviruses suggests NS1′ may derive from ribosomal frameshifting. Virol. J. 6:14–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mason PW. 1989. Maturation of Japanese encephalitis virus glycoproteins produced by infected mammalian and mosquito cells. Virology 169:354–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Macdonald J, Tonry J, Hall RA, Williams B, Palacios G, Ashok MS, Jabado O, Clark D, Tesh RB, Briese T, Lipkin WI. 2005. NS1 protein secretion during the acute phase of West Nile virus infection. J. Virol. 79:13924–13933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mackenzie JM, Jones MK, Young PR. 1996. Immunolocalization of the dengue virus nonstructural glycoprotein NS1 suggests a role in viral RNA replication. Virology 220:232–240 [DOI] [PubMed] [Google Scholar]
- 29. Gillespie LK, Hoenen A, Morgan G, Mackenzie JM. 2010. The endoplasmic reticulum provides the membrane platform for biogenesis of the flavivirus replication complex. J. Virol. 84:10438–10447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Adams SC, Broom AK, Sammels LM, Hartnett AC, Howard MJ, Coelen RJ, Mackenzie JS, Hall RA. 1995. Glycosylation and antigenic variation among Kunjin virus isolates. Virology 206:49–56 [DOI] [PubMed] [Google Scholar]