Abstract
The human parainfluenza virus (hPIV) hemagglutinin-neuraminidase (HN) protein binds (H) oligosaccharide receptors that contain N-acetylneuraminic acid (Neu5Ac) and cleaves (N) Neu5Ac from these oligosaccharides. In order to determine if one of HN′s two functions is predominant, we measured the affinity of H for its ligands by a solid-phase binding assay with two glycoprotein substrates and by surface plasmon resonance with three monovalent glycans. We compared the dissociation constant (Kd) values from these experiments with previously determined Michaelis-Menten constants (Kms) for the enzyme activity. We found that glycoprotein substrates and monovalent glycans containing Neu5Acα2-3Galβ1-4GlcNAc bind HN with Kd values in the 10 to 100 μM range. Km values for HN were previously determined to be on the order of 1 mM (M. M. Tappert, D. F. Smith, and G. M. Air, J. Virol. 85:12146–12159, 2011). A Km value greater than the Kd value indicates that cleavage occurs faster than the dissociation of binding and will dominate under N-permissive conditions. We propose, therefore, that HN is a neuraminidase that can hold its substrate long enough to act as a binding protein. The N activity can therefore regulate binding by reducing virus-receptor interactions when the concentration of receptor is high.
INTRODUCTION
The human parainfluenza viruses (hPIVs) are paramyxoviruses that cause croup, bronchitis, and pneumonia in children under age 6 years. The hPIVs have two envelope glycoproteins: the hemagglutinin-neuraminidase (HN) and the fusion protein (F). HN binds to (H) and cleaves (N) N-acetylneuraminic acid (Neu5Ac; sialic acid) on target cells and, upon binding, activates the fusion protein to assist in viral entry (1–3).
HN, like influenza virus hemagglutinin, is known to be a receptor-binding protein. Both of these proteins confer the ability to bind to sialic acids on red blood cells (RBCs), and the inhibition of cell adsorption by monoclonal antibodies that also block hemagglutination suggests that hemagglutination corresponds to receptor binding (4). While the physiological receptor(s) for hPIV has not yet been identified, the binding specificity of the hPIVs in vitro is known. hPIVs bind to structures containing the motif Neu5Acα2-3Galβ1-4GlcNAc on a glycan array or on a thin-layer chromatography plate spotted with gangliosides (5, 6). hPIV type 1 (hPIV1) and hPIV2 tolerate modifications to the motif, including sulfation of Gal, fucosylation of GlcNAc (sialyl-Lewisx), and extension with additional lactosamines or branched glycan structures, while hPIV3 requires at least one extra Gal for binding and tolerates fucosylation but not sulfation or branching (5, 7). Binding may be limited to motifs on N-glycans or glycolipids, because inhibition of glycolipid formation or N-glycosylation renders COS-7 cells resistant to Newcastle disease virus (NDV) infection, while inhibitors of O-glycosylation do not (8).
We previously found that HN can cleave the same motifs that it binds and, in the case of hPIV2 and hPIV3, some nonreceptor glycans as well. Identical glycans are more susceptible to N when fixed to a surface (e.g., attached to a glycan array or glycoprotein or lipid on an RBC membrane) than when attached to a glycoprotein in solution. Our findings suggest that N can be activated by interaction with substrate on a surface, perhaps through a conformational change in the HN protein (7).
Because HN can both bind and destroy the same set of receptor structures, alterations in the balance between H and N activities are detrimental to the virus. hPIV3 produced in receptor-depleted cells, a growth condition that mimics increased N, generates escape mutants with an increased affinity of H for the ligand that offsets the higher N activity (9). The T193A and H552Q mutants of hPIV3, which have high H (measured as hemadsorption) and wild-type N (measured as cleavage of fluorescent substrate) activity, require exogenous neuraminidase for optimal growth in an airway epithelium model, again suggesting that N must increase to balance the increase in H (10). Although it was originally thought that the different pH optima for binding (pH 7) and neuraminidase activity (approximately pH 5) indicated that the former occurs at the cell surface, while the latter occurs intracellularly (11), these findings suggest that N must also act at the cell surface.
H-N imbalances may cause detrimental effects on the virus by causing abnormal fusion with the target cell. The high-H hPIV3 mutants with the H552Q and T193A mutations both demonstrate a combination of high fusogenicity and poor growth (10, 12), suggesting that a mismatch in binding and cleavage activity leads to suboptimal fusion triggering. One possible explanation is that fusion is triggered before the virus reaches the proper distance from the cell surface for fusion peptide insertion. Alternatively, excessive fusogenicity might lead to a multiplicity of infection higher than that which is optimal for virus production.
The question of whether HN balances its binding and cleavage functions by partially separating them into two active sites is a subject of much debate. Many experiments yield results consistent with a single site. For example, monoclonal antibodies raised against hPIV1 HN inhibit both H and N or neither H nor N, suggesting that the two activities use the same site or two sites that are structurally connected (13). In wild-type hPIV3, the N-site inhibitor zanamivir blocks H activity, fusion, and plaque formation (14) and competes with red blood cell binding (15), suggesting that there is no second site available for binding if the N site is occupied. Only one sialic acid per HN monomer is observed in the crystal structure of wild-type hPIV3 HN, and it is located at the N site (16). On the other hand, a pH 6.3 crystal structure of NDV HN shows sialic acid binding in the N site and also at the dimer interface (17). There is some evidence for this second site in certain hPIV mutants; sialic acid can be modeled into the putative second site in the hPIV3 H552Q mutant (18). H552Q is at the dimer interface and confers the ability to bind RBCs in the presence of N-site inhibitors (18, 19), suggesting that the increased H avidity is the result of a secondary sialic acid binding site in this region. A similar second site may also exist in hPIV1, although its function is limited by a nearby N glycan in the wild-type virus (20).
Several key aspects of HN binding and activity in hPIV are still unknown, including the binding affinity and avidity of candidate receptors and how the binding of HN differs upon N cleavage of sialic acid and during any intermediate transition state(s) of the enzyme. We now report on the quantification of the binding of H to potential glycoprotein and small-molecule ligands, which we previously used as cleavage substrates, using a colorimetric binding assay and surface plasmon resonance. We then compared the dissociation constant (Kd) values to the enzymatic Michaelis-Menten constant (Km) and found that Km was greater than Kd. We propose that N acts as a regulator of binding by reducing high concentrations of receptors, thus preventing unwanted fusion events that might occur in the presence of excess binding.
MATERIALS AND METHODS
Virus culture and purification.
hPIV1 strain C35 was from ATCC. hPIV2 strain Oklahoma/3955/2005 and hPIV3 strain Oklahoma/410/2009 were obtained from the virology laboratory at the University of Oklahoma Children's Hospital. All viruses were grown in LLC-MK2 cells, purified by sucrose gradient centrifugation, and assayed for hemagglutination titer as previously described (5, 7). Total viral protein and total HN in a virus preparation were determined by gel electrophoresis and comparison of Coomassie blue-stained band intensities with the intensity of a bovine serum albumin (BSA) standard, as previously described (7).
Expression and preparation of purified monomeric HN.
Gene constructs were designed as follows: coding sequences for an N-terminal honeybee mellitin secretion signal and a C-terminal 6-histidine (His) tag were added to the sequence of the globular domain of hPIV1 strain Oklahoma/4409/2010 (GenBank accession number JN089925; amino acids 119 to 575 [21]), hPIV3 strain Oklahoma/410/2009 (GenBank accession number JF912197; amino acids 125 to 572 [17]), or hPIV2 strain Oklahoma/283/2009 (GenBank accession number JF912195; amino acids 103 to 571, indicated by sequence alignment with hPIV1 and hPIV3). Each construct was codon optimized for expression in the Spodoptera frugiperda cell line Sf9, synthesized, and ligated into the ampicillin resistance plasmid pMA between 5′ SalI and 3′ NotI restriction sites by GeneArt, Inc. pMA plasmid containing the recombinant HN gene was amplified in OneShot MAX Efficiency DH5α-T1R competent Escherichia coli (Invitrogen) following the protocol included with the cells. Amplified plasmid was purified using a plasmid midikit (Qiagen). The HN gene was sequentially digested with SalI and NotI (Invitrogen), purified by electrophoresis, and eluted from the gel using a QIAquick gel extraction kit. The excised gene was ligated into pFastBac (Invitrogen) between the SalI and NotI sites with T4 DNA ligase for 30 min at 37°C (Invitrogen). Recombinant pFastBac-HN was amplified in OneShot MAX Efficiency DH5α-T1R competent E. coli, and the successful integration of the HN gene was verified by SalI and NotI digestion. Bacmid was generated from pFastBac-HN using MAX Efficiency DH10Bac competent E. coli (Invitrogen) following the company's protocols. Bacmid DNA was purified using a Qiagen large construct kit. Proper insertion of the HN gene into the bacmid was verified by PCR using the following primers: hPIV1 fwd, 5′-ACCATCTCCGGTTGCGTGCG-3′; hPIV1 rev, 5′-CGTTCACAGCGTCGGGGGAC-3′; hPIV2 fwd, 5′-GAAGTTCCTCGTGAACGTG-3′; hPIV2 rev, 5′-GCGGAGGAATGGGATGAT-3′; hPIV3 fwd, 5′-ATGAAGTTCCTCGTGAACG-3′; and hPIV3 rev, 5′-ATCTCGGTCTTGAACAGCA-3′. PCR was performed in an MJ Research programmable thermal cycler programmed as follows: 94°C for 5 min; 30 cycles of 94°C for 1 min, 50°C for 1 min, and 72°C for 1 min; and ending with 72°C for 5 min. Agarose gel visualization of a band at the appropriate ∼1,800-kb size was considered confirmation of successful insertion.
Purified hPIV1, -2, or -3 HN bacmid DNA was transfected into Sf9 cells as follows: cells were plated in 6-well tissue culture plates (Falcon) at a density expected to yield 70 to 80% confluence and allowed to adhere for 1 h at 27°C in BaculoGold medium (BD) without antibiotics. Cells were transfected using Cellfectin II reagent (Invitrogen) following the protocols included with the reagent. Following transfection, cells were maintained in BaculoGold growth medium (BD). After 72 h, the neuraminidase (N) activity of the culture supernatant was determined by the fluorescence method of Potier et al. (22) adapted for 96-well microplates, as previously described (7). Supernatant with N activity was collected as the passage 1 (P1) stock and used to infect Sf9 cells to generate a P2 stock. P2 stock was used to infect Sf9 cells to generate HN protein using 500 μl P2 supernatant per T175 flask of Sf9 cells.
To purify HN, ≥200 ml culture medium from Sf9 cells infected with P2 stock was centrifuged at 1,000 × g for 5 min in a GS-6R tabletop centrifuge (Beckman) to remove cell debris and then purified on 5 ml of a His-Select nickel-nitrilotriacetic acid (NTA) affinity gel (Sigma) in a 1-cm-diameter column following the protocol included with the affinity resin. Eluted fractions with N activity were pooled, concentrated in a 10,000-molecular-weight-cutoff Amicon ultracentrifugal filter (5-ml volume in a 15-ml conical tube; Millipore) at 2,000 × g, washed with CaMgBBS (0.15 M NaCl, 0.25 mM CaCl2, 0.8 mM MgCl2, 20 mM H3BO3, 0.13 mM Na2B4O7, pH 7.2) to remove imidazole, and concentrated to a final volume of approximately 100 μl. The amount of HN present in each sample was quantified by SDS-PAGE as previously described (7). Briefly, the intensity of the stained HN band was compared to the intensities of known amounts of BSA on the same gel. N activity was determined using the substrates 2′-(4-methylumbelliferyl)-α-d-N-acetylneuraminic acid (MUN), 3′-N-acetylneuraminyllactose (3′NANL), bovine fetuin (bFetuin), bFetuin digested with proteinase K (bFetuin peptides), and human α1-acid glycoprotein (hAGP) as previously described (7).
The oligomeric state of the purified HN was determined by fast protein liquid chromatography (FPLC) over a Superdex 200 column. To calibrate the column, a mixture of 400 μg each of goat IgG (Sigma), bovine serum albumin (Sigma), hAGP (Sigma), trypsin (Worthington), and fungal proteinase K (Invitrogen) was made in FPLC running buffer (50 mM Tris, 3 mM NaN3, 150 mM NaCl, pH 8). The mixture was run on a UPC-900 FPLC (Amersham Biosciences) at 4°C and a flow rate of 0.5 ml/min with a 2.02-ml loop emptying wash, and 0.5-ml fractions were collected with a Frac-920 fraction collector (General Electric). Each fraction was subjected to 9% SDS-PAGE with Coomassie blue staining. hPIV1, -2, or -3 HN was then diluted to a 1-ml volume with FPLC running buffer and subjected to FPLC under the same conditions. Each fraction was assayed for N activity, and fractions with N activity were subjected to 9% SDS-PAGE with Coomassie blue staining.
Preparation of biotinylated glycoprotein ligands.
hAGP was from Sigma. bFetuin was purified from fetal calf serum (Sigma) by the method of Spiro (23). hAGP was dissolved in phosphate-buffered saline (PBS) to a concentration of 30 mg/ml (0.75 mM), and bFetuin was dissolved to a concentration of 40 mg/ml (0.83 mM). Both proteins were biotinylated for 30 min at room temperature with a 20-fold molar excess of EZ-Link sulfo-NHS-LC-biotin (sulfosuccinimidyl-6-[biotin-amido]hexanoate, 10 mM stock solution; Thermo Scientific). Biotinylated proteins were centrifuged at 2,000 × g in a GS-6R tabletop centrifuge (Beckman) in a 10,000-molecular-weight-cutoff Amicon ultracentrifugal filter (Millipore) to reduce the PBS and unreacted biotin, washed with CaMgBBS, and concentrated to a volume of ≤0.5 ml. Successful biotinylation was confirmed by 9% SDS-PAGE, blotting onto a polyvinylidene difluoride (PVDF) membrane, blocking the blot with 2% dry milk in rinse buffer (0.14 M NaCl, 3 mM KH2PO4, 8 mM Na2HPO4, 2.5 mM KCl, 0.05% Tween 20), binding to a 1:1,000 dilution of alkaline phosphatase-coupled streptavidin (streptavidin-AP; Promega) in antibody buffer (10% supplemented calf serum [HyClone] in rinse buffer), and detecting with 5-bromo-4-chloro-3′-indolylphosphate–nitro blue tetrazolium chloride (BCIP-NBT; Sigma). The final protein concentration was determined using a Bio-Rad protein assay with BSA (Pierce) as the standard.
Quantification of glycoprotein ligand binding to immobilized virions.
A total of 32 hemagglutination units (HAU) of hPIV1, -2, or -3 in 50 μl CaMgBBS was bound to each well of a 96-well flat-bottom enzyme-linked immunosorbent assay (ELISA) plate (Microlon; Greiner Bio-One) for 16 h at 4°C. The plate was then blocked with 2% BSA (Sigma) in CaMgBBS for 1 h at 37°C and washed three times with CaMgBBS in an ELx405 auto plate washer (BioTek Instruments). After the wash, 50-μl 2-fold dilutions of biotinylated bFetuin or hAGP in 1% BSA–CaMgBBS were made in rows 1 and 2 of the plate. Fifty microliters of a 1:1,000 dilution of goat anti-hPIV1 (US Biologicals), guinea pig anti-hPIV2, or guinea pig anti-hPIV3 (BEI Resources) in 1% BSA–CaMgBBS was added to each well of row 3 of the plate as a control for total virus binding, and 50 μl 1% BSA–CaMgBBS was added to rows 4 and 5. The plate was incubated for 1 h at room temperature and then washed as before. After the wash, 50 μl of a 1:1,000 dilution of streptavidin-AP was added to each well of rows 1, 2, and 4. Fifty microliters of alkaline phosphatase-coupled anti-goat or anti-guinea pig serum (Sigma) was added to each well of row 3. The plate was again incubated for 1 h at room temperature and then washed. Ten microliters of 1% BSA–CaMgBBS was added to each well of row 5, and 2-fold dilutions of streptavidin-AP were made. Fifty microliters of p-nitrophenylphosphate (pNPP; 1 tablet in 10 ml; Sigma) in diethanolamine buffer (10% [vol/vol] diethanolamine, pH 9.8) was added to each well. The assay was allowed to develop at room temperature for 30 min, and then it was read at 405 nm in a SpectraMax M2 microplate reader using SoftMax Pro software. In this setup, rows 1 and 2 measure the ligand bound to virus, row 3 is a control for the amount of virus bound to the plate, row 4 is a control for the streptavidin-AP background, and row 5 is a control for the relationship between the absorbance and the amount of streptavidin-AP present. All experiments were repeated three times. Binding was also measured at pH 5.5, substituting 1% BSA in 50 mM NaCH3COO−, 4 mM CaCl2 for 1% BSA in CaMgBBS. In the pH 5.5 experiments, all binding steps were performed at 4°C to minimize neuraminidase activity. In a separate experiment, each glycoprotein ligand was bound to a blocked ELISA plate without virus for 1 h at 37°C, and the plate was washed, developed with streptavidin-AP as described above, and read to determine the nonspecific binding of protein to the plate. The nonspecific binding of the protein to the plate and the streptavidin-AP background were subtracted from each virus-ligand binding curve.
Analysis of glycoprotein ligand binding curves.
Subtracted binding curves were first fit to the Hill equation using Prism v.4 software to determine whether ligand is bound by multiple interacting sites. The Hill equation is as follows: Y = (Bmax · xh)/(xh · Kdh), where Y is the absorbance minus background, x is the concentration of ligand, Bmax is the maximal binding in absorbance units, Kd is the ligand concentration that gives half-maximal binding, and h is the Hill constant. All curves for which the model failed or yielded a value of h that was near 1 were then fit to two hyperbolic binding equations, as follows, to determine whether binding is mediated by a single site (model 1) or by two independent sites with different Kd values (model 2). The equation for one-site binding is Y = (Bmax · x)/(Kd + x), and the equation for two-site binding is Y = [(Bmax1 · x)/(Kd1 + x)] + [(Bmax2 · x)/(Kd2 + x)], where Bmax is the maximal binding of a single site in absorbance units; Kd is the ligand concentration that gives half-maximal binding at a single site; Bmax1 and Bmax2 are the maximal binding of sites 1 and 2, respectively; and Kd1 and Kd2 are the ligand concentrations that give half-maximal binding at sites 1 and 2, respectively. All Bmax and Kd values were constrained to be >0. The two hyperbolic binding models were compared using the extra-sum-of-squares F test in Prism v.4, according to the following equation F = [(SS1 − SS2)/(DF1 − DF2)]/(SS2/DF2), where SS1 and SS2 are the sum-of-squares values for models 1 and 2, respectively, and DF1 and DF2 are the degrees of freedom of models 1 and 2, respectively. The simpler model was considered a statistically significant better fit when P was <0.05 or when the two-site model failed, as seen by 95% confidence intervals that spanned orders of magnitude.
Surface plasmon resonance.
Biotinylated Galβ1-4GlcNAcβ1-3Galβ1-4GlcNAc (dilactosamine [DiLN]), Neu5Acα2-3Galβ1-4GlcNAcβ1-3Galβ1-4GlcNAc (3′-sialyl-dilactosamine [3′SiaDiLN]), Neu5Acα2-3Galβ1-4GlcNAcβ1-3Galβ1-3GlcNAc, and Neu5Acα2-3Galβ1-4Glc (3′-N-acetylneuraminyllactose [NANL]) were obtained from the Consortium for Functional Glycomics Core D (The Scripps Research Institute, San Diego, CA). All surface plasmon resonance experiments were performed on a Biacore 3000 instrument (General Electric) running v.3.2 control software. Each glycan was dissolved to a concentration of 0.5 μg/ml in CaMgBBS with 0.05% surfactant P20 (General Electric). Glycans were captured on a streptavidin sensor chip (General Electric) as 10 μl of 0.01 μM glycan at a flow rate of 2 μl/min, using degassed CaMgBBS-P20 as running buffer. DiLN was captured on flow cell 1 as a negative-control surface, 3′SiaDiLN was captured on flow cell 2, Neu5Acα2-3Galβ1-4GlcNAcβ1-3Galβ1-3GlcNAc was captured on flow cell 3, and NANL was captured on flow cell 4. Solutions of 10, 7.5, 5, 2.5, 1.25, and 0.625 μM of hPIV1, -2, or -3 HN were made in CaMgBBS-P20, and 5 μl of each dilution was injected over the sensor chip at 5 μl/min. All experiments were performed twice. A 50-μl wash was used to regenerate the chip between each binding. All binding experiments were performed at room temperature. Sensorgrams were recorded in the Fc2-1, Fc3-1, and Fc4-1 modes and processed using BIAEvaluation v.3.2 RC1 software. All baselines were defined as 0, wash steps and air spikes were removed, and successive bindings were overlaid to generate a composite curve for the concentration series. The association constant (Ka) and dissociation constant (Kd) were then determined from the association and dissociation phases of the curve using the Langmuir 1:1 binding model. The model fit was evaluated by visual assessment, residuals, and chi-square values.
Use of photographic processing software in construction of figures.
Adobe Photoshop CS3 software was used to crop duplicate lanes from the SDS-polyacrylamide gel in Fig. 2A. In all other figures, Photoshop was used only to assemble data into a final image file.
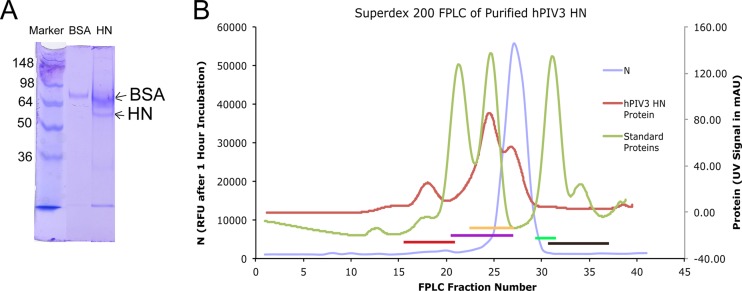
Fig 2.
(A) Partially purified HN after Ni-NTA affinity chromatography. Lane 1, SeeBlue Plus2 standards; lane 2, BSA standard for comparison; lane 3, PIV3 HN sample. The upper band was determined to be BSA by mass spectrometry. The band immediately below was confirmed to be HN by mass spectrometry. All lower bands were also determined to be HN fragments by mass spectrometry and likely represent degradation products. Numbers on the left are molecular masses (in kilodaltons). (B) HN (detected by neuraminidase activity; blue trace) elutes from FPLC after IgG (red bar), BSA (purple bar), and hAGP (yellow bar) but before trypsin (green bar) and proteinase K (brown bar), consistent with a monomer with a molecular mass of ∼45 kDa. Results for hPIV3 are shown; results for hPIV1 were similar, while for hPIV2, the N activity eluted earlier, indicating a dimer. mAU, milli-absorbance units.
RESULTS
hPIV binding to glycoprotein ligands.
In our previous study, we quantified the enzyme activity of HN on hPIV1, -2, and -3 virions and obtained Km values (7). In order to determine the relative strengths of H and N and test for the presence of a second binding site, we quantified the binding of immobilized hPIV virions to soluble hAGP and bFetuin, which are 40- to 50-kDa glycoproteins with highly branched sialylated glycans that were previously shown to be substrates for hPIV N (7).
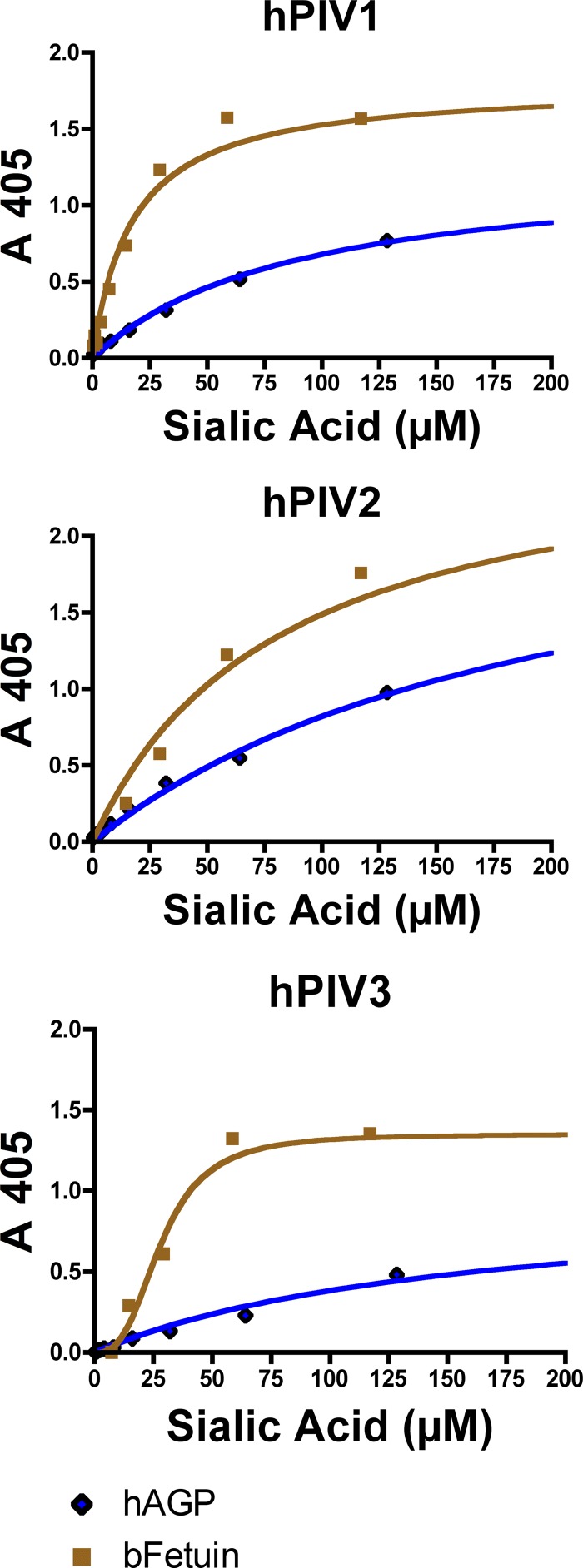
hPIV1, -2, and -3 virions all bound hAGP and bFetuin in a colorimetric binding assay at pH 7 (Fig. 1). The binding curves were fitted to three different models of binding site-ligand interaction: a model for interacting binding sites (the Hill equation), a model for two different kinds of independent binding sites, and a model for one binding site. The Hill equation models interacting binding sites, where an h value of ≈1 implies that all binding sites are independent and an h value not equal to 1 indicates cooperativity among sites. For all binding experiments except the experiment with hPIV3-fetuin, the Hill model yielded an h value near 1. For hPIV3 binding fetuin, h was equal to 3 ± 0.5.
Fig 1.
Binding of hPIV1, -2, and -3 HN (as whole virus) to biotinylated glycoprotein ligands at pH 7. In these experiments, the virus was immobilized and the glycoprotein ligand was in solution. Binding curves from one representative experiment are shown. Note that all three viruses exhibit a higher affinity for bFetuin than hAGP.
Virus-ligand binding curves, except for the curve for PIV3-fetuin, which gave a sigmoidal binding curve, were fit to two hyperbolic binding equations. The one-site binding equation assumes that there is only one kind of binding site per protein unit, while the two-site binding equation assumes that there are two kinds of binding sites, each with its own dissociation constant. In all of the 15 comparisons between the one-site and two-site binding models, the simpler one-site model was the best fit for the data. The two-site model either failed to be fitted or gave 95% confidence limits orders of magnitude wide.
The Kd values calculated from the one-site hyperbolic binding models (hPIV1 and -2 and hPIV3 binding hAGP) or the Hill model (hPIV3 binding fetuin) are shown in Table 1. At pH 7 and room temperature, all three viruses bound bFetuin with at least a 3-fold higher affinity (3-fold lower Kd) than hAGP. Kd ranged from 7.7 μM sialic acid for hPIV1 binding bFetuin to 218 μM sialic acid for hPIV2 binding hAGP. Binding at pH 5.5 was not detectable using this assay, and preliminary experiments showed that binding at 37°C was not significantly different from that at room temperature (see data at www.functionalglycomics.org under Glycan Array:1033).
Table 1.
Kd values for hPIV binding to glycoprotein ligands determined from colorimetric binding assaya
| Virus | Ligand | Kd (μM cleavable sialic acidb) |
|---|---|---|
| hPIV1 | hAGP | 90.5 ± 10.6 |
| bFetuin | 20.6 ± 2.8 | |
| hPIV2 | hAGP | 218.2 ± 10.9 |
| bFetuin | 7.7 ± 1.8 | |
| hPIV3 | hAGP | 124.7 ± 30.2 |
| bFetuin | 33.7 ± 6.7 |
Kd values were calculated from the experiments whose results are shown in Fig. 1.
Cleavable sialic acid was used for comparison with Km values.
Recombinant hPIV HN binding to monovalent ligands.
Monovalent interactions between a glycan with one sialic acid (such as NANL) and a single HN binding site are too low in affinity to resist washing steps, and these ligands are therefore unsuitable for the 96-well colorimetric binding assay. However, binding is detectable on the glycan array, where fixation of the ligands to a solid support at high density allows an effectively multivalent interaction with intact virions. In our previous study, we noted that monovalent, small-molecule substrates differed from glycoprotein substrates in their neuraminidase susceptibility (7), and we therefore wished to determine whether differences in binding existed as well. To determine the binding of HN to monovalent ligands, we expressed soluble HN in insect cells and measured its interaction with ligand using surface plasmon resonance. In surface plasmon resonance, the low-affinity monovalent interactions are detectable because no washing is required when measuring binding.
Sf9 cell culture medium had measurable neuraminidase activity on MUN substrate at 4 days posttransfection with bacmid DNA and at 3 days postinfection with P1 or P2 recombinant baculovirus stock. Partial purification of hPIV1, -2, or -3 HN by Ni-NTA affinity column and concentration gave a protein preparation that separated into two bands on SDS-PAGE, an upper band at ∼70 kDa and a lower band at ∼45 kDa (Fig. 2A). Mass spectrometry of the two bands confirmed that the upper band is BSA and the lower band is HN. HN from hPIV1 and -3, measured by N activity, eluted from a Superdex 200 column slightly after hAGP but before trypsin (Fig. 2B), which is the position expected for a monomer. HN from hPIV2 eluted slightly before hAGP, as would be expected of a dimer (data not shown). Despite its monomeric or dimeric state, in comparison to a tetramer state on the virus (24), purified hPIV2 and -3 HN exhibited neuraminidase activity with Km values similar to those of the HN on intact virions of the same strain. The kcat values for purified HN monomers or dimers were larger than the kcat values for virion HN for glycoprotein substrates but not for small-molecule substrates (Table 2).
Table 2.
Km and kcat of purified HNa
| Virus | Substrate | Purified HN |
Whole virusb |
||
|---|---|---|---|---|---|
| Km (mM cleavable sialic acid, pH 5.5, 37°C) | kcat (s−1) | Km (mM cleavable sialic acid) | kcat (s−1) | ||
| hPIV2 | MUN | 0.44 ± 0.07 | 49 ± 15 | 0.18 ± 0.04 | 57 ± 3.4 |
| NANL | 0.50 ± 0.03 | 4.2 ± 1.1 | 0.47 ± 0.06 | 10 ± 1.7 | |
| bFetuin | 0.34 ± 0.08 | 8.2 + 2.2 | 0.40 + 0.07 | 1.5 ± 0.08 | |
| bFetuin peptides | 0.43 ± 0.11 | 10 ± 1.3 | 1.22 ± 0.24 | 8.1 ± 0.69 | |
| hAGP | 0.98 ± 0.16 | 16 ± 2.2 | 0.64 ± 0.04 | 4.76 + 0.72 | |
| hPIV3 | MUN | 4.9 ± 0.49 | 16 ± 1.5 | 8.2 ± 0.67 | 12 ± 2.3 |
| NANL | —c | — | — | — | |
| bFetuin | — | — | — | — | |
| bFetuin peptides | — | — | — | — | |
| hAGP | 1.4 ± 0.31 | 3.5 + 0.62 | 2.5 + 0.5 | 1.36 ± 0.22 | |
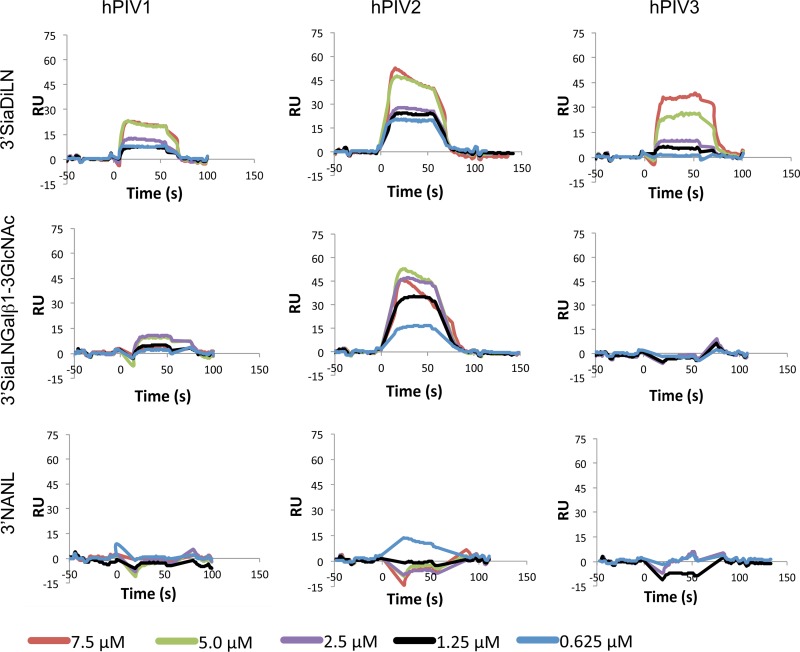
The binding of soluble hPIV1, -2, and -3 HN to three ligands was investigated by surface plasmon resonance (Biacore) using immobilized biotinylated glycans and flowing purified HN over the chip. Neu5Acα2-3Galβ1-4GlcNAcβ1-3Galβ1-4GlcNAc (3′SiaDiLN) and Neu5Acα2-3Galβ1-4GlcNAcβ1-3Galβ1-3GlcNAc (3′SiaLNGalβ1-3GlcNAc) are known to be ligands for all three viruses on the Consortium for Functional Glycomics Core H glycan array, while 3′-N-acetylneuraminyllactose (NANL) is a cleavage substrate for hPIV1 and -2 and a ligand for hPIV1 on the glycan array (5, 7). On the Biacore instrument, immobilized 3′SiaDiLN bound to all three viruses, 3′SiaLNGalβ1-3GlcNAc bound only to hPIV2, and 3′NANL bound to none of the three viruses. The Kd values for all quantifiable bindings were in the range of 10 to 80 μM HN, except for 3′SiaDiLN binding to hPIV3, which exhibited a Kd in the mM range (Table 3). All three HNs bound their immobilized ligands quickly following injection, dissociated rapidly and completely with washing, and did not require a regeneration solution to completely elute from the sensor surface (Fig. 3). Binding experiments at pH 5 were not attempted, lest neuraminidase digest the glycans immobilized on the sensor chip.
Table 3.
Soluble HN binding monovalent ligands determined by surface plasmon resonancea
| Virus | Ligand | kon (1/M · s) | SE kon (1/M · s) | koff (1/s) | SE koff (1/s) | Kd (μM HN) | χ2 |
|---|---|---|---|---|---|---|---|
| hPIV1 | 3′SiaDiLN | 6.2 × 103 | 1.82 × 103 | 0.50 | 0.06 | 79 | 1.8 |
| 3′SiaLNGalβ1-3GlcNAc | —b | — | — | — | — | — | |
| 3′NANL | — | — | — | — | — | — | |
| hPIV2 | 3′SiaDiLN | 1.2 × 104 | 1.1 × 103 | 0.34 | 5.1 × 10−3 | 28 | 3.8 |
| 3′SiaLNGalβ1-3GlcNAc | 1.1 × 104 | 1.7 × 103 | 0.13 | 8.0 × 10−3 | 10 | 1.4 | |
| 3′NANL | — | — | — | — | — | — | |
| hPIV3 | 3′SiaDiLN | 51 | 123 | 0.22 | 8.5 × 10−3 | 4300 | 1.4 |
| 3′SiaLNGalβ1-3GlcNAc | — | — | — | — | — | — | |
| 3′NANL | — | — | — | — | — | — |
3′SiaDiLN, Neu5Acα2-3Galβ1-4GlcNAcβ1-3Galβ1-4GlcNAc; 3′SiaLNGalβ1-3GlcNAc, Neu5Acα2-3Galβ1-4GlcNAcβ1-3Galβ1-3GlcNAc; 3′NANL, Neu5Acα2-3Galβ1-4Glc; kon, on-rate constant; koff, off-rate constant.
—, binding was not measurable.
Fig 3.
Surface plasmon resonance sensorgrams for hPIV1, hPIV2, and hPIV3 soluble HN binding to monovalent ligands. Note that binding occurs rapidly and dissociates rapidly, as indicated by the steepness of the curves. Binding was not measurable for hPIV2 on NANL and hPIV3 on 3′SiaLNGalβ1-3GlcNAc or NANL. RU, relative units.
DISCUSSION
hPIV HN binds glycoprotein ligands in a manner consistent with a single binding site.
We tested our whole-virus binding data using models for (i) multiple interacting active sites, (ii) two types of binding sites with separate Kd values, and (iii) a single type of active site. The one-site and two-site hyperbolic models are related and therefore can be compared by the F test. The F test determines whether the more complicated model (the two-site model) is a better fit for the data than the simpler model (the one-site model) using a sum-of-squares analysis.
Each HN tetramer has four N sites (site I) and two dimer interfaces, each of which could contain a binding-only site (site II). In the crystal structure of NDV HN, it was noted that three active-site residues (R174, Y526, and E547) are each connected to a site II component by movable loops, such that even though their nominal distance is 12 Å from site II at pH 6.5, any change in either site could indirectly affect the other by moving the loop (17). It is therefore expected that if hPIV HN has a site II, its binding will not be independent of site I and binding curves will exhibit cooperativity. However, five of the six virus-glycoprotein ligand pairs tested either failed to fit the interacting-sites model or fit the model with an h value of ≈1, indicating no cooperativity. These findings suggest either that there is a single binding site or that sites I and II are completely independent of one another. They also indicate that there is no cooperativity among the N sites in an HN tetramer in these virus-ligand pairs or that each tetramer binds only one molecule of ligand. In contrast, cooperative N activity has been demonstrated in some strains of NDV (25). hPIV3 does exhibit cooperative binding to bFetuin in the colorimetric binding assay, although there is no measurable N activity with bFetuin. The glycans on bFetuin (26, 27) may be packed more densely than those of hAGP, allowing multiple interactions between an HN oligomer and a single bFetuin molecule, or their triantennary structure may be less accessible to the binding site, thus requiring cooperation among the monomers for adequate binding. Models of the hPIV3 dimer interface suggest that a site II could accommodate only one sialic acid on one branch of a glycan ligand (18), and therefore, it is unlikely that the cooperative binding is the result of discrimination between the triantennary glycans of bFetuin and the tetraantennary glycans of hAGP (28) by a site II.
Virus-ligand binding curves that did not exhibit cooperativity were fit to two hyperbolic binding models. The one-site model implies that there is only one kind of binding site in each protein, while the two-site model allows two independent but different binding sites. In two-site fits of the virion to glycoprotein ligand binding curves, Kd1 was equal to Kd2, indicating that the two sites are in fact identical. Furthermore, the Kd values for intact virions and purified HN are on the same order of magnitude, even though HN on the virion is a fully assembled tetramer that might have an intersubunit second site, while purified HN is monomeric and unlikely to have a second site because it lacks a dimer interface. If the tetramer had a second binding site, the apparent affinity for ligand would likely be higher than that of the monomer, because the observed binding would be the sum of the values for the two sites. The region at the dimer interface that has been proposed to be a second sialic acid binding site in hPIV3 has been implicated in fusion (10), but our data do not support the idea that this site is involved in receptor binding.
Why do the hPIVs lack the second receptor-binding site that appears to be present in NDV? One possible explanation is that the overall N activity of two-sited HNs, such as those of NDV (25) or Sendai virus (29), measured as the amount of sialic acid released from 3′NANL per unit time, is much higher than the N activity of hPIV (19). If the balance between H and N activity is critical, then the mutation that caused the N activity reduction in hPIV relative to that in the ancestral virus required a corresponding reduction in H activity. Perhaps the evolutionary loss of site II provided this reduction in H.
HN is primarily a neuraminidase that holds its substrate long enough for apparent binding.
In our previous study, we determined the Michaelis-Menten constants for hPIV1, -2, and -3 virions on small-molecule and glycoprotein substrates (7), and we have now shown that Km values for whole hPIV and soluble HN are similar: ∼0.5 mM for NANL and bFetuin and ∼1 to 2 mM for hAGP, in those cases where cleavage was measurable. We can now compare binding and cleavage efficiencies. For glycoprotein substrates/ligands, hPIV1, -2, and -3 virions all show that Km was greater than Kd (Table 4). For monovalent substrates/ligands, due to low binding or low activity, the best comparison that we can make is Km using NANL (MUN for hPIV3) and Kd using 3′SiaDiLN, and Km was greater than Kd for hPIV1 and -2 soluble HN (Table 4, last column). The finding that Km was greater than Kd for the same or a similar substrate when both binding and cleavage were measured at the same temperature and at their optimal pH indicates that the affinity of HN for the products of the enzymatic reaction is lower than the affinity for the intact ligand and the interaction of HN with receptor will proceed to cleavage before the binding complex dissociates, as long as the chemical environment is N permissive. When the hPIV3 substrate/ligand is monovalent, Kd is high at 4.3 mM and Km is nearly equal to Kd, which indicates that the virus has little affinity for the ligand; hPIV3 may not use glycans with only a monovalent binding motif as receptors. hPIV3 exhibits cooperative binding to bFetuin, unlike all other tested virus-ligand binding pairs, in which ligand interacts with the binding sites on the tetramer independently. This difference in binding may contribute to bFetuin's resistance to hPIV3 N. On the whole, the data suggest that HN is primarily a neuraminidase but holds its ligand long enough to have apparent binding activity.
Table 4.
Comparison of Km and Kd valuesa
| Virus | Substrate | Substrate type | Km (μM sialic acid, pH 5.5, RTb) | Kd (μM sialic acid, pH 7, RT) | Km/Kd | Km/Kd (NANL or MUN/3′SiaDiLN)c |
|---|---|---|---|---|---|---|
| hPIV1 (whole) | bFetuin | Glycoprotein | 460 ± 140 | 21 ± 2.8 | 23 | |
| hPIV1 (whole) | hAGP | Glycoprotein | 2,000 ± 130 | 91 ± 11 | 22 | |
| hPIV1 (whole) | NANL | Monovalent | 530 ± 50 | |||
| hPIV1 (HN) | 3′SiaDiLN | Monovalent | 79 | 7 | ||
| hPIV2 (whole) | bFetuin | Glycoprotein | 400 ± 70 | 7.7 ± 1.8 | 52 | |
| hAGP | Glycoprotein | 640 ± 40 | 218 ± 11 | 3 | ||
| hPIV2 (HN) | NANL | Monovalent | 440 ± 70 | |||
| hPIV2 (HN) | 3′SiaDiLN | Monovalent | 28 | 16 | ||
| hPIV2 (HN) | 3′SiaLNGalβ1-3GlcNAc | Monovalent | 10 | |||
| hPIV3 (whole) | bFetuin | Glycoprotein | 34 ± 7 | |||
| hPIV3 (whole) | hAGP | Glycoprotein | 250 ± 50 | 125 ± 30 | 2 | |
| hPIV3 (HN) | MUN | Monovalent | 4,900 ± 490 | |||
| hPIV3 (HN) | 3′SiaDiLN | 4,300 | 1.1 |
Each Km or Kd is measured at the same temperature and at optimal pH for that function. Blank cells indicate that Km or Kd was not obtained due to low activity.
RT, room temperature.
Because MUN cannot be tested by surface plasmon resonance and 3′NANL showed no binding, Km/Kd ratios for HN binding monovalent substrate or ligand were calculated using a cleavage substrate and its most chemically similar ligand with measurable binding.
The structure of HN is consistent with the idea that this protein is primarily a neuraminidase. HN shares its basic structure, a tetramer of six-bladed β propellers with an active site at the center of each subunit, with the influenza virus neuraminidase (30, 31). Neuraminidases of some recent H3N2 influenza viruses with a mutation in the neuraminidase active site have been found to bind red blood cells in an oseltamivir-sensitive manner (32). A structural and enzymatic study of the mutant neuraminidase showed that it avidly bound 3′-sialylated glycans but did not cleave them (33), and the NeuAc2-3 binding by mutant neuraminidase can be used by the virus to enter cells (34). Thus, the hPIV HN is intermediate between active influenza virus neuraminidase and the mutant that binds substrate but shows very low hydrolysis.
The finding that Km was greater than Kd for N and H is consistent with the hypothesis that N acts to limit H activity in the presence of high-density sialic acid receptors and with the known pH sensitivity of HN.
A Km greater than Kd also implies that more ligand than is necessary for observable binding is necessary to reach the maximal cleavage rate. A target cell or membrane microdomain with a high density of sialic acid-containing receptors would have two characteristics critical for HN regulation: the presence of acidic sialyl groups would lower the local pH to bring it closer to the pH 5 optimum for N, and the effective sialic acid concentration would be closer to Km than Kd. In this environment, HN would be expected to act as a neuraminidase and destroy receptors. In this case, N acts to reduce the receptor density, thus preventing the excessive fusogenicity that has proved detrimental to virus replication in model systems (10, 12).
A target cell or membrane microdomain with a low density of sialic acid-containing receptors, in contrast, would present HN with both the nearly neutral pH environment that favors binding and an effective sialic acid concentration closer to Kd than Km, both of which would favor the binding function of HN. In addition, with a low receptor density, perhaps only one or two monomers of a tetramer can engage a receptor and binding is insufficient to trigger the conformational changes that activate N (7). The virus will then remain bound to its receptor long enough to trigger fusion and guarantee successful entry into the cell. In spite of the dominance of N over H in the mechanism of HN action, N is inhibited both by the higher local pH and by the lower concentration of receptors, allowing the critical life cycle step of target cell binding to occur.
ACKNOWLEDGMENTS
This work was supported by the Oklahoma Center for the Advancement of Science and Technology (OCAST), grant HR09-001. Biotinylated glycans were supplied by the Consortium for Functional Glycomics Core D, funded by NIGMS grant GM62116.
The following reagents were obtained through the NIH Biodefense and Emerging Infections Research Resources Repository, NIAID, NIH: polyclonal anti-human parainfluenza virus 2 Greer (antiserum, guinea pig; NR-3231) and polyclonal anti-human parainfluenza virus 3 NIH 47885 (antiserum, guinea pig; NR-3235).
We thank Paul Cook at the University of Oklahoma for assistance with enzymatic and binding data interpretation.
Footnotes
Published ahead of print 5 June 2013
REFERENCES
- 1. Iorio RM, Glickman RL, Sheehan JP. 1992. Inhibition of fusion by neutralizing monoclonal antibodies to the haemagglutinin-neuraminidase glycoprotein of Newcastle disease virus. J. Gen. Virol. 73(Pt 5):1167–1176 [DOI] [PubMed] [Google Scholar]
- 2. Moscona A, Peluso RW. 1991. Fusion properties of cells persistently infected with human parainfluenza virus type 3: participation of hemagglutinin-neuraminidase in membrane fusion. J. Virol. 65:2773–2777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Portner A, Scroggs RA, Marx PS, Kingsbury DW. 1975. A temperature-sensitive mutant of Sendai virus with an altered hemagglutinin-neuraminidase polypeptide: consequences for virus assembly and cytopathology. Virology 67:179–187 [DOI] [PubMed] [Google Scholar]
- 4. Miura N, Uchida T, Okada Y. 1982. HVJ (Sendai virus)-induced envelope fusion and cell fusion are blocked by monoclonal anti-HN protein antibody that does not inhibit hemagglutination activity of HVJ. Exp. Cell Res. 141:409–420 [DOI] [PubMed] [Google Scholar]
- 5. Amonsen M, Smith DF, Cummings RD, Air GM. 2007. Human parainfluenza viruses hPIV1 and hPIV3 bind oligosaccharides with alpha2-3-linked sialic acids that are distinct from those bound by H5 avian influenza virus hemagglutinin. J. Virol. 81:8341–8345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Suzuki T, Portner A, Scroggs RA, Uchikawa M, Koyama N, Matsuo K, Suzuki Y, Takimoto T. 2001. Receptor specificities of human respiroviruses. J. Virol. 75:4604–4613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tappert MM, Smith DF, Air GM. 2011. Fixation of oligosaccharides to a surface may increase the susceptibility to human parainfluenza virus 1, 2, or 3 hemagglutinin-neuraminidase. J. Virol. 85:12146–12159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ferreira L, Villar E, Munoz-Barroso I. 2004. Gangliosides and N-glycoproteins function as Newcastle disease virus receptors. Int. J. Biochem. Cell Biol. 36:2344–2356 [DOI] [PubMed] [Google Scholar]
- 9. Murrell M, Porotto M, Weber T, Greengard O, Moscona A. 2003. Mutations in human parainfluenza virus type 3 hemagglutinin-neuraminidase causing increased receptor binding activity and resistance to the transition state sialic acid analog 4-GU-DANA (zanamivir). J. Virol. 77:309–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Palermo LM, Porotto M, Yokoyama CC, Palmer SG, Mungall BA, Greengard O, Niewiesk S, Moscona A. 2009. Human parainfluenza virus infection of the airway epithelium: viral hemagglutinin-neuraminidase regulates fusion protein activation and modulates infectivity. J. Virol. 83:6900–6908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Merz DC, Prehm P, Scheid A, Choppin PW. 1981. Inhibition of the neuraminidase of paramyxoviruses by halide ions: a possible means of modulating the two activities of the HN protein. Virology 112:296–305 [DOI] [PubMed] [Google Scholar]
- 12. Porotto M, Murrell M, Greengard O, Doctor L, Moscona A. 2005. Influence of the human parainfluenza virus 3 attachment protein's neuraminidase activity on its capacity to activate the fusion protein. J. Virol. 79:2383–2392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yewdell J, Gerhard W. 1982. Delineation of four antigenic sites on a paramyxovirus glycoprotein via which monoclonal antibodies mediate distinct antiviral activities. J. Immunol. 128:2670–2675 [PubMed] [Google Scholar]
- 14. Greengard O, Poltoratskaia N, Leikina E, Zimmerberg J, Moscona A. 2000. The anti-influenza virus agent 4-GU-DANA (zanamivir) inhibits cell fusion mediated by human parainfluenza virus and influenza virus HA. J. Virol. 74:11108–11114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Porotto M, Fornabaio M, Greengard O, Murrell MT, Kellogg GE, Moscona A. 2006. Paramyxovirus receptor-binding molecules: engagement of one site on the hemagglutinin-neuraminidase protein modulates activity at the second site. J. Virol. 80:1204–1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lawrence MC, Borg NA, Streltsov VA, Pilling PA, Epa VC, Varghese JN, McKimm-Breschkin JL, Colman PM. 2004. Structure of the haemagglutinin-neuraminidase from human parainfluenza virus type III. J. Mol. Biol. 335:1343–1357 [DOI] [PubMed] [Google Scholar]
- 17. Zaitsev V, von Itzstein M, Groves D, Kiefel M, Takimoto T, Portner A, Taylor G. 2004. Second sialic acid binding site in Newcastle disease virus hemagglutinin-neuraminidase: implications for fusion. J. Virol. 78:3733–3741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Porotto M, Fornabaio M, Kellogg GE, Moscona A. 2007. A second receptor binding site on human parainfluenza virus type 3 hemagglutinin-neuraminidase contributes to activation of the fusion mechanism. J. Virol. 81:3216–3228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bousse T, Takimoto T. 2006. Mutation at residue 523 creates a second receptor binding site on human parainfluenza virus type 1 hemagglutinin-neuraminidase protein. J. Virol. 80:9009–9016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Alymova IV, Taylor G, Mishin VP, Watanabe M, Murti KG, Boyd K, Chand P, Babu YS, Portner A. 2008. Loss of the N-linked glycan at residue 173 of human parainfluenza virus type 1 hemagglutinin-neuraminidase exposes a second receptor-binding site. J. Virol. 82:8400–8410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang ZM, Tong LL, Grant D, Cihlar T. 2001. Expression and characterization of soluble human parainfluenza virus type 1 hemagglutinin-neuraminidase glycoprotein. J. Virol. Methods 98:53–61 [DOI] [PubMed] [Google Scholar]
- 22. Potier M, Mameli L, Belisle M, Dallaire L, Melancon SB. 1979. Fluorometric assay of neuraminidase with a sodium (4-methylumbelliferyl-alpha-d-N-acetylneuraminate) substrate. Anal. Biochem. 94:287–296 [DOI] [PubMed] [Google Scholar]
- 23. Spiro RG. 1960. Studies on fetuin, a glycoprotein of fetal serum. I. Isolation, chemical composition, and physiochemical properties. J. Biol. Chem. 235:2860–2869 [PubMed] [Google Scholar]
- 24. Thompson SD, Laver WG, Murti KG, Portner A. 1988. Isolation of a biologically active soluble form of the hemagglutinin-neuraminidase protein of Sendai virus. J. Virol. 62:4653–4660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mahon PJ, Deng R, Mirza AM, Iorio RM. 1995. Cooperative neuraminidase activity in a paramyxovirus. Virology 213:241–244 [DOI] [PubMed] [Google Scholar]
- 26. Green ED, Adelt G, Baenziger JU, Wilson S, Van Halbeek H. 1988. The asparagine-linked oligosaccharides on bovine fetuin. Structural analysis of N-glycanase-released oligosaccharides by 500-megahertz 1H NMR spectroscopy. J. Biol. Chem. 263:18253–18268 [PubMed] [Google Scholar]
- 27. Spiro RG, Bhoyroo VD. 1974. Structure of the O-glycosidically linked carbohydrate units of fetuin. J. Biol. Chem. 249:5704–5717 [PubMed] [Google Scholar]
- 28. Hermentin P, Witzel R, Doenges R, Bauer R, Haupt H, Patel T, Parekh RB, Brazel D. 1992. The mapping by high-pH anion-exchange chromatography with pulsed amperometric detection and capillary electrophoresis of the carbohydrate moieties of human plasma alpha 1-acid glycoprotein. Anal. Biochem. 206:419–429 [DOI] [PubMed] [Google Scholar]
- 29. Portner A. 1981. The HN glycoprotein of Sendai virus: analysis of site(s) involved in hemagglutinating and neuraminidase activities. Virology 115:375–384 [DOI] [PubMed] [Google Scholar]
- 30. Colman PM, Varghese JN, Laver WG. 1983. Structure of the catalytic and antigenic sites in influenza virus neuraminidase. Nature 303:41–44 [DOI] [PubMed] [Google Scholar]
- 31. Crennell S, Takimoto T, Portner A, Taylor G. 2000. Crystal structure of the multifunctional paramyxovirus hemagglutinin-neuraminidase. Nat. Struct. Biol. 7:1068–1074 [DOI] [PubMed] [Google Scholar]
- 32. Lin YP, Gregory V, Collins P, Kloess J, Wharton S, Cattle N, Lackenby A, Daniels R, Hay A. 2010. Neuraminidase receptor binding variants of human influenza A(H3N2) viruses resulting from substitution of aspartic acid 151 in the catalytic site: a role in virus attachment? J. Virol. 84:6769–6781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhu X, McBride R, Nycholat CM, Yu W, Paulson JC, Wilson IA. 2012. Influenza virus neuraminidases with reduced enzymatic activity that avidly bind sialic acid receptors. J. Virol. 86:13371–13383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gulati S, Smith DF, Cummings RD, Couch RB, Griesemer SB, St George K, Webster RG, Air GM. 2013. Human H3N2 influenza viruses isolated from 1968 to 2012 show varying preference for receptor substructures with no apparent consequences for disease or spread. PLoS One 8:e66325. 10.1371/journal.pone.0066325 [DOI] [PMC free article] [PubMed] [Google Scholar]