Abstract
GB virus B (GBV-B; family Flaviviridae, genus Hepacivirus) has been studied in New World primates as a model for human hepatitis C virus infection, but the distribution of GBV-B and its relatives in nature has remained obscure. Here, we report the discovery of a novel and highly divergent GBV-B-like virus in an Old World monkey, the black-and-white colobus (Colobus guereza), in Uganda. The new virus, guereza hepacivirus (GHV), clusters phylogenetically with GBV-B and recently described hepaciviruses infecting African bats and North American rodents, and it shows evidence of ancient recombination with these other hepaciviruses. Direct sequencing of reverse-transcribed RNA from blood plasma from three of nine colobus monkeys yielded near-complete GHV genomes, comprising two distinct viral variants. The viruses contain an exceptionally long nonstructural 5A (NS5A) gene, approximately half of which codes for a protein with no discernible homology to known proteins. Computational structure-based analyses indicate that the amino terminus of the GHV NS5A protein may serve a zinc-binding function, similar to the NS5A of other viruses within the family Flaviviridae. However, the 521-amino-acid carboxy terminus is intrinsically disordered, reflecting an unusual degree of structural plasticity and polyfunctionality. These findings shed new light on the natural history and evolution of the hepaciviruses and on the extent of structural variation within the Flaviviridae.
INTRODUCTION
GB virus B (GBV-B), together with hepatitis C virus (HCV), belongs to the genus Hepacivirus within the family Flaviviridae (1). This family also includes the genera Pestivirus and Flavivirus and the recently accepted genus Pegivirus, which contains the GBV-A, -C, and -D viruses (1) as well as newly described viruses in bats, horses, and rodents (2–5). GBV-B was first described in 1967 in tamarins (Saguinus spp.) and other New World monkeys after their inoculation with acute-phase plasma from a physician (“G.B.”) with unexplained hepatitis (6). Subsequent analysis of acute-phase tamarin plasma led to the identification of two novel RNA viruses, GBV-A and GBV-B (7). While GBV-A was shown to be an indigenous tamarin virus that does not cause hepatitis, GBV-B was directly associated with the development of acute hepatitis in tamarins (8). Attempts to detect GBV-B in the initial human inoculum as well as in additional tamarins or other New World monkeys failed; thus, the natural host and evolutionary origin of GBV-B have remained unclear (1, 9–11).
Because GBV-B is closely related to HCV, experimental infection of New World primates with this virus has been proposed as a surrogate animal model for HCV infection, currently only possible in chimpanzees (Pan troglodytes) (12). Experimental infection of tamarins with GBV-B usually results in acute, self-resolving infection with subacute hepatitis (13). However, persistent infection has been described in marmosets, including chronic and progressive hepatitis C-like disease (14). The first nonprimate hepaciviruses (NPHV) were recently identified in domestic dogs and horses (11, 15). These viruses cluster phylogenetically with the hepatitis C viruses, with molecular dating estimates suggesting a common ancestor no more recent than 500 to 1,000 years before the present (ybp) (11, 15). Other hepaciviruses distantly related to GBV-B have been identified in three rodent species from the Southwestern United States, deer mice (Peromyscus maniculatus), desert wood rats (Neotoma lepida), and hispid pocket mice (Chaetodipus hispidus) (4), as well as in two African bat species, the striped leaf-nosed bat (Hipposideros vittatus) and the large-eared free-tailed bat (Otomops martiensseni) (5). To our knowledge, however, no hepaciviruses have yet been identified in wild nonhuman primates (NHPs).
Here we report the discovery and characterization of the first hepacivirus infecting a wild nonhuman primate, the black-and-white colobus (Colobus guereza), an Old World monkey, in Kibale National Park, Uganda. This virus, guereza hepacivirus (GHV), shares common ancestry with GBV-B, rodent hepaciviruses (RHV), and one of three recently discovered bat hepaciviruses (BHV), and it shows evidence of ancient recombination with these viruses. Notably, GHV contains an unusual nonstructural 5A (NS5A) protein that is approximately twice the length of any other known NS5A protein within the Flaviviridae and that exhibits unique structural features, including an exceptionally long intrinsically disordered region at the carboxy terminus. These findings shed new light on the host range, natural history, and evolution of the hepaciviruses. In addition, the unique features of the NS5A protein of GHV substantially expand our understanding of the extent of structural and functional variation within the Flaviviridae.
MATERIALS AND METHODS
Sampling and high-throughput sequencing.
Black-and-white colobus (BWC) monkeys were sampled from Kibale National Park, Uganda, a 795-km2 forested park in Western Uganda (0°13′–0°41″N, 30°19′–30°32″E) known for its exceptional primate diversity and biomass (16). As part of a long-term study of primate ecology and health (17), nine animals were immobilized and sampled as previously described (18). All animal protocols received prior approval from the Uganda National Council for Science and Technology, the Uganda Wildlife Authority, and the University of Wisconsin Animal Care and Use Committee, and all samples were shipped in accordance with international laws under Ugandan CITES permit no. 002290.
One milliliter of blood plasma from each animal was filtered (0.45-μm pore) to remove residual host cells, and viral RNA was isolated using the Qiagen QIAamp MinElute virus spin kit (Qiagen, Hilden, Germany), omitting carrier RNA. After DNase treatment, RNA was random hexamer primed, subjected to double-stranded cDNA synthesis, and prepared for sequencing on the Illumina MiSeq using the Nextera DNA sample preparation kit (Illumina, San Diego, CA) as previously described (19). Sequence data were analyzed using CLC Genomics Workbench 5.5 (CLC bio, Aarhus, Denmark). Briefly, low-quality reads (Phred quality score below 30) and short reads (<100 bp) were removed, and the remaining reads were subjected to de novo assembly. Assembled contiguous sequences (contigs) were queried against the GenBank database using the basic local alignment search tools blastn and blastx (20).
Phylogenetic and evolutionary analyses.
Translated protein sequences for complete NS3 helicase or NS5A RNA-dependent RNA polymerase (RdRp) motifs of 40 viruses available in GenBank were included in phylogenetic analyses to represent known major clades within the Flaviviridae and the maximum diversity within each clade. Sequence alignments were generated using MAAFT (21) across the four Flaviviradae genera using TranslatorX (22). Neighbor-joining trees (23) were constructed from translated amino acid alignments using the computer program MEGA5 (24), with 1,000 bootstrap replicates of the data to assess the statistical confidence of phylogenetic groupings.
Time to the most recent common ancestor (TMRCA) analyses were conducted using BEAST v1.6.2 (25), with a relaxed molecular clock and an uncorrelated log normal rate distribution, a Yule tree prior, the HKY nucleotide substitution model with gamma distributed rates, an estimated proportion of invariable sites, and two alignments of HCV, NPHV, and GBV-B NS5B sequences (nucleotides [nt] 8200 to 8800, based on HCV-H reference strain numbering). The first alignment was composed of 597 bp and included all three codon positions (123cdp), while the second alignment was composed of 398 bp containing only the first and second codon positions (12cdp) to account for site saturation at the wobble position. TMRCAs were inferred using previously determined HCV-1 and HCV-6 evolutionary rates (1.0 × 10−3 and 1.72 × 10−4 nucleotide substitutions per site per year, respectively). Fifty million Monte Carlo Markov chains (MCMC) were used in each run, and chain convergence and mixing, effective sample sizes (ESS) (all of which were greater than 400), and Bayes factors were determined using the program Tracer v1.5. Maximum clade credibility (MCC) trees were obtained using TreeAnnotator after a burn-in of the first 1,000 trees. MCC trees were viewed using the program FigTree v1.3.1.
Recombination within GHV and between GHV and the other hepaciviruses was assessed using the statistical recombination analysis methods available in the computer package RDP3 (26). Synonymous (dS) and nonsynonymous (dN) substitution rates of codon-aligned GHV sequences were calculated using SNAP (27). Single nucleotide polymorphisms (SNPs) above a minimum variant threshold of 5% and intrahost sequence diversity (πS and πN) were quantified as previously described (28). Amino acid similarity between GHV and related hepaciviruses was plotted across codon-aligned genomes by the sliding-window method implemented in SimPlot v3.5.1 (29).
Protein structure and function analysis of NS5A.
Three-dimensional (3D) models representing NS5A tertiary protein structure were derived using a protein structure modeling ab initio approach based on the iterative implementation of the threading assembly refinement (I-TASSER) method (30, 31). Full-atomic 3D models from the 95 N-terminal residues (residues 1 to 95) and 470 C-terminal residues (residues 414 to 883) of NS5A, encompassing GHV-1 (variant from animal BWC08) polyprotein positions 1862 to 1956 and 2275 to 2744 (denoted as T-211 and T-216, respectively), were generated using the I-TASSER software package v1.1 (30, 32). Accuracy assessments of 3D models were based on two scoring functions: the confidence (C) score and the template modeling (TM) score (31, 32). The top-ranked 3D model from each NS5A region was then selected for further modeling refinements and function analysis. Overall stereochemical quality of 3D models was assessed with PROCHECK v3.5 (33) after additional refinements to remove atomic clashes were carried out on the top-ranked 3D models of T-211 and T-216 using WHATIF v8.0 (34). Secondary structure (3 states: coil, helix, and strand) of the 95-amino-acid (aa) N-terminal and 470-aa C-terminal NS5A regions were determined by computer prediction using PSIPRED v2.6 (35).
Sequence-based analyses of NS5A to identify intrinsically disordered regions (IDRs) and sites within IDRs with capacity to undergo disorder-to-order transitions for binding interactions were conducted with ANCHOR v1.0 (36). Identification of possible binding partners involved in protein-protein or substrate-protein interactions at predicted binding sites, as determined by the ANCHOR algorithm (37), was based on a database of functional sites conforming to the constraints of linear motifs (LMs) (38). LMs obtained from the Eukaryotic Linear Motif server (39) and from the Calmodulin Target Database server (40) were also included in the database of LMs (servers last accessed on 8 March 2012).
Additional function associations annotated to the GHV-1 NS5A protein were determined by performing structure-based function analysis on 3D models T-211 and T-216. Functional predictions from 3D models were performed using a template-matching method (41) that searches for and identifies proteins with significant structure matching to a query template in the Protein Data Bank (PDB). The known functional characteristics (based on Gene Ontology [GO] terms; http://sbkb.org/) and molecular interaction pathways (Kyoto Encyclopedia of Genes and Genomes [KEGG] pathways; http://www.genome.jp/kegg/) of the corresponding matching PDB templates (matches with E values of <0.10) were then associated to NS5A.
The total personal supercomputer computational resources consumed to run the I-TASSER suite programs to generate 3D models of the NS5A 470-aa and 95-aa fragments were 34.7 and 6.3 h, respectively. Runs were conducted in parallel using a total of 28 central processing units (CPU) for a total maximum of 520 CPU h and a peak memory of 16 GB. The data required a total storage space of 47.2 GB.
RESULTS
Discovery and genomic characterization of guereza hepacivirus (GHV).
Deep sequencing of RNA from blood plasma of nine black-and-white colobus monkeys from Kibale National Park, Uganda, resulted in 1,633,000 to 2,232,000 trimmed-paired-end reads per sample. In three animals (BWC04, BWC05, and BWC08), de novo assembly revealed an RNA virus with genomic architecture matching known viruses in the family Flaviviridae. Sequences covering the entire coding region as well as partial 5′ and 3′ untranslated regions (UTRs) were acquired for viruses from all three animals, with an average coverage depth between 464× and 2,220× (GenBank sequence accession no. KC551800 to KC551802). Pairwise nucleotide-level comparisons among the three variants revealed that two variants were highly similar to each other (98% identity [ID] between the variants from animals BWC05 and BWC08) but divergent from the third variant (85% average ID between the variant from animal BWC04 and the variants from animals BWC05 and BWC08). A query against the GenBank database revealed that the two viruses were most closely related to viruses in the genus Hepacivirus. Across the coding genome, the new viruses shared limited nucleotide identity with other hepaciviruses: HCV (43%), NPHV (43%), RHV (47%), GBV-B (48%), and BHV (50%). To indicate host species of origin (Colobus guereza), we designate the new viral variants GHV (guereza hepacivirus). Based on the established nomenclature (42), we classified the three GHV variants into two subtypes: GHV-1 (from animals BWC05 and BWC08) and GHV-2 (from animal BWC04).
GHV encodes a 3,336-aa (GHV-1) or 3,334-aa (GHV-2) polyprotein and 5′ and 3′ UTR sequences of at least 270 nt and 134 nt, respectively. Notably, the GHV coding sequence is over 1 kb longer than those of GBV-B, NPHV or HCV, largely due to an unusually long NS5A gene. Overall, the 5′ UTR of GHV had limited homology to the UTRs of other hepaciviruses. However, a 99-bp region adjacent to the start of the polyprotein showed structures analogous to stem-loops IIId, IIIe, and IIIf in the HCV 5′ UTR (data not shown). The remaining 5′ UTR returned no matches after searching against the GenBank sequence databases. A canonical microRNA 122 (miRNA-122) binding site (CACUCC), also present near the 5′ UTR N terminus of HCV, GBV-B, NPHV, and RHV (15), was detected in both GHV-1 and GHV-2 5′ UTR sequences at nucleotides 11 to 18, potentially indicating hepatotropicity of these viruses. The longest 3′ UTR, recovered from GHV-2, had a length of 199 nt and did not share homology with 3′ UTRs of other hepaciviruses. The three stem-loop structures present in the 98X region of HCV were replaced by a single stem-loop with 45 bp in the stem and a bulge of 5 bases. This singular stem-loop also includes a short 6-nt poly(C) tract 46 nucleotides downstream of the stop codon and is thus comparable to the 10-nt poly(C) tract observed in RHV (4).
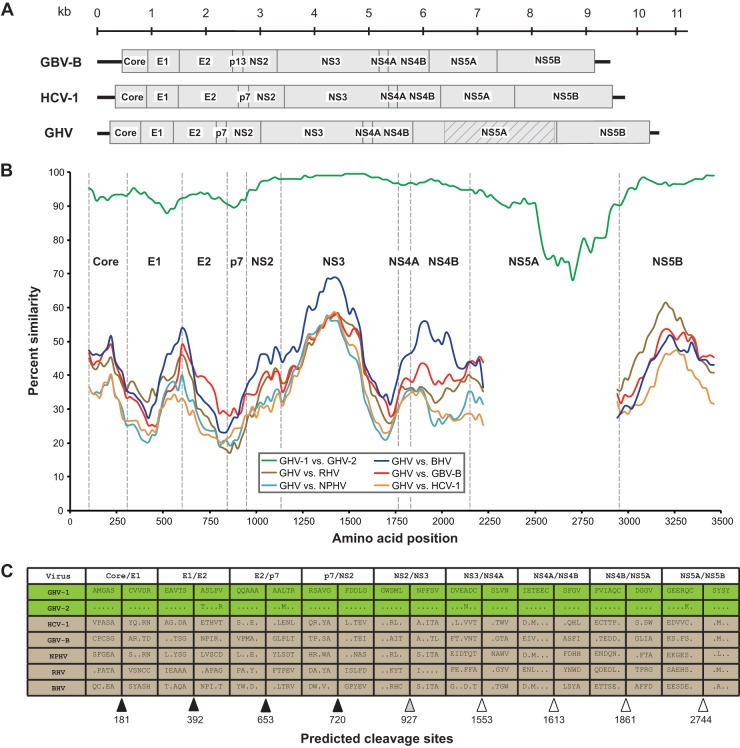
GHV polyprotein cleavage sites were predicted based on an alignment with members of the Hepacivirus and Pegivirus genera and through manual (43, 44) and in silico (45, 46) signalase and NS3-NS4A protease cleavage site prediction. Similar to HCV, GBV-B, NPHV, BHV, and RHV, predicted cleavage sites on the GHV polyprotein resulted in 10 viral proteins representing the typical Hepacivirus genome organization (Fig. 1A and C). Within the core, no canonical ribosomal slippery site was identified. However, a possible alternative reading frame protein (ARFP), which overlaps much of the HCV ARFP/F protein (47), was identified for GHV, with a potential AUG start codon 82 nt downstream of the polyprotein initiation site and a coding capacity of 136 aa. Previous studies have demonstrated initiation of ARFP translation from AUG start codons located downstream of this polyprotein initiation site (47). The GHV E1 and E2 proteins each contained four predicted N-glycosylation sites, similar in abundance to sites predicted for GBV-B (3 and 6 sites, respectively), RHV (2 and 4 sites), and BHV (2 and 5 sites), but fewer than the number of sites in HCV (6 and 11 sites) and NPHV (4 and 10 sites). Of GHV's predicted N-glycosylation sites, two in E1 and all four in E2 were conserved with GBV-B, while two in each protein were conserved with RHV and BHV. Such N-glycosylation sites are important for the proper folding of the Hepacivirus envelope proteins, and they have been highlighted to support evolutionary distinctions among hepaciviruses (1, 11).
Fig 1.
Genome organization, amino acid similarity, and polyprotein cleavage sites of hepaciviruses HCV-1, GBV-B, and GHV. (A) The genome organization of the novel GHV is shown in comparison to those of HCV-1 and GBV-B. Boxes represent mature proteins and are drawn to scale. The shaded area within GHV NS5A denotes a region without homology to known proteins. Black lines adjacent to core and NS5B proteins represent untranslated regions (UTRs). (B) Sliding-window similarity plots across aligned coding regions. Dashed vertical lines indicate start positions of inferred viral proteins. (C) Amino acid sequences of GHV and related viruses adjacent to predicted protease cleavage sites. Proposed cleavage sites for signalase (black triangles), NS2-NS3 protease (gray triangle), and NS3-4A protease (white triangles) are indicated. Amino acid positions of cleavage sites in relation to GHV-1 are included below the triangles.
Amino acids essential for NS2 (His-855 and Cys-895, with reference to GHV-1) and NS3 (His-983, Asp-1007, and Ser-1066, with reference to GHV-1) protease function are conserved between GHV-1 and GHV-2 and with GBV-B and HCV (1). Of note, as in HCV, GHV appears to encode a p7-like protein, rather than the p13 protein characteristic of GBV-B (48). The HCV and GHV p7 are both 63 amino acids in length and consist of two transmembrane domains connected by a cytoplasmic loop. Unlike HCV, the GHV cytoplasmic loop contains three instead of two positively charged amino acids, resembling the p7 protein of bovine viral diarrhea virus (BVDV) (49) and the p7 homolog of GBV-B, comprising the C-terminal half of p13 (48).
The GHV NS5A protein (882 to 883 aa) is approximately twice the length of any other known NS5A within the Flaviviridae. By comparison, the next largest known NS5A, belonging to a pestivirus, BVDV-1, comprises 496 aa. While the first 175 aa of the N terminus and the last 45 aa of the C terminus aligned convincingly with the NS5A of GBV-B, the intervening 662- to 663-aa region aligned poorly, or not at all, with the corresponding 191 aa of the GBV-B NS5A. BLAST comparisons of the GHV NS5A C-terminal ∼690 aa yielded no detectable homology to currently described nucleotide or protein sequences within the GenBank databases. Proposed NS3-4A serine protease cleavage sites for GBV-B and GHV (based on sequence alignment) do not completely adhere to the canonical form reported for HCV (44, 50). Therefore, we cannot exclude the existence of cryptic NS3-4A serine protease cleavage sites within this region of the GHV NS5A. However, apparent homology between GHV and GBV-B in N- and C-terminal regions of NS5A suggests that the entire sequence is expressed as a single protein.
Sliding-window analyses of amino acid similarities between GHV and selected hepaciviruses are shown in Fig. 1B. Across the region from core to NS4B, GHV is most similar to BHV. However, in NS5B GHV shares the highest sequence similarity with RHV. This pattern of discordant similarity suggests recombination; however, formal analysis of recombination using RDP3 proved inconclusive. Among the hepaciviruses compared, the amino acid similarity was highest in the NS3 helicase domain and across the NS5B RNA-dependent RNA polymerase (RdRp) motifs. Between GHV-1 and GHV-2, the amino acid similarity was lowest in the NS5A protein, particularly in the aforementioned region, with no detectable homology to any sequence in GenBank (Fig. 1).
Phylogenetic and genetic diversity analyses.
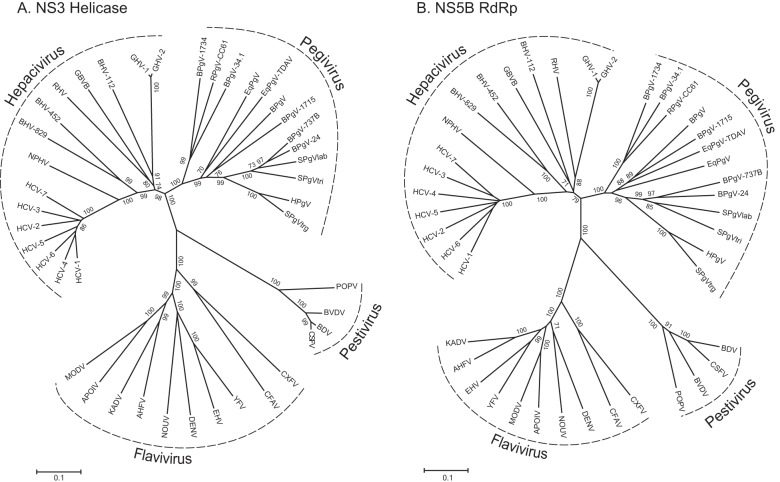
Phylogenies constructed from NS3 helicase and NS5B RdRp alignments (Fig. 2A and B) yielded topologies consistent with established relationships among the Flaviviridae (1, 11). Together, these trees supported the grouping of GHV within the Hepacivirus genus and the shared common ancestry of GHV, GBV-B, RHV, and BHV-112. Consistent with the sliding-window similarity analysis described above, the phylogenetic position of GHV was discordant between the two trees, with GHV sharing a most recent common ancestor with BHV-112 in the NS3 helicase phylogeny but with RHV in the NS5B RdRp phylogeny.
Fig 2.
Phylogenetic analyses of conserved regions in the NS3 helicase (motifs I to VI) (A) and NS5B RdRp (B) genes of GHV aligned with representative members of the family Flaviviridae. Trees were constructed using neighbor-joining analysis of amino acid alignments with 1,000 bootstrap replicates; only bootstrap values of >70% are shown. Regions included in the analyses corresponded to positions 3667 to 4470 (helicase domain of NS3) and 7711 to 8550 (RdRp in NS5B; numbered according to the AF011751 HCV genotype 1a reference sequence). GenBank accession numbers of sequences used in the analyses are as follows: GHV-1 from BWC08, KC551800; GHV-1 from BWC05, KC551801; GHV-2 from BWC04, KC551802; GBV-B, NC_001655; HCV-1, NC_004102; HCV-2, NC_009823; HCV-3, NC_009824; HCV-4, NC_009825; HCV-5, NC_009826; HCV-6, NC_009827; HCV-7, EF108306; NPHV, JF744991; YFV, NC_002031; EHV, DQ859060; DENV, NC_001477; NOUV, EU159426; AHFV, NC_004355; KADV, DQ235146; APOIV, NC_003676; MODV, NC_003635; CXFV, NC_008604; CFAV, NC_001564; KRV, NC_005064; NAKV, GQ165809; POPV, EF100713; BVDV-1, NC_001461; BDV-1a, NC_003679; CSFV, NC_002657; BPgV, GU566735; SPgVlab, NC_001837; SPgVtri, AF023425; SPgVtrg, AF070476; HPgV, NC_001710; BPgV-1715, KC796088; BPgV-1734, KC796087; BPgV-24, KC796082; BPgV-34.1, KC796093; BPgV-737B, KC796081; EqPgV, NC_020902; EqPgV-TDAV, KC145265; RPgV-CC61, KC815311; RHV, KC815310; BHV-829, KC796074; BHV-112, KC796077; and BHV-452, KC796090.
To investigate the timing of GHV's divergence from the other hepaciviruses, we estimated the time to most recent common ancestor (TMRCA) for all seven HCV genotypes, NPHV, GBV-B, and GHV. The relaxed molecular clock used in this analysis was based on substitution rates previously used to determine divergence times for HCV (HCV-1, 1 × 10−3; HCV-6, 1.7 × 10−4) (51, 52) and that approximate the substitution rates proposed for GBV-B during chronic infection of tamarins (1.9 × 10−3 in year 1 and 1 × 10−3 in year 2) (53). Mean TMRCA values for the GHV/GBV-B clade were 692 years before present (ybp) (95% highest posterior density [hpd], 249 to 1,739 ybp) and 1,705 ybp (95% hpd, 480 to 5,248 ybp), based on HCV-1 and HCV-6 substitution rates, respectively. This is similar to the timing of the HCV/NPHV split (624 ybp [95% hpd, 232 to 1,487 ybp] and 1,550 ypb [95% hpd, 518 to 4,667 ybp]), based on HCV-1 and HCV-6 substitution rates, respectively. However, it is more recent than the root of Hepacivirus clade (985 ybp [95% hpd, 395 to 2,356 ybp] and 2,613 ypb [95% hpd, 887 to 7,750 ybp]), based on HCV-1 and HCV-6 substitution rates, respectively. TMRCA estimates using only first and second codon positions were proportionally similar but approximately half as ancient in all cases (data not shown). Because of uncertainties associated with extrapolation of short-term evolutionary rates to deeper time scales (54) as well as potential differences in GHV substitution rates compared to HCV and GBV-B, these dates should be regarded as minimum estimates.
GHV subtypes were further characterized by assessing both intra- and interhost genetic diversity. Intrahost genetic variability was low and consisted of 35 to 99 single nucleotide polymorphisms (SNPs) across the GHV open reading frame (ORF). This amount of variability is higher than that for dengue virus but considerably lower than that for HCV (55, 56). The majority of polymorphisms detected were present at frequencies below 10%, indicating that many of these SNPs may be selectively neutral. Across the coding genome, intrahost synonymous nucleotide diversity (πS) was significantly higher than intrahost nonsynonymous nucleotide diversity (πN) (P < 0.01), with πS exceeding πN by a ratio of 77:1 and with the highest intrahost diversity in NS5A (Table 1).
Table 1.
Mean synonymous and nonsynonymous intrahost viral genetic diversity in regions of the GHV coding genome
| Region | Mean ± SE intrahost genetic diversitya |
|
|---|---|---|
| πS | πN | |
| Core | 0.00401 ± 0.00089 | 0.00000 ± 0.00000*** |
| E1 | 0.00436 ± 0.00283 | 0.00029 ± 0.00029 |
| E2 | 0.00182 ± 0.00026 | 0.00000 ± 0.00000*** |
| p7 | 0.00408 ± 0.00207 | 0.00000 ± 0.00000* |
| NS2 | 0.00662 ± 0.00295 | 0.00000 ± 0.00000* |
| NS3 | 0.00483 ± 0.00233 | 0.00012 ± 0.00006* |
| NS4A | 0.01048 ± 0.00605 | 0.00000 ± 0.00000 |
| NS4B | 0.01022 ± 0.00153 | 0.00008 ± 0.00008*** |
| NS5A | 0.00699 ± 0.00319 | 0.00030 ± 0.00016* |
| NS5B | 0.00551 ± 0.00317 | 0.00002 ± 0.00002 |
| All | 0.00542 ± 0.00201 | 0.00007 ± 0.00004** |
Synonymous (πS) and nonsynonymous (πN) intrahost diversity was determined for each viral gene as well as for the entire coding genome (All) and represents averaged values for GHV-1 and GHV-2. z tests of the hypothesis show that πS = πN: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Consonant with this pattern, most nucleotide differences between GHV-1 and GHV-2 consensus sequences occurred at synonymous sites with low dN/dS ratios (0.046), comparable to published values for NPHV (structural genes, 0.057; nonstructural genes, 0.03) and human Pegivirus (structural genes, 0.063; nonstructural genes, 0.029) (15). The region of lowest GHV intersubtype nucleotide diversity and lowest dN/dS was NS3 (11.5% difference; dN/dS ratio, 0.011), and the region of highest GHV intersubtype nucleotide diversity and highest dN/dS ratio was NS5A (19.3% difference; dN/dS ratio, 0.089).
NS5A protein structure and inferred functions.
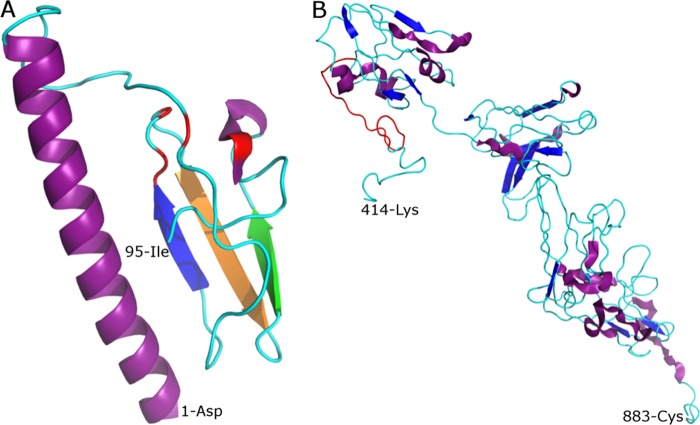
Three approaches (primary sequence comparison as well as secondary and tertiary structure comparisons) were used to identify proteins with similar structure and presumably similar function to the GHV NS5A. All tertiary structure-based analyses, initiated from computer-generated 3D models of NS5A, were found to be either of good or satisfactory stereochemical quality (Table 2). Sequence alignments revealed that the 95 N-terminal residues of NS5A contain a zinc-binding motif, C17CxCx20C, which is conserved in NS5A of hepaciviruses and other related viruses (57). Secondary structure predictions using PSIPRED (35) indicated a topology consisting of two α-helices and three β-strands across this region, and tertiary modeling (model T-211) (Fig. 3) supported high structural similarity between the GHV and HCV NS5A zinc-binding motifs (root mean square deviation [RMSD], <2 Å) (Table 3), with predicted β-strands arranging to form an antiparallel β-sheet (57). The absence of homologous sequences in GenBank precluded similar sequence-based analyses of the remaining portion of the GHV NS5A protein.
Table 2.
Accuracy and Ramachandran plot statistics of NS5A 3D modelsa
| Parameter | Result for: |
|
|---|---|---|
| T-211 | T-216 | |
| Accuracy of modelsb | ||
| C score | −1.70 | −3.10 |
| TM score | 0.52 ± 0.15 | 0.37 ± 0.12 |
| Estimated RMSD (Å) | 7.2 ± 4.2 | 14.9 ± 3.6 |
| Stereochemical quality, no. of residues (%)c | ||
| Most favored regions (A, B, L) | 68 (93.2) | 278 (74.1) |
| Additional allowed regions (a, b, l, p) | 5 (6.8) | 62 (16.5) |
| Generously allowed regions (∼a, ∼b, ∼l, ∼p) | 0 (0.0) | 15 (4.0) |
| Disallowed regions (XX) | 0 (0.0) | 20 (5.3) |
| Non-Gly and non-Pro residues | 73 (100.0) | 375 (100.0) |
| End residues (excluding Gly and Pro) | 2 | 2 |
| Gly residues | 14 | 36 |
| Pro residues | 6 | 57 |
| Total no. of residues | 95 | 470 |
3D models were generated using the I-TASSER method (32). Briefly, to excise continuous fragments from structural alignments, 5 threading programs were sequentially implemented against the Protein Data Bank (PDB) to select the best templates matching the query sequence. Template selection by each method was restricted to 20 matches. Assembly of continuous fragments was performed by running up to 15 Monte Carlo simulations. Full-atomic 3D models (n = 5) were generated after energy minimization refinements of assembled structures.
The accuracy of predicted models was evaluated by confidence (C) scores. The C score is a confidence score for estimating the quality of predicted models. This score is typically in the range of −5 to 2, where a high value signifies a model with a high confidence and vice versa. A TM score of >0.5 indicates a model of correct topology, and a TM score of <0.17 means a random similarity. Also shown is the estimated root mean square deviation (RMSD) resolution of predicted models in angstroms.
Stereochemical quality assessment of residue spatial geometries, as defined by Ramachandran plot, was conducted with the PROCHECK program v3.5 (33).
Fig 3.
Three-dimensional models of the GHV-1 NS5A protein. The ab initio-generated models are based on the 95-aa N-terminal (T-211) and 470-aa C-terminal (T-216) sequences of the black-and-white colobus 08 (BWC08) variant. (A) T-211. Shown is the antiparallel β-sheet consisting of three β-strands colored in blue, orange, and green, which, respectively, resemble strands B1, B2, and B3 described previously for the crystal structure (PDB, 1zh1) of the HCV NS5A zinc-binding site region (64). The putative conserved cysteine residues involved in zinc coordination (C44, C62, C64, and C86) are colored red. Also shown in purple are the long and short α-helix structures (the amino-terminal helix being a putative conserved anchor component of NS5A). (B) T-216. The α-helix and β-strand secondary structure elements are shown in purple and blue, respectively. The longest segment in the NS5A region found to have significant structural homology to RHD of the IκBα p65 protein is colored red.
Table 3.
Template matches from representative structures in the PDB and statistics of sequence alignment
| Parameter | Result for: |
|
|---|---|---|
| T-211 | T-216 | |
| Template matchesa | ||
| PDB entry of matched template | 1zh1 | 1ikn |
| Name of PDB entry | Structure of zinc-binding domain of HCV NS5A | IκBα/NF-κB complex |
| UniProt ID of PDB entry | Q9WMX2 | Q04207 |
| E valueb | 4.46 × 10−4 | 0.059 |
| Alignment statistics | ||
| Sequence length of GVH-1 NS5A, aa | 95 | 470 |
| Sequence length of matched template (PDB), aa | 163 (1zh1A) | 283 (1iknA) |
| Sequence identity (%) | 16.84 | 23.7 |
| Structural similarity (%)c | 98.2 | 64.9 |
| Longest continuous segment length, aad | 54 | 18 |
| Total no. of residues fittede | 54 | 41 |
| RMSD (Å) | 1.63 | 2.96 |
Reverse-template comparisons of NS5A 3D structures versus structures in PDB were done by submitting T-211 and T-216 templates to the ProFunc server (http://www.ebi.ac.uk/thornton-srv/databases/profunc/; accessed on 3 June 2012). Shown is the PDB template match (based on E value scores) for each NS5A structure template. Structural and sequence statistics are also shown based on alignments from query and matched PDB (chain A) templates.
Matches are categorized as follows: certain (E value, <1.00E−06), probable (1.00E−06 < E value < 0.01), possible (0.01 < E value < 0.10), or long shots (0.10 < E value < 10.0). Details on scoring functions and methods are described elsewhere (41).
The structural similarity shows the percentage of all residues that lie in structurally fittable segments (i.e., segments consisting of at least 7 consecutive residues that, when structurally superposed on equivalent C-α positions, give a root mean squaxyre deviation (RMSD) of <3.0 Å.
Length of longest fittable segment.
Number of residue pairs from the two structures that could be superposed on the C-α atoms to give an RMSD of <3.0 Å.
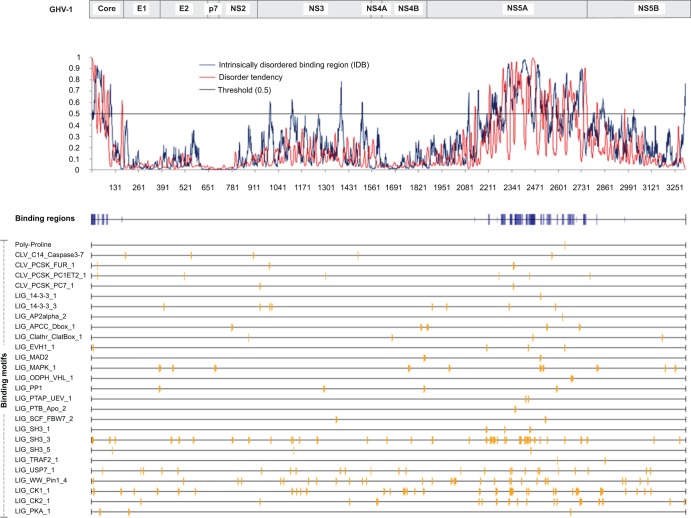
The HCV NS5A protein is known to have intrinsically disordered regions (IDRs) that are relevant to many of its inferred functions, including host regulatory and signaling processes (58–60). In silico analysis of the GHV-1 polyprotein predicted the presence of intrinsically disordered regions within the core and NS5A proteins (Fig. 4). In particular, 70.8% of NS5A's 521-aa C terminus (residues 2223 to 2743) was predicted to be intrinsically disordered, including several continuous disordered stretches of >20 aa (residues 2223 to 2260, 2304 to 2457, 2468 to 2512, 2565 to 2588, 2628 to 2656, and 2677 to 2743). Disorder in the HCV core and NS5A proteins has been observed previously, but the length of the disordered region in the GHV NS5A is unprecedented among known flaviviruses (60, 61).
Fig 4.
Disorder tendency and binding regions of GHV-1 proteins. Profiles of a general disorder prediction (red) using the IUPRED method and of disordered binding regions (blue) using the ANCHOR method (see Materials and Methods) are shown. Blue bars indicate predicted binding regions. Shading is directly proportional to the prediction score (lighter shading represents a lower score and vice versa) and is based on a threshold of 0.5 (black). Matching motifs are also indicated with yellow bars and are mapped according to their position within NS5A. Only a selected set of 27 binding motifs (n = 94, in total) is shown.
Further analysis of the GHV NS5A detected IDRs with tendency to undergo substrate-induced disorder-to-order transitions, suggesting that the GHV NS5A, like that of HCV, may have the capacity to bind transiently to a range of substrates, including class 1, 3, and 5 Src homology 3 (SH3) protein domains (58–60) (Fig. 4). Congruently, IDRs exhibiting class II polyproline motifs PxxPx[KR] and PxxPxx[KR] (known as PP2.1 and PP2.2, respectively) were identified in the GHV NS5A C terminus. In HCV, these motifs are known to bind several SH3 domains, thus mediating protein-protein interactions in many signaling processes in the cytoplasm (58). While the PP2.2 motif was observed in both GHV-1 variants (2637-PPPPMVR-2643) and in GHV-2 (2636-PPPPMVR-2642), the PP2.1 motif was only present in GHV-2 (2421-PSSPTR-2426) and in the GHV-1 variant from animal BWC05 (2487-PEQPVR-2492).
Three-dimensional modeling of the 470 C-terminal residues of NS5A (model T-216) (Fig. 3), which includes the longest IDRs detected (Fig. 4), indicated that regions with a structural helix or strands were few and separated by long stretches of coiled residues. Concordantly, the PSIPRED (35) secondary structure profile for this region indicated a low percentage of residues in strand (10%) or helix (20%) conformation. Additionally, this model revealed structural modules that were similar to crystal structure models of the IκBα p65 subunit, which is known to interact with a variety of nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB)/Rel transcription factors (62). Most of these modules (the largest comprising an 18-aa-long stretch, residues 436 to 454) matched the N-terminus Rel homology domain of the IκBα p65 protein with significant structural similarity (RMSD < 3 Å) (Table 3). In relation to the HCV NS5A, the parallel presence of IDRs and the apparent conservation of N- and C-terminal functional motifs in GHV lend additional support to the hypothesis that the putative NS5A sequence expresses a single protein product.
DISCUSSION
The detection of GHV in the black-and-white colobus represents the first documented natural infection of a nonhuman primate with a Hepacivirus and expands the known host range of this globally important viral genus to Old World monkeys. The recent discovery of hepaciviruses infecting dogs, horses, rodents, and bats (4, 5, 11, 15) suggests that the evolution of this viral genus has involved several host shifts across diverse mammalian families. Phylogenetic analyses of GHV using NS3 helicase and NS5B RdRp amino acid sequence alignments confidently support the shared common ancestry among GHV, GBV-B, RHV, and BHV-112 within the Flaviviridae. This lineage's root near the origin of the Hepacivirus clade and the inclusion in this clade of viruses infecting both New and Old World monkeys suggest an early divergence of GHV from the other major Hepacivirus lineage containing HCV and the NPHVs, at least 1,000 to 1,500 ybp. Similarity plot analysis demonstrates the greatest amino acid identity between GHV and BHV-112 across most of their genomes, possibly reflecting the East African origins of these two viruses. However, GHV is more similar to RHV in NS5B. This pattern is reflected in topological discordance between phylogenies constructed from NS3 helicase and NS5B RdRp amino acid alignments. Together, these analyses suggest that cross-species transmission and ancient recombination have played important roles in the evolution of GHV and related hepaciviruses.
Genomic analyses of the two GHV subtypes showed moderate sequence divergence, with the majority of changes occurring at synonymous sites, suggesting an overall pattern of purifying selection. Analysis of intrahost variability supports this conclusion, in that the majority of polymorphisms within infected hosts were selectively neutral and overall synonymous nucleotide diversity significantly exceeded nonsynonymous nucleotide diversity across the viral genomes. These observations suggest that GHV is adapted to its host; in this light, we note that no animals appeared clinically ill at the time of sampling, nor have any been observed with overt clinical signs subsequently. While we are unable to rule out disease association definitively or to ascertain tissue tropism, the presence of an miRNA-122 binding site in the GHV 5′ UTR is suggestive of hepatotropism. For HCV, two such sites have been identified in the 5′ UTR with functions essential for replication in hepatocytes (63). Similarly, the importance of this binding site for hepaciviruses in general has been proposed based on its detection in the 5′ UTRs of RHV, NPHV, and GBV-B (4).
GHV contains features in the NS5A gene that substantially broaden our understanding of structural and functional variability within the Flaviviridae. The N-terminal amphipathic α-helix and structural organization within the GHV NS5A zinc-binding motif resemble the N-terminal membrane anchor and zinc binding regions of the NS5A in hepaciviruses and pestiviruses (57, 64). The degree of structural similarity at these regions suggests that, like the HCV NS5A, the GHV NS5A may perform functions involving cellular membrane association and zinc-binding coordination (e.g., RNA replication) (57, 65). However, the C-terminal half of GHV's unprecedentedly long NS5A protein is intrinsically disordered. IDRs can experience transient substrate-induced disorder-to-order conformations to provide low-affinity binding motifs that stabilize interactions mediated by canonical high-affinity binding sites. Accordingly, the extensive repertoire of binding substrates for HCV's NS5A protein has been largely attributed to intrinsic disorder at functionally active IDRs in domains II and III (59, 60, 66).
Interestingly, a region encompassing a 154-aa-long IDR within the GHV-1 NS5A (residues 2304 to 2457 in the polyprotein) exhibits structural similarity to the Rel homology domain of the transcription factor IκBα p65, which has been shown to interact with a variety of NF-κB/Rel transcription factors (62). This observation suggests that GHV NS5A may be involved in modulating key regulators involved in expression of proinflammatory, immunomodulatory, and antiapoptotic genes (67). Moreover, the GHV NS5A protein contains a tumor necrosis factor (TNF) receptor-associated factor 2 (TRAF2) binding motif (733-SFQE-736; polyprotein positions 2594 to 2597). For HCV, the NS5A-TRAF2 protein complex has been shown to inhibit TNF-induced NF-κB activation, thereby increasing cellular resistance to apoptotic stimuli (68), perhaps modulating host responses to pathogens, persistence of viral infection, and disease severity (69). An inhibitory effect by NS5A-protein complexes on TNF-induced NF-κB activation has also been reported for BVDV, albeit mediated through NS5A-binding interactions with different host immunomodulatory proteins (70).
The presence of the class II polyproline binding motif PP2.2 in the disordered region of all GHV variants provides further support for a potential role of GHV NS5A in fine-tuning a variety of cellular signaling pathways. This motif is also highly conserved in the NS5A of HCV and has been demonstrated to mediate interactions with SH3 protein domains (58, 71). Highly disordered regions at the polyproline-containing SH3 binding domain in HCV NS5A transiently adopt α-helical structures to form a noncanonical SH3-binding motif, which, in addition to the class II canonical SH3-binding motifs, is necessary for “bridging integrator protein 1” (Bin1)-SH3 binding (58). The PP2.1 and PP2.2 motifs found in GHV might also play important roles in the binding of other SH3 proteins, such as Src family kinases (e.g., Lyn and Fyn), which modulate activity of proteins that regulate several signaling pathways (72). The NS5A functional predictions proposed herein for GHV await experimental confirmation. Nevertheless, available data suggest that GHV encodes an 883- or 882-aa-long NS5A protein: (i) structure-based functional associations are compatible between the HCV and GHV NS5A proteins, (ii) canonical, HCV-like NS3/4A protease cleavage sites are absent within the GHV NS5A, and (iii) a possible NS3/4A protease cleavage site exists at the proposed C terminus of GHV's NS5A, including the “P1” Cys residue conserved among the hepaciviruses, pestiviruses, and pegiviruses.
Research on HCV has been limited by the lack of a suitable animal model. The chimpanzee model uniquely permits studies of HCV infectivity and pathogenesis (73) but has considerable drawbacks, including expense, availability, biocontainment, and ethical considerations (13). GBV-B has been advocated as an NHP surrogate model for HCV infection, but the natural host of this virus is still unknown, and GBV-B can only infect New World monkeys. Old World monkeys are the most commonly used NHPs in biomedical research and have served as well-established animal models for human disease (e.g., simian immunodeficiency virus [SIV] infection of macaques [Macaca sp.] or African green monkeys [Chlorocebus sp.] mimicking HIV infection in humans). The fact that GHV was found in an Old World primate host raises the possibility that it might infect other Old World monkeys and could lead to a useful animal model for human viral hepatitis, especially considering the putative liver tropism of GHV. Furthermore, although GHV shares a common ancestor with GBV-B, RHV, and BHV-112, it exhibits important structural and proposed functional similarities to HCV, such as the presence of a possible ARFP-F homolog within the core protein, the putative expression of a p7 protein, and the occurrence of IDRs in both core and NS5A proteins. GHV therefore contains unique combinations of structural and functional features that hold promise for expanding our understanding of the basic biology of the hepaciviruses. Examination of the ability of GHV to cause clinical disease is clearly in order, as are focused attempts to discover related viruses in other animals.
ACKNOWLEDGMENTS
We are grateful to the Uganda Wildlife Authority, the Uganda National Council for Science and Technology, and Makerere University Biological Field Station for granting permission to conduct this research, J. Byaruhanga, P. Katurama, A. Nyamwija, J. Rusoke, A. Mbabazi, and P. Omeja for assistance in the field, and L. Kilby for assistance with permitting and logistics. We also thank C. A. Lynberg, Centers for Disease Control and Prevention, IT Research & Development, for providing access to personal supercomputer computational resources to run 3D modeling predictions.
This work was funded by NIH grant TW009237 as part of the joint NIH-NSF Ecology of Infectious Disease program and the UK Economic and Social Research Council, through grants P51OD011106 and P51RR000167 to the Wisconsin National Primate Research Center, and by the University of Wisconsin School of Medicine and Public Health Wisconsin Partnership Program through the Wisconsin Center for Infectious Disease (WisCID).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Published ahead of print 5 June 2013
REFERENCES
- 1. Stapleton J, Foung S, Muerhoff A, Bukh J, Simmonds P. 2011. The GB viruses: a review and proposed classification of GBV-A, GBV-C (HGV), and GBV-D in genus Pegivirus within the family Flaviviridae. J. Gen. Virol. 92:233–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chandriani S, Skewes-Cox P, Zhong W, Ganem DE, Divers TJ, Blaricum AJV, Tennant BC, Kistler AL. 2013. Identification of a previously undescribed divergent virus from the Flaviviridae family in an outbreak of equine serum hepatitis. Proc. Natl. Acad. Sci. U. S. A. 110:E1407–E1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kapoor A, Simmonds P, Cullen JM, Scheel T, Medina JL, Giannitti F, Nishiuchi E, Brock KV, Burbelo PD, Rice CM, Lipkin WI. 2013. Identification of a pegivirus (GBV-like virus) that infects horses. J. Virol. 87:7185–7190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kapoor A, Simmonds P, Scheel TK, Hjelle B, Cullen JM, Burbelo PD, Chauhan LV, Duraisamy R, Sanchez Leon M, Jain K, Vandegrift KJ, Calisher CH, Rice CM, Lipkin WI. 2013. Identification of rodent homologs of hepatitis C virus and pegiviruses. mBio 4:e00216–13. 10.1128/mBio.00216-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Quan PL, Firth C, Conte JM, Williams SH, Zambrana-Torrelio CM, Anthony SJ, Ellison JA, Gilbert AT, Kuzmin IV, Niezgoda M, Osinubi MO, Recuenco S, Markotter W, Breiman RF, Kalemba L, Malekani J, Lindblade KA, Rostal MK, Ojeda-Flores R, Suzan G, Davis LB, Blau DM, Ogunkoya AB, Alvarez Castillo DA, Moran D, Ngam S, Akaibe D, Agwanda B, Briese T, Epstein JH, Daszak P, Rupprecht CE, Holmes EC, Lipkin WI. 2013. Bats are a major natural reservoir for hepaciviruses and pegiviruses. Proc. Natl. Acad. Sci. U. S. A. 110:8194–8199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Deinhardt F, Holmes A, Capps R, Popper H. 1967. Studies on the transmission of human viral hepatitis to marmoset monkeys. I. Transmission of disease, serial passages, and description of liver lesions. J. Exp. Med. 125:673–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Simons J, Pilot-Matias T, Leary T, Dawson G, Desai S, Schlauder G, Muerhoff A, Erker J, Buijk S, Chalmers M, Van Sant CL, Mushahwar IK. 1995. Identification of two flavivirus-like genomes in the GB hepatitis agent. Proc. Natl. Acad. Sci. U. S. A. 92:3401–3405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schaluder G, Dawson G, Simons J, Pilot-Matias T, Gutierrez R, Heynen C, Knigge M, Kurpiewski G, Buijk S, Leary T, Muerhoff AS, Desai SM, Mushahwar IK. 1995. Molecular and serologic analysis in the transmission of the GB hepatitis agents. J. Med. Virol. 46:81–90 [DOI] [PubMed] [Google Scholar]
- 9. Bukh J, Apgar C. 1997. Five new or recently discovered (GBV-A) virus species are indigenous to New World monkeys and may constitute a separate genus of the Flaviviridae. Virology 229:429–436 [DOI] [PubMed] [Google Scholar]
- 10. Bukh J, Apgar C, Govindarajan S, Purcell R. 2001. Host range studies of GB virus-B hepatitis agent, the closest relative of hepatitis C virus, in New World monkeys and chimpanzees. J. Med. Virol. 65:694–697 [DOI] [PubMed] [Google Scholar]
- 11. Kapoor A, Simmonds P, Gerold G, Qaisar N, Jain K, Henriquez J, Firth C, Hirschberg D, Rice C, Shields S, Lipkin W. 2011. Characterization of a canine homolog of hepatitis C virus. Proc. Natl. Acad. Sci. U. S. A. 108:11608–11613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bright H, Carroll A, Watts P, Fenton R. 2004. Development of a GB virus B marmoset model and its validation with a novel series of hepatitis C virus NS3 protease inhibitors. J. Virol. 78:2062–2071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Akari H, Iwasaki Y, Yoshida T, Iijima S. 2009. Non-human primate surrogate model of hepatitis C virus infection. Microbiol. Immunol. 53:53–57 [DOI] [PubMed] [Google Scholar]
- 14. Iwasaki Y, Mori K, Ishii K, Maki N, Iijima S, Yoshida T, Okabayashi S, Katakai Y, Lee Y, Saito A, Fukai H, Kimura N, Ageyama N, Yoshizaki S, Suzuki T, Yasutomi Y, Miyamura T, Kannagi M, Akari H. 2011. Long-term persistent GBV-B infection and development of a chronic and progressive hepatitis C-like disease in marmosets. Front. Microbiol. 2:240. 10.3389/fmicb.2011.00240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Burbelo PD, Dubovi EJ, Simmonds P, Medina JL, Henriquez JA, Mishra N, Wagner J, Tokarz R, Cullen JM, Iadarola MJ, Rice CM, Lipkin WI, Kapoor A. 2012. Serology-enabled discovery of genetically diverse hepaciviruses in a new host. J. Virol. 86:6171–6178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Struhsaker T. 1997. Ecology of an African rain forest: logging in Kibale and the conflict between conservation and exploitation. University Press of Florida, Gainesville, FL [Google Scholar]
- 17. Goldberg TL, Paige SB, Chapman CA. 2012. The Kibale EcoHealth Project: exploring connections among human health, animal health, and landscape dynamics in western Uganda, p 452–465 In Aguirre AA, Daszak P, Ostfeld RS. (ed), New directions in conservation medicine: applied cases of ecological health. Oxford University Press, New York, NY [Google Scholar]
- 18. Lauck M, Hyeroba D, Tumukunde A, Weny G, Lank SM, Chapman CA, O'Connor DH, Friedrich TC, Goldberg TL. 2011. Novel, divergent simian hemorrhagic fever viruses in a wild Ugandan red colobus monkey discovered using direct pyrosequencing. PLoS One 6:e19056. 10.1371/journal.pone.0019056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lauck M, Sibley SD, Hyeroba D, Tumukunde A, Weny G, Chapman CA, Ting N, Switzer WM, Kuhn JH, Friedrich TC, O'Connor DH, Goldberg TL. 2013. Exceptional simian hemorrhagic fever virus diversity in a wild African primate community. J. Virol. 87:688–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Altschul S, Gish W, Miller W, Myers E, Lipman D. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 21. Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059–3066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Abascal F, Zardoya R, Telford M. 2010. TranslatorX: multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res. 38:W7–W13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425 [DOI] [PubMed] [Google Scholar]
- 24. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Drummond AJ, Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7:214. 10.1186/1471-2148-7-214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Martin DP, Lemey P, Lott M, Moulton V, Posada D, Lefeuvre P. 2010. RDP3: a flexible and fast computer program for analyzing recombination. Bioinformatics 26:2462–2463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Korber B. 2000. HIV signature and sequence variation analysis, p 55–72 In Rodrigo AG, Learn GH. (ed), Computational analysis of HIV molecular sequences. Kluwer Academic Publishers, Dordrecht, Netherlands [Google Scholar]
- 28. Hughes A, Becker E, Lauck M, Karl J, Braasch A, O'Connor D, O'Connor S. 2012. SIV genome-wide pyrosequencing provides a comprehensive and unbiased view of variation within and outside CD8 T lymphocyte epitopes. PLoS One 7:e47818. 10.1371/journal.pone.0047818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lole K, Bollinger R, Paranjape R, Gadkari D, Kulkarni S, Novak N, Ingersoll R, Sheppard H, Ray S. 1999. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 73:152–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wu S, Skolnick J, Zhang Y. 2007. Ab initio modeling of small proteins by iterative TASSER simulations. BMC Biol. 5:17. 10.1186/1741-7007-5-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang Y. 2007. Template-based modeling and free modeling by I-TASSER in CASP7. Proteins 69(Suppl 8):108–117 [DOI] [PubMed] [Google Scholar]
- 32. Roy A, Kucukural A, Zhang Y. 2010. I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5:725–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Laskowski R, MacAurther M, Moss D, Thornton J. 1993. Procheck—a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 26:47–60 [Google Scholar]
- 34. Vriend G. 1990. WHAT IF: a molecular modeling and drug design program. J. Mol. Graph. 8:29, 52–56 [DOI] [PubMed] [Google Scholar]
- 35. Jones D. 1999. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292:195–202 [DOI] [PubMed] [Google Scholar]
- 36. Dosztanyi Z, Meszaros B, Simon I. 2009. ANCHOR: web server for predicting protein binding regions in disordered proteins. Bioinformatics 25:2745–2746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Meszaros B, Simon I, Dosztanyi Z. 2009. Prediction of protein binding regions in disordered proteins. PLoS Comput. Biol. 5:e1000376. 10.1371/journal.pcbi.1000376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fuxreiter M, Tompa P, Simon I. 2007. Local structural disorder imparts plasticity on linear motifs. Bioinformatics 23:950–956 [DOI] [PubMed] [Google Scholar]
- 39. Gould C, Diella F, Via A, Puntervoll P, Gemund C, Chabanis-Davidson S, Michael S, Sayadi A, Bryne J, Chica C, Seiler M, Davey N, Haslam N, Weatheritt R, Budd A, Hughes T, Pas J, Rychlewski L, Trave G, Aasland R, Helmer-Citterich M, Linding R, Gibson T. 2010. ELM: the status of the 2010 eukaryotic linear motif resource. Nucleic Acids Res. 38:D167–D180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yap K, Kim J, Truong K, Sherman M, Yuan T, Ikura M. 2000. Calmodulin target database. J. Struct. Funct. Genomics 1:8–14 [DOI] [PubMed] [Google Scholar]
- 41. Laskowski R, Watson J, Thornton J. 2005. Protein function prediction using local 3D templates. J. Mol. Biol. 351:614–626 [DOI] [PubMed] [Google Scholar]
- 42. Nakano T, Lau GMG, Lau GML, Sugiyama M, Mizokami M. 2012. An updated analysis of hepatitis C virus genotypes and subtypes based on the complete coding region. Liver Int. 32:339–345 [DOI] [PubMed] [Google Scholar]
- 43. Nielsen H, Engelbrecht J, Brunak S, von Heijne G. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1–6 [DOI] [PubMed] [Google Scholar]
- 44. Shiryaev SA, Thomsen ER, Cieplak P, Chudin E, Cheltsov AV, Chee MS, Kozlov IA, Strongin AY. 2012. New details of HCV NS3/4A proteinase functionality revealed by a high-throughput cleavage assay. PLoS One 7:e35759. 10.1371/journal.pone.0035759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Petersen T, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8:785–786 [DOI] [PubMed] [Google Scholar]
- 46. Reynolds SM, Kall L, Riffle ME, Bilmes JA, Noble WS. 2008. Transmembrane topology and signal peptide prediction using dynamic Bayesian networks. PLoS Comput. Biol. 4:e1000213. 10.1371/journal.pcbi.1000213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Branch AD, Stump DD, Gutierrez JA, Eng F, Walewski JL. 2005. The hepatitis C virus alternate reading frame (ARF) and its family of novel products: the alternate reading frame protein/F-protein, the double-frameshift protein, and others. Semin. Liver Dis. 25:105–117 [DOI] [PubMed] [Google Scholar]
- 48. Ghibaudo D, Cohen L, Penin F, Martin A. 2004. Characterization of GB virus B polyprotein processing reveals the existence of a novel 13-kDa protein with partial homology to hepatitis C virus p7 protein. J. Biol. Chem. 279:24965–24975 [DOI] [PubMed] [Google Scholar]
- 49. Takikawa S, Engle R, Emerson S, Purcell R, St Claire M, Bukh J. 2006. Functional analyses of GB virus B p13 protein: development of a recombinant GB virus B hepatitis virus with a p7 protein. Proc. Natl. Acad. Sci. U. S. A. 103:3345–3350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sbardellati A, Scarselli E, Amati V, Falcinelli S, Kekule AS, Traboni C. 2000. Processing of GB virus B non-structural proteins in cultured cells requires both NS3 protease and NS4A cofactor. J. Gen. Virol. 81:2183–2188 [DOI] [PubMed] [Google Scholar]
- 51. Magiorkinis G, Magiorkinis E, Paraskevis D, Ho SY, Shapiro B, Pybus OG, Allain JP, Hatzakis A. 2009. The global spread of hepatitis C virus 1a and 1b: a phylodynamic and phylogeographic analysis. PLoS Med. 6:e1000198. 10.1371/journal.pmed.1000198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pybus OG, Barnes E, Taggart R, Lemey P, Markov PV, Rasachak B, Syhavong B, Phetsouvanah R, Sheridan I, Humphreys IS, Lu L, Newton PN, Klenerman P. 2009. Genetic history of hepatitis C virus in East Asia. J. Virol. 83:1071–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Takikawa S, Engle R, Faulk K, Emerson S, Purcell R, Bukh J. 2010. Molecular evolution of GB virus B hepatitis virus during acute resolving and persistent infections in experimentally infected tamarins. J. Gen. Virol. 91:727–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Worobey M, Telfer P, Souquiere S, Hunter M, Coleman CA, Metzger MJ, Reed P, Makuwa M, Hearn G, Honarvar S, Roques P, Apetrei C, Kazanji M, Marx PA. 2010. Island biogeography reveals the deep history of SIV. Science 329:1487. 10.1126/science.1193550 [DOI] [PubMed] [Google Scholar]
- 55. Lauck M, Alvarado-Mora M, Becker E, Bhattacharya D, Striker R, Hughes A, Carrilho F, O'Connor D, Pinho J. 2012. Analysis of hepatitis C virus intrahost diversity across the coding region by ultradeep pyrosequencing. J. Virol. 86:3952–3960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Parameswaran P, Charlebois P, Tellez Y, Nunez A, Ryan EM, Malboeuf CM, Levin JZ, Lennon NJ, Balmaseda A, Harris E, Henn MR. 2012. Genome-wide patterns of intrahuman dengue virus diversity reveal associations with viral phylogenetic clade and interhost diversity. J. Virol. 86:8546–8558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tellinghuisen T, Paulson M, Rice C. 2006. The NS5A protein of bovine viral diarrhea virus contains an essential zinc-binding site similar to that of the hepatitis C virus NS5A protein. J. Virol. 80:7450–7458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Feuerstein S, Solyom Z, Aladag A, Favier A, Schwarten M, Hoffmann S, Willbold D, Brutscher B. 2012. Transient structure and SH3 interaction sites in an intrinsically disordered fragment of the hepatitis C virus protein NS5A. J. Mol. Biol. 420:310–323 [DOI] [PubMed] [Google Scholar]
- 59. Gupta G, Qin H, Song J. 2012. Intrinsically unstructured domain 3 of hepatitis C virus NS5A forms a “fuzzy complex” with VAPB-MSP domain which carries ALS-causing mutations. PLoS One 7:e39261. 10.1371/journal.pone.0039261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hanoulle X, Badillo A, Verdegem D, Penin F, Lippens G. 2010. The domain 2 of the HCV NS5A protein is intrinsically unstructured. Protein Pept. Lett. 17:1012–1018 [DOI] [PubMed] [Google Scholar]
- 61. Ivanyi-Nagy R, Lavergne JP, Gabus C, Ficheux D, Darlix JL. 2008. RNA chaperoning and intrinsic disorder in the core proteins of Flaviviridae. Nucleic Acids Res. 36:712–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Beg A, Baldwin A. 1993. The IkB proteins: multifunctional regulators of Rel/NF-kB transcription factors. Genes Dev. 7:2064–2070 [DOI] [PubMed] [Google Scholar]
- 63. Li Y, Masaki T, Yamane D, McGivern DR, Lemon SM. 2013. Competing and noncompeting activities of miR-122 and the 5′ exonuclease Xrn1 in regulation of hepatitis C virus replication. Proc. Natl. Acad. Sci. U. S. A. 110:1881–1886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Tellinghuisen T, Marcotrigiano J, Rice C. 2005. Structure of the zinc-binding domain of an essential component of the hepatitis C virus replicase. Nature 435:374–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Penin F, Brass V, Appel N, Ramboarina S, Montserret R, Ficheux D, Blum H, Bartenschlager R, Moradpour D. 2004. Structure and function of the membrane anchor domain of hepatitis C virus nonstructural protein 5A. J. Biol. Chem. 279:40835–40843 [DOI] [PubMed] [Google Scholar]
- 66. Yamasaki L, Arcuri H, Jardim A, Bittar C, de Carvalho-Mello I, Rahal P. 2012. New insights regarding HCV-NS5A structure/function and indication of genotypic differences. Virol. J. 9:14. 10.1186/1743-422X-9-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Pahl H. 1999. Activators and target genes of Rel/NF-kappaB transcription factors. Oncogene 18:6853–6866 [DOI] [PubMed] [Google Scholar]
- 68. Park K, Choi S, Lee S, Hwang S, Lai M. 2002. Nonstructural 5A protein of hepatitis C virus modulates tumor necrosis factor alpha-stimulated nuclear factor kappa B activation. J. Biol. Chem. 277:13122–13128 [DOI] [PubMed] [Google Scholar]
- 69. Rahman M, McFadden G. 2011. Modulation of NF-kappaB signalling by microbial pathogens. Nat. Rev. Microbiol. 9:291–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Zahoor M, Yamane D, Mohamed Y, Suda Y, Kobayashi K, Kato K, Tohya Y, Akashi H. 2010. Bovine viral diarrhea virus non-structural protein 5A interacts with NIK- and IKKbeta-binding protein. J. Gen. Virol. 91:1939–1948 [DOI] [PubMed] [Google Scholar]
- 71. Nanda S, Herion D, Liang T. 2006. The SH3 binding motif of hepatitis C virus NS5A protein interacts with Bin1 and is important for apoptosis and infectivity. Gastroenterology 130:794–809 [DOI] [PubMed] [Google Scholar]
- 72. Macdonald A, Harris M. 2004. Hepatitis C virus NS5A: tales of a promiscuous protein. J. Gen. Virol. 85:2485–2502 [DOI] [PubMed] [Google Scholar]
- 73. Bukh J, Meuleman P, Tellier R, Engle RE, Feinstone SM, Eder G, Satterfield WC, Govindarajan S, Krawczynski K, Miller RH, Leroux-Roels G, Purcell RH. 2010. Challenge pools of hepatitis C virus genotypes 1–6 prototype strains: replication fitness and pathogenicity in chimpanzees and human liver-chimeric mouse models. J. Infect. Dis. 201:1381–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]