Abstract
Nonhuman primate-simian immunodeficiency virus (SIV) models are powerful tools for studying the pathogenesis of human immunodeficiency virus type 1 (HIV-1) in the brain. Our laboratory recently isolated a neuropathogenic viral swarm, SIVsmH804E, a derivative of SIVsmE543-3, which was the result of sequential intravenous passages of viruses isolated from the brains of rhesus macaques with SIV encephalitis. Animals infected with SIVsmH804E or its precursor (SIVsmH783Br) developed SIV meningitis and/or encephalitis at high frequencies. Since we observed macaques with a combination of meningitis and encephalitis, as well as animals in which meningitis or encephalitis was the dominant component, we hypothesized that distinct mechanisms could be driving the two pathological states. Therefore, we assessed viral populations in the meninges and the brain parenchyma by laser capture microdissection. Viral RNAs were isolated from representative areas of the meninges, brain parenchyma, terminal plasma, and cerebrospinal fluid (CSF) and from the inoculum, and the SIV envelope fragment was amplified by PCR. Phylogenetic analysis of envelope sequences from the conventional progressors revealed compartmentalization of viral populations between the meninges and the parenchyma. In one of these animals, viral populations in meninges were closely related to those from CSF and shared signature truncations in the cytoplasmic domain of gp41, consistent with a common origin. Apart from magnetic resonance imaging (MRI) and positron-emission tomography (PET) imaging, CSF is the most accessible assess to the central nervous system for HIV-1-infected patients. However, our results suggest that the virus in the CSF may not always be representative of viral populations in the brain and that caution should be applied in extrapolating between the properties of viruses in these two compartments.
INTRODUCTION
Prior to the development of highly active antiretroviral therapy (HAART), a significant proportion of human immunodeficiency virus type 1 (HIV-1)-infected individuals developed a range of motor and cognitive symptoms together with behavioral symptoms, such as muscle weakness, impairment of short- and long-term memory, social withdrawal, and change of personality, that are collectively referred to as HIV-1-associated dementia (HAD) or HIV encephalopathy. The most severe forms of HAD can lead to a mute and vegetative state which is lethal. HAD in many individuals is associated with HIV-1 encephalitis (HIVE), which is characterized by the presence of HIV-expressing multinucleated giant cells (MNGC) in the brain. However, the development and application of HAART for use by HIV-1-infected individuals have decreased the incidence of progression to HAD/HIVE dramatically, at least in developed countries, where HIV-1-infected individuals have access to and can afford therapy. Despite effective suppression of systemic viral replication by HAART, to near or under detectable levels in the plasma, there has been an increased number of patients with HIV-1-associated neurocognitive disorder (HAND), also referred to as minor cognitive and motor disorder (MCMD) (1). Unlike HAD, the progression of HAND/MCMD is more gradual, and symptoms are minor, making it difficult to differentiate from non-HIV-related cognitive and motor symptoms. Therefore, the reported prevalence is extremely variable depending upon the study and criteria used for diagnosis (15 to 50%) (1, 2). Several models for the pathogenesis of HAND/MCMD arising during HAART treatment have been suggested, such as incomplete suppression of viral replication in the brain due to poor penetration of drugs to the central nervous system (CNS) (3–5), effects of aging on the brain (6–8), and CNS toxicity of long-term application of antiretroviral therapy (ART) (9, 10), but the driving mechanism(s) of progression to HAND during HAART has not yet been determined.
Nonhuman primate-simian immunodeficiency virus (SIV) models are powerful tools for studying the pathogenesis of HIV-1. Infection of macaques with certain strains of SIV can result in CNS disorders, such as meningitis and encephalitis, that are associated with characteristic multinucleated giant cells. The rhesus macaque-SIV model allows accessibility to samples of any part of the brain at any stage of disease progression to evaluate the mechanisms of viral pathogenicity and accessibility of HAART drugs to the brain (11–13). Our laboratory recently isolated a neuropathogenic viral swarm, SIVsmH783Br, a derivative of SIVsmE543-3, by sequential intravenous passages of viruses isolated from the brains of rhesus macaques with SIV encephalitis (SIVE) (14). An additional passage was performed to further adapt SIVsmH783Br in this study; we isolated SIVsmH804E from the brain of a SIVsmH783Br-infected animal that developed severe SIV encephalitis. Animals infected with SIVsmH804E or its precursor (SIVsmH783Br) developed SIV meningitis and/or encephalitis at high frequencies.
Our previous report demonstrated the compartmentalization of viral populations between plasma, cerebrospinal fluid (CSF), and the brain of the animal infected with SIVsmH631Br, a precursor of SIVsmH783Br and SIVsmH804E (15). This observation was consistent with the majority of studies of viral populations in the CNS of HIV-infected patients (16–18), although at least one report suggested that viral populations could redistribute to systemic tissues (19). It has also been reported that viral populations are compartmentalized within different regions of the brain from the same individual (20, 21). However, not much analysis has been done on the viral populations in the meninges, which may contain specific sequences that play an important role in the development of meningitis. Since in our preliminary studies we observed compartmentalization between virus populations from the brain and the CSF, suggestive of different sources for these populations (15), we wished to investigate whether the meninges were a potential source of CSF virus. In addition, pathological findings were somewhat variable among our study animals; thus, we observed macaques with meningoencephalitis as well as animals in whom meningitis or encephalitis was the predominant, if not exclusive, feature in the multiple CNS samples we examined. From these observations, we hypothesized that distinct mechanisms could be driving the two pathological states. To address these two related questions, we focused on studying viral populations in the brain parenchyma and the meninges by using laser capture microdissection (LCM) to assess whether there was evidence of compartmentalization between these two regions of the CNS.
MATERIALS AND METHODS
Animals and viruses.
The four rhesus macaques that form the basis of this study were selected from a pool of 12 macaques inoculated with either of two closely related neuropathogenic isolates: SIVsmH783Br and SIVsmE804E. These two isolates cumulatively resulted in SIV encephalitis and/or meningitis in over 70% of inoculated animals. SIVsmH783Br was isolated from the brain of RhH783, a macaque inoculated with SIVsmH631Br that developed SIVE (15). Briefly, the fresh brain from RhH783 was collected at the time of necropsy and homogenized. The supernatant of the homogenized brain was incubated with fresh rhesus macaque peripheral blood mononuclear cells (PBMC) to isolate the viral stock, SIVsmH783Br. Five hundred 50% tissue culture infective doses (TCID50) of SIVsmH783Br was inoculated intravenously into six Indian-origin rhesus macaques (SIV, simian retrovirus [SRV], and simian T cell leukemia virus type 1 [STLV-1] seronegative) for in vivo analysis (14, 22). These macaques did not express any of the major histocompatibility complex (MHC) alleles known to be restrictive for SIVmac239. They included a mixture of TRIM5α genotypes, although only one was homozygous restrictive (TRIMTFP/TFP) (23). The viral swarm, SIVsm804E, was isolated from the brain of one of these SIVsmH783Br-inoculated macaques, RhH804, and was inoculated intravenously into an additional six TRIMTFP/Q (moderately susceptible) rhesus macaques. Only one of these macaques expressed an MHC allele associated with restriction of SIVmac239. Macaques chosen for the present study included three inoculated with SIVsmH783Br (RhDBTN, Rh802, and Rh804), expressing TRIMTFP/Q, TRIMCypA/Q, and TRIMTFP/Q, and one inoculated with SIVsmH804E (Rh817), expressing TRIMTFP/Q. None of these four macaques expressed MHC-I alleles known to be restrictive for SIVmac239. All animals were housed in accordance with American Association for Accreditation of Laboratory Animal Care standards. The investigators adhered to the Guide for the Care and Use of Laboratory Animals (24) and to NIAID Animal Care and Use Committee-approved protocols.
ISH to identify SIV-expressing cells.
Formalin-fixed, paraffin-embedded brain sections were assayed for SIV RNA expression by in situ hybridization (ISH) as previously described (25). Briefly, the sections were hybridized overnight at 45°C with either a sense or an antisense digoxigenin-UTP-labeled SIVmac239 riboprobe that spanned the entire genome. The hybridized sections were blocked with 3% normal sheep and horse sera in 0.1 M Tris, pH 7.4, and then incubated with sheep anti-digoxigenin–alkaline phosphatase (Roche Molecular Biochemicals) and nitroblue tetrazolium–5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (BCIP; Vector Laboratories), followed by counterstaining with Nuclear Fast Red solution (Sigma). ISH-stained tissues were visualized and photographed with a Zeiss Axio Imager Z1 microscope (Zeiss).
Isolation of viral RNA and sequencing.
CSF and plasma were collected from animals at the time of necropsy and cryopreserved. The viral RNA was extracted using a QIAamp Viral RNA minikit (Qiagen Inc., Santa Clara, CA). All animals used in this study were perfused with saline prior to sample collection to avoid blood contamination in the tissues. Fresh brain tissue was collected from the frontal, parietal, and temporal lobes, the cerebellum, and the midbrain during necropsy. Either the left or right hemisphere was used for laser capture studies, and the opposite hemisphere was used for histopathology. Fresh tissues were embedded in OCT compound and cryopreserved for LCM. Three sections of brain tissue (8 μm thick) were made from the same tissue block. The first section was stained with KK45 (NIH AIDS Repository Program) antibody to confirm the presence of viral protein. The third section was stained with hematoxylin and eosin to determine the delineation between the meninges and the brain parenchyma. The middle section was placed on a membrane slide (Leica, Buffalo Grove, IL) to cut out areas of meninges and the brain parenchyma by use of a Leica AMS-LMD (Leica, Buffalo Grove, IL) microscope. This allowed us to verify the presence of SIV-infected cells in the serial sections without need for fixation of the section used for LCM, since that would interfere with PCR amplification of the 3-kb Env gene fragment. The same procedure was performed with several tissue blocks from the areas of the brain listed above for each of the four animals. Multiple pieces consisting of meninges were laser excised and pooled in a single tube, and a similar procedure was followed for the parenchyma. In addition, two individual perivascular lesions (PVL), with only a single piece of tissue per tube, were collected from the brain of animal Rh804. Total RNA was extracted from each sample by use of an Arcturus PicoPure RNA isolation kit (Arcturus, Mountain View, CA) and then treated with DNase I (Invitrogen, Grand Island, NY) to digest residual genomic DNA. The viral RNA was reverse transcribed using a Thermoscript reverse transcriptase PCR (RT-PCR) system (Invitrogen) and the primer 9341-R (CATCATCCACATCATCCATG). The envelope DNA fragment was amplified by PCR with Platinum Taq DNA polymerase (Invitrogen) and primers 6463-F (GGTGTTGCTATCATTGTCAGC) and 9341-R and then subcloned into the pCR4-TOPO vector by use of a TOPO TA cloning kit (Invitrogen) for sequencing. A minimum of 9 clones were selected for the two inocula (SIVsmH783Br and SIVsmH804E) and for each sample from the four monkeys, and the complete envelope gene was sequenced.
Phylogenetic analysis of sequences.
The sequences were aligned using MAFFT (26, 27), and the alignments were viewed and manually corrected, where necessary, by using BioEdit (http:www.mbio.ncsu.edu/bioedit/bioedit.html). Columns containing gaps in the alignment were stripped prior to phylogenetic analyses. Trees were built using the maximum likelihood method, with the general time-reversible model of evolution and gamma distribution of rate variation across sites, using the MEGA software package (28). Trees were rendered with FigTree (http://tree.bio.ed.ac.uk/software/figtree/), with rooting on the SIVsmE543-3 sequence (accession number U72748.2). We used the LANL HIV Database Highlighter tool (http://www.hiv.lanl.gov/content/sequence/HIGHLIGHT/highlighter_top.html), which was designed to detect intrapatient recombination of HIV-1 sequences, to rule out recombination between viruses within a sample.
Construction of env chimeras.
Viral RNAs were extracted from the brain parenchyma and meninges of animal RhH804 as described above. A DNA fragment including part of vpx, vpr, and most of the envelope gene was amplified by RT-PCR with Herculase II Fusion DNA polymerase (Stratagene, Santa Clara, CA), using the primers Bst-F (GAAGAGGCCTTCGAATGGCTAAACAG) and 9341-R. The amplified DNA fragments were digested with the BstBI and NdeI restriction enzymes (New England BioLabs, Ipswich, MA), substituted for the corresponding region of the full-length SIVsmE543-3 clone, and evaluated for production of virus by transfection of 293T cells.
Viral replication in PBMC and MDMs.
Virus replication of the clones obtained above was evaluated in rhesus macaque PBMC and monocyte-derived macrophages (MDMs) as described elsewhere (14, 22). PBMC from SIV-naive, healthy rhesus macaques were separated from whole blood, stimulated with 5 μg of phytohemagglutinin (PHA) per ml and 10% interleukin-2 (IL-2) (Advanced Biotechnologies, Columbia, MD) for 3 days, and maintained in RPMI 1640 medium containing 10% fetal calf serum (FCS) and 10% IL-2. Rhesus macaque MDMs were obtained from rhesus macaque PBMC as previously described. Briefly, fresh PBMC were incubated with anti-nonhuman-primate CD14 magnetic beads (Miltenyi Biotec, Auburn, CA) and positively selected with MACS separation columns (Miltenyi Biotec). A total of 3 × 105 cells per well of CD14-positive cells were cultured in a 46-well plastic plate for 4 days in RPMI 1640 containing 10% FCS, 10% human serum type AB (Sigma, St. Louis, MO), and 20 ng/ml of macrophage colony-stimulating factor (R&D Systems, Minneapolis, MN). Wells were washed two times with Hanks balanced salt solution (HBSS) and cultured in fresh medium for three additional days. PBMC (5 × 105/well) were dispensed into a 46-well plastic plate and then inoculated with each chimeric virus at a multiplicity of infection (MOI) of 0.01, using the spinoculation method (29). MDMs were incubated with virus at an MOI of 0.01 for 1 h and then washed twice with HBSS and cultured in fresh medium. Virion-associated reverse transcriptase (RT) activity of the culture supernatant was monitored periodically.
Nucleotide sequence accession numbers.
The nucleotide sequences generated in this study have been deposited in GenBank under accession numbers KF198990 to KF199058 (RhDBTN tree), KF199059 to KF199135 (Rh802 tree), KF198820 to KF198938 (Rh804 tree), and KF198939 to KF198989 (Rh817 tree).
RESULTS
In vivo passage of SIV from the brain.
To develop a reproducible rhesus macaque model of neuro-AIDS, our group has been conducting sequential intravenous rhesus macaque passages of viruses isolated from the brains of rhesus macaques with SIVE, and we recently isolated the neuropathogenic viral swarm SIVsmH783Br (14). An additional passage was conducted with the virus isolated from a SIVsmH783Br-infected animal, designated SIVsmH804E, by intravenous inoculation into a group of six rhesus macaques. Although this study is still in progress, with one animal surviving, three of these animals developed SIVE, similar to the incidence of SIVE observed using the previous passage of virus, i.e., SIVsmH783Br (data not shown). The SIVsmH783Br and SIVsmH804E isolates used in this study were approximately 1.7% divergent in envelope sequence from the original parental SIVsmE543-3 isolate and are quite closely related to one another (0.6% divergent), despite differing by one animal passage.
A representative group of 4 animals that developed SIVE (RhDBTN, Rh802, Rh804, and Rh817) was selected prospectively from the pool of both SIVsmH783Br- and SIVsmH804E-infected animals for subsequent analysis by laser capture microdissection (Table 1). Our criteria for selection were as follows: (i) pathological evidence of SIV encephalitis, with or without involvement of the meninges; (ii) detection of SIV expression by in situ hybridization; and (iii) properly cryopreserved brain tissues. The last criterion eliminated any retrospective samples. Two of these monkeys, RhDBTN and Rh802, progressed rapidly, developing neurological symptoms (tremors and motor weakness) necessitating euthanasia at 13 and 18 weeks postinoculation, respectively. The other two monkeys, Rh804 and Rh817, progressed more slowly and were sacrificed at 43 and 47 weeks postinoculation, respectively, also due to neurological symptoms. Histopathologic examination of the brain tissue revealed that animals RhDBTN, Rh802, and Rh804 had SIV-induced meningoencephalitis, whereas Rh817 had only histopathologic evidence of encephalitis (Table 1) upon examination of multiple tissue sections. As observed previously, lesions were characterized by perivascular accumulations of SIV-expressing macrophages and multinucleated giant cells admixed with lymphocytes as well as glial nodules.
Table 1.
Summary of study animalsa
| Animal | Inoculum | Meningitis | Encephalitis | CSF viral load at death (log10 copies) | Plasma viral load at death (log10 copies) | Time of survival (wk) |
|---|---|---|---|---|---|---|
| DBTN | SIVsm783br | Yes | Yes | 6 | 4 | 13 |
| H802 | SIVsm783br | Yes | Yes | 6 | 6 | 18 |
| H804 | SIVsm783br | Yes | Yes | 7 | 7 | 43 |
| H817 | SIVsm804E | No | Yes | 3 | 4 | 47 |
The animals indicated with shading were rapid progressors.
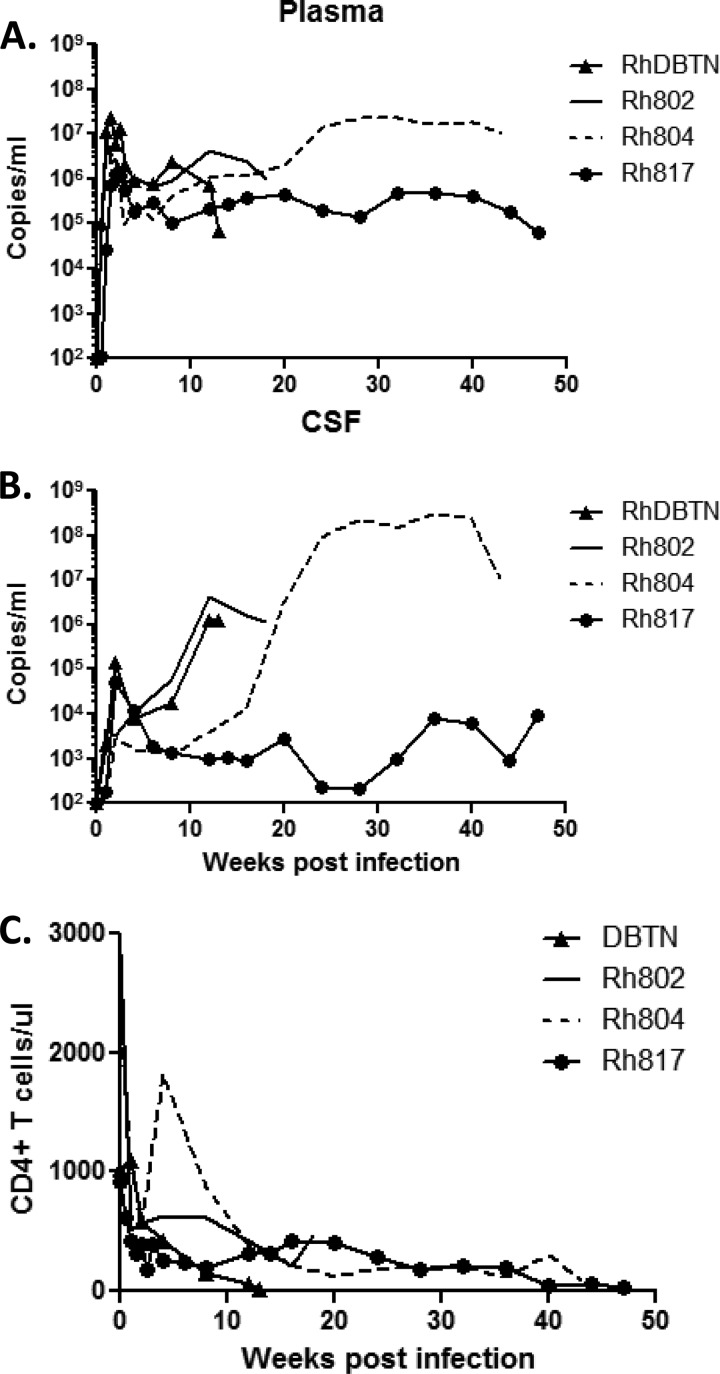
We previously reported that progression to encephalitis correlates with the CSF viral RNA load, with a threshold of 104 viral RNA copies/μl of CSF, but does not correlate with the plasma viral RNA load in SIVsmH783Br-infected animals. Each of the animals in the present study exhibited high peak plasma viral RNA levels and maintained robust viremia throughout the course of infection (Fig. 1A). Three animals, RhDBTN, Rh802, and Rh804, described in the previous study, each had a CSF viral RNA load at the time of necropsy of >104 copies/μl (14). In contrast, the plasma viral load for Rh817 (infected with SIVsmH804E) was approximately 1 log lower throughout the course of infection (Fig. 1A). The peak CSF viral RNA load was comparable to that of one of the rapid progressors, RhDBTN, but the levels decreased to close to the detection limit by 28 weeks postinfection. However, CSF viral RNA loads increased terminally to 9 × 103 copies/ml, which is very close to the CSF threshold we reported previously (Fig. 1B) (14). Flow cytometric analysis of circulating CD4+ T lymphocytes showed that CD4+ T cells were either depleted or decreased to <500 cells/μl of blood by week 12 in all animals. CD4+ T cell counts continued to decline and eventually resulted in severe depletion in the two conventional progressors by the time of necropsy (Fig. 1B).
Fig 1.
Plasma (A) and CSF (B) viral RNA loads and numbers of circulating CD4 T cells (C) of SIVsmH783Br- and SIVsmH804E-infected animals. Solid lines, RhH802; broken lines, RhH804; closed circles, RhH817; closed triangles, RhDBTN.
Detection of SIV expression in the brain.
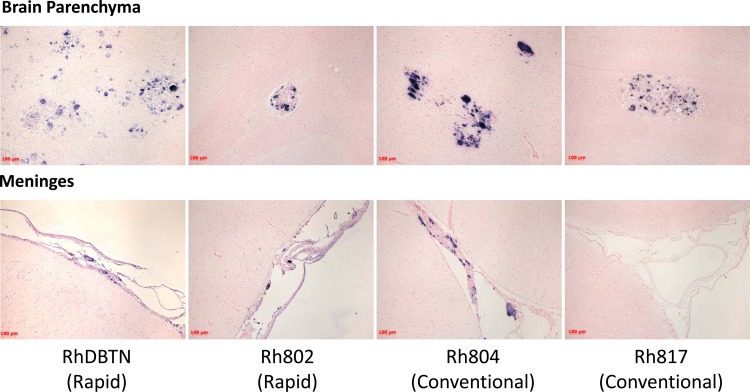
To confirm the site of viral replication in the brain, SIV-specific ISH was conducted to detect actively transcribed viral RNA. Three animals, RhDBTN, Rh802, and Rh804, had strong signals of SIV-positive cells in both the meninges and the brain parenchyma (Fig. 2). This result was consistent with the presence of MNGC in both compartments, as observed by hematoxylin and eosin staining, consistent with SIV meningoencephalitis (Table 1). On the other hand, SIV-positive cells were observed only in the brain parenchyma for animal Rh817 (Fig. 2). This result was also consistent with the lack of inflammation and MNGC in the meninges of this macaque, as observed by hematoxylin and eosin staining (Table 1). The brain tissues were also evaluated by confocal microscopy using SIV-specific ISH and immunohistochemistry for CD3, HAM56, and DAPI (4′,6-diamidino-2-phenylindole) to confirm the phenotypes of the SIV-positive cells in the brain. Consistent with our previous report on the ancestral viral swarm SIVsmH631Br-infected animals (15), SIV RNA signals predominantly colocalized with HAM56, indicating that macrophages are the main target of SIVsmH783Br and SIVsmH804E in both the brain parenchyma and meninges (data not shown).
Fig 2.
SIV-specific in situ hybridization of brain tissue. The upper panels show representative areas of the brain parenchyma with characteristic lesions for the four study animals. The lower panels show representative areas of meninges from the same four animals. The sets of upper and lower panels show images for animals RhDBTN, RhH802, RhH804, and RhH817, from left to right. Multinucleated giant cells expressing SIV RNA were observed in the meninges of all animals but Rh817.
Laser capture microdissection and phylogenetic analysis of viral populations.
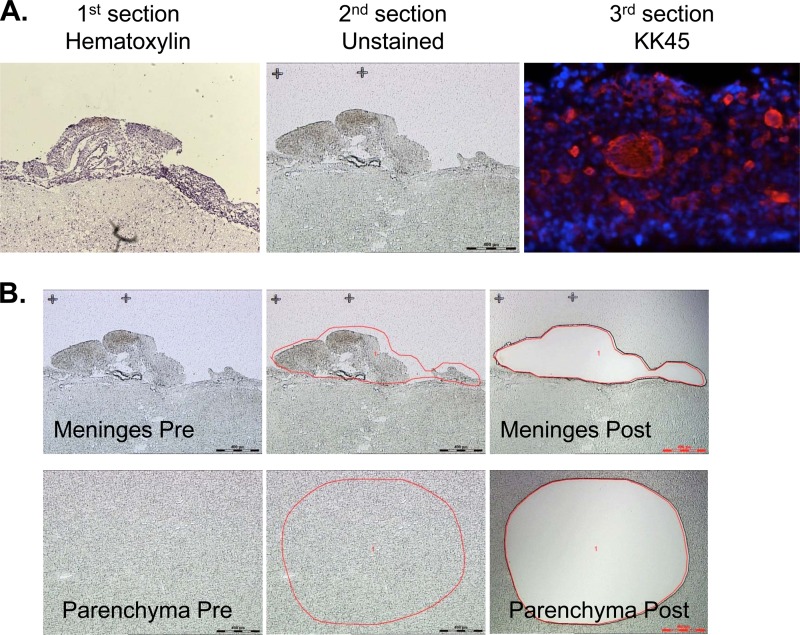
To avoid contamination and to isolate the specific sites where the virus was actively replicating, we utilized laser capture microdissection to sample these two compartments. The cryopreserved brain sections from RhDBTN, Rh802, Rh804, and Rh817 were placed on membrane slides. The meninges and the brain parenchyma were then isolated using a laser (Fig. 3). Laser capture microdissection is widely used in the study of HIV-1, including studies of HIVE. However, most of these studies amplify relatively small fragments (<500 nucleotides [nt]) of viral DNA (30–33). To obtain viral sequences that were representative of actively expressed virus, we chose to amplify the entire envelope sequence from viral RNA. This strategy made it necessary to used unfixed tissues in order to preserve RNA for subsequent PCR amplification. The viral RNAs from these two compartments were extracted, and the whole envelope sequences were successfully amplified by RT-PCR. For comparative purposes, the whole envelope sequence was also amplified in parallel from plasma and CSF samples collected at the time of necropsy and from the original inocula (see Table 2 for numbers of clones). The minute sample sizes of laser-captured samples precluded the use of limiting dilution PCR. However, recombination, which can be a problem in bulk PCR such as that used in this study, was not detected by analysis with the LANL HIV Database Highlighter tool (http://www.hiv.lanl.gov/content/sequence/HIGHLIGHT/highlighter_top.html) (data not shown). Of the 299 envelope sequences, the majority (78%) encoded intact open reading frames. The majority of nonsynonymous substitutions were observed within the V1/V2 region, with a scattering of changes in other variable regions. There was no consistent loss or gain of potential N-linked glycosylation sites (PNGs) specific to any particular compartment. Phylogenetic trees were generated with envelope sequences derived from each animal.
Fig 3.
Schematic images of laser capture microdissection sections. (A) Images of hematoxylin- and eosin-stained, unstained, and KK45-stained serial sections of the RhH804 brain, from left to right. (B) Sequential images of meninges (upper panels, from left to right) and the brain parenchyma (lower panels, from left to right) captured during the microdissection procedure.
Table 2.
Summary of envelope clones and prevalence of premature stop codons
| Animal or virus | Tissue or fluid | Total no. of clones | No. of clones with premature stop codon | No. of clones with gp41 truncations |
|---|---|---|---|---|
| SIVsm783Br inoculum | 12 | 0 | 0 | |
| RhDBTN | Plasma | 20 | 9 | 0 |
| CSF | 15 | 3 | 0 | |
| Parenchyma | 9 | 0 | 0 | |
| Meninges | 11 | 0 | 0 | |
| Rh802 | Plasma | 10 | 1 | 0 |
| CSF | 10 | 0 | 0 | |
| Parenchyma | 25 | 0 | 0 | |
| Meninges | 19 | 1 | 0 | |
| Rh804 | Plasma | 11 | 0 | 0 |
| CSF | 12 | 2 | 4 | |
| Parenchyma | 15 | 1 | 0 | |
| PVL-A | 20 | 6 | 0 | |
| PVL-B | 15 | 2 | 0 | |
| Meninges | 33 | 17 | 9 | |
| SIVsmH804E inoculum | 10 | 1 | 1 | |
| Rh817 | Plasma | 10 | 0 | 0 |
| CSF | 9 | 5 | 0 | |
| Parenchyma | 11 | 3 | 0 | |
| Meninges | 10 | 0 | 0 |
Evaluation of compartmentalization in rapid-progressor macaques.
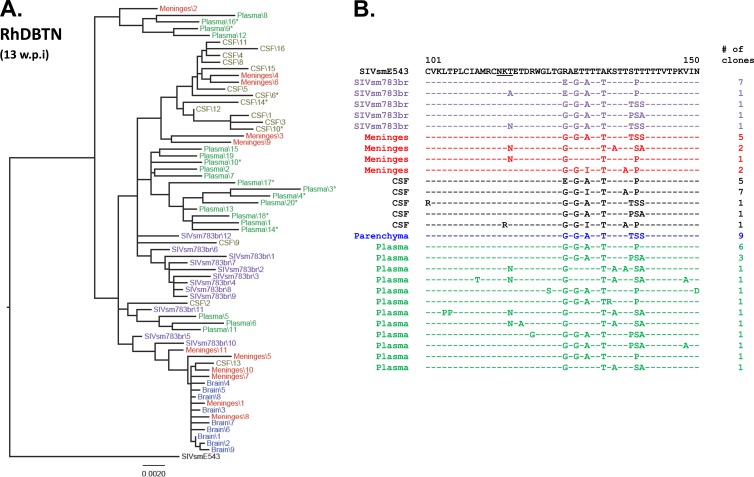
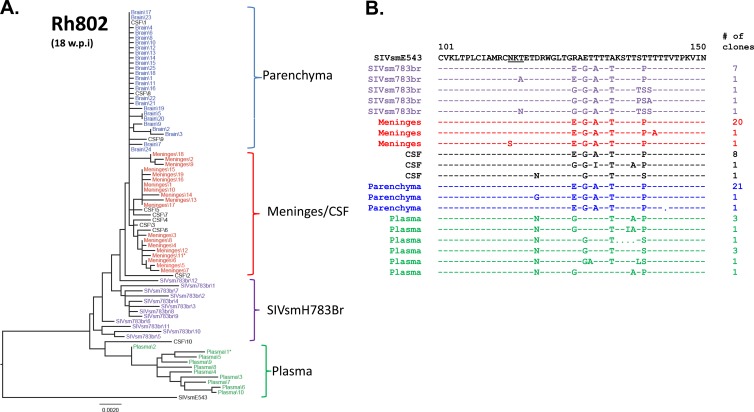
In a previous study, we examined CSF and plasma of a rapid-progressor macaque that developed SIVE and found intermingling of sequences from plasma and CSF, in contrast to the compartmentalization we saw in two conventional progressors from the same study (15). However, the prior study did not examine sequences derived from the brain. The present study allowed us to expand our observations to the brain and meninges, in addition to the plasma and CSF, of two additional rapid progressors. The first animal, RhDBTN, which was inoculated with SIVsmH783Br, progressed rapidly and developed meningoencephalitis by 13 weeks postinfection. Unlike typical rapid progressors, this animal had evidence of seroconversion by Western blot analysis (data not shown). Although the inoculum, plasma, CSF, and brain parenchymal variants formed separate main clusters, some clones from each compartment were intermingled among clones from other compartments (Fig. 4A). Thus, some envelope sequences derived from all four compartments were randomly dispersed throughout the tree, without evidence of compartmentalization. Most nonsynonymous mutations were observed in the V1/V2 region of the envelope, but there were no obvious signature amino acid substitutions to indicate compartmentalization in this animal (Fig. 4B). For example, a T116N substitution that shifted a PNG was observed in the plasma and meninges, though not in the brain parenchyma. The other animal that progressed rapidly, Rh802, was also infected with SIVsmH783Br and developed neurologic disease by week 18 postinoculation. Similar to RhDBTN, this animal showed weak seroconversion to SIV proteins, which is somewhat atypical of rapid progressors. Env sequences from Rh802 also showed a low degree of compartmentalization. Viral populations from meninges and the brain parenchyma formed separate clusters, but the brain parenchymal populations were highly homogeneous, and the degree of divergence between these two compartments was within 0.002 substitution per site. The viral population from plasma was most divergent in this animal and formed a distinct cluster with the longest branches (Fig. 5A). Examination of nonsynonymous mutations in the V1/V2 region of the virus confirmed the phylogenetic analysis; meningeal, brain parenchymal, and CSF populations were relatively homogeneous, whereas plasma populations contained several unique amino acid substitutions (Fig. 5B). For example, plasma viruses showed a D119N substitution and also contained populations of variants which shared the same amino acid sequence with the original SIVsmE543 isolate at positions 126, 128, and 130 (Fig. 5B).
Fig 4.
Phylogenetic analysis of envelope sequences amplified from plasma and CNS of RhDBTN. The figure shows a phylogenetic analysis of envelope nucleotide sequences (A) and a representative amino acid alignment 5′ of the V1/V2 region of the envelope (B). The black letters at the top indicate the original SIVsmE543-3 isolate, purple indicates the inoculum, blue indicates the brain parenchyma, red indicates meninges, black indicates CSF, and green indicates plasma. *, presence of a premature stop codon. The scale bar indicates the number of substitutions per site. w.p.i, weeks postinfection.
Fig 5.
Phylogenetic analysis of envelope sequences amplified from plasma and CNS of RhH802. The figures shows a phylogenetic analysis of envelope nucleotide sequences (A) and a representative amino acid alignment of the V1/V2 region of the envelope (B). The black letters at the top indicate the original SIVsmE543-3 isolate, purple indicates the inoculum, blue indicates the brain parenchyma, red indicates meninges, black indicates CSF, and green indicates plasma. *, presence of a premature stop codon. The scale bar indicates the number of substitutions per site.
Evaluation of compartmentalization in conventional-progressor macaques.
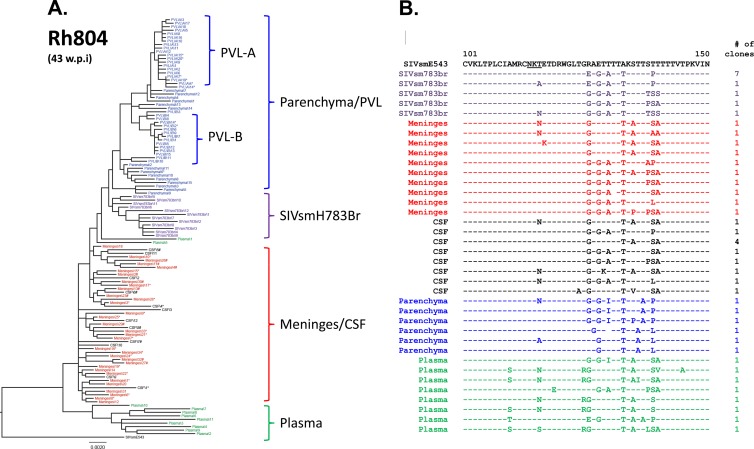
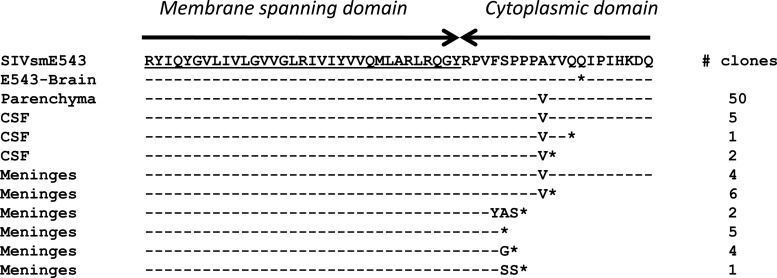
A different picture emerged when we examined sequences cloned from the two conventional-progressor macaques, Rh804 and Rh817. Clones from tissues of animal Rh804, which progressed to meningoencephalitis by week 43 postinoculation, showed a higher level of divergence from the inoculum, with clear compartmentalization between the brain and plasma and also within the CNS. The brain parenchyma, meninges/CSF, inoculum, and plasma formed four distinct clusters. Unlike the case for the two rapid progressors, the meningeal and brain parenchymal populations clearly formed separate clusters in the phylogenetic tree, as shown in Fig. 6A. The plasma isolates were most divergent compared with the variants from the rest of the compartments, as observed in Rh802. Interestingly, the CSF and meningeal isolates intermingled with each other, forming one cluster, suggesting that they share an origin (Fig. 6A). Amino acid alignments of the V1/V2 region also suggested that meningeal and CSF clones were closely related to each other but distinct from the clones from the brain parenchyma (Fig. 6B). The T116N substitution that shifted a PNG observed in clones from RhDBTN was also seen in multiple compartments in Rh804. In addition to cloning from multiple pooled regions of the brain parenchyma, we also cloned envelopes from two circumscribed perivascular lesions (PVL-A and PVL-B). Clones from each of these lesions clustered separately but fell within the larger cluster of clones from the parenchyma. This confirmed that while there might be regional differences in sequence, clones from the parenchyma of the brain still remained distinct from meningeal clones. In addition, there were signature truncations in the cytoplasmic domain of gp41 that were found in both meningeal (80%) and CSF (30%) clones but not in the 50 clones from the brain parenchyma that we analyzed. This truncation was caused by a mixture of insertions and duplications which introduced frameshifts resulting in the introduction of a premature stop codon at a similar site in the cytoplasmic domain, but at slightly different locations (Fig. 7). The presence of these signature truncations also suggests that viruses from meninges and the CSF have a common origin.
Fig 6.
Phylogenetic analysis of envelope sequences amplified from plasma and CNS of RhH804. The figure shows a phylogenetic analysis of envelope nucleotide sequences (A) and a representative amino acid alignment of the V1/V2 region of the envelope (B). The black letters at the top indicate the original SIVsmE543-3 isolate, purple indicates the inoculum, blue indicates the brain parenchyma, red indicates meninges, black indicates CSF, and green indicates plasma. *, presence of a premature stop codon in the envelope; #, truncation of the gp41 cytoplasmic domain. The scale bar indicates the number of substitutions per site.
Fig 7.
Amino acid alignment of sequences from the meninges and brain parenchyma at the area of gp41 cytoplasmic tail truncations, with the original SIVsmE543-3 sequence as a reference. An amino acid alignment for isolates from the brain, CSF, and meninges is shown, with the number of homologous isolates indicated on the right. The E543-Brain sequence was isolated from the monkey infected with SIVsmE543-3 reported in our previous study. *, stop codon.
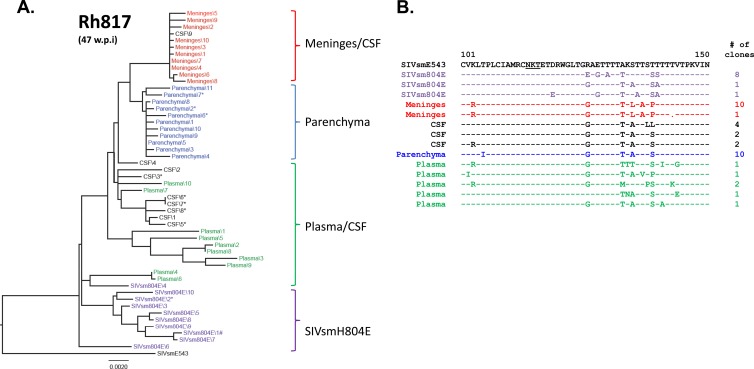
To confirm our observation of compartmentalization in this conventional-progressor macaque, viral populations in another conventional-progressor macaque, Rh817, were also assessed. Although we could not observe cells actively producing SIV RNA in meninges of this animal by in situ hybridization, the viral envelope fragment was successfully amplified by PCR, suggesting that there are SIV-infected cells that actively expressing a low level of SIV RNA but do not cause any observable histopathology in the meninges. Consistent with the observations on animal Rh804, viral populations were compartmentalized between the meninges and the brain parenchyma in animal Rh817. The inoculum, meninges, and brain parenchyma formed distinct clusters, except for in the plasma cluster, where some variants intermingled with the CSF and inoculum clusters. One of the CSF clones fell within the meningeal cluster, but the majority of CSF clones grouped with the plasma clones. This is perhaps not too surprising considering the low level of CSF viral load in this animal relative to the plasma viral load. The possibility that these CSF clones were actually due to low-level contamination of the CSF with plasma cannot be eliminated from consideration. Premature truncations of gp41 were not observed in any of the clones from this animal (Fig. 8A). The compartmentalization of the viral populations between meninges and the brain parenchyma in Rh817 is also supported by the amino acid alignment of the V1/V2 region, with meningeal variants containing the unique amino acid substitutions S135L, T137A, and T139P (Fig. 8B). None of the clones from any of the compartments showed the shifting or loss of PNGs (i.e., T119N) observed in the other animals.
Fig 8.
Phylogenetic analysis of envelope sequences amplified from plasma and CNS of RhH817. The figure shows a phylogenetic analysis of envelope nucleotide sequences (A) and a representative amino acid alignment of the V1/V2 region of the envelope (B). The black letters at the top indicate the original SIVsmE543-3 isolate, purple indicates the inoculum, blue indicates the brain parenchyma, red indicates meninges, black indicates CSF, and green indicates plasma. *, presence of premature stop codons. The scale bar indicates the number of substitutions per site.
Replication kinetics of Env chimeras from the CNS of Rh804.
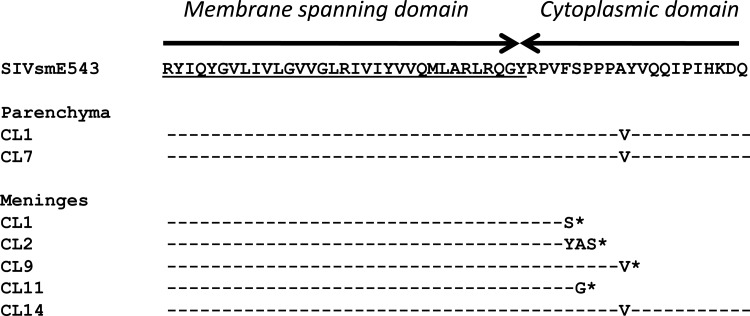
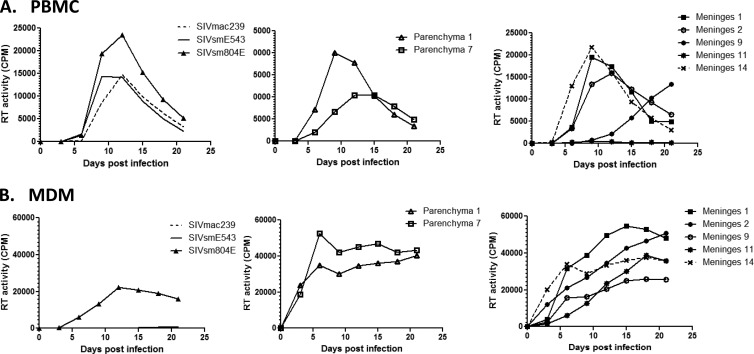
We observed a high frequency of stop codons introduced by substitution, insertion, or duplication of nucleotides throughout the envelope sequences of meningeal and CSF isolates from Rh804 (Table 2). Some of these were located in gp120, presumably resulting in nonfunctional Env proteins. This was particularly observed among clones derived from the meninges. However, some of the meningeal clones had premature truncations within the cytoplasmic domain of gp41, without any other inactivating mutations. In order to assess the biological significance of gp41 truncation on these clones, DNA fragments containing a part of vpx, the whole vpr gene, and most of the envelope gene were amplified from the brain parenchyma and meninges of animal Rh804 and substituted for the corresponding region of SIVsmE543-3 to develop chimeric clones. Two clones were made from the brain parenchyma, and five clones were made from the meninges. Four of five clones from the meninges contained the gp41 truncation in various forms, and the remaining clone encoded an intact transmembrane (TM) glycoprotein (Fig. 9). Following production of virus stocks in 293T cells, the infectivity of these clones was assessed in PBMC (Fig. 10A) and MDMs (Fig. 10B). As controls, we measured the replication of the pathogenic molecular clone SIVmac239, the original SIVsmE543-3 clones, and the viral isolate from the brain of Rh804, i.e., SIVsmH804E. Each of the chimeric viruses replicated at relatively high levels in PBMC compared to the levels of SIVmac239 and SIVsmE543-3, with an approximately 1.6-fold higher peak RT value. SIVmac239 is known to be a T-tropic virus that does not replicate in macrophages in vitro, which is consistent with the lack of replication in MDMs observed in this study. Our previous report suggested that SIVsmE543-3 replicates in macrophages, but we did not observe signs of replication in three independent attempts in this study. One possible explanation is a difference in viral susceptibility of the host, since the degree of viral replication in macrophages is significantly influenced by the donor (22). On the other hand, SIVsmH804E replicated to high levels in MDMs. This result indicates that SIVsmH804E is a dual-tropic SIV that can infect and replicate in both T lymphocytes and macrophages. The two chimeras with envelopes from the brain parenchymal clones that lack premature TM truncations both replicated in PBMC and MDMs. Parenchymal clone 1 showed better replication in PBMC, with an earlier onset of replication and an approximately 2-fold higher peak RT value than that of parenchymal clone 7. On the other hand, parenchymal clone 7 showed slightly better replication in MDMs than parenchymal clone 1. Each of the chimeric viruses derived from the meninges was infectious in MDMs, regardless of whether or not they had a prematurely truncated TM glycoprotein. Meningeal clone 14, which contained the envelope without truncation of the TM glycoprotein, replicated in both PBMC and MDMs, and the replication kinetics was similar to that of parenchymal clone 7. Three chimeric viruses (meningeal clones 1, 2, and 9) that contained envelopes with a truncated gp41 cytoplasmic tail in various forms also replicated in PBMC and MDMs, although one of these (meningeal clone 9) had a delayed replication peak in PBMC. One clone (meningeal clone 11) replicated only in MDMs. In general, the chimeric virus isolates with gp41 truncations replicated at a level comparable to that of the clones without the TM truncation. This result indicates that truncation at gp41 is not lethal to the virus but failed to delineate any clear biological difference between viruses expressing a truncated versus intact TM glycoprotein.
Fig 9.
Amino acid alignment of chimeric virus sequences around the gp41 cytoplasmic domain, with the original SIVsmE543-3 sequence as a reference. Two chimeras were constructed from the brain parenchymal isolates, and five clones were constructed from the meningeal isolates. *, stop codon.
Fig 10.
Replication kinetics of chimeric viruses expressing envelopes from the RhH804 CNS. (A) Replication kinetics of chimeric viruses on PBMC. (B) Replication kinetics of chimeric viruses on MDMs. Broken lines, SIVmac239; solid lines, SIVsmE543-3; closed triangles, SIVsmH804E; open triangles, parenchymal clone 1; open squares, parenchymal clone 7; closed squares, meningeal clone 1; open circles, meningeal clone 2; closed circles, meningeal clone 9; stars, meningeal clone 11; broken lines with crosses, meningeal clone 14. The figure shown is representative of four independent experiments.
DISCUSSION
Nonhuman primate-SIV models are useful for elucidating mechanisms responsible for the pathogenesis of HIV-1 in the brain for HAART-treated/untreated HIV-1-infected individuals. We previously reported that the viral isolate SIVsmH783Br, which has been passaged sequentially through rhesus macaques, induces neurological symptoms at a high frequency (14). In the present study, we conducted an additional in vivo passage in an attempt to further adapt SIVsmH783Br to replication in the brain, and we isolated SIVsmH804E from one of the six animals infected with SIVsmH783Br. A group of six animals infected with SIVsmH804E showed a disease course and rate of induction of neuropathogenesis comparable to those of SIVsmH783Br-infected animals, suggesting that an additional passage did not have much influence on increasing the neuropathogenicity of SIVsmH783Br. The use of laser capture microdissection allowed us to explore viral populations in the CNS of animals with SIVE, confirming the compartmentalization of SIV sequences in the CNS compared to systemic circulation. Additionally, we observed a compartmentalization of virus in the brain parenchyma versus meninges in those animals that developed SIVE during chronic infection, consistent with distinct selective pressures in these two “compartments.”
Previous studies in our lab have demonstrated that the patterns and severity of pathological lesions in SIV-infected macaques which progress rapidly versus those that survive for longer periods are distinct (25). Therefore, we divided our study animals into rapid and conventional progressors based upon the rate of disease progression. Those animals that progressed rapidly showed neurological symptoms with high viral RNA loads in CSF within 3 to 5 months after infection, and they progressed to severe meningoencephalitis. On the other hand, conventional progressors developed neurological symptoms at a later stage of the disease course, typically within 10 months to 2 years after infection. Unlike rapid progressors, where inflammation was readily observed in both the meninges and brain parenchyma, either meningitis or encephalitis was the predominant feature in a subset of the conventional progressors (14). We studied one of these animals, in whom encephalitis was the dominant pathological finding (Rh817). Although we were unable to observe SIV expression in the meninges of this animal, we were still able to amplify viral RNA from laser capture samples, and these sequences were distinct from those in the brain parenchyma. The results from the phylogenetic analysis of viral populations in meninges and the brain parenchyma clearly indicate that they are distinct from each other in conventional progressors. This result suggests that the progression to meningitis and encephalitis is induced by two distinct viral populations, suggesting that two pathological states progress in parallel. The driving mechanism(s) of this compartmentalization is still unclear, but based on the result that the V1/V2 region of the viral envelope sequence was most divergent between the two compartments, we could speculate that a selective pressure such as cell tropism, the microenvironment, or immunological pressure might have contributed to this compartmentalization (34–36).
It has previously been reported that there is less compartmentalization within the CNS of rapid progressors, whereas conventional progressors show clear compartmentalization (37). Indeed, our observations are consistent with this, as there was no or less compartmentalization between meninges and the brain parenchyma in rapid progressors, but they were clearly compartmentalized in conventional progressors. It is still unclear why rapid progressors showed less compartmentalization in the CNS, but possible explanations include the following: (i) animals were sacrificed at relatively short times after infection, and viral populations in the CNS did not have sufficient time to diversify; (ii) catastrophic destruction of the immune system by the rapid progression of disease caused less selective pressure in the CNS; and (iii) there was a loss of the barrier between compartments due to a breakdown of the blood-brain barrier (BBB). Further assessments are required to answer this question in future studies.
The choroid plexus produces a large amount of CSF each day to exclude metabolites produced in the CNS. There is evidence that cells in the choroid plexus can be infected by HIV-1 and produce viruses which can be disseminated to the CNS (38, 39). However, the results of phylogenetic analyses in the present study revealed that the CSF viral population was most closely related to that of the meninges in at least one of the study animals. This result suggests that the choroid plexus might not be the sole source of viral populations in the CSF and that the meninges may play a major role in production of virus. This speculation is supported by the fact that those monkeys with high CSF viral RNA loads always had evidence of meningitis, whereas the monkey in whom encephalitis was the predominant feature exhibited 3 to 4 log lower viral RNA loads in the CSF. The other evidence that the CSF population is closely related to populations in meninges is that they shared unique truncations of the cytoplasmic domain of gp41 in this animal. Although the patterns of insertion and duplications were variable among the isolates, these unique truncations were observed at similar positions in the N terminus of the cytoplasmic domain in 80% of meningeal clones and 30% of CSF clones but were never identified in the brain parenchymal or plasma clones.
It has previously been reported that truncation of the cytoplasmic domain of gp41 increases the fusion activity of the virion by efficient exposure of fusion epitopes followed by binding of gp120 and the CD4 receptor, which may also increase the sensitivity to neutralizing antibodies (40, 41). This suggests that these truncations result from viral adaptation to the unique microenvironment in the meninges or perhaps to differences in the macrophage populations between the brain parenchyma and meninges, which show distinct phenotypes and properties (42, 43). In addition, we also found a high frequency of stop codons at random positions along the envelope in meningeal and CSF clones (Table 2). Premature stop codons were observed to some degree in all the animals, as would be expected from PCR error and the expected presence of some inactive viruses in tissues (Table 2). However, the frequency was increased in the meninges and CSF from Rh804 and, to some degree, in other tissues from this animal. This suggests the possibility of the influence of some unknown restriction factor, similar to but distinct from the G-to-A hypermutation induced by APOBEC3G (44). The presence of premature stop codons in gp120 is lethal to the virus, whereas stop codons at certain regions, such as the cytoplasmic domain of gp41, are not. Therefore, the survival of isolates with gp41 truncations may not be a specific adaptation to the microenvironment of the meninges but rather just represent the survivors of some unknown selective pressure. On the other hand, although we identified these gp41 truncations in only one of the two conventional progressors in this study, we previously reported the presence of similar truncations in isolates from the brain of the original SIVsmE543-infected animal (45). In addition, other studies using well-characterized neurovirulent SIV clones demonstrated that a similar truncation at the TM domain of gp41 is responsible for CD4 independence (46, 47). These observations suggest that those gp41 truncations may play important roles in the pathogenesis of SIV in the CNS. The viral replication kinetics of envelope chimeras in PBMC and MDMs revealed that the viral isolates with truncated gp41 were replication competent, and the truncations did not appear to influence macrophage tropism. Although we could not identify any significance of truncations relative to replication efficiency or cell tropism, further analyses are needed to fully understand their biological significance.
Based upon the present study, we have clearly shown that meningitis and encephalitis in conventional progressors with SIVE are associated with two distinct viral populations. It is not clear if these viral populations diverged from one another through the course of infection or whether they were initially established by distinct virus populations. CSF populations in at least one of the animals we examined were most closely related to the population in the meninges. CSF is the most easily accessible CNS sample from HIV-1-infected patients. Although our model may not directly reflect HIV-1 infection in the human CNS due to its more rapid course, our results suggest that viral populations in the CSF may not necessarily be representative of the populations in the brain parenchyma, where most of the damage to motor and memory functions occurs.
ACKNOWLEDGMENTS
We thank Heather Cronise, Joanne Swerczek, and Richard Herbert at NIHAC for excellent care of the study animals and Owen Schwartz and Lilly Koo of the Confocal Laboratory, Research Technologies Branch, NIAID, for assistance with laser capture microdissection techniques.
This work was supported by the intramural research program of NIAID, NIH, and by a JSPS Research Fellowship for Japanese Biomedical and Behavioral Researchers at NIH.
Footnotes
Published ahead of print 29 May 2013
REFERENCES
- 1. Heaton RK, Clifford DB, Franklin DR, Jr, Woods SP, Ake C, Vaida F, Ellis RJ, Letendre SL, Marcotte TD, Atkinson JH, Rivera-Mindt M, Vigil OR, Taylor MJ, Collier AC, Marra CM, Gelman BB, McArthur JC, Morgello S, Simpson DM, McCutchan JA, Abramson I, Gamst A, Fennema-Notestine C, Jernigan TL, Wong J, Grant I, CHARTER Group 2010. HIV-associated neurocognitive disorders persist in the era of potent antiretroviral therapy: CHARTER study. Neurology 75:2087–2096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cysique LA, Maruff P, Brew BJ. 2004. Prevalence and pattern of neuropsychological impairment in human immunodeficiency virus-infected/acquired immunodeficiency syndrome (HIV/AIDS) patients across pre- and post-highly active antiretroviral therapy eras: a combined study of two cohorts. J. Neurovirol. 10:350–357 [DOI] [PubMed] [Google Scholar]
- 3. Cusini A, Vernazza PL, Yerly S, Decosterd LA, Ledergerber B, Fux CA, Rohrbach J, Widmer N, Hirschel B, Gaudenz R, Cavassini M, Klimkait T, Zenger F, Gutmann C, Opravil M, Gunthard HF, Swiss HIV Cohort Study 2013. Higher CNS penetration-effectiveness of long-term combination antiretroviral therapy is associated with better HIV-1 viral suppression in cerebrospinal fluid. J. Acquir. Immune Defic. Syndr. 62:28–35 [DOI] [PubMed] [Google Scholar]
- 4. Garvey L, Winston A, Walsh J, Post F, Porter K, Gazzard B, Fisher M, Leen C, Pillay D, Hill T, Johnson M, Gilson R, Anderson J, Easterbrook P, Bansi L, Orkin C, Ainsworth J, Palfreeman A, Gompels M, Phillips AN, Sabin CA, UK Collaborative HIV Cohort (CHIC) Study 2011. Antiretroviral therapy CNS penetration and HIV-1-associated CNS disease. Neurology 76:693–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Letendre S, Marquie-Beck J, Capparelli E, Best B, Clifford D, Collier AC, Gelman BB, McArthur JC, McCutchan JA, Morgello S, Simpson D, Grant I, Ellis RJCHARTER Group 2008. Validation of the CNS penetration-effectiveness rank for quantifying antiretroviral penetration into the central nervous system. Arch. Neurol. 65:65–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ances BM, Vaida F, Yeh MJ, Liang CL, Buxton RB, Letendre S, McCutchan JA, Ellis RJ. 2010. HIV infection and aging independently affect brain function as measured by functional magnetic resonance imaging. J. Infect. Dis. 201:336–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cysique LA, Maruff P, Bain MP, Wright E, Brew BJ. 2011. HIV and age do not substantially interact in HIV-associated neurocognitive impairment. J. Neuropsychiatry Clin. Neurosci. 23:83–89 [DOI] [PubMed] [Google Scholar]
- 8. Ernst T, Chang L. 2004. Effect of aging on brain metabolism in antiretroviral-naive HIV patients. AIDS 18(Suppl 1):S61–S67 [PubMed] [Google Scholar]
- 9. Pettersen JA, Jones G, Worthington C, Krentz HB, Keppler OT, Hoke A, Gill MJ, Power C. 2006. Sensory neuropathy in human immunodeficiency virus/acquired immunodeficiency syndrome patients: protease inhibitor-mediated neurotoxicity. Ann. Neurol. 59:816–824 [DOI] [PubMed] [Google Scholar]
- 10. Rihs TA, Begley K, Smith DE, Sarangapany J, Callaghan A, Kelly M, Post JJ, Gold J. 2006. Efavirenz and chronic neuropsychiatric symptoms: a cross-sectional case control study. HIV Med. 7:544–548 [DOI] [PubMed] [Google Scholar]
- 11. Burudi EM, Fox HS. 2001. Simian immunodeficiency virus model of HIV-induced central nervous system dysfunction. Adv. Virus Res. 56:435–468 [DOI] [PubMed] [Google Scholar]
- 12. Strickland SL, Gray RR, Lamers SL, Burdo TH, Huenink E, Nolan DJ, Nowlin B, Alvarez X, Midkiff CC, Goodenow MM, Williams K, Salemi M. 2012. Efficient transmission and persistence of low-frequency SIVmac251 variants in CD8-depleted rhesus macaques with different neuropathology. J. Gen. Virol. 93:925–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zink MC, Amedee AM, Mankowski JL, Craig L, Didier P, Carter DL, Munoz A, Murphey-Corb M, Clements JE. 1997. Pathogenesis of SIV encephalitis. Selection and replication of neurovirulent SIV. Am. J. Pathol. 151:793–803 [PMC free article] [PubMed] [Google Scholar]
- 14. Dang Q, Whitted S, Goeken RM, Brenchley JM, Matsuda K, Brown CR, Lafont BA, Starost MF, Iyengar R, Plishka RJ, Buckler-White A, Hirsch VM. 2012. Development of neurological disease is associated with increased immune activation in simian immunodeficiency virus-infected macaques. J. Virol. 86:13795–13799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dang Q, Goeken RM, Brown CR, Plishka RJ, Buckler-White A, Byrum R, Foley BT, Hirsch VM. 2008. Adaptive evolution of simian immunodeficiency viruses isolated from 2 conventional-progressor macaques with encephalitis. J. Infect. Dis. 197:1695–1700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Caragounis EC, Gisslen M, Lindh M, Nordborg C, Westergren S, Hagberg L, Svennerholm B. 2008. Comparison of HIV-1 pol and env sequences of blood, CSF, brain and spleen isolates collected ante-mortem and post-mortem. Acta Neurol. Scand. 117:108–116 [DOI] [PubMed] [Google Scholar]
- 17. Pillai SK, Pond SL, Liu Y, Good BM, Strain MC, Ellis RJ, Letendre S, Smith DM, Gunthard HF, Grant I, Marcotte TD, McCutchan JA, Richman DD, Wong JK. 2006. Genetic attributes of cerebrospinal fluid-derived HIV-1 env. Brain 129:1872–1883 [DOI] [PubMed] [Google Scholar]
- 18. Schnell G, Price RW, Swanstrom R, Spudich S. 2010. Compartmentalization and clonal amplification of HIV-1 variants in the cerebrospinal fluid during primary infection. J. Virol. 84:2395–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lamers SL, Gray RR, Salemi M, Huysentruyt LC, McGrath MS. 2011. HIV-1 phylogenetic analysis shows HIV-1 transits through the meninges to brain and peripheral tissues. Infect. Genet. Evol. 11:31–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chang J, Jozwiak R, Wang B, Ng T, Ge YC, Bolton W, Dwyer DE, Randle C, Osborn R, Cunningham AL, Saksena NK. 1998. Unique HIV type 1 V3 region sequences derived from six different regions of brain: region-specific evolution within host-determined quasispecies. AIDS Res. Hum. Retroviruses 14:25–30 [DOI] [PubMed] [Google Scholar]
- 21. Shapshak P, Segal DM, Crandall KA, Fujimura RK, Zhang BT, Xin KQ, Okuda K, Petito CK, Eisdorfer C, Goodkin K. 1999. Independent evolution of HIV type 1 in different brain regions. AIDS Res. Hum. Retroviruses 15:811–820 [DOI] [PubMed] [Google Scholar]
- 22. Kuwata T, Dehghani H, Brown CR, Plishka R, Buckler-White A, Igarashi T, Mattapallil J, Roederer M, Hirsch VM. 2006. Infectious molecular clones from a simian immunodeficiency virus-infected rapid-progressor (RP) macaque: evidence of differential selection of RP-specific envelope mutations in vitro and in vivo. J. Virol. 80:1463–1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kirmaier A, Wu F, Newman RM, Hall LR, Morgan JS, O'Connor S, Marx PA, Meythaler M, Goldstein S, Buckler-White A, Kaur A, Hirsch VM, Johnson WE. 2010. TRIM5 suppresses cross-species transmission of a primate immunodeficiency virus and selects for emergence of resistant variants in the new species. PLoS Biol. 8:e1000462. 10.1371/journal.pbio.1000462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. National Research Council 1996. Guide for the care and use of laboratory animals. National Academies Press, Washington, DC [Google Scholar]
- 25. Brown CR, Czapiga M, Kabat J, Dang Q, Ourmanov I, Nishimura Y, Martin MA, Hirsch VM. 2007. Unique pathology in simian immunodeficiency virus-infected rapid progressor macaques is consistent with a pathogenesis distinct from that of classical AIDS. J. Virol. 81:5594–5606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Katoh K, Asimenos G, Toh H. 2009. Multiple alignment of DNA sequences with MAFFT. Methods Mol. Biol. 537:39–64 [DOI] [PubMed] [Google Scholar]
- 27. Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059–3066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. O'Doherty U, Swiggard WJ, Malim MH. 2000. Human immunodeficiency virus type 1 spinoculation enhances infection through virus binding. J. Virol. 74:10074–10080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Churchill MJ, Gorry PR, Cowley D, Lal L, Sonza S, Purcell DF, Thompson KA, Gabuzda D, McArthur JC, Pardo CA, Wesselingh SL. 2006. Use of laser capture microdissection to detect integrated HIV-1 DNA in macrophages and astrocytes from autopsy brain tissues. J. Neurovirol. 12:146–152 [DOI] [PubMed] [Google Scholar]
- 31. Marras D, Bruggeman LA, Gao F, Tanji N, Mansukhani MM, Cara A, Ross MD, Gusella GL, Benson G, D'Agati VD, Hahn BH, Klotman ME, Klotman PE. 2002. Replication and compartmentalization of HIV-1 in kidney epithelium of patients with HIV-associated nephropathy. Nat. Med. 8:522–526 [DOI] [PubMed] [Google Scholar]
- 32. Thompson KA, Varrone JJ, Jankovic-Karasoulos T, Wesselingh SL, McLean CA. 2009. Cell-specific temporal infection of the brain in a simian immunodeficiency virus model of human immunodeficiency virus encephalitis. J. Neurovirol. 15:300–311 [DOI] [PubMed] [Google Scholar]
- 33. Torres-Munoz JE, Nunez M, Petito CK. 2008. Successful application of hyperbranched multidisplacement genomic amplification to detect HIV-1 sequences in single neurons removed from autopsy brain sections by laser capture microdissection. J. Mol. Diagn. 10:317–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Laird ME, Igarashi T, Martin MA, Desrosiers RC. 2008. Importance of the V1/V2 loop region of simian-human immunodeficiency virus envelope glycoprotein gp120 in determining the strain specificity of the neutralizing antibody response. J. Virol. 82:11054–11065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rossi F, Querido B, Nimmagadda M, Cocklin S, Navas-Martin S, Martin-Garcia J. 2008. The V1-V3 region of a brain-derived HIV-1 envelope glycoprotein determines macrophage tropism, low CD4 dependence, increased fusogenicity and altered sensitivity to entry inhibitors. Retrovirology 5:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Walter BL, Wehrly K, Swanstrom R, Platt E, Kabat D, Chesebro B. 2005. Role of low CD4 levels in the influence of human immunodeficiency virus type 1 envelope V1 and V2 regions on entry and spread in macrophages. J. Virol. 79:4828–4837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chen MF, Westmoreland S, Ryzhova EV, Martin-Garcia J, Soldan SS, Lackner A, Gonzalez-Scarano F. 2006. Simian immunodeficiency virus envelope compartmentalizes in brain regions independent of neuropathology. J. Neurovirol. 12:73–89 [DOI] [PubMed] [Google Scholar]
- 38. Chen H, Wood C, Petito CK. 2000. Comparisons of HIV-1 viral sequences in brain, choroid plexus and spleen: potential role of choroid plexus in the pathogenesis of HIV encephalitis. J. Neurovirol. 6:498–506 [DOI] [PubMed] [Google Scholar]
- 39. Petito CK, Chen H, Mastri AR, Torres-Munoz J, Roberts B, Wood C. 1999. HIV infection of choroid plexus in AIDS and asymptomatic HIV-infected patients suggests that the choroid plexus may be a reservoir of productive infection. J. Neurovirol. 5:670–677 [DOI] [PubMed] [Google Scholar]
- 40. Edwards TG, Wyss S, Reeves JD, Zolla-Pazner S, Hoxie JA, Doms RW, Baribaud F. 2002. Truncation of the cytoplasmic domain induces exposure of conserved regions in the ectodomain of human immunodeficiency virus type 1 envelope protein. J. Virol. 76:2683–2691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wyss S, Dimitrov AS, Baribaud F, Edwards TG, Blumenthal R, Hoxie JA. 2005. Regulation of human immunodeficiency virus type 1 envelope glycoprotein fusion by a membrane-interactive domain in the gp41 cytoplasmic tail. J. Virol. 79:12231–12241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chinnery HR, Ruitenberg MJ, McMenamin PG. 2010. Novel characterization of monocyte-derived cell populations in the meninges and choroid plexus and their rates of replenishment in bone marrow chimeric mice. J. Neuropathol. Exp. Neurol. 69:896–909 [DOI] [PubMed] [Google Scholar]
- 43. Polfliet MMJ, van de Veerdonk F, Dopp EA, van Kesteren-Hendrikx EML, van Rooijen N, Dijkstra CD, van den Berg TK. 2002. The role of perivascular and meningeal macrophages in experimental allergic encephalomyelitis. J. Neuroimmunol. 122:1–8 [DOI] [PubMed] [Google Scholar]
- 44. Depboylu C, Eiden LE, Weihe E. 2007. Increased APOBEC3G expression is associated with extensive G-to-A hypermutation in viral DNA in rhesus macaque brain during lentiviral infection. J. Neuropathol. Exp. Neurol. 66:901–912 [DOI] [PubMed] [Google Scholar]
- 45. Hirsch V, Adger-Johnson D, Campbell B, Goldstein S, Brown C, Elkins WR, Montefiori DC. 1997. A molecularly cloned, pathogenic, neutralization-resistant simian immunodeficiency virus, SIVsmE543-3. J. Virol. 71:1608–1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bonavia A, Bullock BT, Gisselman KM, Margulies BJ, Clements JE. 2005. A single amino acid change and truncated TM are sufficient for simian immunodeficiency virus to enter cells using CCR5 in a CD4-independent pathway. Virology 341:12–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Overholser ED, Babas T, Zink MC, Barber SA, Clements JE. 2005. CD4-independent entry and replication of simian immunodeficiency virus in primary rhesus macaque astrocytes are regulated by the transmembrane protein. J. Virol. 79:4944–4951 [DOI] [PMC free article] [PubMed] [Google Scholar]