Abstract
Histo-blood group antigens (HBGAs) have been suggested to be receptors or coreceptors for human noroviruses (HuNoVs) expressed on the intestinal epithelium. We isolated an enteric bacterium strain (SENG-6), closely related to Enterobacter cloacae, bearing HBGA-like substances from a fecal sample of a healthy individual by using a biopanning technique with anti-HBGA antibodies. The binding capacities of four genotypes of norovirus-like particles (NoVLPs) to Enterobacter sp. SENG-6 cells were confirmed by enzyme-linked immunosorbent assay (ELISA). Transmission electron microscopy demonstrated that NoVLPs bound mainly to extracellular polymeric substances (EPS) of Enterobacter sp. SENG-6, where the HBGA-like substances were localized. EPS that contained HBGA-like substances extracted from Enterobacter sp. SENG-6 was shown by enzyme-linked immunosorbent assay (ELISA) to be capable of binding to NoVLPs of a GI.1 wild-type strain (8fIIa) and a GII.6 strain that can recognize A antigen but not to an NoVLP GI.1 mutant strain (W375A) that loses the ability to bind to A antigen. Enzymatic cleavage of terminal N-acetyl-galactosamine residues in the bacterial EPS weakened bacterial EPS binding to the GI.1 wild-type strain (8fIIa). These results indicate that A-like substances in the bacterial EPS play a key role in binding to NoVLPs. Since the specific binding of HuNoVs to HBGA-positive enteric bacteria is likely to affect the transmission and infection processes of HuNoVs in their hosts and in the environment, further studies of human enteric bacteria and their binding capacity to HuNoVs will provide a new scientific platform for understanding interactions between two types of microbes that were previously regarded as biologically unrelated.
INTRODUCTION
Human noroviruses (HuNoVs) are major causative agents of nonbacterial acute gastroenteritis in humans, which constitute a substantial disease burden worldwide (1). Noroviruses belong to the genus Norovirus in the family Caliciviridae. The genus Norovirus is divided into five genogroups, i.e., GI, GII, GIII, GIV, and GV, and the strains in each genogroup can be further divided into genotypes (2). GI, GII, and GIV NoVs, which include at least 15 genotypes, 18 genotypes, and 1 genotype, respectively (3), infect humans of all age groups, causing symptoms such as nausea, vomiting, diarrhea, abdominal cramps, headache, and fever (4). Human-to-human infection is the main transmission route of HuNoVs, but contaminated water and sea products such as oysters are reported to be sources or vehicles of infection (5, 6) because of environmental contamination with domestic wastewater (7, 8).
The lack of tissue cells for replicating HuNoVs has impeded the study of the life cycle of this important human pathogen. One major finding related to productive infections with HuNoVs is the interaction with histo-blood group antigens (HBGAs), which have been proposed to be receptors or coreceptors of human small intestinal epithelial cells for HuNoVs (9, 10). HBGAs comprise ABH and Lewis antigens, which are structurally related oligosaccharides, and each HuNoV genotype or strain has its own HBGA recognition pattern profile (11–13). For example, virus-like particles (VLPs) of Norwalk virus (NV/68), a genotype 1 strain in genogroup I (GI.1) and the prototype strain of norovirus, bind to HBGAs in saliva from secretor-positive individuals and preferentially bind to H type 1, Lewis b (Leb), and type A carbohydrates (11, 14). Furthermore, VLPs of GII.4 (r104) can recognize a broader range of blood group carbohydrates than other genotypes (12) although the ligand binding patterns have changed over time (15). The importance of the HBGA recognition pattern for HuNoV infections has been emphasized because GII.4 strains are the most prevalent etiological agents of infectious diseases caused by norovirus, probably because of their broad HBGA recognition profile.
In this study, we focused on a group of human enteric bacteria that produce HBGA-positive extracellular polymeric substances (EPS). EPS comprise organic macromolecules such as polysaccharides, proteins, nucleic acids, lipids, and other polymeric compounds located on or outside the cell surface (16). Humans possess immunoglobulin M (IgM) antibodies against nonself HBGAs in the blood, which is attributable to the presence of enteric bacteria with blood group activity (17). This led us to speculate that human enteric bacteria may capture HuNoV particles via specific interactions with HBGA-like bacterial substances. To elucidate the specific interaction between HuNoV particles and HBGA-like bacterial substances, we screened blood group-active human enteric bacteria from human feces using a biopanning technique with anti-HBGA antibodies. We tested the binding capacity of four genotypes to norovirus-like particles (NoVLPs) for bacterial cells using enzyme-linked immunosorbent assays (ELISAs). NoVLP binding to bacterial cells was observed by transmission electron microscopy (TEM), and the localization of HBGA-like bacterial substances was analyzed by immuno-TEM. EPS, surface-retained organic matter (SOM), and lipopolysaccharide (LPS) were extracted from bacterial cells, and the interactions between HBGA-like substances in the extracted bacterial polymers and NoVLPs were examined by ELISA. The specific interactions between HBGA-like substances and NoVLPs were evaluated further by the quartz crystal microbalance (QCM) method.
MATERIALS AND METHODS
Isolation of human enteric bacteria bearing HBGA-like substances.
We screened enteric bacteria bearing HBGA-like substances from human feces using anti-HBGA antibodies. Fifty microliters of anti-blood group A, B, or O(H) mouse monoclonal antibodies (sc-69951, sc-69952, and sc-52372, respectively; Santa Cruz Biotechnology Inc., USA) was added to each well of an ELISA plate and kept at room temperature (RT) for 1 h to coat the well. The wells were washed two times with 0.1 M phosphate-buffered saline (PBS), and a diluted human fecal suspension derived from a healthy adult was added to the wells. After incubation at RT for 1 h, the wells were washed two times with PBS and 2% brilliant green bile broth (Kanto Chemical Co., Inc., Japan), Escherichia coli broth (Nihon Pharmaceutical Co., Ltd., Japan), Enterobacteriaceae enrichment mannitol broth (Nihon Pharmaceutical Co., Ltd.), or enterobacteria enrichment broth-Mossel (Nihon Pharmaceutical Co., Ltd.) was added to each well. After overnight incubation at 37°C under microaerobic conditions, the enriched cells were cultured further by streaking onto an agar plate containing the same medium, and several colonies were selected randomly. The selected colonies were cultured again by streaking onto the same agar plates, and a single colony was selected. The selected cells were cultured in the same culture broth, collected by centrifugation, and suspended in PBS. Tenfold dilution series were prepared with PBS, and the optical densities at 600 nm (OD600) of the diluted cell suspensions were measured using a spectrophotometer. The presence of HBGA-like substances on the cell surface in the diluted cell suspensions was detected on the basis of their aggregation activities with anti-blood group A, B, or O(H) antibody-coated beads from a blood typing kit (ABO sphia; Kamakura Techno-Science Inc., Japan). The names of the antibodies used in the blood typing kit were not disclosed, but they could detect at least type 1 and type 2 HBGAs because human blood, saliva, and sperm could be used as samples. The minimum OD600 in the 10-fold dilution series that gave a positive result was determined for each isolated strain. The isolated bacterial strains were identified by sequencing of the 16S rRNA gene by targeting its 1,466 nucleotides using the primers 27F, 530F, 907R, and 1492R (18).
Assay for HBGA binding activity of NoVLPs.
NoVLP genotypes GI.7 (accession number AB758449), GII.3 (AB758450), GII.4 (AB668028), and GII.6 (AB758451) were prepared according to a published method (19). These genotypes were selected because GII.3, GII.4, and GII.6 are the most prevalent GII noroviruses (20–22), while GI.7 is also detected frequently in epidemiological studies (23). The HBGA binding profile of each genotype (Table 1) was tested as follows. Fifty microliters of NoVLP suspension was added to each well of an ELISA plate and kept at 4°C for 24 h to coat the well. Triplicate wells were used for each sample. The wells were washed two times with PBS and blocked with 5% bovine serum albumin (BSA) in PBS. After incubation at RT for 2 h, the wells were washed two times with PBS, and 50 μl of biotin-conjugated type 1 A (GLT 01-084; GlycoTech, MD, USA), B (GLT 01-085; GlycoTech), H (GLT 01-037; GlycoTech), or Lewis b carbohydrate (GLT 01-042; GlycoTech) (diluted to 1:10 with PBS containing 5% BSA; Eve Bio-Science Co., Ltd., Japan) was added to the wells. Plates were incubated at RT for 2 h. The wells were washed two times with PBS, and 50 μl of horseradish peroxidase (HRP)-conjugated streptavidin (518-59941; Thermo Scientific, Japan) diluted 1:10 with PBS containing 5% BSA was inoculated into each well. After incubation at RT for 1 h, the wells were washed four times with PBS, and the bound HRP-conjugated streptavidin was measured by staining with o-phenylenediamine dihydrochloride (P-7288; Sigma-Aldrich, Japan) and H2O2 in citrate-phosphate buffer for 30 min. The coloring reaction was stopped with 2 M H2SO4. The absorbance at 490 nm was measured using a plate reader (Arvo MX; PerkinElmer, Japan).
Table 1.
Binding profiles of norovirus-like particles prepared in this study
| Genotype (GenBank accession no.) | Histo-blood group type 1 antigen bindinga |
|||
|---|---|---|---|---|
| A | B | H | Lewis b | |
| GI.7 (AB758449) | + | − | − | − |
| GII.3 (AB758450) | + | − | − | − |
| GII.4 (AB668028) | + | + | + | + |
| GII.6 (AB758451) | + | + | + | + |
The absence (−) and presence (+) of binding are indicated.
Assay to measure the binding activity of NoVLP to bacterial cells.
NoVLP binding to isolated bacterial cells was investigated by ELISA and TEM. The isolated bacterial strain Enterobacter sp. strain SENG-6 was incubated in Luria-Bertani medium at 37°C for 18 h, and the number of cells suspended in PBS (pH 6.5) was adjusted to 109 cells/ml. Staphylococcus epidermidis (ATCC 35984), an HBGA-negative strain, was incubated in Reasoner's 2A medium at 37°C for 18 h, and the cell number was adjusted to 109 cells/ml. One milliliter of the cell suspension was centrifuged at 3,000 × g for 5 min, resuspended with 400 μl of NoVLPs (GI.7, GII.3, GII.4, or GII.6) in PBS (109 to 1010 particles/ml), and incubated at RT for 1 h. NoVLPs suspended in PBS (without bacterial cells) were used as controls. The mixture was filtered using a mixed cellulose acetate membrane filter (0.20-μm pore size and 13-mm diameter) (13CP020AN; Advantec, Toyo Roshi Kaisha Ltd., Japan) to recover unbound NoVLPs. The recovered NoVLPs were detected using an ELISA as follows. Fifty microliters of NoVLP suspension was added to each well of an ELISA plate and kept at RT for 1 h to coat the well. Triplicate wells were used for each sample. The wells were then washed two times with PBS and blocked with 5% BSA in PBS. After incubation at RT for 2 h, the wells were washed two times with PBS, and 50 μl of anti-NoVLP rabbit antibody (diluted 1:100 with PBS containing 5% BSA; Eve Bio-Science Co., Ltd., Japan) was added to the wells (the anti-NoVLP rabbit antibodies were purified using protein A from the antisera of rabbits immunized separately with each genotype). The plates were incubated at RT for 1 h. The wells were washed two times with PBS, and 50 μl of HRP-conjugated anti-rabbit IgG antibody (SAB-300; Stressgen Bioreagents, Belgium) diluted 1:500 with PBS containing 5% BSA was inoculated into each well. After incubation at RT for 1 h, the wells were washed four times with PBS, and the bound HRP-conjugated antibodies were measured as described previously.
TEM imaging of NoVLP binding to bacterial cells was performed as follows. Enterobacter sp. SENG-6 was incubated at 37°C for 18 h, as described previously, and the number of cells suspended in M9 broth was adjusted to 109 cells/ml. One milliliter of the cell suspension was centrifuged at 3,000 × g for 5 min, and the resulting cell pellet was resuspended in M9 broth (a washing step). The washing step was repeated two times. The cell suspension (109 cells/ml) was centrifuged at 3,000 × g for 5 min, resuspended with 100 μl of GII.6 NoVLPs in PBS (1011 to 1012 particles/ml), and incubated at 4°C for 1 h. The washing step with M9 broth was repeated two times, and the washed cells were placed on collodion membrane-coated copper grids, negatively stained with 1% phosphotungstic acid, and observed using a transmission electron microscope (JEM-1400; JEOL Ltd., Japan).
Immuno-TEM observation for localizing HBGA-like substances.
The localizations of HBGA-like substances were analyzed by immuno-TEM. Enterobacter sp. SENG-6 was incubated at 37°C for 18 h, as described previously. Two milliliters of cell suspension was centrifuged at 3,000 × g for 5 min, resuspended in 2 ml of PBS containing 4% paraformaldehyde and 0.1% glutaraldehyde, and incubated at 4°C for 2 h. The fixed cells were collected by centrifugation at 12,000 × g for 5 min. The resulting cell pellet was washed by soaking in 2 ml of PBS at 4°C for 10 min. This washing step was repeated two times. The cell pellet was dehydrated by soaking in 2 ml of increasing concentrations of ethanol (70, 80, 90, and 95%) and then mixed gently using a rotator at 4°C for 15 min. The following dehydration and infiltration reactions were also conducted on a rotator at 4°C. For complete dehydration, the pellet was soaked in 2 ml of 100% ethanol for 20 min. This final dehydration step was repeated two times. Next, the cell pellet was infiltrated with a mixture of 2 ml of LR White resin (medium grade; London Resin Company, Ltd., England) and 100% ethanol (1:2) for 12 h, followed by a mixture of 2 ml of LR White resin and 100% ethanol (2:1) for 12 h. The solution was replaced with 2 ml of pure LR White resin for 1 h, followed by another 2 ml of pure LR White resin for 12 h. The cell pellet in the pure LR White resin was polymerized using an UV polymerizer (TUV-100; Dosaka EM Co., Ltd., Japan) at 4°C for 3 days. Ultrathin sections (70 nm thick) were prepared from the embedded cell pellet using a diamond knife on an ultramicrotome (RMC MTXL; Boeckeler Instruments Inc., USA) and then placed on nickel grids. The ultrathin sections on grids were washed by floating the grid upside-down on a drop of PBS at RT for 1 min. The following incubations were also conducted by floating the grid upside-down on drops of reagent at RT. The sections were blocked with 4% BSA in PBS for 5 min and incubated with anti-blood group A, B, or O(H) mouse monoclonal antibody (sc-69951, sc-69952, and sc-52372, respectively; Santa Cruz Biotechnology, Inc.) diluted 1:30 with PBS containing 1% BSA for 2 h, followed by washing with PBS (six washes of 1 min each). In a competitive adsorption test, an ultrathin section was incubated with 100 μg/ml of soybean agglutinin prior to incubation with anti-blood group A antibody (sc-69951; Santa Cruz Biotechnology, Inc.). The bound primary antibodies were localized by incubating the sections on anti-mouse IgM antibody gold conjugate (10-nm particles) (ab39613; Abcam, Japan) diluted 1:10 with PBS containing 1% BSA for 1 h, followed by washing with PBS (six washes of 1 min each). The sections were fixed with 2% glutaraldehyde in PBS for 15 min, followed by washing with deionized distilled water (DDW; seven washes of 1 min each). Finally, the sections were stained with 5% uranyl acetate, followed by washing with 50% ethanol for 1 min and DDW (four washes of 1 min each). Stained sections were observed by TEM. A grid was also prepared without the incubation with primary antibodies and used as a control.
Assay of NoVLP binding to HBGA-like substances in bacterial polymeric substances.
Based on a hypothesis that HBGA-like substances were polysaccharides in EPS or LPS, we extracted EPS and LPS from Enterobacter sp. SENG-6. Bacterial EPS was also extracted from S. epidermidis ATCC 35984. EPS was extracted according to the method of Liu and Fang (24) with some modifications. This procedure was used because Pan et al. found that the application of this method to biofilms of algae-bacteria produced a higher yield of carbohydrate in EPS, whereas the protein yield was lower than that with other extraction methods (25). The bacterial strains were incubated at 37°C for 18 to 20 h in 400 ml of each medium, as described previously. The cell suspension (1010 to 1011 cells/ml) was transferred to two 225-ml centrifuge tubes and centrifuged at 3,000 × g for 5 min. The resulting cell pellet was resuspended in 40 ml of PBS using a vortex mixer for 2 min to displace EPS from the cells. No chemicals were used in this first step of EPS recovery. Therefore, the destruction of bacterial cells and the recovery of cellular proteins and genomic DNA components were minimized. Eighty milliliters of the cell suspension was transferred to two 50-ml centrifuge tubes and centrifuged at 9,000 ×g for 5 min. The supernatant containing EPS was collected and filtered using a mixed cellulose ester membrane filter (0.20-μm pore size and 25-mm diameter) (25AS020AS; Advantec, Toyo Roshi Kaisha, Ltd.) to remove bacterial cells. Eighty milliliters of the filtrate was purified with mixed cellulose dialysis tubing (molecular mass cutoff of 3,500 to 5,000 Da) (131204; Spectrum Laboratories, Inc., USA) in DDW at 4°C for 24 h. The purified solution was lyophilized at −50°C for 48 h, and the resulting pellet was dissolved in 4 ml of DDW.
The remaining bacterial cells in the EPS extract from Enterobacter sp. SENG-6 were used to extract SOM, which is mainly composed of LPS, according to Takaara et al. (26). The cell pellet was lyophilized using a vacuum freeze dryer, and the lyophilized cells were suspended in 4 ml of autoclaved distilled water at 65°C. Next, 4 ml of 90% phenol (65°C) was added. After vigorous stirring with a vortex mixer, the samples were stirred for 15 min at 65°C and left for 15 min on ice. The cooled samples were centrifuged at 4,000 × g for 30 min at 2°C. Components of the cell-associated hydrophilic substances, which comprised mainly LPS and cellular RNA (27), were fractionated in the aqueous phase. The aqueous phase (4 ml) was then mixed with 4 ml of 90% phenol at 65°C. The samples were cooled on ice and centrifuged at 4,000 × g for 30 min at 2°C. The aqueous phase was collected and treated with 100 mg/ml RNase A (Sigma-Aldrich) for 6 h at 37°C to eliminate RNA components. Next, RNase A molecules were removed by phenol-chloroform precipitation, and organic substances in the supernatant were purified further by ethanol precipitation. The degradation of RNA molecules by the RNase A treatment was confirmed visibly by agarose gel (1.5%) electrophoresis, ethidium bromide staining, and UV light excitation. The pellet collected after ethanol precipitation was suspended in DDW and stored as an SOM fraction at −80°C until further analysis. In addition to SOM, LPS of Enterobacter sp. SENG-6 was extracted separately using an LPS Extraction Kit (17141; Cosmo Bio Co., Ltd., Tokyo, Japan). The endotoxin units of SOM and LPS were quantified using an EndoLISA Endotoxin Detection Kit (609033; J. K. International, Tokyo, Japan) to confirm the presence of lipid A in the extracted bacterial substances.
NoVLP binding to the extracted EPS, SOM, and LPS was analyzed by ELISA. Fifty microliters of NoVLP GII.6 suspension in PBS (109 to 1010 particles/ml) was added to each well of an ELISA plate and maintained at RT for 1 h to coat the well. Triplicate wells were used for each sample. The wells were washed two times with PBS and blocked with 5% BSA in PBS. After incubation at RT for 2 h, the wells were washed two times with PBS, and 50 μl of the extracted EPS, SOM, or LPS was applied to the wells. After incubation at RT for 1 h, the wells were washed two times with PBS, and 50 μl of anti-blood group A, B, or O(H) mouse monoclonal antibody (sc-69951, sc-69952, or sc-52372, respectively; Santa Cruz Biotechnology, Inc., USA) diluted 1:100 with PBS containing 5% BSA was applied to the wells. After incubation at RT for 1 h, the wells were washed two times with PBS, and 50 μl of HRP-conjugated anti-mouse IgM antibody (A90-101P; Bethyl Laboratories, USA) diluted 1:500 with PBS containing 5% BSA was inoculated into each well. After incubation at RT for 1 h, the wells were washed four times with PBS, and the bound HRP-conjugated antibodies were measured as described previously. Wells without addition of EPS were used as controls.
In order to prove the specific binding of NoVLPs to the bacterial EPS, NoVLPs from the GI.1 wild-type strain 8fIIa (GenBank accession number M87661) and its mutant strain (W375A), kindly provided by Mary K. Estes, Baylor College of Medicine, were tested against the EPS extracted from Enterobacter sp. SENG-6. The mutant strain (W375A) does not recognize A and O(H) antigens (28). Furthermore, the extracted EPS treated with α-N-acetyl-galactosaminidase, which specifically catalyzes the hydrolysis of α-linked d-N-acetyl-galactosamine residues from oligosaccharides, was also used in ELISAs for detecting A antigen. The enzyme treatment was performed as follows. One hundred microliters of reaction mixture contained 88 μl of extracted EPS, 10 μl of 10× G7 reaction buffer, and 1 μl of α-N-acetyl-galactosaminidase to yield a final concentration of 0.2 U/ml (New England BioLabs, Inc., Japan), and 1 μl of 10 mg/ml BSA. One microliter of DDW was added instead of α-N-acetyl-galactosaminidase to control EPS. The tubes were incubated at 37°C for 24 h.
Specific interaction between NoVLPs and HBGA-like substances.
The quartz crystal microbalance (QCM) method was used to evaluate the specific interaction between GII.6 NoVLPs and HBGA-like substances in the extracted EPS. The principle of QCM is the same as an electric scale, where the frequency of an energized quartz crystal changes relative to the amount of substances accumulated on the quartz crystal surface. Therefore, the specific interaction between ligand and receptor molecules is detected when the ligand molecules are inoculated onto the quartz crystal surface where the receptor molecules are immobilized. Fifty microliters of NoVLP suspension in PBS at a concentration of 20 μg of total protein/ml was placed on a sensor chip and incubated at RT for 1 h. The sensor chip was blocked with 0.1% BSA in PBS. After incubation at RT for 1 h, the chip was washed with DDW and installed in the reactor of a QCM instrument (Single-Q0500; BioLab/AsOne Co., Ltd., Japan). The reactor was filled with 500 μl of PBS and stirred at 300 rpm during measurements. When the frequency of the chip stabilized, a 3-μl aliquot of 1 mg of total organic carbon (TOC)/ml of the extracted EPS in PBS was inoculated into the reactor consecutively to produce a total of 24 μl, and the change in the frequency of the chip (ΔF) was recorded. This adsorption experiment was performed three times, and the adsorption isotherms were obtained. As controls, adsorption isotherms were obtained without NoVLPs immobilized on the sensor chip. The difference between the ΔF values of the test and control conditions (ΔΔF) was used to calculate the equilibrium binding constant. The molecular weight of the HBGA-like EPS from Enterobacter sp. SENG-6 was estimated to calculate the apparent equilibrium binding constant of the HBGA-like EPS from Enterobacter sp. SENG-6 and GII.6 NoVLPs according to the Langmuir adsorption isotherm. In brief, the extracted EPS was fractionated using an ultrafiltration device (molecular mass cutoff of 300 kDa) (Nanosep centrifugal device; Pall Corporation, USA), and the filtrate was collected. The amount of A-like substances in the filtrate was measured by ELISA using anti-blood group A antibody, as described above.
Nucleotide sequence accession numbers.
The partial 16S rRNA gene sequence of Enterobacter sp. SENG-6 was deposited in the GenBank/EMBL/DDBJ database under accession number AB758448. The accession numbers for the capsid genes of the HuNoV genotypes of GI.7, GII.3, GII.4, and GII.6 were deposited under accession numbers AB758449, AB758450, AB668028, and AB758451, respectively.
RESULTS
Isolation of human enteric bacteria bearing HBGA-like substances.
We screened blood group-active enteric bacteria from human feces using a biopanning technique with anti-HBGA antibodies. Of 35 isolated strains, 21 screened bacterial strains had blood group activity with A, B, or O(H) (Table 2). The phylogenetic relationships of the 16S rRNA gene sequences of the 21 isolated strains and related enteric bacteria are shown in Fig. 1. These enteric bacteria were Gram negative and were genetically related to Enterobacter cloacae (ATCC 13047T), Shigella flexneri (ATCC 29903T), or Escherichia fergusonii (ATCC 35469T). From these, we isolated a bacterium (designated strain SENG-6) that was closely related to Enterobacter cloacae (ATCC 13047T) with a 16S rRNA gene sequence similarity of 99.9%. Strain SENG-6 had A, B, and O(H) blood group activities. Twelve of 15 strains that were closely related to S. flexneri (ATCC 29903T), sharing 99.3 to 99.9% of its 16S rRNA gene sequence, had both A and O(H) blood group activities (e.g., NMCA1-7 and NMC20B1-4), whereas two other strains (MURA1-1 and MUR18B1-4) with similarities of 99.5 and 99.6%, respectively, had only A blood group activity, while NMC20B1-6 with a similarity of 99.5% had only O(H) blood activity. All five strains that were closely related to E. fergusonii (ATCC 35469T), sharing 99.4 to 99.8% of its 16S rRNA gene sequence, had both A and O(H) blood group activities.
Table 2.
Blood group activities of isolated and reference bacterial strains used in this study
| Strain | Growth mediuma | Blood group activity |
OD600b | Genetically similar strain based on 16S rRNA gene analysis |
|||
|---|---|---|---|---|---|---|---|
| A | B | O(H) | Organism (strain no.) | % Similarity | |||
| SENG-6 | BGLB | + | + | + | 0.068 | E. cloacae (ATCC 13047T) | 99.9 |
| NMCA1-7 | BGLB | + | − | + | 0.920 | S. flexneri (ATCC 29903T) | 99.6 |
| NMCA1-8 | BGLB | + | − | + | 0.082 | E. fergusonii (ATCC 35469T) | 99.8 |
| NMC18B1-3 | BGLB | + | − | + | 0.091 | E. fergusonii (ATCC 35469T) | 99.4 |
| NMC18B1-6 | BGLB | + | − | + | 0.058 | E. fergusonii (ATCC 35469T) | 99.5 |
| NMC20B1-4 | BGLB | + | − | + | 0.821 | S. flexneri (ATCC 29903T) | 99.5 |
| NMC20B1-6 | BGLB | − | − | + | 0.302 | S. flexneri (ATCC 29903T) | 99.5 |
| KKA1-1 | EC | + | − | + | 0.465 | E. fergusonii (ATCC 35469T) | 99.8 |
| KKB1-1 | EC | + | − | + | 0.502 | E. fergusonii (ATCC 35469T) | 99.5 |
| TZWH1-1 | EEM | + | − | + | 0.502 | S. flexneri (ATCC 29903T) | 99.6 |
| TZWH1-2 | EEM | + | − | + | 0.089 | S. flexneri (ATCC 29903T) | 99.6 |
| TZWH2-2 | EEM | + | − | + | 0.515 | S. flexneri (ATCC 29903T) | 99.3 |
| TZWH2-3 | EEM | + | − | + | 0.570 | S. flexneri (ATCC 29903T) | 99.6 |
| TZWH2-4 | EEM | + | − | + | 0.568 | S. flexneri (ATCC 29903T) | 99.4 |
| TZWA1-2 | EEM | + | − | + | 0.532 | S. flexneri (ATCC 29903T) | 99.6 |
| TZWA1-3 | EEM | + | − | + | 0.504 | S. flexneri (ATCC 29903T) | 99.6 |
| TZWA1-4 | EEM | + | − | + | 0.257 | S. flexneri (ATCC 29903T) | 99.4 |
| MURA1-1 | Mossel | + | − | − | 0.407 | S. flexneri (ATCC 29903T) | 99.5 |
| MURA1-6 | Mossel | + | − | + | 0.428 | S. flexneri (ATCC 29903T) | 99.9 |
| MUR18B1-4 | Mossel | + | − | − | 0.367 | S. flexneri (ATCC 29903T) | 99.6 |
| MUR20B1-4 | Mossel | + | − | + | 0.263 | S. flexneri (ATCC 29903T) | 99.7 |
| ATCC 35984c | R2A | − | − | − | − | − | |
BGLB, 2% brilliant green bile broth; EC, E. coli broth; EEM, Enterobacteriaceae enrichment mannitol broth; Mossel, enterobacteria enrichment broth-Mossel; R2A, Reasoner's 2A.
Minimum value with an HBGA-positive result.
Staphylococcus epidermidis ATCC 35984 was used as the HBGA-negative strain, and it gave an HBGA-negative result, even at an OD600 of 1.357.
Fig 1.
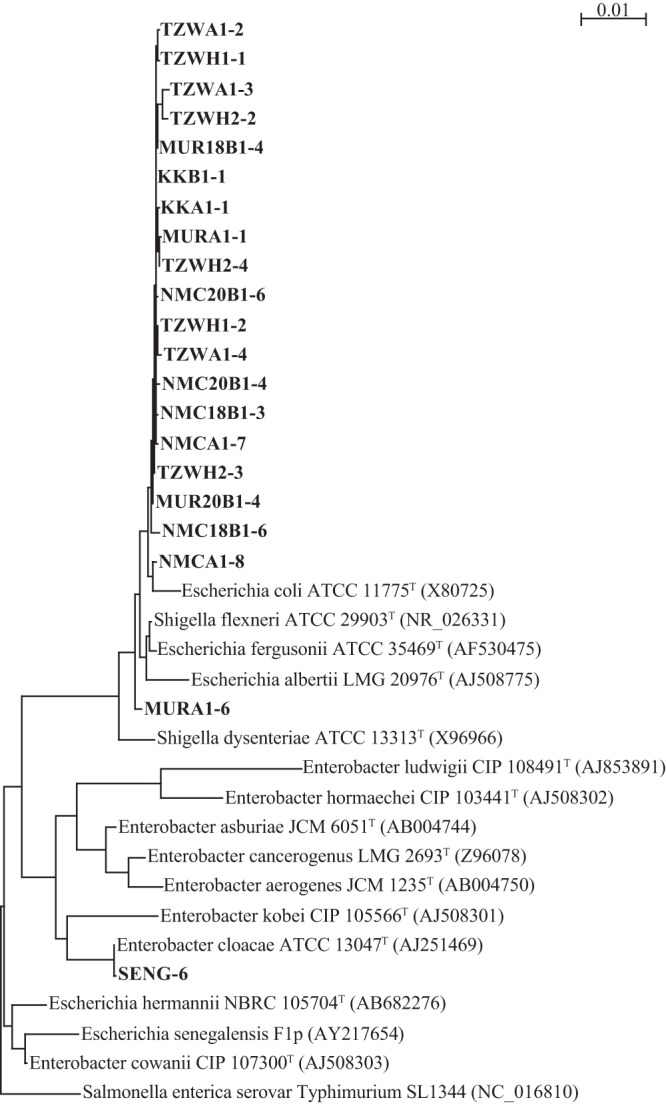
Phylogenetic relationships of 16S rRNA gene sequences of isolated bacterial strains and related enteric bacteria. The phylogenetic tree was constructed by the neighbor-joining method with 1,000 bootstrap replicates using ClustalX (version 2.1). Bold type indicates the bacterial strains isolated in this study; Salmonella enterica serovar Typhimurium SL1344 was used as an outgroup. The scale bar represents the number of substitutions per site.
Strains SENG-6 and NMC18B1-6 produced HBGA-positive results at the minimum optical density (OD600) of 0.068 and 0.058, respectively (Table 2), which showed that these strains carried a high abundance of HBGA-like substances. SENG-6 possessed A, B, and O(H) blood group activities; therefore, this strain was used in further experiments. Staphylococcus epidermidis (ATCC 35984) was used as the HBGA-negative strain because it had an HBGA-negative result, even at an OD600 of 1.357 (Table 2).
NoVLP binding to isolated bacterial cells.
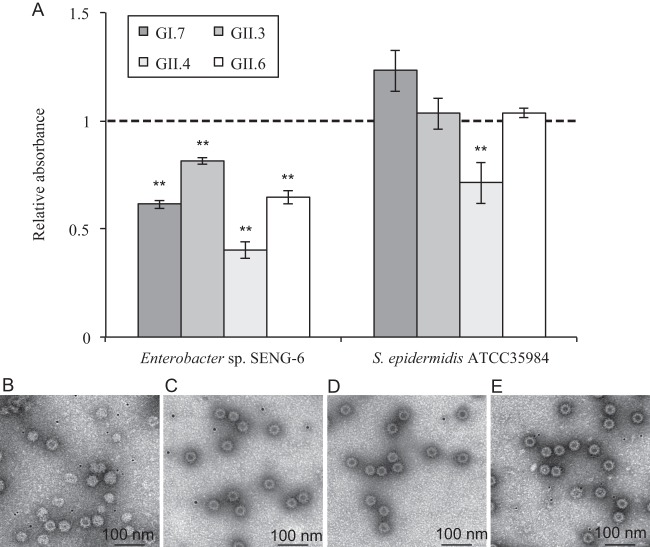
The blood group activity test suggested that HBGA-like substances were present in the bacterial outer components, including the LPS of Gram-negative bacteria and/or EPS, including the capsule. The binding of NoVLP genotypes GI.7, GII.3, GII.4, and GII.6 to Enterobacter sp. SENG-6 and S. epidermidis ATCC 35984 was tested. Each NoVLP genotype was mixed with each strain, and unbound NoVLPs were detected by ELISA (Fig. 2). NoVLPs from each genotype were also mixed with PBS (without bacterial cells), and used as negative controls. All NoVLP genotypes tested bound significantly to Enterobacter sp. SENG-6 (P < 0.01). While GII.4 NoVLPs bound to S. epidermidis ATCC 35984 (Fig. 2A), it was previously shown that this genotype recognizes a broad range of ligands other than HBGAs, such as those containing sialic acid (29–31).
Fig 2.
NoVLP binding to bacterial cells. (A) Cells of Enterobacter sp. SENG-6 and Staphylococcus epidermidis (ATCC 35984) were mixed with each NoVLP genotype (GI.7, GII.3, GII.4, and GII.6) in PBS. Unbound NoVLPs were recovered by filtration and detected by ELISA. Suspended NoVLPs in PBS that were not mixed with bacterial cells were filtered and used as controls. The results were expressed as the absorbance relative to the control; therefore, values of <1 (dashed line) indicate the significant binding of NoVLPs to bacterial cells. The error bars represent the standard deviations of triplicate independent measurements. **, P < 0.01 (t test). (B) GI.7 NoVLPs. (C) GII.3 NoVLPs. (D) GII.4 NoVLPs. (E) GII.6 NoVLPs. Black dots in the viewing fields in panels B to E are avidin-conjugated gold nanoparticles (average diameter, 5 nm), which were added to estimate the diameter of NoVLPs.
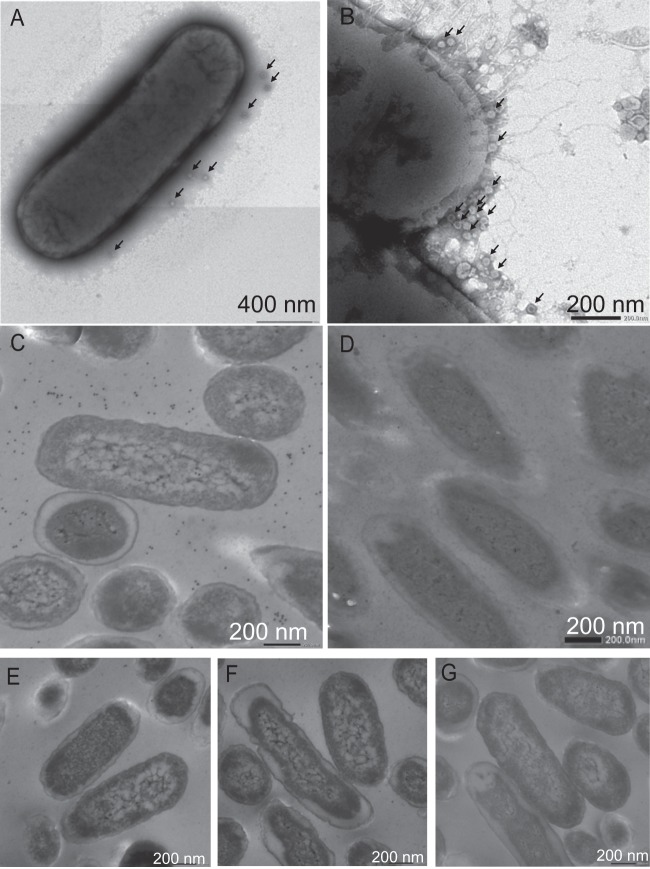
The direct visualization with TEM confirmed that NoVLP genotypes GI.7 and GII.6 bound to Enterobacter sp. SENG-6 (Fig. 3A and B). The majority of these NoVLPs were observed to bind mainly to the EPS of Enterobacter sp. SENG-6 (Fig. 3A and B), whereas a few NoVLPs were observed in the region adjacent to bacterial cells. However, it was not determined by this technique whether NoVLPs bound to the LPS of Enterobacter sp. SENG-6 because most LPS components were covered by EPS, which would hamper the interaction between NoVLPs and LPS.
Fig 3.
Binding of NoVLPs to bacterial cells and localization of HBGA-like substances of Enterobacter sp. SENG-6. Enterobacter sp. SENG-6 was reacted with GI.7 (A) or GII.6 (B) NoVLPs in PBS. NoVLPs are indicated by arrows in both panels. For the localization of HBGA-like substance, ultrathin sections of Enterobacter sp. SENG-6 were labeled with immunogold nanoparticles after being reacted with anti-blood group A, B, and O(H) antibodies (primary antibodies) and observed by TEM. Ultrathin sections without primary antibodies were used as controls. (C) A-like substances. (D) A-like substances after pretreatment with soybean agglutinin, a lectin specific to N-acetyl-galactosamine. (E) B-like substances. (F) O(H)-like substances. (G) Primary antibody-negative control.
Localization of HBGA-like substances in bacterial cell components.
Ultrathin sections of Enterobacter sp. SENG-6 were examined by immuno-TEM to determine the localization of HBGA-like substances. Enterobacter sp. SENG-6 was labeled with gold nanoparticles carrying anti-mouse IgM after reacting with anti-blood group A, B, or O(H) antibody (used as the primary antibody). A number of gold nanoparticles were observed in the extracellular spaces of Enterobacter sp. SENG-6 (Fig. 3C), which had reacted with anti-A antibody. The scattered occurrence of A-like substances outside bacterial cells suggests that these antigens were present at the terminals of each extracellular polymer although this needs to be confirmed by the structural analysis of EPS. Figure 3D shows that the number of gold nanoparticles in the ultrathin section was reduced dramatically by pretreatment of the ultrathin section with soybean agglutinin, which is a lectin specific to N-acetyl-galactosamine. This demonstrated that anti-A antibodies labeled with gold nanoparticles in the ultrathin section shown in Fig. 3C specifically bound to A-like substances that contained N-acetyl-galactosamine. In contrast, few B- and O(H)-like substances were observed in Enterobacter sp. SENG-6 (Fig. 3E and F, respectively) although the B and O(H) blood group activities of this bacterial strain were confirmed using a blood group typing kit (Table 2). The failure to detect B- and O(H)-like substances in the immuno-TEM images may be attributable to a subtle difference in the recognition of the antibodies used in the blood group typing kit and immuno-TEM or insufficient amounts of B- and O(H)-like substances in immuno-TEM although it is difficult to compare the relative amounts of each HBGA-like substance. Gold nanoparticles did not accumulate at the edges of the cell surfaces (Fig. 3C), which showed that only limited amounts of HBGA-like substances were present in the LPS of Enterobacter sp. SENG-6, even if blood type-active substances were present.
NoVLP binding to HBGA-like substances in bacterial EPS, SOM, and LPS.
TEM observations showed that NoVLPs were captured mainly in the bacterial EPS where the A-like substances were present, whereas LPS had no or only a small number of specific binding sites for NoVLPs (Fig. 3). To further elucidate the localization of HBGA-like substances, EPS, SOM, and LPS were extracted separately from Enterobacter sp. SENG-6 and tested to detect HBGA-like substances by ELISA using GII.6 NoVLPs. GII.6 is one of the best genotypes for the binding assay because this genotype was shown to bind to A, B, and O(H) antigens (Table 1). The bacterial EPS were also extracted from S. epidermidis ATCC 35984 and used in the same ELISA. SOM and LPS of S. epidermidis ATCC 35984 were not extracted because S. epidermidis ATCC 35984 is Gram positive.
Figure 4A shows that A- and B-like substances were detected in the extracted EPS of Enterobacter sp. SENG-6. B-like substances were not detected in immuno-TEM (Fig. 3E), but the extraction and concentration of EPS allowed them to be detected by ELISA. However, no significant signals for A-, B-, and O(H)-like substances were detected in SOM and LPS of Enterobacter sp. SENG-6. Likewise, no signals for A-, B-, and O(H)-like substances were detected in the EPS of S. epidermidis ATCC 35984 (Fig. 4A). Enterobacter sp. SENG-6 was positive for all A, B, and O(H) blood group antigens (Table 2), whereas O(H)-like substances were not detected by ELISA, which may be caused by the difference in the recognition of anti-O(H) antibodies used in the blood typing kit and ELISA.
Fig 4.
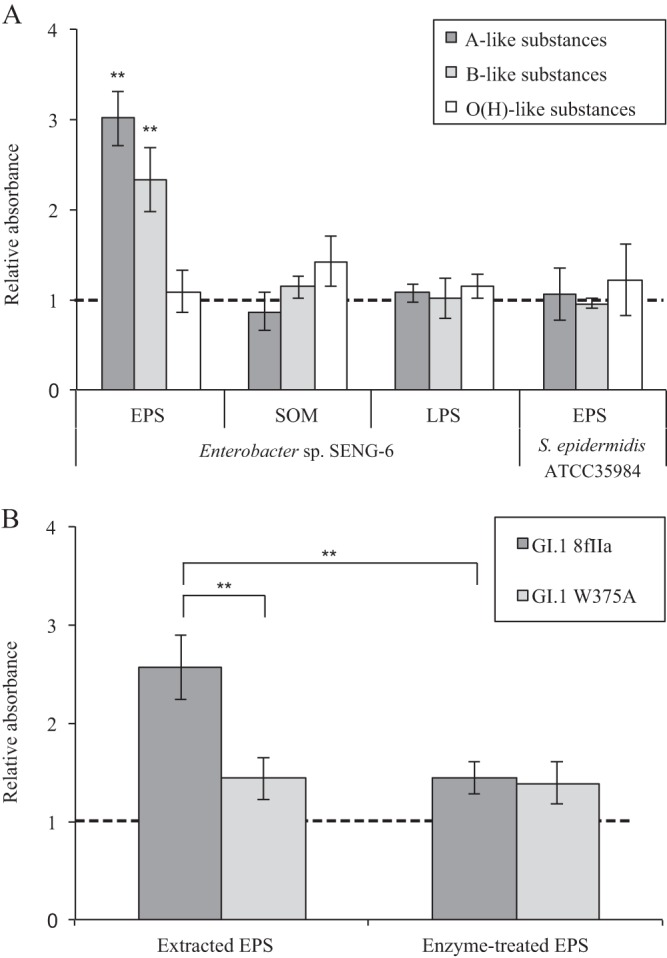
Detection of HBGA-like substances in extracted bacterial polymers. (A) The wells on an ELISA plate were coated with GII.6 NoVLPs, and EPS, SOM, and LPS extracted from Enterobacter sp. SENG-6 and EPS extracted from Staphylococcus epidermidis (ATCC 35984) were inoculated into the wells. We used ELISAs to measure A-, B-, and O(H)-like substances in the EPS, SOM, or LPS which bound to NoVLPs. A well without the incubation of bacterial polymeric substances was used as a control. (B) The wells were coated with GI.1 NoVLPs (strain 8fIIa or the W375A mutant), and the EPS extracted from Enterobacter sp. SENG-6 or the EPS treated with α-N-acetyl-galactosaminidase was reacted. A-like substances which bound to the NoVLPs were detected by ELISA. All results are expressed as the absorbance relative to the control; therefore, values of >1 (dashed line) indicate the presence of A-, B-, or O(H)-like substances. The error bars represent the standard deviations of triplicate independent measurements. *, P < 0.05; **, P < 0.01 (t test).
The binding between GII.6 NoVLPs and EPS of Enterobacter sp. SENG-6 containing A-like substances is shown clearly in Fig. 4A although it is not clear whether the A-like substances in the EPS of Enterobacter sp. SENG-6 are the main binding sites for GII.6 NoVLPs. To test the function of A-like substances in EPS as the binding site for NoVLPs, we performed ELISAs using the NoVLPs of GI.1 wild-type (8fIIa) and mutant (W375A) strains. The GI.1 wild-type strain (8fIIa) can recognize A antigen, whereas the mutant strain (W375A) has lost its ability to bind to A antigen because of a point mutation from a tryptophan to an alanine residue at position 375 (28). Figure 4B shows that A-like substances in the EPS of Enterobacter sp. SENG-6 were detected when NoVLPs of the GI.1 wild-type strain (8fIIa) were immobilized in ELISA, but they were not detected when NoVLPs of the GI.1 mutant strain (W375A) were immobilized. Furthermore, the enzymatic treatment of EPS with N-acetyl-galactosaminidase, which can release the terminal N-acetyl-galactosamine from A antigen, weakened the interaction between EPS and NoVLPs of the GI.1 wild-type strain (8fIIa) (Fig. 4B). These results show that A-like substances in the EPS of Enterobacter sp. SENG-6 are the binding sites of NoVLPs. The A antigen has also been found in HBGA-like substances in the digestive tissues of oysters (32), and the critical role of the terminal N-acetyl-galactosamine residue in the strain-dependent accumulation of HuNoVs in oysters has been discussed previously (29, 30). Bacterial A-like substances can also affect the genotype-dependent fate of HuNoVs in the environment because each HuNoV genotype has its own HBGA binding profile (12).
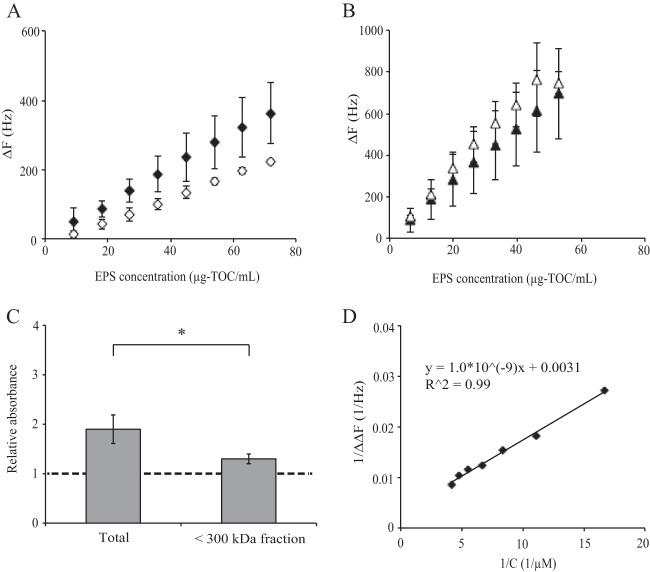
The adsorption isotherms of the extracted EPS were determined for GII.6 NoVLPs, which were immobilized on a sensor chip (Fig. 5A and B). The ΔF values for the EPS of Enterobacter sp. SENG-6 with GII.6 NoVLPs were higher than those under the NoVLP-negative condition, and this difference was statistically significant according to a Mann-Whitney U test performed for each EPS concentration (Fig. 5A). In contrast, the ΔF values for the EPS extracted from S. epidermidis ATCC 35984 were not different from those under the NoVLP-negative condition (Fig. 5B). These results indicated that the EPS extracted from Enterobacter sp. SENG-6 can interact specifically with NoVLPs. In order to estimate the molecular mass of HBGA-positive EPS from Enterobacter sp. SENG-6, the amount of A-like substances in the EPS extracted from Enterobacter sp. SENG-6 was determined before and after ultrafiltration with a molecular mass cutoff of 300 kDa. As a result, the amount of A-like substances was significantly reduced by ultrafiltration (Fig. 5C), which suggested that the majority of A-like substances existed in EPS with a molecular mass of >300 kDa. If we assume that A-like antigens dominated the EPS and that the EPS carrying the HBGA-like substances had a molecular mass of 300 kDa, the reciprocal plots of ΔΔF (the difference between ΔF with NoVLPs and that without NoVLPs) (Fig. 5A) and EPS concentration is obtained (Fig. 5D), which gives an apparent equilibrium binding constant of 3.1 × 106 M−1 according to the Langmuir adsorption isotherm. Since the molecular mass of HBGA-positive EPS is expected to be larger than 300 kDa (Fig. 5C), the apparent equilibrium binding constant for the HBGA-positive EPS of Enterobacter sp. SENG-6 and GII.6 NoVLPs must be >3.1 × 106 M−1.
Fig 5.
Binding avidity between NoVLPs and bacterial EPS. The adsorption isotherm between GII.6 NoVLPs and EPS from Enterobacter sp. SENG-6 (A) and Staphylococcus epidermidis (ATCC 35984) (B) was measured using the QCM method. GII.6 NoVLPs were immobilized on a sensor chip, and various amounts of extracted EPS were inoculated. The change in frequency of the sensor chip was measured, which depended on the amount of EPS bound to GII.6 NoVLPs. The error bars represent the standard deviations of triplicate independent measurements. Filled symbols in panels A and B are ΔF values in the presence of NoVLPs, while open symbols are those in the absence of NoVLPs. (C) Detection of A-like substances in EPS extracted from Enterobacter sp. SENG-6 before and after ultrafiltration. The wells of an ELISA plate were coated with GII.6 NoVLPs, and the wells were inoculated with EPS extracted from Enterobacter sp. SENG-6 before or after ultrafiltration with a molecular mass cutoff of 300 kDa. A well without EPS was used as a control. The results are expressed as the absorbance relative to the control. The error bars represent the standard deviations. *, P < 0.05 (t test). (D) Reciprocal plots of ΔΔF and the concentration of EPS. ΔΔF is the difference between the ΔF value in the presence of NoVLPs and that in the absence of NoVLPs.
DISCUSSION
Blood group activities of human enteric bacteria.
The presence of blood group-active enteric bacteria was reported many years ago. Springer et al. tested 282 Gram-negative bacterial strains from genera such as Escherichia, Salmonella, and Klebsiella, which were isolated from clinical specimens, and approximately 50% of the strains inhibited anti-blood group A, B, and/or O(H) agglutinins. Of the tested strains belonging to Escherichia, some strains had a single aggregation activity [A, B, or O(H)], whereas others had multiple aggregation activities [ABO(H), AO(H), AB, or BO(H)] (17). In the present study, 21 blood group-active strains were isolated from human feces using a biopanning technique with anti-HBGA antibodies (Table 2). The isolated bacterial strains must represent only a small fraction of the actual population of HBGA-positive bacteria. We analyzed only culturable strains in a fecal sample from a single individual under microaerobic conditions in this study, and it is recognized that the majority of enteric bacteria in human feces are likely to be dead, viable but not culturable (33), or obligate anaerobes.
The blood group activities of various widely distributed Gram-negative bacteria were reported initially by Springer et al. (17), as described above, and these blood group activities of enteric bacteria were attributed to the presence of HBGA-like O antigen in LPS. The structural basis of the HBGA-like O antigen was clarified recently. For example, Anderson et al. showed that the O-antigen polysaccharide of E. coli O86 contained a B-type trisaccharide where alpha-l-fucose and beta-d-galactose formed part of the O-antigen backbone, while alpha-d-galactose was conjugated to beta-d-galactose (34). Salmonella enterica subsp. enterica serovar Milwaukee O:43 (group U) also has human blood group B activity with the O polysaccharide (35). Therefore, we initially postulated that HBGA-like substances were present in the LPS of the isolated strains. However, immuno-TEM imaging (Fig. 3) and ELISAs (Fig. 4) clearly showed that HBGA-like substances were localized mainly in the bacterial EPS. Only Klebsiella sp. was suggested to have blood group-active substances in its capsular polysaccharide although this was not proved (17). This is the first study to show that human enteric bacteria possess HBGA-like substances in their EPS and that they can capture HuNoVLPs.
Interaction between bacterial substances and NoVLPs.
Four genotypes of NoVLPs (GI.7, GII.3, GII.4, and GII.6) were used in the binding assay to HBGA-positive (Enterobacter sp. SENG-6) and HBGA-negative (S. epidermidis ATCC 35984) cells, and only GII.4 bound to both strains (Fig. 2A). It has been shown that NoV GII.4 may bind to multiple carbohydrate moieties, including alpha-d-Gal, alpha-d-Man/alpha-d-Glc, and alpha-l-Fuc, in addition to GalNAc, GlcNAc, and sialic acid (30). Although extracellular polymeric substances of planktonic S. epidermidis cells have not been well analyzed so far, the broad binding capacity of NoV GII.4 is likely to contribute to the binding to HBGA-negative cells of S. epidermidis ATCC 35984 observed in the present study. This binding property of NoV GII.4 did not allow us to use this genotype in the binding assays described in the legends of Fig. 3 to 5 because it was necessary to analyze the specific interaction between bacterial HBGAs and NoVLPs. In that context, NoV GII.6 was available for those binding assays in the present study because this genotype can recognize all HBGAs tested (Table 1) but seemed not to have a binding spectrum as broad as that of GII.4, as judged by the binding assay results shown in Fig. 2. Further analysis of the binding spectrum of multiple genotypes of HuNoVs other than GII.4 may provide a clue for better understanding the genotype-specific lifestyle of HuNoVs.
Significance of the blood group-active enteric bacteria: a hypothesis.
The binding is likely to affect at least two aspects of the ecology of HuNoVs, particularly the fate of HuNoVs in their hosts after HuNoVs enter the human intestine and encounter large numbers of enteric bacteria. In this regard, there is increasing understanding of the roles played by the intestinal microbiota in viral infections. Using antibiotic-treated and germ-free mice, Kane et al. showed that bacterial LPS plays a key role in the infection of mouse mammary tumor virus (MMTV) (36). MMTV-bound bacterial LPS was recognized by Toll-like receptor 4, and it affected the induction of the inhibitory cytokine interleukin-10. However, this enhanced viral titers, and MMTV overcame the immune response. Kuss et al. found that antibiotic-treated mice were less susceptible to poliovirus infection and demonstrated that exposure to bacteria or their N-acetyl-glucosamine-containing surface polysaccharides, including LPS and peptidoglycan, enhanced the infectivity of poliovirus (37). These results suggest that antibiotic-mediated microbiota depletion reduces virus infections and that viruses may exploit intestinal microbes for replication and transmission. Therefore, it is likely that the commensal microbiota in the human intestine will also affect the likelihood of infection with human enteric viruses, including HuNoVs, in the intestine. A higher level of blood group-active EPS in the mucosa of the intestine (jejunum and ileum) probably provides HuNoVs with a greater opportunity for infecting intestinal cells because norovirus particles may be retained in the intestine, provided that norovirus-binding bacteria are present although it is also possible that bacterial HBGA will be an inhibitor of HuNoV binding to human HBGA on intestinal cells. The abundance of blood group-active EPS could also affect rotavirus infection, another major gastroenteritis virus, because spike protein VP8 of human rotavirus recognizes HBGAs (38, 39). Thus, enteric virus infections may be characterized on the basis of the bacterial community composition of the human intestine, which varies considerably with age, lifestyle, and season. Understanding the role of the intestinal microbiota in viral transmission and pathogenesis may lead to the development of new antiviral strategies although the role of bacterial HBGAs in the infections of intestinal mucosal cells with HuNoVs remains largely speculative at this moment.
Another important implication concerning HBGA-positive bacterial EPS for the HuNoV life cycle is the transport of viral particles from acute gastroenteritis patients to aquatic environments. Enteric viruses are known to be associated mainly with particles in environmental waters (40). Various environmental materials can capture human enteric viruses, including activated sludge (41), estuarine sediment (41, 42), river sediment (43), and soils (41, 44). da Silva et al. showed that the GI and GII noroviruses found in wastewater samples were associated mainly with particles measuring <180 μm (45). Sano et al. demonstrated that indigenous noroviruses were not present in the permeates of mixed liquor and treated wastewater filtered using a microfiltration membrane (pore size, 0.1 μm) (46). Gentry et al. showed that an unexpectedly large number of HuNoVs were present in the fraction collected using 63- and 200-μm-mesh plankton nets, which suggested the binding of HuNoV particles to phytoplankton and zooplankton (47). Viral particles associated with environmental materials are physically more stable than freely moving viral particles (48). Viruses associated with organic or inorganic particles are protected from inactivation by chlorine (49), UV (40), and salinity (osmotic shock) (50) because these exogenous stresses are impeded by the entities that capture the viral particles. Furthermore, enteric viruses associated with environmental suspended solids may be transported in a different manner from freely moving virions (51, 52). Thus, HuNoV particles associated with HBGA-positive EPS that is attached to or disengaged from enteric bacterial cells must have different fates from free viral particles. Since the proteolytic virucidal activity of some bacterial species has been reported (53), the association of HuNoV particles with HBGA-positive EPS detached from bacterial cells may have an advantage in terms of the environmental persistency of the virus. These hypotheses about the impact of bacterial HBGAs on the environmental persistency of HuNoVs largely remain to be proved and are challenging issues in further studies.
Avidity of the blood group-active bacterial EPS with HuNoV particles.
Thus, the effect of the blood group-active bacterial EPS on the fate of HuNoVs is totally dependent on the binding avidity of EPS to HuNoV particles. If we assume that all the components in the extracted EPS are blood group active and that they have a molecular mass of >300 kDa (Fig. 5C), we can estimate the apparent equilibrium binding constant according to the Langmuir adsorption isotherm as >3.1 × 106 M−1. This value is comparable with that obtained for the interactions between ß-galactose-specific lectins and lactose-carrying glycopolystyrenes, which have binding constants of 106 to 107 M−1 (54). The interactions between enteric viruses and environmental adsorbents were previously shown to have no binding specificity (41–45, 47), whereas HuNoV binding to bacterial HBGAs is regarded as specific, as illustrated by human HBGAs and HuNoVs (12). In addition to enthalpy-driven adsorption via an array of hydrogen binding routes (28), the entropy effect caused by multiple binding sites also contributes to the increased binding avidity although structural analyses of bacterial EPS are essential to obtain an insight into the true value of the binding constant. The specific binding capacity of blood group-active enteric bacteria with HuNoVs provides a new scientific platform for understanding the interactions between two types of microbes that were previously regarded as biologically unrelated.
Conclusions.
Enterobacter sp. SENG-6, an isolated strain from a fecal sample of a healthy person, exhibited blood group activity. We observed A-like substances in its extracellular region. EPS extracted from Enterobacter sp. SENG-6 captured norovirus particles, specifically via A-like substances, with an avidity comparable to the avidities between multi-glycoconjugate polymers and lectins. Further studies of human enteric bacteria capable of binding to HuNoVs are warranted to gain new insights into the biological role of HBGA-positive bacterial EPS in the transmission and infection of HuNoVs in the environment and are likely to provide a clue for the development of novel prevention strategies.
ACKNOWLEDGMENTS
We are very grateful to Mary Estes, Baylor College of Medicine, TX, for providing the NoVLP GI.1 wild-type strain (8fIIa) and the mutant strain (W375A). We also thank Natsumi Ushijima, Support Section for Education and Research, Graduate School of Dental Medicine, Hokkaido University, for her technical assistance during TEM observations. We thank Rie Nomachi, Kanae Kawamura, and Megumi Tazawa, Division of Environmental Engineering, Faculty of Engineering, Hokkaido University, for their technical support during NoVLP production and bacterial cultivation.
This study was supported by the Japan Science and Technology Agency through Core Research for Evolutionary Science and Technology and the Japan Society for the Promotion of Science through Grant-in-Aid for Young Scientist (A) (22686049), Challenging Exploratory Research (21659157), and Research Fellowships for Young Scientists (23-4434).
Footnotes
Published ahead of print 26 June 2013
REFERENCES
- 1.Lopman BA, Reacher MH, Vipond IB, Hill D, Perry C, Halladay T, Brown DW, Edmunds WJ, Sarangi J. 2004. Epidemiology and cost of nosocomial gastroenteritis, Avon, England, 2002–2003. Emerg. Infect. Dis. 10:1827–1834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zheng DP, Ando T, Frankhauser RL, Beard RS, Glass RI, Monroe SS. 2006. Norovirus classification and proposed strain nomenclature. Virology 346:312–323 [DOI] [PubMed] [Google Scholar]
- 3.Kageyama T, Shinohara M, Uchida K, Fukushi S, Hoshino FB, Kojima S, Takai R, Oka T, Takeda N, Katayama K. 2004. Coexistence of multiple genotypes including newly identified genotypes in outbreaks of gastroenteritis due to norovirus in Japan. J. Clin. Microbiol. 42:2988–2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Green KY. 2007. Caliciviridae: the noroviruses, p 949–979 In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE. (ed), Fields virology, 5th ed, vol 1 Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 5.Le Guyader FS, Bon F, DeMedici D, Parnaudeau S, Bertone A, Crudeli S, Doyle A, Zidane M, Suffredini E, Kohli E, Maddalo F, Monini M, Gallay A, Pommepuy M, Pothier P, Ruggeri FM. 2006. Detection of multiple noroviruses associated with an international gastroenteritis outbreaks linked to oyster consumption. J. Clin. Microbiol. 44:3878–3882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nenonen NP, Hannoun C, Larsson CU, Bergstrom T. 2012. Marked genomic diversity of norovirus genogroup I strains in a waterborne outbreak. Appl. Environ. Microbiol. 78:1846–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ueki Y, Sano D, Watanabe T, Akiyama K, Omura T. 2005. Norovirus pathway in water environment estimated by genetic analysis of strains from patients of gastroenteritis, sewage, treated wastewater, river water and oysters. Water Res. 39:4271–4280 [DOI] [PubMed] [Google Scholar]
- 8.Perez-Sautu U, Sano D, Guix S, Kasimir G, Pinto RM, Bosch A. 2012. Human norovirus occurrence and diversity in the Llobregat river catchment, Spain. Environ. Microbiol. 14:494–502 [DOI] [PubMed] [Google Scholar]
- 9.Donaldson EF, Lindesmith LC, LoBue AD, Baric RS. 2010. Viral shape-shifting: norovirus evasion of the human immune system. Nat. Rev. Microbiol. 8:231–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tan M, Jiang X. 2010. Norovirus gastroenteritis, carbohydrate receptors, and animal models. PLoS Pathog. 6:e1000983. 10.1371/journal.ppat.1000983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hutson AM, Atmar RL, Estes MK. 2004. Norovirus disease: changing epidemiology and host susceptibility factors. Trends Microbiol. 12:279–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shirato H, Ogawa S, Ito H, Sato T, Kameyama A, Narimatsu H, Xiaofan Z, Miyamura T, Wakita T, Ishii K, Takeda N. 2008. Noroviruses distinguish between type 1 and type 2 histo-blood group antigens for binding. J. Virol. 82:10756–10767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kubota T, Kumagai A, Ito H, Furukawa S, Someya Y. 2012. Structural basis for the recognition of Lewis antigens by genogroup I norovirus. J. Virol. 86:11138–11150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harrington PR, Lindesmith L, Yount B, Moe CL, Baric RS. 2002. Binding of Norwalk virus-like particles to ABH histo-blood group antigens is blocked by antisera from infected human volunteers or experimentally vaccinated mice. J. Virol. 76:12335–12343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lindesmith LC, Donaldson EF, LoBue AD, Cannon JL, Zheng D-P, Vinje J, Baric RS. 2008. Mechanisms of GII.4 norovirus persistence in human populations. PLoS Med. 5:e31. 10.1371/journal.pmed.0050031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neyens E, Baeyens J, Dewil R, De heyder B. 2004. Advanced sludge treatment affects extracellular polymeric substances to improve activated sludge dewatering. J. Hazard. Mater. 106:83–92 [DOI] [PubMed] [Google Scholar]
- 17.Springer GF, Williamson P, Brandes WC. 1961. Blood group activity of gram-negative bacteria. J. Exp. Med. 113:1077–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lane DJ. 1991. 16S/23S rRNA sequencing, p 115–175 In Stackebrant E, Goodfellow M. (ed), Nucleic acid techniques in bacterial systematics. John Wiley & Sons Ltd., London, United Kingdom [Google Scholar]
- 19.Kitamoto N, Tanaka T, Natori K, Takeda N, Nakata S, Jiang X, Estes MK. 2002. Cross-reactivity among several recombinant calicivirus virus-like particles (VLPs) with monoclonal antibodies obtained from mice immunized orally with one type of VLP. J. Clin. Microbiol. 40:2459–2465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iritani N, Kaida A, Kubo H, Abe N, Goto K, Ogura H, Seto Y. 2010. Molecular epidemiology of noroviruses detected in seasonal outbreaks of acute nonbacterial gastroenteritis in Osaka City, Japan, from 1996–1997 to 2008–2009. J. Med. Virol. 82:2097–2105 [DOI] [PubMed] [Google Scholar]
- 21.Han TH, Kim CH, Chung JY, Park SH, Hwang ES. 2011. Emergence of norovirus GII-4/2008 variant and recombinant strains in Seoul, Korea. Arch. Virol. 156:323–329 [DOI] [PubMed] [Google Scholar]
- 22.Zeng M, Xu X, Zhu C, Chen J, Zhu Q, Lin S, Jie Y, Shu X, Chinese Pediatric Study Group of Norovirus Diarrhea 2012. Clinical and molecular epidemiology of norovirus infection in childhood diarrhea in China. J. Med. Virol. 84:145–151 [DOI] [PubMed] [Google Scholar]
- 23.Medici MC, Martinelli M, Abelli LA, Ruggeri FM, Di Bartolo I, Arcangeletti MC, Pinardi F, De Conto F, Izzi G, Bernasconi S, Chezzi C, Dettori G. 2006. Molecular epidemiology of norovirus infections in sporadic cases of viral gastroenteritis among children in Northern Italy. J. Med. Virol. 78:1486–1492 [DOI] [PubMed] [Google Scholar]
- 24.Liu H, Fang HH. 2002. Extraction of extracellular polymeric substances (EPS) of sludges. J. Biotechnol. 95:249–256 [DOI] [PubMed] [Google Scholar]
- 25.Pan X, Liu J, Zhang D, Chen X, Li L, Song W, Yang J. 2010. A comparison of five extraction methods for extracellular polymeric substances (EPS) from biofilm by using three-dimensional excitation-emission matrix (3DEEM) fluorescence spectroscopy. Water SA 36:111–116 [Google Scholar]
- 26.Takaara T, Sano D, Masago Y, Omura T. 2010. Surface-retained organic matter of Microcystis aeruginosa inhibiting coagulation with polyaluminum chloride in drinking water treatment. Water Res. 44:3781–3786 [DOI] [PubMed] [Google Scholar]
- 27.Westphal O, Jann K. 1965. Bacterial lipopolysaccharide, p 83–91 In Whistler RL, Wolfrom ML. (ed), Methods in carbohydrate chemistry. Academic Press Inc., New York, NY [Google Scholar]
- 28.Choi JM, Hutson AM, Estes MK, Prasad BVV. 2008. Atomic resolution structural characterization of recognition of histo-blood group antigens by Norwalk virus. Proc. Natl. Acad. Sci. U. S. A. 105:9175–9180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maalouf H, Zakhour M, Le Pendu J, Saux JC, Atmar RL, Le Guyader FS. 2010. Distribution in tissue and seasonal variation of norovirus genogroup I and II ligands in oysters. Appl. Environ. Microbiol. 76:5621–5630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maalouf H, Schaeffer J, Parnaudeau S, Le Pendu J, Atmar RL, Crawford SE, Le Guyader FS. 2011. Strain-dependent norovirus bioaccumulation in oysters. Appl. Environ. Microbiol. 77:3189–3196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Esseili MA, Wang Q, Saif LJ. 2011. Binding of human GII.4 norovirus virus-like particles to carbohydrates of romaine lettuce leaf cell wall materials. Appl. Environ. Microbiol. 78:786–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Le Guyader FS, Loisy F, Atmar RL, Hutson AM, Estes MK, Ruvoen-Clouet N, Pommepuy M, Le Pendu J. 2006. Norwalk virus-specific binding to oyster digestive tissues. Emerg. Infect. Dis. 12:931–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.George I, Crop P, Servais P. 2002. Fecal coliform removal in wastewater treatment plants studies by plate counts and enzymatic methods. Water Res. 36:2607–2617 [DOI] [PubMed] [Google Scholar]
- 34.Andersson M, Carlin N, Leontein K, Lindquist U, Slettengren K. 1989. Structural studies of the O-antigenic polysaccharide of Escherichia coli O86, which possesses blood-group B activity. Carbohydr. Res. 185:211–223 [DOI] [PubMed] [Google Scholar]
- 35.Perry MB, Maclean LL. 1992. Structural characterization of the O-polysaccharide of the lipopolysaccharide produced by Salmonella milwaukee O:43 (group U) which possesses human blood group B activity. Biochem. Cell Biol. 70:49–55 [DOI] [PubMed] [Google Scholar]
- 36.Kane M, Case LK, Kopaskie K, Kozlova A, MacDearmid C, Chervonsky AV, Golovkina TV. 2011. Successful transmission of a retrovirus depends on the commensal microbiota. Science 334:245–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuss SK, Best GT, Etheredge CA, Pruijssers AJ, Frierson JM, Hooper LV, Dermody TS, Pfeiffer JK. 2011. Intestinal microbiota promote enteric virus replication and systemic pathogenesis. Science 334:249–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu L, Crawford SE, Czako R, Cortes-Penfield NW, Smith DF, Le Pendu J, Estes MK, Prasad BVV. 2012. Cell attachment protein VP8* of a human rotavirus specifically interacts with A-type histo-blood group antigen. Nature 485:256–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang P, Xia M, Tan M, Zhong W, Wei C, Wang L, Morrow A, Jiang X. 2012. Spike protein VP8* of human rotavirus recognize histo-blood group antigens in a type-specific manner. J. Virol. 86:4833–4843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Templeton MR, Andrews RC, Hofmann R. 2005. Inactivation of particle-associated viral surrogates by ultraviolet light. Water Res. 39:3487–3500 [DOI] [PubMed] [Google Scholar]
- 41.Gerba CP, Goyal SM, Hurst CJ, Labelle RL. 1980. Type and strain dependence of enterovirus adsorption to activated sludge, soils and estuarine sediments. Water Res. 14:1197–1198 [Google Scholar]
- 42.LaBelle RL, Gerba CP. 1980. Influence of estuarine sediment on virus survival under field conditions. Appl. Environ. Microbiol. 39:749–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miura T, Masago Y, Sano D, Omura T. 2011. Development of an effective method for recovery of viral genomic RNA from environmental silty sediments for quantitative molecular detection. Appl. Environ. Microbiol. 77:3975–3981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meschke JS, Sobsey MD. 1998. Comparative adsorption of Norwalk virus, poliovirus 1 and F+ RNA coliphage MS2 to soils suspended in treated wastewater. Water Sci. Technol. 38:187–189 [PubMed] [Google Scholar]
- 45.da Silva AK, Le Guyader FS, Le Saux JC, Pommepuy M, Montgomery MA, Elimelech M. 2008. Norovirus removal and particle association in a waste stabilization pond. Environ. Sci. Technol. 42:9151–9157 [DOI] [PubMed] [Google Scholar]
- 46.Sano D, Ueki Y, Watanabe T, Omura T. 2006. Membrane separation of indigenous noroviruses from sewage sludge and treated wastewater. Water Sci. Technol. 54:77–82 [DOI] [PubMed] [Google Scholar]
- 47.Gentry J, Vinje J, Guadagnoli D, Lipp EK. 2009. Norovirus distribution within an estuarine environment. Appl. Environ. Microbiol. 75:5474–5480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fong TT, Lipp EK. 2005. Enteric viruses of humans and animals in aquatic environments: health risks, detection, and potential water quality assessment tools. Microbiol. Mol. Biol. Rev. 69:357–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hejkal TW, Wellings FM, LaRock PA, Lewis AL. 1979. Survival of poliovirus within organic solids during chlorination. Appl. Environ. Microbiol. 38:114–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smith EM, Gerba CP, Melnick JL. 1978. Role of sediment in the persistence of enteroviruses in the estuarine environment. Appl. Environ. Microbiol. 35:685–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shen C, Phanikumar MS, Fong TT, Aslam I, McElmurry SP, Molloy SL, Rose JB. 2008. Evaluating bacteriophage P22 as a tracer in a complex surface water system: The Grand River, Michigan. Environ. Sci. Technol. 42:2426–2431 [DOI] [PubMed] [Google Scholar]
- 52.Walshe GE, Pang L, Flury M, Close ME, Flintoft M. 2010. Effects of pH, ionic strength, dissolved organic matter, and flow rate on the co-transport of MS2 bacteriophages with kaolinite in gravel aquifer media. Water Res. 44:1255–1269 [DOI] [PubMed] [Google Scholar]
- 53.Cliver DO, Herrmann JE. 1972. Proteolytic and microbial inactivation of enteroviruses. Water Res. 6:797–805 [Google Scholar]
- 54.Matsuura K, Tsuchida A, Okahata Y, Akaike T, Kobayashi K. 1998. A quartz-crystal microbalance study of adsorption behaviors of artificial glycoconjugate polymers onto chemically modified gold surfaces and their interactions with lectins. Bull. Chem. Soc. Jpn. 71:2073–2977 [Google Scholar]