Abstract
Parainfluenza virus 5 (PIV5) is a promising viral vector for vaccine development. PIV5 is safe, stable, efficacious, cost-effective to produce and, most interestingly, it overcomes preexisting antivector immunity. We have recently reported that PIV5 expressing the hemagglutinin (HA) from highly pathogenic avian influenza (HPAI) virus H5N1 (PIV5-H5) provides sterilizing immunity against lethal doses of HPAI H5N1 infection in mice. It is thought that induction of apoptosis can lead to enhanced antigen presentation. Previously, we have shown that deleting the SH gene and the conserved C terminus of the V gene in PIV5 results in mutant viruses (PIV5ΔSH and PIV5VΔC) that enhance induction of apoptosis. In this study, we inserted the HA gene of H5N1 into PIV5ΔSH (PIV5ΔSH-H5) or PIV5VΔC (PIV5VΔC-H5) and compared their efficacies as vaccine candidates to PIV5-H5. We have found that PIV5ΔSH-H5 induced the highest levels of anti-HA antibodies, the strongest T cell responses, and the best protection against an H5N1 lethal challenge in mice. These results suggest that PIV5ΔSH is a better vaccine vector than wild-type PIV5.
INTRODUCTION
Parainfluenza virus 5 (PIV5), a nonsegmented negative-sense single-stranded RNA virus, is a member of the genus Rubulavirus of the family Paramyxoviridae (1). Several characteristics of PIV5 make it an attractive vaccine candidate vector. First, PIV5 is thought to be a contributing factor for kennel cough (2–6), and kennel cough vaccines containing live PIV5 have been used in dogs over 30 years without any safety concern for dogs or humans (7, 8). Second, PIV5 can be produced in high titers in many cell lines, including Vero cells, which have been approved for vaccine production. In a laboratory setting, it can be easily grown to >108 PFU/ml (1) and has been mass-produced for the veterinary vaccine market (7, 8). Third, PIV5 can infect both contemporary laboratory human cell lines and primary human cells (9). Fourth, in recent studies, a single dose of a live recombinant PIV5 expressing the HA gene (rPIV5-H5) from the highly pathogenic avian influenza (HPAI) virus H5N1 subtype provided sterilizing immunity against a lethal dose of influenza A virus H5N1 infection in mice (10, 11). PIV5 expressing NP, an internal protein of influenza virus, protected against lethal influenza virus challenge in mice, as well (12). The levels of protection afforded by PIV5-based H5N1 vaccine candidates in mice are unprecedented. In contrast, a vaccinia virus expressing NP did not provide any protection against the challenge (13) and an adenovirus containing NP provides 80% protection against the lethal H1N1 challenge, but the mice lost ca. 30% weight (14). Fifth, PIV5 vaccination has the advantage of needle-free intranasal delivery. Finally, preexisting anti-PIV5 immunity does not negatively affect the immunogenicity of a PIV5-based vaccine (15).
PIV5 encodes eight known viral proteins (1). The nucleocapsid protein (NP), phosphoprotein (P), and large RNA polymerase (L) protein are important for transcription and replication of the viral RNA genome. P and L form the viral RNA-dependent RNA polymerase (1). The V protein plays important roles in viral pathogenesis, as well as in regulating viral RNA synthesis (1, 16). Recombinant PIV5 lacking the conserved C terminus of the V protein (PIV5VΔC) induces apoptosis in infected cells via an intrinsic pathway (17). The fusion (F) protein, a glycoprotein, mediates both cell-to-cell and virus-to-cell fusion in a pH-independent manner that is essential for virus entry into cells. The hemagglutinin-neuraminidase (HN), another viral glycoprotein, is also involved in virus entry and release from the host cells. The matrix (M) protein plays an important role in virus assembly and budding (18, 19). The small hydrophobic (SH) protein is a 44-residue hydrophobic integral membrane protein (20). Recombinant PIV5 without SH (PIV5ΔSH) induces apoptosis in L929 cells through a tumor necrosis factor alpha (TNF-α)-mediated extrinsic apoptotic pathway (21–23).
Virus infection generally elicits a protective host immune response that resists reinfection and is the foundation of vaccinology. The ability of foreign antigens, such as viral proteins, to be recognized by the host immune system in part determines their effectiveness as a vaccine antigen. Apoptotic cells are a source of antigens for professional antigen-presenting cells, such as dendritic cells. It is thought that the apoptotic pathway activated by virus infection may also play a role in antigen presentation and that various apoptotic pathways may affect antigen presentation differently. Unlike most paramyxoviruses, PIV5 can productively infect many cell types with little or no detectable cytopathic effect (CPE) over a long period of time (22). The ability of PIV5 to grow productively without inducing CPE suggests that PIV5 likely encodes antiapoptosis mechanisms to prevent infected cells from undergoing cell death. Recombinant PIV5 viruses lacking SH (PIV5ΔSH) induce apoptosis via a TNF-α-mediated extrinsic pathway, suggesting that SH plays an essential role in blocking TNF-α-mediated apoptosis (23). PIV5 lacking the conserved C terminus (PIV5VΔC) induces apoptosis in infected cells via an intrinsic apoptotic pathway in which endoplasmic reticulum stress likely plays an important role (17). We hypothesize that mutant PIV5 viruses that induce apoptosis will be better vectors for delivering foreign antigens, such as H5N1 proteins, than wild-type PIV5. We tested this hypothesis by expressing the HA protein from H5N1 (H5-HA) using mutant PIV5 (PIV5ΔSH-H5 and PIV5VΔC-H5) viruses and examined the efficacies in comparison to PIV5-H5.
MATERIALS AND METHODS
Cells.
MDBK and Vero cells were maintained in Dulbecco modified Eagle medium (DMEM) containing 10% fetal bovine serum (FBS), 100 IU of penicillin/ml, and 100 μg of streptomycin/ml. BHK and BSR-T7 cells were maintained in DMEM containing 10% FBS and 10% tryptose phosphate broth (TPB). G418 (400 μg/ml) was added to BSR-T7 cells. All cells were incubated at 37°C and 5% CO2. Virus-infected cells were grown in DMEM containing 2% FBS. Plaque assays were performed using BHK cells.
Mice.
Six- to eight-week-old female BALB/c mice (Charles River Labs, Frederick, MD) were used for all studies. Mice were anesthetized via intraperitoneal administration of Avertin (2,2,2-tribromoethanol) prior to all intranasal vaccinations and influenza virus challenges. Mouse HPAI infections were performed in enhanced BSL3 facilities in HEPA filtered isolators according to guidelines approved by the institutional biosafety program at the University of Georgia and for use of Select Agents approved by the Centers for Disease Control and Prevention (CDC), Atlanta, GA. All animal studies were conducted under guidelines approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Georgia.
Influenza viruses.
Highly pathogenic A/Vietnam/1203/2004 (H5N1; kindly provided by Richard Webby, St. Jude Children's Research Hospital, Memphis, TN) was propagated in the allantoic cavity of embryonated hen eggs at 37°C for 24 h. rgA/VN-PR8 (H5N1; kindly provided by Ruben Donis, CDC) were propagated in the allantoic cavity of embryonated hen eggs at 37°C for 48 to 72 h. All viruses were divided into aliquots and stored at −80°C. All experiments using live, highly pathogenic A/Vietnam/1203/2004 were reviewed and approved by the institutional biosafety program at the University of Georgia and were conducted in enhanced biosafety level 3 (BSL3+) containment according to guidelines for the use of select agents approved by the CDC.
Construction of plasmids containing full-length PIV5 genomes and HA.
To generate a plasmid containing H5-HA insertion between the HN and L genes in the PIV5ΔSH genome (PIV5ΔSH-H5), the plasmid pPIV5ΔSH containing the full-length genome of PIV5 lacking the SH gene was used (21). To generate a plasmid containing H5-HA insertion between the HN and L genes in the PIV5VΔC genome (PIV5VΔC-H5), the plasmid pPIV5VΔC containing the full-length genome of PIV5 lacking the conserved cysteine-rich domain of the V gene was used (24). The plasmid containing the H5-HA gene without cleavage site was used as a DNA template for PCR amplification using appropriate oligonucleotide primer pairs as described previously (10). Sequences of the primers for the H5-HA gene, PIV5ΔSH-H5, and PIV5VΔC-H5 are available on request.
Virus rescue and sequencing.
The rescue of infectious recombinant PIV5 was performed as described previously (25). Briefly, the plasmids PIV5ΔSH-H5 or PIV5VΔC-H5 containing H5-HA in the mutant PIV5 genome, and three helper plasmids pPIV5-NP, pPIV5-P, and pPIV5-L encoding NP, P, and L proteins, were cotransfected into BSR-T7 cells at 95% confluence in 6-cm plates with JetPrime (Polyplus-Transfection, Inc, New York, NY). The amounts of plasmids used were as follows: 5 μg of full-length PIV5ΔSH-H5 or PIV5VΔC-H5 plasmids, 1 μg of pPIV5-NP, 0.3 μg of pPIV5-P, and 1.5 μg of pPIV5-L. After 72 h of incubation at 37°C, the media were harvested, and cell debris was pelleted by low-speed centrifugation (3,000 rpm, 10 min). Plaque assays were used to obtain single clones of recombinant viruses. The full-length genomes of a single plaque-purified clone of PIV5ΔSH-H5 or two clones of PIV5VΔC-H5 viruses (PIV5VΔC-H5-3 and PIV5VΔC-H5-4) were sequenced as described previously (10). Total RNAs from the media of PIV5ΔSH-H5 or PIV5VΔC-H5 virus-infected Vero cells were purified using the viral RNA extraction kit (Qiagen, Inc., Valencia, CA). cDNAs were prepared using random hexamers, and aliquots of the cDNA were then amplified using appropriate oligonucleotide primer pairs. PCR products were sequenced.
Growth of recombinant PIV5-HA viruses in vitro and in vivo.
Vero or MDBK cells in six-well plates were infected with PIV5, PIV5-H5, PIV5ΔSH-H5, PIV5ΔSH, PIV5VΔC-H5-3, PIV5VΔC-H5-4, or PIV5VΔC at a multiplicity of infection (MOI) of 0.1. The cells were then washed with phosphate-buffered saline (PBS) and maintained in DMEM–2% FBS. Media were collected at 24-h intervals. The titers of viruses were determined using plaque assay with BHK cells.
To analyze the growth of viruses in mice, 6-week-old wild-type BALB/c mice were infected with 105 PFU of PIV5, PIV5-H5, PIV5ΔSH-H5, PIV5VΔC-H5-3, or PIV5VΔC-H5-4 in a 50-μl volume intranasally. Mice were euthanized 3 days postinfection, and the lungs were collected to determine virus titers.
Detection of viral protein expression.
An immunofluorescence assay was used to detect expression of viral proteins. MDBK cells in 24-well plates were mock infected or infected with PIV5, PIV5-H5, PIV5ΔSH-H5, PIV5VΔC-H5-3, or PIV5VΔC-H5-4 at an MOI of 0.1. At 2 days postinfection (dpi), the cells were washed with PBS and then fixed in 0.5% formaldehyde. The cells were permeabilized in PBS–0.1% saponin solution and then incubated for 30 min with monoclonal anti-PIV5-V/P or anti-H5-HA antibody. The cells were washed with PBS–1% bovine serum albumin (BSA) and then incubated with fluorescein isothiocyanate (FITC)-labeled goat anti-mouse antibody. The cells were incubated for 30 min, washed, and photographed using a fluorescence microscope (Advanced Microscopy Group).
Expression levels of H5-HA in virus-infected cells were compared using flow cytometry. MDBK cells in six-well plates were mock infected or infected with PIV5, PIV5-H5, PIV5ΔSH-H5, PIV5VΔC-H5-3, or PIV5VΔC-H5-4 at an MOI of 5. The cells were collected at 2 dpi and fixed with 0.5% formaldehyde for 1 h. The fixed cells were resuspended in FBS-DMEM (50:50) and then permeabilized in 70% ethanol overnight. The cells were washed once with PBS and then incubated with mouse anti-H5-HA antibody in PBS–1% BSA for 1 h at 4°C. The cells were stained with anti-mouse antibody labeled with phycoerythrin for 1 h at 4°C in the dark and then washed once with PBS–1% BSA. The fluorescence intensity was measured using a flow cytometer (BD LSR II).
ELISA.
H5-HA-specific serum antibody titers were measured using an IgG enzyme-linked immunosorbent assay (ELISA). For the generation of immune serum, mice were vaccinated with 103 PFU of PIV5, PIV5-H5, PIV5ΔSH-H5, PIV5VΔC-H5-3, or PIV5VΔC-H5-4 intranasally, and blood samples collected on day 21 postvaccination. Immulon 2HB 96-well microtiter plates (Thermo Lab Systems) were coated with 1 μg of recombinant H5-HA protein (BEI Resource)/ml and incubated at 4°C overnight. Assays were performed according to the manufacturer's instructions (KPL, Inc.). Serial dilution of serum samples from PIV5-, PIV5-H5-, PIV5ΔSH-H5-, PIV5VΔC-H5-3-, or PIV5VΔC-H5-4-inoculated mice were added into coated plates. The goat anti-mouse IgG conjugated to AP (KPL, Inc) was added, and the plates were developed. The optical density (OD) was measured at 405 nm on a Bio-Tek Powerwave XS plate reader. The IgG titer was determined to be the lowest serum dilution with an OD greater than that of the mean of naive serum plus two standard deviations.
IFN-γ ELISPOT assay.
To measure the T cell responses in the spleens of vaccinated mice, a gamma interferon (IFN-γ) enzyme-linked immunospot assay (ELISPOT) assay was performed. Mice were vaccinated using PBS or 103 PFU of PIV5, PIV5-H5, or PIV5ΔSH-H5 intranasally. At day 21 postvaccination, the mice were sacrificed, and the spleens were collected. The spleens were homogenized and washed with Hanks balanced salt solution. Gey's solution was added to remove the red blood cells. Splenocytes in complete tumor medium were added to 96-well plates (BD Biosciences). The cells were mock stimulated or stimulated with H5N1 HA protein (BEI Resource), Ebola GP peptide P2 (EYLFEVDNL) as an irrelevant peptide, or phorbol myristate acetate (PMA)-ionomycin. Cultures were incubated at 37°C and in 5% CO2 for 48 h. The splenocytes were removed, and the plates were washed and incubated with antibody and substrate according to the manufacturer's instructions (BD Biosciences). The spots were counted using an AID ViruSpot Reader (Cell Technology, Inc.). The results are presented as mean numbers of cytokine-secreting cells subtracted from the total number of mock-stimulated cells per 106 splenocytes.
Influenza virus challenge of PIV5-vaccinated mice.
All animal experiments were carried out strictly according to the protocol approved by the IACUC. Mice were immunized with a single dose of 103 PFU of PIV5, PIV5-H5, PIV5ΔSH-H5, PIV5VΔC-H5-3, or PIV5VΔC-H5-4 or with 2,000 PFU of rgA/VN-PR8 intranasally. At day 21 postvaccination, the mice were challenged with 10 50% lethal doses (LD50) of H5N1 HPAI. After challenge, the mice were monitored for weight loss and survival. Mice were scored based upon clinical signs of infection (ruffled fur, hunched posture, and/or dyspnea = 1 point each; <25% weight loss = 1 point; 25 to 35% weight loss = 2 points; >35% weight loss = 3 points; neurological symptoms = 3 points). Animals were humanely euthanized upon reaching 3 points.
RESULTS
Generation of PIV5 mutants expressing HA.
We have generated PIV5 mutants expressing H5-HA (collective called PIV5-HA) (Fig. 1). Although we have found that insertion between the SH and HN junction results in the most efficacious vaccine in case of PIV5 expressing H5-HA (10), we have used the HN and L gene junction as the preferred site for the present study for two reasons. One is that the junction between the HN and L gene is the most distal to the leader sequence, the de facto promoter for transcription; thus, the insertion would have the least impact on PIV5 viral gene expression. The second reason was that it was easier to measure an improvement in a suboptimal vector to determine whether modification of PIV5 could improve efficacy. We generated full-length PIV5 genomes with appropriate mutations and insertion of H5-HA and recovered infectious virus as before (24). As a part of routine analysis of the recombinant viruses, we plaque purified and sequenced entire genome of the viruses. Although PIV5ΔSH-H5 (ZL128) had the exact input sequences, there were mutations in the genome of PIV5VΔC-H5 viruses. We sequenced many rPIV5VΔC-H5 clones, and they all contained mutations at different locations besides the intended mutations within the V/P gene to give rise to VΔC within the V/P gene. We decided to pick two clones of recovered viruses with different mutations besides the intended mutations that abrogate expression of the C terminus of the V protein to test. The rationale was that if all mutants had the same effects and the common feature was the lack of the conserved C terminus of V, it would likely be the determining factor in the phenotype. Two plaque-purified clones of PIV5VΔC-H5 containing mutation sites in the L gene were used for all further experiments; PIV5VΔC-H5-4 contained a Y1490S mutation, and PIV5VΔC-H5-3 contained the same mutation site plus an extra E1658D mutation (Fig. 1).
Fig 1.
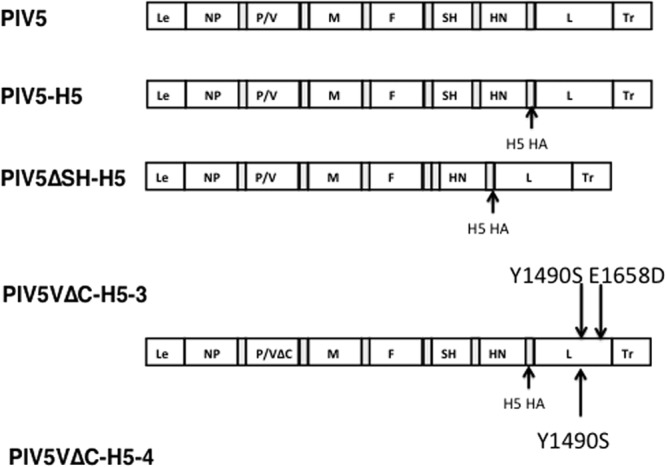
Schematics of recombinant PIV5 expressing H5-HA. PIV5 genome contains seven genes in the order of 3′-NP-V/P-M-F-SH-HN-l-5′ with leader (Le) and trailer (Tr) regions located at the ends of the genome. PIV5ΔSH lacking the SH gene and PIV5VΔC lacking the conserved C-terminal of the V protein have been described before (21, 24). The H5N1 HA gene was inserted into PIV5, PIV5ΔSH, or PIV5VΔC between the HN and L genes. The additional mutation sites within two clones of PIV5VΔC-H5 genome were as indicated.
Expression of H5-HA from PIV5-HA virus-infected cells was confirmed by using immunofluorescence (Fig. 2A). The HA protein was only detected in PIV5-H5-, PIV5ΔSH-H5-, or PIV5VΔC-H5-infected MDBK cells but not in mock- or PIV5-infected MDBK cells (Fig. 2A). Expression levels of HA of infected cells were further quantified by using flow cytometry (Fig. 2B). PIV5ΔSH-H5 produced the highest levels of HA expression, PIV5VΔC-H5-3 and PIV5-H5 produced similar higher levels of HA, and PIV5VΔC-H5-4 produced the lowest levels of H5-HA expression in MDBK cells.
Fig 2.
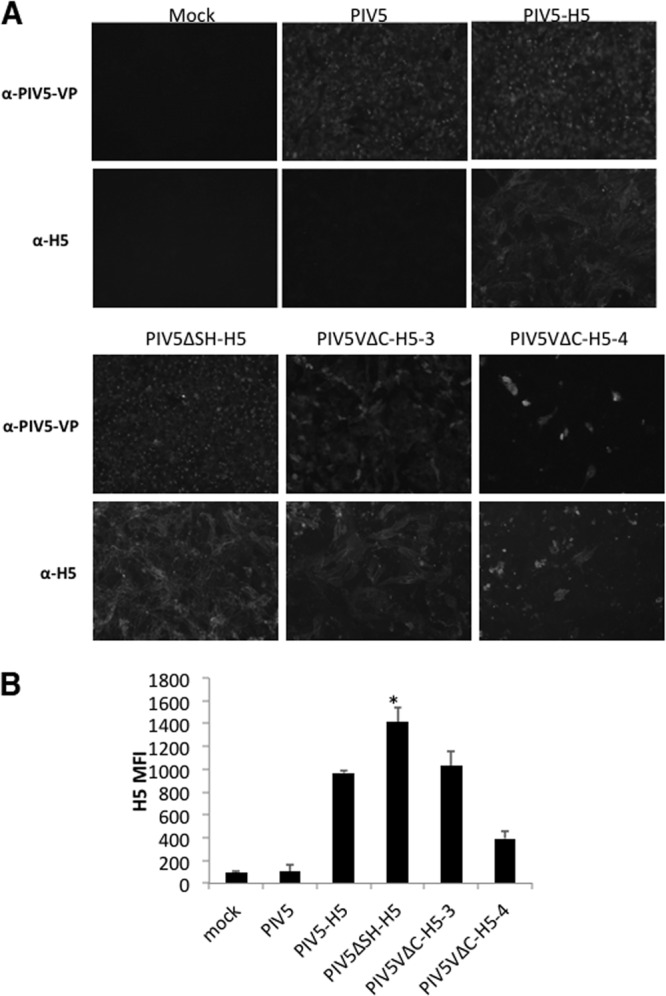
Confirmation of H5-HA expression. (A) HA expression in PIV5-HA virus-infected MDBK cells. MDBK cells were mock infected or infected with PIV5, PIV5-H5, PIV5ΔSH-H5, PIV5VΔC-H5-3, or PIV5VΔC-H5-4 at an MOI of 0.1. At 2 dpi, the cells were fixed, permeabilized, and then incubated with monoclonal anti-PIV5-V/P or anti-H5N1-H5 antibodies. The cells were photographed using a fluorescence microscope (Advanced Microscopy Group). (B) Comparison of H5-HA protein expression levels in virus-infected MDBK cells by flow cytometer. MDBK cells were mock infected or infected with PIV5, PIV5-H5, PIV5ΔSH-H5, PIV5VΔC-H5-3, or PIV5VΔC-H5-4 at an MOI of 5. The mean fluorescence intensity (MFI) of H5-HA was examined by flow cytometer at 2 dpi. *, P < 0.05 between PIV5ΔSH-H5 and PIV5-H5.
Analysis of PIV5-HA viruses in vitro and in vivo.
To compare the growth of PIV5-HA viruses in vitro, multiple-step growth curves were performed using MDBK or Vero cells. For PIV5ΔSH-based vaccine candidates, MDBK cells, the best cell line for growing PIV5, were used. PIV5, PIV5-H5, PIV5ΔSH-H5, and PIV5ΔSH replicated similarly in MDBK cells (Fig. 3A). For PIV5VΔC-based vaccine candidates, Vero cells, which lack IFN genes, were used to avoid the complication of IFN since PIV5VΔC induces higher levels of IFN expression in IFN-competent cells. In Vero cells, PIV5, PIV5-H5, PIV5VΔC-H5-3, and PIV5VΔC replicated with similar kinetics. PIV5VΔC-H5-4 replicated slower than the other viruses, with a peak titer that was >1 log lower (Fig. 3B).
Fig 3.
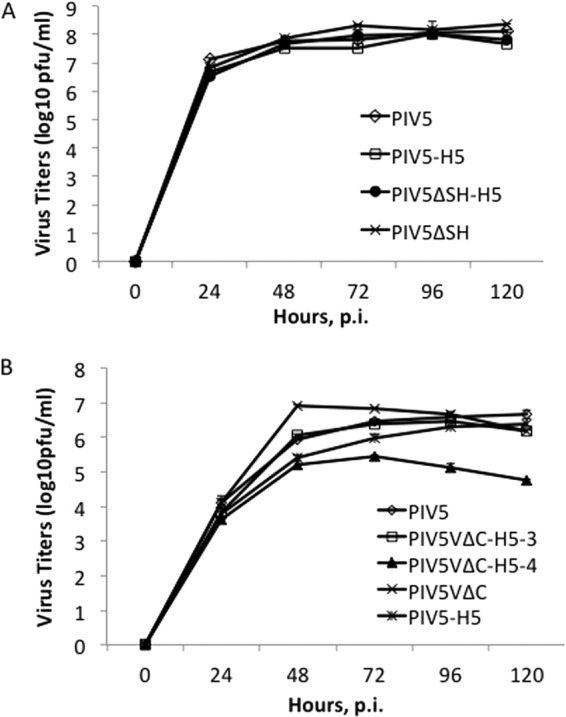
Growth of PIV5-HA viruses in vitro. (A) MBDK cells were infected with PIV5, PIV5-H5, PIV5ΔSH-H5, or PIV5ΔSH. (B) Vero cells were infected with PIV5, PIV5-H5, PIV5VΔC-H5-3, PIV5VΔC-H5-4, or PIV5VΔC at an MOI of 0.1. Media were collected at 24-h intervals, and virus titers were determined by plaque assay on BHK cells. p.i., postinfection.
To examine the growth of PIV5-HA viruses in vivo, BALB/c mice were vaccinated with 105 PFU of PIV5, PIV5-H5, PIV5ΔSH-H5, PIV5VΔC-H5-3, or PIV5VΔC-H5-4 intranasally. Titers of virus in the lungs of vaccinated mice were determined at 3 days postinfection (Fig. 4). The titers in the lungs of PIV5- and PIV5-H5-vaccinated mice were similar. PIV5ΔSH-H5- and PIV5VΔC-H5-vaccinated mice had lower titers of viruses in the lungs.
Fig 4.
Growth of PIV5-HA viruses in vitro and in vivo. Mice were vaccinated with 105 PFU of PIV5, PIV5-H5, PIV5ΔSH-H5, PIV5VΔC-H5-3, or PIV5VΔC-H5-4 intranasally. Mice were euthanized on day 3 postvaccination to determine lung virus titers.
Immune responses to PIV5-HA virus inoculation in mice.
To investigate HA antibody production among PIV5-HA viruses in vivo, mice were vaccinated with PIV5, PIV5-H5, PIV5ΔSH-H5, PIV5VΔC-H5-3, or PIV5VΔC-H5-4 intranasally. At day 21 postvaccination, blood samples were collected and sera were prepared. PIV5ΔSH-H5 induced the highest levels of anti-H5-HA antibodies, PIV5VΔC-H5-3 and PIV5-H5 induced similar levels of antibodies, and PIV5VΔC-H5-4 produced the lowest levels of antibodies (Fig. 5A).
Fig 5.
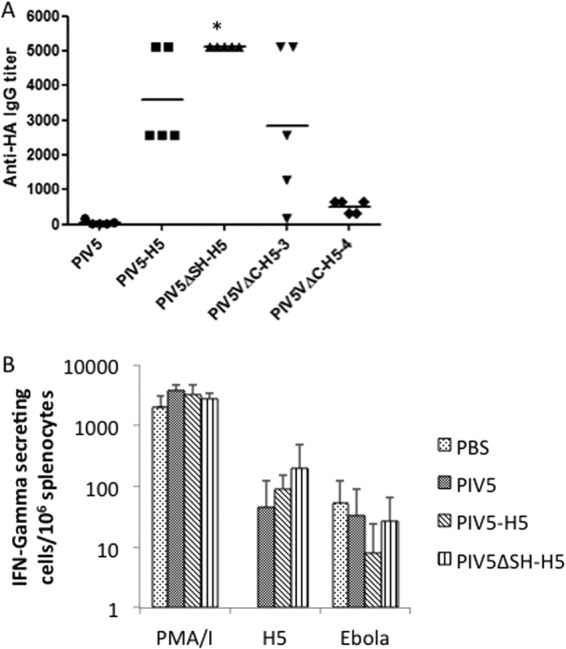
Induction of humoral and cellular responses by PIV5-HA viruses. (A) HA-antibody levels in mice induced by PIV5-HA viruses. Mice were vaccinated with 103 PFU of PIV5, PIV5-H5, PIV5ΔSH-H5, PIV5VΔC-H5-3, or PIV5VΔC-H5-4 intranasally. At day 21 postvaccination, blood samples were collected, sera were prepared, and an ELISA was performed to detect H5-HA-specific IgG. *, P < 0.05 between PIV5ΔSH-H5 and PIV5-H5. (B). T cell response in mice induced by PIV5-HA viruses. Mice were vaccinated with PBS or 103 PFU of PIV5, PIV5-H5, or PIV5ΔSH-H5 intranasally. At day 21 postvaccination, the mice were sacrificed, and the spleens were collected. Splenocytes were restimulated with HA, Ebola GP P2 as a negative control, or PMA-ionomycin as a positive control. The results are presented as the mean number of cytokine-secreting cells subtracted from the number of mock-stimulated cells per 106 splenocytes.
Differences in T cell responses induced by PIV5-H5 and PIV5ΔSH-H5 were examined using an IFN-γ ELISPOT assay. Mice were vaccinated with PIV5, PIV5-H5, or PIV5ΔSH-H5 intranasally. At day 21 postvaccination, mice were euthanized and splenocytes were obtained for IFN-γ ELISPOT assays. Compared to PIV5-vaccinated mice, PIV5-H5- and PIV5ΔSH-H5-vaccinated mice induced higher levels of H5-HA-specific T cell responses. PIV5ΔSH-H5-vaccinated mice induced higher levels of T cell responses than PIV5-H5-vaccinated mice, although the difference was not statistically significant.
Determining efficacy of PIV5-HA viruses against H5N1 HPAI challenge in mice.
To examine whether PIV5ΔSH-H5 or PIV5VΔC-H5 viruses could provide better protection against H5N1 HPAI challenge, mice were immunized with a single dose of PIV5, PIV5-H5, PIV5ΔSH-H5, PIV5VΔC-H5-3, or PIV5VΔC-H5-4 or with 2,000 PFU of rgA/VN-PR8 intranasally. At day 21 postvaccination, mice were challenged with 10 LD50 H5N1 HPAI. All PIV5-immunized mice lost body weight and succumbed to the infection. In contrast, 90% of mice vaccinated with PIV5VΔC-H5-4 and 100% of mice vaccinated with PIV5-H5, PIV5ΔSH-H5, or PIV5VΔC-H5-3 survived challenge. PIV5ΔSH-H5-vaccinated mice lost the least amount of body weight and recovered by 4 days postchallenge, indicating that PIV5ΔSH-H5 provided the best protection (Fig. 6). We have performed this immunization-challenge regime three times and, each time, PIV5ΔSH-H5 outperformed PIV5-H5 in reducing morbidity (lower percentage of weight loss) and providing complete protection (100% survival). Unfortunately, the results from PIV5VΔC-H5 immunization and challenge were variable, and it is inconclusive whether PIV5VΔC-H5 is better than PIV5-H5.
Fig 6.
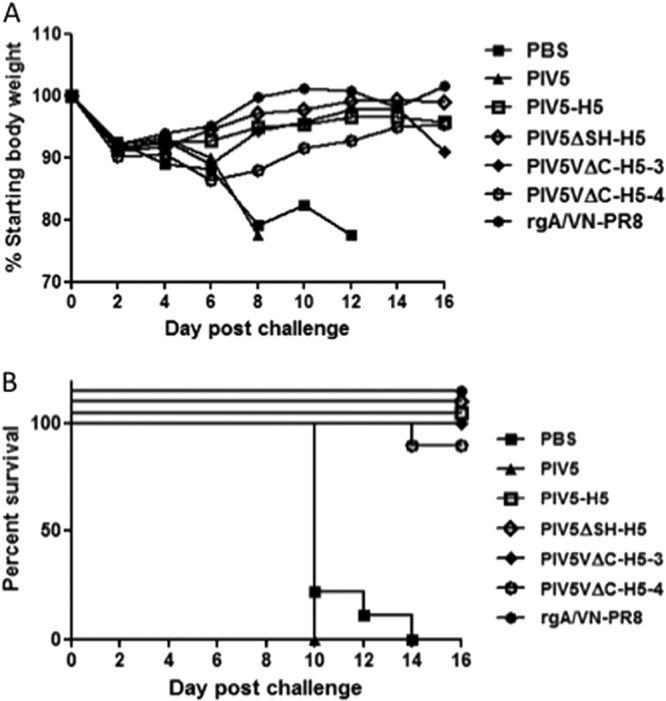
Protection of PIV5-HA viruses against H5N1 HPAI challenge. (A). Mice were vaccinated with PBS (n = 9), 103 PFU of PIV5 (n = 7), PIV5-H5 (n = 10), PIV5ΔSH-H5 (n = 8), PIV5VΔC-H5-3 (n = 10), or PIV5VΔC-H5-4 (n = 10) or with 2,000 PFU of rgA/VN-PR8 (n = 10). At day 21 postvaccination, the mice were challenged with 10 LD50 H5N1 HPAI. Weight loss (A) and survival (B) were monitored for 16 days after H5N1 challenge. The P value of peak weight loss between PIV5-H5 and PIV5ΔSH-H5 is 0.28.
DISCUSSION
Influenza A virus continues to cause significant morbidity and mortality each year. Influenza A viruses are classified by two major surface glycoproteins, hemagglutinin (HA) and neuraminidase (NA). There are 17 HA and 9 NA subtypes, differing by ≥30% in protein homology, which are used to categorize influenza A virus into subtypes (e.g., H1N1, H3N2, H5N1, etc.). Point mutations in the antibody-binding sites of these surface glycoproteins allow viruses to evade antibody-mediated immunity and reinfect humans and animals (antigenic drift). When different influenza A virus subtypes infect the same host, the exchange of gene segments can occur, often resulting in a new virus with a unique combination of viral genome (antigenic shift), which may give rise to pandemics. Vaccines that match circulating virus strains are needed for optimal protection. Current seasonal influenza vaccines consist of two influenza A viruses (H1N1 and H3N2) and one or two influenza B viruses. The development of an influenza vaccine providing broadly cross-protective immunity will be of great advantage. Recently, PIV5-based influenza virus vaccines have been generated and found to be highly effective in animals (10–12). In the present study, the efficacy of a PIV5-based vaccine was further improved by modifying the PIV5 vector. The modified PIV5 vector, one lacking SH, has the potential to be an excellent vector for developing a cross-protective universal influenza vaccine because of its increasing efficacy.
Even though all past links of PIV5 and human diseases have been proven false and humans have been likely exposed to PIV5 without any illness, it is not possible to exclude PIV5 from causing any known or unknown human diseases (i.e., it is impossible to prove negative). PIV5ΔSH has already been shown to be attenuated in mice. By inserting the H5-HA, PIV5ΔSH-H5 should be even further attenuated, thus reducing any potential safety risk of using it as viral vector.
Although PIV5VΔC-H5 afforded similar protection against a lethal challenge compared to PIV5ΔSH-H5, several characteristics of this mutated construct make it less ideal for use as a vaccine candidate. Previously, when rPIV5VΔC was cultures in Vero cells, an IFN-defective cell line, viruses with mutations that regained expression of the V protein, as well as mutations at other sites, emerged (24). This is likely due to the way that the deletion of the C terminus was made and the importance of the V protein in virus replication. Because the V and P mRNAs are both transcribed from the same V/P gene, it is impossible to delete the entire coding sequence for V from the PIV5 genome without also deleting the N terminus of the P protein, an essential part of viral RNA polymerase complex. Instead, multiple point mutations were introduced within the region of the V/P gene that is critical for transcribing both mRNAs to force the V/P gene only make the P mRNA and not the V mRNA (24, 26). At the time of the first publication of rPIV5VΔC, only the V/P gene region was sequenced due to technical difficulties in sequencing the entire viral RNA genome. Thus, it is not clear whether the original rPIV5VΔC harbored additional mutations in other regions of the genome. PIV5VΔC is likely a mixed population, and thus it is difficult to determine the predominant populations within a given stock at a given time. This problem can be further exacerbated by the lower dose used for in vivo infection. We speculate this may be the reason that PIV5VΔC-H5 clones behaved inconsistently in vivo as a vaccine candidate. Because of the rapid rise of mutations in the genome beyond the V/P gene, the stability of PIV5VΔC is a concern; thus, PIV5VΔC is not a good vector for vaccine development.
It is interesting that PIV5VΔC-H5-4 did not grow as well as the others, especially PIV5VΔC-H5-3, in Vero cells. PIV5-VΔC-H5-3 had two mutations in L (Y1490S and E1650D) and PIV5VΔC-H5-4 had only one mutation in L (Y1490S). It is possible that additional L-E1650D mutation enhances virus replication since the mutation is in the L protein, the major component of viral RNA-dependent RNA polymerase. Previously, it has been reported that the V protein of PIV5 plays a role in viral RNA synthesis (16). Thus, the emerging of mutations within the L gene is consistent with the role of V in viral RNA synthesis. Further studies of these potential compensational mutations may lead to a better understanding of the interactions between V and L, as well as the synthesis of viral RNA.
The mechanism of increased immunity afforded by PIV5ΔSH is not known. Although PIV5ΔSH-H5 gave rise to a higher level of HA expression in infected cells than did PIV5-H5, the replication of PIV5ΔSH-H5 was less robust in the lungs of immunized mice than that of PIV5-H5, suggesting that antigen levels per se may not be the critical factor in explaining the better protection afforded by PIV5ΔSH-H5. In tissue culture cells, PIV5ΔSH induces higher level of apoptosis than wild-type virus. It is possible that even though the induction of apoptosis by PIV5ΔSH-H5 results in faster clearance by host (lower titers of virus in lungs of infected mice comparing to PIV5-H5 in Fig. 4), the infected cells that undergo apoptosis are better sources of antigens for the immune system to recognize. Further studies are needed.
ACKNOWLEDGMENTS
We are grateful to Ruben Donis and Richard Webby for providing the VN-H5N1-PR8/CDC-RG (H5N1; rgA/VN-PR8) and A/Vietnam/1203/2004 viruses, respectively. We appreciate helpful discussions and technical assistance from all of the members of Biao He's laboratory.
This study was supported by grants from the National Institute of Allergy and Infectious Disease (R01AI070847) to B.H.
Footnotes
Published ahead of print 26 June 2013
REFERENCES
- 1.Lamb RA, Kolakofsky D. 2001. Paramyxoviridae: the viruses and their replication, p 1487–1531 In Knipe DM, Howley PM. (ed), Fields virology, 4th ed. Lippincott/The Williams & Wilkins Co, Philadelphia, PA [Google Scholar]
- 2.Azetaka M, Konishi S. 1988. Kennel cough complex: confirmation and analysis of the outbreak in Japan. Nippon Juigaku Zasshi 50:851–858 [DOI] [PubMed] [Google Scholar]
- 3.Binn LN, Eddy GA, Lazar EC, Helms J, Murnane T. 1967. Viruses recovered from laboratory dogs with respiratory disease. Proc. Soc. Exp. Biol. Med. 126:140–145 [DOI] [PubMed] [Google Scholar]
- 4.Cornwell HJ, McCandlish IA, Thompson H, Laird HM, Wright NG. 1976. Isolation of parainfluenza virus SV5 from dogs with respiratory disease. Vet. Rec. 98:301–302 [DOI] [PubMed] [Google Scholar]
- 5.McCandlish IA, Thompson H, Cornwell HJ, Wright NG. 1978. A study of dogs with kennel cough. Vet. Rec. 102:293–301 [DOI] [PubMed] [Google Scholar]
- 6.Rosenberg FJ, Lief FS, Todd JD, Reif JS. 1971. Studies of canine respiratory viruses. I. Experimental infection of dogs with an SV5-like canine parainfluenza agent. Am. J. Epidemiol. 94:147–165 [DOI] [PubMed] [Google Scholar]
- 7.Chladek DW, Williams JM, Gerber DL, Harris LL, Murdock FM. 1981. Canine parainfluenza-Bordetella bronchiseptica vaccine immunogenicity. Am. J. Vet. Res. 42:266–270 [PubMed] [Google Scholar]
- 8.Kontor EJ, Wegrzyn RJ, Goodnow RA. 1981. Canine infectious tracheobronchitis: effects of an intranasal live canine parainfluenza-Bordetella bronchiseptica vaccine on viral shedding and clinical tracheobronchitis (kennel cough). Am. J. Vet. Res. 42:1694–1698 [PubMed] [Google Scholar]
- 9.Tompkins SM, Lin Y, Leser GP, Kramer KA, Haas DL, Howerth EW, Xu J, Kennett MJ, Durbin RK, Durbin JE, Tripp R, Lamb RA, He B. 2007. Recombinant parainfluenza virus 5 (PIV5) expressing the influenza A virus hemagglutinin provides immunity in mice to influenza A virus challenge. Virology 362:139–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Z, Mooney AJ, Gabbard JD, Gao X, Xu P, Place RJ, Hogan RJ, Tompkins SM, He B. 2013. Recombinant parainfluenza virus 5 expressing hemagglutinin of influenza A virus H5N1 protected mice against lethal highly pathogenic avian influenza virus H5N1 challenge. J. Virol. 87:354–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mooney AJ, Li Z, Gabbard JD, He B, Tompkins SM. 2013. Recombinant parainfluenza virus 5 vaccine encoding the influenza virus hemagglutinin protects against H5N1 highly pathogenic avian influenza virus infection following intranasal or intramuscular vaccination of BALB/c mice. J. Virol. 87:363–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Z, Gabbard JD, Mooney A, Gao X, Chen Z, Place RJ, Tompkins SM, He B. 2013. Single dose vaccination of a recombinant parainfluenza virus 5 expressing NP from H5N1 provides broad immunity against influenza A viruses. J. Virol. 87:5985–5993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lawson CM, Bennink JR, Restifo NP, Yewdell JW, Murphy BR. 1994. Primary pulmonary cytotoxic T lymphocytes induced by immunization with a vaccinia virus recombinant expressing influenza A virus nucleoprotein peptide do not protect mice against challenge. J. Virol. 68:3505–3511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Price GE, Soboleski MR, Lo CY, Misplon JA, Quirion MR, Houser KV, Pearce MB, Pappas C, Tumpey TM, Epstein SL. 2010. Single-dose mucosal immunization with a candidate universal influenza vaccine provides rapid protection from virulent H5N1, H3N2 and H1N1 viruses. PLoS One 5:e13162. 10.1371/journal.pone.0013162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen Z, Xu P, Salyards GW, Harvey SB, Rada B, Fu ZF, He B. 2012. Evaluating a parainfluenza virus 5-based vaccine in a host with pre-existing immunity against parainfluenza virus 5. PLoS One 7:e50144. 10.1371/journal.pone.0050144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin Y, Horvath F, Aligo JA, Wilson R, He B. 2005. The role of simian virus 5 V protein on viral RNA synthesis. Virology 338:270–280 [DOI] [PubMed] [Google Scholar]
- 17.Sun M, Rothermel TA, Shuman L, Aligo JA, Xu S, Lin Y, Lamb RA, He B. 2004. Conserved cysteine-rich domain of paramyxovirus simian virus 5 V protein plays an important role in blocking apoptosis. J. Virol. 78:5068–5078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmitt AP, He B, Lamb RA. 1999. Involvement of the cytoplasmic domain of the hemagglutinin-neuraminidase protein in assembly of the paramyxovirus simian virus 5. J. Virol. 73:8703–8712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmitt AP, Leser GP, Waning DL, Lamb RA. 2002. Requirements for budding of paramyxovirus simian virus 5 virus-like particles. J. Virol. 76:3952–3964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hiebert SW, Richardson CD, Lamb RA. 1988. Cell surface expression and orientation in membranes of the 44 amino acid SH protein of simian virus 5. J. Virol. 62:2347–2357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He B, Leser GP, Paterson RG, Lamb RA. 1998. The paramyxovirus SV5 small hydrophobic (SH) protein is not essential for virus growth in tissue culture cells. Virology 250:30–40 [DOI] [PubMed] [Google Scholar]
- 22.He B, Lin GY, Durbin JE, Durbin RK, Lamb RA. 2001. The SH integral membrane protein of the paramyxovirus simian virus 5 is required to block apoptosis in MDBK cells. J. Virol. 75:4068–4079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin Y, Bright AC, Rothermel TA, He B. 2003. Induction of apoptosis by paramyxovirus simian virus 5 lacking a small hydrophobic gene. J. Virol. 77:3371–3383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He B, Paterson RG, Stock N, Durbin JE, Durbin RK, Goodbourn S, Randall RE, Lamb RA. 2002. Recovery of paramyxovirus simian virus 5 with a V protein lacking the conserved cysteine-rich domain: the multifunctional V protein blocks both interferon-beta induction and interferon signaling. Virology 303:15–32 [DOI] [PubMed] [Google Scholar]
- 25.He B, Paterson RG, Ward CD, Lamb RA. 1997. Recovery of infectious SV5 from cloned DNA and expression of a foreign gene. Virology 237:249–260 [DOI] [PubMed] [Google Scholar]
- 26.Thomas SM, Lamb RA, Paterson RG. 1988. Two mRNAs that differ by two nontemplated nucleotides encode the amino coterminal proteins P and V of the paramyxovirus SV5. Cell 54:891–902 [DOI] [PMC free article] [PubMed] [Google Scholar]


