Abstract
The HIV-1 latent reservoir represents an important source of genetic diversity that could contribute to viral evolution and multidrug resistance following latent virus reactivation. This could occur by superinfection of a latently infected cell. We asked whether latent viruses might be reactivated when their host cells are superinfected, and if so, whether they could contribute to the generation of recombinant viruses. Using populations of latently infected Jurkat cells, we found that latent viruses were efficiently reactivated upon superinfection. Pathways leading to latent virus reactivation via superinfection might include gp120-CD4/CXCR4-induced signaling, modulation of the cellular environment by Nef, and/or the activity of Tat produced upon superinfection. Using a range of antiviral compounds and genetic approaches, we show that gp120 and Nef are not required for latent virus reactivation by superinfection, but this process depends on production of functional Tat by the superinfecting virus. In a primary cell model of latency in unstimulated CD4 T cells, superinfection also led to latent virus reactivation. Drug-resistant latent viruses were also reactivated following superinfection in Jurkat cells and were able to undergo recombination with the superinfecting virus. Under drug-selective pressure, this generated multidrug-resistant recombinants that were identified by unique restriction digestion band patterns and by population-level sequencing. During conditions of poor drug adherence, treatment interruption or treatment failure, or in drug-impermeable sanctuary sites, reactivation of latent viruses by superinfection or other means could provide for the emergence or spread of replicatively fit viruses in the face of strong selective pressures.
INTRODUCTION
Latently infected resting CD4 T cells represent the major barrier to curing HIV-1 infection. In all HIV-1-infected individuals, a latent reservoir is established very early after acute infection (1, 2). Latent viruses are replication-competent proviruses that are integrated into cellular chromosomal DNA and do not produce viral particles but can lead to virus production following appropriate activation signals. Multiple mechanisms are involved in both the establishment and maintenance of latency, and these generally act to suppress viral transcription or to limit gene expression from any viral transcripts produced (recent reviews include references 3, 4, 5, 6, and 7). Since latent viruses are nonreplicating, they are not affected by host immune responses or by antiretroviral therapy. Reactivation and depletion of the latent reservoir are currently a major goal of the field, and multiple clinical trials have been carried out or are under way (8, 9). Nonetheless, eradication of latent reservoirs is far from being achieved at present.
Treatment failure can occur when viruses develop resistance to one or more of the drugs in a treatment regimen. Since viruses can be continually deposited into the latent reservoir during periods of low-level viremia or during treatment failure and can exit the latent reservoir when their host cell is activated (10), latency provides a means for the archiving and reemergence of sequences representing the history of a patient's quasispecies. When drug-resistant viruses are present, they are also archived in the latent reservoir (11–15). Since viral rebound from latently infected cells occurs upon treatment interruption or treatment failure (16), previously existing drug-resistant viruses that are present in the latent reservoir preclude patients from being treated with that drug or drug combination in the future.
Cells multiply infected with HIV-1 in vivo have been previously reported. This is especially common in secondary lymphoid tissues (17), where the majority of lymphocytes reside, and splenocytes have been reported to harbor 3 to 4 proviruses on average, with some cells containing up to 8 proviruses (18). In addition, 5 to 25% of infected lymphocytes in peripheral blood were reported to carry multiple proviruses (19). Multiply infected cells can arise from one of two general mechanisms, namely, by simultaneous infection by several viruses or by sequential infection. Cell-to-cell transmission has been shown to lead to the simultaneous transfer of multiple virions across virological synapses in a process referred to as multiploid inheritance (20), leading to multiple integrated, transcriptionally active proviruses (21). In addition, the formation of polysynapses can lead to simultaneous transmission of virions from one infected cell to multiple target cells (22). By locally increasing the multiplicity of infection, polysynapses might contribute to the generation of multiply infected cells by both cell-to-cell transmission as well as by superinfection. Superinfection, whether by cell-to-cell transmission or cell-free infection, leads to the generation of multiply infected cells via sequential infection (23).
The extreme genetic diversity of HIV-1 is a result of the high rate of nucleotide misincorporation and the propensity for template switching by the viral reverse transcriptase (RT). Retroviruses package two genomic RNA molecules into each viral particle, and RT switches between these two templates several times during the process of reverse transcription (reviewed in references 24 and 25). During infection of Jurkat T cells, an average of 7 to 8 template switches per virus were reported to occur at essentially random locations, and an average of 30 template switches per virus were reported in macrophages (26). Multiply infected cells can produce heterozygous virions, which can result in the formation of recombinant viruses when RT switches between nonidentical templates during reverse transcription. The large number (>50) and high prevalence of circulating recombinant forms (CRFs) demonstrate the evolutionary success of HIV-1 recombinants on a global scale (27). Recombination within individual patients has been documented in numerous studies (reviewed in reference 25), and recombination involving drug-resistant viruses provides a mechanism for the spread of drug resistance throughout a patient's quasispecies (28–30).
Since the latent reservoir represents an archive of the history of a patient's quasispecies, including viruses with any previously existing drug resistance mutations (11–15), latent viruses represent an important source for the further generation of genetic diversity under selective pressure. This could occur following superinfection of a latently infected cell. Superinfection of latently infected cells could occur either during treatment interruption or failure, during periods of low-level viremia, or in compartmentalized sites of viral replication such as sanctuary sites that might result from poor drug penetration. Although this process is likely to be rare, the combination of the high multiplicity of infection that is common in vivo coupled with the potentially strong selective advantage of any resulting recombinant viruses renders this an important process (31, 32). In fact, it has been suggested that superinfection might modulate levels of latency for many viruses, including HIV (33), and it has been suggested that latent viruses might contribute to HIV-1 recombination in vivo (25, 34).
In this study, we asked whether latent viruses would be reactivated upon superinfection of their host cells, and if so, whether they could contribute to the generation of recombinant viruses. Using cell line and primary cell models of HIV-1 latency, we found that superinfection reactivated latent viruses and that this process required Tat production from the superinfecting virus. We also found that reactivated, drug-resistant latent viruses could contribute to the development of multidrug resistance via recombination with superinfecting viruses.
MATERIALS AND METHODS
Cell lines, viruses, and antiviral compounds.
Jurkat (clone E6-1) and HeLa-tat-III (HeLa-tat) cells were obtained through the NIH AIDS Research and Reference Reagent Program. Jurkat cells were maintained in RPMI 1640 medium (Invitrogen) supplemented with 10% fetal bovine serum (FBS), 1% l-glutamine, and 1% penicillin-streptomycin. HeLa-tat and 293T cells were maintained in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen) supplemented with 10% FBS, 1% l-glutamine, and 1% penicillin-streptomycin. pNL4-3-ΔE-EGFP (expressing enhanced green fluorescent protein) was obtained through the NIH AIDS Research and Reference Reagent Program, while pBR-NL4-3-IRES-EGFP (containing an internal ribosome entry site), pBR-NL4-3-IRES-dsRed, and pBR-NL4-3-IRES-dsRed-nef-stop were kind gifts from J. Münch and F. Kirchhoff (35). The following constructs were created by either site-directed mutagenesis or cloning: pNL4-3-ΔE-EGFP-tat(H13L), pNL4-3-ΔE-EGFP-tat(H13L)-RT(ΔSbfI/K103N), pBR-NL4-3-IRES-dsRed-RT(M184V/ΔMboI), pBR-NL4-3-IRES-dsRed-tat(H13L), and pBR-NL4-3-IRES-dsRed-tat(C22G). The nucleotide changes introduced are as follows: tat H13L, CAT to TTA; tat C22G, TGT to GGA; ΔSbfI, CCTGCAGG to CCTGCTGG; RT K103N, AAA to AAC; RT M184V, ATG to GTG; ΔMboI, GATC to GTTC. Replication-competent reporter viruses were produced by transfection of ∼9 × 106 293T cells with 25 μg of plasmid DNA using Lipofectamine 2000 (Invitrogen). The viruses used in Fig. 3 [pBR-NL4-3-IRES-dsRed-tat(wt/H13L/C22G), where “wt” represents “wild type”] were produced under the same conditions, except by transfection of HeLa-tat cells. Pseudoviruses were produced by cotransfection of 293T cells with 6.25 μg pVPack-VSV-G (Stratagene)—a vesicular stomatitis virus G protein (VSV-G) envelope-encoding construct—in combination with 18.75 μg of pNL4-3-ΔE-EGFP derivatives, as described above. All transfections were carried out using Opti-MEM medium (Invitrogen) supplemented with 2.5% FBS. Virus-containing supernatants were harvested at 48 h posttransfection, clarified by centrifugation for 5 min at 470 × g, and passed through a 0.45-μm-pore filter. All viruses were then treated with 50 U/ml benzonase (Sigma) in the presence of added benzonase buffer (10× = 500 mM Tris-HCl [pH 8.0], 10 mM MgCl2, and 1 mg/ml bovine serum albumin [BSA]) at 37°C for 20 min to digest residual plasmid DNA. Viral titers were determined by enzyme-linked immunosorbent assay (ELISA) for viral capsid (p24), using a Vironostika HIV-1 antigen (Ag) kit (bioMérieux). The RT inhibitor efavirenz (EFV), the integrase inhibitor raltegravir (RAL), and the protease (PR) inhibitor darunavir (DRV) were obtained through the NIH AIDS Research and Reference Reagent Program.
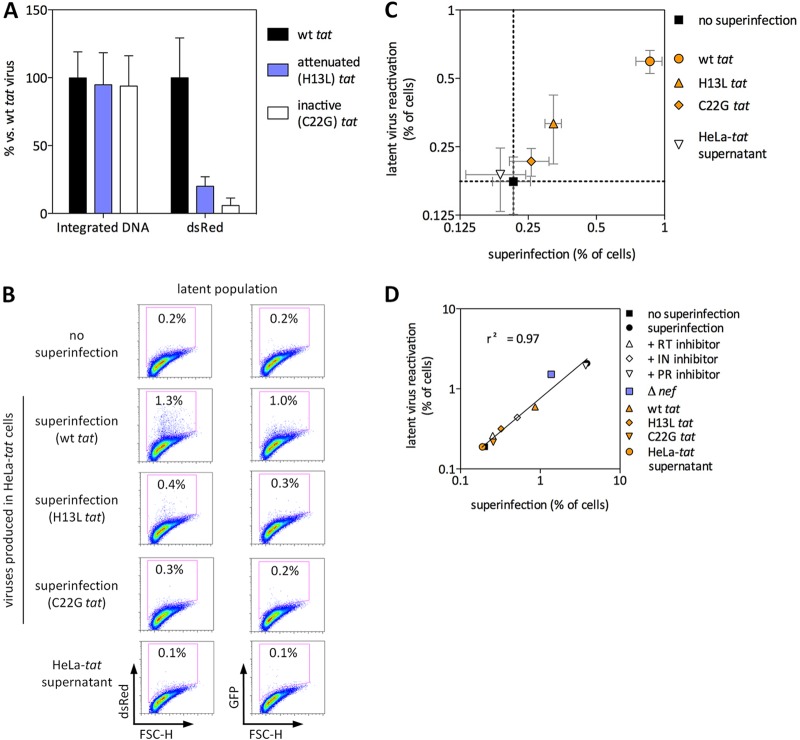
Fig 3.
Latent virus reactivation via superinfection requires expression of functional Tat by the superinfecting virus. (A) wt, attenuated (H13L), or inactivated (C22G) tat viruses were produced by transfection of HeLa-tat cells and were then used to infect Jurkat cells in the presence of 1 μM DRV to prevent reinfection. Levels of integrated viral DNA were measured by Alu-gag qPCR, while infectivity was determined by flow cytometry for virus-encoded dsRed. (B and C) Latently infected Jurkat cells were superinfected with NL4-3-dsRed (wt or H13L or C22G tat) or were treated with HeLa-tat supernatant as a control for any secreted Tat present in the superinfecting virus inoculum. FSC-H, forward scatter height. Representative results are shown in panel B, and the results of three independent experiments performed in triplicate are shown in panel C. (The axes in panel C are log2 rather than log10.) dsRed indicates superinfecting virus, and GFP represents latent virus. (D) Summary of the superinfection experiments shown in Fig. 1 to 3. Linear regression analysis was used to test for any correlation between superinfection and latent virus reactivation. Error bars represent SEM (n = 3 for error bar calculations).
Jurkat cell latency model.
Populations of latently infected cells were established as described previously (36). Briefly, Jurkat cells were infected with VSV-G-pseudotyped NL4-3-ΔE-EGFP-tat(H13L) or NL4-3-ΔE-EGFP-tat(H13L)-RT(ΔSbfI/K103N) and were cultured for up to 2 months. At various time points, samples were treated for 24 h with tumor necrosis factor α (TNF-α) (20 ng/ml) to reactivate latent viruses, before being fixed in 2% paraformaldehyde for 20 min. Flow cytometry was performed using a FACSCalibur or LSRFortessa cell analyzer (Becton, Dickinson), and data were analyzed with either FCS Express or FlowJo software. Live cells were gated by forward and side scatter properties, single cells were then gated based on forward and side scatter width and height, and levels of EGFP were then measured.
Primary cell latency model.
A previously described primary cell latency model (37) was used with minor modifications. Peripheral blood mononuclear cells (PBMC) were isolated from blood of HIV-1-negative donors by Ficoll-Hypaque density gradient centrifugation and were immediately processed to isolate unstimulated CD4 T cells using a Dynabeads untouched human CD4 T-cell isolation kit (Invitrogen). Isolated cells were stained with anti-CD3-phycoerythrin (PE), anti-CD4-e450, and anti-CD69-fluorescein isothiocyanate (FITC) and acquired on an LSRFortessa cell analyzer. Isolated CD4 T cells were cultured overnight in RPMI supplemented with 10% FBS, 1% l-glutamine, and 1% penicillin-streptomycin, in the absence of interleukin-2 (IL-2). The following day, CD4 T cells (∼0.5 million) were infected with NL4-3-IRES-dsRed (wt or H13L tat) by spinoculation in 5-ml polystyrene tubes at 1,200 × g for 2 h at 25°C, using 200 ng p24 per million cells (50% virus-containing supernatant and 50% supplemented RPMI, by volume). Immediately after spinoculation, cells were resuspended in supplemented RPMI in the presence of 1 μM DRV and then cultured in 96-well round-bottom plates for 3 days. On day 3 postinfection (p.i.), samples of uninfected or infected cells were incubated with anti-CD3/CD28 magnetic beads (Invitrogen), using 1 bead per cell in the presence of 10 μM RAL, to reactivate postintegration latent viruses. Cells were fixed 48 or 72 h later and used for flow cytometry.
Superinfection. (i) Jurkat cells.
A total of 2 × 105 Jurkat cells latently infected with NL4-3-ΔE-EGFP-tat(H13L), or uninfected Jurkat cells were infected with NL4-3-IRES-dsRed (or its tat/nef mutant derivatives: 120 ng p24 was used for each virus in Fig. 1 and 2, while 90 ng p24 was used for the viruses in Fig. 3. A wide range of superinfecting virus levels were used, as indicated, in Fig. 4. Similarly, 2 × 105 Jurkat cells latently infected with NL4-3-ΔE-EGFP-tat(H13L)-RT(ΔSbfI/K103N) or uninfected Jurkat cells were infected with 300 ng p24 of NL4-3-IRES-dsRed-RT(M184V/ΔMboI). Infections were by spinoculation at 1,500 × g for 2 h at 37°C in the presence of EFV, RAL, or DRV as required (1 μM each). Following spinoculation, cells were allowed to rest for 1 h at 37°C and then were resuspended in supplemented RPMI containing EFV, RAL, or DRV as required. At 72 h p.i., cells were fixed and analyzed by flow cytometry.
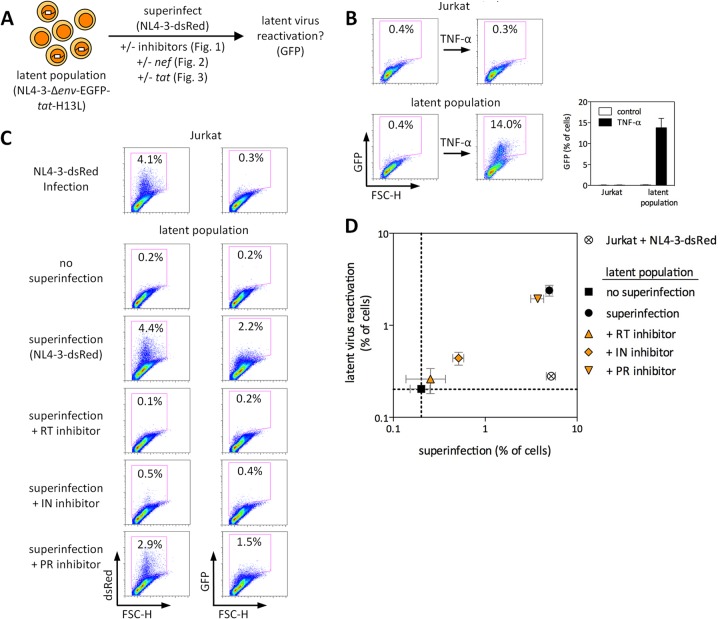
Fig 1.
Superinfection of latently infected cells reactivates latent HIV-1 and requires gene expression but not gp120-CD4/CXCR4 signaling from the superinfecting virus. (A) Schematic representation of the experimental design. (B) Characterization of the latently infected Jurkat cell populations used in Fig. 1 to 4. FSC-H, forward scatter height. Results of three independent experiments performed in duplicate are shown. (C and D) Uninfected or latently infected Jurkat cells were superinfected with NL4-3-dsRed in the presence or absence of 1 μM RT inhibitor EFV, IN inhibitor RAL, or PR inhibitor DRV. Representative results are shown in panel C, and the results of three independent experiments performed in triplicate are shown in panel D. dsRed represents superinfecting virus, and GFP represents latent virus. Error bars represent standard errors of the mean (SEM [n = 3 for error bar calculations]).
Fig 2.
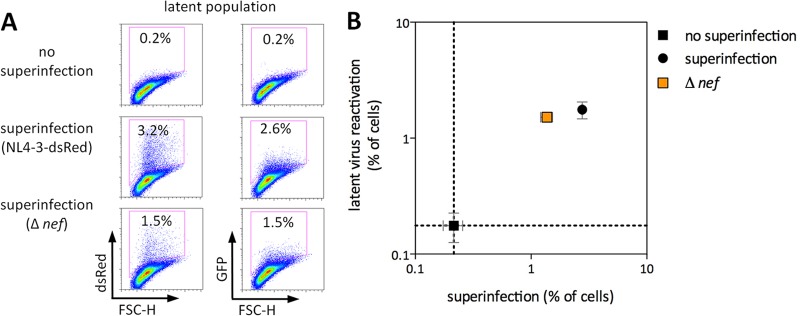
Superinfecting virus Nef is not required for latent virus reactivation. Uninfected or latently infected Jurkat cells were superinfected with NL4-3-dsRed or NL4-3-dsRed-Δnef. Representative results are shown in panel A, and the results of three independent experiments performed in triplicate are shown in panel B. dsRed represents superinfecting virus, and GFP represents latent virus. FSC-H, forward scatter height. Error bars represent SEM (n = 3 for error bar calculations).
Fig 4.
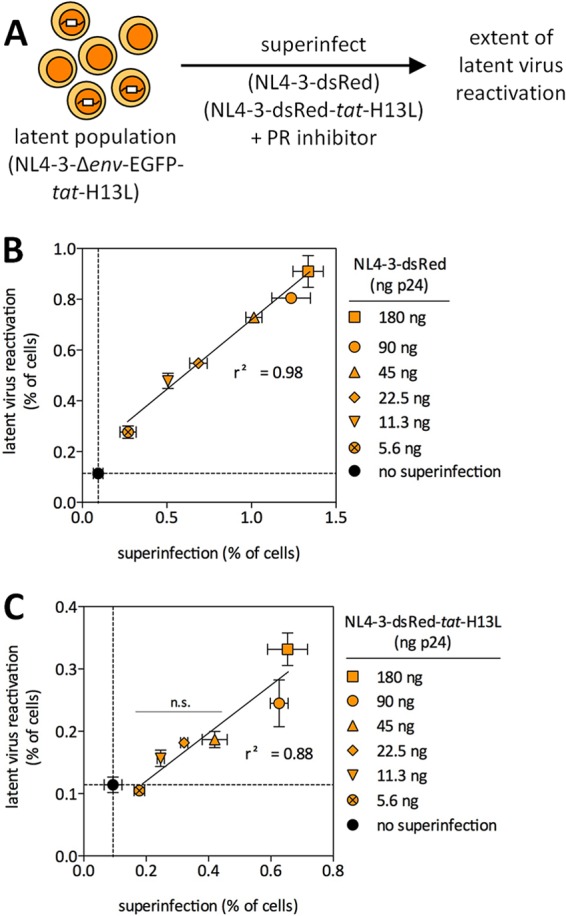
Efficiency of latent virus reactivation following superinfection. (A) Schematic representation of the experimental design. (B and C) Latently infected cells were superinfected with a wide range of NL4-3-dsRed viruses, encoding wt or H13L Tat. The indicated superinfection inoculum levels (ng p24) are per 75,000 cells. DRV (1 μM) was added immediately after superinfection to limit viruses to a single round of replication. Linear regression analysis was used to test for any correlation between superinfection and latent virus reactivation. One-way ANOVA indicated that the reactivation of latent viruses was statistically significant (P < 0.0001) for both wt and H13L tat superinfections. Dunnett's multiple-comparison posttest was used to determine which conditions led to a statistically significant latent virus reactivation, compared to mock-superinfected latently infected cells (“no superinfection”). All conditions were statistically significant in panel B, while the two highest superinfection input levels were statistically significant in panel C. Error bars represent SEM (n = 3 for error bar calculations). The results of three independent experiments performed in duplicate are shown.
(ii) Primary cells.
Latently infected CD4 T cells (3 days p.i.) were superinfected by spinoculation with NL4-3-IRES-GFP (or supplemented RPMI for controls; 1,200 × g for 2 h at 25°C), using 200 ng p24 per million cells. Cells were cultured in supplemented RPMI (in the absence of IL-2) plus 1 μM DRV for 3 days before fixation and flow cytometry.
PCR. Integrated HIV-1 DNA.
A total of 2 × 105 Jurkat cells were infected with 90 ng p24 of NL4-3-IRES-dsRed (or its tat mutant derivatives) by spinoculation as described above, and 1 μM DRV was added to prevent reinfection. Cellular DNA was extracted 48 h p.i. using a DNeasy blood and tissue kit (Qiagen). A previously described nested Alu-gag PCR (38) was used with the following modifications. The first-round reaction (performed in both the presence and the absence of an Alu-specific primer) was performed using undiluted samples (65 ng DNA) and 1:4 dilutions of each sample (16.25 ng DNA from infected Jurkat cells diluted with DNA from uninfected Jurkat cells [65 ng DNA total]) in the presence of 2 mM MgCl2 and 200 μM deoxynucleoside triphosphates (dNTPs) in a total volume of 20 μl, using the primers Alu-F (5′-GCCTCCCAAAGTGCTGGGATTACAG-3′) and gag-R (5′-GTTCCTGCTATGTCACTTCC-3′). The cycling conditions were 95°C for 2 min, followed by 20 cycles of 95°C for 15 s, 50°C for 15 s, and 72°C for 3.5 min. Nine microliters of the resulting first-round product was used as the template for the second-round nested reaction in the presence of 5 mM MgCl2 (final concentration, including carryover from the first round) and 200 μM added dNTPs, in a total volume of 20 μl. The second-round primers were LTR-F (long terminal repeat forward) (5′-TTAAGCCTCAATAAAGCTTGCC-3′) and LTR-R (long terminal repeat reverse) (5′-GTTCGGGCGCCACTGCTAGA-3′), and only the “wild-type” probe (38) was used. The second-round cycling conditions were 50°C for 2 min, 95°C for 1 min, and 45 cycles of 95°C for 15 s and 60°C for 30 s, using Platinum quantitative PCR (qPCR) SuperMix-UDG (Invitrogen) on a Corbett Rotor-Gene 6000 thermocycler. To generate a standard curve for relative quantification of integrated DNA, Alu-gag PCR was first performed on a 2-fold dilution series of DNA from infected Jurkat cells (diluted with DNA from uninfected Jurkat cells). Samples were normalized to their β-globin contents as described previously (39).
RT-PCR for viral genomic RNA.
Viral RNA was extracted from supernatants of infected cells using a QIAamp viral RNA minikit (Qiagen). RT-PCR was performed using a SuperScript III one-step RT-PCR kit (Invitrogen) and 6 μl viral RNA template, with primers Recomb-F (5′-AATGGATGGCCCAAAAGTTAAACA-3′ [corresponding to HXB2 coordinates 2140 to 2163] and Recomb-R 5′-CTGTTAATTGTTTCACATCATTAGTGTGGG-3′ [corresponding to HXB2 coordinates 3174 to 3203]) in a total volume of 30 μl. The cycling conditions were 55°C for 15 min, 94°C for 2 min, and 40 cycles of 94°C for 20 s, 60°C for 20 s, and 68°C for 1 min. Products were visualized on 1% agarose Tris-acetate-EDTA (TAE) gels.
Identification of recombinant viruses.
To analyze recombination by restriction digestion, 5 μl of each RT-PCR product (containing amplified viral genomic RNA) was double digested with both SbfI and MboI (New England BioLabs) in a total volume of 15 μl for 15 min at 37°C. Products were then visualized on 1% agarose TAE gels (1 h at 125 V), and band patterns were compared to digests of plasmids representing wt, latent, or superinfecting viruses. To analyze recombination by sequencing, RT-PCR products containing amplified viral genomic RNA were sequenced by standard methods using the primers Recomb-F and Recomb-R (see above). All chromatograms were visually inspected, and chromatogram peak intensities at relevant nucleotide positions were used to calculate the relative proportion of each virus in the population. Results from forward and reverse sequence reads were averaged for each position. As described in further detail in the Results section, estimates for the percentage of recombinant viruses in each population are conservative.
Statistical analyses.
Unpaired two-tailed t tests, one-way analysis of variance (ANOVA), and linear regression analysis were performed with GraphPad Prism 5.0.
RESULTS
Superinfection of latently infected cells reactivates latent HIV-1.
To determine whether superinfection of latently infected cells would reactivate latent viruses, we first used a Jurkat-based model of HIV-1 latency establishment and reactivation that we have previously described (36). This model takes advantage of viruses encoding the tat H13L variant that attenuates Tat activity by decreasing its interaction with P-TEFb. When H13L Tat is present at sufficient levels, it appears to be fully functional, supporting viral gene expression at levels comparable to those of wild-type tat viruses (40). However, lower concentrations of H13L Tat lead to more rapid shutdown of viral transcription compared to wt (40). This facilitates the entry into latency, since a low Tat threshold is reached sooner, allowing the establishment of latency (41–43). H13L tat viruses have been extensively characterized in the context of HIV-1 latency in both Jurkat and primary cells and represent genuinely latent viruses that behave similarly to latent viruses that encode wt Tat (36, 40, 44–47). The use of H13L tat thus represents an experimentally useful tool to increase the number and frequency of latently infected cells available for study.
In our Jurkat latency model, a population of latent GFP reporter viruses representing diverse integration sites is established. Culturing these cells for several weeks gives rise to a heterogeneous population of cells harboring latent proviruses, with no virus-producing cells present. In the latent populations used here, ∼14% of cells harbored TNF-α-inducible latent viruses (Fig. 1A and B). Uninfected Jurkat cells or latently infected Jurkat cells that encode viral EGFP were then infected with the replication-competent reporter virus NL4-3-dsRed (where dsRed is expressed from an internal ribosome entry site [IRES] from nef transcripts [35]). Superinfection of latently infected cells, in the absence of any antiviral inhibitors, led to reactivation of latent virus, as demonstrated by the increased percentage of EGFP-positive cells following superinfection (Fig. 1C and D).
Interaction of gp120 with CD4 and CXCR4 is not required for latent virus reactivation.
We next wished to characterize the mechanism(s) by which superinfection can reactivate latent viruses. In resting or suboptimally activated CD4 T cells, interaction of gp120 with CD4 and either CCR5 or CXCR4 can lead to induction of Ca2+ and NFAT (nuclear factor of activated T cells)—an important transcription factor involved in HIV-1 transcription—in the absence of full cellular activation (48–52). Additionally, HIV-1 envelope was reported to induce viral replication from resting cells of HIV-1-infected patients (53). Latently infected cells were superinfected in the presence of inhibitory levels (1 μM) of the RT inhibitor efavirenz (EFV). Blocking productive superinfection at reverse transcription, which is downstream of the gp120-CD4/CXCR4 interaction, resulted in no increase in latent virus gene expression compared to that of latently infected cells that were not superinfected (P = 0.75) (Fig. 1C and D).
Reactivation of latent viruses by superinfection requires gene expression of the superinfecting virus.
Next, latently infected cells were superinfected in the presence of the integrase (IN) inhibitor raltegravir (RAL), which prevents integration and thus productive viral gene expression, or in the presence of the protease (PR) inhibitor darunavir (DRV), which acts after integration and viral gene expression. As shown in Fig. 1C to D, latent virus reactivation required gene expression of the superinfecting virus. It is noteworthy that superinfection in the presence of an integrase inhibitor led to a minor and borderline significant (P = 0.05) increase in latent virus reactivation. This could be due to incomplete inhibition of viral replication in the presence of 1 μM RAL, which might be explained by the comparatively poor inhibitory capacity of this drug during a single round of viral replication (54). Alternatively, low-level gene expression from unintegrated viral DNA might explain this observation (55).
Nef is not required for latent virus reactivation via superinfection.
HIV-1 Nef modulates numerous cellular pathways, including several related to T-cell activation. Recent studies suggest that Nef lowers the activation threshold for CD4 T cells. This implies that when cells encounter activation signals in the presence of Nef, greater induction of transcription factors, including NF-κB and NFAT, as well as greater Ca2+ release and IL-2 production, can result (reviewed in references 56 and 57). Furthermore, it has been reported that Nef alone is sufficient to upregulate numerous cellular genes involved in LTR-driven transcription, including NFAT and many other transcription factors, as well as CDK9 and other factors involved in the elongation of viral transcripts (58). Expression of Nef from unintegrated DNA can also modulate T-cell activation pathways (59). We therefore wished to determine whether Nef might be contributing to the reactivation of latent viruses that we observed in Fig. 1. Latently infected cells were superinfected with a replication-competent reporter virus containing two stop codons near the start of nef (Δnef virus). Consistent with the enhancement of infectivity associated with Nef, superinfection with Δnef virus resulted in fewer dsRed-positive cells compared to superinfection with Nef-encoding virus. However, latent viruses were reactivated at least as efficiently in the absence of Nef as with wt virus (Fig. 2), excluding a requirement for Nef in the reactivation of latent viruses by superinfection in our model.
Latent virus reactivation via superinfection requires expression of functional Tat by the superinfecting virus.
It is reasonable to hypothesize that production of Tat by superinfecting viruses might be required for reactivation of latent viruses. Accordingly, we produced replication-competent reporter viruses that encode functional (wt), attenuated (H13L), or transactivation-negative (C22G) Tat. Infection of Jurkat cells with these viruses resulted in equivalent levels of integrated DNA, whereas the percentage of cells positive for Tat-dependent viral reporter gene expression followed the expected pattern of wt > H13L > C22G (Fig. 3A). This demonstrates that these viruses are equally functional for all steps up to and including integration and that any differences in latent virus reactivation following superinfection would be due to different Tat activities and not due to any defect in superinfection. These viruses were then used to superinfect latently infected cells (Fig. 3B and C). While superinfection with wt tat virus efficiently reactivated latent viruses, superinfection with attenuated tat virus resulted in a modest but statistically insignificant reactivation of latent virus. Latent virus reactivation was not detectable when transactivation-negative tat virus was used for superinfection. Of note, the level of superinfection achieved with wt tat virus here is lower than that in comparable infections shown in Fig. 1 and 2. This is due to the use of HeLa-tat cells for production of the viruses used here (necessary for the production of C22G tat viruses and used for H13L and wt tat viruses to ensure consistency) as opposed to virus production in 293T cells, used elsewhere in this study. Since HeLa-tat transfection produced relatively low viral titers, lower viral inputs were used for the subsequent superinfections. However, all comparisons made with these viruses are internal (i.e., wt versus H13L versus C22G virus, all produced in HeLa-tat cells), and all were used at equal p24 infection levels. As a control for any secreted Tat that might result from the use of HeLa-tat cells for virus production, latently infected cells were incubated directly with HeLa-tat supernatant; no latent virus reactivation was observed in this case (Fig. 3B and C).
Together, the results of Fig. 1 to 3 show that reactivation of latent viruses by superinfection requires gene expression of the superinfecting virus and, specifically, production of functional Tat. Linear regression analysis reveals a strong positive correlation (r2 = 0.97) between the extent of superinfection and the extent of latent virus reactivation (Fig. 3D).
Efficiency of latent virus reactivation following superinfection.
Based on the above results, we reasoned that latent viruses would be reactivated by superinfection across a wide range of infection rates and that superinfection with viruses encoding H13L Tat might also reactivate latent viruses if higher superinfection rates were achieved. Therefore, latently infected cells were superinfected with either wt or H13L tat viruses (both produced in 293T cells) using a 32-fold range of superinfecting virus inocula (Fig. 4A). DRV (1 μM) was added to limit infection to a single round. As expected, latent viruses were reactivated across a wide range of superinfecting virus input levels when wt tat viruses were used (Fig. 4B), and there was a strong positive correlation (r2 = 0.98) between the extent of superinfection and the extent of latent virus reactivation. One-way ANOVA and Dunnett's multiple-comparison posttest indicated that the reactivation of latent viruses was statistically significant (P < 0.0001) for all superinfection input levels. Interestingly, latent viruses were also reactivated when H13L tat viruses were used for superinfection (Fig. 4C) (r2 = 0.88). One-way ANOVA and Dunnett's multiple-comparison posttest indicated that the reactivation of latent viruses was statistically significant (P < 0.0001) at the two highest superinfection input levels, while a nonsignificant reactivation was observed for lower superinfection levels. This also provides additional support for our above finding that latent virus reactivation following superinfection requires Tat function.
Latent HIV-1 can be reactivated by superinfection in unstimulated primary CD4 T cells.
To test our hypothesis in a more physiologically relevant system, we used a primary cell model of HIV-1 latency that involves direct infection of unstimulated CD4 T cells. In this model, latency is established in multiple CD4 T-cell subsets, including naive, central memory, and transitional memory cells (37). Furthermore, CD4 T cells are cultured in the absence of cytokines, such as IL-2, and are infected shortly after isolation, preserving the in vivo distribution of CD4 T-cell subsets. The authors of this model also showed that infection of freshly isolated CD4 T cells gave near-identical results compared to infection of more extensively purified resting memory cells, which is likely because activated CD4 T cells in peripheral blood are present at only low frequency. Additionally, the latent viruses generated in this model respond to reactivating compounds with the same patterns observed for ex vivo-treated patient cells (37). Although some productively infected cells were also generated, spreading infection was prevented by a protease inhibitor.
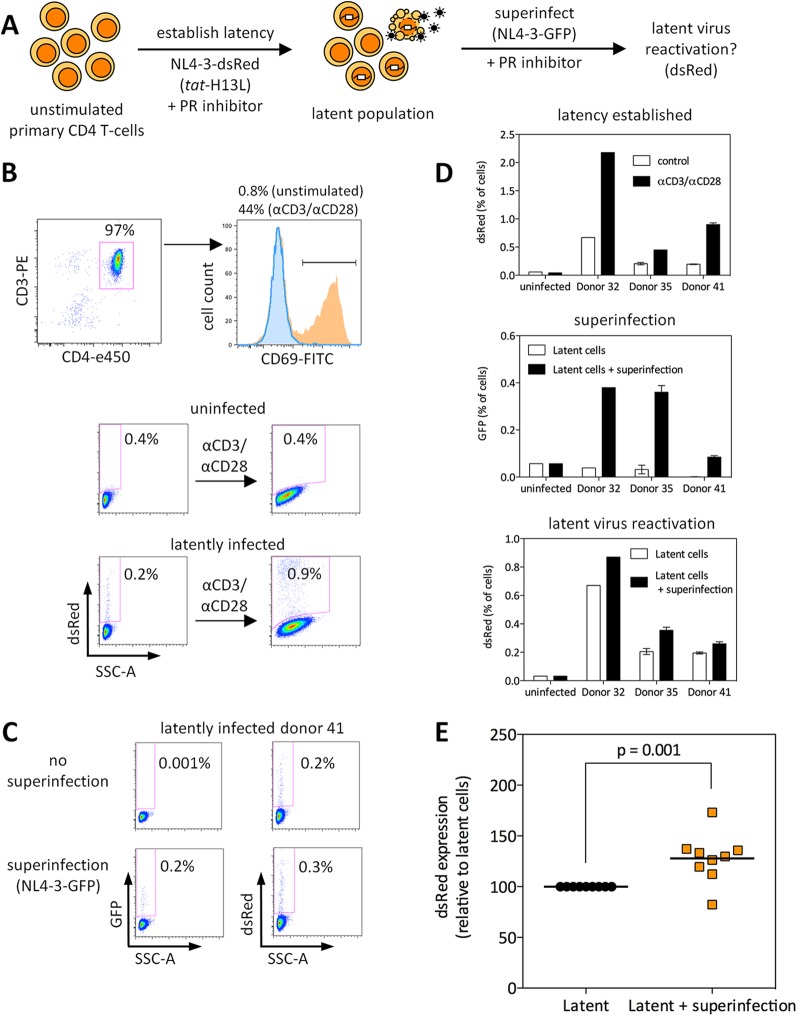
We first assessed the purity and activation status of CD4 T cells isolated from multiple donors. The vast majority (typically ∼97%) of cells expressed both CD3 and CD4, but they did not express the activation marker CD69 (<1%) (Fig. 5B). Additionally, these cells were small and were nondividing, consistent with their being resting CD4 T cells. Isolated CD4 T cells from individual donors were infected with replication-competent dsRed-encoding reporter viruses, in the presence of 1 μM protease inhibitor DRV to prevent spreading infection. Across a wide range of infecting virus levels (25 ng to 400 ng p24 per million cells), infection with attenuated (H13L) tat virus resulted in higher levels of silencing than infection with wt tat virus, and postintegration, latent viruses were reactivated in all cases by treatment with anti-CD3/CD28 beads in the presence of 10 μM RAL (data not shown). Since the ratio of silent to active viruses was greater for attenuated tat virus infections (Fig. 5B) (data not shown), we used an attenuated tat reporter virus to address our hypothesis in this primary cell model (Fig. 5A).
Fig 5.
Superinfection of latently infected primary CD4 T cells leads to reactivation of latent virus. (A) Schematic representation of the experimental approach. (B) Isolated CD4 T cells were stained with anti-CD3-PE, anti-CD4-e450, and anti-CD69-FITC to determine purity and activation status; one representative donor is shown. In the histogram, the portion outlined and shaded blue depicts freshly isolated CD4 T cells, and the portion shaded orange depicts CD4 T cells incubated for 24 h with anti-CD3/CD28 beads (1:1 ratio) to induce T-cell activation. Latency was established using NL4-3-dsRed (tat H13L) and is shown for one of nine donors. SSC-A, side scatter area. (C) Superinfection of latently infected cells led to latent virus reactivation; dsRed represents latent virus, and GFP represents superinfecting virus. One of nine donors is shown. (D) Three days after infection, cells were incubated with anti-CD3/CD28 beads (1:1 ratio) to determine the number of latent viruses (top panel) or were superinfected by spinoculation with GFP-expressing virus (or spinoculated with RPMI only as a control [middle panel]). The extent of latent virus reactivation was determined 3 days after superinfection (bottom panel). (E) The results from nine individual donors are shown. The percentage of dsRed-expressing cells (in the absence of superinfection) for each donor was set at 100%, and the percentage of dsRed-expressing cells following superinfection with GFP virus is depicted.
Next, latency was established in primary cells of multiple donors, and these cells were superinfected with a GFP reporter virus (in the presence of 1 μM DRV) to assess latent virus reactivation (Fig. 5B to D). The results of Fig. 5D to E show that superinfection led to a modest but significant reactivation of latent viruses in cells from nine individual donors (mean ∼30% increase in virus-expressing cells; P = 0.001). Due to the low overall infection rates achievable in unstimulated primary CD4 T cells, higher rates of superinfection and latent virus reactivation might be unlikely to occur. Nonetheless, these data indicate that, similarly to Jurkat cells, latent viruses can be reactivated from unstimulated primary CD4 T cells by superinfection.
Drug-resistant latent viruses are reactivated by superinfection with other drug-resistant viruses.
We next wished to determine whether reactivated latent viruses could contribute to the generation of recombinants. Latent Jurkat populations were established, in which the latent virus encodes the RT drug resistance mutation K103N, which confers resistance to nonnucleoside reverse transcriptase inhibitors (NNRTIs), including EFV (Fig. 6B). To facilitate the identification of potential recombinants, the latent viruses also contained a noncoding restriction site change that removes an SbfI site (ΔSbfI) located ∼20 nucleotides from the K103N mutation. K103N latent populations were superinfected with a dsRed-encoding drug-resistant reporter virus that encodes the RT mutation M184V, which provides for resistance to nucleoside RT inhibitors (NRTIs), including emtricitabine (FTC). The superinfecting virus also contained a noncoding restriction site change that removed an MboI site (ΔMboI) located ∼20 nucleotides from the M184V mutation (Fig. 6A). As demonstrated in Fig. 6C to D, superinfection of K103N latent populations with M184V virus led to reactivation of drug-resistant latent viruses.
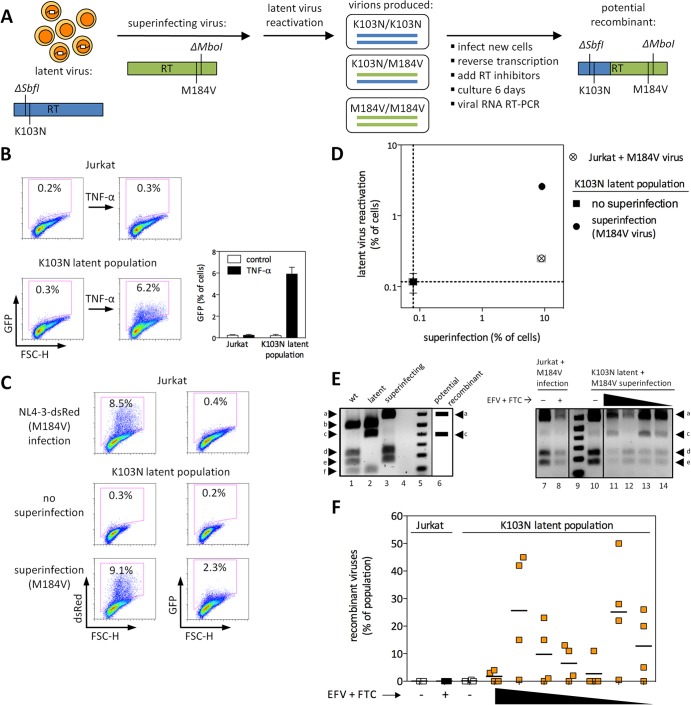
Fig 6.
Drug-resistant latent viruses are reactivated by superinfection, can recombine with superinfecting viruses, and can contribute to the development of multidrug-resistant recombinants. (A) Schematic representation of the experimental design. (B) Characterization of the drug-resistant latent virus population used in these experiments. FSC-H, forward scatter height. Results of three independent experiments performed in duplicate are shown. (C and D) Uninfected or latently infected Jurkat cells were superinfected with NL4-3-dsRed-RT(M184V/ΔMboI). Representative results are shown in panel C, and the results from three independent experiments performed in triplicate are shown in panel D. dsRed represents superinfecting virus, and GFP represents latent virus. Error bars represent SEM (n = 3 for error bar calculations). (E) (Left) Plasmids representing wt, latent, or superinfecting viruses (lanes 1 to 3) were double digested with MboI and SbfI and run on agarose gels. Lane 4, empty; lane 5, DNA ladder. The presence of bands a and c in the same lane would indicate a recombinant virus derived from both the latent and superinfecting viruses (lane 6). (Right) RT-PCR products from supernatants of one representative experiment, as depicted in panel A, were double digested with MboI and SbfI. Lanes 7 and 8, infection of Jurkat cells; lane 9, DNA ladder; lanes 10 to 14, superinfection of latently infected cells. As described in the Results section, recombinant viruses are represented in lanes 11, 13, and 14, but not lane 12. Lanes 7 to 14 are from the same experiment and were run on the same gel. (F) Results of sequence analysis from two independent experiments performed in duplicate are shown. The recombinant virus is ΔSbfI K103N M184V ΔMboI on the same genomic RNA. The highest drug concentrations applied were 20 nM EFV plus 16 μM FTC, while the lowest drug concentrations were 2.5 nM EFV plus 250 nM FTC.
Reactivated latent viruses can recombine with superinfecting viruses, which can contribute to the development of multidrug-resistant recombinants.
To determine whether recombination would occur between reactivated latent viruses and superinfecting viruses, supernatants of superinfected latent cells (which are expected to include some heterozygous virions [ΔSbfI K103N/M184V ΔMboI]) were collected 3 days after superinfection and used for new infections (Fig. 6A). After 20 h (a time sufficient for the completion of reverse transcription but prior to the next round of viral replication [39]), EFV and FTC were added together at a range of concentrations to select for potential recombinants. This will select for only a small fraction of recombinant viruses: i.e., only those in which recombination occurred between amino acid positions 103 and 184 of RT and in the correct orientation to maintain both resistance mutations. Following addition of RT inhibitors, cultures were maintained for 6 days, and ∼1 kb of RT was then amplified by RT-PCR from supernatant viral genomic RNA.
Two approaches were used for the identification of recombinant viruses. First, RT amplicons were subject to double-restriction enzyme digestion with SbfI and MboI, making use of the noncoding restriction site changes introduced into each virus. As shown in Fig. 6E (left), recombinant but not parental viruses are expected to result in both bands a and c. Recombinant viruses were detected in many but not all biological replicates across a range of drug-selective pressures, with representative results shown in Fig. 6E (right). Lanes 11, 13, and 14 (showing bands a, c, d, and e) represent a mixture of recombinant and superinfecting viruses. In contrast, only superinfecting virus was present following infection of Jurkat cells (lanes 7 and 8) or of latent populations with no drug-selective pressure (lane 10). Comparison of the intensities of band a across different lanes gives an approximation of the overall level of virus present, since both superinfecting and recombinant viruses contribute to band a. (For example, lane 12 represents a lower level of virus than lane 11, 13, or 14, consistent with the absence of recombinant viruses despite drug-selective pressure in that sample).
Second, bulk sequencing was used to estimate the proportion of recombinants in each sample. Recombinant viruses were only considered to be present when mathematically necessary. For example, if a population was 40% ΔSbfI K103N and 90% M184V ΔMboI, then at least 30% of the population must be recombinant viruses (where ΔSbfI, K103N, and M184V ΔMboI are on the same genomic RNA). If all mutations were present at <50%, the population was not considered to contain recombinants. The presence of the ΔSbfI and ΔMboI mutations additionally confirms that recombinants are genuine, as opposed to the de novo acquisition of resistance mutations by either parental virus. As shown in Fig. 6F, multidrug-resistant recombinant viruses resulted from superinfection of latently infected cells in many but not all biological replicates. Since levels of recombinants increase with longer times in culture (in the presence of EFV plus FTC), the specific percentages depicted are not themselves important. Rather, the presence versus absence of recombinants should be noted. No recombinants were detected following infection of Jurkat cells or following superinfection of latently infected cells in the absence of drug-selective pressure. Together, these results demonstrate that latent viruses can serve as a source for recombination, which might contribute to the emergence of multidrug-resistant recombinants.
DISCUSSION
Latent virus reactivation might occur by superinfection of latently infected cells (33). Although it has been suggested that latent viruses might contribute to recombination in vivo (25, 34), this process has not been experimentally characterized. In this study, we asked whether latent viruses would be reactivated when their host cells are superinfected, and if so, whether they could contribute to the generation of recombinants. We demonstrated that superinfection of latently infected cells led to latent virus reactivation (Fig. 1), although we found no evidence for gp120 (Fig. 1) or Nef (Fig. 2) being required for this process in our Jurkat latency model. Inhibitors targeting different stages of viral replication were used to demonstrate that latent virus reactivation required gene expression of the superinfecting virus (Fig. 1), and use of functional, attenuated, or inactivated tat viruses demonstrated that latent virus reactivation required the activity of newly expressed Tat by the superinfecting virus (Fig. 3). Superinfection using wt or H13L tat viruses led to reactivation of latent viruses across a wide range of superinfection levels (Fig. 4). Superinfection of latently infected primary CD4 T cells also led to reactivation of latent viruses (Fig. 5). Finally, drug-resistant latent viruses were subject to reactivation by superinfection in Jurkat cells (Fig. 6A to D), which led to the generation of multidrug-resistant recombinants under selective pressure (Fig. 6E and F).
Lentiviruses, including HIV-1 have evolved various strategies to downregulate cell surface CD4, which might serve to impair immune recognition of infected cells and/or to limit cellular superinfection in a phenomenon referred to as superinfection immunity (35, 60, 61). Although CD4 downregulation can decrease superinfection rates (35), this effect is not absolute, and others have observed minimal interference with superinfection (26). Regardless of the magnitude of superinfection immunity, we were interested in superinfection of latently infected cells. Since latent viruses express little or no viral gene products, neither CD4 downregulation nor superinfection immunity would be expected.
Most clinically relevant latent viruses are found in resting CD4 T cells. This implies that superinfection of latently infected cells in vivo would require infection of resting cells, which is much less efficient than infection of activated cells. Nonetheless, infection of resting cells does occur both in vitro and in vivo (reviewed in references 3 and 62). Furthermore, infection of phenotypically resting CD4 T cells is enhanced in chemokine- or cytokine-rich environments, such as secondary lymphoid tissues (22, 63–67) where the majority of lymphocytes—including multiply infected cells—reside, and several studies have reported that pretreatment of resting CD4 T cells with various chemokines increases subsequent infection rates (68–70).
Reactivation of latent viruses by superinfection (Fig. 1 to 4) results in cells expressing two genetically distinct viral genomes. Notably, HIV-1 has a much higher effective rate of recombination than some other retroviruses, such as murine leukemia virus (MLV). This is not due to higher rates of RT template switching, but rather to higher rates of heterozygous genomic RNA dimerization and packaging (25, 71, 72). The segregation of HIV-1 but not MLV genomic RNA molecules into assembling virions is effectively a random process, and there is now direct physical evidence that heterozygous HIV-1 virions are produced according to a Hardy-Weinberg equilibrium (73). Furthermore, it has been estimated that nearly all HIV-1 virions undergo recombination during reverse transcription, as opposed to only a subpopulation of viruses (26, 72, 74). It has also been suggested that superinfection contributes to recombination to a much greater extent than does cell-to-cell transmission, on the assumption that multiple infection by cell-to-cell transmission involves genetically identical virions (23). Regardless of the pathways of infection through which recombinant viruses arise, their evolutionary success is apparent given the global abundance of CRFs. More direct examples of the success of recombinants are shown by studies in which rhesus macaques were inoculated with two simian immunodeficiency virus (SIV) strains, each deleted in one or more accessory genes. In these studies, recombinants emerged as the dominant quasispecies in most macaques (31, 32). In settings of highly active antiretroviral therapy (HAART), the selective advantages of multidrug-resistant viruses might be even greater than for these accessory gene-deleted lentiviruses.
Previous studies have examined superinfection of cell lines harboring defective or latent viruses, although not in the same context as explored here. The authors of one study infected the U1 and ACH-2 cell lines (which harbor latent or defective proviruses) or their parental cell lines with HIV-1 that was pseudotyped with an amphotropic MLV envelope (75). In that study, superinfection was used as a tool to uncover cellular determinants of viral latency in U1 and ACH-2 cells and provided useful insights into HIV-1 latency at a time when little was known in that regard. However, latent virus reactivation and subsequent recombination were not addressed. A second study demonstrated recombination when a cell line chronically infected with a Vpr-deleted provirus was superinfected with other accessory gene-deleted viruses (76). More recently, our group has demonstrated recombination following superinfection of a cell line chronically infected with a multidrug-resistant virus, although the cell line carried an envelope-defective virus rather than a latent virus (77). Another study recently demonstrated that coinfection with distinct reporter viruses led to higher than expected levels of double-positive cells and that this required Tat (78). This likely indicates that Tat expression from one coinfecting virus is sufficient to drive expression of both viral genomes, potentially limiting the earliest events involved in the establishment of latency. In the present study, we have used populations of both Jurkat cells and unstimulated primary CD4 T cells, each harboring latent viral genomes across diverse integration sites. Of note, the latent viruses used here encode H13L Tat that attenuates its activity by decreasing Tat–P-TEFb interactions. This is reminiscent of the enrichment of attenuated tat viruses that was identified in resting CD4 T cells of patients on suppressive therapy, where those tat mutations also caused decreased affinity for P-TEFb (79). Finally, one study showed that coinfection of a T-cell line with two distinct drug-resistant viruses led to the emergence of multidrug-resistant recombinants following passage in cell culture under selective conditions (80).
While many of the same mechanisms appear to govern the establishment and maintenance of latency in Jurkat cells and primary cells (3, 81), their intracellular environments exhibit important differences. Thus, our examination of the effect of gp120 on latent virus reactivation (Fig. 1) might not be applicable to latency in primary resting cells, whose activation state is unlikely to be represented by Jurkat cells. Similarly, this issue might apply to our examination of the role of Nef in latent virus reactivation (Fig. 2). If anything, however, our results might suggest a modest inhibitory effect of Nef on latent virus reactivation in Jurkat cells (Fig. 2B and 3D; compare the Δnef data point to the linear regression line), although this is purely speculative. These differences notwithstanding, our results indicate that latent viruses can be reactivated by superinfection in both Jurkat cells and primary CD4 T cells. It is worth discussing our use of an NRTI-resistant virus in the recombination experiments presented in Fig. 6, since some NRTI resistance mutations alter recombination rates. However, the M184V mutation used here has only a minor effect on RT template-switching rates (82). Additionally, it has been shown that recombination occurs at similar frequencies in both Jurkat cells and primary cells (26).
Recombination is expected to occur whenever there is ongoing replication. While two recent studies have provided evidence for ongoing HIV-1 replication during suppressive HAART (83, 84), the general consensus is that ongoing replication does not occur in most HAART-treated patients (85–88). Nonetheless residual viremia is present in most HAART-treated individuals, which likely arises from reactivation of latent viruses, and a recent study demonstrated that residual viremia during long-term suppressive HAART was infectious (89). This suggests that new rounds of replication could occur during periods of low drug adherence or treatment interruption or even during adherent treatment if the residual virus was drug resistant. Superinfection of latently infected cells might be expected to occur regularly in untreated patients and could also occur during HAART as a result of infectious residual viremia, regardless of whether the residual viremia originated from activation of individual latent viruses, low-level ongoing replication, or viral rebound following treatment failure. As demonstrated here, reactivated latent viruses are capable of undergoing recombination. Recombination is widely acknowledged to increase viral evolution in individual patients (24, 25, 34, 90, 91), often though not always accelerating the emergence of multidrug resistance (92–94). Since all viral quasispecies, including drug-resistant viruses, can be latently archived (11–15), reactivation of latent viruses by superinfection or other means could provide for the emergence or spread of replicatively fit viruses in the face of strong selective pressures.
ACKNOWLEDGMENTS
This work was supported by grants from the Canadian Institutes of Health Research (CIHR). D.A.D. is the recipient of a doctoral research award from CIHR. Work by D.A.D. was performed in partial fulfillment of the requirements for the Ph.D. degree from the Faculty of Graduate Studies and Research, McGill University, Montréal, Québec, Canada.
We thank Maureen Oliveira, Susan Colby-Germinario, and Cesar Collazos for providing technical assistance. We also thank Christian Young of the Lady Davis Institute flow cytometry facility for aid with flow cytometry analysis. pBR-NL4-3-IRES-dsRed and derivatives were kindly provided by J. Münch and F. Kirchhoff.
Footnotes
Published ahead of print 26 June 2013
REFERENCES
- 1.Chun TW, Engel D, Berrey MM, Shea T, Corey L, Fauci AS. 1998. Early establishment of a pool of latently infected, resting CD4(+) T cells during primary HIV-1 infection. Proc. Natl. Acad. Sci. U. S. A. 95:8869–8873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Archin NM, Vaidya NK, Kuruc JD, Liberty AL, Wiegand A, Kearney MF, Cohen MS, Coffin JM, Bosch RJ, Gay CL, Eron JJ, Margolis DM, Perelson AS. 2012. Immediate antiviral therapy appears to restrict resting CD4+ cell HIV-1 infection without accelerating the decay of latent infection. Proc. Natl. Acad. Sci. U. S. A. 109:9523–9528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Donahue DA, Wainberg MA. 2013. Cellular and molecular mechanisms involved in the establishment of HIV-1 latency. Retrovirology 10:11. 10.1186/1742-4690-10-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hakre S, Chávez L, Shirakawa K, Verdin E. 2012. HIV latency: experimental systems and molecular models. FEMS Microbiol. Rev. 36:706–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siliciano RF, Greene WC. 2011. HIV latency. Cold Spring Harb. Perspect. Med. 1:a007096. 10.1101/cshperspect.a007096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mbonye U, Karn J. 2011. Control of HIV latency by epigenetic and non-epigenetic mechanisms. Curr. HIV Res. 9:554–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abbas W, Herbein G. 2012. Molecular understanding of HIV-1 latency. Adv. Virol. 2012:574967. 10.1155/2012/574967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geeraert L, Kraus G, Pomerantz RJ. 2008. Hide-and-seek: the challenge of viral persistence in HIV-1 infection. Annu. Rev. Med. 59:487–501 [DOI] [PubMed] [Google Scholar]
- 9.Chun T-W, Fauci AS. 2012. HIV reservoirs: pathogenesis and obstacles to viral eradication and cure. AIDS 26:1261–1268 [DOI] [PubMed] [Google Scholar]
- 10.Han Y, Wind-Rotolo M, Yang H-C, Siliciano JD, Siliciano RF. 2007. Experimental approaches to the study of HIV-1 latency. Nat. Rev. Microbiol. 5:95–106 [DOI] [PubMed] [Google Scholar]
- 11.Persaud D, Pierson T, Ruff C, Finzi D, Chadwick KR, Margolick JB, Ruff A, Hutton N, Ray S, Siliciano RF. 2000. A stable latent reservoir for HIV-1 in resting CD4(+) T lymphocytes in infected children. J. Clin. Invest. 105:995–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruff CT, Ray SC, Kwon P, Zinn R, Pendleton A, Hutton N, Ashworth R, Gange S, Quinn TC, Siliciano RF, Persaud D. 2002. Persistence of wild-type virus and lack of temporal structure in the latent reservoir for human immunodeficiency virus type 1 in pediatric patients with extensive antiretroviral exposure. J. Virol. 76:9481–9492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Monie D, Simmons RP, Nettles RE, Kieffer TL, Zhou Y, Zhang H, Karmon S, Ingersoll R, Chadwick K, Zhang H, Margolick JB, Quinn TC, Ray SC, Wind-Rotolo M, Miller M, Persaud D, Siliciano RF. 2005. A novel assay allows genotyping of the latent reservoir for human immunodeficiency virus type 1 in the resting CD4+ T cells of viremic patients. J. Virol. 79:5185–5202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lambotte O, Chaix M-L, Gubler B, Nasreddine N, Wallon C, Goujard C, Rouzioux C, Taoufik Y, Delfraissy J-F. 2004. The lymphocyte HIV reservoir in patients on long-term HAART is a memory of virus evolution. AIDS 18:1147–1158 [DOI] [PubMed] [Google Scholar]
- 15.Verhofstede C, Noë A, Demecheleer E, De Cabooter N, Van Wanzeele F, Van Der Gucht B, Vogelaers D, Plum J. 2004. Drug-resistant variants that evolve during nonsuppressive therapy persist in HIV-1-infected peripheral blood mononuclear cells after long-term highly active antiretroviral therapy. J. Acquir. Immune Defic. Syndr. 35:473–483 [DOI] [PubMed] [Google Scholar]
- 16.Joos B, Fischer M, Kuster H, Pillai SK, Wong JK, Böni J, Hirschel B, Weber R, Trkola A, Günthard HF, Swiss HIV Cohort Study 2008. HIV rebounds from latently infected cells, rather than from continuing low-level replication. Proc. Natl. Acad. Sci. U. S. A. 105:16725–16730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gratton S, Cheynier R, Dumaurier MJ, Oksenhendler E, Wain-Hobson S. 2000. Highly restricted spread of HIV-1 and multiply infected cells within splenic germinal centers. Proc. Natl. Acad. Sci. U. S. A. 97:14566–14571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jung A, Maier R, Vartanian J-P, Bocharov G, Jung V, Fischer U, Meese E, Wain-Hobson S, Meyerhans A. 2002. Recombination: multiply infected spleen cells in HIV patients. Nature 418:144. 10.1038/418144a [DOI] [PubMed] [Google Scholar]
- 19.Josefsson L, King MS, Makitalo B, Brännström J, Shao W, Maldarelli F, Kearney MF, Hu W-S, Chen J, Gaines H, Mellors JW, Albert J, Coffin JM, Palmer SE. 2011. Majority of CD4+ T cells from peripheral blood of HIV-1-infected individuals contain only one HIV DNA molecule. Proc. Natl. Acad. Sci. U. S. A. 108:11199–11204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Del Portillo A, Tripodi J, Najfeld V, Wodarz D, Levy DN, Chen BK. 2011. Multiploid inheritance of HIV-1 during cell-to-cell infection. J. Virol. 85:7169–7176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Russell RA, Martin N, Mitar I, Jones E, Sattentau QJ. 27 May 2013. Multiple proviral integration events after virological synapse-mediated HIV-1 spread. Virology [Epub ahead of print.] 10.1016/j.virol.2013.05.005 [DOI] [PubMed] [Google Scholar]
- 22.Rudnicka D, Feldmann J, Porrot F, Wietgrefe S, Guadagnini S, Prévost M-C, Estaquier J, Haase AT, Sol-Foulon N, Schwartz O. 2009. Simultaneous cell-to-cell transmission of human immunodeficiency virus to multiple targets through polysynapses. J. Virol. 83:6234–6246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dixit NM, Perelson AS. 2004. Multiplicity of human immunodeficiency virus infections in lymphoid tissue. J. Virol. 78:8942–8945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smyth RP, Davenport MP, Mak J. 2012. The origin of genetic diversity in HIV-1. Virus Res. 169:415–429 [DOI] [PubMed] [Google Scholar]
- 25.Onafuwa-Nuga A, Telesnitsky A. 2009. The remarkable frequency of human immunodeficiency virus type 1 genetic recombination. Microbiol. Mol. Biol. Rev. 73:451–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levy DN, Aldrovandi GM, Kutsch O, Shaw GM. 2004. Dynamics of HIV-1 recombination in its natural target cells. Proc. Natl. Acad. Sci. U. S. A. 101:4204–4209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hemelaar J. 2013. Implications of HIV diversity for the HIV-1 pandemic. J. Infect. 66:391–400 [DOI] [PubMed] [Google Scholar]
- 28.Günthard HF, Leigh-Brown AJ, D'Aquila RT, Johnson VA, Kuritzkes DR, Richman DD, Wong JK, D'Aquila RT. 1999. Higher selection pressure from antiretroviral drugs in vivo results in increased evolutionary distance in HIV-1 pol. Virology 259:154–165 [DOI] [PubMed] [Google Scholar]
- 29.Nora T, Charpentier C, Tenaillon O, Hoede C, Clavel F, Hance AJ. 2007. Contribution of recombination to the evolution of human immunodeficiency viruses expressing resistance to antiretroviral treatment. J. Virol. 81:7620–7628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ray N, Harrison JE, Blackburn LA, Martin JN, Deeks SG, Doms RW. 2007. Clinical resistance to enfuvirtide does not affect susceptibility of human immunodeficiency virus type 1 to other classes of entry inhibitors. J. Virol. 81:3240–3250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wooley DP, Smith RA, Czajak S, Desrosiers RC. 1997. Direct demonstration of retroviral recombination in a rhesus monkey. J. Virol. 71:9650–9653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim E-Y, Busch M, Abel K, Fritts L, Bustamante P, Stanton J, Lu D, Wu S, Glowczwskie J, Rourke T, Bogdan D, Piatak M, Lifson JD, Desrosiers RC, Wolinsky S, Miller CJ. 2005. Retroviral recombination in vivo: viral replication patterns and genetic structure of simian immunodeficiency virus (SIV) populations in rhesus macaques after simultaneous or sequential intravaginal inoculation with SIVmac239Δvpx/Δvpr and SIVmac239Δnef. J. Virol. 79:4886–4895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berngruber TW, Weissing FJ, Gandon S. 2010. Inhibition of superinfection and the evolution of viral latency. J. Virol. 84:10200–10208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Charpentier C, Nora T, Tenaillon O, Clavel F, Hance AJ. 2006. Extensive recombination among human immunodeficiency virus type 1 quasispecies makes an important contribution to viral diversity in individual patients. J. Virol. 80:2472–2482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wildum S, Schindler M, Münch J, Kirchhoff F. 2006. Contribution of Vpu, Env, and Nef to CD4 down-modulation and resistance of human immunodeficiency virus type 1-infected T cells to superinfection. J. Virol. 80:8047–8059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Donahue DA, Kuhl BD, Sloan RD, Wainberg MA. 2012. The viral protein Tat can inhibit the establishment of HIV-1 latency. J. Virol. 86:3253–3263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lassen KG, Hebbeler AM, Bhattacharyya D, Lobritz MA, Greene WC. 2012. A flexible model of HIV-1 latency permitting evaluation of many primary CD4 T-cell reservoirs. PLoS One 7:e30176. 10.1371/journal.pone.0030176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu JJ, Wu TL, Liszewski MK, Dai J, Swiggard WJ, Baytop C, Frank I, Levine BL, Yang W, Theodosopoulos T, O'Doherty U. 2008. A more precise HIV integration assay designed to detect small differences finds lower levels of integrated DNA in HAART treated patients. Virology 379:78–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Donahue DA, Sloan RD, Kuhl BD, Bar-Magen T, Schader SM, Wainberg MA. 2010. Stage-dependent inhibition of HIV-1 replication by antiretroviral drugs in cell culture. Antimicrob. Agents Chemother. 54:1047–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pearson R, Kim YK, Hokello J, Lassen K, Friedman J, Tyagi M, Karn J. 2008. Epigenetic silencing of human immunodeficiency virus (HIV) transcription by formation of restrictive chromatin structures at the viral long terminal repeat drives the progressive entry of HIV into latency. J. Virol. 82:12291–12303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weinberger LS, Burnett JC, Toettcher JE, Arkin AP, Schaffer DV. 2005. Stochastic gene expression in a lentiviral positive-feedback loop: HIV-1 Tat fluctuations drive phenotypic diversity. Cell 122:169–182 [DOI] [PubMed] [Google Scholar]
- 42.Weinberger LS, Dar RD, Simpson ML. 2008. Transient-mediated fate determination in a transcriptional circuit of HIV. Nat. Genet. 40:466–470 [DOI] [PubMed] [Google Scholar]
- 43.Singh A, Weinberger LS. 2009. Stochastic gene expression as a molecular switch for viral latency. Curr. Opin. Microbiol. 12:460–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tyagi M, Karn J. 2007. CBF-1 promotes transcriptional silencing during the establishment of HIV-1 latency. EMBO J. 26:4985–4995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tyagi M, Pearson RJ, Karn J. 2010. Establishment of HIV latency in primary CD4+ cells is due to epigenetic transcriptional silencing and P-TEFb restriction. J. Virol. 84:6425–6437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim YK, Mbonye U, Hokello J, Karn J. 2011. T-cell receptor signaling enhances transcriptional elongation from latent HIV proviruses by activating P-TEFb through an ERK-dependent pathway. J. Mol. Biol. 410:896–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mbonye UR, Gokulrangan G, Datt M, Dobrowolski C, Cooper M, Chance MR, Karn J. 2013. Phosphorylation of CDK9 at Ser175 enhances HIV transcription and is a marker of activated P-TEFb in CD4(+) T lymphocytes. PLoS Pathog. 9:e1003338. 10.1371/journal.ppat.1003338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cicala C, Arthos J, Censoplano N, Cruz C, Chung E, Martinelli E, Lempicki RA, Natarajan V, VanRyk D, Daucher M, Fauci AS. 2006. HIV-1 gp120 induces NFAT nuclear translocation in resting CD4+ T-cells. Virology 345:105–114 [DOI] [PubMed] [Google Scholar]
- 49.Cicala C, Arthos J, Selig SM, Dennis G, Hosack DA, Van Ryk D, Spangler ML, Steenbeke TD, Khazanie P, Gupta N, Yang J, Daucher M, Lempicki RA, Fauci AS. 2002. HIV envelope induces a cascade of cell signals in non-proliferating target cells that favor virus replication. Proc. Natl. Acad. Sci. U. S. A. 99:9380–9385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Missé D, Gajardo J, Oblet C, Religa A, Riquet N, Mathieu D, Yssel H, Veas F. 2005. Soluble HIV-1 gp120 enhances HIV-1 replication in non-dividing CD4+ T cells, mediated via cell signaling and Tat cofactor overexpression. AIDS 19:897–905 [DOI] [PubMed] [Google Scholar]
- 51.Davis CB, Dikic I, Unutmaz D, Hill CM, Arthos J, Siani MA, Thompson DA, Schlessinger J, Littman DR. 1997. Signal transduction due to HIV-1 envelope interactions with chemokine receptors CXCR4 or CCR5. J. Exp. Med. 186:1793–1798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vasiliver-Shamis G, Dustin ML, Hioe CE. 2010. HIV-1 Virological synapse is not simply a copycat of the immunological synapse. Viruses 2:1239–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kinter AL, Umscheid CA, Arthos J, Cicala C, Lin Y, Jackson R, Donoghue E, Ehler L, Adelsberger J, Rabin RL, Fauci AS. 2003. HIV envelope induces virus expression from resting CD4+ T cells isolated from HIV-infected individuals in the absence of markers of cellular activation or apoptosis. J. Immunol. 170:2449–2455 [DOI] [PubMed] [Google Scholar]
- 54.Shen L, Peterson S, Sedaghat AR, McMahon MA, Callender M, Zhang H, Zhou Y, Pitt E, Anderson KS, Acosta EP, Siliciano RF. 2008. Dose-response curve slope sets class-specific limits on inhibitory potential of anti-HIV drugs. Nat. Med. 14:762–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sloan RD, Wainberg MA. 2011. The role of unintegrated DNA in HIV infection. Retrovirology 8:52. 10.1186/1742-4690-8-52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Abraham L, Fackler OT. 2012. HIV-1 Nef: a multifaceted modulator of T cell receptor signaling. Cell Commun. Signal. 10:39. 10.1186/1478-811X-10-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Landi A, Iannucci V, Nuffel AV, Meuwissen P, Verhasselt B. 2011. One protein to rule them all: modulation of cell surface receptors and molecules by HIV Nef. Curr. HIV Res. 9:496–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Simmons A, Aluvihare V, McMichael A. 2001. Nef triggers a transcriptional program in T cells imitating single-signal T cell activation and inducing HIV virulence mediators. Immunity 14:763–777 [DOI] [PubMed] [Google Scholar]
- 59.Wu Y, Marsh JW. 2001. Selective transcription and modulation of resting T cell activity by preintegrated HIV DNA. Science 293:1503–1506 [DOI] [PubMed] [Google Scholar]
- 60.Geleziunas R, Bour S, Wainberg MA. 1994. Cell surface down-modulation of CD4 after infection by HIV-1. FASEB J. 8:593–600 [DOI] [PubMed] [Google Scholar]
- 61.Michel N, Allespach I, Venzke S, Fackler OT, Keppler OT. 2005. The Nef protein of human immunodeficiency virus establishes superinfection immunity by a dual strategy to downregulate cell-surface CCR5 and CD4. Curr. Biol. 15:714–723 [DOI] [PubMed] [Google Scholar]
- 62.Vatakis DN, Nixon CC, Zack JA. 2010. Quiescent T cells and HIV: an unresolved relationship. Immunol. Res. 48:110–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li Q, Duan L, Estes JD, Ma Z-M, Rourke T, Wang Y, Reilly C, Carlis J, Miller CJ, Haase AT. 2005. Peak SIV replication in resting memory CD4+ T cells depletes gut lamina propria CD4+ T cells. Nature 434:1148–1152 [DOI] [PubMed] [Google Scholar]
- 64.Unutmaz D, KewalRamani VN, Marmon S, Littman DR. 1999. Cytokine signals are sufficient for HIV-1 infection of resting human T lymphocytes. J. Exp. Med. 189:1735–1746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kinter A, Moorthy A, Jackson R, Fauci AS. 2003. Productive HIV infection of resting CD4+ T cells: role of lymphoid tissue microenvironment and effect of immunomodulating agents. AIDS Res. Hum. Retroviruses 19:847–856 [DOI] [PubMed] [Google Scholar]
- 66.Eckstein DA, Penn ML, Korin YD, Scripture-Adams DD, Zack JA, Kreisberg JF, Roederer M, Sherman MP, Chin PS, Goldsmith MA. 2001. HIV-1 actively replicates in naive CD4(+) T cells residing within human lymphoid tissues. Immunity 15:671–682 [DOI] [PubMed] [Google Scholar]
- 67.Weissman D, Daucher J, Barker T, Adelsberger J, Baseler M, Fauci AS. 1996. Cytokine regulation of HIV replication induced by dendritic cell-CD4-positive T cell interactions. AIDS Res. Hum. Retroviruses 12:759–767 [DOI] [PubMed] [Google Scholar]
- 68.Saleh S, Solomon A, Wightman F, Xhilaga M, Cameron PU, Lewin SR. 2007. CCR7 ligands CCL19 and CCL21 increase permissiveness of resting memory CD4+ T cells to HIV-1 infection: a novel model of HIV-1 latency. Blood 110:4161–4164 [DOI] [PubMed] [Google Scholar]
- 69.Cameron PU, Saleh S, Sallmann G, Solomon A, Wightman F, Evans VA, Boucher G, Haddad EK, Sékaly R-P, Harman AN, Anderson JL, Jones KL, Mak J, Cunningham AL, Jaworowski A, Lewin SR. 2010. Establishment of HIV-1 latency in resting CD4+ T cells depends on chemokine-induced changes in the actin cytoskeleton. Proc. Natl. Acad. Sci. U. S. A. 107:16934–16939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Saleh S, Wightman F, Ramanayake S, Alexander M, Kumar N, Khoury G, Pereira C, Purcell D, Cameron PU, Lewin SR. 2011. Expression and reactivation of HIV in a chemokine induced model of HIV latency in primary resting CD4+ T cells. Retrovirology 8:80. 10.1186/1742-4690-8-80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhuang J, Mukherjee S, Ron Y, Dougherty JP. 2006. High rate of genetic recombination in murine leukemia virus: implications for influencing proviral ploidy. J. Virol. 80:6706–6711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Onafuwa A, An W, Robson ND, Telesnitsky A. 2003. Human immunodeficiency virus type 1 genetic recombination is more frequent than that of Moloney murine leukemia virus despite similar template switching rates. J. Virol. 77:4577–4587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen J, Nikolaitchik O, Singh J, Wright A, Bencsics CE, Coffin JM, Ni N, Lockett S, Pathak VK, Hu W-S. 2009. High efficiency of HIV-1 genomic RNA packaging and heterozygote formation revealed by single virion analysis. Proc. Natl. Acad. Sci. U. S. A. 106:13535–13540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rhodes T, Wargo H, Hu W-S. 2003. High rates of human immunodeficiency virus type 1 recombination: near-random segregation of markers one kilobase apart in one round of viral replication. J. Virol. 77:11193–11200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen BK, Saksela K, Andino R, Baltimore D. 1994. Distinct modes of human immunodeficiency virus type 1 proviral latency revealed by superinfection of nonproductively infected cell lines with recombinant luciferase-encoding viruses. J. Virol. 68:654–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kishi M, Tokunaga K, Zheng YH, Bahmani MK, Kakinuma M, Nonoyama M, Lai PK, Ikuta K. 1995. Superinfection of a defective human immunodeficiency virus type 1 provirus-carrying T cell clone with vif or vpu mutants gives cytopathic virus particles by homologous recombination. AIDS Res. Hum. Retroviruses 11:45–53 [DOI] [PubMed] [Google Scholar]
- 77.Quan Y, Liang C, Brenner BG, Wainberg MA. 2009. Multidrug-resistant variants of HIV type 1 (HIV-1) can exist in cells as defective quasispecies and be rescued by superinfection with other defective HIV-1 variants. J. Infect. Dis. 200:1479–1483 [DOI] [PubMed] [Google Scholar]
- 78.Brégnard C, Pacini G, Danos O, Basmaciogullari S. 2012. Suboptimal provirus expression explains apparent nonrandom cell coinfection with HIV-1. J. Virol. 86:8810–8820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yukl S, Pillai S, Li P, Chang K, Pasutti W, Ahlgren C, Havlir D, Strain M, Günthard H, Richman D, Rice AP, Daar E, Little S, Wong JK. 2009. Latently-infected CD4+ T cells are enriched for HIV-1 Tat variants with impaired transactivation activity. Virology 387:98–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Moutouh L, Corbeil J, Richman DD. 1996. Recombination leads to the rapid emergence of HIV-1 dually resistant mutants under selective drug pressure. Proc. Natl. Acad. Sci. U. S. A. 93:6106–6111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dahabieh MS, Ooms M, Simon V, Sadowski I. 2013. A double-fluorescent HIV-1 reporter shows that the majority of integrated HIV-1 is latent shortly after infection. J. Virol. 87:4716–4727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nikolenko GN, Svarovskaia ES, Delviks KA, Pathak VK. 2004. Antiretroviral drug resistance mutations in human immunodeficiency virus type 1 reverse transcriptase increase template-switching frequency. J. Virol. 78:8761–8770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Buzón MJ, Massanella M, Llibre JM, Esteve A, Dahl V, Puertas MC, Gatell JM, Domingo P, Paredes R, Sharkey M, Palmer S, Stevenson M, Clotet B, Blanco J, Martinez-Picado J. 2010. HIV-1 replication and immune dynamics are affected by raltegravir intensification of HAART-suppressed subjects. Nat. Med. 16:460–465 [DOI] [PubMed] [Google Scholar]
- 84.Buzón MJ, Codoñer FM, Frost SDW, Pou C, Puertas MC, Massanella M, Dalmau J, Llibre JM, Stevenson M, Blanco J, Clotet B, Paredes R, Martinez-Picado J. 2011. Deep molecular characterization of HIV-1 dynamics under suppressive HAART. PLoS Pathog. 7:e1002314. 10.1371/journal.ppat.1002314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Eisele E, Siliciano RF. 2012. Redefining the viral reservoirs that prevent HIV-1 eradication. Immunity 37:377–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Johnston R, Barré-Sinoussi F. 2012. Controversies in HIV cure research. J. Int. AIDS Soc. 15:16. 10.1186/1758-2652-15-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Maldarelli F. 2011. Targeting viral reservoirs: ability of antiretroviral therapy to stop viral replication. Curr. Opin. HIV AIDS 6:49–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Evering TH, Mehandru S, Racz P, Tenner-Racz K, Poles MA, Figueroa A, Mohri H, Markowitz M. 2012. Absence of HIV-1 evolution in the gut-associated lymphoid tissue from patients on combination antiviral therapy initiated during primary infection. PLoS Pathog. 8:e1002506. 10.1371/journal.ppat.1002506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sahu GK, Sarria JC, Cloyd MW. 2010. Recovery of replication-competent residual HIV-1 from plasma of a patient receiving prolonged, suppressive highly active antiretroviral therapy. J. Virol. 84:8348–8352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bocharov G, Ford NJ, Edwards J, Breinig T, Wain-Hobson S, Meyerhans A. 2005. A genetic-algorithm approach to simulating human immunodeficiency virus evolution reveals the strong impact of multiply infected cells and recombination. J. Gen. Virol. 86:3109–3118 [DOI] [PubMed] [Google Scholar]
- 91.Vijay NNV, Vasantika Ajmani R, Perelson AS, Dixit NM. 2008. Recombination increases human immunodeficiency virus fitness, but not necessarily diversity. J. Gen. Virol. 89:1467–1477 [DOI] [PubMed] [Google Scholar]
- 92.Althaus CL, Bonhoeffer S. 2005. Stochastic interplay between mutation and recombination during the acquisition of drug resistance mutations in human immunodeficiency virus type 1. J. Virol. 79:13572–13578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Arora P, Dixit NM. 2009. Timing the emergence of resistance to anti-HIV drugs with large genetic barriers. PLoS Comput. Biol. 5:e1000305. 10.1371/journal.pcbi.1000305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Carvajal-Rodríguez A, Crandall KA, Posada D. 2007. Recombination favors the evolution of drug resistance in HIV-1 during antiretroviral therapy. Infect. Genet. Evol. 7:476–483 [DOI] [PMC free article] [PubMed] [Google Scholar]