Abstract
A novel human Middle East respiratory syndrome coronavirus (MERS-CoV) caused outbreaks of severe acute respiratory syndrome (SARS)-like illness with a high mortality rate, raising concerns of its pandemic potential. Dipeptidyl peptidase-4 (DPP4) was recently identified as its receptor. Here we showed that residues 377 to 662 in the S protein of MERS-CoV specifically bound to DPP4-expressing cells and soluble DPP4 protein and induced significant neutralizing antibody responses, suggesting that this region contains the receptor-binding domain (RBD), which has a potential to be developed as a MERS-CoV vaccine.
TEXT
As of 17 June 2013, WHO had been informed of 64 laboratory-confirmed cases of infection with a novel human coronavirus (hCoV), Middle East respiratory syndrome (MERS) CoV (MERS-CoV; previously known as hCoV-Erasmus Medical Center [hCoV-EMC]) (1, 2), including 38 deaths (3). Human-to-human transmission of MERS-CoV has occurred in household, work environment, and health care settings (4, 5), raising concerns of its potential to cause a pandemic similar to that caused by severe acute respiratory syndrome (SARS) coronavirus (SARS-CoV) in 2002 to 2003 (6).
In 2003, Farzan and colleagues successfully identified the receptor of SARS-CoV, angiotensin-converting enzyme 2 (ACE2) (7), and a 193-amino-acid fragment in the spike (S) protein (residues 318 to 510) as the receptor-binding domain (RBD) (8). We found that SARS-CoV S-RBD contains a critical neutralizing site (9) which induces potent neutralizing antibodies and protection against SARS-CoV infection in an animal model (10).
Since MERS-CoV is genetically related to SARS-CoV (1), we compared their S protein sequences and predicted that the RBD of MERS-CoV might be located in the region spanning residues 377 to 662 of the S1 subunit (Fig. 1). Using the Swiss-Model Workplace homology modeling server (11) and basing our work on the X-ray crystal structure of the SARS-CoV S-RBD (Protein Data Bank [PDB] identification no. 2DD8) (12), we predicted the conformational structure of the region consisting of residues 377 to 662 in the S1 subunit of the MERS-CoV S protein (13). We noticed that the SARS-CoV S-RBD and the predicted MERS-CoV S-RBD possessed similar core structures but had an extended secondary structure consisting predominantly of the receptor-binding motifs (RBM) (12, 14). The extended region in MERS-CoV S-RBD is much longer than that in SARS-CoV S-RBD, suggesting that MERS-CoV and SARS-CoV use different receptors. Indeed, it has been proven that dipeptidyl peptidase-4 (DPP4; also known as CD26) is the functional receptor of MERS-CoV (15).
Fig 1.
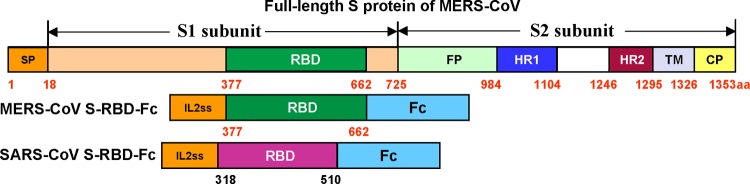
Schematic representation of MERS-CoV S protein and constructs of MERS-CoV and SARS-CoV S-RBD-Fc proteins. SP, signal peptide; RBD, receptor-binding domain; FP, fusion peptide; HR1, heptad repeat 1; HR2, heptad repeat 2; TM, transmembrane domain; CP, cytoplasmic domain.
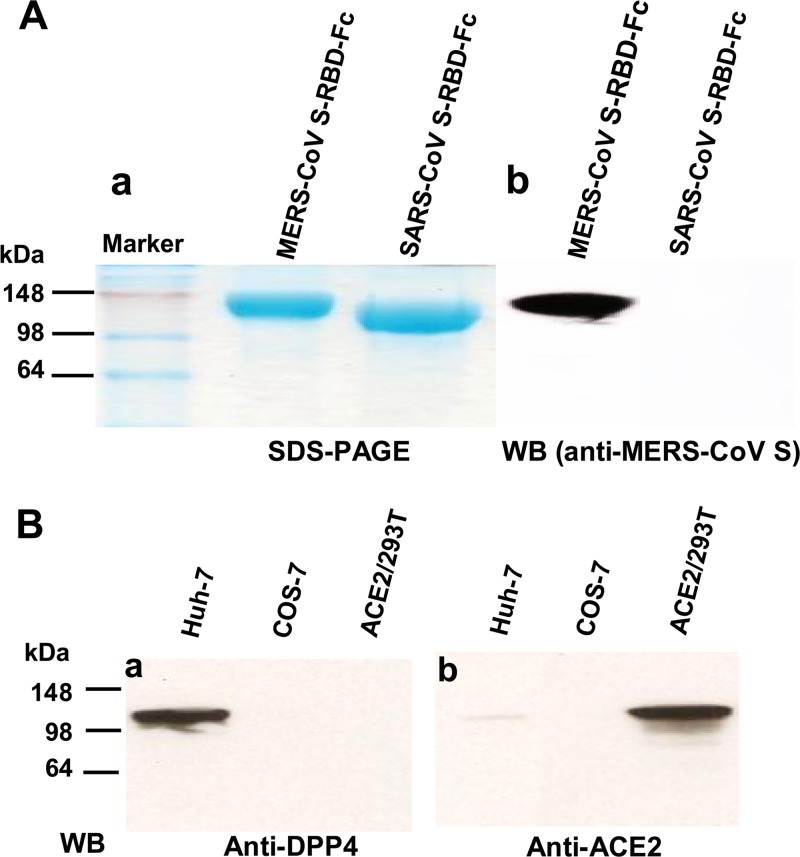
We then constructed MERS-CoV S-RBD based on the synthesized codon-optimized MERS-CoV S sequences (GenBank accession no. AFS88936.1) and fused it to Fc of human IgG using pFUSE-hIgG1-Fc2 expression vector (here named Fc) (InvivoGen, San Diego, CA). The SARS-CoV S-RBD-Fc was constructed by fusing RBD of codon-optimized SARS-CoV S sequence into the Fc vector referred to above as a control (Fig. 1) (16). The S-RBD-Fc proteins were expressed in 293T cell culture supernatant and purified by protein A affinity chromatography (GE Healthcare, Piscataway, NJ) (17). We found that both MERS-CoV and SARS-CoV S-RBD-Fc proteins were highly purified from transfected culture supernatants (Fig. 2A, panel a). MERS-CoV S-RBD-Fc could be recognized by an MERS-CoV S-specific polyclonal antibody (1:1,000), while SARS-CoV S-RBD-Fc could not react with this antibody, as detected by Western blotting (Fig. 2A, panel b).
Fig 2.
Characterization of MERS-CoV S-RBD-Fc and detection of DPP4 expression in cells. (A) SDS-PAGE and Western blot analysis of MERS-CoV S-RBD-Fc. The protein molecular mass markers (kDa) are indicated on the left. SARS-CoV S-RBD-Fc was included as the control. MERS-CoV S-specific polyclonal antibodies were used for Western blot (WB) analysis. (B) Detection of expression of MERS-CoV's receptor DPP4 and SARS-CoV's receptor ACE2 in Huh-7, COS-7, and ACE2/293T cells by SDS-PAGE and Western blot (WB) analysis. Anti-DPP4 (a) and anti-ACE2 (b) monoclonal antibodies (MAbs) (R&D Systems, Minneapolis, MN) at 1 μg/ml were applied for the detection.
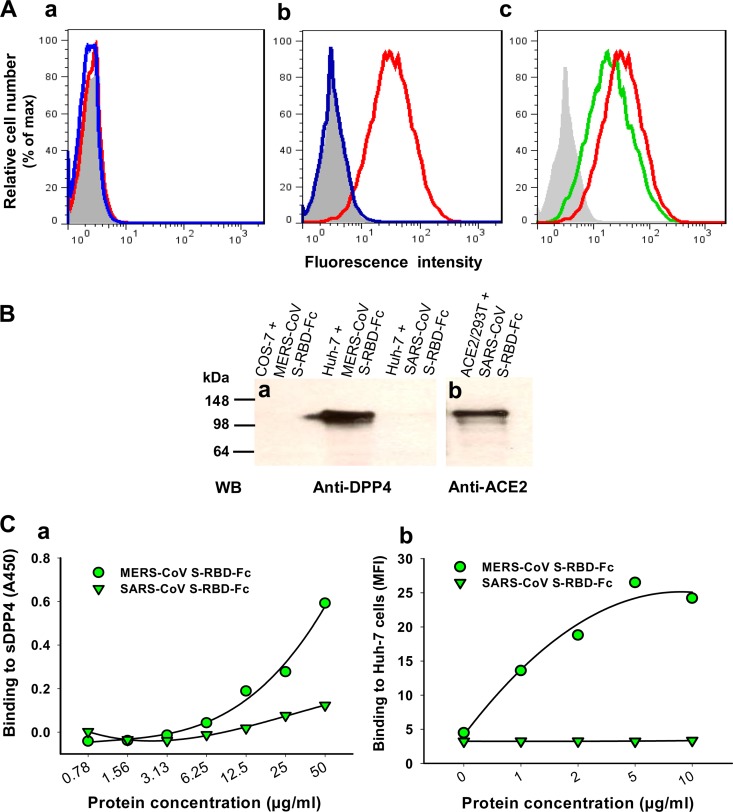
Using analysis performed by Western blotting, we found that DPP4 was highly expressed in Huh-7 cells but not in COS-7 cells (ATCC, Manassas, VA) and ACE2/293T cells (293T expressing SARS-CoV receptor ACE2) (Fig. 2B, panel a). However, ACE2 was highly expressed in ACE2/293T cells but not in COS-7 cells. There was a weak expression of ACE2 in Huh-7 cells (Fig. 2B, panel b). We then tested the binding of MERS-CoV S-RBD to DPP4-expressing Huh-7 cells as well as blockage of this binding using flow cytometry. Huh-7 and COS-7 (2 × 105) cells (control) were incubated with MERS-CoV and SARS-CoV S-RBD-Fc (5 μg/ml), respectively, at 37°C for 10 min and then continued to incubate at 4°C for 30 min. After washes, cells were incubated with DyLight-488-labeled goat anti-human IgG antibody at 4°C for 30 min before analysis. The competitive binding assay was performed by incubating soluble human DPP4 (sDPP4; 5 μg/ml) with MERS-CoV S-RBD-Fc (5 μg/ml) followed by the same procedure as described above. MERS-CoV S-RBD-Fc protein failed to interact with COS-7 cells, which have no DPP4 expression (Fig. 3A, panel a), but it strongly bound Huh-7 cells expressing the DPP4 receptor. No binding of SARS-CoV S-RBD-Fc with Huh-7 cells was revealed (Fig. 3A, panel b). The binding of MERS-CoV S-RBD-Fc to Huh-7 cells could be blocked by sDPP4 (Fig. 3A, panel c), suggesting that residues 377 to 662 of MERS-CoV S protein might contain the RBD.
Fig 3.
Specific binding of MERS-CoV S-RBD to DPP4. (A) Flow cytometric analysis of MERS-CoV S-RBD binding to DPP4-expressing Huh-7 cells and blockage of the binding with DPP4. (a) The MERS-CoV S-RBD-Fc did not bind to COS-7 cells that express no DPP4 (red line). Mock-incubated COS-7 cells (gray shading) and SARS-CoV S-RBD-Fc (blue line) were used as controls. (b) MERS-CoV S-RBD-Fc (red line) bound strongly to Huh-7 cells that express a high level of DPP4, while SARS-CoV S-RBD-Fc did not bind to Huh-7 cells (blue line). Mock-incubated Huh-7 cells were used as a control (gray shading). (c) Binding of MERS-CoV S-RBD-Fc to Huh-7 cells (red line) was blocked by sDPP4 (green line). Mock-incubated Huh-7 cells were used as a control (gray shading). (B) Coimmunoprecipitation analysis of interactions between MERS-CoV S-RBD-Fc and DPP4. The interactions between MERS-CoV S-RBD-Fc and DPP4 in the Huh-7 cell lysates and between SARS-CoV S-RBD-Fc and ACE2 in the ACE2/293T cell lysates were detected by coimmunoprecipitation analysis, using anti-DPP4 (a) and anti-ACE2 (b) MAbs at 1 μg/ml, respectively. COS-7 cells expressing neither DPP4 nor ACE2 were used as a control. (C) Detection of dose-dependent binding of MERS-CoV S-RBD-Fc to sDPP4 (2 μg/ml) by ELISA (a) and of DPP4-expressing Huh-7 cells by flow cytometric analysis (median fluorescence intensity [MFI]) (b). SARS-CoV S-RBD-Fc was included as a control.
Subsequently, we conducted a coimmunoprecipitation assay to analyze the interactions between MERS-CoV S-RBD-Fc and DPP4 in the Huh-7 cell lysates and between SARS-CoV S-RBD-Fc and ACE2 in the ACE2/293T cell lysates. The Huh-7 cell lysates (5 × 107/ml in 1 ml lysis buffer containing 0.3% N-decyl-β-d-maltopyranoside–phosphate-buffered saline [PBS]) were incubated with MERS-CoV S-RBD-Fc protein (20 μg) plus protein A Sepharose beads (50% [vol/vol], 200 μl) at 4°C for 1 h (15). After being washed with lysis buffer and PBS, beads were subjected to SDS-PAGE and Western blot analysis for detection of DPP4 using anti-DPP4 MAb. A clear band (corresponding to the size of DPP4 at ∼110 kDa) was detected from the beads that were preincubated with the mixture of MERS-CoV S-RBD-Fc and Huh-7 cell lysates but was not detected from the beads preincubated with the mixture of MERS-CoV S-RBD-Fc and COS-7 or the mixture of SARS-CoV S-RBD-Fc and Huh-7 cell lysates (Fig. 3B, panel a). A band of similar size (∼120 kDa) corresponding to ACE2 was detected from the beads preincubated with the mixture of SARS-CoV S-RBD-Fc and ACE2/293T cell lysates by the use of anti-ACE2 MAb (Fig. 3B, panel b). Using an enzyme-linked immunosorbent assay (ELISA), we further detected the binding of MERS-CoV S-RBD-Fc protein to sDPP4 in a dose-dependent manner and no significant binding of SARS-CoV S-RBD-Fc with sDPP4 (Fig. 3C, panel a). These results were consistent with those detected by flow cytometry, which showed the dose-dependent binding of MERS-CoV S-RBD-Fc protein to DPP4-expressing Huh-7 cells (Fig. 3C, panel b). The data presented above confirm the binding specificity of MERS-CoV S-RBD-Fc to its receptor DPP4.
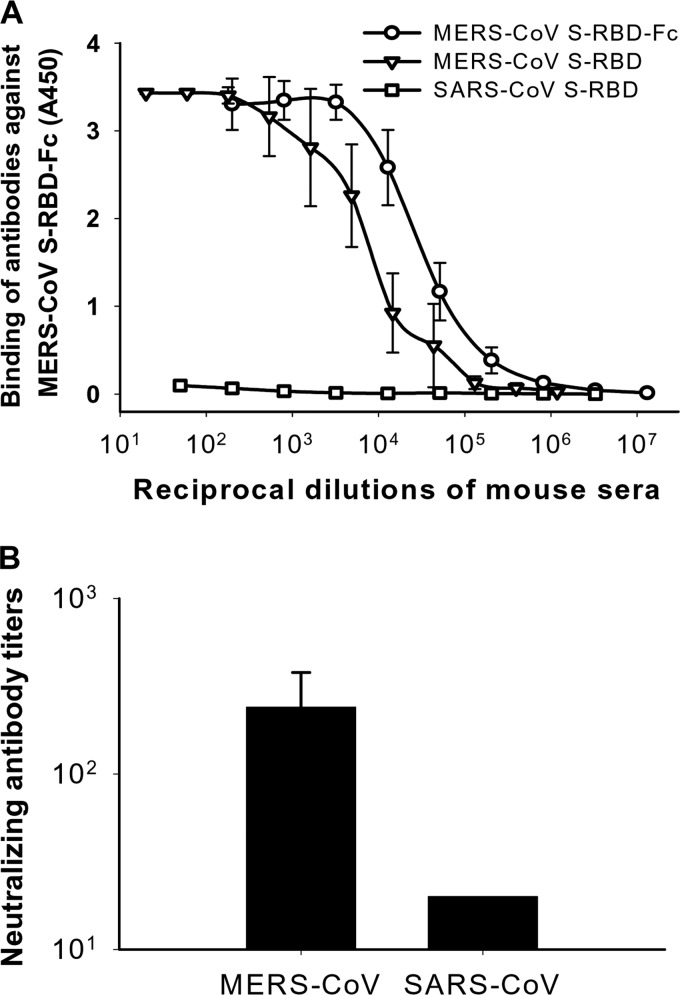
To detect the immunogenicity of MERS-CoV S-RBD-Fc, we used the purified protein to immunize BALB/c mice in the presence of Montanide ISA 51 adjuvant (Seppic, Fairfield, NJ) and collected mouse sera 10 days after the second vaccination to detect antibody responses and neutralizing activity against MERS-CoV and SARS-CoV. MERS-CoV S-RBD protein without Fc was constructed by fusing MERS-CoV S-RBD plus a 6×His tag and stop codon into the Fc vector and purified from 293T cell culture supernatant by the use of nickel-nitrilotriacetic acid (Ni-NTA) Superflow resin (Qiagen, Valencia, CA) (17, 18). The antibodies reacted strongly with MERS-CoV S-RBD with or without Fc, but they had only background binding to SARS-CoV S-RBD (18) (Fig. 4A). The mouse antisera against MERS-CoV S-RBD could effectively neutralize MERS-CoV infection in cell cultures in vitro with a neutralizing antibody titer of 1:240 ± 139, a level of neutralizing antibodies similar to that detected in MERS-CoV-infected patients (19), while, at the same time, these antisera neutralized SARS-CoV infection only marginally, with a low titer (<1:40) (Fig. 4B). These data suggest that MERS-CoV S-RBD could induce significant neutralizing antibody responses to MERS-CoV. However, MERS-CoV S-RBD-specific antibodies did not cross-react with or cross-neutralize SARS-CoV or vice versa (20).
Fig 4.
Immunogenicity of MERS-CoV S-RBD-Fc and neutralization of MERS-CoV and SARS-CoV by antibodies against MERS-CoV S-RBD-Fc. (A) Binding of antibodies to MERS-CoV S-RBD-Fc, MERS-CoV S-RBD, and SARS-CoV S-RBD in sera of mice immunized with MERS-CoV S-RBD-Fc detected by ELISA. (B) Neutralization of MERS-CoV and SARS-CoV infection in Vero E6 and FRhK4 cells by antisera of mice immunized with MERS-CoV S-RBD-Fc. The titers were determined as the highest serum dilutions that completely prevented cytopathic effect (CPE) in at least 50% of the wells and are expressed as means ± standard deviations.
Notably, the titers (∼1:240) of MERS-CoV neutralizing antibody elicited by MERS-CoV S-RBD-Fc protein were relatively lower than those of SARS-CoV neutralizing antibody induced by SARS-CoV S-RBD-Fc (≥1:1,000) (9, 16), suggesting that the immunogenicity of the MERS-CoV S-RBD is not as strong as that of SARS-CoV S-RBD. Nevertheless, previous studies in S-based SARS vaccines have revealed that mean neutralizing antibody titers as low as 1:284 could protect vaccinated animals against SARS-CoV challenge (21). The neutralizing antibody responses induced by MERS-CoV S-RBD-Fc protein may be strong enough to protect vaccinated animals against MERS-CoV infection. Therefore, further evaluation of the in vivo protective effect of MERS-CoV S-RBD-based vaccine is essential.
In conclusion, we found that a 286-amino-acid fragment (residues 377 to 662) of the MERS-CoV S protein contains the viral RBD, which mediates binding of MERS-CoV to its receptor DPP4 and serves as a critical target for the development of vaccines to prevent MERS-CoV infection and combat any future pandemic of this SARS-like disease.
ACKNOWLEDGMENTS
This study was supported in part by grants from an intramural fund of the New York Blood Center (NYB000068) and the National Program of China (2011CB504706, 2012CB519001).
We thank Charles M. Rice at The Rockefeller University for providing Huh-7 cells.
Footnotes
Published ahead of print 3 July 2013
REFERENCES
- 1.Zaki AM, van BS, Bestebroer TM, Osterhaus AD, Fouchier RA. 2012. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 367:1814–1820 [DOI] [PubMed] [Google Scholar]
- 2.de Groot RJ, Baker SC, Baric RS, Brown CS, Drosten C, Enjuanes L, Fouchier RA, Galiano M, Gorbalenya AE, Memish Z, Perlman S, Poon LL, Snijder EJ, Stephens GM, Woo PC, Zaki AM, Zambon M, Ziebuhr J. 2013. Middle East respiratory syndrome coronavirus (MERS-CoV); announcement of the coronavirus study group. J. Virol. 87:7790–7792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization 17 June 2013. Middle East respiratory syndrome coronavirus (MERS-CoV)—update. World Health Organization, Geneva, Switzerland: http://www.who.int/csr/don/2013_06_17/en/index.html [Google Scholar]
- 4.The Health Protection Agency (HPA) UK Novel Coronavirus Investigation Team 2013. Evidence of person-to-person transmission within a family cluster of novel coronavirus infections, United Kingdom, February 2013. Euro Surveill. 18:20427. [DOI] [PubMed] [Google Scholar]
- 5.Memish ZA, Zumla AI, Al-Hakeem RF, Al-Rabeeah AA, Stephens GM. 2013. Family cluster of Middle East respiratory syndrome coronavirus infections. N. Engl. J. Med. 368:2487–2494 [DOI] [PubMed] [Google Scholar]
- 6.Peiris JS, Lai ST, Poon LL, Guan Y, Yam LY, Lim W, Nicholls J, Yee WK, Yan WW, Cheung MT, Cheng VC, Chan KH, Tsang DN, Yung RW, Ng TK, Yuen KY. 2003. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 361:1319–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, Berne MA, Somasundaran M, Sullivan JL, Luzuriaga K, Greenough TC, Choe H, Farzan M. 2003. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426:450–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wong SK, Li W, Moore MJ, Choe H, Farzan M. 2004. A 193-amino acid fragment of the SARS coronavirus S protein efficiently binds angiotensin-converting enzyme 2. J. Biol. Chem. 279:3197–3201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He Y, Lu H, Siddiqui P, Zhou Y, Jiang S. 2005. Receptor-binding domain of severe acute respiratory syndrome coronavirus spike protein contains multiple conformation-dependent epitopes that induce highly potent neutralizing antibodies. J. Immunol. 174:4908–4915 [DOI] [PubMed] [Google Scholar]
- 10.Du L, He Y, Zhou Y, Liu S, Zheng BJ, Jiang S. 2009. The spike protein of SARS-CoV: a target for vaccine and therapeutic development. Nat. Rev. Microbiol. 7:226–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arnold K, Bordoli L, Kopp J, Schwede T. 2006. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22:195–201 [DOI] [PubMed] [Google Scholar]
- 12.Prabakaran P, Gan J, Feng Y, Zhu Z, Choudhry V, Xiao X, Ji X, Dimitrov DS. 2006. Structure of severe acute respiratory syndrome coronavirus receptor-binding domain complexed with neutralizing antibody. J. Biol. Chem. 281:15829–15836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang S, Lu L, Du L, Debnath AK. 2013. A predicted receptor-binding and critical neutralizing domain in S protein of the novel human coronavirus HCoV-EMC. J. Infect. 66:464–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li F, Li W, Farzan M, Harrison SC. 2005. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science 309:1864–1868 [DOI] [PubMed] [Google Scholar]
- 15.Raj VS, Mou H, Smits SL, Dekkers DH, Muller MA, Dijkman R, Muth D, Demmers JA, Zaki A, Fouchier RA, Thiel V, Drosten C, Rottier PJ, Osterhaus AD, Bosch BJ, Haagmans BL. 2013. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature 495:251–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Du L, Zhao G, He Y, Guo Y, Zheng BJ, Jiang S, Zhou Y. 2007. Receptor-binding domain of SARS-CoV spike protein induces long-term protective immunity in an animal model. Vaccine 25:2832–2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Du L, Zhao G, Sun S, Zhang X, Zhou X, Guo Y, Li Y, Zhou Y, Jiang S. 2013. A critical HA1 neutralizing domain of H5N1 influenza in an optimal conformation induces strong cross-protection. PLoS One 8:e53568. 10.1371/journal.pone.0053568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Du L, Zhao G, Chan CC, Sun S, Chen M, Liu Z, Guo H, He Y, Zhou Y, Zheng BJ, Jiang S. 2009. Recombinant receptor-binding domain of SARS-CoV spike protein expressed in mammalian, insect and E. coli cells elicits potent neutralizing antibody and protective immunity. Virology 393:144–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gierer S, Bertram S, Kaup F, Wrensch F, Heurich A, Kramer-Kuhl A, Welsch K, Winkler M, Meyer B, Drosten C, Dittmer U, von HT, Simmons G, Hofmann H, Pohlmann S. 2013. The spike-protein of the emerging betacoronavirus EMC uses a novel coronavirus receptor for entry, can be activated by TMPRSS2 and is targeted by neutralizing antibodies. J. Virol. 87:5502–5511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Du L, Ma C, Jiang S. 2013. Antibodies induced by receptor-binding domain in spike protein of SARS-CoV do not cross-neutralize the novel human coronavirus hCoV-EMC. J. Infect. S0163-4453(13)00111–4. 10.1016/j.jinf.2013.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bisht H, Roberts A, Vogel L, Bukreyev A, Collins PL, Murphy BR, Subbarao K, Moss B. 2004. Severe acute respiratory syndrome coronavirus spike protein expressed by attenuated vaccinia virus protectively immunizes mice. Proc. Natl. Acad. Sci. U. S. A. 101:6641–6646 [DOI] [PMC free article] [PubMed] [Google Scholar]