Abstract
Human immunodeficiency virus type 1 (HIV-1) infection is a significant global public health problem for which development of an effective prophylactic vaccine remains a high scientific priority. Many concepts for a vaccine are focused on induction of appropriate titers of broadly neutralizing antibodies (bNAbs) against the viral envelope (Env) glycoproteins gp120 and gp41, but no immunogen has yet accomplished this goal in animals or humans. One approach to induction of bNAbs is to design soluble, trimeric mimics of the native viral Env trimer. Here, we describe structural studies by negative-stain electron microscopy of several variants of soluble Env trimers based on the KNH1144 subtype A sequence. These Env trimers are fully cleaved between the gp120 and gp41 components and stabilized by specific amino acid substitutions. We also illustrate the structural consequences of deletion of the V1/V2 and V3 variable loops from gp120 and the membrane-proximal external region (MPER) from gp41. All of these variants adopt a trimeric configuration that appropriately mimics native Env spikes, including the CD4 receptor-binding site and the epitope for the VRC PG04 bNAb. These cleaved, soluble trimer designs can be adapted for use with multiple different env genes for both vaccine and structural studies.
INTRODUCTION
The mature human immunodeficiency virus type 1 (HIV-1) envelope (Env) glycoprotein trimer is comprised of three copies of a noncovalently linked gp120/gp41 heterodimer that arises from cleavage of the viral gp160 precursor protein. The env sequence is highly variable as the gene evolves under selective pressures created by several factors, including neutralizing antibodies. The resulting amino acid variability, in addition to an extensively glycosylated surface, creates a formidable barrier for antibody recognition and broad neutralization. Thus, the envelope glycoproteins represent a challenging, moving target for the humoral immune system (1). While broadly neutralizing antibodies (bNAbs) have been isolated from a subset of HIV-1-infected individuals, they play at most a limited role in controlling viral replication as they only arise around 1 to 2 years postinfection (2–5). However, experimental data in animal models suggest that vaccine-induced bNAbs, if present at appropriate titers, would protect people from infection (6–12). Accordingly, many research groups are now trying to design Env-based antigens that might be able to induce bNAbs when used as vaccine immunogens.
While the link between the design and structure of an antigen and the resulting immune response is poorly understood, it is important that Env-based immunogens are carefully characterized at the molecular level using a range of biochemical, biophysical, and structural tools (13) in order for such correlations to be made. One line of approach to an Env vaccine involves testing the hypothesis that presentation of a “native” (i.e., trimeric) form of Env to the immune system is important for generation of a suitable immune response, including the induction of bNAbs. Thus, the immunogen should present many of the complex quaternary epitopes recognized by bNAbs and be glycosylated in a way that mimics the native structure of Env spikes. For practical reasons that aid in production and purification, the immunogen should also be soluble (i.e., secreted from cells) and stable. These requirements are quite challenging, as there is currently no atomic resolution structure of an Env trimer from which to rationally construct such an antigen. Nonetheless, various strategies have been used to create soluble Env trimers with the general goal of mimicking the native spikes on the virus surface (14–20). Most of these approaches have involved eliminating the cleavage site between gp120 and gp41 and adding a trimerization motif (21, 22) to create a more stable trimer. Here, we describe structural characterization of soluble trimers that are made by a different approach which involves retaining the cleavage site but in the presence of trimer-stabilizing substitutions. These soluble trimers are generally designated SOSIP gp140s as they contain an introduced disulfide bond (SOS) between residues A501 and T605 (in the JRFL or HXB2 numbering) that covalently links gp120 and the gp41 ectodomain (gp41ECTO) and a point substitution, I559P (IP), in gp41ECTO to stabilize intratrimer interactions (17, 18).
Recently, we described various structural studies on complexes between bNAbs and SOSIP gp140 trimers based on two different subtype A sequences, KNH1144 and BG505 (23–27). Here, we present the results of additional structural studies on variants of the KNH1144 SOSIP gp140 trimers that contain or lack the V1/V2 and V3 variable loops of gp120 and/or the membrane-proximal external region (MPER) of gp41ECTO. The various trimer forms have been studied both free and as complexes with soluble CD4 (sCD4) and the antigen binding fragment (Fab) of the CD4 binding site-directed (CD4bs) bNAb, VRC PG04. Our results demonstrate that the SOSIP gp140 trimers adopt similar morphologies to the virion-associated Env complexes in the unliganded and CD4-liganded conformations, irrespective of the presence of the V1/V2 and V3 loops and the MPER. The soluble SOSIP trimers, therefore, structurally mimic virally expressed, membrane-embedded Env but can be made as soluble and stable recombinant proteins (28). The MPER-deleted variant, SOSIP.664 gp140, in particular has improved biophysical and solubility properties that make it an attractive candidate for structural and immunogenicity studies, as described further in the accompanying article (29).
MATERIALS AND METHODS
SOSIP constructs.
Construct design, expression, and purification were undertaken as described in the accompanying article (29). The nomenclature SOSIP.681 is used for the full-length SOSIP gp140 where the KNH1144 clade A sequence terminates at residue 681. A 17-residue, C-terminal truncation of this construct produces SOSIP.664, which lacks the MPER. Replacement of the V1/V2 and V3 loops in each of these constructs with GGG linkers produces the SOSIP.681.ΔV1V2V3 and SOSIP.664.ΔV1V2V3 constructs, respectively.
SOSIP complex formation and purification.
The various SOSIP constructs were purified using Superose 6 (GE Healthcare Life Sciences) size exclusion chromatography (SEC) either unliganded or in complex with VRC PG04 Fab. For complex formation, ligands were added in 6× molar excess to the SOSIP gp140 protomer and incubated on ice for 15 min prior to purification. Complexes with soluble CD4 (sCD4) were incubated for 4 h on ice and not subjected to further SEC purification, as these complexes would dissociate during the purification, possibly due to dilution of the samples as they flowed over the column. Comparable samples consisting of the SOSIP.664 construct were purified in a buffer containing 20 mM Tris-HCl, 150 mM sodium chloride (pH 8.0), whereas purification of samples containing the SOSIP.681 construct was performed in the same buffer supplemented with 0.025% (wt/vol) CYMAL-7 to avoid any aggregation due to the hydrophobic MPER. Eluting fractions containing the desired components as verified by SDS-PAGE and blue native (BN)-PAGE were pooled and subjected to structural characterization.
Electron microscopy.
Samples possessing CYMAL-7 were treated with Bio-Beads (Bio-Rad, CA) for 2 h prior to grid preparation to remove excess detergent. Negatively stained grids were prepared as previously described (23). Briefly, ∼0.1 mg/ml of the purified SOSIP gp140 (liganded or free) was applied to a freshly glow-discharged, carbon-coated grid and stained with 2% uranyl formate. Grids were viewed using a FEI Tecnai TF20 electron microscope operating at a magnification of 100,000× and an accelerating voltage of 120 kV. Images were acquired on a Gatan 4,096-by-4,096 charge-coupled device (CCD) camera in 5° increments from 0 to 55° tilt angles at a defocus range of −600 to −720 nm and a dose of less than 16 e−/Å2 using the Leginon system (30). The tilt angles provided additional particle orientations to improve the image reconstructions. The pixel size of the CCD camera was calibrated at this magnification to be 1.09 Å using a two-dimensional (2D) catalase crystal with known unit cell parameters.
Data collection and image reconstruction.
Particles were automatically selected from micrographs with DoG Picker (31). Contrast transfer function (CTF) estimations for the untilted and tilted micrographs were determined with ctffind3 and ctftilt (32). Particles were binned in groups of 4 (80-by-80 boxes), and reference-free 2D class averages were calculated using the Sparx package (33). Class averages and their associated particles were discarded based on their morphology and quality. In particular, particles that were too small (Fabs or sCD4 alone) and too large (aggregates) to be Env trimer complexes were removed at this point. Image reconstructions were generated using the angular reconstitution method to minimize model bias. For the SOSIP.681 gp140 constructs, the micelle region had to be masked out for the image reconstruction to succeed, as follows: (i) align all raw particles to the reference-free class averages, (ii) mask out the micelle from the class averages and the raw particles using the e2proc2d.py procedure of EMAN2 (34), (iii) recalculate reference-free 2D class averages, (iv) generate 40 ab initio models from the micelle-excluded, reference-free 2D class averages using the EMAN2 package, (v) refine each model against the kept reference-free 2D class averages with the Sparx package (33), (vi) select the best model and refine against the micelle-excluded raw particles with the Sparx package, and (vii) apply the alignment parameters of the micelle-excluded raw particles to the unmasked particles (note that the micelle-excluded particles are in the same orientation as the unmasked particles) and back-project using the sxrecons3d_n.py routine in Sparx to generate an image reconstruction (33, 34). For liganded SOSIP trimers, a similar procedure was used. The ab initio model exhibiting Fab or sCD4-like density served as the initial model for iterative image reconstruction against the CTF-corrected particles using Sparx (33). For unliganded trimers, a comparison of projections from the model to the reference-free class averages was used as the criterion to identify the initial model for iterative image reconstruction. The presence of micelles in the final image reconstructions of SOSIP.681 gp140 trimers supports each image reconstruction, as the micelles were excluded from the image reconstruction procedures and only included for the final back-projection step. This process is akin to an omit map in X-ray crystallography and should be applicable to any system. The resolutions of the final image reconstructions, as determined by a Fourier shell correlation (FSC) of 0.5, ranged between 14 and 20 Å and are listed in Table 1. For comparisons between reconstructions, maps were always filtered to the lowest resolution within the set.
Table 1.
Image reconstruction detailsa
| Complex (SOSIP) | No. of particles | Resolution (Å)b | EMDB accession no. |
|---|---|---|---|
| 681 | 5,543 | 18 | EMD-5700 |
| 681 (ΔV1/V2, V3) | 10,838 | 17 | EMD-5701 |
| 681 + VRC PG04 | 14,325 | 15 | EMD-5702 |
| 681 (ΔV1/V2, V3) + VRC PG04 | 9,248 | 18 | EMD-5703 |
| 681 + sCD4 | 4,506 | 22 | EMD-5704 |
| 664c | 15,248 | 14 | EMD-5705 |
| 664 + VRC PG04 | 12,236 | 15 | EMD-5706 |
| 664 (ΔV1/V2, V3) + VRC PG04 | 11,958 | 19 | EMD-5707 |
| 664 + sCD4 | 16,704 | 16 | EMD-5708 |
| 664 (ΔV1/V2, V3) + sCD4 | 12,877 | 20 | EMD-5709 |
| HIV BaL Envd | 4,7421 | 25 | EMD-5022 |
| HIV BaL Env + sCD4e | NAf | 24 | EMD-5455 |
| HIV BaL Env + VRC03e | NA | 23 | EMD-5458 |
Shown are the images reconstructed, the number of particles used to generate them, their resolution, and the EMDB accession codes. Also tabulated are the image reconstructions and their respective resolutions reported earlier (15, 24, 40).
Resolution based on a Fourier shell correlation (FSC) of 0.5.
See reference 15.
See reference 24.
See reference 40.
NA, not available.
The contour levels of the VRC PG04-liganded image reconstructions were selected at the highest threshold to fully accommodate the crystal structure of the VRC PG04 Fab component. Thus, we used the molecular envelope of the Fab crystal structure as a standard. The contour levels of the unliganded image reconstructions were selected according to the quality of their fit to their corresponding liganded image reconstructions to maximize volume overlap. This procedure normalizes the thresholds of all reconstructions to one another and that of the VRC PG04 Fab.
Model fitting and enantiomer detection of image reconstructions.
The model fitting and enantiomer detection procedures have been described previously (35, 36). The correct enantiomer of the image reconstruction was determined by fitting the crystal structures of HIV-1 gp120 liganded to the VRC PG04 Fab (Protein Data Bank [PDB] entry 3SE9), HIV-1 gp120 liganded to sCD4 (PDB entry 1GC1), and gp120 core structure (gp120core) (PDB entry 3TIH) into each enantiomer filtered to the same resolution (Table 1) using the program Molrep with C3 symmetry (37). The structures fit the same-handed enantiomer with a higher correlation coefficient than the opposite enantiomer. The V1/V2 (PDB entry 3U2S) and V3 loops were manually docked into the image reconstructions using the start and end residues of each structure as the anchor points for connecting to the computationally docked gp120core. Docking was performed in UCSF Chimera.
Map segmentation.
Maps were segmented using the Split Map function of the Color Zone tool in UCSF Chimera (38). Briefly, the gp120core structure (PDB entry 3TIH) was superimposed onto the different EM-fitted gp120 structures for consistency. This procedure was done to eliminate any differences between the gp120 constructs that were used in the different crystallization experiments. A 12-Å mask radius was used around each atom of the docked crystal structure, and the volume was segmented using the Split Map function. The same procedure was used for segmenting the regions of the manually fitted V1/V2 and V3 loops.
Protein structure accession numbers.
The EM reconstructions have been deposited in the Electron Microscopy Data Bank (EMDB) under accession code no. EMD-5700, EMD-5701, EMD-5702, EMD-5703, EMD-5704, EMD-5705, EMD-5706, EMD-5707, EMD-5708, and EMD-5709.
RESULTS
Soluble SOSIP gp140 trimers resemble Env spikes on the surface of HIV-1.
Here, we present comparisons of SOSIP gp140 trimer reconstructions based on the subtype A sequence KNH1144 that was terminated at either residue 681 or 664 in gp41ECTO. These two trimers are designated SOSIP.681 and SOSIP.664 gp140s (Fig. 1). The rationale for the deletion of the MPER from the SOSIP.664 construct, together with biophysical and antigenicity data, is described in the accompanying article (29).
Fig 1.
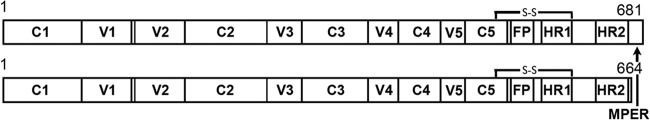
Schematic representation of the SOSIP constructs used in this study. The gp120 constant domains are labeled C1 to C5, while the variable loops are labeled V1 to V5. A disulfide bond (SOS) stabilizes the gp120-gp41 interaction. The fusion peptide is labeled FP, while the heptad repeat 1 (HR1), heptad repeat 2 (HR2), and the membrane-proximal region (MPER) are also indicated.
The image reconstruction of the SOSIP.681 gp140 trimers can be segmented into three regions: (i) a “body” that describes the molecular envelope for the three gp120cores, (ii) a “waist” that describes the molecular envelope connecting the “body” to the “stem,” and (iii) a stem that describes what is likely the C-terminal region of gp41ECTO (Fig. 2) along with the molecular envelope for a micelle presumed to have formed from the detergent CYMAL-7, which was added to prevent aggregation of the SOSIP.681 trimers into rosettes, via the hydrophobic MPER (see Fig. S1 and S2 in the supplemental material). Comparison of the EM image reconstruction of the soluble SOSIP.681 trimers using single-particle analysis with the published cryo-electron tomography (cryo-ET) reconstruction of viral surface Env (39) (EMD-5022) reveals that both reconstructions (Fig. 2) are similar, considering the differences in their reported resolutions (Table 1) and distinct methods used to obtain them. When viewed down the 3-fold axis of symmetry of the image reconstruction with the viral membrane distal to the viewer, the two image reconstructions are of comparable sizes and both exhibit morphologies similar to a three-bladed fan. A side view of the image reconstructions reveals a bulbous density attributed to the gp120core narrowing to a “waist” at the putative gp120-gp41 interaction site. The HIV-1 viral membrane bilayer region observed in the cryo-ET image reconstruction is recapitulated as a micelle in the detergent-solubilized SOSIP.681 gp140 single-particle reconstruction. Nonetheless, some minor differences can also be seen between the two image reconstructions. For example, the virion-associated Env is somewhat more elongated, is wider, and has more of a twist to the gp120 portion. These variations may arise from genuine differences between soluble and virion-associated forms of Env trimers or from image reconstruction artifacts, such as the missing wedge that is present in cryo-ET data or stain artifacts that may occur in negative-stain EM experiments. Overall, we conclude that, at the limited resolution of these image reconstructions, the soluble SOSIP gp140 trimers are very similar in appearance to Env spikes present on HIV-1 virus particles.
Fig 2.
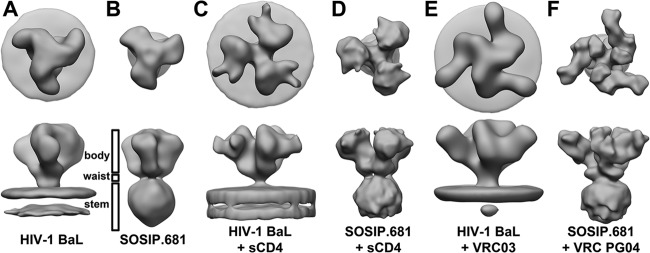
Comparison of SOSIP constructs to HIV-decorated Env trimers. Shown is the surface representation of the SOSIP image reconstructions compared to the published HIV-1 surface Env trimers. (Top view) Looking down the 3-fold axis of each trimer (with the viral membrane in the distance). (Side view) Looking parallel to the viral membrane at the bottom of the figure. (A) Unliganded HIV-1 BaL (EMD-5022); (B) SOSIP.681 filtered to 25-Å resolution; (C) HIV-1 BaL liganded to sCD4 (EMD-5455); (D) SOSIP.681 liganded to sCD4 filtered to 25-Å resolution; (E) HIV-1 BaL liganded to VRC03 (EMD-5458); (F) SOSIP.681 liganded to VRC PG04 filtered to 15-Å resolution. Labeled are the body, waist, and stem of the SOSIP.681 image reconstructions (see the text).
We further characterized the detergent-solubilized SOSIP.681 gp140 trimers by generating image reconstructions in complex with either sCD4 or VRC PG04 and compared them to the published structures of viral surface Env complexes with sCD4 and VRC03, which recognizes a similar epitope to VRC PG04 (40, 41) (EMD-5455 and EMD-5458) (Fig. 2C to F). While sCD4 is not clearly distinguished in the EM reconstruction here, the structure of the trimer adopts an opened phenotype similar to that of viral surface Env. The binding of sCD4 to either form of Env induces nearly indistinguishable conformational changes in the trimers, specifically a 15 to 20° rotation out of plane and a 45 to 50° rotation around an axis parallel to the 3-fold axis (see Fig. S3E and S4A in the supplemental material). These results suggest that the stabilizing mutations introduced in the SOSIP trimers do not prevent previously observed conformational changes from occurring. Binding of VRC03 to Env and VRC PG04 (PGV04) to SOSIP.681 produces similar image reconstructions but without the large conformational changes that occur upon CD4 binding. Notably, more definition is observed in the PGV04 image reconstruction allowing unambiguous docking to the VRC PG04 Fab crystal structure (see Fig. S3C).
V1/V2 and V3 variable loops are located at the apex of the SOSIP gp140 trimer.
To identify the locations of the V1/V2 and V3 loops in the SOSIP gp140 trimer and to distinguish whether they interact via a cis (same protomer) or trans (adjacent protomer) mechanism on the membrane-distal surface of gp120, we generated an image reconstruction of SOSIP.681.ΔV1V2V3 gp140 trimers and compared it to SOSIP.681. In the SOSIP.681.ΔV1V2V3 gp140 construct, each of the V1/V2 and V3 loops was replaced with a polyglycine (GGG) sequence, as described in the accompanying article (29). The image reconstructions yield similar molecular envelopes, and subsequent fitted gp120cores therefore allow us to identify the locations of the V1/V2 and V3 loops (Fig. 3A and B and 4A and B). Rigid-body fitting of the gp120core crystal structure into each image reconstruction supports an interpretation of the EM data (see Fig. S3 in the supplemental material) in which the V1/V2 loops localize to the apex of the SOSIP gp140 trimers, while V3 forms a juxtaposed bump that protrudes from the outer domain of the gp120core. The density that we ascribe to V3 appears to primarily associate with the gp120 protomer from which it emanates in a cis mode. The V1/V2 density for all the protomers forms a continuous cap on the trimer, suggestive of a cis interaction between these domains at the periphery of the trimer and a trans interaction near the axis of symmetry. Given the limited resolution of our image reconstruction (Table 1), we cannot make a definitive conclusion about the quaternary interactions and precise extent of the V1/V2 and V3 loops, but our observations on the SOSIP gp140 trimer are consistent with those drawn from several previous studies of Env (28, 42–44).
Fig 3.
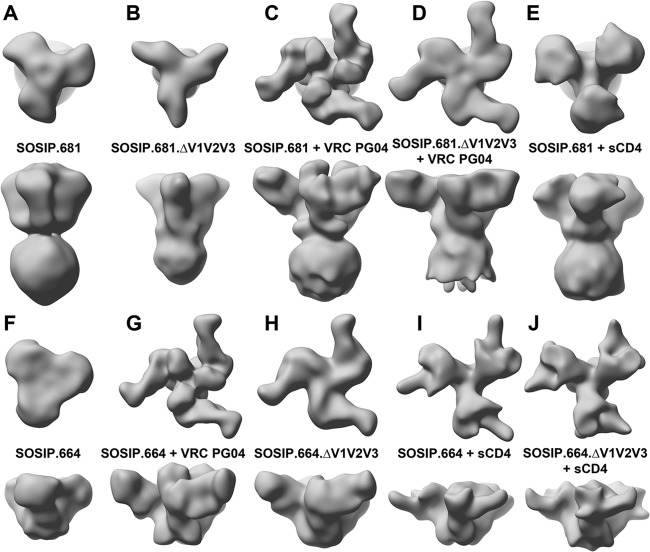
Comparison of SOSIP image reconstructions. Shown is the surface representation of image reconstructions for the various liganded and unliganded SOSIP.681 and SOSIP.664 low-pass filtered to 22-Å resolution. Shown are the top and side views of (A) SOSIP.681, (B) SOSIP.681.ΔV1V2V3, (C) SOSIP.681 liganded to VRC PG04, (D) SOSIP.681.ΔV1V2V3 liganded to VRC PG04, (E) SOSIP.681 liganded to sCD4, (F) SOSIP.664, (G) SOSIP.664 liganded to VRC PG04, (H) SOSIP.664.ΔV1V2V3 liganded to VRC PG04, (I) SOSIP.664 liganded to sCD4, and (J) SOSIP.664.ΔV1V2V3 liganded to sCD4.
Fig 4.
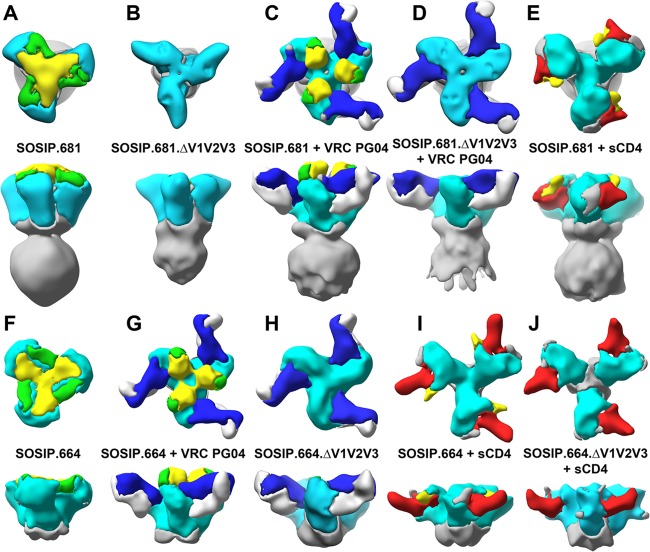
Comparison of segmented SOSIP image reconstructions. Surface representation of image reconstructions segmented using the docked crystal structures of gp120core, V1/V2 and V3 loops, gp120core liganded to VRC PG04, and gp120core liganded to sCD4. The image layout is identical to Fig. 3. The coloring is as follows: gp120core, cyan; V1/V2, yellow; V3, green; VRC PG04 heavy chain, blue; VRC PG04 light chain, white; and gp41 and termini of gp120, gray. See Materials and Methods for details.
Comparison of the positions and orientations of the gp120core crystal structure fitted into each model (see Materials and Methods) did not identify any significant difference between the positioning or orientation of the gp120core protomers in the two image reconstructions. This outcome contrasts with earlier cryo-ET reconstructions of HIV-1 particles bearing Env trimers lacking the V1/V2 loop, where the gp120cores appeared to adopt multiple orientations that significantly deviated from their orientation in full-length Env trimmers (28). The discrepant outcomes may arise because of the trimer-stabilizing changes engineered into the SOSIP gp140 constructs, different Env sequences, soluble versus membrane-bound trimers, and/or the presence of the V3 loops in the virion-associated trimers compared to their absence from the soluble trimmers (28).
VRC PG04 bNAb binds the SOSIP gp140 trimer irrespective of the presence of the MPER and the V1/V2 and V3 loops.
Recent structural studies of viral membrane Env spikes showed that, while the VRC01 and VRC03 bNAbs recognized their epitopes in the CD4 binding site, they did not trigger the conformational changes that are induced by CD4 binding that open up the trimer (40). Atomic resolution studies of VRC01-like Fab-gp120 complexes reveal that VRC01, VRC03, and VRC PG04 all share a highly similar binding interface with gp120 (41). Thus, we sought to characterize how VRC PG04 bound to SOSIP KNH1144 gp140 trimers (Fig. 3C and D and G and H). In the liganded image reconstruction, there is stronger definition of the V1/V2 regions at the trimer apex, which appears in the form of a triumvirate of towers (Fig. 3C and G). These well-defined image reconstructions allowed unambiguous docking of the VRC PG04 Fab-gp120 X-ray structure (PDB entry 3SE9). Binding of VRC PG04 causes a small (∼17°) clockwise rotation of the SOSIP.681 gp120cores around the 3-fold axis (Fig. 3; see Fig. S3C and S4B in the supplemental material) and translation closer to the symmetry axis relative to the gp120 positions in the unliganded SOSIP.681 reconstruction (Fig. 3C).
The “waist” (i.e., the molecular envelope connecting the gp120 “body” with its associated micelle “stem”) becomes enlarged in the liganded image reconstruction. This outcome is accompanied by a change in the envelope near the base of the “body” (Fig. 3A and C). Therefore, the interaction between SOSIP.681 gp140 trimers and VRC PG04 may not simply be restricted to a small rigid-body motion of the gp120cores but may also involve a rigid body or conformational change in the gp140 waist region. Such differences, which have not been described in similar studies performed on envelope-embedded Env proteins (40), may be attributed to the improvement in the resolution of our image reconstructions or authentic differences between membrane-embedded Env and our soluble constructs.
To investigate whether the observed triumvirate of towers assumed to be the V1/V2 and closely associated V3 loops in the SOSIP.681-VRC PG04 complex play a structural role, we generated additional image reconstructions of SOSIP.681.ΔV1V2V3 liganded to VRC PG04 (Fig. 3D). Comparison of the liganded and unliganded SOSIP.681.ΔV1V2V3 image reconstructions showed that the gp120cores undergo similar rotations and translations in both cases (see Fig. S3 in the supplemental material). Hence, the interaction between VRC PG04 and the SOSIP.681 gp140 trimers is not dependent on the presence of the V1/V2 and V3 loops. An ∼8° clockwise rotation in the plane of the paper (i.e., perpendicular to the 3-fold axis) can be seen in the gp120cores when comparing the side views of the liganded SOSIP.681 and SOSIP.681.ΔV1V2V3. We attribute this difference to deletion of the V1/V2 and V3 regions (Fig. 3D).
Next, we generated an image reconstruction of SOSIP.664 gp140 trimers liganded to VRC PG04 to study the influence of the MPER on this interaction. A comparison to the VRC PG04-liganded SOSIP.681 image reconstruction indicates that the gp120cores rotate slightly more in the MPER-deleted trimers, while moving closer to the axis of symmetry (Fig. 3G; see Fig. S3 and S4B in the supplemental material). Hence, the interaction between the MPER and the gp120core, whether direct or indirect, slightly restricts the degree to which the gp120core can rotate when bound to VRC PG04. The towers seen with the SOSIP.681 gp140 trimers are also formed with SOSIP.664 (i.e., in the absence of the MPER), but their closer proximity to one another reinforces the conclusion that the MPER may play a small role in the overall architecture of the SOSIP gp140 trimer (Fig. 3C and G). Alternatively, the presence of detergent and/or micelle in the SOSIP.681 image reconstruction may be responsible for the observed difference. We are currently investigating this possibility.
For a final study, we generated an image reconstruction of the SOSIP.664.ΔV1V2V3 gp140 trimers bound to VRC PG04 (Table 1 and Fig. 3H). A comparison to the image reconstruction of the liganded SOSIP.664 gp140 (i.e., with intact variable loops) indicates that the gp120cores now rotate by approximately an additional 10° clockwise around the axis perpendicular to the 3-fold axis (see Fig. S4C in the supplemental material). These results, in combination with those from the SOSIP.681 gp140 studies described above, indicate that the overall interactions of VRC PG04 with the SOSIP gp140 trimers at this level of resolution are only slightly modified when the V1/V2 and V3 loops and MPER domains are absent.
Binding of sCD4 results in an opening of the SOSIP trimers independent of the V1/V2 and V3 loops.
Binding of sCD4 to Env has been shown to induce a large conformational change in both the position and orientation of the gp120cores with respect to one another (39). Preliminary image analysis also indicated that the SOSIP trimer was capable of binding sCD4 (29, 45). To further investigate the effect of sCD4 on our SOSIP constructs, we generated image reconstructions of SOSIP.681, SOSIP.664, and SOSIP.664.ΔV1V2V3 liganded to sCD4 (Fig. 3E, I, and J). Analysis of all liganded and unliganded reconstructions reveals that sCD4 is able to open up all variants of SOSIP regardless of whether the V1/V2 and V3 or the MPER regions are present. This observation suggests that binding of sCD4 most likely alters the interaction between the gp120core and gp41 that leads to the open conformation of Env.
Cross talk between gp41 and gp120.
Collectively, our reconstructions presented here and elsewhere suggest that the interactions between gp120 and gp41, and possibly the conformation of gp41 itself, differ when the V1/V2 and V3 loops are deleted from SOSIP Env trimers (Fig. 3), as is particularly evident in the image reconstructions of MPER-containing SOSIP trimers. Upon deletion of the V1/V2 and V3 loops, the molecular envelope defining the “waist” become less discrete, while the size of the micelle in the image reconstruction becomes reduced, as observed in comparisons of the unliganded SOSIP.681 to SOSIP.681.ΔV1V2V3 and VRC PG04-liganded SOSIP.681 to SOSIP.681.ΔV1V2V3 (Fig. 3A to D). Thus, small conformational changes may arise upon deletion of these loops. We note that unliganded SOSIP.664 does not exhibit as prominent a V1/V2/V3 density as the unliganded SOSIP.681 image reconstruction, but the corresponding PG VRC04-liganded reconstructions are very similar in V1/V2/V3 (Fig. 3). This observation supports earlier biochemical studies suggesting that cross talk exists between the V1/V2 and V3 and MPER and that modification of either region could affect the binding of antibodies to the other (46, 47). These findings are also consistent with a recent study illustrating that an MPER binding antibody can cause the trimer to adopt an open conformation, similar to the CD4-bound state (48).
While it appears that the presence of the MPER may result in subtle changes in the trimer, SOSIP.664 retains a similar shape and reactivity with VRC PG04 and other broadly neutralizing antibodies (23, 25, 27), as well as sCD4. Thus, the SOSIP mutations allow removal of the transmembrane domain and MPER without compromising the structural or antigenic profile of the trimer.
DISCUSSION
Currently, there is a tremendous drive to develop immunogens for vaccine development that mimic the native HIV-1 Env. Here we describe a series of structural studies using negative-stain electron microscopy that evaluate and characterize soluble versions of HIV-1 Env trimers in the presence or absence of ligands that bind to the CD4 binding site. These studies are focused on understanding how two specific regions of the Env, namely, the combined V1/V2 and V3 loops and MPER, affect the architecture and organization of Env in the absence and presence of ligands.
Our studies illustrate that SOSIP.681, a stabilized and soluble Env trimer possessing a disulfide between gp120/gp41 and a single point mutation, I559P, folds up into a quaternary structure that is similar to that described for native virally displayed HIV-1 Env (39). Moreover, we demonstrate that SOSIP.681 adopts conformations that are virtually indistinguishable from the virally displayed HIV-1 Env in the presence of sCD4 and VRC PG04 (40) (Fig. 2). Thus, SOSIP.681 appears to be a suitable platform for immunogen design. We also demonstrate that while the V1/V2 and V3 and MPER have subtle effects on the organization of the trimer, their deletion does not alter the ability of the SOSIP trimer to bind sCD4 or VRC PG04. Removal of MPER from SOSIP.681 to produce SOSIP.664 results in a minor rotational as well as positional shift in the gp120cores, whereas deletion of the V1/V2 and V3 loops from SOSIP.681 does not alter the orientation or position of the gp120cores relative to one another (see Fig. S3 in the supplemental material). This observation, along with our docking of available gp120 crystal structures, suggests that non-MPER gp41 plays an important role in the organization and position of gp120s in the trimer, likely via an extensive interaction surface. Additionally, we have further shown that these two regions play a limited role in how sCD4 or CD4bs antibodies (VRC PG04) affect the organization of SOSIP. For example, the gp120cores of SOSIP.664.ΔV1V2V3 undergo a similar conformational change as that of SOSIP.681 upon binding to sCD4 or VRC PG04 (Fig. 2 and 3). This result also suggests that the conformational changes observed in Env that alter the gp120-gp41 interaction can occur independently of these two regions. Conversely, while deletion of the V1/V2 and V3 loops does not alter the overall organization of Env, the micelles associated with the SOSIP.681 image reconstructions appear to be more intimately associated with the “body” of SOSIP when V1/V2 and V3 have been removed; hence, the “waist” of this construct appears less defined.
The increased definition in our EM reconstructions at the apex of the trimer as a result of VRC PG04 binding may be due to decreased flexibility in the V1/V2 loops induced by the ligand. Recent biochemical studies that show CD4bs antibodies, such as VRC01, VRC03, and VRC PG04, inhibit binding of CD4-induced (CD4i) antibodies, such as 17b (49). Docking the crystal structure of the gp120core liganded to CD4 and 17b Fab (PDB entry 1RZK) into our VRC PG04 liganded SOSIP.681 and SOSIP.664 image reconstructions clearly demonstrates that 17b Fab would clash with the V1/V2 and V3 towers when VRC PG04 binds to Env (see Fig. S5 in the supplemental material). In the CD4 liganded state, these towers appear to become orthogonal to the Env 3-fold axis of symmetry or change conformation such that they no longer clash with the CD4i antibodies (Fig. 4E and I). Our studies also demonstrate that the V1/V2 and V3 loops of a gp120 protomer predominantly interact in cis but also in trans near the 3-fold axis of symmetry.
While we observe that the MPER has some subtle structural effect on the gp120s, this region can be added or removed without affecting the major antigenic sites on gp120, including the CD4 binding site, as well as the N-linked glycans at N332 and N160 (23, 25–27). This result demonstrates the increased utility of the SOSIP trimer as it is soluble, does not aggregate, and is easier to express and purify. Because of this ease of handling, this platform can be readily modified by mutation to display epitopes designed to elicit particular bNAbs and provide the quaternary restraints necessary to guide an appropriate immune response. Additionally, due to the homogeneity achieved during purification, the SOSIP trimer is ideally suited for further structural investigation of bNAb complexes. In conclusion, we have systematically dissected the structural roles of the HIV-1 Env V1/V2 and V3 loops and the MPER and demonstrated that the soluble SOSIP gp140 trimer has a very similar appearance to virion-associated Env at the resolution reported here. Furthermore, the two forms of Env undergo similar conformational changes in the presence of CD4 or the CD4bs antibody VRC PG04. The stability provided to the soluble trimer by the SOSIP mutations should have a beneficial effect for immunogenicity, as it will reduce the exposure of epitopes that may elicit nonneutralizing antibodies. Studies to explore the antigenicity and immunogenicity of a SOSIP.664 gp140 construct based on a different subtype A sequence, BG505, are now in progress, as are higher-resolution structural explorations.
Supplementary Material
ACKNOWLEDGMENTS
We thank Y. Hua, L. Kong, and R. S. Stanfield for technical assistance and W. C. Koff and D. R. Burton for valuable discussions.
This work was supported by NIH grants HIVRAD P01 AI82362, R37 AI36082, and R01 AI84817, the International AIDS Vaccine Initiative Neutralizing Antibody Center, Scripps CHAVI-ID (UM1 AI100663), the University of California, San Diego Center for AIDS Research (CFAR), an NIH-funded program (P30 AI036214) supported by the NIH Institutes and Centers NIAID, NCI, NIMH, NIDA, NICHD, NHLBI, and NIA, a Vidi grant from the Netherlands Organization for Scientific Research (R.W.S.), a Starting Investigator Grant from the European Research Council (R.W.S.), and a Canadian Institutes of Health Research fellowship (J.-P.J.). The three-dimensional reconstructions were conducted at the National Resource for Automated Molecular Microscopy, which is supported by NIH through the National Center for Research Resources' P41 program (RR017573).
Images were generated using the UCSF Chimera package.
Footnotes
Published ahead of print 3 July 2013
This article is contribution 24023 from The Scripps Research Institute.
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.01222-13.
REFERENCES
- 1.Karlsson Hedestam GB, Fouchier RAM, Phogat S, Burton DR, Sodroski J, Wyatt RT. 2008. The challenges of eliciting neutralizing antibodies to HIV-1 and to influenza virus. Nat. Rev. Microbiol. 6:143–155 [DOI] [PubMed] [Google Scholar]
- 2.Simek MD, Rida W, Priddy FH, Pung P, Carrow E, Laufer DS, Lehrman JK, Boaz M, Tarragona-Fiol T, Miiro G, Birungi J, Pozniak A, McPhee DA, Manigart O, Karita E, Inwoley A, Jaoko W, Dehovitz J, Bekker L-G, Pitisuttithum P, Paris R, Walker LM, Poignard P, Wrin T, Fast PE, Burton DR, Koff WC. 2009. Human immunodeficiency virus type 1 elite neutralizers: individuals with broad and potent neutralizing activity identified by using a high-throughput neutralization assay together with an analytical selection algorithm. J. Virol. 83:7337–7348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doria-Rose NA, Klein RM, Manion MM, O'Dell S, Phogat A, Chakrabarti B, Hallahan CW, Migueles SA, Wrammert J, Ahmed R, Nason M, Wyatt RT, Mascola JR, Connors M. 2009. Frequency and phenotype of human immunodeficiency virus envelope-specific B cells from patients with broadly cross-neutralizing antibodies. J. Virol. 83:188–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sather DN, Armann J, Ching LK, Mavrantoni A, Sellhorn G, Caldwell Z, Yu X, Wood B, Self S, Kalams S, Stamatatos L. 2009. Factors associated with the development of cross-reactive neutralizing antibodies during human immunodeficiency virus type 1 infection. J. Virol. 83:757–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stamatatos L, Morris L, Burton DR, Mascola JR. 2009. Neutralizing antibodies generated during natural HIV-1 infection: good news for an HIV-1 vaccine? Nat. Med. 15:866–870 [DOI] [PubMed] [Google Scholar]
- 6.Burton DR, Poignard P, Stanfield RL, Wilson IA. 2012. Broadly neutralizing antibodies present new prospects to counter highly antigenically diverse viruses. Science 337:183–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conley AJ, Kessler JA, Boots LJ, McKenna PM, Schleif WA, Emini EA, Mark GE, Katinger H, Cobb EK, Lunceford SM, Rouse SR, Murthy KK. 1996. The consequence of passive administration of an anti-human immunodeficiency virus type 1 neutralizing monoclonal antibody before challenge of chimpanzees with a primary virus isolate. J. Virol. 70:6751–6758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hessell AJ, Poignard P, Hunter M, Hangartner L, Tehrani DM, Bleeker WK, Parren PWHI, Marx PA, Burton DR. 2009. Effective, low-titer antibody protection against low-dose repeated mucosal SHIV challenge in macaques. Nat. Med. 15:951–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hessell AJ, Rakasz EG, Poignard P, Hangartner L, Landucci G, Forthal DN, Koff WC, Watkins DI, Burton DR. 2009. Broadly neutralizing human anti-HIV antibody 2G12 is effective in protection against mucosal SHIV challenge even at low serum neutralizing titers. PLoS Pathog. 5:e1000433. 10.1371/journal.ppat.1000433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mascola JR. 2003. Defining the protective antibody response for HIV-1. Curr. Mol. Med. 3:209–216 [DOI] [PubMed] [Google Scholar]
- 11.Klein F, Halper-Stromberg A, Horwitz JA, Gruell H, Scheid JF, Bournazos S, Mouquet H, Spatz LA, Diskin R, Abadir A, Zang T, Dorner M, Billerbeck E, Labitt RN, Gaebler C, Marcovecchio PM, Incesu R-B, Eisenreich TR, Bieniasz PD, Seaman MS, Bjorkman PJ, Ravetch JV, Ploss A, Nussenzweig MC. 2012. HIV therapy by a combination of broadly neutralizing antibodies in humanized mice. Nature 492:118–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hessell AJ, Rakasz EG, Tehrani DM, Huber M, Weisgrau KL, Landucci G, Forthal DN, Koff WC, Poignard P, Watkins DI, Burton DR. 2010. Broadly neutralizing monoclonal antibodies 2F5 and 4E10 directed against the human immunodeficiency virus type 1 gp41 membrane-proximal external region protect against mucosal challenge by simian-human immunodeficiency virus SHIVBa-L. J. Virol. 84:1302–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schief WR, Ban Y-EA, Stamatatos L. 2009. Challenges for structure-based HIV vaccine design. Curr. Opin. HIV AIDS 4:431–440 [DOI] [PubMed] [Google Scholar]
- 14.Wyatt R. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280:1884–1888 [DOI] [PubMed] [Google Scholar]
- 15.Depetris RS, Julien J-P, Khayat R, Lee JH, Pejchal R, Katpally U, Cocco N, Kachare M, Massi E, David KB, Cupo A, Marozsan AJ, Olson WC, Ward AB, Wilson IA, Sanders RW, Moore JP. 2012. Partial enzymatic deglycosylation preserves the structure of cleaved recombinant HIV-1 envelope glycoprotein trimers. J. Biol. Chem. 287:24239–24254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma B-J, Alam SM, Go EP, Lu X, Desaire H, Tomaras GD, Bowman C, Sutherland LL, Scearce RM, Santra S, Letvin NL, Kepler TB, Liao H-X, Haynes BF. 2011. Envelope deglycosylation enhances antigenicity of HIV-1 gp41 epitopes for both broad neutralizing antibodies and their unmutated ancestor antibodies. PLoS Pathog. 7:e1002200. 10.1371/journal.ppat.1002200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Binley JM, Sanders RW, Clas B, Schuelke N, Master A, Guo Y, Kajumo F, Anselma DJ, Maddon PJ, Olson WC, Moore JP. 2000. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J. Virol. 74:627–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanders RW, Vesanen M, Schuelke N, Master A, Schiffner L, Kalyanaraman R, Paluch M, Berkhout B, Maddon PJ, Olson WC, Lu M, Moore JP. 2002. Stabilization of the soluble, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J. Virol. 76:8875–8889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dey AK, David KB, Lu M, Moore JP. 2009. Biochemical and biophysical comparison of cleaved and uncleaved soluble, trimeric HIV-1 envelope glycoproteins. Virology 385:275–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beddows S, Franti M, Dey AK, Kirschner M, Iyer SPN, Fisch DC, Ketas T, Yuste E, Desrosiers RC, Klasse P-J, Maddon PJ, Olson WC, Moore JP. 2007. A comparative immunogenicity study in rabbits of disulfide-stabilized, proteolytically cleaved, soluble trimeric human immunodeficiency virus type 1 gp140, trimeric cleavage-defective gp140 and monomeric gp120. Virology 360:329–340 [DOI] [PubMed] [Google Scholar]
- 21.Pancera M, Lebowitz J, Schön A, Zhu P, Freire E, Kwong PD, Roux KH, Sodroski J, Wyatt R. 2005. Soluble mimetics of human immunodeficiency virus type 1 viral spikes produced by replacement of the native trimerization domain with a heterologous trimerization motif: characterization and ligand binding analysis. J. Virol. 79:9954–9969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang X, Lee J, Mahony EM, Kwong PD, Wyatt R, Sodroski J. 2002. Highly stable trimers formed by human immunodeficiency virus type 1 envelope glycoproteins fused with the trimeric motif of T4 bacteriophage fibritin. J. Virol. 76:4634–4642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pejchal R, Doores KJ, Walker LM, Khayat R, Huang P-S, Wang S-K, Stanfield RL, Julien J-P, Ramos A, Crispin M, Depetris R, Katpally U, Marozsan A, Cupo A, Maloveste S, Liu Y, McBride R, Ito Y, Sanders RW, Ogohara C, Paulson JC, Feizi T, Scanlan CN, Wong C-H, Moore JP, Olson WC, Ward AB, Poignard P, Schief WR, Burton DR, Wilson IA. 2011. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science 334:1097–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harris A, Borgnia MJ, Shi D, Bartesaghi A, He H, Pejchal R, Kang YK, Depetris R, Marozsan AJ, Sanders RW, Klasse P-J, Milne JLS, Wilson IA, Olson WC, Moore JP, Subramaniam S. 2011. Trimeric HIV-1 glycoprotein gp140 immunogens and native HIV-1 envelope glycoproteins display the same closed and open quaternary molecular architectures. Proc. Natl. Acad. Sci. U. S. A. 108:11440–11445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Julien J-P, Lee JH, Cupo A, Murin CD, Derking R, Hoffenberg S, Caulfield MJ, King CR, Marozsan AJ, Klasse P-J, Sanders RW, Moore JP, Wilson IA, Ward AB. 2013. Asymmetric recognition of the HIV-1 trimer by broadly neutralizing antibody PG9. Proc. Natl. Acad. Sci. U. S. A. 110:4351–4356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Julien J-P, Sok D, Khayat R, Lee JH, Doores KJ, Walker LM, Ramos A, Diwanji DC, Pejchal R, Cupo A, Katpally U, Depetris RS, Stanfield RL, McBride R, Marozsan AJ, Paulson JC, Sanders RW, Moore JP, Burton DR, Poignard P, Ward AB, Wilson IA. 2013. Broadly neutralizing antibody PGT121 allosterically modulates CD4 binding via recognition of the HIV-1 gp120 V3 base and multiple surrounding glycans. PLoS Pathog. 9:e1003342. 10.1371/journal.ppat.1003342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kong L, Lee JH, Doores KJ, Murin CD. 26 May 2013. Supersite of immune vulnerability on the glycosylated face of HIV-1 envelope glycoprotein gp120. Nat. Struct. Mol. Biol. [Epub ahead of print.] 10.1038/nsmb.2594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu G, Liu J, Taylor KA, Roux KH. 2011. Structural comparison of HIV-1 envelope spikes with and without the V1/V2 loop. J. Virol. 85:2741–2750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klasse P-J, Depetris RS, Pejchal R, Julien J-P, Khayat R, Lee JH, Marozsan AJ, Cupo A, Cocco N, Korzun J, Yasmeen A, Ward AB, Wilson IA, Sanders RW, Moore JP. 2013. Influences on trimerization and aggregation of soluble, cleaved HIV-1 SOSIP envelope glycoprotein. J. Virol. 87:9873–9855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, Carragher B. 2005. Automated molecular microscopy: the new Leginon system. J. Struct. Biol. 151:41–60 [DOI] [PubMed] [Google Scholar]
- 31.Voss NR, Yoshioka CK, Radermacher M, Potter CS, Carragher B. 2009. DoG Picker and TiltPicker: software tools to facilitate particle selection in single particle electron microscopy. J. Struct. Biol. 166:205–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mindell JA, Grigorieff N. 2003. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 142:334–347 [DOI] [PubMed] [Google Scholar]
- 33.Hohn M, Tang G, Goodyear G, Baldwin PR, Huang Z, Penczek PA, Yang C, Glaeser RM, Adams PD, Ludtke SJ. 2007. SPARX, a new environment for cryo-EM image processing. J. Struct. Biol. 157:47–55 [DOI] [PubMed] [Google Scholar]
- 34.Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. 2007. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 157:38–46 [DOI] [PubMed] [Google Scholar]
- 35.Dreyfus C, Laursen NS, Kwaks T, Zuijdgeest D, Khayat R, Ekiert DC, Lee JH, Metlagel Z, Bujny MV, Jongeneelen M, van der Vlugt R, Lamrani M, Korse HJWM, Geelen E, Sahin Ö, Sieuwerts M, Brakenhoff JPJ, Vogels R, Li OTW, Poon LLM, Peiris M, Koudstaal W, Ward AB, Wilson IA, Goudsmit J, Friesen RHE. 2012. Highly conserved protective epitopes on influenza B viruses. Science 337:1343–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khayat R, Lander GC, Johnson JE. 2010. An automated procedure for detecting protein folds from sub-nanometer resolution electron density. J. Struct. Biol. 170:513–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vagin A, Teplyakov A. 2010. Molecular replacement with MOLREP. Acta Crystallogr. D Biol. Crystallogr. 66:22–25 [DOI] [PubMed] [Google Scholar]
- 38.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605–1612 [DOI] [PubMed] [Google Scholar]
- 39.Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. 2008. Molecular architecture of native HIV-1 gp120 trimers. Nature 455:109–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tran EEH, Borgnia MJ, Kuybeda O, Schauder DM, Bartesaghi A, Frank GA, Sapiro G, Milne JLS, Subramaniam S. 2012. Structural mechanism of trimeric HIV-1 envelope glycoprotein activation. PLoS Pathog. 8:e1002797. 10.1371/journal.ppat.1002797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu X, Zhou T, Zhu J, Zhang B, Georgiev I, Wang C, Chen X, Longo NS, Louder M, McKee K, O'Dell S, Perfetto S, Schmidt SD, Shi W, Wu L, Yang Y, Yang ZY, Yang Z, Zhang Z, Bonsignori M, Crump JA, Kapiga SH, Sam NE, Haynes BF, Simek M, Burton DR, Koff WC, Doria-Rose NA, Connors M, NISC Comparative Sequencing Program. Mullikin JC, Nabel GJ, Roederer M, Shapiro L, Kwong PD, Mascola JR. 2011. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science 333:1593–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.White TA, Bartesaghi A, Borgnia MJ, Meyerson JR, de la Cruz MJV, Bess JW, Nandwani R, Hoxie JA, Lifson JD, Milne JLS, Subramaniam S. 2010. Molecular architectures of trimeric SIV and HIV-1 envelope glycoproteins on intact viruses: strain-dependent variation in quaternary structure. PLoS Pathog. 6:e1001249. 10.1371/journal.ppat.1001249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mao Y, Wang L, Gu C, Herschhorn A, Xiang S-H, Haim H, Yang X, Sodroski J. 2012. Subunit organization of the membrane-bound HIV-1 envelope glycoprotein trimer. Nat. Struct. Mol. Biol. 19:893–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu J, Chen I, Kwang J. 2005. Characterization of a previously unidentified viral protein in porcine circovirus type 2-infected cells and its role in virus-induced apoptosis. J. Virol. 79:8262–8274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kang YK, Andjelic S, Binley JM, Crooks ET, Franti M, Iyer SPN, Donovan GP, Dey AK, Zhu P, Roux KH, Durso RJ, Parsons TF, Maddon PJ, Moore JP, Olson WC. 2009. Structural and immunogenicity studies of a cleaved, stabilized envelope trimer derived from subtype A HIV-1. Vaccine 27:5120–5132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu L, Cimbro R, Lusso P, Berger EA. 2011. Intraprotomer masking of third variable loop (V3) epitopes by the first and second variable loops (V1V2) within the native HIV-1 envelope glycoprotein trimer. Proc. Natl. Acad. Sci. U. S. A. 108:20148–20153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ringe R, Bhattacharya J. 2012. Association of enhanced HIV-1 neutralization by a single Y681H substitution in gp41 with increased gp120-CD4 interaction and macrophage infectivity. PLoS One 7:e37157. 10.1371/journal.pone.0037157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harris AK, Bartesaghi A, Milne JLS, Subramaniam S. 2013. HIV-1 envelope glycoprotein trimers display open quaternary conformation when bound to the gp41 MPER-directed broadly neutralizing antibody Z13e1. J. Virol. 87:7191–7196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Falkowska E, Ramos A, Feng Y, Zhou T, Moquin S, Walker LM, Wu X, Seaman MS, Wrin T, Kwong PD, Wyatt RT, Mascola JR, Poignard P, Burton DR. 2012. PGV04, an HIV-1 gp120 CD4 binding site antibody, is broad and potent in neutralization but does not induce conformational changes characteristic of CD4. J. Virol. 86:4394–4403 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.