Abstract
Since 1998, 9 of the 26 serotypes of bluetongue virus (BTV) have spread throughout Europe, and serotype 8 has suddenly emerged in northern Europe, causing considerable economic losses, direct (mortality and morbidity) but also indirect, due to restriction in animal movements. Therefore, many new types of vaccines, particularly subunit vaccines, with improved safety and efficacy for a broad range of BTV serotypes are currently being developed by different laboratories. Here we exploited a reverse genetics-based replication-deficient BTV serotype 1 (BTV-1) (disabled infectious single cycle [DISC]) strain to generate a series of DISC vaccine strains. Cattle and sheep were vaccinated with these viruses either singly or in cocktail form as a multivalent vaccine candidate. All vaccinated animals were seroconverted and developed neutralizing antibody responses to their respective serotypes. After challenge with the virulent strains at 21 days postvaccination, vaccinated animals showed neither any clinical reaction nor viremia. Further, there was no interference with protection with a multivalent preparation of six distinct DISC viruses. These data indicate that a very-rapid-response vaccine could be developed based on which serotypes are circulating in the population at the time of an outbreak.
INTRODUCTION
Vaccination is one of the most effective approaches for controlling infectious viral diseases known to date. Extensive knowledge of the basic biology of viruses at the molecular level coupled with recent technology developments has resulted in a number of newly designed vaccines for both human and animal viral diseases. However, the generation of effective vaccines for viruses with multiple distinct serotypes remains laborious and highly challenging. The insect-borne bluetongue virus (BTV) consists of 26 serologically distinct viral serotypes (1). BTV is the causative agent of bluetongue (BT) disease of ruminants (sheep, goats, and cattle), with sheep being the most susceptible host with the highest mortality rate. BTV is endemic in both tropical and subtropical countries of the world, and it was considered exotic in Europe prior to 1998. However, several outbreaks in Europe of a number of BTV serotypes, which caused significant losses in European livestock and agriculture, have since been reported.
BTV belongs to the Orbivirus genus in the Reoviridae family, and like other members of the family, BTV is a nonenveloped icosahedral particle. BTV possesses a complex double-capsid structure consisting of seven structural proteins (VP1 to VP7) and a genome of 10 double-stranded RNA (dsRNA) segments. The outer capsid is made up of two major proteins, the larger ∼110-kDa VP2 protein and the 60-kDa VP5 protein. VP2 is a highly variable, serotype-determining protein, and it binds to the cellular receptor. VP5 is less variable and is a membrane penetration protein. These two proteins loosely interact with each other, and both are directly attached to the surface layer of the inner capsid (termed the core), which consists of the remaining five structural proteins and the viral genome. The core surface layer is made up of multiple copies of a single major protein, VP7. VP3 forms an inner scaffolding layer for the VP7 layer, which in turn surrounds the three minor proteins VP1 (polymerase), VP4 (capping enzyme), and VP6 (helicase) in addition to the genomic dsRNAs. In addition, four nonstructural proteins (NS1 to NS4) are synthesized in virus-infected cells. Both core proteins and NS proteins, unlike the outer capsid proteins, are highly conserved among BTV serotypes (2).
Although vaccination has been an effective approach to control BTV spread, currently available vaccines are associated with undesirable side effects. There are two types of BTV vaccines commercially available, namely, conventional live-attenuated and chemically inactivated vaccines. Although both types of vaccine can protect against BTV infection, problems such as incomplete protection, association with teratogenic effects, and incomplete attenuation have been reported (3, 4). Consequently, there are many current efforts to develop new types of vaccines with improved safety and efficacy for a broad range of BTV serotypes (5–11). Most of these efforts concentrate on the development of subunit vaccines.
Recently, we exploited a BTV reverse genetics technology to develop replication-deficient BTV serotypes based on the introduction of a lethal mutation in one of the genes essential for replication, i.e., the gene encoding the viral helicase VP6 protein (12). We have demonstrated that the VP6 deletion viruses (disabled infectious single cycle [DISC]) could replicate only in a VP6-complementing cell line but were excellent at inducing protective neutralizing antibody responses in vaccinated animals. As BTV genome segments reassort readily among different serotypes, it was possible to utilize the VP6 DISC virus strains to generate alternate serotypes by exchanging the two RNA segments that encode the two outer capsid proteins of a different serotype.
In this report, we have extended this approach to the generation of a series of monovalent disabled BTV serotypes, including the recent European serotypes that have caused serious disease in animals. The immunogenicity of each of the disabled virus strains was then assessed in the animal hosts. Furthermore, due to the precedent set by three polyvalent, attenuated live virus vaccines (each containing five serotypes) currently in use in South Africa to control bluetongue disease, we tested the DISC viruses in sheep as a multivalent vaccine candidate. The ratio of specific serotypes in each preparation was calculated to prevent interference between strains with respect to immunity due to cross-reactivity. Our data show clearly that there was no interference with protection with a multivalent preparation of six distinct serotypes of DISC viruses and that full protection was conferred against all six serotypes. The ease of effective DISC vaccine development will facilitate the inclusion of new serotypes in existing vaccination programs. In this manner, a very-rapid-response vaccine could be developed based on which serotypes are circulating in the population at the time of an outbreak.
MATERIALS AND METHODS
Cells and viruses.
BSR cells (BHK-21 subclone) were maintained in Dulbecco modified Eagle medium (DMEM; Sigma-Aldrich) supplemented with 5% (vol/vol) fetal bovine serum (FBS; Invitrogen). The stable BSR9 (BSR-VP6) cell line was grown in DMEM–5% FBS supplemented with 7.5 g/ml of puromycin (Sigma-Aldrich). BTV serotype stocks were generated by infecting BSR cells and harvested at 100% cytopathic effect. The challenge strains (BTV serotype 2 [BTV-2]/SAD2001/01, BTV-4/MOR2009/0, and BTV-8/DE2008) had been isolated and grown in embryonated chicken eggs and/or cell culture (passage levels E1/BHK2/KC1, KC1, and VERO2, respectively). The inoculated doses were confirmed by titration on mammalian Vero and insect KC (derived from Culicoides) cells.
Rescue of reassortant disabled viruses in a complementing cell line.
Segments S2 (VP2) and S6 (VP5) from BTV-2, -4, -8, -10, -13, -21, and -24 were obtained using a sequence-independent system as has been previously described (13, 14). Briefly, dsRNAs from purified core particles were ligated to a self-annealing primer before reverse transcription-PCR (RT-PCR) was performed using a specific primer. cDNAs amplified from segments S2 and S6 of the above-mentioned serotypes were cloned into pUC19 and fully sequenced (Source Bioscience) before insertion of the T7 promoter at the 5′ end and insertion of a unique restriction enzyme site that generates the correct end of the segment at the 3′ end. Modified S9 segments with either a deletion in the coding sequence of VP6 or a deletion and insertion of the enhanced green fluorescent protein (EGFP) gene were described previously (12, 15).
For synthesis of uncapped T7 transcripts for segments S2 and S6, a RiboMAX T7 large-scale RNA production system (Promega) or a TranscriptAid T7 high-yield transcription kit (Thermo Scientific) was used according to the manufacturer's instructions. Reassortant viruses were rescued from confluent monolayers of complementing cells (BSR9) transfected with six plasmids that drive the expression of BTV proteins VP1, VP3, VP4, VP6, NS1, and NS2 as described previously (16) prior to transfection with seven T7-derived single-stranded RNAs (ssRNAs) from BTV-1, one of the modified S9 (S9Δ or S9Δ-EGFP) ssRNAs, and a combination of S2 and S6 ssRNAs from a particular serotype.
Reassortant defective viruses were rescued and grown in the BSR9 complementing cell line as described previously (15). Titration of all defective viruses was performed in the BSR9 cell line, and the virus titer was determined by plaque assay or by determination of the 50% tissue culture infective dose (TCID50).
Vaccination and virus challenge.
For the monovalent vaccine test, 18 BTV-naive heifers between 9 and 18 months of age were used for the animal experiment. Three vaccine groups of three heifers each were inoculated with one of the reassortant disabled viruses (BTV-2D, -4D, and -8D) using two doses administered 21 days apart. Nine received a lysate from BSR9 cells and were kept as controls. Vaccine identity and dose were confirmed with a TaqVet European BTV typing kit (LSI, France). The type-specific quantification cycle (Cq) values for the reassortant viruses were 16.5, 15.2, and 15.1, respectively. Injection sites were examined daily for 1 week after each immunization to detect local reactions.
Clinical surveillance based on the Central Veterinary Institute (CVI) score (17) and measurements of rectal temperature were conducted daily from 4 days before the first vaccination (V1) until the end of the challenge experiment (day 63). A rectal temperature of an individual animal that was more than 1 degree (°C) higher than the average rectal temperature during the 3 days before challenge was considered to represent the presence of fever. An increase of more than 1 degree over the baseline temperature was awarded 1 point on the CVI score sheet, an increase of more than 1.5 degrees 2 points, and an increase of more than 2 degrees 3 points.
Serum samples were taken at 4, 5, 7, 10, 12, 14, and 21 days after both the first and second vaccinations. On day 42 after the first vaccination, the vaccine groups and three controls each were challenged with a virulent BTV strain of the same serotype as the vaccine. Each animal was subcutaneously (s.c.) injected with 4 × 105 TCID50 of virus.
Whole-blood samples (with EDTA anticoagulant) were taken at 0, 3 to 7, 9, 12, 14, 17, 19, and 21 days postchallenge (dpc), and serum samples were collected at 0 and 21 dpc.
For cocktail vaccine testing, sheep were used and experiments were performed under the guidelines of the European Community (86/609) and were approved by the Committee on the Ethics of Animal Experiments of the CVI (permit 2012-029). Twenty-seven female Blessumer sheep that were 6 to 24 months old and free of BTV and BTV antibodies were commercially sourced from the same flock of a Dutch farm. The sheep were randomly allocated to three groups of six animals and one group of nine animals. At 0 days postvaccination (0 dpv) and 21 dpv, groups of six sheep were subcutaneously (s.c.) vaccinated in the neck with 2 ml of multivalent DISC vaccine. The group of nine sheep served as controls and were similarly injected with an equivalent dose of lysate from BSR9 cells. At 35 dpv, control sheep were randomly divided into groups corresponding to the three groups of six vaccinated sheep. On day 42, these newly formed groups of six vaccinated and three control sheep were challenged subcutaneously with 4 × 1 ml of 105 TCID50/ml of either virulent BTV-2 or BTV-4 or BTV-8. Virus was injected subcutaneously to sheep between the shoulder blades to the left and right of the spinal cord. Three weeks after challenge at 63 dpv and 21 dpc, the trial was finalized.
Body temperature was recorded daily in periods of injections. Fever was defined as the average temperature plus 2 times the standard deviation. Clinical signs were daily recorded according to the clinical score by clinical surveillance based on the CVI score table for BTV-8 animal trials (17). Samples of EDTA blood and serum were frequently collected until the end of the experiment.
Serum neutralization assay.
For detection of a neutralizing antibody response in vaccinated animals, a standard serum neutralization assay of BTV was used as described previously (12). Briefly, serum samples were serially diluted 1:2 and added to confluent monolayers of BSR cells in 96-well plates. About 100 PFU of each BTV serotype were added per well and incubated for 3 days. All dilutions were performed in triplicate in each experiment. The neutralizing titers were defined as the highest dilution of sera allowing complete neutralization of the virus.
BT competition ELISA and early-detection ELISA.
Serum samples were analyzed with the ID Screen Bluetongue Competition assay and ID Screen Bluetongue Early Detection (ID VET, Montpellier, France) according to the manufacturer's instructions. In addition to the kit controls, a 2-fold dilution series of an anti-BTV antibody-positive reference serum (CIRAD, Montpellier, France) was included for the competition assay as the working standard in each assay to monitor the performance of the enzyme-linked immunosorbent assay (ELISA) over time. Results are expressed as percentages of negativity (% Negativity) compared to the negative kit control results and converted to a positive (percent sample-to-positive ratio [% S/P] > 30), uncertain (% S/P ≤ 30 but > 25), or negative (% S/P ≤ 25) result according to cutoff values previously determined (18).
RNA extraction and RT-qPCR.
RNA extraction from the EDTA blood samples was performed according to Vandenbussche et al. (19). The triplex quantitative RT-PCR (RT-qPCR) assay included primers and probes for a pan-BTV/segment 5 (S5)-specific reaction and for an internal control (IC) and external control (EC) as described before (19) and was performed on a LightCycler 480 instrument (Roche Diagnostics, Mannheim, Germany). For this assay, crossing point (Cp) values < 40.0 were classified as positive, Cp values = 40.0 and < 45.0 were classified as uncertain, and Cp values > 45.0 were considered negative. The acceptance criteria for the IC were set at a Cp value of 28.9 and for the EC at a Cp value of 33.6 for blood samples.
RESULTS
Rescue of seven disabled BTV serotypes by exchanging the two RNA segments responsible for serotype determination.
As the first step in the development of a new-generation BTV vaccine, a previously described sequence-independent strategy (13, 14) was used to clone segments S2 and S6 (encoding VP2 and VP5, respectively) from six different BTV serotypes (BTV-2, -4, -10, -13, -21, and -24). Subsequently, the resulting clones of all the segments were fully sequenced. In order to generate T7 transcripts with the exact ends for both segments S2 and S6 of each of the six serotypes, the T7 promoter and a unique restriction site at the end of each segment were introduced as described previously (13).
Reassortant disabled viruses were rescued in BSR-VP6-complementing cells (BSR9) that were transfected with 10 RNA transcripts, 7 of which were from BTV-1 (S1, S3, S4, S5, S7, S8, and S10), mutated segment S9 of BTV-10 that was incapable of the correct expression of the essential viral protein VP6 (15), and the S2 and S6 segments from one of the serotypes. The ability of the VP6-expressing cell line (15) to constitutively express VP6 under conditions of selective pressure was used to overcome the lack of a functional VP6 in the rescued system. Each rescued virus (namely, BTV-2D, -4D, -10D, -13D, -21D, and -24D) was propagated further, genomic dsRNAs were isolated, and their gel electrophoresis profiles were compared to that of the wild-type BTV-1 (Fig. 1A). Samples of genomic dsRNAs of BTV-1D and BTV-8D, generated previously (12), were also included as positive controls.
Fig 1.
Characterization of reassortant disabled viruses generated by reverse genetics. (A) Left panel: the genomic profile of purified dsRNA from cells infected with the reassortant BTV-2D was analyzed by nondenaturing PAGE. BTV-1 and BTV-2 profiles were included as a control. Right panel: details of the genomic profile of dsRNA from each of the reassortant disabled viruses. Arrowheads indicate differences in the mobility of segments S2 and S6. (B) Replication in normal (upper panel) or complementing (lower panel) cells of the reassortant disabled BTV strains. Monolayers of normal BSR or complementing BSR9 cell lines were infected at a multiplicity of infection (MOI) of 0.1, and samples were collected as indicated. Cells and supernatant were harvested, and the total titer was determined by TCID50. Total titers at the indicated time points are expressed as TCID50/ml.
To confirm that these reassortant disabled viruses were incapable of growth in normal cells, to exclude any recombination or reversion, and to determine the efficiency of their growth in complementary cells, the growth kinetics of each virus was evaluated by infecting both cell lines at a low multiplicity of infection (MOI) and measuring the virus titers at 18, 24, and 48 h postinfection. The results obtained demonstrated that these viruses were not able to grow in normal BSR cells since no infectious virus particles could be detected 18 h postinfection (Fig. 1B, upper panel). However, each virus was capable of replication in the complementary cells, with titers ranging from 1.3 ×104 (BTV-10D) to 1.7 × 106 (BTV-2D) at 48 h postinfection (Fig. 1B, lower panel).
Monoserotype vaccine strains protect cattle from virulent virus challenge.
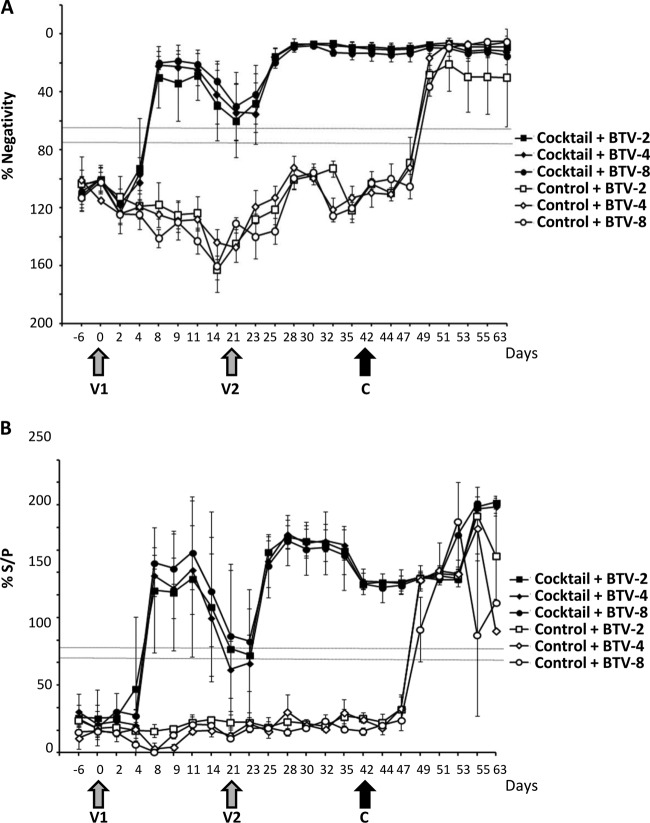
In a previous report, we demonstrated that two BTV-1D and BTV-8D viruses were able to elicit neutralizing antibody responses in sheep that conferred full protection against homologous virulent virus challenges. Since BTV-8 had caused severe BT disease not only in sheep but also in cattle during the BTV-8 outbreaks in Europe (20–22), it was important to assess the protective efficacy of the BTV-8D vaccine strain in cattle. For comparison, two new vaccine strains, BTV-2D and -4D, were also assessed in cattle in parallel as monovalent vaccines. BTV-naive heifers were segregated into 3 groups, and each group was vaccinated once and then again 21 days later with one of the three vaccine strains as described in Materials and Methods. As a control, 3 groups of 3 heifers each were inoculated with a complementing BSR9 cell lysate. No major side effects were observed after each vaccination, indicating that the vaccines were well tolerated. When the humoral immune response to each vaccine was assessed with the competitive ELISA (cELISA), it was clear that the response was fairly weak after the first vaccination (V1). However, all vaccinated heifers had seroconverted by 5 days after the second vaccination (V2) when assessed by the specific cELISA (Fig. 2A). The result of the early-detection ELISA demonstrated a more pronounced IgM response, especially in the BTV-4D and -8D groups (Fig. 2B), where 5 of 6 animals seroconverted at 7 days postvaccination (dpv), with the sixth animal seroconverting at 10 dpv. Although this seroconversion was transient (all animals were again negative at 21 dpv), the animals in these 2 groups responded more quickly to the second vaccination, with 5 of the 6 animals becoming positive at 5 days postrevaccination (dprv) (day 26). The IgM response in the BTV-2D was more heterogeneous and weak. None of the control group animals presented specific antibodies against BTV and became BTV positive only after virulent virus challenges.
Fig 2.
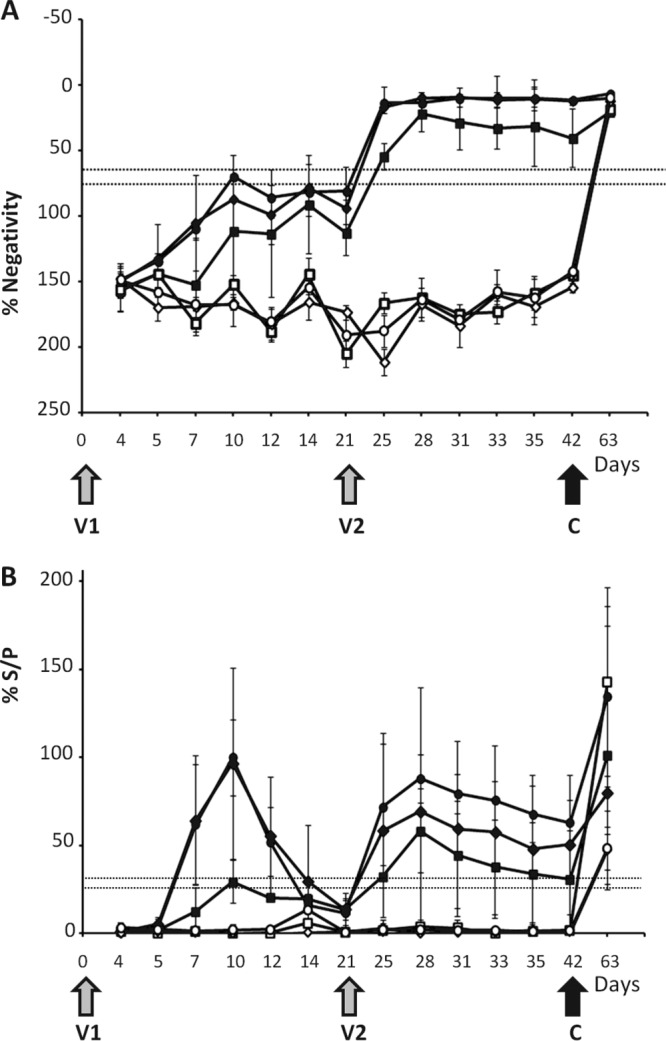
Detection of group-specific antibody response in sera of vaccinated cattle by ELISA. (A) Antibodies directed against the VP7 protein of BTV were detected using a competition ELISA kit. The data points represent the mean antibody responses of the animals in each group (vaccinated with the BTV2D, -4D, or -8D strain or the control) with standard deviations. Results are expressed as percentages of negativity (% Negativity) compared to the negative control included in the kit and were classified into positive (≤65%), uncertain (>65% but ≤75%), and negative (>75%) groups. Gray arrows indicate the first and second vaccination times (V1 and V2), and the black arrow indicates the time of challenge with homologous virulent virus serotypes (C). The samples were as follows (symbol): 2D plus BTV-2 (■), 4D plus BTV-4 (⧫), 8D plus BTV-8 (●), control plus BTV-2 (☐), control plus BTV-4 (◊), and control plus BTV-8 (○). (B) Early detection of IgG and IgM antibodies after the first vaccination. The data represent the mean antibody responses of the animals in each group with standard deviations. The samples were evaluated by the sample-to-positive ratio expressed as a percentage (% S/P) and were classified into positive (% S/P > 30), uncertain (% S/P ≤ 30 but > 25), and negative (% S/P ≤ 25) groups. The samples were as follows (symbol): 2D plus BTV-2 (■), 4D plus BTV-4 (⧫), 8D plus BTV-8 (●), control plus BTV-2 (☐), control plus BTV-4 (◊), and control plus BTV-8 (○).
Serum neutralization tests (SNT) were undertaken on all serum samples on the day of challenge using BTV-2, BTV-4, and BTV-8 to determine if the heifer had demonstrated a response prior to virulent virus challenge. Sera from all vaccinated heifers had neutralizing antibody (NA) specific to the serotype of the vaccine virus, and the NA titers ranged from 16 to 32. In comparison, the control heifer vaccinated with BSR9 lysate did not have any detectable NA.
Viremia levels in both the vaccinated and control groups were determined from day 3 onward after challenge. The protection conferred by the vaccines was assessed by BTV-specific real-time qRT-PCR to detect virus replication. No BTV replication was detectable in any of the vaccinated heifers (Fig. 3). In contrast, all animals in control groups became BTV positive in the real-time RT-PCR after the virulent virus challenges. These results indicated that the disabled strains were not able to replicate in vaccinated animals, as no genomic RNA were detectable during the vaccination period. These data support the idea of the safety of these vaccine candidate strains. More importantly, these data also indicated that these defective viruses were able to elicit strong immune responses that protected all vaccinated animals from homologous virulent virus challenges. Overall, these results complement our results presented in a previous report using a different animal host where all vaccinated sheep were protected by a monovalent vaccine for BTV-1 and -8 (12).
Fig 3.

Virus replication in vaccinated cattle challenged with a virulent strain. Detection of genomic BTV RNA was performed using a real-time RT-PCR assay in postchallenged samples from cattle vaccinated with the monoserotype vaccines. The samples were as follows (symbol): 2D plus BTV-2 (■), 4D plus BTV-4 (⧫), 8D plus BTV-8 (●), control plus BTV-2 (☐), control plus BTV-4 (◊), and control plus BTV-8 (○). The data points represent the means and standard deviations of RNA copy numbers of the animals in each group.
A multivalent cocktail vaccine can protect sheep against multiple serotypes.
Simultaneous circulation of two or more serotypes of BTV emphasizes the necessity of a vaccine that protects against several serotypes. Working toward the development of a multiserotype vaccine, we decided to test a cocktail mixture of six disabled viruses, including serotypes BTV-1D, -2D, -4D, -8D, 13D, and -21D. Some of the serotypes included in the vaccine represent important strains circulating in Europe in the last few years.
Three groups of six sheep were inoculated with the cocktail vaccine in a prime and boost protocol. One group of nine sheep received an equivalent dose of cell lysate and served as the control group. Blood samples were collected at regular intervals over the experimental period to determine the serological response to the vaccination and for detection of the virus replication. In parallel, body temperatures, as a sign of disease, of all groups were recorded routinely.
The presence of BTV antibody in vaccinated sheep was monitored based on VP7, the group-specific antigen, using two different ELISA methods, early detection and cELISA (Fig. 4). All vaccinated sheep showed seroconversion at 8 dpv. This initial response was transient in 4 of the 18 animals after the first vaccination and gave titers below the threshold (negative) prior to the second vaccination (day 21); this trend was evident in both ELISA methods used. However, all animals were seroconverted at 4 dprv (day 25) and remained positive until the end of the trial. The control sheep vaccinated with the BSR9 lysate did not show a response to VP7 until 7 days postchallenge with virulent BTV.
Fig 4.
Detection of group-specific antibody response in sera of sheep vaccinated with multivalent vaccines by ELISA. (A) Antibodies directed against the VP7 protein of BTV were detected using a competition ELISA kit. The data points represent the mean antibody responses of the animals in each group (vaccinated with the vaccine cocktail and control) with standard deviations. Results are expressed as percentages of negativity (% Negativity) compared to the negative control included in the kit and were classified into positive (≤65), uncertain (>65 but ≤75), and negative (>75) groups. The samples were as follows (symbol): 2D plus BTV-2 (■), 4D plus BTV-4 (⧫), 8D plus BTV-8 (●), control plus BTV-2 (☐), control plus BTV-4 (◊), and control plus BTV-8 (○). Arrows indicate first and second vaccination times (V1 and V2), and the black arrow indicates the time of challenge with homologous virulent virus serotypes (C). (B) Early detection of IgG and IgM antibodies after the first vaccination. The data represent the mean antibody responses of the animals in each group (vaccinated with the vaccine cocktail and control) with standard deviations. The samples were as follows (symbol): 2D plus BTV-2 (■), 4D plus BTV-4 (⧫), 8D plus BTV-8 (●), control plus BTV-2 (☐), control plus BTV-4 (◊), and control plus BTV-8 (○). The samples were evaluated by the sample-to-positive ratio expressed as a percentage (% S/P) and were classified into positive (% S/P > 30), uncertain (% S/P ≤ 30 but > 25), and negative (% S/P ≤ 25) groups.
The serum samples from the control and vaccinated animals collected on the day of the challenge were also tested by SNT using BTV-1, -2, -4, -8, -13, and -21 to determine if the vaccinated animals elicited serotype-specific NA. All animals vaccinated with the cocktail vaccine had neutralizing antibodies against all the homologous serotypes included in the cocktail (Fig. 5). The NA titers ranged between 16 and 128 for all serotypes tested, and there was no apparent interference with respect to the ability of the animal to respond to each of the vaccine strains present in the cocktail (Fig. 5). This result indicates that the cocktail vaccine would be able to protect animals against all the different serotypes included in the cocktail vaccine.
Fig 5.
Neutralizing antibody response of vaccinated sheep. Samples of sera from vaccinated (gray columns) and control (open columns) animals were taken 21 days after the second vaccination and tested for neutralizing activity against all BTV serotypes included in the vaccine (BTV-1, -2, -4, -8, -13, and -21) as indicated. The neutralizing titer in each animal was determined as the maximum dilution of serum that showed complete protection.
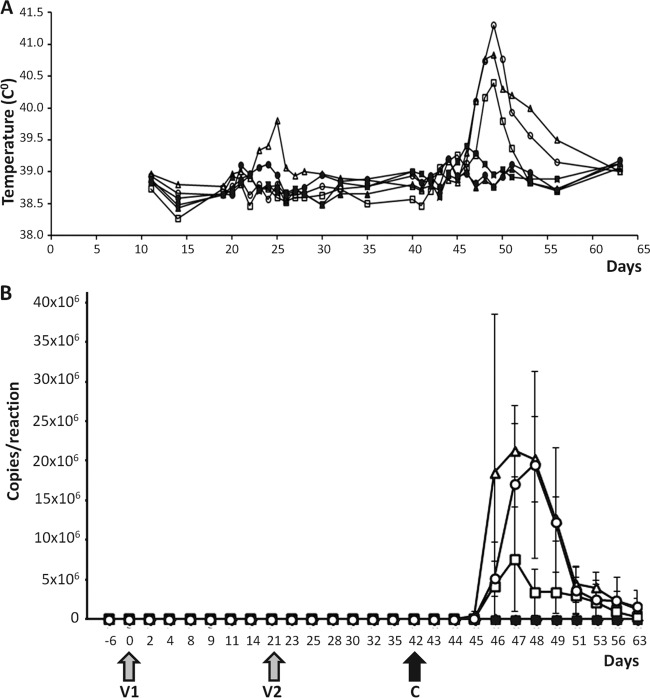
Moreover, the recorded temperatures in all groups showed no major differences between control and vaccinated animals before challenge (Fig. 6A). The only side effect from vaccination included one animal in a control group that had a high temperature after the second inoculation with cell lysate (control group) that lasted for a very short period of time. After challenge, vaccinated sheep did not develop fever, whereas control sheep had fever for several days (2 to 3 days, 4 to 12 days, and 4 to 6 days for BTV-2, -4, and -8, respectively), with a peak at 7 days postchallenge (day 49) for all control sheep. Three vaccinated sheep showed a slightly increased rate of breathing, mild depression, and less appetite after challenge with virulent BTV-2 for more than 1 day. All other vaccinated sheep showed no clinical signs or only very mild signs, such as less appetite for 1 day. Unvaccinated sheep showed severe clinical signs after challenge for several days based on the CVI clinical score (17). Challenge with BTV-2 resulted in increased breathing, mild nasal discharges, less appetite, and mild depression. These clinical signs lasted for a longer period than for the sheep challenged with BTV-4 and -8. The sheep challenged with BTV-4 or -8 showed very severe clinical signs, including no appetite, depression, increased breathing, painful feet, loss of body weight, and high fever. One of three control animals in both groups died or was euthanized for ethical reasons due to clinical signs associated with bluetongue disease.
Fig 6.
Clinical signs and viremia in sheep vaccinated with the multivalent cocktail vaccine. (A) Body temperature as a sign of BT disease was recorded at different time points for all animals in each group. (B) The presence of genomic RNA in blood from all animals was determined by real-time RT-PCR at the indicated times during the experiment. V1, V2, and C represent the times of the first and second vaccinations and challenge, respectively. The samples were as follows (symbol): 2D plus BTV-2 (■), 4D plus BTV-4 (⧫), 8D plus BTV-8 (●), control plus BTV-2 (☐), control plus BTV-4 (◊), and control plus BTV-8 (○).
The safety and protection conferred by the cocktail vaccine were analyzed by real-time RT-PCR in blood samples taken routinely (Fig. 6B). Following both vaccinations, traces of the vaccine RNA were detected in all animals with very low copy numbers (67% showed less than 1,500 copies per reaction, with a maximum of 7,300). This detection was transient, with the results for 50% of the animals becoming negative at 4 dpv. Two animals remained borderline positive until 11 dpv. The detection period upon revaccination was significantly shorter, as the animals were borderline positive only at 2 dprv, with the exception of 1 animal which was positive until 4 dprv. None of the samples from vaccinated animals that were challenged with BTV-2, BTV-4, or BTV-8 exhibited any virus replication. In comparison, all control animals showed virus replication as expected (Fig. 6B).
DISCUSSION
Early attempts at vaccination against BTV used serum from infected sheep that had survived the disease (23). This mild strain of virus was serially propagated in sheep. This vaccination was used for many years in South Africa despite the fact that the vaccine was found not to be safe and did not provide adequate immunity. Subsequently, live-attenuated virus vaccines of multiple BTV serotypes were developed by serial passage in embryonated chicken eggs. Because of the number of serotypes of BTV circulating, multivalent vaccination has generally been used in areas of endemicity (24; reviewed in reference 25). Despite the success of attenuated vaccines in areas of endemicity, the use of such vaccines has some drawbacks. Teratologic effects as a result of vaccination with attenuated BTV are well documented (26). Furthermore, viremia following vaccination both in laboratory experiments and in the field has been sufficient for the vaccine strain to be transmitted (27).
In this report, we extended our previous work on the development of a new generation of vaccines against BTV based on the generation of disabled viruses that cannot replicate in animals or normal cells but can replicate only in a particular complementary cell line. We combined the genome segment reassortment capabilities of the virus to replace the outer capsid proteins (encoded by S2 and S6) of a defective BTV-1 (BTV-1D) strain using the reverse genetics system. This allowed us to rapidly generate new vaccine strains of various serotypes that still retained the deficiency in virus replication in normal cells and can be propagated only on specific complementing cells. Moreover, after 8 serial passages, neither any reversion to virulence nor any cytopathic effect in normal cells was detectable, indicating that these viruses are stable and potentially excellent vaccine candidates.
Initially, we tested three of these vaccine strains (BTV-2D, BTV-4D, and BTV-8D) in cattle since infected cattle generally have prolonged viremia mostly without any BT disease symptoms (except the recent scenario of BTV-8 in European cattle) and, therefore, cattle are considered the major reservoir for the virus replication. When groups of cattle were vaccinated with one of the vaccine strains (BTV-2D, BTV-4D, or BTV-8D) in a two-dose protocol and challenged with a homologous virulent strain, all developed a positive immune response that inhibited challenge virus replication, as no BTV genome could be detected by highly sensitive real-time PCR and there was no clinical sign of BTV disease in any of the vaccinated animals. The data demonstrated that these defective virus strains are very effective at inducing protective immunity in vaccinated cattle in a manner similar to that seen in a previous sheep study (12). Further, a protocol consisting of two doses administered 3 weeks apart seems to be adequate to confer complete protection. These data are comparable to those achieved with inactivated vaccines in cattle (28–30).
In some areas of endemicity such as in South Africa, multiple serotypes cocirculate each season with high epidemic potential; in fact, the circulation of two or more highly virulent virus serotypes (e.g., BTV-1 and BTV-8) had been observed in Europe (31). Therefore, immunization of susceptible animals with a multivalent vaccine that can confer protection against multiple serotypes is ideal for the prevention of BTV outbreak. The disabled reassortant virus strains of six different serotypes that we describe in this report showed great promise as suitable vaccines with all the requisite properties for animal vaccination. Indeed, when these vaccine strains were mixed and the cocktail was inoculated to groups of sheep, all animals were seroconverted in a manner similar to that seen in a study of single vaccine strains reported previously (12). All vaccinated animals also developed neutralizing antibodies which inhibited the replication of challenged viruses, although traces of the vaccine virus RNAs were detectable following vaccination. This presence of RNA after vaccination corroborates recently published data where vaccine RNA could be detected at up to 9 dpv in sheep (32) and up to 7 dpv in cattle (33). However, none of the animals showed any clinical signs of disease after the virulent virus challenge and no trace of infectious virus replication from the challenged virus strains that could be detected by real-time PCR. These results demonstrate that the multivalent cocktail vaccine afforded complete protection in sheep against each virulent serotype and that there was no evidence of interference between the serotypes present in the cocktail. These results are very similar to those of previous vaccine efficacy studies performed in sheep with cocktails of virus-like particles (VLPs) (2 or multiple serotypes), where VLPs of multiple BTV serotypes were shown to be completely protective against homologous virulent virus challenges with no interference between serotypes (34–36). These results are also consistent with the commercially available live attenuated and inactivated virus vaccines, where limited interference was observed in the cocktail BTV vaccines.
Altogether, our data demonstrated that disabled BTV viruses did not replicate either in sheep or in cattle but did confer complete protection against virulent virus challenges in both type of animals. Thus, these DISC virus vaccines are safe as subunit vaccines but express viral proteins at the natural sites of viral infection in animal hosts in a manner similar to that seen with the natural viruses. Another advantage of this DISC virus strain is that a single strain can be used as the backbone of other serotypes by reassortment, facilitating rapid development of highly efficacious vaccine strains. Another significant aspect of these vaccines is that these DISC strains can be used as a multivalent cocktail vaccine that did not show any interference with respect to immune response between different strains and conferred protection against all homologous virus infection.
In conclusion, our data clearly showed that the reverse genetics technology allows the generation of vaccines in a rapid and reliable manner against the threat of an outbreak of bluetongue disease regardless of the serotype involved. Emerging serotypes can therefore be rapidly prepared from genomic material to vaccine for a safe product and a robust immune response.
ACKNOWLEDGMENTS
This study was funded by the European Union (FP7, ORBIVAC, grant 245266) and partly by the Biotechnology and Biological Sciences Research Council (BBSRC), United Kingdom.
We are thankful to P. P. C. Mertens (Pirbright Institute, United Kingdom) for kindly providing the BTV challenge virus strains.
Footnotes
Published ahead of print 3 July 2013
REFERENCES
- 1.Maan NS, Maan S, Belaganahalli MN, Ostlund EN, Johnson DJ, Nomikou K, Mertens PP. 2012. Identification and differentiation of the twenty six bluetongue virus serotypes by RT-PCR amplification of the serotype-specific genome segment 2. PLoS One 7:e32601. 10.1371/journal.pone.0032601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roy P, Marshall JJ, French TJ. 1990. Structure of the bluetongue virus genome and its encoded proteins. Curr. Top. Microbiol. Immunol. 162:43–87 [DOI] [PubMed] [Google Scholar]
- 3.Veronesi E, Darpel KE, Hamblin C, Carpenter S, Takamatsu HH, Anthony SJ, Elliott H, Mertens PP, Mellor PS. 2010. Viraemia and clinical disease in Dorset Poll sheep following vaccination with live attenuated bluetongue virus vaccines serotypes 16 and 4. Vaccine 28:1397–1403 [DOI] [PubMed] [Google Scholar]
- 4.Batten CA, Maan S, Shaw AE, Maan NS, Mertens PP. 2008. A European field strain of bluetongue virus derived from two parental vaccine strains by genome segment reassortment. Virus Res. 137:56–63 [DOI] [PubMed] [Google Scholar]
- 5.Boone JD, Balasuriya UB, Karaca K, Audonnet JC, Yao J, He L, Nordgren R, Monaco F, Savini G, Gardner IA, Maclachlan NJ. 2007. Recombinant canarypox virus vaccine co-expressing genes encoding the VP2 and VP5 outer capsid proteins of bluetongue virus induces high level protection in sheep. Vaccine 25:672–678 [DOI] [PubMed] [Google Scholar]
- 6.Perrin A, Albina E, Breard E, Sailleau C, Prome S, Grillet C, Kwiatek O, Russo P, Thiery R, Zientara S, Cetre-Sossah C. 2007. Recombinant capripoxviruses expressing proteins of bluetongue virus: evaluation of immune responses and protection in small ruminants. Vaccine 25:6774–6783 [DOI] [PubMed] [Google Scholar]
- 7.Franceschi V, Capocefalo A, Calvo-Pinilla E, Redaelli M, Mucignat-Caretta C, Mertens P, Ortego J, Donofrio G. 2011. Immunization of knock-out alpha/beta interferon receptor mice against lethal bluetongue infection with a BoHV-4-based vector expressing BTV-8 VP2 antigen. Vaccine 29:3074–3082 [DOI] [PubMed] [Google Scholar]
- 8.Calvo-Pinilla E, Navasa N, Anguita J, Ortego J. 2012. Multiserotype protection elicited by a combinatorial prime-boost vaccination strategy against bluetongue virus. PLoS One 7:e34735. 10.1371/journal.pone.0034735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Noad R, Roy P. 2003. Virus-like particles as immunogens. Trends Microbiol. 11:438–444 [DOI] [PubMed] [Google Scholar]
- 10.Roy P, Noad R. 2009. Virus-like particles as a vaccine delivery system: myths and facts. Adv. Exp. Med. Biol. 655:145–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Noad R, Roy P. 2009. Bluetongue vaccines. Vaccine 27(Suppl. 4):D86–D89 [DOI] [PubMed] [Google Scholar]
- 12.Matsuo E, Celma CC, Boyce M, Viarouge C, Sailleau C, Dubois E, Breard E, Thiery R, Zientara S, Roy P. 2011. Generation of replication-defective virus-based vaccines that confer full protection in sheep against virulent bluetongue virus challenge. J. Virol. 85:10213–10221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boyce M, Celma CC, Roy P. 2008. Development of reverse genetics systems for bluetongue virus: recovery of infectious virus from synthetic RNA transcripts. J. Virol. 82:8339–8348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maan S, Rao S, Maan NS, Anthony SJ, Attoui H, Samuel AR, Mertens PP. 2007. Rapid cDNA synthesis and sequencing techniques for the genetic study of bluetongue and other dsRNA viruses. J. Virol. Methods 143:132–139 [DOI] [PubMed] [Google Scholar]
- 15.Matsuo E, Roy P. 2009. Bluetongue virus VP6 acts early in the replication cycle and can form the basis of chimeric virus formation. J. Virol. 83:8842–8848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsuo E, Roy P. 2013. Minimum requirements for bluetongue virus primary replication in vivo. J. Virol. 87:882–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Rijn PA, Geurts Y, van der Spek AN, Veldman D, van Gennip RG. 2012. Bluetongue virus serotype 6 in Europe in 2008—emergence and disappearance of an unexpected non-virulent BTV. Vet. Microbiol. 158:23–32 [DOI] [PubMed] [Google Scholar]
- 18.Vandenbussche F, Vanbinst T, Verheyden B, Van Dessel W, Demeestere L, Houdart P, Bertels G, Praet N, Berkvens D, Mintiens K, Goris N, De Clercq K. 2008. Evaluation of antibody-ELISA and real-time RT-PCR for the diagnosis and profiling of bluetongue virus serotype 8 during the epidemic in Belgium in 2006. Vet. Microbiol. 129:15–27 [DOI] [PubMed] [Google Scholar]
- 19.Vandenbussche F, Vandemeulebroucke E, De Clercq K. 2010. Simultaneous detection of bluetongue virus RNA, internal control GAPDH mRNA, and external control synthetic RNA by multiplex real-time PCR. Methods Mol. Biol. 630:97–108 [DOI] [PubMed] [Google Scholar]
- 20.Maan S, Maan NS, Ross-smith N, Batten CA, Shaw AE, Anthony SJ, Samuel AR, Darpel KE, Veronesi E, Oura CA, Singh KP, Nomikou K, Potgieter AC, Attoui H, van Rooij E, van Rijn P, De Clercq K, Vandenbussche F, Zientara S, Breard E, Sailleau C, Beer M, Hoffman B, Mellor PS, Mertens PP. 2008. Sequence analysis of bluetongue virus serotype 8 from the Netherlands 2006 and comparison to other European strains. Virology 377:308–318 [DOI] [PubMed] [Google Scholar]
- 21.Mintiens K, Meroc E, Mellor PS, Staubach C, Gerbier G, Elbers AR, Hendrickx G, De Clercq K. 2008. Possible routes of introduction of bluetongue virus serotype 8 into the epicentre of the 2006 epidemic in north-western Europe. Prev. Vet. Med. 87:131–144 [DOI] [PubMed] [Google Scholar]
- 22.Purse BV, Mellor PS, Rogers DJ, Samuel AR, Mertens PP, Baylis M. 2005. Climate change and the recent emergence of bluetongue in Europe. Nat. Rev. Microbiol. 3:171–181 [DOI] [PubMed] [Google Scholar]
- 23.Theiler A. 1908. The immunization of mules with polyvalent serum and virus, p 192–213 In Transvaal department of agricultural report in veterinary bacteriology, 1906-1907. Transvaal Department of Agriculture, Pretoria, South Africa [Google Scholar]
- 24.Alexander RA, Haig DA, Adelaar TF. 1947. The attenuation of bluetongue virus by serial passage through fertile eggs. Onderstepoort J. Vet. Sci. Anim. Ind. 21:231–241 [PubMed] [Google Scholar]
- 25.Verwoerd DW. 2012. History of Orbivirus research in South Africa. J. S. Afr. Vet. Assoc. 83:E1–E6 [DOI] [PubMed] [Google Scholar]
- 26.Savini G, Lorusso A, Paladini C, Migliaccio P, Di Gennaro A, Di Provvido A, Scacchia M, Monaco F. 2012. Bluetongue serotype 2 and 9 modified live vaccine viruses as causative agents of abortion in livestock: a retrospective analysis in Italy. Transbound. Emerg. Dis. [Epub ahead of print.] 10.1111/tbed.12004 [DOI] [PubMed] [Google Scholar]
- 27.Elia G, Savini G, Decaro N, Martella V, Teodori L, Casaccia C, Di Gialleonardo L, Lorusso E, Caporale V, Buonavoglia C. 2008. Use of real-time RT-PCR as a rapid molecular approach for differentiation of field and vaccine strains of bluetongue virus serotypes 2 and 9. Mol. Cell. Probes 22:38–46 [DOI] [PubMed] [Google Scholar]
- 28.Schwartz-Cornil I, Mertens PP, Contreras V, Hemati B, Pascale F, Breard E, Mellor PS, MacLachlan NJ, Zientara S. 2008. Bluetongue virus: virology, pathogenesis and immunity. Vet. Res. 39:46. 10.1051/vetres:2008023 [DOI] [PubMed] [Google Scholar]
- 29.Savini G, Hamers C, Conte A, Migliaccio P, Bonfini B, Teodori L, Di Ventura M, Hudelet P, Schumacher C, Caporale V. 2009. Assessment of efficacy of a bivalent BTV-2 and BTV-4 inactivated vaccine by vaccination and challenge in cattle. Vet. Microbiol. 133:1–8 [DOI] [PubMed] [Google Scholar]
- 30.Wäckerlin R, Eschbaumer M, Konig P, Hoffmann B, Beer M. 2010. Evaluation of humoral response and protective efficacy of three inactivated vaccines against bluetongue virus serotype 8 one year after vaccination of sheep and cattle. Vaccine 28:4348–4355 [DOI] [PubMed] [Google Scholar]
- 31.Wilson AJ, Mellor PS. 2009. Bluetongue in Europe: past, present and future. Philos. Trans. R. Soc. Lond. B Biol. Sci. 364:2669–2681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Steinrigl A, Revilla-Fernandez S, Eichinger M, Koefer J, Winter P. 2010. Bluetongue virus RNA detection by RT-qPCR in blood samples of sheep vaccinated with a commercially available inactivated BTV-8 vaccine. Vaccine 28:5573–5581 [DOI] [PubMed] [Google Scholar]
- 33.De Leeuw I, Garigliany M, Bertels G, Willems T, Desmecht D, De Clercq K. 2013. Bluetongue virus RNA detection by real-time RT-PCR in post-vaccination samples from cattle. Transbound. Emerg. Dis. [Epub ahead of print.] 10.1111/tbed.12100 [DOI] [PubMed] [Google Scholar]
- 34.Roy P, Bishop DHL, LeBlois H, Erasmus BJ. 1994. Long-lasting protection of sheep against bluetongue challenge after vaccination with virus-like particles: evidence for homologous and partial heterologous protection. Vaccine 12:805–811 [DOI] [PubMed] [Google Scholar]
- 35.Pérez de Diego AC, Athmaram TN, Stewart M, Rodriguez-Sanchez B, Sanchez-Vizcaino JM, Noad R, Roy P. 2011. Characterization of protection afforded by a bivalent virus-like particle vaccine against bluetongue virus serotypes 1 and 4 in sheep. PLoS One 6:e26666. 10.1371/journal.pone.0026666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stewart M, Dubois E, Sailleau C, Breard E, Viarouge C, Desprat A, Thiery R, Zientara S, Roy P. 2013. Bluetongue virus serotype 8 virus-like particles protect sheep against virulent virus infection as a single or multi-serotype cocktail immunogen. Vaccine 31:553–558 [DOI] [PubMed] [Google Scholar]