Abstract
We have generated hexon-modified adenovirus serotype 5 (Ad5) vectors that are not neutralized by Ad5-specific neutralizing antibodies in mice. These vectors are attractive for the advancement of vaccine products because of their potential for inducing robust antigen-specific immune responses in people with prior exposure to Ad5. However, hexon-modified Ad5 vectors displayed an approximate 10-fold growth defect in complementing cells, making potential vaccine costs unacceptably high. Replacing hypervariable regions (HVRs) 1, 2, 4, and 5 with the equivalent HVRs from Ad43 was sufficient to avoid Ad5 preexisting immunity and retain full vaccine potential. However, the resulting vector displayed the same growth defect as the hexon-modified vector carrying all 9 HVRs from Ad43. The growth defect is likely due to a defect in capsid assembly, since DNA replication and late protein accumulation were normal in these vectors. We determined that the hexon-modified vectors have a 32°C cold-sensitive phenotype and selected revertants that restored vector productivity. Genome sequencing identified a single base change resulting in a threonine-to-methionine amino acid substitution at the position equivalent to residue 342 of the wild-type protein. This mutation has a suppressor phenotype (SP), since cloning it into our Ad5 vector containing all nine hypervariable regions from Ad43, Ad5.H(43m-43), increased yields over the version without the SP mutation. This growth improvement was also shown for an Ad5-based hexon-modified vector that carried the hexon hypervariable regions of Ad48, indicating that the SP mutation may have broad applicability for improving the productivity of different hexon-modified vectors.
INTRODUCTION
Adenovirus vectors are considered a leading viral vector platform for vaccines because of their robust immunogenicity and manufacturing feasibility. The most potent adenovirus vectors for use as vaccines are based on adenovirus serotype 5 (Ad5) (1–6). Ad5 vectors can be grown to very high yields in bioreactors and can be purified efficiently with reasonable cost-of-goods estimates for vaccines. Adenovirus-based vectors are capable of generating robust and protective T cell and antibody responses in animal models (7–17), and clinical data conclusively show that Ad5 vectors can induce potent CD8+ and CD4+ T cell and antibody responses in vaccinated volunteers (18–25). Most encouragingly, the protective capacity of a DNA prime-Ad5 boost regimen expressing two malaria antigens demonstrated sterile protection from malaria in 27% of test subjects (26). However, the high prevalence of Ad5-specific neutralizing antibodies (NAb) in human populations, especially in sub-Saharan Africa, has the potential to limit the effectiveness of Ad5-based vaccines (23, 27–30).
Hexon is the most abundant adenoviral structural protein, and studies show that it is the major target for NAb in vivo (23, 31, 32). These NAbs target the nine hypervariable regions that form the exposed surface of the hexon protein (31, 33). We and others have shown that it is possible to replace the nine hypervariable regions of the Ad5 hexon with those derived from group C (31) or group D (34, 35) serotypes. The Ad5-based hexon-modified vectors that contain subgroup D hypervariable regions (Ad43 or Ad48) induced robust transgene-specific immune responses that were unaffected by Ad5-specific NAb in murine and nonhuman primate vaccine models. In addition to their role as the primary determinants of NAb, the Ad5 hexon HVRs play a critical role in vector tropism. In particular, the Ad5 HVR residues interact with scavenger receptors on Kupffer cells (36–39) and with coagulation factors, such as factor X (40–42). Thus, Ad vectors with modifications in the hexon HVRs have potential utility in targeted gene delivery.
A major impediment to the advancement of hexon-modified vectors for vaccines is their reduced growth characteristics in complementing cells, relative to those of unmodified vectors (34). We identified a mutation in the hexon that restores productivity to hexon-modified vectors while retaining their immune potential as vaccines and their ability to avoid preexisting Ad5 NAb. This new vector base will be of particular value in developing nations, which are very sensitive to the cost of goods and have high percentages of seropositivity to Ad5 among their populations.
MATERIALS AND METHODS
Cells and viruses.
293 cells were obtained from the American Type Culture Collection (ATCC, Manassas, VA) and maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS). 293-ORF6 cells express both Ad5 early region 1 (E1) and E4 gene products (43). They support the growth of E1/E4-deleted vectors based on Ad5 as well as alternative adenovirus serotypes (43–46). Adherent 293-ORF6 cells were maintained in DMEM supplemented with 10% FBS, and suspension 293-ORF6 cells were grown in shaker flasks and maintained in serum-free 293SFM II medium with l-glutamine (Life Technologies, NY). Adenovirus vectors (referred to here as adenovectors) were generated by using our plasmid-based adenovirus vector construction system as previously described (35). Briefly, the recombinant Ad5 genomes (containing a transgene expression cassette and hexon modification) were liberated from plasmids by digestion with PacI restriction endonuclease, transfected into monolayer 293 or 293-ORF6 cells, and cell lysates were serially passaged every 3 days until cytopathic effect (CPE) was observed. The CPE lysates were further expanded, analyzed to determine vector titer, particle concentration, and vector genomic integrity by PCR, and used to seed a production run in suspension 293-ORF6 cells in shaker flasks using serum-free media. Suspension 293-ORF6 cells (1 × 106 cells/ml) were infected with recombinant adenovectors at a multiplicity of infection (MOI) of 20 focus-forming units (FFU)/cell and then incubated at 37°C in 5% CO2 on a shaker at 125 rpm. At 18 to 24 h postinfection, the infected cells were fed with an equal volume of medium. At 72 h postinfection, infected cells were pelleted, resuspended in Benzonase buffer (25 mM Tris[pH 8.0], 10 mM NaCl, 5 mM MgCl2, 0.0025% Tween 80), and then frozen and thawed for three cycles. Benzonase (EMD Biosciences, Darmstadt, Germany) was added to the cell lysate at a concentration of 1 unit/2 × 104 cells and incubated at room temperature overnight. Adenovirus vectors were purified by three rounds of continuous cesium chloride (CsCl) isopycnic gradient centrifugation following standard procedures. Briefly, the lysate was layered on top of a 1.33-g/ml CsCl solution and then centrifuged at 66,900 × g for a minimum of 16 h at 4°C. Visible bands were collected from the third equilibrium gradient and dialyzed against final formulation buffer, and the purified vectors were stored at −80°C. Physical particle units (PU) were determined by absorbance at 260 nm (nM) following disruption of the capsid with sodium dodecyl sulfate (SDS).
The Ad5 hexon in the H(43s-43) vector replaces Ad5 HVRs 1, 2, 4, 5, and 7 to 9 with the corresponding HVRs from Ad43; HVRs 3 and 6 are not modified in this vector. The Ad5 hexon in the H(48-48) vector replaces Ad5 HVRs 1 to 9 with the corresponding HVRs from Ad48. The specific modifications to the Ad5 hexon in the H(43m-43) (35), H(43s-43) and H(48-48) vector are provided in Table 1. All of the Plasmodium yoelii circumsporozoite protein (PyCSP)-expressing vectors have E1 and E3 deleted, except for AdPyCSP.11D and AdPyCSP.H(43m-43)0.11D, which have E1, E3, and E4 deleted.
Table 1.
Specific amino acid substitutions in the hexon protein of Ad5 in H(43m-43), H(43s-43), and H(48-48) backbones
| Hexon protein | H(43m-43) |
H(43s-43) |
H(48-48) |
|||
|---|---|---|---|---|---|---|
| Ad5 aaa inserted | Ad43 aa inserted | Ad5 aa deleted | Ad43 aa inserted | Ad5 aa deleted | Ad48 aa inserted | |
| HVR 1 | 136–165 | 136–153 | 136–165 | 136–153 | 136–165 | 136–150 |
| HVR 2 | 190–192 | 178–184 | 190–192 | 178–184 | 190–192 | 173–185 |
| HVR 3 | 212–218 | 204–209 | —b | —b | 212–220 | 203–210 |
| HVR 4 | 252–258 | 244–251 | 252–258 | 244–251 | 248–258 | 238–251 |
| HVR 5 | 271–279 | 264–271 | 270–279 | 263–271 | 268–281 | 261–277 |
| HVR 6 | 305–310 | 279–302 | —b | —b | 305–310 | 301–306 |
| HVR 7 | 418–428 | 410–420 | 418–428 | 410–420 | 418–428 | 414–424 |
| HVR 8 | 435–436 | —c | 435–436 | —c | 431–438 | 427–434 |
| HVR 9 | 440–451 | 430–440 | 440–451 | 430–440 | 443–551 | 439–446 |
aa, amino acids.
This HVR is not present in Ad43, so no additional amino acids are inserted.
This HVR is not modified.
In vitro vector growth assay.
293 cells were seeded in 6-well plates at ∼70% confluence (8.5 × 105 cells/well) and grown in a 5% CO2 incubator at 37°C. After 24 h, cells were infected in triplicate with hexon-modified vectors with or without the suppressor phenotype (SP) mutation or control vectors at an MOI of 10 focus-forming units (FFU)/cell. Infections were performed in a volume of 200 μl of medium at either 32°C or 37°C. Plates were gently rocked every 15 min to ensure good coverage of the monolayer. At the end of the 60-min incubation, virus was aspirated from the cells, and the cells were washed with phosphate-buffered saline (PBS), overlaid with 3 ml of prewarmed DMEM plus 10% FBS, and incubated in a 5% CO2 incubator at either 32°C or 37°C. At various time points postinfection (24, 48, and/or 72 h) the infected cells were scraped into 15-ml polypropylene tubes and frozen and thawed three times to lyse infected cells and release vector particles. The number of viral particles was measured by the FFU assay as follows. 293 or 293-ORF6 cells were seeded in collagen-coated 6-well plates at (1.2 ×106 cells/well) and grown in a 5% CO2 incubator at 37°C. After 24 h, cells were infected with 10-fold dilutions of the viral lysates as described above. Twenty-four hours postinfection, cells were washed with PBS, fixed for 10 min with ice-cold methanol, and assayed for infection by immunofluorescence using monoclonal antibody 38-2 against the Ad5 DNA-binding protein (DBP) (47) conjugated to fluorescein isothiocyanate (FITC). Ten random fields of DBP-positive cells (FFU) were counted from wells containing between 4 and 50 fluorescent cells using a Nikon Eclipse TE300 inverted microscope. The average number of FFU per field was used to calculate the number of FFU/well, the total number of FFU in the cell lysate and the number of FFU/cell. Statistical analysis was performed using unpaired, two-tailed t tests. P values of less than 0.05 were considered significant.
Viral genome replication assay.
A total of 1.1 × 106 cells 293 cells were cultured per well in 6-well plates and incubated overnight at 37°C, 5% CO2. Cells were infected in triplicate with 10 FFU/cell of AdPyCSP, AdPyCSP.H(43m-43), and AdPyCSP.H(43m-43)sp. Infected-cell cultures were incubated at 37°C or 32°C in 5.0% CO2 for 1 h; then, the infection medium was aspirated and cells were overlaid with 2 ml of fresh medium. Cells were placed back in the incubator at 37°C or 32°C with 5.0% CO2 for virus replication. Mock-infected and infected cells were harvested at 0, 6, 12, 24, and 48 h postinfection. Cell pellets were resuspended in 1 ml of PBS. A 200-μl portion of cell suspension was used for DNA extraction using a QIAamp DNA minikit (Qiagen) following the manufacturer's instructions. Isolated DNA was further diluted 1:100 in water. Triplicate quantitative PCRs (qPCRs) were performed using 20 μl of diluted sample, 2× TaqMan universal PCR master mix (Life Technologies, Grand Island, NY), 300 nM forward primer (Ad5s3830 [ATTGTGACTGACTTTGCTTTCCTG]), 300 nM reverse primer (Ad5a3902 [GCCAAAAGAGCCGTCAACTT]), and 250 nM probe (Ad5s3866 [6FAM-AGCAGTGCAGCTTCCCGTTCATCC-MGBNFQ]) in a 50-μl reaction. Thermal cycling was performed on a ViiA 7 instrument using conditions recommended by the manufacturer for a 96-well standard curve assay. Primers and probes align with the polypeptide IX (pIX) reading frame of the Ad5 genome. The control plasmid pAGE1(C.GFP)E3(10X)BR4 concentration was measured by spectrophotometry and diluted to 1 × 108 copies per μl. Further 10-fold dilutions were used as a standard curve for genome quantification, with a range of 2 × 109 to 2 × 102. 293 cells contain multiple copies of the pIX reading frame in their genomes; this is also amplified during the qPCRs. To calculate viral genome copy number per cell, the quantity of pIX from the mock-infected sample at the corresponding temperature and time point was subtracted from the quantity of the infected sample pIX. This value was multiplied by the reciprocal of the dilution factor and divided by 1.1 × 106 cells.
Immunoblot analysis.
For immunoblot analysis, whole-cell lysates from infected cells were electrophoresed on 4 to 12% bis-Tris NuPAGE gels (Novex by Life Technologies, Grand Island, NY), electroblotted to polyvinylidene difluoride (PVDF) membranes (0.2-μm pore size; Life Technologies, Grand Island, NY), blocked with TBST (10 mM Tris [pH 8], 150 mM NaCl, 0.05% Tween 20) containing 5% milk overnight at 4°C, and incubated with (1:10,000) anti-Ad5 rabbit serum. The rabbit serum was obtained by immunizing a New Zealand White rabbit with 1 × 1010 PU AdNull (two 0.5-ml injections into the right quadriceps at an interval of 8 weeks), and the serum was collected 6 weeks after the second administration (48). The immunoblots were washed in TBST and incubated with (1:10,000) donkey anti-rabbit immunoglobulin G (IgG) conjugated to horseradish peroxidase (Millipore, Temecula, CA) for 60 min at room temperature followed by washing in TBST and detection by enhanced chemiluminescence (ECL Prime; GE Life Sciences, Pittsburgh, PA).
Immunizations.
Female 5- to 8-week-old BALB/c AnNCr mice (Harlan, Indianapolis, IN) were immunized with 1 × 108 PU of adenovirus vectors expressing PyCSP in a 0.1-ml volume by intramuscular (IM) immunization (injections into tibialis anterior muscles with a 29 1/2-gauge needle). Vector administrations to generate preexisting Ad5-neutralizing antibodies were delivered by IM injection into the left gastrocnemius muscle (1 × 1010 PU) followed by a booster immunization with the same vector (1 × 1010 PU) given 4 weeks later. Ad5 NAb were generated using an Ad5 vector expressing luciferase, Ad5L, and hexon-specific Ad5 NAb were generated using a chimeric Ad5 vector containing an Ad35 fiber, Ad5L.F(35).
Neutralizing-antibody assay.
Our NAb assay is based on the capacity of antibodies to neutralize adenovectors that express luciferase (49) and is described elsewhere (35). Briefly, BALB/c mice were immunized with two doses of adenovector to generate Ad5-specific NAb. Three or four weeks after the last administration of adenovector, serum was collected, heat inactivated at 56°C for 60 min, serially diluted, and mixed with the luciferase-expressing adenovectors. After incubation at room temperature for 30 min, the virus/serum mixture was used to infect A549 cells (ATCC, Manassas, VA) seeded on 96-well plates. Luciferase activity from individual wells was measured using a 96-well luminometer, and the highest dilution of serum that resulted in 90% inhibition of luciferase activity was determined.
Immunological assays.
Cellular immune responses were assessed by intracellular cytokine staining (ICS) assays (6 mice/group, assayed individually) as described previously (48), using splenocytes harvested from immunized or control mice as effector cells. Targets were major histocompatibility complex (MHC)-matched A20.2J cells pulsed with synthetic peptides. A total of 1 × 106 splenocytes/mouse were stained for viability, CD3, CD8, and gamma interferon (IFN-γ). Splenocytes plus peptide-loaded A20 cells were incubated for 2 h, Golgi inhibited, incubated another 6 to 16 h, and then stained and fixed. All available cells were acquired (LSRII; BD Biosciences) and then analyzed with FlowJo (Mac OSX version 8.8.6). Data are presented as mean percent cytokine producers of total CD3+ CD8+ T cells. Statistical analysis of ICS data was performed using one-way analysis of variance (ANOVA) followed by Bonferroni's mean comparison test. P values of less than 0.05 were considered significant.
RESULTS
Growth and immune avoidance of different Ad5 hexon-modified vectors.
We previously reported that an Ad5-based hexon-modified vector containing substitutions in all nine HVRs [AdCSP.H(43m-43)] induced robust T cell responses in mice with preexisting Ad5 immunity (35). This vector also exhibited reduced growth in 293-ORF6 production cells (Fig. 1). We hypothesized that the growth defect and the immune evasion phenotypes were the result of different HVR substitutions and that by modifying only a subset of the nine HVRs, we could develop a hexon-modified vector that avoids preexisting Ad5 immunity and grows well.
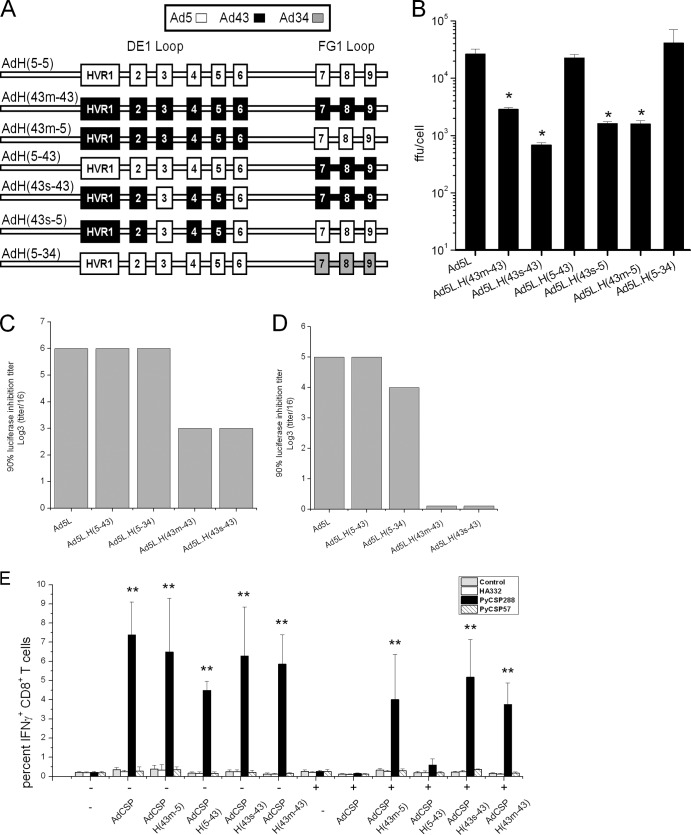
Fig 1.
Ad5 vectors carrying DE1 hypervariable loop substitutions are reduced for growth and avoid Ad5-specific NAb in vitro and in vivo. (A) Schematic representation of Ad5 vectors carrying HVR substitutions in the hexon DE1 and FG1 hypervariable loops. The nomenclature of the vectors consists of “AdH,” which represents the Ad5 hexon, followed by a parenthetical indication of the serotype from which the DE1 and FG1 loop HVRs are derived. (B) Cells were infected with the indicated CsCl-purified vectors at an MOI of 10 FFU/cell. Seventy-two hours later, recombinant vector was harvested from the cells by three cycles of freeze-thawing, and vector yield was determined with a standard FFU assay. Data are means ± SEM (n = 3). *, P < 0.05 compared with Ad5L. (C and D) Mice were immunized with two administrations (1 ×1010 PU each) of adenovector with an interval of 1 month. The serum obtained 3 weeks after the last immunization was pooled and diluted 1:16, and then 3-fold serial dilutions were tested for neutralizing activity for the indicated vectors. Two different adenovectors were used to immunize mice and generate serum, an Ad5 vector [Ad5L] (C) and an Ad5 vector with an Ad35 fiber replacement [Ad5L.F(35)] (D). (E) Naive BALB/c mice (−) or mice preimmunized with two injections (1 ×1010 PU each) of Ad5Null (+) were immunized with the indicated vectors expressing PyCSP. PyCSP-specific T cell responses were assessed by ICS 4 weeks after immunization. Targets were MHC-matched A20.2J cells pulsed with synthetic peptides representing the immunodominant CD8+ T cell epitope (PyCSP280-288) or the CD4+ T cell epitope with nested subdominant CD8+ T cell epitope (PyCSP57-70) from PyCSP or a defined CD8+ T cell epitope for the influenza virus hemagglutinin antigen (HA332). Error bars indicate standard errors of the means (6 per group). **, P < 0.01 compared with naive mice.
In an attempt to decouple the immune evasion from the growth defect phenotypes, we generated a set of adenovectors with various HVR substitutions (Fig. 1A) and tested them for growth and immune evasion. All of these vectors had particle-to-active-particle ratios less than 50 (35). We tested the growth of these vectors by infecting 293-ORF6 cells at an MOI of 10 FFU/cell, and the vector yields were determined 72 h after infection (Fig. 1B). A vector that contained all nine HVRs from Ad43 [Ad5L.H(43m-43)] displayed a 10-fold reduction in growth relative to the unmodified vector [Ad5L]. This growth defect was also observed with vectors that contained HVRs 1 to 6 from Ad43 [Ad5L.H(43m-5)], HVRs 1, 2, 4, and 5 from Ad43 [Ad5L.H(43s-5)], and HVRs 1, 2, 4, 5, 7, 8, and 9 from Ad43 [Ad5L.H(43s-43)]. Vectors that maintain Ad5 HVRs 1 to 5 but carry Ad43 or Ad34 substitutions in HVRs 7, 8, and 9 [Ad5L.H(5-43) and Ad5L.H(5-34)] grew as well as vectors containing the wild-type Ad5 hexon. These results indicate that replacement of Ad5 HVRs 1, 2, 4, and 5 with the comparable HVR sequences from Ad43 has deleterious effects on vector growth and that HVRs 7, 8, and 9 can be modified with no effect on vector yield.
Next we tested these vectors to determine if avoidance of NAb cosegregated with the vector growth phenotype. Mice were immunized with two administrations (1 × 1010 PU each) of Ad5 vector at an interval of 1 month, and the serum from these mice was tested for neutralizing activity against an unmodified Ad5 vector that expresses luciferase [Ad5L] or the chimeric hexon-modified luciferase vectors shown in Fig. 1A. This serum efficiently neutralized the Ad5 vector and the hexon-modified vectors that maintain Ad5 HVRs 1 to 5 but carry Ad43 or Ad34 substitutions in HVRs 7, 8, and 9 [Ad5L.H(5-43) and Ad5L.H(5-34)]. The serum was less effective at neutralizing the hexon-modified vector containing all nine Ad43 HVRs [Ad5L.H(43m-43)] and a vector that contained HVRs 1, 2, 4, 5, and 7 to 9 from Ad43 [Ad5L.H(43s-43)] (Fig. 1C).
To determine if the residual neutralizing activity observed in the hexon-modified vectors was due to NAb specific for the Ad5 fiber, we generated NAb from mice using an Ad5 vector containing an Ad35 fiber. Serum from these mice neutralized the Ad5, Ad5L.H(5-43), and Ad5L.H(5-34) vectors but did not neutralize Ad5L.H(43m-43) and Ad5L.H(43s-43) (Fig. 1D). These data indicate that neutralizing antibodies specific for both the hexon HVRs and the fiber are functional in vitro and that the hexon-specific NAb preferentially target HVRs 1, 2, 4, and 5 or a subset of these.
To assess whether the same HVRs are important for in vivo neutralization, we generated Ad5-specific NAb responses in mice by injecting them twice (with a four-week interval) with 1 × 1010 PU of an Ad5 vector expressing no antigen (Ad5Null), and then 4 weeks later we immunized them with 1 × 108 PU of adenovectors containing different HVRs and expressing PyCSP. In the absence of Ad5-specific NAb, all of the hexon-modified vectors performed similarly (Fig. 1E), inducing approximately 6% of CD8+ T cells to express IFN-γ following stimulation with antigen-presenting cells (APCs) loaded with a peptide encoding the dominant PyCSP CD8+ epitope. This confirmed that hexon-modified adenovectors maintain their immune potential. Mice that received two injections of Ad5Null had high levels of serum Ad5-specific NAb (titers > 2,048; data not shown). When these mice were immunized with vectors containing the wild-type Ad5 hexon [AdCSP] or HVRs 1 to 6 from Ad5 and HVRs 7 to 9 from Ad43 [AdCSP.H(5-43)], they did not produce PyCSP-specific CD8+ T cell responses that were significantly different from those in naive mice. Vectors with Ad43 substitutions in HVRs 1 to 6 [AdCSP.H(43m-5), HVRs 1 to 9 [Ad5L.H(43m-43)], and HVRs 1, 2, 4, 5, and 7 to 9 [Ad5L.H(43s-43)] were capable of inducing robust CD8+ T cell responses even in the presence of Ad5 NAb. These results suggest that the hexon-specific NAb that function in vivo target Ad5 HVRs 1, 2, 4, and 5 or a subset of these.
Taken together, these results indicate that modification of HVRs 7 to 9 does not have a deleterious effect on vector growth but also does not enable these vectors to avoid Ad5 NAb. On the other hand, vectors with modification of HVRs 1, 2, 4, and 5 avoid Ad5 NAb in vitro and in vivo, but these vectors are subject to a growth disadvantages relative to unmodified Ad5-based vectors.
Cold-sensitive phenotype of the hexon-modified vector.
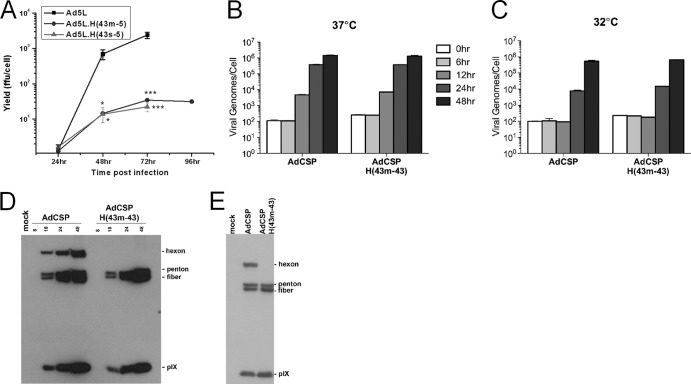
The growth reduction that we observed with hexon-modified vectors may be due to disruption of structural interactions within the viral capsid, as the HVR loops that reside on the surface of the virion interact with each other to stabilize the trimeric structure of the hexon (50–52) and interact with neighboring hexon trimers or other capsid components (53–57). We reasoned that if there was a structural change in the capsid of the hexon-modified vectors, we might be able to demonstrate reduced stability of the vector when exposed to destabilizing forces. We performed experiments to determine whether these chimeric vectors were less stable than unmodified vector by analyzing their stability following incubation at 37°C or 42°C and by exposing the vectors to multiple freeze-thaw cycles (data not shown). None of these stresses could distinguish the hexon-modified from the nonmodified vectors in a loss-of-titer assay. However, we did uncover a cold-sensitive phenotype when growing these vectors at 32°C. At this temperature, the hexon-modified vectors showed a 100-fold growth defect relative to an unmodified Ad5 vector (Fig. 2A). Growth of the Ad5L.H(43m-5) vector reached a plateau between 72 and 96 h after infection, indicating that the reduction in viral yield was not due to a delay in growth. We observed similar differences in the growth of AdCSP versus AdCSP.H(43m-43) and AdCSP.H(43s-43), indicating that the transgene did not contribute to the cold-sensitive growth defect.
Fig 2.
Cold-sensitive phenotype of hexon-modified vectors. 293 cells were infected with the indicated vectors carrying substitutions in the Ad5 hexon DE1 hypervariable loop at an MOI of 10 FFU/cell. (A) Infected cells were incubated at 32°C. At the indicated time points, recombinant vector was harvested from the cells by three cycles of freeze-thawing, and vector yield was determined using a standard FFU assay. Data are means ± SEM (n = 2). *, P < 0.05, and ***, P < 0.001, relative to Ad5L. To assess viral DNA replication, infected cells were incubated at 37°C (B) and 32°C (C) and harvested at various time points after infection. Viral DNA was extracted and analyzed by qPCR. Genome copy numbers per cell were calculated as the difference between the sample and mock cells, divided by the number of cells infected. Error bars indicate standard errors of the means (n = 3). To assess late adenovirus protein accumulation, infected cells were incubated at 37°C (D) and 32°C (E) and harvested at various time points (above each lane, in hours) after infection. Infected cell lysates were obtained, and immunoblots were probed with anti-adenovirus polyclonal antisera.
Our hypothesis that the reduced growth of AdH(43m-43) vectors is due to changes in structural interactions within the viral capsids predicted that these vectors would replicate their DNA and accumulate late proteins normally. To test this, we compared viral DNA replication and late protein accumulation between an unmodified vector (AdCSP) and a hexon-modified vector that displayed the growth defect [AdCSP.H(43m-43)]. We quantified viral genome copy numbers at various time points following infection of 293 cells by qPCR (Fig. 2B and C). At 37°C, viral DNA replication was apparent at 12 h and increased at 24 and 48 h after infection with both vectors. At 32°C, DNA replication was delayed substantially for both vectors, but by 48 h after infection, replication was similar to that seen at 37°C. We did not observe differences between the hexon-modified and unmodified vectors at any time point.
To determine if the hexon-modified vector displayed a defect in late protein accumulation, we infected 293 cells with AdCSP and AdCSP.H(43m-43) at an MOI of 10 FFU/cell at 32°C and 37°C. Cell lysates were prepared at various times after infection, and immunoblots were probed with an antibody that detected hexon, penton, fiber, and pIX proteins (Fig. 2D and E). When vectors were grown at 37°C, penton, fiber, and pIX were observed 18 h after infection, accumulated to higher levels at 24 and 48 h, and were indistinguishable between cells infected with the unmodified and hexon-modified vector (Fig. 2D). At 32°C we observed similar accumulations of penton, fiber, and pIX at 48 h postinfection for both infections (Fig. 2E). The rabbit antisera recognized hexon from the unmodified but not the hexon-modified vector, indicating that the hexon-specific antibodies that were detected in the immunoblot assay were specific for the HVR residues.
Selection of a hexon-modified vector with improved growth.
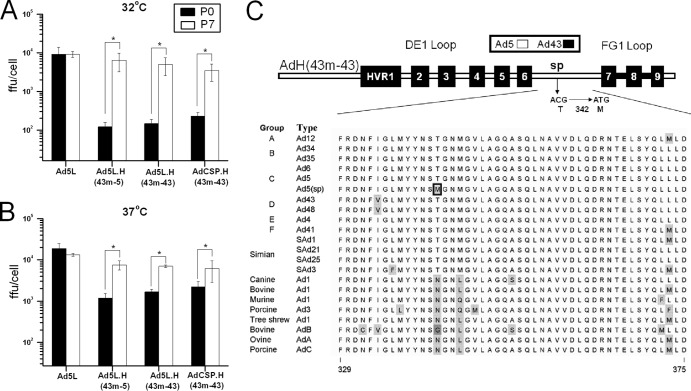
The dramatic growth defect that we observed with the hexon-modified vectors that carried HVR 1, 2, 4, and 5 from Ad43 allowed us to select for a vector with improved growth. We serially passaged three cold-sensitive hexon-modified vectors, Ad5L.H (43m-5), Ad5L.H(43m-43), and AdCSP.H(43m-43), at 32°C in 293 cells. The growth of these vectors at 32°C and 37°C was determined using material from passage 0 (P0) and passage 7 (P7). At 32°C, we saw dramatic increases in the growth properties of each of the hexon-modified vector P7 stocks relative to the P0 stocks (Fig. 3A). Substantial differences in titers were also observed between the P0 and P7 stocks when the growth was analyzed at 37°C (Fig. 3B). These differences in the growth of the P0 and P7 stocks were not seen with the unmodified Ad5L control vector. These results suggested that we had selected mutants with improved growth, which subsequently became the dominant genetic entity in the population.
Fig 3.
Selection of revertant vectors with improved growth following serial passaging at 32°C. Control and hexon-modified vectors were serially passaged seven times in 293 cells at 32°C. The growth of vectors from the original vector stock (passage 0) and from passage 7 were determined by infecting 293 cells at an MOI of 10 FFU/cell and incubating them at 32°C (A) and 37°C (B). Infected cells were harvested 72 h after infection, and virus yield was determined by using the FFU assay. Error bars indicate standard errors of the means (n = 3). *, P < 0.05. (C) Schematic representation of the SP mutation in a highly conserved region between HVR 6 and HVR 7. The position of the SP mutation is indicated. An amino acid alignment of the region between HVR 6 and HVR7 (AA329 to AA375) is shown (amino acid numbers are based on the Ad5 hexon sequence). The boxed methionine indicates the location of the SP mutation in the AdH(43m-43) vector. Shaded amino acids indicate residues that are not conserved relative to Ad5. Serotypes from each of the human adenovirus groups as well as nonhuman primate and nonhuman species are represented.
To identify the mutation(s) that were associated with the improved growth of the P7 stocks, we sequenced hexon, pIX, pIIIa, and 100K. 100K mediates trimerization of the hexon during capsid assembly (58, 59), and hexon HVR4 is thought to interact with pIX (53, 54) and pIIIa (53, 54). No alterations were found in pIX, IIIa, or 100K. However, we did identify a single nucleotide substitution (cytosine to thymine) at a position equivalent to nucleotide position 1025 in the wild-type Ad5 hexon gene in two of the three P7 stocks, AdH(43m-43) and AdH(43m-5) (Table 2). This mutation was predicted to result in a threonine (T)-to-methionine (M) substitution at the position corresponding to amino acid 342 (T342M) in the wild-type Ad5 hexon. We hypothesized that this mutation was responsible for the suppressor phenotype (SP) of the P7 viruses. The mutation occurred in a highly conserved region between the DE1 and FG1 loops of the hexon (Fig. 3C). The threonine is completely conserved in all human and simian adenovirus species. However, nonprimate adenoviruses typically have an asparagine or glycine residue at this position.
Table 2.
Nucleotides at position 1025a in the Ad5 hexon gene
| Vector | Nucleotide in: |
|
|---|---|---|
| Passage 0 | Passage 7 | |
| Ad5L.H(5-5) | C | C |
| Ad5L.H(43m-43) | C | T |
| Ad5L.H(43m-5) | C | T |
| AdCSP.H(43m-43) | C | C |
Nucleotide position with respect to the wild-type Ad5 hexon gene.
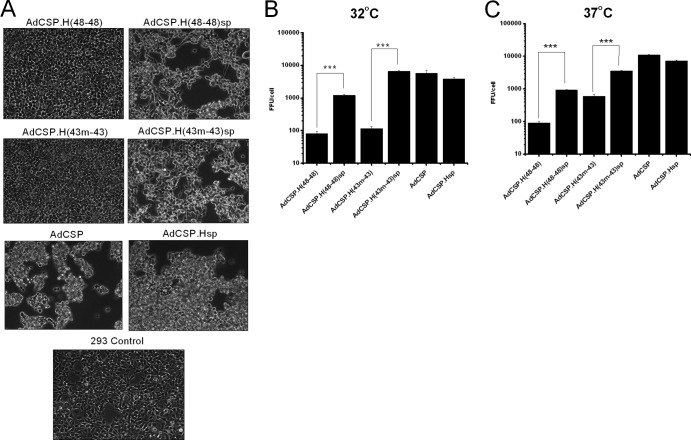
To determine if the reverted phenotype was a direct consequence of the T342M hexon mutation and not due to unidentified second-site mutations, we constructed a new vector, AdCSP.H(43m-43sp), based on the hexon-modified vector backbone and containing the T342M mutation. The hexon gene in this new vector is identical to the revertant selected and analyzed for Fig. 3. We then tested this new vector for its growth potential in 293 cells. Adenovirus vectors grown in monolayer cultures of complementing cells induce a cellular cytopathic effect (CPE), which is an indicator of vector replication. We observed CPE 72 h after infection with an Ad5 vector containing a wild-type hexon gene (AdCSP) but not with the vector containing the HVR residues of Ad43 [AdCSP.H(43m-43)]. As predicted, the T342M mutation restored the growth of the defective hexon-modified vector as determined by CPE observation at 72 h postinfection (Fig. 4A). Moreover, this T342M mutation fully restored the growth of the hexon-modified vector at 32°C, representing an 80-fold increase in vector productivity over the original hexon-modified vector (Fig. 4B). Similarly, we observed a 6-fold increase in vector yields at 37°C, approaching the productivity of Ad5 vectors carrying wild-type hexons (Fig. 4C). It was formally possible that another mutation in an adenovirus gene contributed to the enhanced growth observed with AdCSP.H(43m-43)sp and the passage 7 material from Ad5L.H(43m-43). To test this, we plaque purified and expanded the Ad5L.H(43m-43) (P7) material to passage 11, purified the vector by three rounds of CsCl isopycnic gradient centrifugation, and obtained the full-length sequence. Sequencing revealed a single nucleotide change at a position equivalent to nucleotide 1025 in the wild-type hexon but did not detect any additional mutations. These results indicate that a single amino acid substitution at hexon position 342 substantially increased the yield of a hexon-modified vector.
Fig 4.
The SP mutation in the hexon rescues the growth of the hexon-modified vectors. The SP mutation was cloned into the genomes of two different hexon-modified vectors and a vector with an unmodified hexon gene. The growth phenotype of these vectors was analyzed following infection of 293 cells at 32°C or at 37°C. 293 cells were infected with the indicated vectors with or without SP mutation at an MOI of 10 FFU/cell. (A) Images of cells after infection with the indicated vectors and incubation at 32°C for 72 h. The growth of each vector was assessed 72 h after infection from cell lysates of cells incubated at 32°C (B) and 37°C (C) following infection. Error bars indicate standard errors of the means (n = 3). ***, P < 0.001.
To determine if the SP mutation would improve the growth of a vector containing a wild-type hexon or a vector containing different substitutions in the Ad5 hexon HVRs, we built the SP mutation into a vector with a wild-type hexon (AdCSP.Hsp) and a different hexon-modified vector containing all nine HVRs sequences derived from Ad48 [AdCSP.H(48-48)sp]. These vectors were then tested for growth along with controls carrying the same hexon sequences but without the SP mutation. The SP mutation increased productivity of the AdCSP.H(48-48) vector when tested at both 32°C and 37°C but did not increase the yields of a vector carrying the wild-type Ad5 hexon (Fig. 4). This result indicates that the SP mutation can improve the growth of different hexon-modified vectors.
To determine if the SP mutation had any effect on the immunogenicity of hexon-modified Ad5 vectors, we compared induction of T cell responses in mice immunized with unmodified, hexon-modified, and hexon-modified SP vectors expressing PyCSP. BALB/c mice were immunized with 1 × 108 PU of PyCSP-expressing adenovectors, and antigen-specific T cell responses were evaluated by ICS two weeks postimmunization. All of the PyCSP expressing vectors induced robust PyCSP-specific IFN-γ+ CD8+ T cell responses. There were no significant differences in CSP-specific T cell responses among the mice immunized with the E1-, E3-deleted vectors [AdCSP, AdCSP.H(43m-43), AdCSP.H(43m-43)sp, AdCSP.H(48-48), and AdCSP.H(48-48)sp] (Fig. 5). In addition, we observed comparable T cell responses among mice immunized with unmodified or hexon-modified E1-, E3-, E4-deleted (11D) vectors.
Fig 5.
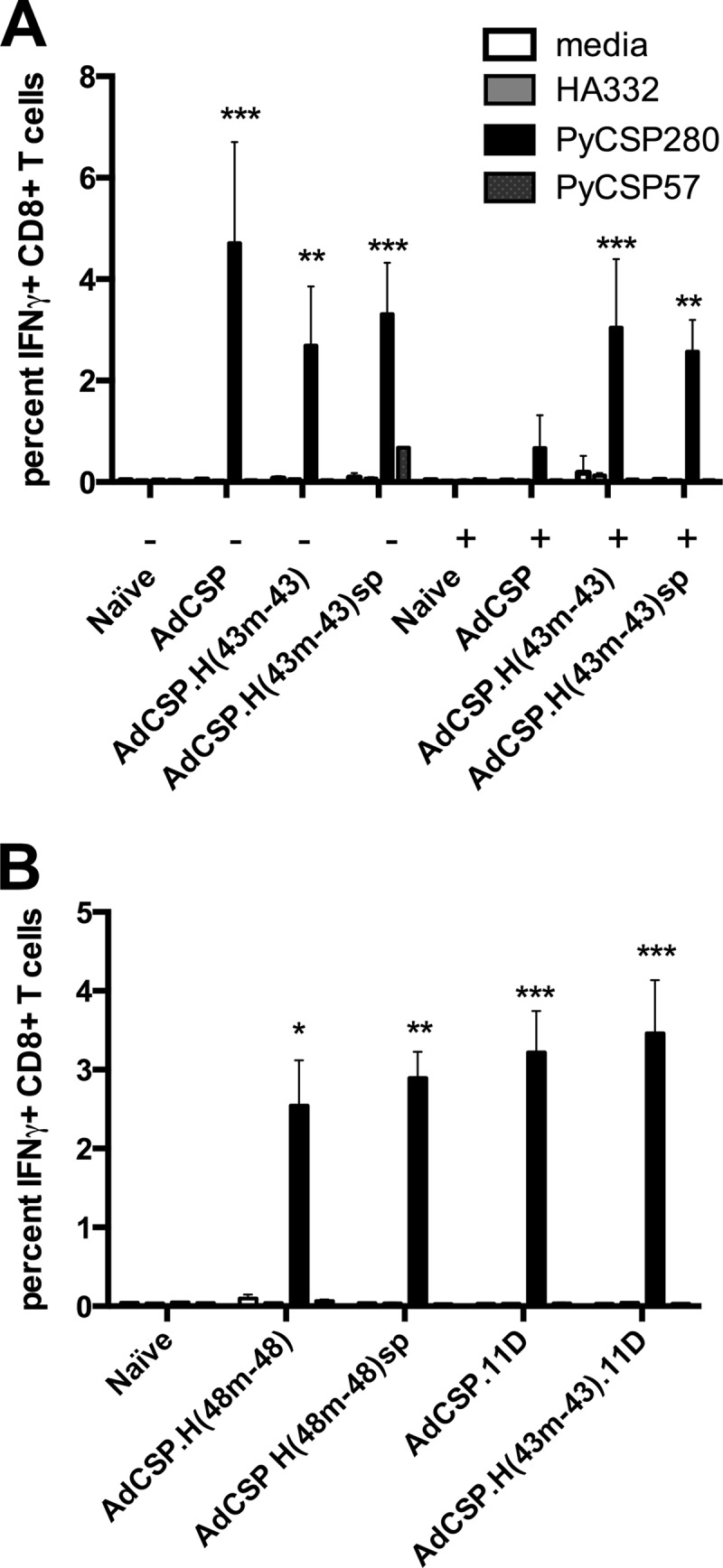
The SP mutation does not affect immunogenicity of hexon-modified Ad vectors. PyCSP-specific T cell responses were assessed by ICS 2 weeks after immunization with PyCSP expressing vectors. Targets were MHC-matched A20.2J cells pulsed with synthetic peptides, as indicated. (A) BALB/c mice were preimmunized with two injections (1 ×1010 PU each) of Ad5Null vector administered at an interval of 4 weeks. Four weeks later, naive mice (−) and the preimmunized mice (+) were immunized with AdCSP or PyCSP-expressing Ad5 hexon-modified vectors containing HVRs from Ad43 with or without the SP mutation, as indicated. (B) Naive mice were immunized with E1-, E3-deleted, PyCSP-expressing, Ad5 hexon-modified vectors containing HVRs from Ad48 with or without the SP mutation, an E1-, E3-, E4-deleted vector (11D) with a wild-type hexon, and an E1-, E3-, E4-deleted hexon-modified vector containing the HVRs from Ad43, as indicated. Error bars indicate standard deviations of the means (6 per group). *, P < 0.05, **, P < 0.01, and ***, P < 0.001, compared with the value for naive mice.
To determine the effect of preexisting Ad5 NAb on the immunogenicity of these vectors, mice were preimmunized with two administrations (1 × 1010 PU each) of Ad5Null to generate high levels of preexisting Ad5 NAb. As previously described (34, 35), the unmodified vector [AdCSP] was substantially reduced in its capacity to induce PyCSP-specific T cell responses under these conditions. In contrast, both the hexon-modified [AdCSP.H(43m-43)] and the hexon-modified vector with the SP mutation [AdCSP.H(43m-43sp)] induced significantly higher PyCSP-specific T cell responses in preimmunized mice (Fig. 5A). Thus, hexon-modified vectors carrying the SP mutation retain their full vaccine potential and avoid preexisting Ad5 NAb in vivo.
The ability of the SP mutation to increase vector productivity is not due to an apparent change in capsid composition or particle-to-FFU ratios. All of the vectors containing the SP mutation produced to date have particle-to-FFU ratios of <50. The protein composition of vectors carrying the SP mutation, as assessed by silver-stained gels, was identical to those of wild-type Ad5 and a hexon-modified vector without the SP mutation (data not shown). Also, there was no difference among unmodified, hexon-modified and hexon-modified SP vectors with respect to viral DNA replication and late protein accumulation (data not shown).
DISCUSSION
Preexisting Ad5 NAb which are prevalent in human populations (23, 27–30) have been shown to inhibit vaccines based on Ad5 (18, 20). The serotype-specific NAb that are most potent at inhibiting the immunogenicity of adenovirus vectors bind to the HVR residues (23, 31, 32, 60). We developed a hexon-modified Ad5 vector, termed AdH(43m-43), in which all nine HVRs in the Ad5 hexon were replaced with those from the rare serotype Ad43. This hexon-modified vector was capable of inducing robust antigen-specific immune responses in mice with high Ad5 NAb titers (35), suggesting that this vector may be capable of circumventing preexisting Ad5 NAb in humans. Thus, hexon-modified Ad5 vectors have potential utility in vaccine and therapeutic applications. However, we observed a substantial (10-fold) growth defect with this vector. The poor growth of hexon-modified Ad5 vectors would likely increase the cost of goods for products utilizing this technology, an issue that is especially relevant in developing countries. Our discovery of a mutation that improved the growth of hexon-modified Ad vectors will facilitate the advancement of this technology for malaria and other vaccine applications.
We hypothesized that the growth defect was due to altered interactions between the Ad43 HVR residues and the other Ad5 capsid components and that this change in protein-protein interactions may affect the efficiency of virus assembly or the stability of the capsids. Our finding that adenovirus DNA replication and late protein synthesis is normal in hexon-modified vectors supports this hypothesis. We could not identify conditions that affected the stability of the Ad5L.H(43m-43) capsids to a greater degree than unmodified Ad5 vectors. However, this vector displayed a cold-sensitive phenotype, showing 100-fold-reduced growth in 293 cells at 32°C. Since cold-sensitive mutations are thought to reflect a change in the ability of a protein to interact with another viral or cellular macromolecule (61), these results suggested that protein-protein interactions important for the assembly of capsids may be affected by the HVR substitutions and that growth at suboptimal temperature magnifies the effect.
Mutations in adenovirus that severely affect viral growth can sometimes be rescued by serial propagation of the defective virus in cell culture. The growth defect results in selection of revertants with improved growth characteristics (62). Since growth of Ad5L.H(43m-5) was reduced 100-fold at 32°C, these conditions were ideal for selection of a revertant. By passaging this vector seven times in 293 cells at 32°C, we selected a revertant with vastly improved growth characteristics. The improvement in growth was associated with and caused by a second-site mutation, a T-to-M substitution at amino acid position 342 in the hexon (relative to the wild-type Ad5 hexon sequence). This mutation was termed SP, for “suppressor mutation,” and it occurred in a highly conserved region between HVR6 and HVR7. The growth-rescuing effect of the SP mutation was not limited to the AdH(43m-5)] vector. Incorporation of the SP mutation rescued the growth defects of other vectors containing different Ad43 HVR substitutions as well as a vector containing all nine HVRs from Ad48. These findings indicate that the SP mutation may have broad applications to other hexon-modified vectors with growth defects. It would be interesting to compare the structures of a hexon-modified vector, a hexon-modified vector carrying the SP mutation, and a vector carrying the wild-type hexon to determine if there are key changes in the structure of the hexon or adenovirus capsid which are induced by the substitution of HVR residues and corrected by the revertant. We also selected a vector with improved growth that did not have the T-to-M substitution at amino acid position 342. Sequencing of the hexon, pIX, pIIIa, and 100K did not reveal any mutations, suggesting that the improved growth of this vector was due to alterations in other adenovirus proteins. Full genome sequencing of this vector may reveal or confirm interactions between the hexon and other minor capsid proteins (53–57).
Our work also identified a subset of HVRs that are critical for immune avoidance and contribute to the growth defect we observed with AdH(43m-43)-based vectors. Using a set of vectors with different HVR substitutions, we determined that HVRs 1, 2, 4, and 5 or a subset of these were the key determinants for Ad5 NAb in vitro and in vivo. Substitution of only these HVRs with those derived from Ad43 conferred on the hybrid vector the capacity to avoid Ad5-specific NAb responses in mice but also reduced vector growth in complementing cells. Substitution of Ad5 HVRs 7, 8, and 9 with Ad43 HVRs had minimal impact on the immune avoidance phenotype and did not reduce vector yields in complementing cells. Earlier published studies showed that Ad5 vectors with modifications to only HVR1 (63) or HVR5 (64) avoided Ad5 NAb in vitro and could induce transgene-specific immune responses in animals previously exposed to Ad5. However, more recent work indicates that Ad5-, Ad3-, and Ad7-specific NAb target multiple HVRs (65, 66). Ad5-based hexon-modified vectors carrying HVRs 1 to 3, HVR 4, or HVR 5 of Ad48 underperformed relative to an Ad5 vector containing all HVRs of Ad48, indicating that modification to multiple HVRs is necessary for full immune avoidance in vivo (65). Our results extend these findings and demonstrate that Ad5 HVRs 1, 2, 4, and 5, or a subset of these, contribute substantially to immune avoidance, whereas HVRs 7 to 9 do not, at least in BALB/c mice.
Recently, the U.S. Navy demonstrated that a DNA prime-Ad5 boost regimen expressing two malaria antigens induced robust T cell responses and protected 4/15 volunteers from experimental challenge by the bites of infected mosquitoes. Of those 15 volunteers, 5 had high titers of Ad5 NAb (>500) and 10 had low titers (<500). Interestingly, all of the protected individuals were in the low-titer group (26). Although this result lacks statistical significance because of the small numbers of volunteers in the trial, the trend suggests that in addition to blunting vaccine-induced immune responses, Ad5 NAb may also prevent Ad5-based vaccine efficacy. Three large trials now have been conducted using Ad5 in the HIV system, and none have shown evidence of efficacy. In all the studies, the vector was potently immunogenic. In one study (STEP), there was a suggestion of increased HIV infection in the vaccinated group that may have been associated with preexisting neutralizing antibodies to Ad5 (19). The technology described in this paper may provide a means of limiting the impact of antibody responses to adenovirus vectors such as Ad5 and possibly those derived from other subgroups or species. These data support the advancement of novel adenovirus vectors that are as immunogenic as Ad5 but are not inhibited by highly prevalent Ad5-specific NAb. The Ad5 hexon-modified vectors described here possess these functional characteristics and are a promising platform for vaccine development.
ACKNOWLEDGMENTS
We thank Rena Cohen for help with the preparation of the manuscript, Chris Lazarski for help with figures and statistics, Grant Liao for assistance with neutralizing antibody assays, Holly Torano for assistance with immunoblotting, Jennifer Tseng for assistance with plaque purification of the passage 9 material, and Randy Osborn for animal vector administrations and animal care.
This work was supported by Advanced Technology Small Business Innovation Research grant 1R43 AI077309-01 from the National Institutes of Health, National Institute for Allergy and Infectious Diseases.
Footnotes
Published ahead of print 3 July 2013
REFERENCES
- 1.Lemckert AA, Sumida SM, Holterman L, Vogels R, Truitt DM, Lynch DM, Nanda A, Ewald BA, Gorgone DA, Lifton MA, Goudsmit J, Havenga MJ, Barouch DH. 2005. Immunogenicity of heterologous prime-boost regimens involving recombinant adenovirus serotype 11 (Ad11) and Ad35 vaccine vectors in the presence of anti-ad5 immunity. J. Virol. 79:9694–9701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shiver JW, Emini EA. 2004. Recent advances in the development of HIV-1 vaccines using replication-incompetent adenovirus vectors. Annu. Rev. Med. 55:355–372 [DOI] [PubMed] [Google Scholar]
- 3.Ophorst OJ, Radosevic K, Havenga MJ, Pau MG, Holterman L, Berkhout B, Goudsmit J, Tsuji M. 2006. Immunogenicity and protection of a recombinant human adenovirus serotype 35-based malaria vaccine against Plasmodium yoelii in mice. Infect. Immun. 74:313–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barouch DH, Pau MG, Custers JH, Koudstaal W, Kostense S, Havenga MJ, Truitt DM, Sumida SM, Kishko MG, Arthur JC, Korioth-Schmitz B, Newberg MH, Gorgone DA, Lifton MA, Panicali DL, Nabel GJ, Letvin NL, Goudsmit J. 2004. Immunogenicity of recombinant adenovirus serotype 35 vaccine in the presence of pre-existing anti-Ad5 immunity. J. Immunol. 172:6290–6297 [DOI] [PubMed] [Google Scholar]
- 5.Ophorst OJ, Kostense S, Goudsmit J, De Swart RL, Verhaagh S, Zakhartchouk A, Van Meijer M, Sprangers M, Van Amerongen G, Yuksel S, Osterhaus AD, Havenga MJ. 2004. An adenoviral type 5 vector carrying a type 35 fiber as a vaccine vehicle: DC targeting, cross neutralization, and immunogenicity. Vaccine 22:3035–3044 [DOI] [PubMed] [Google Scholar]
- 6.Hensley SE, Cun AS, Giles-Davis W, Li Y, Xiang Z, Lasaro MO, Williams BR, Silverman RH, Ertl HC. 2007. Type I interferon inhibits antibody responses induced by a chimpanzee adenovirus vector. Mol. Ther. 15:393–403 [DOI] [PubMed] [Google Scholar]
- 7.Jacobs SC, Stephenson JR, Wilkinson GW. 1992. High-level expression of the tick-borne encephalitis virus NS1 protein by using an adenovirus-based vector: protection elicited in a murine model. J. Virol. 66:2086–2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiang ZQ, Yang Y, Wilson JM, Ertl HC. 1996. A replication-defective human adenovirus recombinant serves as a highly efficacious vaccine carrier. Virology 219:220–227 [DOI] [PubMed] [Google Scholar]
- 9.Lubeck MD, Natuk R, Myagkikh M, Kalyan N, Aldrich K, Sinangil F, Alipanah S, Murthy SC, Chanda PK, Nigida SM, Jr, Markham PD, Zolla-Pazner S, Steimer K, Wade M, Reitz MS, Jr, Arthur LO, Mizutani S, Davis A, Hung PP, Gallo RC, Eichberg J, Robert-Guroff M. 1997. Long-term protection of chimpanzees against high-dose HIV-1 challenge induced by immunization. Nat. Med. 3:651–658 [DOI] [PubMed] [Google Scholar]
- 10.Rodrigues EG, Zavala F, Eichinger D, Wilson JM, Tsuji M. 1997. Single immunizing dose of recombinant adenovirus efficiently induces CD8+ T cell-mediated protective immunity against malaria. J. Immunol. 158:1268–1274 [PubMed] [Google Scholar]
- 11.Sullivan NJ, Geisbert TW, Geisbert JB, Xu L, Yang ZY, Roederer M, Koup RA, Jahrling PB, Nabel GJ. 2003. Accelerated vaccination for Ebola virus haemorrhagic fever in non-human primates. Nature 424:681–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shiver JW, Fu TM, Chen L, Casimiro DR, Davies ME, Evans RK, Zhang ZQ, Simon AJ, Trigona WL, Dubey SA, Huang L, Harris VA, Long RS, Liang X, Handt L, Schleif WA, Zhu L, Freed DC, Persaud NV, Guan L, Punt KS, Tang A, Chen M, Wilson KA, Collins KB, Heidecker GJ, Fernandez VR, Perry HC, Joyce JG, Grimm KM, Cook JC, Keller PM, Kresock DS, Mach H, Troutman RD, Isopi LA, Williams DM, Xu Z, Bohannon KE, Volkin DB, Montefiori DC, Miura A, Krivulka GR, Lifton MA, Kuroda MJ, Schmitz JE, Letvin NL, Caulfield MJ, Bett AJ, Youil R, Kaslow DC, Emini EA. 2002. Replication-incompetent adenoviral vaccine vector elicits effective anti-immunodeficiency-virus immunity. Nature 415:331–335 [DOI] [PubMed] [Google Scholar]
- 13.Gilbert SC, Schneider J, Hannan CM, Hu JT, Plebanski M, Sinden R, Hill AV. 2002. Enhanced CD8 T cell immunogenicity and protective efficacy in a mouse malaria model using a recombinant adenoviral vaccine in heterologous prime-boost immunisation regimes. Vaccine 20:1039–1045 [DOI] [PubMed] [Google Scholar]
- 14.Tan Y, Hackett NR, Boyer JL, Crystal RG. 2003. Protective immunity evoked against anthrax lethal toxin after a single intramuscular administration of an adenovirus-based vaccine encoding humanized protective antigen. Hum. Gene Ther. 14:1673–1682 [DOI] [PubMed] [Google Scholar]
- 15.Wang J, Thorson L, Stokes RW, Santosuosso M, Huygen K, Zganiacz A, Hitt M, Xing Z. 2004. Single mucosal, but not parenteral, immunization with recombinant adenoviral-based vaccine provides potent protection from pulmonary tuberculosis. J. Immunol. 173:6357–6365 [DOI] [PubMed] [Google Scholar]
- 16.Epstein SL, Kong WP, Misplon JA, Lo CY, Tumpey TM, Xu L, Nabel GJ. 2005. Protection against multiple influenza A subtypes by vaccination with highly conserved nucleoprotein. Vaccine 23:5404–5410 [DOI] [PubMed] [Google Scholar]
- 17.Worgall S, Krause A, Rivara M, Hee KK, Vintayen EV, Hackett NR, Roelvink PW, Bruder JT, Wickham TJ, Kovesdi I, Crystal RG. 2005. Protection against P. aeruginosa with an adenovirus vector containing an OprF epitope in the capsid. J. Clin. Invest. 115:1281–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Catanzaro AT, Koup RA, Roederer M, Bailer RT, Enama ME, Moodie Z, Gu L, Martin JE, Novik L, Chakrabarti BK, Butman BT, Gall JG, King CR, Andrews CA, Sheets R, Gomez PL, Mascola JR, Nabel GJ, Graham BS. 2006. Phase 1 safety and immunogenicity evaluation of a multiclade HIV-1 candidate vaccine delivered by a replication-defective recombinant adenovirus vector. J. Infect. Dis. 194:1638–1649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buchbinder SP, Mehrotra DV, Duerr A, Fitzgerald DW, Mogg R, Li D, Gilbert PB, Lama JR, Marmor M, Del Rio C, McElrath MJ, Casimiro DR, Gottesdiener KM, Chodakewitz JA, Corey L, Robertson MN. 2008. Efficacy assessment of a cell-mediated immunity HIV-1 vaccine (the Step Study): a double-blind, randomised, placebo-controlled, test-of-concept trial. Lancet 372:1881–1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McElrath MJ, De Rosa SC, Moodie Z, Dubey S, Kierstead L, Janes H, Defawe OD, Carter DK, Hural J, Akondy R, Buchbinder SP, Robertson MN, Mehrotra DV, Self SG, Corey L, Shiver JW, Casimiro DR. 2008. HIV-1 vaccine-induced immunity in the test-of-concept Step Study: a case-cohort analysis. Lancet 372:1894–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abel B, Tameris M, Mansoor N, Gelderbloem S, Hughes J, Abrahams D, Makhethe L, Erasmus M, de Kock M, van der Merwe L, Hawkridge A, Veldsman A, Hatherill M, Schirru G, Pau MG, Hendriks J, Weverling GJ, Goudsmit J, Sizemore D, McClain JB, Goetz M, Gearhart J, Mahomed H, Hussey GD, Sadoff JC, Hanekom WA. 2010. The novel tuberculosis vaccine, AERAS-402, induces robust and polyfunctional CD4+ and CD8+ T cells in adults. Am. J. Respir. Crit. Care Med. 181:1407–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sedegah M, Kim Y, Peters B, McGrath S, Ganeshan H, Lejano J, Abot E, Banania G, Belmonte M, Sayo R, Farooq F, Doolan DL, Regis D, Tamminga C, Chuang I, Bruder JT, King CR, Ockenhouse CF, Faber B, Remarque E, Hollingdale MR, Richie TL, Sette A. Identification and localization of minimal MHC-restricted CD8+ T cell epitopes within the Plasmodium falciparum AMA1 protein. Malar. J. 9:241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sumida SM, Truitt DM, Lemckert AA, Vogels R, Custers JH, Addo MM, Lockman S, Peter T, Peyerl FW, Kishko MG, Jackson SS, Gorgone DA, Lifton MA, Essex M, Walker BD, Goudsmit J, Havenga MJ, Barouch DH. 2005. Neutralizing antibodies to adenovirus serotype 5 vaccine vectors are directed primarily against the adenovirus hexon protein. J. Immunol. 174:7179–7185 [DOI] [PubMed] [Google Scholar]
- 24.Tamminga C, Sedegah M, Regis D, Chuang I, Epstein JE, Spring M, Mendoza-Silveiras J, McGrath S, Maiolatesi S, Reyes S, Steinbeiss V, Fedders C, Smith K, House B, Ganeshan H, Lejano J, Abot E, Banania GJ, Sayo R, Farooq F, Belmonte M, Murphy J, Komisar J, Williams J, Shi M, Brambilla D, Manohar N, Richie NO, Wood C, Limbach K, Patterson NB, Bruder JT, Doolan DL, King CR, Diggs C, Soisson L, Carucci D, Levine G, Dutta S, Hollingdale MR, Ockenhouse CF, Richie TL. 2011. Adenovirus-5-vectored P. falciparum vaccine expressing CSP and AMA1. Part B: safety, immunogenicity and protective efficacy of the CSP component. PLoS One 6:e25868. 10.1371/journal.pone.0025868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sedegah M, Tamminga C, McGrath S, House B, Ganeshan H, Lejano J, Abot E, Banania GJ, Sayo R, Farooq F, Belmonte M, Manohar N, Richie NO, Wood C, Long CA, Regis D, Williams FT, Shi M, Chuang I, Spring M, Epstein JE, Mendoza-Silveiras J, Limbach K, Patterson NB, Bruder JT, Doolan DL, King CR, Soisson L, Diggs C, Carucci D, Dutta S, Hollingdale MR, Ockenhouse CF, Richie TL. 2011. Adenovirus 5-vectored P. falciparum vaccine expressing CSP and AMA1. Part A: safety and immunogenicity in seronegative adults. PLoS One 6:e24586. 10.1371/journal.pone.0024586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chuang I, Sedegah M, Cicatelli S, Spring M, Polhemus M, Tamminga C, Patterson N, Guerrero M, Bennett JW, McGrath S, Ganeshan H, Belmonte M, Farooq F, Abot E, Banania JG, Huang J, Newcomer R, Rein L, Litilit D, Richie NO, Wood C, Murphy J, Sauerwein R, Hermsen CC, McCoy AJ, Kamau E, Cummings J, Komisar J, Sutamihardja A, Shi M, Epstein JE, Maiolatesi S, Tosh D, Limbach K, Angov E, Bergmann-Leitner E, Bruder JT, Doolan DL, King CR, Carucci D, Dutta S, Soisson L, Diggs C, Hollingdale MR, Ockenhouse CF, Richie TL. DNA prime/adenovirus boost malaria vaccine encoding P. falciparum CSP and AMA1 induces sterile protection associated with cell-mediated immunity. PLoS One 8:e55571. 10.1371/journal.pone.0055571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vogels R, Zuijdgeest D, van Rijnsoever R, Hartkoorn E, Damen I, de Bethune MP, Kostense S, Penders G, Helmus N, Koudstaal W, Cecchini M, Wetterwald A, Sprangers M, Lemckert A, Ophorst O, Koel B, van Meerendonk M, Quax P, Panitti L, Grimbergen J, Bout A, Goudsmit J, Havenga M. 2003. Replication-deficient human adenovirus type 35 vectors for gene transfer and vaccination: efficient human cell infection and bypass of preexisting adenovirus immunity. J. Virol. 77:8263–8271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheng C, Gall JG, Kong WP, Sheets RL, Gomez PL, King CR, Nabel GJ. 2007. Mechanism of Ad5 vaccine immunity and toxicity: fiber shaft targeting of dendritic cells. PLoS Pathog. 3:e25. 10.1371/journal.ppat.0030025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thorner AR, Vogels R, Kaspers J, Weverling GJ, Holterman L, Lemckert AA, Dilraj A, McNally LM, Jeena PM, Jepsen S, Abbink P, Nanda A, Swanson PE, Bates AT, O'Brien KL, Havenga MJ, Goudsmit J, Barouch DH. 2006. Age dependence of adenovirus-specific neutralizing antibody titers in individuals from sub-Saharan Africa. J. Clin. Microbiol. 44:3781–3783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kostense S, Koudstaal W, Sprangers M, Weverling GJ, Penders G, Helmus N, Vogels R, Bakker M, Berkhout B, Havenga M, Goudsmit J. 2004. Adenovirus types 5 and 35 seroprevalence in AIDS risk groups supports type 35 as a vaccine vector. AIDS 18:1213–1216 [DOI] [PubMed] [Google Scholar]
- 31.Gall JG, Crystal RG, Falck-Pedersen E. 1998. Construction and characterization of hexon-chimeric adenoviruses: specification of adenovirus serotype. J. Virol. 72:10260–10264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gall J, Kass-Eisler A, Leinwand L, Falck-Pedersen E. 1996. Adenovirus type 5 and 7 capsid chimera: fiber replacement alters receptor tropism without affecting primary immune neutralization epitopes. J. Virol. 70:2116–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roy S, Shirley PS, McClelland A, Kaleko M. 1998. Circumvention of immunity to the adenovirus major coat protein hexon. J. Virol. 72:6875–6879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roberts DM, Nanda A, Havenga MJ, Abbink P, Lynch DM, Ewald BA, Liu J, Thorner AR, Swanson PE, Gorgone DA, Lifton MA, Lemckert AA, Holterman L, Chen B, Dilraj A, Carville A, Mansfield KG, Goudsmit J, Barouch DH. 2006. Hexon-chimaeric adenovirus serotype 5 vectors circumvent pre-existing anti-vector immunity. Nature 441:239–243 [DOI] [PubMed] [Google Scholar]
- 35.Bruder JT, Semenova E, Chen P, Limbach K, Patterson NB, Stefaniak ME, Konovalova S, Thomas C, Hamilton M, King CR, Richie TL, Doolan DL. 2012. Modification of Ad5 hexon hypervariable regions circumvents pre-existing Ad5 neutralizing antibodies and induces protective immune responses. PLoS One 7:e33920. 10.1371/journal.pone.0033920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alemany R, Suzuki K, Curiel DT. 2000. Blood clearance rates of adenovirus type 5 in mice. J. Gen. Virol. 81:2605–2609 [DOI] [PubMed] [Google Scholar]
- 37.Xu Z, Tian J, Smith JS, Byrnes AP. 2008. Clearance of adenovirus by Kupffer cells is mediated by scavenger receptors, natural antibodies, and complement. J. Virol. 82:11705–11713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haisma HJ, Boesjes M, Beerens AM, van der Strate BW, Curiel DT, Pluddemann A, Gordon S, Bellu AR. 2009. Scavenger receptor A: a new route for adenovirus 5. Mol. Pharm. 6:366–374 [DOI] [PubMed] [Google Scholar]
- 39.Khare R, May SM, Vetrini F, Weaver EA, Palmer D, Rosewell A, Grove N, Ng P, Barry MA. 2011. Generation of a Kupffer cell-evading adenovirus for systemic and liver-directed gene transfer. Mol. Ther. 19:1254–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parker AL, Waddington SN, Nicol CG, Shayakhmetov DM, Buckley SM, Denby L, Kemball-Cook G, Ni S, Lieber A, McVey JH, Nicklin SA, Baker AH. 2006. Multiple vitamin K-dependent coagulation zymogens promote adenovirus-mediated gene delivery to hepatocytes. Blood 108:2554–2561 [DOI] [PubMed] [Google Scholar]
- 41.Shayakhmetov DM, Gaggar A, Ni S, Li ZY, Lieber A. 2005. Adenovirus binding to blood factors results in liver cell infection and hepatotoxicity. J. Virol. 79:7478–7491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kalyuzhniy O, Di Paolo NC, Silvestry M, Hofherr SE, Barry MA, Stewart PL, Shayakhmetov DM. 2008. Adenovirus serotype 5 hexon is critical for virus infection of hepatocytes in vivo. Proc. Natl. Acad. Sci. U. S. A. 105:5483–5488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brough DE, Lizonova A, Hsu C, Kulesa VA, Kovesdi I. 1996. A gene transfer vector-cell line system for complete functional complementation of adenovirus early regions E1 and E4. J. Virol. 70:6497–6501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abrahamsen K, Kong HL, Mastrangeli A, Brough D, Lizonova A, Crystal RG, Falck-Pedersen E. 1997. Construction of an adenovirus type 7a E1A− vector. J. Virol. 71:8946–8951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nan X, Peng B, Hahn TW, Richardson E, Lizonova A, Kovesdi I, Robert-Guroff M. 2003. Development of an Ad7 cosmid system and generation of an Ad7deltaE1deltaE3HIV(MN) env/rev recombinant virus. Gene Ther. 10:326–336 [DOI] [PubMed] [Google Scholar]
- 46.Lemiale F, Haddada H, Nabel GJ, Brough DE, King CR, Gall JG. 2007. Novel adenovirus vaccine vectors based on the enteric-tropic serotype 41. Vaccine 25:2074–2084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cleghon V, Piderit A, Brough DE, Klessig DF. 1993. Phosphorylation of the adenovirus DNA-binding protein and epitope mapping of monoclonal antibodies against it. Virology 197:564–575 [DOI] [PubMed] [Google Scholar]
- 48.Bruder JT, Stefaniak ME, Patterson NB, Chen P, Konovalova S, Limbach K, Campo JJ, Ettyreddy D, Li S, Dubovsky F, Richie TL, King CR, Long CA, Doolan DL. 2010. Adenovectors induce functional antibodies capable of potent inhibition of blood stage malaria parasite growth. Vaccine 28:3201–3210 [DOI] [PubMed] [Google Scholar]
- 49.Sprangers MC, Lakhai W, Koudstaal W, Verhoeven M, Koel BF, Vogels R, Goudsmit J, Havenga MJ, Kostense S. 2003. Quantifying adenovirus-neutralizing antibodies by luciferase transgene detection: addressing preexisting immunity to vaccine and gene therapy vectors. J. Clin. Microbiol. 41:5046–5052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roberts MM, White JL, Grutter MG, Burnett RM. 1986. Three-dimensional structure of the adenovirus major coat protein hexon. Science 232:1148–1151 [DOI] [PubMed] [Google Scholar]
- 51.Rux JJ, Burnett RM. 2000. Type-specific epitope locations revealed by X-ray crystallographic study of adenovirus type 5 hexon. Mol. Ther. 1:18–30 [DOI] [PubMed] [Google Scholar]
- 52.Rux JJ, Kuser PR, Burnett RM. 2003. Structural and phylogenetic analysis of adenovirus hexons by use of high-resolution x-ray crystallographic, molecular modeling, and sequence-based methods. J. Virol. 77:9553–9566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saban SD, Silvestry M, Nemerow GR, Stewart PL. 2006. Visualization of alpha-helices in a 6-angstrom resolution cryoelectron microscopy structure of adenovirus allows refinement of capsid protein assignments. J. Virol. 80:12049–12059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fabry CM, Rosa-Calatrava M, Conway JF, Zubieta C, Cusack S, Ruigrok RW, Schoehn G. 2005. A quasi-atomic model of human adenovirus type 5 capsid. EMBO J. 24:1645–1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu H, Jin L, Koh SB, Atanasov I, Schein S, Wu L, Zhou ZH. 2010. Atomic structure of human adenovirus by cryo-EM reveals interactions among protein networks. Science 329:1038–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reddy VS, Natchiar SK, Stewart PL, Nemerow GR. 2010. Crystal structure of human adenovirus at 3.5 A resolution. Science 329:1071–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nemerow GR, Stewart PL, Reddy VS. 2012. Structure of human adenovirus. Curr. Opin. Virol. 2:115–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cepko CL, Sharp PA. 1982. Assembly of adenovirus major capsid protein is mediated by a nonvirion protein. Cell 31:407–415 [DOI] [PubMed] [Google Scholar]
- 59.Morin N, Boulanger P. 1986. Hexon trimerization occurring in an assembly-defective, 100K temperature-sensitive mutant of adenovirus 2. Virology 152:11–31 [DOI] [PubMed] [Google Scholar]
- 60.Bradley RR, Lynch DM, Iampietro MJ, Borducchi EN, Barouch DH. 2012. Adenovirus serotype 5 neutralizing antibodies target both hexon and fiber following vaccination and natural infection. J. Virol. 86:625–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shenk T, Williams J. 1984. Genetic analysis of adenoviruses. Curr. Top. Microbiol. Immunol. 111:1–39 [DOI] [PubMed] [Google Scholar]
- 62.Schmid SI, Hearing P. 1997. Bipartite structure and functional independence of adenovirus type 5 packaging elements. J. Virol. 71:3375–3384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shiratsuchi T, Rai U, Krause A, Worgall S, Tsuji M. 2010. Replacing adenoviral vector HVR1 with a malaria B cell epitope improves immunogenicity and circumvents preexisting immunity to adenovirus in mice. J. Clin. Invest. 120:3688–3701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Abe S, Okuda K, Ura T, Kondo A, Yoshida A, Yoshizaki S, Mizuguchi H, Klinman D, Shimada M. 2009. Adenovirus type 5 with modified hexons induces robust transgene-specific immune responses in mice with pre-existing immunity against adenovirus type 5. J. Gene Med. 11:570–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bradley RR, Maxfield LF, Lynch DM, Iampietro MJ, Borducchi EN, Barouch DH. 2012. Adenovirus serotype 5-specific neutralizing antibodies target multiple hexon hypervariable regions. J. Virol. 86:1267–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Qiu H, Li X, Tian X, Zhou Z, Xing K, Li H, Tang N, Liu W, Bai P, Zhou R. 2012. Serotype-specific neutralizing antibody epitopes of human adenovirus type 3 (HAdV-3) and HAdV-7 reside in multiple hexon hypervariable regions. J. Virol. 86:7964–7975 [DOI] [PMC free article] [PubMed] [Google Scholar]