Abstract
The relationship between parasitoid wasps and polydnaviruses constitutes one of the few known mutualisms between viruses and eukaryotes. Viral particles are injected with the wasp eggs into parasitized larvae, and the viral genes thus introduced are used to manipulate lepidopteran host physiology. The genome packaged in the particles is composed of 35 double-stranded DNA (dsDNA) circles produced in wasp ovaries by amplification of viral sequences from proviral segments integrated in tandem arrays in the wasp genome. These segments and their flanking regions within the genome of the wasp Cotesia congregata were recently isolated, allowing extensive mapping of amplified sequences. The bracovirus DNAs packaged in the particles were found to be amplified within more than 12 replication units. Strikingly, the nudiviral cluster, the genes of which encode particle structural components, was also amplified, although not encapsidated. Amplification of bracoviral sequences was shown to involve successive head-to-head and tail-to-tail concatemers, which was not expected given the nudiviral origin of bracoviruses.
INTRODUCTION
Polydnaviruses are unique viruses that have evolved a mutualistic life style with tens of thousands of parasitoid wasp species (1–3). They belong to the Polydnaviridae (PDV), divided into two genera, Ichnovirus (IV) and Bracovirus (BV), associated with parasitoid wasps from several subfamilies of Ichneumonidae and Braconidae, respectively (4). These unconventional viruses have an atypical life cycle that requires two separate species: the parasitoid wasp, in which viral particles are produced in specialized cells of the calyx in the ovaries, and the lepidopteran larvae, in which the viral genes packaged in the particles are expressed (5). One original feature of these atypical viruses is that their genomes are divided into two parts, both of which reside permanently in the wasp genome and are transmitted vertically (6–8). The first part of the genome is composed of proviral segments used to produce multiple double-stranded DNA (dsDNA) circles packaged in the particles and injected into the lepidopteran host (5, 9–14). This packaged genome carries virulence genes causing host immune suppression and developmental arrest that are necessary to ensure successful wasp larva development in the host (15, 16). Recently, in silico de novo annotation predicted more packaged genes than previously reported (222 coding DNA sequences or CDS) (17) and numerous gene families. Seven of these gene families encoded proteins containing eukaryotic conserved domains (PTP, VANK, cystatin, RNase T2, BEN, Cys rich, and C-type lectin), one family codes for a P94-like baculovirus protein, and 29 families are specific to BVs (EP1-like, EP2-like, Ser rich, and BV families 1 to 26). However, unlike free viruses, the packaged bracovirus genome does not contain the genes involved in particle production (18), and as a consequence, virions are not produced in parasitized larvae (6–8, 19–22). Indeed, the genes involved in particle production, constituting the second part of the virus genome, reside permanently in the wasp genome (22–24). For example, in the ichneumonid wasp Hyposoter dydimator, associated with the ichnovirus Hyposoter dydimator ichnovirus (HdIV), these genes, referred to as ichnovirus structural-protein-encoding regions (IVSPERs), are clustered in specialized genomic regions amplified during virus particle production. They show no relatedness to other proteins (21). In bracoviruses, they belong to the gene set typical of nudiviruses (which are closely related to baculoviruses) and appear to have originated from a nudiviral genome captured by the common ancestor of bracovirus-associated wasps (7, 8, 25). The nudiviral genes encode proteins of the viral transcriptional machinery (nudiviral RNA polymerase subunits) and of the particle structural components and envelope proteins potentially involved in host cell entry (homologous to baculovirus pif genes [for per os infectivity factors]) (7, 8, 20, 22, 26). Interestingly, half of the nudiviral genes identified are clustered within the genome of the braconid wasp Cotesia congregata in a region referred to as the nudiviral cluster, thought to reflect the original organization of the nudivirus genome captured by the ancestor of bracovirus-associated wasps (7, 8, 23). The other nudiviral genes identified are dispersed in different C. congregata genomic regions containing wasp genes and mobile elements (7, 8).
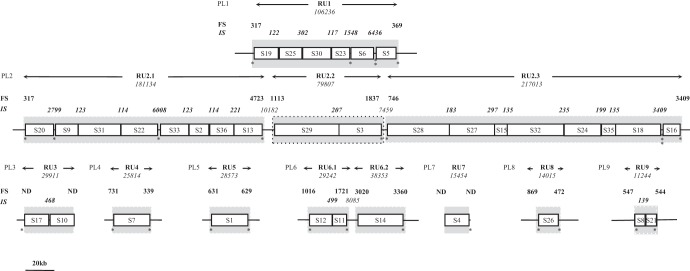
Based on analyses of two braconid parasitoid wasps, C. congregata and Chelonus inanitus, the bracovirus proviral segments were originally thought to be located at a single chromosomal locus in a tandem array referred to as the proviral macrolocus (6, 20, 27). Recent genomic analyses of bracovirus proviral regions of Glyptapanteles flavicoxis, Glyptapanteles indiensis, and C. congregata (GfBV, GiBV, and CcBV, respectively) using bacterial artificial chromosome (BAC) insert sequencing revealed that 68 to 75% of the proviral segments were indeed localized within a 550- to 700-kb-long macrolocus (12, 17, 28). The organization of the macrolocus was found to be conserved within two regions named PL1 and PL2 (for proviral loci 1 and 2), linked by a wasp gene-containing region. In addition, several proviral loci (seven in C. congregata: PL3 to PL9), containing one to three proviral segments, were not localized in close proximity to the macrolocus (Fig. 1). Some of these loci, however, could be present in the same chromosomal region, since CcBV segments S7, S8 (from isolated loci PL4 and PL9, respectively), and S30 (from the macrolocus) were all detected on the short arm of chromosome 5 in a previous study using in situ hybridization on metaphase chromosomes (6).
Fig 1.
Schematic representation of CcBV PL in the genome of the braconid wasp C. congregata with a map of the regions amplified during particle production (RUs). CcBV segments are represented by white boxes in which segment names are indicated (17), separated and flanked by ISs and FSs (black lines). Their lengths (in bp) are indicated above their positions. The numbers in boldface identify ANES indicated in Table 1 (amplified FSs and ISs). The replication units are shown by the gray areas (amplified FSs could not be represented to scale due to space limitations), and their sizes (in bp) are indicated by double arrows. Each asterisk marks the presence of a common AT-rich sequence motif (the sequences are listed in Fig. 5).
Analysis of proviral genomes also confirmed that direct-repeat sequences terminate all bracovirus proviral segments. These repeats, named DRJs (for direct-repeat junctions) are involved in a late step of packaged circle production (18). The process of excision leading to circularization was proposed to occur through juxtaposition of the DRJs of a proviral genome segment, followed by a recombination event involving a site-specific recombinase (27, 29, 30). Accordingly, all the bracovirus viral circles sequenced to date were found to retain a single DRJ corresponding to the recombination of proviral segment extremities (12, 17, 28).
Before their excision, DNAs have to be amplified from proviral segments used as master sequences. In CcBV and Chelonus inanitus BV (CiBV), viral DNA amplification does not occur after separate excision of individual segments, as initially proposed (30, 31). Indeed in C. congregata, we showed that two segments, S8 and S21 (on PL9), were amplified together within a large progenitor molecule (30). This large molecule was detected from the onset of viral particle production during wasp pupal development and 1 day before dsDNA circles are produced (30).
From the dispersed organization of the proviral sequences, we expected either that isolated loci were amplified separately or that large regions of wasp genome separating proviral loci were amplified along with viral DNA. In the present study, we finely mapped the regions amplified during virus particle production onto the proviral form and searched for conserved sequences that might play the role of replication origins. We also tested whether the nudiviral cluster encoding the most abundant particle components was amplified during virus particle production. Finally, we analyzed the structure of the amplified molecule corresponding to a replication unit (i.e., circular/linear, head-to-tail/other concatemers) by Southern blotting. For the first time, we provide a complete overview of bracovirus amplification and reveal unexpected insights into the bracovirus replication mechanism.
MATERIALS AND METHODS
Insects.
The gregarious larval endoparasitoid wasps C. congregata (Hymenoptera, Braconidae, Microgastrinae) were reared on their natural host, the tobacco hornworm, Manduca sexta (Lepidoptera, Sphingidae), as previously described (32).
Females lay their eggs in fourth-instar larvae of the host, and their progeny emerge 10 to 12 days later to spin their cocoons on the back of the caterpillar. Under our rearing conditions, the wasps emerge between 5 and 6 days after cocoon spinning.
Extraction of wasp DNA.
The sex of each individual C. congregata wasp was visually checked twice under a binocular dissecting microscope. We selected adults 1 day after emergence as a source of DNA because amplification of bracovirus DNA is very active at this time (22, 30). DNA was extracted from adult males and females using the DNeasy Blood and Tissue kit (Qiagen, France) according to the manufacturer's instructions.
PCRs.
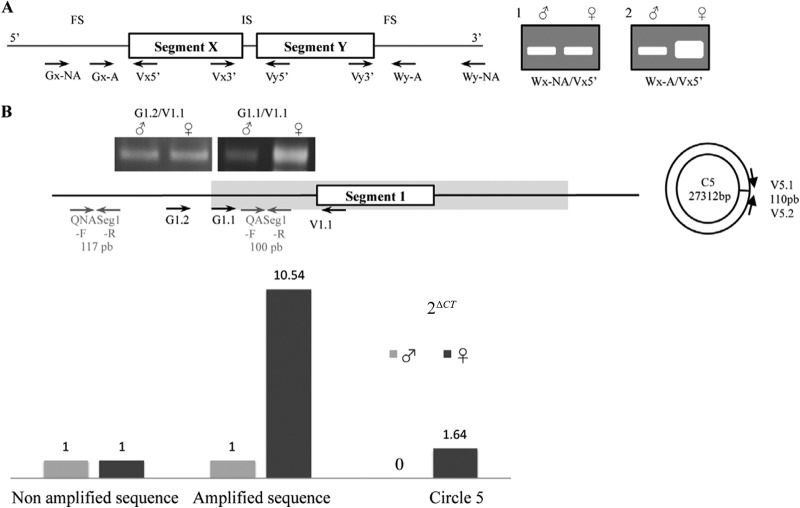
Semiquantitative PCRs were performed for all targeted regions using 10 ng of wasp DNA, specific primers, and the most adapted Taq polymerase, depending on the fragment size amplified. Fragments below 4 kb were amplified using Advantage 2 polymerase (Clontech, France), and long-range PCR fragments were generated using TaKara LA Taq (Clontech, France). PCR conditions were determined to perform the amplification within the exponential phase, and the wasp actin gene was used as an internal control to normalize male and female samples. With Advantage 2 polymerase, after a 4-min denaturation step at 95°C, amplifications were carried out for 25 cycles with the following steps: 60 s at 95°C for denaturation, 60 s for annealing (at the temperature corresponding to the lowest melting temperature [Tm] of the primer pair), and an extension time at 72°C depending on the fragment size (90 s per kb). With LA Taq, after a 4-min denaturation step at 94°C, amplifications were carried out for 14 cycles with the steps 20 s at 98°C for denaturation, 30 s for annealing, and 20 min at 68°C, and then for 16 cycles with the steps 20 s at 98°C for denaturation and 30 s for annealing, with the extension time increased by 15 s in each cycle. PCR products were resolved on an agarose gel stained with ethidium bromide. Amplified DNA band intensities were then estimated using Image J software (http://imagej.nih.gov/ij/). PCR specificity was confirmed by sequencing amplified purified products (Nucleospin extract II; Macherey-Nagel, France) on an ABI 3100-Avant capillary sequencer (BigDye Terminator; Life Technologies, France). Relative quantification was also performed by real-time PCR on PL5 (Fig. 2B and C) using primers designed in the RU5 flanking regions. Relative quantitative PCRs (qPCRs) were carried out on an ABI Prism 7000 Sequence Detection System (Life Technologies, France) in 96-well plates (25 μl per well). The PCR conditions consisted of 40 cycles of 15 s at 94°C and 60 s at 55°C, followed by a dissociation protocol. Serial dilutions of DNA ranging from 0.1 to 10 ng were treated in triplicate in a reaction buffer containing 1× SYBR green mix (Mesa Green qPCR MasterMix; Eurogentec, France) and 300 nM specific primers (Fig. 2B; see Table S1 in the supplemental material). The mean threshold cycle (Ct) values were used to estimate the fold change in viral DNA quantities between amplified and nonamplified sequences by the 2ΔCT method (33).
Fig 2.
Experimental approaches used for mapping sequences amplified in wasp ovaries. (A) Schematic representation of the semiquantitative PCR approach. The boxes represent two proviral segments in tandem array in the wasp genome, and the black line corresponds to FSs and an IS. The arrows indicate primer positions. The letters “V” and “W” correspond to primers localized in proviral segments and their flanking sequences, respectively. A series of W primers were designed at different distances from the proviral segments (FSs) to determine to what extent sequences in 5′ and 3′ flanking regions were amplified (Gx-NA and Gx-A, primers in a flanking region not amplified and amplified, respectively). For 100- to 1,000-bp intersegments, primers were designed close to the segment extremities (Vx3′/Vy5′). The expected results after electrophoresis of PCR products are as follows: for a sequence not amplified in ovaries, the same intensity for the bands obtained from male and female DNA samples (i.e., PCR performed using Wx-NA and Vx5′) (gel 1), and for a sequence amplified in ovaries, a much higher-intensity band obtained from a female sample (i.e., PCR performed using Wx-A and Vx5′) (gel 2). (B) Schematic representation of proviral locus 5, RU5 (gray area), and circle 5 and the experimental results obtained using semiquantitative PCR and relative real-time PCR analyses. For the qPCR assay, we designed a primer pair (QASeg1-F/QASeg1-R; 100-bp PCR product) inside the replication unit previously mapped by semiquantitative PCR (G1.1/V1.1; 816-bp product). Another primer pair was designed in the nonamplified region (QNASeg1-F/QNASeg1-R; 117-bp product) and on the circle 5 (PL1)-specific junction (110-bp product). Measurements based on three replicates indicated a more than 10-fold difference in the amount of amplified DNA in females compared to males. Semiquantitative PCR is sufficient to assess such a difference and allows us to test the amplification states of large regions in a single assay.
Primer design.
Primers were designed based on the sequence of each proviral locus (accession numbers HF586472 to HF586480 for PL1 to PL9). The sequences listed in Table S1 in the supplemental material are longer than the usual primers (25-mers) to compensate for TA bias (70%) and to increase PCR specificity in the context of the numerous duplications characterizing the proviral segments (17).
Replication unit-mapping approach.
A schematic representation of replication unit mapping by a semiquantitative PCR assay using adult male and female genomic DNAs is shown in Fig. 2. Only females produce virus particles, and males are used as controls for proviral DNA normalization (not amplified). By using appropriate primers, the amplification of a bracoviral sequence is visualized in a PCR assay by an intensity of the band in the female sample that is higher than that obtained with male DNA. Two (theoretical) proviral segments, X and Y, separated in the wasp genome by an intersegmental sequence (IS), are represented. To test whether the IS was part of the replication unit, we used two primers located at the 3′ end of the first segment (Vx3′) and the 5′ end of the next segment (Vy5′). To test whether the region flanking a proviral segment (FS) was part of the replication unit, primers were designed at the extremities of the segments (Vx5′ and Vy3′) and in the wasp flanking regions (Gx-A and Gy-A). Primers were then designed further upstream or downstream of the proviral sequence (Wx-NA and Wy-NA) until the region tested was no longer found to be amplified.
Sequence analysis.
To highlight conserved motifs, the replication units were analyzed using MEME (http://meme.sdsc.edu/meme/intro.html) and WebACT (http://www.webact.org/WebACT/home) software (34, 35). The alignment was performed using MUSCLE (http://www.ebi.ac.uk/Tools/msa/muscle/). Secondary-structure motif predictions were based on the consensus and were obtained using mfold software (http://mfold.rna.albany.edu/?q=mfold/).
Southern blotting.
Digested DNA (10 μg genomic DNA and 80 ng virus DNA) was electrophoretically separated on a 0.8% agarose gel and transferred onto a nylon membrane (Biotrans ICN, France), according to the manufacturer's protocol. The filters were incubated for 30 min in hybridization solution (7% SDS, 0.5 M sodium phosphate buffer, pH 7.2, 1 mM EDTA). 32P-labeled probes were prepared by random priming and denatured at 94°C before membrane hybridization at 65°C overnight. The membranes were washed twice in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) containing 0.1% SDS for 20 min at 65°C and twice in 0.2× SSC containing 0.1% SDS for 15 min at 65°C before autoradiography. The EP1 probe corresponds to the HincII 634-bp fragment of EP1 cDNA (32) that is located in segment S8 (positions 28470 to 29104) in proviral locus 9. The immediate downstream probe (IDP) is a 1.2-kb-long HindIII-PstI fragment isolated from a λ phage harboring the EP1 gene (λ121) (27). The sequence is located at bp 23979 to 25203 in PL9 in the S8 5′ flanking sequence and also hybridizes, to a lesser extent, in the S21 3′ flanking sequence (positions 35386 to 35931).
RESULTS
Amplified sequences from the major proviral locus and isolated regions.
Analysis of data obtained in CcBV revealed that the proviral segments are organized into 9 proviral loci (17). The two major proviral loci, PL1 and PL2 (macrolocus), harbor 6 and 18 proviral segments, respectively. The seven other proviral loci are dispersed in the host genome and harbor one to three segments (Fig. 1). Proviral segments of the same locus are separated by intersegmental sequences—not encapsidated in BV particles—ranging in size from 114 bp to 10.1 kb. Previous findings showed that sequences from the two PL9 proviral segments were coamplified along with their corresponding IS (IS9.1) prior to their excision/circularization (30). This observation raised the question of whether coamplification of segments in tandem array with nonencapsidated sequences was a general rule for the whole proviral genome. In order to answer this question, we used a semiquantitative PCR assay to compare the levels of viral DNAs in males, which contain only proviral sequences, and in females, which in addition contain amplified bracovirus DNAs in their ovaries. In this comprehensive analysis, we assessed whether each proviral segment was amplified separately or together with other segments and with intersegmental and flanking sequences. Figure 2 shows a schematic representation of the expected PCR results, depending on primer positions and on whether the tested region is amplified (see Materials and Methods). Semiquantitative and relative real-time PCRs were used to compare the levels of DNA amplification of the same proviral segment and of a nonamplified sequence using male and female wasp DNAs. Both methods were found to be accurate in determining whether DNA was amplified (Fig. 2). The semiquantitative PCR approach was chosen for further analyses because it allowed us to test the levels of amplification of large fragments, thereby enabling convenient assessment of the amplification states of large genomic regions (>700 kb).
Intersegmental sequences of PL1 are amplified.
Most ISs of PL1 were amplified using specific primer pairs designed near the ends of neighboring segments. For the larger intersegmental sequences IS1.4 (1,548 bp) and IS1.5 (6,346 bp), two and three primer pairs, respectively, amplifying overlapping fragments were used to assess the amplification state of the entire sequence (Fig. 3 and Table 1). In both male and female samples, we observed PCR products of the expected sizes. A larger quantity of PCR products was obtained in females than in males as measured using Image J software, indicating that ISs were amplified during replication, along with contiguous segments (Fig. 3). Altogether, these results indicated that the 6 PL1 segments and their ISs were coamplified within the same molecule.
Fig 3.
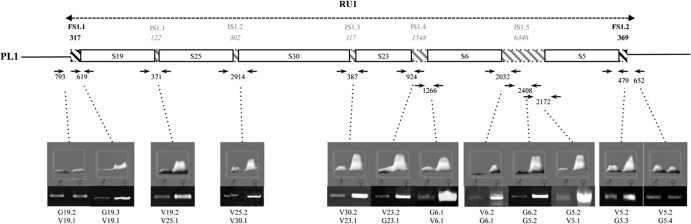
Mapping of PL1 sequences amplified during particle production by semiquantitative PCR. Shown is a PL1 map with the sequences amplified within RU1 indicated inside the double arrow. Intersegments (IS1.1 to IS1.5) are represented by gray-hatched boxes, and their sizes are indicated. The FSs within RU1 are represented by black-hatched boxes. The positions of the primers used are indicated by black arrows, and the numbers in between primer pairs correspond to amplified fragment sizes. The numbers of fragments obtained were compared using gel electrophoresis after PCR amplification had been performed using the same amounts of DNAs extracted from male and female wasps. The intensities of the bands were measured with ImageJ (the results are represented above the electrophoresis gels). The names of the primers used for each PCR are indicated under the electrophoresis gels.
Table 1.
Localization of the regions amplified during virus production on the proviral loci (replication units, flanking sequences, and intersegmental sequences)
| Locus (size [bp]) | RU (size [bp]) | ANES |
||
|---|---|---|---|---|
| Positiona | Min size (bp) | Max size (bp) | ||
| PL1 (146,359) | RU1 (106,236) | FS1.1 (7137–7454) | 317 | 501 |
| IS1.1 (26223–26344) | 122 | NA | ||
| IS1.2 (41570–41871) | 302 | NA | ||
| IS1.3 (63971–64087) | 117 | NA | ||
| IS1.4 (77685–79232) | 1,548 | NA | ||
| IS1.5 (94463–100808) | 6,346 | NA | ||
| FS1.2 (113004–113373) | 369 | 542 | ||
| PL2 (522,749) | RU2.1 (181,134) | FS2.1 (27203–31944) | 4,723 | 5,120 |
| IS2.1 (51094–53892) | 2,799 | NA | ||
| IS2.2 (69843–69965) | 123 | NA | ||
| IS2.3 (100630–100743) | 114 | NA | ||
| IS2.4 (126759–132766) | 6,008 | NA | ||
| IS2.5 (152964–153086) | 123 | NA | ||
| IS2.6 (168062–168175) | 114 | NA | ||
| IS2.7 (185653–185873) | 221 | NA | ||
| FS2.2 (207260–208337) | 1,077 | 1,630 | ||
| RU2.2 (79,807) | FS2.3 (216327–217440) | 1,113 | 2,692 | |
| IS2.8 (263798–264004) | 207 | NA | ||
| FS2.4 (294297–296134) | 1,837 | 3,069 | ||
| RU2.3 (217,013) | FS2.5 (301008–301754) | 746 | 1,546 | |
| IS2.9 (347113–347295) | 183 | NA | ||
| IS2.10 (380464–380760) | 297 | NA | ||
| IS2.11 (389467–389601) | 135 | NA | ||
| IS2.12 (431047–431281) | 235 | NA | ||
| IS2.13 (457384–457582) | 199 | NA | ||
| IS2.14 (468809–468943) | 135 | NA | ||
| IS2.15 (501023–504431) | 3,409 | NA | ||
| FS2.6 (516807–518021) | 1,214 | 2,750 | ||
| PL3 (35,494) | RU3 (29,911) | FS3.1 (ND–5580) | ND | ND |
| IS3.1 (20739–21206) | 468 | NA | ||
| FS3.2 (35493–ND) | ND | ND | ||
| PL4 (66,938) | RU4 (25,814) | FS4.1 (23351–24082) | 731 | 1,561 |
| FS4.2 (48826–49165) | 339 | 1,412 | ||
| PL5 (55,213) | RU5 (28,573) | FS5.1 (3371–4002) | 631 | 752 |
| FS5.2 (31315–31944) | 629 | 718 | ||
| PL6 (143,166) | RU6.1 (29,242) | FS6.1 (16549–17565) | 1,016 | 2,514 |
| IS6.1 (33668–34166) | 499 | NA | ||
| FS6.2 (44070–45791) | 1,721 | 3,020 | ||
| RU6.2 (38,353) | FS6.3 (49133–52153) | 3,020 | 6,794 | |
| FS6.4 (84126–87486) | 3,360 | 4,365 | ||
| PL7(16,348) | RU7 (15,454) | FS7.1 (ND–2) | ND | ND |
| FS7.2 (15455–ND) | ND | ND | ||
| PL8 (155,986) | RU8 (14,015) | FS8.1 (65937–66806) | 869 | 926 |
| FS8.2 (79480–79952) | 472 | 525 | ||
| PL9 (63,782) | RU9 (11,244) | FS9.1 (24687–25234) | 547 | 584 |
| IS9.1 (30267–30405) | 139 | NA | ||
| FS9.2 (35387–35931) | 544 | 582 | ||
ANES positions on the proviral loci. For ISs, the positions indicated are those of the DRJs terminating the proviral segments. Note that for FSs, the positions indicated correspond to the furthest-outside primer tested identifying an amplified region. Min size, FS size estimated from this primer; Max size, size corresponding to the closest primer tested identifying a nonamplified region. NA, not applicable; ND, not determined. The positions refer to sequences that have been deposited in GenBank under accession numbers HF586472 to HF586480 (17).
Short sequences in flanking regions of PL1 are amplified.
To determine whether this amplified molecule terminated at the extremities of the first and last segments of PL1, we assessed whether flanking regions were amplified (Fig. 3 and Table 1). A larger quantity of PCR products was obtained with female than with male DNA using a 5′ primer located 317 bp upstream of the first segment (S19; 619-bp PCR product) (Fig. 3 and Table 1). This result indicated that amplification of the molecule begins upstream of the DRJ core sequence and includes a sequence that we named flanking sequence 1.1 (FS1.1). FS1.1 does not extend beyond 501 bp, since similar amounts of PCR products were obtained using DNA from females and males with a primer localized at this position (793-bp product) (Fig. 3). The same approach indicated that 369 bp of flanking sequence downstream of the last PL1 segment (S5) was amplified during viral replication (FS1.2). Amplified FSs and ISs are not present in the genome packaged in the bracovirus particles, and thus, we propose to refer to these sequences as amplified not encapsidated sequences (ANES). The molecule containing the 6 PL1 segments and their ANES constitutes a replication unit (RU).
The PL2 region is amplified within three replication units.
A similar analysis was performed on the 17 ISs separating the 18 segments of PL2. The results obtained (Table 1) indicated that most ISs were amplified with contiguous segments (summarized in Fig. 1). However, two ISs (joining S13 to S29 and S3 to S28) were not amplified, showing that bracoviral DNA of the PL2 region was not amplified as a single replication unit but within 3 different molecules, namely, RU2.1, RU2.2, and RU2.3. Analysis of RU2.1, RU2.2, and RU2.3 flanking sequences revealed that the sizes of the amplified flanking sequences (5′-3′, respectively) were 4,723 bp to 1,077 bp for RU2.1, 1,113 bp to 1,837 bp for RU2.2, and less than 746 bp to 1,214 bp for RU2.3 (Fig. 1 and Table 1). Altogether, macrolocus proviral segments were amplified within 4 replication units (1 for PL1 and 3 for PL2).
Isolated PLs are generally amplified within a single replication unit.
Using the same approach, we found that the segments located in PL4, PL8, and PL9 were amplified within a single replication unit, along with FSs (Table 1). PL6 segments were amplified within 2 RUs; the first RU encompassed the two proviral segments S12 and S11 and their IS, and the second contained only segment S14 (Fig. 1 and Table 1). PL3 and PL7 (containing 2 and 1 proviral segments, respectively) could not be tested for FS amplification because there are either no or too few sequence data on the FSs of these segments. However, the IS separating the two segments of PL3 was detected as amplified, indicating the locus corresponds to a single RU. Circle 4, for which proviral sequence is contained in PL7, was sequenced from the DNA extracted from virus particles (9); thus, we know that it is amplified, and moreover, the fact that it contains a single segment strongly suggests it constitute only one replication unit.
In summary, we have precisely mapped the amplified molecules onto the proviral form of the viral genome (Fig. 1), and we showed that the 35 CcBV dsDNA circles identified are produced from segments amplified in 12 replication units. These RUs are variable in length, from 11,244 bp (RU9, PL9) to 217,013 bp (RU2.3, PL2). They carry ANES corresponding to FSs and ISs (114 to 6,346 bp long) that represent 3 to 16.6% of each RU (Table 1).
Sequences from the nudiviral cluster are amplified during virus particle production.
Previous studies have shown that the nudiviral genes involved in bracovirus particle production are expressed in the ovaries (7, 8, 20, 22) but not encapsidated in the particles (9, 11). In ichnoviruses, the IVSPERs correspond functionally to the nudiviral cluster; although not encapsidated, they are amplified during ichnovirus particle production (21). We used a PCR approach to assess whether nudiviral genes could be similarly amplified during bracovirus particle production. We observed that for almost all genes of the nudiviral cluster, the amount of PCR product obtained was larger using DNA extracted from females than using DNA from males (Fig. 4). In order to test whether nudiviral genes of the cluster were amplified within the same molecule, we assessed the amplification of the sequences between the different genes by designing primers at the 3′ extremity of one gene and the 5′ extremity of the next gene. Almost all of them were amplified, as well as a 695-bp sequence downstream of the pif-3 gene, but not the Pmv gene located at the 3′ extremity of the cluster (Fig. 4). The sequence of the region upstream of the 38K gene is not available, making it impossible to map the 5′ extremity of the amplified molecule. In contrast to genes present in the cluster, the nudiviral genes isolated in the wasp genome (orf140-like [Fig. 4]; lef-8, odv-e56, and p74 [data not shown]) were not found to be amplified during virus particle production. In conclusion, most of the genes from the nudiviral cluster are specifically amplified during virus particle production within a replication unit more than 14,163 bp long.
Fig 4.
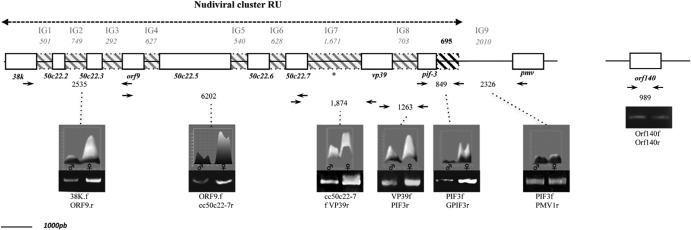
Mapping of amplified sequences from the nudiviral cluster during virus particle production by semiquantitative PCR. Shown is a map of the nudiviral cluster with the sequences amplified within the nudiviral cluster RU indicated inside the double arrow. Intergenic sequences (IG) are shown in gray-hatched boxes, with their lengths. The part of IG9 amplified between the pif-3 gene and the 3′ extremity of the nudiviral cluster RU is indicated as a black-hatched box. The positions of the primers used are indicated by solid arrows, and the numbers between primer pairs correspond to amplified fragment sizes. The amounts of fragments obtained were compared using gel electrophoresis after PCR amplification had been performed using the same amounts of DNAs extracted from male and female wasps. The intensities of the bands were measured with ImageJ (the gel scans are represented above the electrophoresis gels). The names of the primers used for each PCR are indicated under the gels. The asterisk represents the location of the TA motif (the sequence is shown in Fig. 5).
A specific sequence motif is shared by RUs.
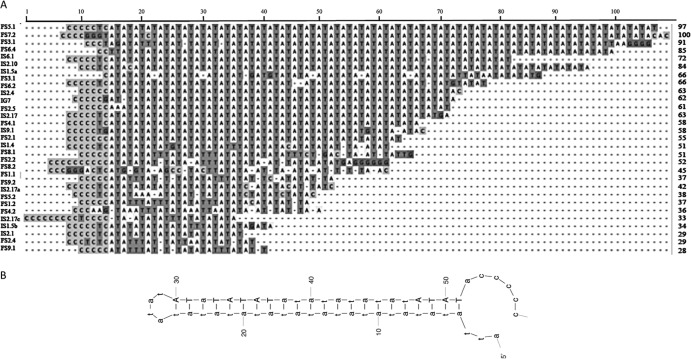
The fact that ANES were amplified along with wasp genomic DNA raised the question of their role in the amplification of the bracoviral segments and the nudiviral cluster. We hypothesized that these sequences possess signals that promote the initiation of DNA amplification in the wasp, such as origins of replication. To identify sequences shared by RUs, we analyzed the 53 CcBV ANES sequences with WebACT and MEME software (34, 35).
We found a conserved common TA-rich sequence motif constituted of a C-rich sequence linked to a TA repeat that is variable in size (up to 100 bp) (Fig. 5A). This motif is present in all RUs, including the nudiviral cluster RU (short common sequence [SCS] localizations are indicated by asterisks in Fig. 1 and 4). The TA-rich stretches can form hairpins (Fig. 5B) that could be involved in genome replication as replication origins.
Fig 5.
(A) Alignment of common TA-rich sequences identified in CcBV ANES. The consensus motif is at the top. The name of each ANES is indicated on the left, and the number of nucleotides of each motif is indicated on the right. The light gray shading represents the C-rich sequences. The T in the TA motif is in dark-gray shading. (B) Secondary-structure motif prediction based on the consensus obtained using mfold software.
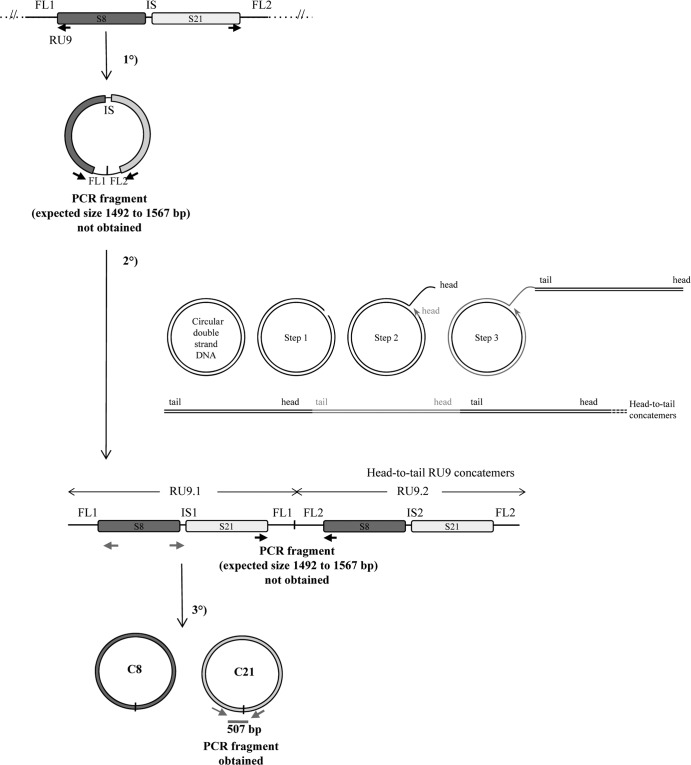
The amplified molecules form successive head-to-head and tail-to-tail concatemers.
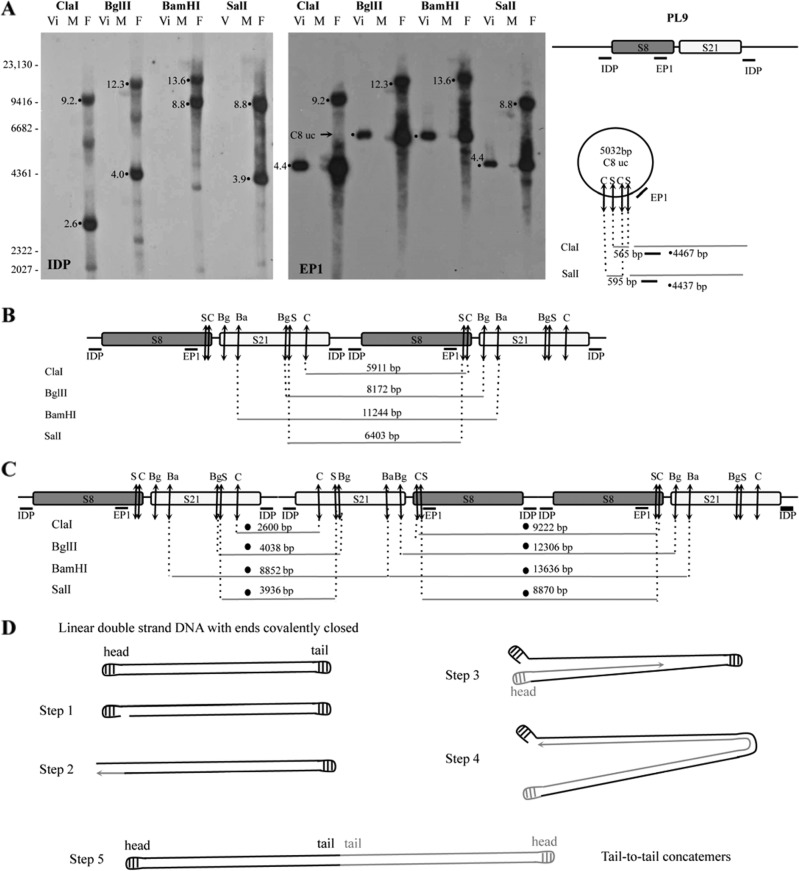
We had proposed initially that proviral segments in tandem array were excised from the chromosome in the calyx cells, forming a large circle, and then amplified by rolling-circle replication (Fig. 6) (30, 36). According to this model, successive concatemers linked head-to-tail are obtained, and head-to-tail junctions on the circle and RU concatemers are predicted, the production of which can be assessed by an inverse PCR using primers designed in opposite orientations at the extremities of the first and last segments of the RU (Fig. 6). We investigated the production of a large master circle and concatemers of RU9 (S21 and S8) by this PCR approach, but we did not obtain the expected PCR fragment (Fig. 6). Using the same DNA sample, the amplification of a specific product was obtained using primers in opposite orientations at the extremities of S21 (Fig. 6). These results strongly suggest that neither a large circle nor rolling-circle concatemers are produced during bracovirus DNA amplification. To confirm these results, we analyzed the amplified molecule by Southern blotting using specific probes that hybridized in a flanking sequence (IDP) and within the segment S8 (EP1) (30) (Fig. 7A). The hybridization patterns obtained using 4 restriction enzymes to digest male, female, and purified-particle DNAs were compared. Interestingly, the sizes of the fragments did not correspond to those expected from a rolling-circle replication model, with production of head-to-tail concatemers confirming the negative PCR results (Fig. 7B). In contrast, a model of linear replication with production of head-to-head and tail-to-tail concatemers fully predicted the patterns of the most intense bands obtained following hybridization of the two probes (Fig. 7C and D). Such a mechanism was previously described in DNA viruses, such as Poxviridae (37, 38).
Fig 6.
Schematic representation of the macrocircle excision and rolling-circle amplification model (tested on RU9). The first step consists of the excision of a large circle from the wasp chromosome, including S8 and S21 sequences and their ANES (FL1-IS-FL2). This molecule is then used as a template for rolling-circle replication. After a nick on one strand of the circular dsDNA molecule (step 1), the 3′ OH end is released to serve as a primer for DNA synthesis by DNA polymerase (step 2 and step 3). Using the other strand as a template, replication proceeds around the circular DNA molecule, displacing the nicked strand as single-stranded DNA (step 3). The molecule produced is a head-to-tail concatemer, which can later be converted to double-stranded circles (C8 and C21 for RU9). This model predicts the production of a junction between ANES flanking sequences (FL1 and FL2) on the template, which is amplified in the concatemer and can be detected by a specific PCR, thus giving rise to an amplified fragment from 1,492 to 1,567 bp long, depending on the actual size of RU9 FL1 and FL2 (Table 1). No PCR fragment was obtained using primers in opposite orientations at the extremities of S8 and S21 (black arrows), while a product of the expected size (507 bp) was obtained using primers (gray arrows) in opposite orientations at the extremities of S21 (assessing the production of the C21 circle junction). These results indicated that the DNA used can be amplified by PCR but that the large circle and concatemers predicted by the model are most probably not produced (unless a particular structure prevents their amplification by PCR) and that bracovirus packaged-sequence amplification is achieved using a different mechanism.
Fig 7.
Southern blot analysis of amplified RU9 molecules in DNA extracted from virus particles (Vi) and female (F) and male (M) wasps undergoing virus replication. The experiment was performed using DNA extracted from purified virus particles (80 ng) and male and female wasps (10 μg) digested with ClaI (C), BglII (Bg), BamHI (Ba), and SalI (S) restriction enzymes and hybridized, as indicated, with IDP localized in RU9 flanking sequences amplified during replication and the ep1 gene probe (EP1) corresponding to a CcBV gene packaged in the particles. (A) Five-hour-exposure autoradiography using IDP and EP1 probes. (B) Map of the amplified RU9 in a model of rolling-circle amplification forming head-to-tail concatemers. (C) Map of RU9 in an alternative model of amplification forming successive head-to-head and tail-to-tail concatemers. The predicted fragments are indicated under the maps, and the corresponding bands are identified in panel A. Note that the fragments that would correspond to RU9 head-to-tail concatemers were not found, whereas all the fragments corresponding to the model of replication implicating head-to-head concatemers were detected and corresponded to the most intense bands of circle C8 uncut (C8 uc) (containing the ep1 gene), whose restriction map is shown on the right in panel A. The less intense bands obtained were those expected from the map of the proviral form (not shown) or corresponded to uncharacterized replication intermediates produced in small amounts or to broken molecules. (D) Linear-replication model (based on poxvirus replication). The linear genome (with covalently closed hairpin termini) replication begins with the introduction of a nick, which exposes a free 3′ OH that serves as a primer for DNA polymerase (step 1). The nascent strand (gray lines) is synthesized from the displaced strand (black lines; step 2), and each forms self-complementary hairpins, which allows leading-strand synthesis to replicate the entire molecule (steps 3 and 4). This process generates a tail-to-tail concatemer (step 5).
DISCUSSION
Recent studies have unraveled the complex organization of BV proviral genomes composed of two distinct components: the “core” genome, which includes nudiviral genes essential for the production of BV particles, and the proviral segments, from which the circular dsDNA molecules packaged in the particles are produced (7, 8, 20, 22). However, our understanding of how proviral segments are amplified in order to provide the large number of circular molecules encapsidated in bracovirus particles is far from complete. It was initially proposed that circles were excised directly from the chromosomal form in the calyx cells (27, 30, 31, 36) and then amplified by rolling-circle replication. However, Southern blot analyses of DNAs from C. congregata females revealed that two sequences from contiguous viral segments were amplified together within the same molecule before individual circles were produced (30). In the parasitoid wasp C. inanitus, a first step of polyploidization of calyx cells was reported, increasing the overall amount of DNA; then, in a second phase, proviral DNA was shown to be selectively amplified by quantitative PCR (36). It was also reported both in C. congregata and C. inanitus that circular viral segments once produced were not further amplified (30, 36). The recent sequencing of genomic regions containing CcBV proviral sequences offered us the opportunity to map DNA molecules amplified during particle production onto the chromosomal regions containing proviral segments (7, 12, 17, 22, 28). Compared to previous analyses performed on a very limited number of segments, we provide here an extensive view of the molecules amplified during particle production.
We determined that the 35 CcBV segments were amplified during particle production in 12 different molecules, each one constituting a different RU containing 1 to 8 segments. Strikingly, ISs and FSs were also amplified within each RU, but they were not encapsidated. However, these ANES do not extend to wasp genomic regions separating proviral loci, and the hypothesis that all bracovirus sequences were amplified within a single molecule was thus clearly excluded (31). ANES are variable in size (114 bp to 6,436 bp) and are mostly constituted of noncoding DNA with only 2 hypothetical genes (ep2-like3 and ep2-like4), located on one large ANES in PL2, that are not expressed during viral-particle production (data not shown). The boundaries of amplified FSs appear to be very precise. Indeed, for several RUs (RU1, RU5, and RU8), the boundaries of amplified regions could be restricted to a region of 40 to 200 bp (Table 1), suggesting that there is a nucleotide start of amplification on the chromosome or that proviral sequences are excised from the chromosome prior to their linear amplification. To our knowledge, the existence of amplified but not encapsidated viral sequences is a unique feature of polydnavirus genomes. This might be explained by the particular life cycle of the symbiotic viruses, which requires that only the virulence genes involved in antagonist relationships with parasitized hosts are packaged in the particles.
All RUs possess a conserved SCS of 28 bp to 100 bp potentially forming a hairpin that could confer replication origin (ori) functions similar to those described in Saccharomyces sp. or in Drosophila for the amplification of chorion genes (39–41). While the biochemical mechanism and factors involved in replication initiation appear to be highly conserved, the DNA sequences at which these events take place are not (41). Indeed, in baculoviruses, ori sequences (named hrs) are closely related within a genome but show very limited homology between different baculoviruses (42–44). In nudiviruses, direct-repeat sequences have been identified and hypothesized to function as an ori (45–48). The SCSs identified in bracoviruses are not similar to sequences from other viruses, but their conservation in both the Glyptapanteles and Cotesia lineages, which separated approximately 17 million years ago (49), in sequences that are not packaged and thus are not involved in host-parasite antagonistic interactions suggests they have an important function, which could be initiation of replication.
We also found that the nudiviral cluster, including several genes that are involved in bracovirus particle production but that are not encapsidated, is amplified during viral replication and thus constitutes an additional RU. Strikingly a similar situation—amplification, but not encapsidation—was described for regions involved in ichnovirus particle production called IVSPERs (21, 24). In the ichneumonid wasp H. dydimator, the three IVSPERs are characterized by high coding sequence densities (exon density, 62.2%), making them atypical compared to the rest of the wasp genome (exon density, 21%). There was also a significant difference in the mean lengths of intergenic sequences between these regions (638 bp) and other portions of the wasp genome (1,669 bp). Moreover, the 40 IVSPER genes in these regions each consist of a single exon, while a large majority of wasp genes are predicted to contain multiple exons. The organization of the nudiviral cluster is comparable to that of IVSPERs, with an exon density of 50%, a mean intergenic-sequence length of 857 bp, and 10 genes each consisting of a single exon (7, 8). Due to these similarities, it could be considered a bracovirus structural-protein-encoding region (BVSPER). The amplification of IVSPERs and BVSPERs in ichnoviruses and bracoviruses, respectively, points to a striking convergence of replication strategies between polydnaviruses, despite their different viral origins (21, 24). It is probably significant that in bracoviruses the BVSPERs encode 38K and VP39, the most abundant proteins of CiBV particles (19, 23). Amplification of the bracovirus structural-protein genes appears to mimic the process described in baculoviruses and other viruses in which late genes are expressed after DNA replication (50, 51). Therefore, amplification might not only increase the copy number, but could also provide access to the viral RNA polymerase and facilitate gene expression. Accordingly, a motif previously characterized in baculovirus genes transcribed by baculovirus RNA polymerase has been found in the promoter of vp39 (7). The presence of this small motif, (A/T/G)TAAG, within 300 nucleotides upstream of the translation start site would lack the specificity necessary to selectively express chromosomally integrated genes but would be sufficient to ensure expression from amplified sequences using viral RNA polymerase, as described for baculovirus late genes (52). In accordance with this hypothesis, it has been shown recently in Microplitis demolitor BV (MdBV) that vp39 and p74 genes coding for structural proteins were preferentially transcribed by the nudivirus-like RNA polymerase. The BVPSER amplification might be required to achieve the production of VP39 at the level required to make bracovirus nucleocapsids (26).
The role of the nudiviral machinery in bracovirus particle production prompted us to hypothesize that amplification of viral sequences might involve a rolling-circle mechanism. Indeed, such a mechanism was described for the baculovirus Autographa californica nucleopolyhedrosis virus (AcMNPV) (53), sharing core genes involved in replication with nudiviruses (the replication mechanism of which is unknown). We identified the presence of fragments corresponding to a succession of head-to-head and tail-to-tail concatemers for RU9. Surprisingly, these types of concatemers are found in the context of linear replication of viral genomes, such as for poxvirus, whose genome is constituted of a closed linear double-stranded DNA that possesses at its extremities TA-rich inverted terminal repeats (ITR) forming a hairpin, in which replication occurs in the cytoplasm (38) (Fig. 7). The production of the head-to-head and tail-to-tail concatemers explains the lack of PCR products expected from a rolling-circle model of replication. This model is also in accord with the lack of nudiviral DNA polymerase detection in transcripts from three braconid species, even when high-throughput sequencing approaches were used (22). Most of the nudiviral genes potentially involved in replication were lacking in braconid wasp ovaries, except a nudiviral helicase and a flap endonuclease we have recently identified by a probabilistic method (22, 54). This led us to propose that bracovirus DNA amplification might involve the wasp replication machinery (7, 8, 22). In accordance with this hypothesis, massive parallel sequencing performed on M. demolitor ovaries has identified transcripts from many cellular DNA polymerases. In addition, DNA polymerase genes from Maverick transposable elements (TE) that are abundant in parasitoid wasp genomes were also detected (22). The contribution of a third partner for bracovirus DNA amplification is also plausible, since domesticated genes from TE have been found to serve in important cellular processes, such as programmed genome rearrangement in ciliates (55). We expect the detailed characterization of bracovirus replication units and the identification of successive head-to-head and tail-to-tail concatemers as replication intermediates will constitute important clues to sort out which genes are actually involved in bracovirus DNA amplification.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the ANR Paratoxose, the University of Tours sequencing platform PPF Genome, and the CNRS Groupement de Recherche on mobile elements (GDR 2157).
We thank Cindy Ménoret and Karine Musset for insect rearing. We thank Elisabeth Huguet for careful reading of the manuscript.
Footnotes
Published ahead of print 26 June 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.00886-13.
REFERENCES
- 1.Stoltz DB, Krell P, Summers MD, Vinson B. 1984. Polydnaviridae—a proposed family of insect viruses with segmented, double-stranded, circular DNA genomes. Intervirology 21:1–4 [DOI] [PubMed] [Google Scholar]
- 2.Strand MR, Burke GR. 2012. Polydnaviruses as symbionts and gene delivery systems. PLoS Pathog. 8:e1002757. 10.1371/journal.ppat.1002757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gundersen-Rindal D, Dupuy C, Huguet CE, Drezen J-M. 2013. Parasitoid polydnaviruses: evolution, pathology and applications. Biocontrol. Sci. Tech. 23:1–61 [Google Scholar]
- 4.Strand MR, Drezen J-M. 2012. Family Polydnaviridae, p 267–278 In King AMQ, Adams MJ, Carsten EB, Lefkowitz EJ. (ed), Virus taxonomy. Ninth report of the International Commitee on Taxonomy of Viruses. Elsevier, London, United Kingdom [Google Scholar]
- 5.Dupuy C, Huguet E, Drezen J-M. 2006. Unfolding the evolutionary story of polydnaviruses. Virus Res. 117:81–89 [DOI] [PubMed] [Google Scholar]
- 6.Belle E, Beckage NE, Rousselet J, Poirié M, Lemeunier F, Drezen J-M. 2002. Visualization of polydnavirus sequences in a parasitoid wasp chromosome. J. Virol. 76:5793–5796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bézier A, Annaheim M, Herbinière J, Wetterwald C, Gyapay G, Bernard-Samain S, Wincker P, Roditi I, Heller M, Belghazi M, Pfister-Wilhem R, Periquet G, Dupuy C, Huguet E, Volkoff A-N, Lanzrein B, Drezen J-M. 2009. Polydnaviruses of braconid wasps derive from an ancestral nudivirus. Science 323:926–930 [DOI] [PubMed] [Google Scholar]
- 8.Bézier A, Herbinière J, Lanzrein B, Drezen J-M. 2009. Polydnavirus hidden face: the genes producing virus particles of parasitic wasps. J. Invertebr. Pathol. 101:194–203 [DOI] [PubMed] [Google Scholar]
- 9.Espagne E, Dupuy C, Huguet E, Cattolico L, Provost B, Martins N, Poirié M, Periquet G, Drezen J-M. 2004. Genome sequence of a polydnavirus: insights into symbiotic virus evolution. Science 306:286–289 [DOI] [PubMed] [Google Scholar]
- 10.Tanaka K, Lapointe R, Barney WE, Makkay AM, Stoltz D, Cusson M, Webb BA. 2007. Shared and species-specific features among ichnovirus genomes. Virology 363:26–35 [DOI] [PubMed] [Google Scholar]
- 11.Webb BA, Strand MR, Dickey SE, Beck MH, Hilgarth RS, Barney WE, Kadash K, Kroemer JA, Lindstrom KG, Rattanadechakul W, Shelby KS, Thoetkiattikul H, Turnbull MW, Witherell AR. 2006. Polydnavirus genomes reflect their dual roles as mutualists and pathogens. Virology 347:160–174 [DOI] [PubMed] [Google Scholar]
- 12.Desjardins CA, Gundersen-Rindal DE, Hostetler JB, Tallon LJ, Fuester RW, Schatz MC, Pedroni MJ, Fadrosh DW, Haas BJ, Toms BS, Chen D, Nene V. 2007. Structure and evolution of a proviral locus of Glyptapanteles indiensis bracovirus. BMC Microbiol. 7:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lapointe R, Tanaka K, Barney WE, Whitfield JB, Banks JC, Béliveau C, Stoltz D, Webb BA, Cusson M. 2007. Genomic and morphological features of a banchine polydnavirus: comparison with bracoviruses and ichnoviruses. J. Virol. 81:6491–6501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Y-F, Gao F, Ye X, Wei S, Shi M, Zheng H, Chen X-X. 2011. Deep sequencing of Cotesia vestalis bracovirus reveals the complexity of a polydnavirus genome. Virology 414:42–50 [DOI] [PubMed] [Google Scholar]
- 15.Beckage NE. 2012. Polydnaviruses as endocrine regulators, p 163–168 In Beckage NE, Drezen J-M. (ed), Parasitoid viruses symbionts and pathogens. Elsevier, San Diego, CA [Google Scholar]
- 16.Strand MR. 2012. Polydnavirus gene products that interact with the host immune system, p 149–162 In Beckage NE, Drezen J-M. (ed), Parasitoid viruses symbionts and pathogens. Elsevier, San Diego, CA [Google Scholar]
- 17.Bézier A, Louis F, Jancek S, Periquet G, Thézé J, Gyapay G, Musset K, Lesobre J, Lenoble P, Dupuy C, Gundersen-Rindal D, Herniou EA, Drezen J-M. Functional Endogenous Viral Elements in the genome of the parasitoid wasp Cotesia congregata: insights into the evolutionary dynamics of bracoviruses. Philos. Trans. R. Soc. B, 10.1098/rstb.2013.0047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dupuy C, Gundersen-Rindal D, Cusson M. 2012. Genomics and replication of polydnaviruses, p 47–62 In Beckage NE, Drezen J-M. (ed), Parasitoid viruses symbionts and pathogens. Elsevier, San Diego, CA [Google Scholar]
- 19.Theilmann DA, Summers MD. 1986. Molecular analysis of Campoletis sonorensis virus DNA in the lepidopteran host Heliothis virescens. J. Gen. Virol. 67:1961–1969 [DOI] [PubMed] [Google Scholar]
- 20.Wetterwald C, Roth T, Kaeslin M, Annaheim M, Wespi G, Heller M, Maser P, Roditi I, Pfister-Wilhelm R, Bézier A, Gyapay G, Drezen J-M, Lanzrein B, Mäser P. 2010. Identification of bracovirus particle proteins and analysis of their transcript levels at the stage of virion formation. J. Gen. Virol. 91:2610–2619 [DOI] [PubMed] [Google Scholar]
- 21.Volkoff A-N, Jouan V, Urbach S, Samain S, Bergoin M, Wincker P, Demettre E, Cousserans F, Provost B, Coulibaly F, Legeai F, Béliveau C, Cusson M, Gyapay G, Drezen J-M. 2010. Analysis of virion structural components reveals vestiges of the ancestral ichnovirus genome. PLoS Pathog. 6:e1000923. 10.1371/journal.ppat.1000923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burke GR, Strand MR. 2012. Deep sequencing identifies viral and wasp genes with potential roles in replication of Microplitis demolitor bracovirus. J. Virol. 86:3293–3306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Drezen J-M, Herniou EA, Bézier A. 2012. Evolutionary progenitors of bracoviruses, p 15–31 In Beckage NE, Drezen J-M. (ed), Parasitoid viruses symbionts and pathogens. Elsevier, San Diego, CA [Google Scholar]
- 24.Volkoff A, Drezen J-M, Cusson M, Webb BA. 2012. The organization of genes encoding ichnovirus structural proteins, p 33–45 In Beckage NE, Drezen J-M. (ed), Parasitoid viruses symbionts and pathogens. Elsevier, San Diego, CA [Google Scholar]
- 25.Thézé J, Bézier A, Periquet G, Drezen J-M, Herniou EA. 2011. Paleozoic origin of insect large dsDNA viruses. Proc. Natl. Acad. Sci. U. S. A. 108:15931–15935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burke GR, Thomas SA, Eum JH, Strand MR. 2013. Mutualistic polydnaviruses share essential replication gene functions with pathogenic ancestors. PLoS Pathog. 9:e1003348. 10.1371/journal.ppat.1003348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Savary S, Beckage N, Tan F, Periquet G, Drezen J-M. 1997. Excision of the polydnavirus chromosomal integrated EP1 sequence of the parasitoid wasp Cotesia congregata (Braconidae, Microgastinae) at potential recombinase binding sites. J. Gen. Virol. 78:3125–3134 [DOI] [PubMed] [Google Scholar]
- 28.Desjardins CA, Gundersen-Rindal DE, Hostetler JB, Tallon LJ, Fadrosh DW, Fuester RW, Pedroni MJ, Haas BJ, Schatz MC, Jones KM, Crabtree J, Forberger H, Nene V. 2008. Comparative genomics of mutualistic viruses of Glyptapanteles parasitic wasps. Genome Biol. 9:R183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Annaheim M, Lanzrein B. 2007. Genome organization of the Chelonus inanitus polydnavirus: excision sites, spacers and abundance of proviral and excised segments. J. Gen. Virol. 88:450–457 [DOI] [PubMed] [Google Scholar]
- 30.Pasquier-Barre F, Dupuy C, Huguet E, Monteiro F, Moreau A, Poirié M, Drezen J-M. 2002. Polydnavirus replication: the EP1 segment of the parasitoid wasp Cotesia congregata is amplified within a larger precursor molecule. J. Gen. Virol. 83:2035–2045 [DOI] [PubMed] [Google Scholar]
- 31.Gruber A, Stettler P, Heiniger P, Lanzrein B. 1996. Polydnavirus DNA of the braconid wasp Chelonus inanitus is integrated in the wasp's genome and excised only in later pupal and adult stages of the female. J. Gen. Virol. 77:2873–2879 [DOI] [PubMed] [Google Scholar]
- 32.Harwood SH, Beckage NE. 1994. Purification and characterization of an early expressed polydnavirus induced protein from the hemolymph of Manduca sexta larvae parasitized by Cotesia congregata. Insect Biochem. Mol. 24:685–698 [Google Scholar]
- 33.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-ΔΔC(T)) method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 34.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. 2009. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37:W202–W208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carver TJ, Rutherford KM, Berriman M, Rajandream M-A, Barrell BG, Parkhill J. 2005. ACT: the Artemis Comparison Tool. Bioinformatics 21:3422–3423 [DOI] [PubMed] [Google Scholar]
- 36.Marti D, Grossniklaus-Bürgin C, Wyder S, Wyler T, Lanzrein B. 2003. Ovary development and polydnavirus morphogenesis in the parasitic wasp Chelonus inanitus. I. Ovary morphogenesis, amplification of viral DNA and ecdysteroid titres. J. Gen. Virol. 84:1141–1150 [DOI] [PubMed] [Google Scholar]
- 37.Rojo G, García-Beato R, Viñuela E, Salas ML, Salas J. 1999. Replication of African swine fever virus DNA in infected cells. Virology 257:524–536 [DOI] [PubMed] [Google Scholar]
- 38.Traktman P. 1996. Poxvirus DNA replication, p 775–798 In DePamphilis ML. (ed), DNA replication in eukaryotic cells. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 39.Calvi BR, Lilly MA, Spradling AC. 1998. Cell cycle control of chorion gene amplification. Gene Dev. 12:734–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Segurado M, De Luis A, Antequera F. 2003. Genome-wide distribution of DNA replication origins at A+T-rich islands in Schizosaccharomyces pombe. EMBO Rep. 4:1048–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cvetic C, Walter JC. 2005. Eukaryotic origins of DNA replication: could you please be more specific? Semin. Cell Dev. Biol. 16:343–353 [DOI] [PubMed] [Google Scholar]
- 42.Carstens EB, Wu Y. 2007. No single homologous repeat region is essential for DNA replication of the baculovirus Autographa californica multiple nucleopolyhedrovirus. J. Gen. Virol. 88:114–122 [DOI] [PubMed] [Google Scholar]
- 43.Okano K, Vanarsdall AL, Mikhailov VS, Rohrmann GF. 2006. Conserved molecular systems of the Baculoviridae. Virology 344:77–87 [DOI] [PubMed] [Google Scholar]
- 44.Vanarsdall AL, Mikhailov VS, Rohrmann GF. 2007. Baculovirus DNA replication and processing. Curr. Drug Targets 8:1096–1102 [DOI] [PubMed] [Google Scholar]
- 45.Burand JP, Kim W, Afonso CL, Tulman ER, Kutish GF, Lu Z, Rock DL. 2012. Analysis of the genome of the sexually transmitted insect virus Helicoverpa zea nudivirus 2. Viruses 4:28–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheng C, Liu S-M, Chow T, Hsiao Y-Y, Wang D, Huang J, Chen H-H. 2002. Analysis of the complete genome sequence of the Hz-1 virus suggests that it is related to members of the Baculoviridae. J. Virol. 76:9024–9034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Y, Kleespies RG, Huger AM, Jehle JA. 2007. The genome of Gryllus bimaculatus nudivirus indicates an ancient diversification of baculovirus-related nonoccluded nudiviruses of insects. J. Virol. 81:5395–5406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Y, Van Oers MM, Crawford AM, Vlak JM, Jehle JA. 2007. Genomic analysis of Oryctes rhinoceros virus reveals genetic relatedness to Heliothis zea virus 1. Arch. Virol. 152:519–531 [DOI] [PubMed] [Google Scholar]
- 49.Murphy N, Banks JC, Whitfield JB, Austin AD. 2008. Phylogeny of the parasitic microgastroid subfamilies (Hymenoptera: Braconidae) based on sequence data from seven genes, with an improved time estimate of the origin of the lineage. Mol. Phylogenet. Evol. 47:378–395 [DOI] [PubMed] [Google Scholar]
- 50.Wang Y, Jehle JA. 2009. Nudiviruses and other large, double-stranded circular DNA viruses of invertebrates: new insights on an old topic. J. Invertebr. Pathol. 101:187–193 [DOI] [PubMed] [Google Scholar]
- 51.Slack J, Arif BM. 2007. The baculoviruses occlusion-derived virus: virion structure and function. Adv. Virus Res. 69:99–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rohrmann GF. 2011. Baculovirus late transcription, p 75–86 In Rohrmann GF. (ed), Baculovirus molecular biology. National Library of Medicine, National Center for Biotechnology Information, Bethesda, MD [Google Scholar]
- 53.Oppenheimer DI, Volkman LE. 1997. Evidence for rolling circle replication of Autographa californica M nucleopolyhedrovirus genomic DNA. Arch. Virol. 142:2107–2113 [DOI] [PubMed] [Google Scholar]
- 54.Herniou E, Huguet E, Thézé J, Bézier A, Periquet G, Drezen J-M. When parasitic wasps hijacked viruses: genomic and functionnal evolution of polydnaviruses. Philos. Trans. R. Soc. B, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baudry C, Malinsky S, Restituito M, Kapusta A, Rosa S, Meyer E, Bétermier M. 2009. PiggyMac, a domesticated piggyBac transposase involved in programmed genome rearrangements in the ciliate Paramecium tetraurelia. Genes Dev. 23:2478–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.