Abstract
Influenza A virus (IAV) remains an important human pathogen largely because of antigenic drift, the rapid emergence of antibody escape mutants that precludes durable vaccination. The most potent neutralizing antibodies interact with cognate epitopes in the globular “head” domain of hemagglutinin (HA), a homotrimeric glycoprotein. The H1 HA possesses five distinct regions defined by a large number of mouse monoclonal antibodies (MAbs), i.e., Ca1, Ca2, Cb, Sa, and Sb. Ca1-Ca2 sites require HA trimerization to attain full antigenicity, consistent with their locations on opposite sides of the trimer interface. Here, we show that full antigenicity of Cb and Sa sites also requires HA trimerization, as revealed by immunofluorescence microscopy of IAV-infected cells and biochemically by pulse-chase radiolabeling experiments. Surprisingly, epitope antigenicity acquired by HA trimerization persists following acid triggering of the globular domains dissociation and even after proteolytic release of monomeric heads from acid-treated HA. Thus, the requirement for HA trimerization by trimer-specific MAbs mapping to the Ca, Cb, and Sa sites is not dependent upon the bridging of adjacent monomers in the native HA trimer. Rather, complete antigenicity of HA (and, by inference, immunogenicity) requires a final folding step that accompanies its trimerization. Once this conformational change occurs, HA trimers themselves would not necessarily be required to induce a highly diverse neutralizing response to epitopes in the globular domain.
INTRODUCTION
The influenza A virus (IAV) hemagglutinin (HA) glycoprotein attaches virions to target cells by binding terminal sialic acid residues on cell surface glycans (1, 2). As a prototypical homotrimeric type I integral membrane protein, HA is synthesized in the endoplasmic reticulum (ER) of infected cells and transported through the Golgi complex (GC) to the plasma membrane (PM), where it is incorporated into budding virions. A variable number (depending on the strain) of N-linked oligosaccharides are added cotranslationally as HA is extruded into the ER through the translocon and subsequently trimmed and modified extensively during HA transport to the cell surface. In addition, the cytoplasmic COOH terminus of HA is palmitoylated during its intracellular transport, likely in an early GC compartment (3).
HA folding begins cotranslationally, as shown by the acquisition of intrachain disulfide bonds and by the binding of monoclonal antibodies (MAbs) specific for discontinuous epitopes within HA to nascent chains (4). Initial folding of HA monomers is likely completed shortly after chain termination (within 1 or 2 min), whereas HA trimerization occurs with a half-life of ∼5 min (5, 6). The localization of the later process appears to be strain (and perhaps subtype) specific, with prototype H2 (7) and H3 (8) HAs trimerizing in the ER while the A/Puerto Rico/8/34 (PR8) H1 HA trimerizes in either the ER-Golgi intermediate compartment or the cis-GC (3, 6).
HA is of great medical importance by virtue of its recognition by neutralizing Abs that provide the bulk of the protection afforded by prior IAV infection or vaccination (9). The variability of HA neutralizing epitopes enables IAV immune escape (10). Most neutralizing Abs are directed to epitopes in the globular “head” domain of HA, the location of the sialic acid binding site. Abs neutralize infectivity by sterically blocking HA binding to its receptor and by interfering with conformational alterations required for HA-mediated fusion of viral and cellular membranes (11). Fusion occurs after the internalization of virions into endosomes, where the acidic pH triggers HA to expose its fusion peptide.
In classical work, now 30 years old, Gerhard and colleagues used a large panel of MAbs to antigenically define PR8 HA (12, 13). This revealed the presence of five overlapping antigenic sites in the globular domain, two highly variable between closely related epidemic strains (strain-specific a [Sa] and Sb) and three that demonstrate somewhat more cross-reactivity (Ca1, Ca2, and Cb). The Sa and Sb sites are located at the very tip of the HA, while Ca and Cb lie further down within the HA spike.
With increasing concern regarding the effectiveness of IAV vaccination and the possibility of generating vaccines that induce broadly neutralizing Abs (14), there is a renewed interest in understanding not only HA antigenicity but also HA immunogenicity. Since virtually all neutralizing Abs are conformation specific, a critical issue is the generation of epitopes during the progressive folding of HA. Biogenesis studies show that HA-specific MAbs are exquisitely sensitive to HA conformational status. The Y8-10C2 MAb (designated Sa11 in original publications) defines an Sa epitope concealed upon HA trimerization (6), making Y8-10C2 a probe for conformational changes, including brief conformational excursions that expose the epitope for Ab binding (15). Conversely, HA trimerization is required to generate the H17-L2 (Ca8) epitope, which is located at the HA trimer interface (6). In IAV PR8-infected cells, Y8-10C2 staining of the secretory compartment is limited to the ER while H17-L2 stains the GC and beyond, consistent with HA trimerization occurring in the GC or during the transit of HA from ER exit sites to the GC. The Sb-specific MAb H28-E23 (Sb9) binds both HA monomers and trimers and stains HA throughout the secretory pathway (6).
The distinct pattern of staining of HA monomer- versus trimerization-dependent MAbs makes possible a very simple screening assay for oligomer discrimination by MAbs specific for the distinct HA antigenic sites. While it is expected that some Ca-specific MAbs, whose escape HA mutants span the trimer interface, are dependent on HA trimerization, it is possible that epitopes recognized by MAbs specific for other sites also require HA trimerization for their formation.
Here, we identify additional HA trimer-specific MAbs and use them to demonstrate that HA antigenicity, and hence folding, is fully completed only upon HA trimerization.
MATERIALS AND METHODS
Cells.
MDCK cells (American Type Culture Collection, Manassas, VA) were maintained in complete medium (Dulbecco's modified Eagle's medium [DMEM] plus GlutaMAX-I [Gibco, Grand Island, NY] supplemented with 4.5 g/liter d-glucose, 110 mg/liter sodium pyruvate, and 7.5% heat-inactivated fetal bovine serum) at 37°C in a humidified atmosphere of 9% carbon dioxide in air.
Virus and viral infections.
IAV PR8 was grown in the allantoic cavity of embryonated chicken eggs and stored as infectious allantoic fluid at −80°C. MDCK monolayers were washed twice with Dulbecco's phosphate-buffered saline (DPBS; Gibco) and incubated with IAV PR8 at 10 infectious doses per cell in AIM medium (minimum essential medium [MEM] plus GlutaMAX-I [Gibco] supplemented with Earle's salts, 0.1% bovine serum albumin [BSA], and 20 mM HEPES [pH 6.6]) at 37°C in a humidified atmosphere of 9% carbon dioxide in air. After 1 h, the infection medium was replaced with complete medium and incubation was continued for an additional 4 h under the same conditions.
Abs.
Mouse anti-HA MAbs were described in previous publications (12, 13, 15–19). We used MAbs as undiluted hybridoma culture fluid supernatants in all of our experiments. The rabbit anti-neuraminidase (NA) polyclonal Abs used were described previously (20). DyLight 488-conjugated donkey anti-mouse IgG (H+L) and DyLight 546-conjugated donkey anti-rabbit IgG (H+L) were from Jackson ImmunoResearch Laboratories, Inc. (West Grove, PA). Horseradish peroxidase (HRP)-conjugated rabbit anti-mouse IgG was purchased from Dako (Carpinteria, CA). HRP-conjugated anti-mouse IgG TrueBlot ULTRA was from eBioscience (San Diego, CA).
Pulse-chase analysis, immunoprecipitation, and immunoblotting.
Confluent monolayers of IAV PR8-infected MDCK cells growing in 75-cm2 culture flasks (Thermo Scientific Nunc, Rochester, NY) were detached with trypsin (Gibco) and washed twice with DPBS. Cells in suspension were pulse-labeled for 2 min with EasyTag [35S]Met (PerkinElmer, Waltham, MA) (1 or 0.2 mCi/ml, depending on whether experiments were performed at 20°C or 37°C, respectively) in Met-free DMEM (Gibco) and then chased in complete medium supplemented with 67 mM l-Met (Sigma-Aldrich, St. Louis, MO). After each chase time point, cell aliquots were removed and washed once with ice-cold DPBS supplemented or not with 20 mM N-ethylmaleimide (NEM; Calbiochem, Billerica, MA). Cells were pelleted and extracted in ice-cold lysis buffer (0.5% Triton X-100, 50 mM Tris-HCl [pH 7.5], 300 mM NaCl, 5 mM EDTA) supplemented with the complete, Mini, EDTA-free protease inhibitor cocktail (Roche Diagnostics, Indianapolis, IN). Equivalent amounts of cell lysates were subjected to immunoprecipitation (IP) as described previously (21). Immunocollected proteins were analyzed by SDS-PAGE (22) and visualized by exposing dried gels to Carestream Kodak BioMax MR (Sigma-Aldrich) films. Gels were stained with Coomassie brilliant blue R prior to drying to ensure that IgG was equally recovered and loaded in all of the lanes. Since the same amount of IgG was added to each sample, this served as a loading control. Immunoblotting studies were performed as described previously (22). Data analysis and quantification were performed with the ImageJ software (http://rsbweb.nih.gov/ij).
Immunofluorescence confocal microscopy.
MDCK cells (1 × 105) growing on 12-mm microscope cover glasses (Marienfeld GmbH & Co. KG, Lauda-Königshofen, Germany) were infected with IAV PR8 as described above in the presence or absence of 10 μM monensin (Sigma-Aldrich). Cells were washed twice with DPBS supplemented with 0.9 mM CaCl2 and 0.43 mM MgCl2 (DPBS-Ca-Mg; Gibco) and then fixed with 3% freshly prepared paraformaldehyde (Electron Microscopy Sciences, Hatfield, PA) in DPBS-Ca-Mg. After permeabilization with 1% Triton X-100, cells were incubated with a mixture of polyclonal anti-NA Abs and the corresponding anti-HA MAb in 5% normal donkey serum (Jackson ImmunoResearch Laboratories, Inc.). Cells were then incubated with fluorescent secondary Abs in balanced salt solution supplemented with 0.1% BSA. Inverted coverslips were mounted on microscope slides with Fluoromount G plus 4′,6-diamidino-2-phenylindole (DAPI; Electron Microscopy Sciences) and analyzed with a TCS SP5 (DMI 6000) confocal microscope system (Leica Microsystems, Deerfield, IL).
Tryptic digestion of recombinant trimerized HA.
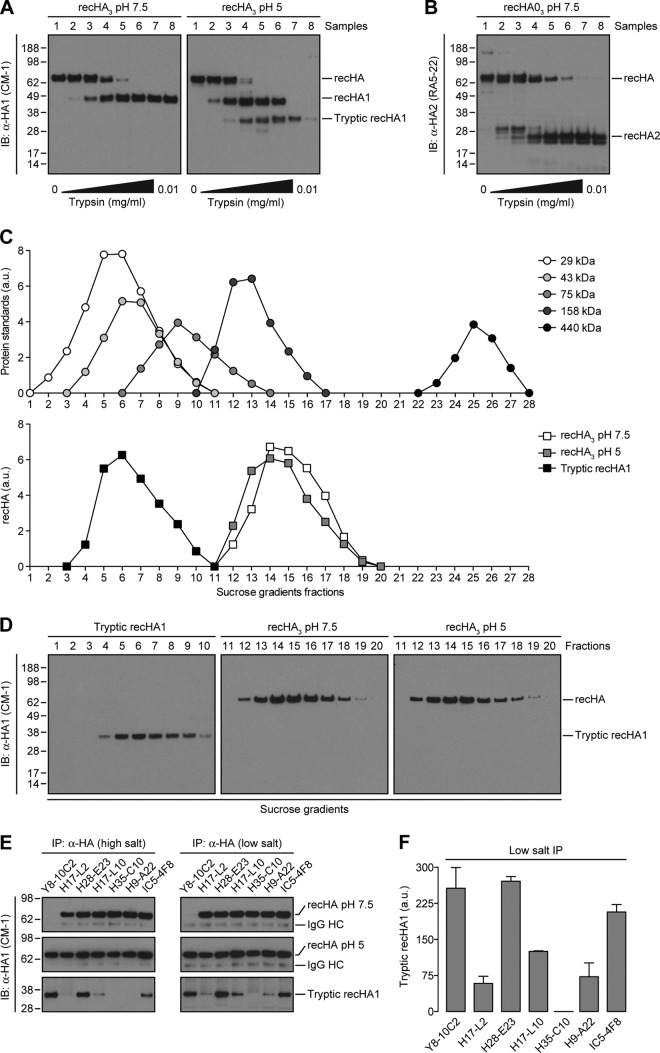
PR8-foldon-His6 recHA (recHA3) (23) in 1% Triton X-100, 50 mM Tris-HCl (pH 7.5), and 100 mM NaCl was incubated at pH 5 (attained by adding 1 M citric acid at pH 1.64) for 15 min at 37°C, returned to neutral pH with 1 M Tris at pH 10.91, and exposed to increasing amounts of trypsin tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK)-treated (Worthington, Lakewood, NJ) for an additional 15 min at 37°C. Digestion was stopped by adding 1 mM phenylmethylsulfonyl fluoride (PMSF) to each sample. Native recHA3, acid-treated recHA3, and the monomeric, tryptic fragment derived from recHA1 (sample 7; see Fig. 5A) were subjected to IP at a high (300 mM) or low (150 mM) concentration of NaCl with specific anti-HA Ca, Cb, Sa, and Sb MAbs.
Fig 5.
HA trimer-specific MAbs to the Ca, Cb, and Sa antigenic sites bind to monomers derived from oligomerized HA. (A and B) Untreated (pH 7.5) and acid-triggered (pH 5) recHA3 were incubated with increasing amounts of trypsin for 15 min at 37°C. Digestion was stopped with 1 mM PMSF. recHA1- (A) or recHA2 (B)-derived fragments were then visualized by immunoblotting (IB) with the anti-HA CM-1 or RA5-22 MAbs, respectively. (C and D) In addition to a cocktail of protein standards, native recHA3, acid-treated recHA3, and the monomeric, tryptic fragments derived from recHA1 (HA tops) (sample 7, A) were fractionated by ultracentrifugation on layered 5 to 25% sucrose gradients as described in Materials and Methods. Sucrose gradient fractions were then analyzed by reducing SDS-PAGE and immunoblotting with the CM-1 MAb (D). The amounts of sedimented standards and recHA proteins in each fraction were quantified by densitometry and expressed in arbitrary units (a.u.) (C). (E) The recHA3 pH 7.5, recHA3 pH 5, and monomeric tryptic recHA1 proteins were subjected to IP at a high (300 mM) or low (150 mM) salt concentration with the HA monomer-specific Y8-10C2 MAb; the HA trimer-reactive H17-L2, H17L-10, H35-C10, and H9-A22 MAbs; or MAbs H28-E23 and IC5-4F8, which recognize both HA species. (F) The amount of immunoprecipitated HA tops was quantified as described in the legend to Fig. 1D. The values to the left of panels A, B, D, and E are molecular sizes in kilodaltons.
Zonal sedimentation analysis on sucrose gradients.
Trimerized recHA (recHA3) and monomeric, tryptic recHA1 fragments were analyzed by ultracentrifugation on layered 5 to 25% (wt/vol) sucrose gradients (9 ml) with a 2-ml 60% sucrose cushion prepared in 50 mM Tris-HCl (pH 7.5) and 100 mM NaCl. Native recHA3, acid-triggered recHA3, and tryptic recHA1 were loaded on the top of separate sucrose gradients and centrifuged in an SW41 rotor (Beckman Coulter Inc., Fullerton, CA) at 35,000 rpm for 16 h at 4°C. Fractions of 0.25 ml were collected from the top of the tube and analyzed by immunoblotting with CM-1, a MAb specific to denatured HA1. The protein standards carbonic anhydrase (29 kDa), ovalbumin (43 kDa), conalbumin (75 kDa), aldolase (158 kDa), and ferritin (440 kDa) (GE Healthcare Biosciences, Pittsburgh, PA) were loaded at 60 μg/gradient and detected by staining nitrocellulose membranes with Ponceau S (Sigma-Aldrich) before immunoblotting. Standards and recHA proteins on sucrose gradient fractions were quantified by densitometry with the ImageJ software.
Surface plasmon resonance.
Steady-state equilibrium binding of MAbs to different recHA proteins was monitored with a ProteOn surface plasmon resonance (SPR) biosensor (Bio-Rad, Hercules, CA) (24, 25). Protein A/G was coupled to GLC sensor chips with amine coupling at 500 resonance units in test flow cells. MAbs in serum-free culture supernatants were captured with protein A/G at 10 μg/ml. Twofold serial dilutions of freshly prepared recHA proteins (starting concentration, 20 μg/ml) were injected at a flow rate of 50 μl/min (contact time, 180 s) for association, and further disassociation was performed over an interval of 600 s at a flow rate of 50 μl/min. Responses from the MAb-coated surface were corrected with a mock surface and a separate injection of buffer only. The anti-CCR5 2D7 MAb was included as a negative control. The Bio-Rad ProteOn manager software (version 3.0) was used for data analysis.
RESULTS
Characterization of nascent HA monomers and trimers via SDS-PAGE.
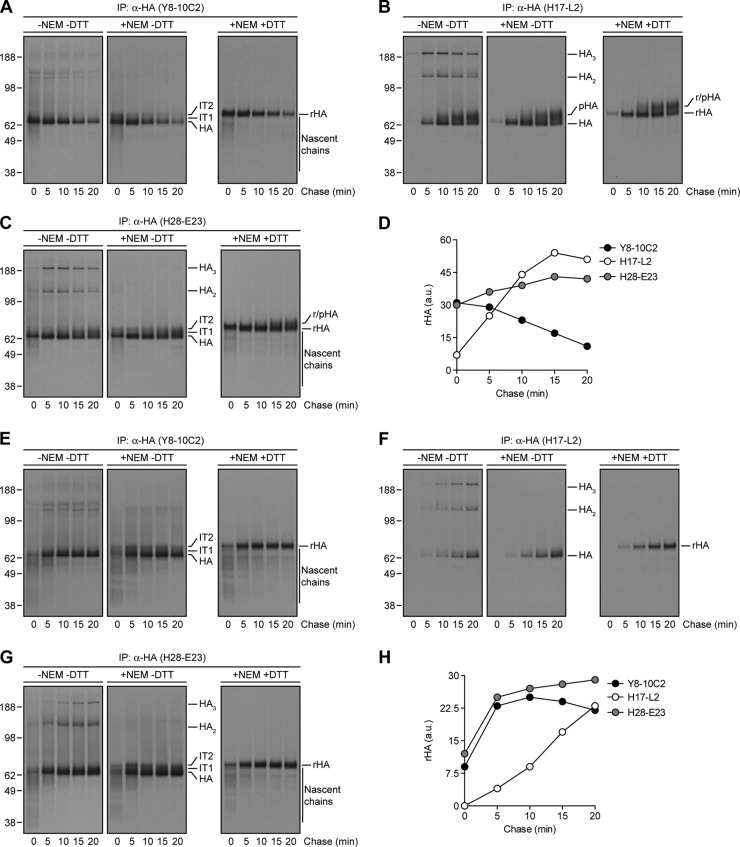
As reported previously (6), after 2 min of labeling of IAV PR8-infected MDCK cells with [35S]Met at 37°C, the anti-HA Y8-10C2 and H17-L2 MAbs recovered distinct biochemical species from Triton X-100 lysates as revealed by SDS-PAGE under reducing and nonreducing conditions (Fig. 1A and B). Under reducing conditions, HA migrated more slowly because of the complete unfolding of domains maintained by disulfide bonds.
Fig 1.
Biochemical characterization of HA maturation, trafficking, and assembly with specific anti-HA MAbs. (A to C) IAV PR8-infected MDCK cells were labeled with [35S]Met for 2 min at 37°C, chased for the indicated times at the same temperature, and lysed with an ice-cold extraction buffer supplemented (+NEM) or not (−NEM) with NEM. Detergent lysates were then precipitated with MAbs specific to HA monomers (Y8-10C2, A), HA trimers (H17-L2, B), or both HA species (H28-E23, C). Immunocollected proteins were boiled in SDS sample buffer supplemented (+DTT) or not (−DTT) with dithiothreitol and analyzed by SDS-PAGE and fluorography. IT1 and IT2, HA folding intermediate species; pHA, processed HA; rHA, reduced HA. (D) The amount of immunoprecipitated rHA was quantified by densitometry and expressed in arbitrary units (a.u.). (E to G) MDCK cells infected with IAV PR8 were radiolabeled and chased at 20°C instead of 37°C. Cell lysates made at the end of each chase time point were subjected to IP with the Y8-10C2 (E), H17-L2 (F), or H28-E23 (G) MAbs. (H) The amount of precipitated rHA was quantified as described for panel D. The values to the left of panels A to C and E to G are molecular sizes in kilodaltons.
Reduced Y8-10C2-reactive HA migrated with a molecular mass (65 kDa) expected for glycosylated HA (termed HA) (Fig. 1A). Immediately after pulse-labeling, Y8-10C2 also recovered a ladder of lower-molecular-mass species that likely represent incomplete nascent chains that cotranslationally fold to create the Y8-10C2 epitope (4). Under nonreducing conditions, we observed several minor higher-molecular-mass species that may represent host proteins bound postlysis to HA via disulfide bonds since they were not present after alkylating free Cys residues by the addition of NEM to the lysis buffer. The addition of NEM also revealed HA with partial intrachain disulfide bonds, termed IT1 and IT2 by Braakman and colleagues (26). Y8-10C2-reactive HA decayed over the chase period because of steric interference of Y8-10C2 access to its epitope coincident with HA trimerization (6, 15) (Fig. 1A and D).
The same experiment demonstrated HA trimerization by a time-dependent increase in the recovery of HA by H17-L2 (Fig. 1B and D). HA exhibited an initial slight increase in mobility likely due to trimming of N-linked oligosaccharides in the early GC, followed by a more substantial decrease in mobility likely due to additions of extra oligosaccharide chains in the distal GC. Under nonreducing conditions, HA trimers migrated as a mixture of monomers, dimers, and trimers. Addition of NEM to the extraction buffer prevented the appearance of denaturation-resistant multimers (all of the samples are boiled in SDS prior to PAGE), indicating that their generation is due to postlysis disulfide bond formation between free Cys residues. Since bromelain cleavage of the COOH terminus of HA prevents the appearance of HA oligomers in nonreducing SDS-PAGE (6), the relevant Cys residues are almost certainly among the three Cys present in the cytoplasmic domain of HA. Using the anti-HA H28-E23 MAb, which recognizes both HA monomers and trimers, to recover HA from detergent lysates gave the expected pattern of HA species in summing the patterns obtained from Y8-10C2 and H17-L2 (Fig. 1C and D).
To analyze HA folding and trimerization with greater detail, we slowed HA biogenesis by performing the pulse-chase protocol at 20°C. This not only increased the amount of incomplete HA chains recovered by Y8-10C2 and H28-E23 but also retarded their resolution into full-length HA (Fig. 1E and G). Completion of HA synthesis required ∼10 min under these conditions, as inferred from the maximal amount of HA recovered by Y8-10C2 (Fig. 1H). At this time, a fairly large fraction of HA had already trimerized, as shown by the kinetics of HA recovery by H17-L2 and H28-E23 (Fig. 1F to H).
Sequential immunoprecipitation clearly demarcates Y8-10C2- versus H17-L2-reactive HA.
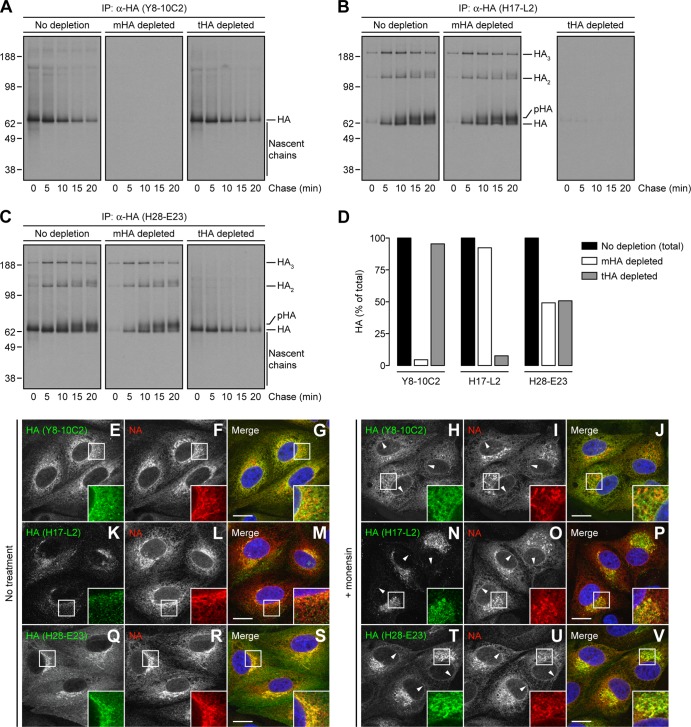
To test the mutual exclusivity of the Y8-10C2 and H17-L2 MAbs to HA, we performed sequential IP, depleting HA species from pulse-chase lysates with one MAb to completion and then analyzing for HA reactive with other MAbs by nonreducing SDS-PAGE. Importantly, we used pulse conditions such that HA monomers and trimers each consisted of ∼50% of the total HA pool. Depletion with Y8-10C2 or H17-L2 completely removed each homologous species without significantly affecting the recovery of the heterologous species qualitatively or quantitatively (Fig. 2A, B, and D). Consistent with this result, each MAb depleted its respective species from HA collected with H28-E23 and reduced the total amount of HA recovered by very close to 50% for each treatment (Fig. 2C and D).
Fig 2.
Specificity of anti-HA MAbs to ER-located HA monomers or trimerized HA lying on the GC. (A to C) IAV PR8-infected MDCK cells were labeled with [35S]Met and chased at 37°C as described in the legend to Fig. 1A to C. HA species were depleted from cell lysates with an irrelevant MAb to the vesicular stomatitis virus N protein (10G-4; no depletion), the HA monomer-specific Y8-10C2 MAb (mHA depleted), or the HA trimer-specific H17-L2 MAb (tHA depleted). After two rounds of depletion, cell extracts were incubated with the Y8-10C2 (A), H17-L2 (B), or H28-E23 (C) MAbs. Immunoprecipitated HA species were analyzed under nonreducing conditions by SDS-PAGE and fluorography. (D) Bar graph representing the percentage of pooled HA-associated radioactivity (from 0 to 20 min) relative to the total HA (100%) in nondepleted cell lysates. (E to V) MDCK cells were infected with IAV PR8 in the absence (no treatment) or presence of 10 μM monensin. Cells were fixed, permeabilized, and incubated with the Y8-10C2 (E to J), H17-L2 (K to P), or H28-E23 (Q to V) MAbs (green channel) and rabbit polyclonal Abs to NA (red channel). DNA was labeled with DAPI (blue channel). Stained cells were examined by confocal fluorescence microscopy. Bars, 10 μm. Arrowheads point to NA colocalization with HA monomers on the nuclear envelope (ER) of cells labeled with Y8-10C2 and H28-E23. The values to the left of panels A to C are molecular sizes in kilodaltons.
Together, these results clearly show the existence of two distinct populations of HA, monomer versus oligomer, each uniquely recognized at high affinity by selected MAbs.
Identification of HA trimerization-dependent Cb and Sa epitopes.
We revisited the subcellular localization of HA monomers and trimers (6) by using confocal microscopy to image MAb binding to fixed and permeabilized cells. We identified the entire secretory pathway by staining cells with rabbit polyclonal Abs raised to a synthetic peptide corresponding to the COOH terminus of NA. These Abs recognize all of the forms of full-length NA, including native tetramers and denatured monomers, in immunoblot assays (20). Immunofluorescence confirmed that the Y8-10C2 and H17-L2 MAbs exhibit complementary patterns, with Y8-10C2 staining the ER (Fig. 2E to G) and H17-L2 staining post-ER structures (GC and PM) (Fig. 2K to M), while H28-E23 stained the entire secretory pathway, giving extensive colocalization with NA (Fig. 2Q to S). We also observed HA trimers on puncta scattered throughout the cytoplasm that did not overlap with ER-associated NA (Fig. 2K to M), which could represent secretory carriers en route to the cell surface or HA in recycling endosomes.
PM staining interfered with intracellular staining because of the tenuity of MDCK cells (Fig. 2Q to S). We therefore treated cells with the H+/Na+ ionophore monensin to slow HA transport through the GC and thus reduce its surface expression (27–29). Monensin altered the morphology of the GC-containing NA (Fig. 2F and I), which failed to stain with Y8-10C2 (Fig. 2H to J) but stained intensely with H17-L2 (Fig. 2N to P) or H28-E23 (Fig. 2T to V). Monensin dramatically redistributed all of the HA trimer-containing structures into perinuclear clusters of membranous vesicles and tubules (Fig. 2N to P). As expected from binding all HA species, H28-E23 staining represented the combined patterns of Y8-10C2 and H17-L2 and extensively colocalized with anti-NA Abs staining throughout the secretory pathway (Fig. 2Q to S and T to V).
We next examined a MAb panel for HA monomer versus trimer binding by immunofluorescence microscopy, scoring HA monomer-specific MAbs to those staining the ER only, HA trimerization-dependent MAbs to those that exclusively stained the GC, and HA monomer/trimerization-dependent MAbs to those exhibiting ER-GC specificity (Table 1). This revealed that, with the exception of Sb-specific MAbs, multiple MAbs specific for the Ca, Cb, and Sa antigenic sites of HA surprisingly stained cells in an HA trimer-specific (GC) pattern.
Table 1.
Immunofluorescence-based screening for anti-HA MAb specificitya
| Anti-HA MAb | |||
|---|---|---|---|
| Original name | Published name | Antigenic site | Staining pattern |
| H2-5B6 | Ca1 | Ca | Monomer/trimer |
| H17-L19 | Ca3 | Ca | Monomer/trimer |
| H18-L9 | Ca5 | Ca | Monomer/trimer |
| H18-S28 | Cx4 | Ca | Monomer/trimer |
| H18-S413 | Cx5 | Ca | Monomer/trimer |
| H33-46 | Cx6 | Ca | Monomer/trimer |
| H33-23 | Cx7 | Ca | Monomer/trimer |
| H28-A2 | Cx8 | Ca | Monomer/trimer |
| Y8-2D1 | Ca2 | Ca | Trimer |
| H17-L10 | Ca6 | Ca | Trimer |
| H17-L2 | Ca8 | Ca | Trimer |
| H3-4C5 | Ca9 | Ca/Sa | Monomer |
| H18-S24 | Cx2 | Ca/Sa | Monomer |
| H35-C9 | Cb2 | Cb | Monomer/trimer |
| H2-5A4 | Cb3 | Cb | Monomer/trimer |
| H18-S415 | Cb5 | Cb | Monomer/trimer |
| H18-S48 | Cb6 | Cb | Monomer/trimer |
| H9-A15 | Cb7 | Cb | Monomer/trimer |
| H20-A15 | Cb8 | Cb | Monomer/trimer |
| H18-S13 | Cb9 | Cb | Monomer/trimer |
| H18-S121 | Cb10 | Cb | Monomer/trimer |
| H18-S112 | Cb11 | Cb | Monomer/trimer |
| H9-D3 | Cb13 | Cb | Monomer/trimer |
| H2-4C2 | Cb14 | Cb | Monomer/trimer |
| H17-L7 | Cb15 | Cb | Monomer/trimer |
| H35-C10 | Cb4 | Cb | Trimer |
| H35-C7 | Cb12 | Cb | Trimer |
| H35-C12 | Cb16 | Cb | Trimer |
| H33-48 | Cb1 | Cb | Monomer |
| PEG-1 | Sa4 | Sa | Monomer/trimer |
| Y8-2C6 | Sa5 | Sa | Monomer/trimer |
| H28-C1 | Sa10 | Sa | Monomer/trimer |
| Y8-4C5 | Sa12 | Sa | Monomer/trimer |
| Y8-3B3 | Sa13 | Sa | Monomer/trimer |
| H2-6A5 | Sa15 | Sa | Monomer/trimer |
| H2-4B3 | Sa1 | Sa | Trimer |
| H16-S19 | Sa2 | Sa | Trimer |
| H16-S53 | Sa3 | Sa | Trimer |
| H2-6A1 | Sa6 | Sa | Trimer |
| H9-A22 | Sa9 | Sa | Trimer |
| H9-B20 | Sa14 | Sa | Trimer |
| Y8-10C2 | Sa11 | Sa | Monomer |
| H18-S21 | Cx1 | Sa | Monomer |
| H35-C6 | Sb1 | Sb | Monomer/trimer |
| H35-C3 | Sb4 | Sb | Monomer/trimer |
| Y8-1C1 | Sb6 | Sb | Monomer/trimer |
| H28-A23 | Sb8 | Sb | Monomer/trimer |
| H28-E23 | Sb9 | Sb | Monomer/trimer |
| H28-D14 | Sb10 | Sb | Monomer/trimer |
| IC5-4F8 | Sb | Monomer/trimer | |
A panel of anti-HA MAbs was analyzed by immunofluorescence microscopy, scoring ER-only staining as HA monomer-specific MAbs, GC-only staining as HA trimer-reactive MAbs, and ER/GC staining as HA monomer/trimerization-dependent MAbs.
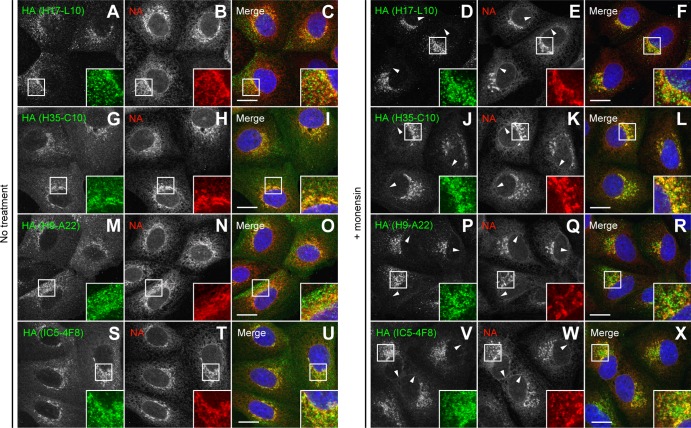
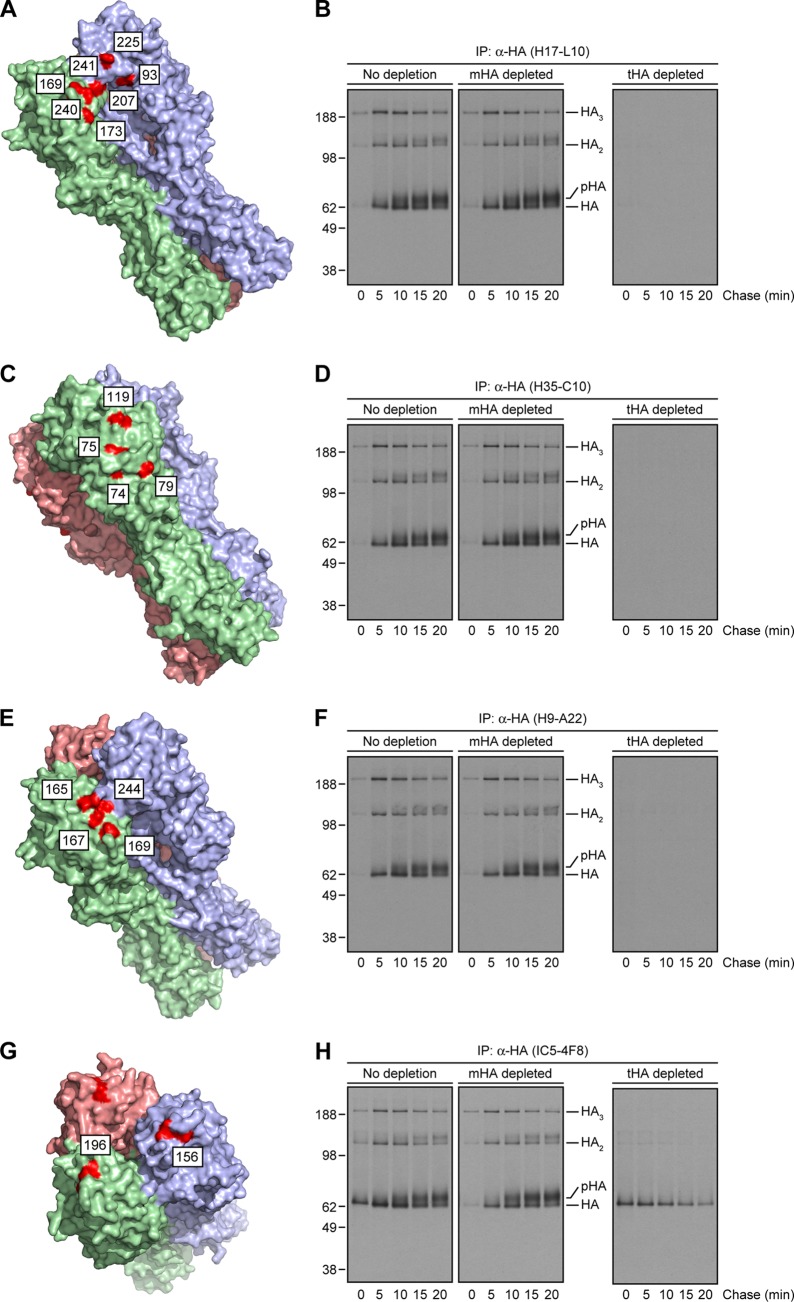
We selected candidate HA trimer-specific MAbs for biochemical analysis, including the HA Ca-specific H17-L10, Cb-specific H35-C10, and Sa-specific H9-A22 MAbs (the staining patterns of these MAbs are shown in Fig. 3A to R). The locations of amino acid substitutions (H3 numbering) that reduce the affinity of these MAbs more than 10-fold are shown in Fig. 4A, C, and E, respectively (12). We performed pulse-chase experiments to examine the HA species recovered by these MAbs in nonreducing gels with extracts that had been depleted of HA monomers or trimers by Y8-10C2 and H17-L2, respectively. In supporting the microscopy data, each MAb demonstrated clear HA trimerization dependence, mimicking the properties of H17-L2 described above (Fig. 2B) in recovering HA only after an ∼5-min chase and from HA monomer- but not trimer-depleted extracts (Fig. 4B, D, and F). As a control, we also characterized the HA Sb-specific IC5-4F8 MAb, which stained both the ER and GC (Fig. 3S to X) and, as predicted, recovered both HA monomers and trimers (Fig. 4G and H).
Fig 3.
Reactivity of various anti-HA MAbs assayed by immunofluorescence microscopy. MDCK cells were infected with IAV PR8 in the absence (no treatment) or presence of 10 μM monensin as described in the legend to Fig. 2E to V. HA was labeled on fixed and permeabilized cells with specific MAbs to the HA Ca (H17-L10, A to F), Cb (H35-C10, G to L), Sa (H9-A22, M to R), and Sb (IC5-4F8, S to X) antigenic sites (green channel). NA was detected with rabbit polyclonal Abs (red channel). DNA was labeled with DAPI (blue channel). Stained cells were examined by confocal fluorescence microscopy. Bars, 10 μm. Arrowheads point to NA colocalization with HA monomers on the nuclear envelope (ER) of cells labeled with IC5-4F8.
Fig 4.
Targeting of the HA Ca, Cb, and Sa antigenic sites by HA trimer-specific MAbs. (A, C, E, and G) PyMOL images of the crystal structure of the IAV PR8 HA trimer (46) (RSCB protein database entry 1RU7) showing amino acid substitutions (red, H3 numbering scheme) on mutants that escape MAbs specific for the HA Ca (A), Cb (C), Sa (E), and Sb (G) antigenic sites. Each of the subunits within the HA oligomer are displayed in light green, pink, and purple. (B, D, F, and H) Detergent extracts from [35S]Met-labeled and chased cells were left untreated or depleted for HA monomers or trimers as described in the legend to Fig. 2A to C before incubation with specific MAbs to the HA Ca (H17-L10, B), Cb (H35-C10, D), Sa (H9-A22, F), and Sb (IC5-4F8, H) antigenic sites. Immunocollected HA species were visualized by SDS-PAGE under nonreducing conditions, followed by fluorography. The values to the left of panels B, D, F, and H are molecular sizes in kilodaltons.
HA trimerization-dependent epitopes are maintained on trimer-derived HA monomers.
Notably, all of the HA trimerization-dependent MAbs identified in the previous section maintain their ability to inhibit hemagglutination mediated by acid-triggered virus (30). Since acid triggering loosens the association of the HA globular domains (30, 31), the binding of HA trimer-specific MAbs may not require bridging between adjacent HA monomers. To examine this possibility, we generated monomeric HA “tops” (composed of the HA globular domain) (31–33) by exposing recombinant purified trimeric HA (recHA3) (23) to pH 5, returning it to neutral pH, and treating it with trypsin. Without acid treatment, recHA3 is converted into HA1 (recHA1, Fig. 5A) and HA2 (recHA2, Fig. 5B) chains by trypsin in a dose-dependent manner, as revealed by immunoblotting with MAbs specific for linear determinants in HA1 (CM-1) or HA2 (RA5-22) (34). Acid exposure, however, increases the sensitivity of HA1 to further digestion, converting HA1 into a trypsin-resistant form that is the only CM-1-reactive specie present in sample 7 (tryptic recHA1, Fig. 5A). We confirmed the oligomeric state of native recHA3, acid-triggered recHA3 (HA trimers with separated globular domains), and HA tops (monomeric HA heads) by zonal sedimentation on 5 to 25% sucrose gradients. As expected, trimerized recHA3 proteins (∼200 kDa) sedimented as discrete peaks immediately following fractions containing aldolase (158 kDa) (Fig. 5C and D). In contrast, monomeric HA tops (∼35 kDa) sedimented on intermediate fractions between those containing carbonic anhydrase (29 kDa) and ovalbumin (43 kDa) (Fig. 5C and D).
We then incubated native recHA3, acid-treated recHA3, or HA tops with various MAbs, detecting immunoreactive HA species recovered on protein A beads by immunoblotting with the CM-1 MAb. The acid-dependent binding of Y8-10C2 demonstrates that recHA3 homogeneously maintains a tight trimeric structure (Fig. 5E). Notably, each of the HA trimerization-dependent MAbs examined (H17-L2, H17-L10, H35-C10, and H9-A22) demonstrated no loss of binding to acid-triggered HA, even when the interaction occurred in a high-salt (300 mM NaCl) buffer (Fig. 5E). Remarkably, three of the four HA trimerization-dependent MAbs examined (H17-L2, H17-L10, and H9-A22) bound the monomeric, tryptic recHA1 fragment (Fig. 5E and F). In contrast to HA monomer-reactive MAbs, however, the binding of each of these MAbs to the HA tops was reduced at 300 mM salt (Fig. 5E), consistent with a decreased affinity for HA tops relative to native HA trimers.
To further examine the interaction of MAbs with native recHA3, acid-triggered recHA3, and fragmented recHA1, we measured Ab on (kon) and off (koff) rate constants by SPR (Table 2). We validated the method by using Y8-10C2, which, as expected (15), demonstrated temperature-dependent binding to native but not acid-treated recHA3 and bound acid-triggered recHA3 with a 10-fold higher affinity than untreated recHA3. SPR also revealed that acid treatment of recHA3 had little effect on the kon and koff values of H28-E23 (HA monomer/trimer binding) or the HA trimer-specific H9-A22 MAb. Moreover, acid treatment of recHA3 increased both the kon and koff values of H17-L10 ∼5-fold while the koff value of H35-C10 decreased ∼2-fold (Table 2).
Table 2.
Kinetic and affinity measurements of MAb binding to recombinant HA proteins by SPRa
| Anti-HA MAb | Kinetic/affinity measurement | recHA3, pH 7.5 | recHA3, pH 5 | Tryptic recHA1 |
|---|---|---|---|---|
| Y8-10C2 (4°C) | kon (1/Ms) | NAb | 3.58E+05 | 2.58E+05 |
| koff (1/s) | NA | 1.99E−05 | 1.69E−04 | |
| KD (1/M) | NA | 5.56E−11 | 6.57E−10 | |
| Y8-10C2 (15°C) | kon (1/Ms) | 1.18E+05 | 3.12E+05 | 1.54E+05 |
| koff (1/s) | 2.48E−04 | 4.07E−05 | 1.91E−04 | |
| KD (1/M) | 2.10E−09 | 1.31E−10 | 1.24E−09 | |
| H28-E23 | kon (1/Ms) | 9.02E+05 | 7.69E+05 | 2.98E+05 |
| koff (1/s) | 2.09E−04 | 1.46E−04 | 3.32E−04 | |
| KD (1/M) | 2.31E−10 | 1.89E−10 | 1.11E−09 | |
| H9-A22 | kon (1/Ms) | 9.07E+05 | 1.04E+06 | 3.35E+05 |
| koff (1/s) | 1.39E−02 | 1.36E−02 | 3.33E−02 | |
| KD (1/M) | 1.54E−08 | 1.30E−08 | 9.92E−08 | |
| H17-L10 | kon (1/Ms) | 1.36E+04 | 6.72E+04 | 6.90E+04 |
| koff (1/s) | 3.56E−03 | 1.67E−02 | 1.34E−02 | |
| KD (1/M) | 2.63E−07 | 2.48E−07 | 1.94E−07 | |
| H35-C10 | kon (1/Ms) | 3.93E+05 | 3.69E+05 | NA |
| koff (1/s) | 4.27E−02 | 1.75E−02 | NA | |
| KD (1/M) | 1.09E−07 | 4.74E−08 | NA |
SPR was performed by coating chip surfaces with various MAbs, followed by injections of 10 μg/ml of native recHA3 (pH 7.5), acid-triggered recHA3 (pH 5), or monomeric tryptic recHA1 (HA tops) at 15°C. For the Y8-10C2 MAb only, the assay was also done at 4°C, where no binding to native recHA3 was observed. Values represent data from two independent experiments.
NA, not applicable.
With HA tops, all of the MAbs demonstrated decreased calculated affinity compared to their binding to native recHA3 or acid-treated recHA3. To some extent, this may reflect underestimation of the HA top concentration, which is needed to calculate the kon value. This does not account for relative kon value differences between different MAbs, however, or the complete lack of binding of H35-C10 which confirms the solution assay data in Fig. 5E. A strength of SPR is that the koff value is independent of the protein concentration. The koff values for all of the MAbs are increased relative to that for acid-treated recHA3, except, rather surprisingly, for H17-L10, which actually decreases slightly. In any event, the data clearly demonstrate that MAbs that are specific for HA monomer, trimer, or both forms bind to a monomeric form of HA with KDs that are in the physiological range.
Taken together, these data conclusively indicate that HA epitopes that require trimerization can persist on monomers generated from HA trimers. Thus, trimerization is required to complete HA folding and not exclusively to align contact residues on adjacent HA monomers.
DISCUSSION
We have explored the relationship between HA oligomerization and fine structure by using MAbs as structural probes. It is important to note the enormous advantages conferred by using antiviral MAbs with neutralizing activity to study the general questions about protein antigenicity and structure. Given that viruses exhibit high mutation rates, it is typically simple to obtain a large panel of escape mutants. With advances in nucleic acid sequencing technology, identification of amino acid substitutions on HA that enable immune escape is now fast and cheap. While not all of the substitutions will represent contact residues for MAbs binding (15), exceptions are likely to be unusual and, in any event, will typically be easy to distinguish by their physical distance from the clustered location of other escape substitutions. In supporting this notion, X-ray crystallography studies on HA escape mutants in conjunction with crystal structures of MAb Fabs in complex with trimerized and monomeric HA allowed Skehel, Knossow, and colleagues to conclusively demonstrate the remarkable coincidence between the locations where anti-HA MAbs bind and the sites of amino acid substitutions on HA variants (35–39). Thus, viral antigens like HA provide a robust, essentially self-reporting system for defining epitope structure.
In a previous publication, we had reported that trimeric PR8 HA migrates as a mixture of monomers, dimers, and trimers in denaturing SDS-PAGE under nonreducing conditions (6). Since we had included the alkylating agent iodoacetamide in the extraction buffer, we concluded that the disulfide bonds in the COOH terminus of HA were not artifactually created postlysis (6). Here, we show that this conclusion is likely erroneous, since replacement of iodoacetamide with NEM prevents oligomerization. This is logical, since the Cys residues implicated in cross-linking of the oligomers are present in the COOH terminus of HA, which should be exposed to the highly reducing environment of the cytosol.
We show that, in addition to HA Ca-specific MAbs, a subset of MAbs specific for both Cb and Sa antigenic sites require HA trimerization for binding and fail to bind HA in the ER. This provides further evidence that PR8 HA trimerization occurs only after export from the ER. While this was highly controversial when first proposed, since the ER was thought to be a general site of oligomer assembly (40, 41), there are now examples of other proteins believed to oligomerize in the GC (42, 43).
Despite the fact that the H17-L10 (Ca) epitope spans the HA trimer interface by escape mutant analysis, we found that this MAb binds to monomeric, trypsin-released HA tops, though with diminished affinity. Although it is formally possible that H17-L10 binding to HA tops is based on the recruitment of two HA monomers to recreate the epitope, this seems unlikely, given (i) the presence of only monomeric HA tops species in solution as assayed by zonal sedimentation analysis on sucrose gradients and (ii) the inability of H17-L10 to bind to HA monomers in pulse-chase experiments. Rather, it is likely that the H17-L10 epitope is created by conformational changes that accompany HA trimerization. Remarkably, and unlike those of the other MAbs tested, the kinetics of H17-L10 binding to acid-treated recHA3 and HA tops are highly similar in SPR, suggesting that while MAb binding may be influenced by mutations in adjacent HA monomers, simultaneous interactions among HA monomers are not necessary to achieve maximal binding energy.
Notably, we show that the HA Cb and Sa epitopes can also require HA trimerization. This might have been expected for the Sa antigenic site, since several critical residues contribute to the HA trimer interface. Nevertheless, as with H17-L10, the H9-A22 (Sa) koff value is not greatly affected by fragmentation, consistent with the achievement of maximal binding energy with monomeric HA derived from trimers. The HA trimerization-dependent binding of the Cb-specific H35-C10 MAb, on the other hand, is surprising, as its epitope is located nearly equidistant from the HA trimer interfaces. Indeed, H35-C10 is the only HA trimer-dependent MAb we studied that does not detectably bind to HA tops, suggesting that its epitope is dependent on subtle conformational elements that require maintained oligomerization. Intriguingly, DuBois and colleagues (44) demonstrated that the HA Cb antigenic site is subject to considerable conformational flexibility, consistent with the idea that H35-C10 detects a Cb conformation on the HA tops that is unfavorable to association with this MAb.
The capacity of HA trimerization-dependent MAbs to bind HA trimer-derived monomers explains our previous observation that these MAbs maintain binding to the HA of acid-triggered virions as measured by hemagglutination inhibition (30). It is important to note that a subset of HA trimerization-dependent MAbs will actually bridge adjacent HA monomers. Although we have yet to clearly identify such MAbs among our panel of anti-IAV PR8 Abs, X-ray crystallography studies have already revealed an example among known MAbs to IAV H3N2 (35).
It will be of great interest to extend these findings to MAbs specific for the stem region of HA that exhibit virus-neutralizing activity. Such Abs offer great promise for thwarting HA antigenic drift. Krammer and colleagues (45) recently reported that the inclusion of a trimerization domain in recombinant soluble HAs is necessary to create the epitopes for several representative HA stem-reactive MAbs. The capacity of such MAbs to distinguish HA biosynthetic intermediates has not, however, been reported to date.
The most important conclusion stemming from these findings is that complete HA antigenicity (and presumably immunogenicity) requires subtle conformational effects that accompany HA trimerization and can largely persist after HA trimers partially (acid treatment) or even completely (proteolytic degradation) dissociate. Thus, although it is clearly established that folding of HA monomers or even isolated globular domains is sufficient to generate a significant fraction of epitopes, we have identified a number of MAbs specific for the HA Ca, Cb, and Sa antigenic sites whose epitopes are created (or completed) only upon HA trimerization. Consequently, judging the degree of folding of HA in vaccines produced from virus or by recombinant means requires a diverse panel comprising multiple MAbs specific for each HA antigenic site. The good news is that if these findings extend to contemporary HA strains, dissociation of trimerized HA in vaccine preparations does not necessarily lead to major losses in the immunogenicity of the globular head domains.
ACKNOWLEDGMENTS
We thank Glennys Reynoso for providing outstanding technical assistance and Joseph Newland and Thomas M. Kristie for expert biochemical advice and insight.
This work was supported by the Division of Intramural Research of the National Institute of Allergy and Infectious Diseases.
Footnotes
Published ahead of print 3 July 2013
REFERENCES
- 1.Hers JF. 1966. Disturbances of the ciliated epithelium due to influenza virus. Am. Rev. Respir. Dis. 93(Suppl):162–177 [DOI] [PubMed] [Google Scholar]
- 2.Skehel JJ, Wiley DC. 2000. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 69:531–569 [DOI] [PubMed] [Google Scholar]
- 3.Russ G, Bennink JR, Bachi T, Yewdell JW. 1991. Influenza virus hemagglutinin trimers and monomers maintain distinct biochemical modifications and intracellular distribution in brefeldin A-treated cells. Cell Regul. 2:549–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Han Y, David A, Liu B, Magadan JG, Bennink JR, Yewdell JW, Qian SB. 2012. Monitoring cotranslational protein folding in mammalian cells at codon resolution. Proc. Natl. Acad. Sci. U. S. A. 109:12467–12472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Copeland CS, Zimmer KP, Wagner KR, Healey GA, Mellman I, Helenius A. 1988. Folding, trimerization, and transport are sequential events in the biogenesis of influenza virus hemagglutinin. Cell 53:197–209 [DOI] [PubMed] [Google Scholar]
- 6.Yewdell JW, Yellen A, Bachi T. 1988. Monoclonal antibodies localize events in the folding, assembly, and intracellular transport of the influenza virus hemagglutinin glycoprotein. Cell 52:843–852 [DOI] [PubMed] [Google Scholar]
- 7.Gething MJ, McCammon K, Sambrook J. 1986. Expression of wild-type and mutant forms of influenza hemagglutinin: the role of folding in intracellular transport. Cell 46:939–950 [DOI] [PubMed] [Google Scholar]
- 8.Copeland CS, Doms RW, Bolzau EM, Webster RG, Helenius A. 1986. Assembly of influenza hemagglutinin trimers and its role in intracellular transport. J. Cell Biol. 103:1179–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Couch RB, Kasel JA. 1983. Immunity to influenza in man. Annu. Rev. Microbiol. 37:529–549 [DOI] [PubMed] [Google Scholar]
- 10.Webster RG, Laver WG, Kilbourne ED. 1975. Antigenic variation of influenza viruses, p 269–314 In Kilbourne ED. (ed), The influenza viruses and influenza. Academic Press, New York, NY [Google Scholar]
- 11.Lambkin R, Dimmock NJ. 1995. All rabbits immunized with type A influenza virions have a serum haemagglutination-inhibition antibody response biased to a single epitope in antigenic site B. J. Gen. Virol. 76(Pt 4):889–897 [DOI] [PubMed] [Google Scholar]
- 12.Caton AJ, Brownlee GG, Yewdell JW, Gerhard W. 1982. The antigenic structure of the influenza virus A/PR/8/34 hemagglutinin (H1 subtype). Cell 31:417–427 [DOI] [PubMed] [Google Scholar]
- 13.Gerhard W, Yewdell J, Frankel ME, Webster R. 1981. Antigenic structure of influenza virus haemagglutinin defined by hybridoma antibodies. Nature 290:713–717 [DOI] [PubMed] [Google Scholar]
- 14.Yewdell JW. 2011. Viva la revolucion: rethinking influenza a virus antigenic drift. Curr. Opin. Virol. 1:177–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yewdell JW, Taylor A, Yellen A, Caton A, Gerhard W, Bachi T. 1993. Mutations in or near the fusion peptide of the influenza virus hemagglutinin affect an antigenic site in the globular region. J. Virol. 67:933–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yewdell JW, Caton AJ, Gerhard W. 1986. Selection of influenza A virus adsorptive mutants by growth in the presence of a mixture of monoclonal antihemagglutinin antibodies. J. Virol. 57:623–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yewdell JW, Frank E, Gerhard W. 1981. Expression of influenza A virus internal antigens on the surface of infected P815 cells. J. Immunol. 126:1814–1819 [PubMed] [Google Scholar]
- 18.Yewdell JW, Gerhard W. 1981. Antigenic characterization of viruses by monoclonal antibodies. Annu. Rev. Microbiol. 35:185–206 [DOI] [PubMed] [Google Scholar]
- 19.Yewdell JW, Webster RG, Gerhard WU. 1979. Antigenic variation in three distinct determinants of an influenza type A haemagglutinin molecule. Nature 279:246–248 [DOI] [PubMed] [Google Scholar]
- 20.Dolan BP, Li L, Takeda K, Bennink JR, Yewdell JW. 2010. Defective ribosomal products are the major source of antigenic peptides endogenously generated from influenza A virus neuraminidase. J. Immunol. 184:1419–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bonifacino JS, Dell'Angelica EC. 2001. Immunoprecipitation. Curr. Protoc. Cell Biol. Chapter 7:Unit 7.2. [DOI] [PubMed] [Google Scholar]
- 22.Magadan JG, Perez-Victoria FJ, Sougrat R, Ye Y, Strebel K, Bonifacino JS. 2010. Multilayered mechanism of CD4 downregulation by HIV-1 Vpu involving distinct ER retention and ERAD targeting steps. PLoS Pathog. 6:e1000869. 10.1371/journal.ppat.1000869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stevens J, Corper AL, Basler CF, Taubenberger JK, Palese P, Wilson IA. 2004. Structure of the uncleaved human H1 hemagglutinin from the extinct 1918 influenza virus. Science 303:1866–1870 [DOI] [PubMed] [Google Scholar]
- 24.Khurana S, Suguitan AL, Jr, Rivera Y, Simmons CP, Lanzavecchia A, Sallusto F, Manischewitz J, King LR, Subbarao K, Golding H. 2009. Antigenic fingerprinting of H5N1 avian influenza using convalescent sera and monoclonal antibodies reveals potential vaccine and diagnostic targets. PLoS Med. 6:e1000049. 10.1371/journal.pmed.1000049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khurana S, Verma N, Yewdell JW, Hilbert AK, Castellino F, Lattanzi M, Del Giudice G, Rappuoli R, Golding H. 2011. MF59 adjuvant enhances diversity and affinity of antibody-mediated immune response to pandemic influenza vaccines. Sci. Transl. Med. 3:85ra48. 10.1126/scitranslmed.3002336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Braakman I, Hoover-Litty H, Wagner KR, Helenius A. 1991. Folding of influenza hemagglutinin in the endoplasmic reticulum. J. Cell Biol. 114:401–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Griffiths G, Quinn P, Warren G. 1983. Dissection of the Golgi complex. I Monensin inhibits the transport of viral membrane proteins from medial to trans Golgi cisternae in baby hamster kidney cells infected with Semliki Forest virus. J. Cell Biol. 96:835–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Strous GJ, Lodish HF. 1980. Intracellular transport of secretory and membrane proteins in hepatoma cells infected by vesicular stomatitis virus. Cell 22:709–717 [DOI] [PubMed] [Google Scholar]
- 29.Tartakoff AM. 1983. Perturbation of vesicular traffic with the carboxylic ionophore monensin. Cell 32:1026–1028 [DOI] [PubMed] [Google Scholar]
- 30.Yewdell JW, Gerhard W, Bachi T. 1983. Monoclonal anti-hemagglutinin antibodies detect irreversible antigenic alterations that coincide with the acid activation of influenza virus A/PR/834-mediated hemolysis. J. Virol. 48:239–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Skehel J, Bayley PM, Brown EB, Martin SR, Waterfield MD, White JM, Wilson IA, Wiley DC. 1982. Changes in the conformation of influenza virus hemagglutinin at the pH optimum of virus-mediated membrane fusion. Proc. Natl. Acad. Sci. U. S. A. 79:968–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ruigrok RW, Aitken A, Calder LJ, Martin SR, Skehel JJ, Wharton SA, Weis W, Wiley DC. 1988. Studies on the structure of the influenza virus haemagglutinin at the pH of membrane fusion. J. Gen. Virol. 69(Pt 11):2785–2795 [DOI] [PubMed] [Google Scholar]
- 33.Wharton SA, Ruigrok RW, Martin SR, Skehel JJ, Bayley PM, Weis W, Wiley DC. 1988. Conformational aspects of the acid-induced fusion mechanism of influenza virus hemagglutinin. Circular dichroism and fluorescence studies. J. Biol. Chem. 263:4474–4480 [PubMed] [Google Scholar]
- 34.Yewdell JW. 2010. Monoclonal antibodies specific for discontinuous epitopes direct refolding of influenza A virus hemagglutinin. Mol. Immunol. 47:1132–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barbey-Martin C, Gigant B, Bizebard T, Calder LJ, Wharton SA, Skehel JJ, Knossow M. 2002. An antibody that prevents the hemagglutinin low pH fusogenic transition. Virology 294:70–74 [DOI] [PubMed] [Google Scholar]
- 36.Bizebard T, Gigant B, Rigolet P, Rasmussen B, Diat O, Bosecke P, Wharton SA, Skehel JJ, Knossow M. 1995. Structure of influenza virus haemagglutinin complexed with a neutralizing antibody. Nature 376:92–94 [DOI] [PubMed] [Google Scholar]
- 37.Fleury D, Barrere B, Bizebard T, Daniels RS, Skehel JJ, Knossow M. 1999. A complex of influenza hemagglutinin with a neutralizing antibody that binds outside the virus receptor binding site. Nat. Struct. Biol. 6:530–534 [DOI] [PubMed] [Google Scholar]
- 38.Fleury D, Wharton SA, Skehel JJ, Knossow M, Bizebard T. 1998. Antigen distortion allows influenza virus to escape neutralization. Nat. Struct. Biol. 5:119–123 [DOI] [PubMed] [Google Scholar]
- 39.Knossow M, Daniels RS, Douglas AR, Skehel JJ, Wiley DC. 1984. Three-dimensional structure of an antigenic mutant of the influenza virus haemagglutinin. Nature 311:678–680 [DOI] [PubMed] [Google Scholar]
- 40.Doms RW, Lamb RA, Rose JK, Helenius A. 1993. Folding and assembly of viral membrane proteins. Virology 193:545–562 [DOI] [PubMed] [Google Scholar]
- 41.Hurtley SM, Helenius A. 1989. Protein oligomerization in the endoplasmic reticulum. Annu. Rev. Cell Biol. 5:277–307 [DOI] [PubMed] [Google Scholar]
- 42.Locker JK, Opstelten DJ, Ericsson M, Horzinek MC, Rottier PJ. 1995. Oligomerization of a trans-Golgi/trans-Golgi network retained protein occurs in the Golgi complex and may be part of its retention. J. Biol. Chem. 270:8815–8821 [DOI] [PubMed] [Google Scholar]
- 43.Musil LS, Goodenough DA. 1993. Multisubunit assembly of an integral plasma membrane channel protein, gap junction connexin43, occurs after exit from the ER. Cell 74:1065–1077 [DOI] [PubMed] [Google Scholar]
- 44.DuBois RM, Aguilar-Yanez JM, Mendoza-Ochoa GI, Oropeza-Almazan Y, Schultz-Cherry S, Alvarez MM, White SW, Russell CJ. 2011. The receptor-binding domain of influenza virus hemagglutinin produced in Escherichia coli folds into its native, immunogenic structure. J. Virol. 85:865–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krammer F, Margine I, Tan GS, Pica N, Krause JC, Palese P. 2012. A carboxy-terminal trimerization domain stabilizes conformational epitopes on the stalk domain of soluble recombinant hemagglutinin substrates. PLoS One 7:e43603. 10.1371/journal.pone.0043603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gamblin SJ, Haire LF, Russell RJ, Stevens DJ, Xiao B, Ha Y, Vasisht N, Steinhauer DA, Daniels RS, Elliot A, Wiley DC, Skehel JJ. 2004. The structure and receptor binding properties of the 1918 influenza hemagglutinin. Science 303:1838–1842 [DOI] [PubMed] [Google Scholar]