Abstract
CD4+ T cells rather than macrophages are the principal cells infected by human immunodeficiency virus type 1 (HIV-1) and simian immunodeficiency virus (SIV) in vivo. Macrophage tropism has been linked to the ability to enter cells through CCR5 in conjunction with limiting CD4 levels, which are much lower on macrophages than on T cells. We recently reported that rhesus macaques (RM) experimentally depleted of CD4+ T cells before SIV infection exhibit extensive macrophage infection as well as high chronic viral loads and rapid progression to AIDS. Here we show that early-time-point and control Envs were strictly CD4 dependent but that, by day 42 postinfection, plasma virus of CD4+ T cell-depleted RM was dominated by Envs that mediate efficient infection using RM CCR5 independently of CD4. Early-time-point and control RM Envs were resistant to neutralization by SIV-positive (SIV+) plasma but became sensitive if preincubated with sCD4. In contrast, CD4-independent Envs were highly sensitive to SIV+ plasma neutralization. However, plasma from SIV-infected CD4+ T cell-depleted animals lacked this CD4-inducible neutralizing activity and failed to neutralize any Envs regardless of sCD4 pre-exposure status. Enhanced sensitivity of CD4-independent Envs from day 42 CD4+ T cell-depleted RM was also seen with monoclonal antibodies that target both known CD4-inducible and other Env epitopes. CD4 independence and neutralization sensitivity were both conferred by Env amino acid changes E84K and D470N that arose independently in multiple animals, with the latter introducing a potential N-linked glycosylation site within a predicted CD4-binding pocket of gp120. Thus, the absence of CD4 T cells results in failure to produce antibodies that neutralize CD4-independent Envs and CD4-pretriggered control Envs. In the absence of this constraint and with a relative paucity of CD4+ target cells, widespread macrophage infection occurs in vivo accompanied by emergence of variants carrying structural changes that enable entry independently of CD4.
INTRODUCTION
CD4+ T cells rather than macrophages are the primary target cells infected by human immunodeficiency virus type 1 (HIV-1) and simian immunodeficiency virus (SIV) in vivo (1). CD4+ T cells and macrophages both express the principal entry coreceptor CCR5, but macrophages express much lower levels of CD4, and macrophage tropism is associated with the ability of Env to enter using CCR5 in conjunction with very-low-to-absent CD4 (2–6). The two-step entry process that requires CD4 binding prior to coreceptor engagement is known to render virus more resistant to antibody neutralization (7–10), suggesting that immune forces may limit emergence of macrophage-tropic variants in vivo. Indeed, infection of macaques using cloned CD4-independent or macrophage-tropic SIV results in either transient, nonprogressive infection or dissemination to the central nervous system (CNS), without immunodeficiency (11, 12). In the CNS, unlike in the systemic compartment, myeloid lineage cells are the dominant targets of infection and macrophage-tropic viruses are common, where a paucity of CD4+ T cell targets combined with the immune-privileged nature of the CNS is thought to contribute to the unique tropism pattern in the brain (1, 13–17). Macrophage-tropic variants have also been reported in some studies to arise during very late-stage disease (18) or following rapid profound virus-induced CD4+ T cell depletion (19, 20). However, the forces that maintain strict CD4 dependence and regulate tropism in the systemic compartment have not been explicitly demonstrated in vivo and neither have mechanisms that might enable widespread systemic macrophage infection.
Recently we showed that extensive macrophage infection developed in rhesus macaques (RM) experimentally depleted of CD4+ T cells from the blood and lymph nodes prior to infection with SIVmac251, and variants capable of using human CCR5 in the absence of CD4 emerged in plasma during chronic infection (21). Depleted animals also experienced high chronic viral load and progressed rapidly to AIDS. Thus, CD4+ T cell depletion prior to infection resulted in adaptation of SIV to decreased CD4 dependence and macrophage tropism in vivo and provides a model in which the forces that regulate tropism during infection can be elucidated.
In this study, we showed that very efficient CD4-independent use of rhesus macaque CCR5 arose in CD4+ T cell-depleted macaques during the postpeak phase of infection and was associated with sensitivity to neutralization by control SIV+ plasma but not by autologous plasma. A key distinguishing feature was the presence of antibody activity in control RM plasma, but not CD4+ T cell-depleted RM plasma, that neutralized control Envs if preincubated with sCD4 but not in the absence of sCD4 exposure. In the absence of this CD4-inducible neutralization activity, and with a paucity of CD4+ T cell targets in CD4+ T cell-depleted animals, circulating SIV Envs acquired 2 amino acid changes in gp120 that impart CD4-independent entry through CCR5. Thus, CD4+ T cells contribute to the production of antibodies targeted to conserved Env conformations that normally are induced only by CD4 engagement. These antibodies were associated with strict CD4 dependence of Env, maintenance of CD4+ T cell targeting, and restrained tropism for CD4-low macrophages in vivo.
MATERIALS AND METHODS
Ethics statement.
All animal experimentation was conducted following guidelines established by the Animal Welfare Act and the NIH for housing and care of laboratory animals and was performed in accordance with institutional regulations after review and approval by the Institutional Animal Care and Use Committees (IACUC) at the Yerkes National Primate Research Center (YNPRC) or the Tulane National Primate Research Center (TNPRC). Studies were also reviewed and approved by the University of Pennsylvania IACUC.
SIVMac envelope clones, mutagenesis, and pseudotyped virus.
The env genes from day 11 and day 42 SIV-infected rhesus macaque plasma were PCR amplified using a procedure for endpoint diluted single genomes as previously described (21). Mutations were introduced into SIV envelopes using a QuikChange II XL site-directed mutagenesis kit (Agilent Inc., Santa Clara, CA) and verified by sequencing. SIVmac239 and SIVmac251.6 Env clones were used as reference controls. Luciferase-expressing pseudotyped viruses carrying SIV Envs on an HIV-1 backbone were generated as previously described (22) and were treated with DNase prior to use in infection.
Virus infection and receptor function assays.
Human 293T cells were maintained in Dulbecco's modified Eagle medium (DMEM) containing 10% fetal bovine serum (D10 media). Entry of pseudotyped viruses was assayed in 293T target cells expressing CD4 and CCR5 or CCR5 alone. Target cells were transfected with plasmids encoding rhesus macaque CCR5 with or without rhesus macaque CD4, using pcDNA 3.1 as a “filler” plasmid (23). Cells transfected with pcDNA 3.1 only were used as a negative control. Target cells (2 × 104 per well in 96-well plates) were infected with pseudotyped viruses (20 ng p24 antigen) by spinoculation at 1,200 × g for 2 h. Cells were then incubated for 72 h at 37°C, and infection was quantified by measuring luciferase content in cell lysates as previously described (23). All data represent a minimum of 3 independent replicate experiments.
Env structural mapping.
The SIV Env core structure (24) was visualized with Jmol (25), and predicted CD4-binding residues were highlighted based on homology with HIV as previously described (24, 26). Residues highlighted were as follows: the HIV-CD4 direct-contact model (more stringent) included residues 107, 293 to 295, 297, 381, 384, 386, 387, 438 to 443, 468 to 472, 479, and 482 to 484; and the HIV-CD4 loss-of-solvent-accessibility model (less stringent) in addition to the above included residues 105, 106, 108, 272, 292, 296, 380, 481, and 485 to 487. Residue 84 was additionally highlighted.
Neutralization assays.
Monoclonal antibodies (MAb) 7D3, 36D5, 17A11, 171C2, and 35C11 have been previously described (27). Plasma from day 11 and day 56 animals in this study or pooled plasma from two chronically SIVmac251-infected macaques (kindly provided by P. Marx) was heat inactivated at 56°C for 1 h. Soluble CD4-183 (sCD4; Pharmacia, Inc.) was obtained from the NIH AIDS Reference Reagents Program. Neutralization and sCD4 exposure assays were performed as previously described (9, 28) with modifications. Pseudotyped virus was mixed with sCD4 in D10 medium to achieve concentrations of virus of 0.8 ng/μl of viral p24 antigen and 50 ng/μl sCD4 and incubated at 37°C for 1 h. Aliquots (25 μl) of the virus-sCD4 mixture were then transferred to wells of a 96-well V-bottom plate, and MAb or plasma was added to achieve final concentrations as noted. The mixture was incubated at 37°C for an additional 1 h, after which 25 μl was added to target cells (293T cells expressing rmCD4 and rmCCR5) plated in 96-well flat-bottom plates (2 × 104 cells in 100 μl medium). Cells were subject to spinoculation as described above, incubated for 72 h, and lysed for measurement of luciferase activity as an indication of infection. Neutralization was determined based on the percentage of infection in the absence of antibody (using virus incubated with or without sCD4, as appropriate, for the no-antibody treatment). Neutralization data represent a minimum of at least three independent replicate experiments.
Plasma binding antibody.
Binding antibodies to SIV Env protein were assessed by an enzyme-linked immunosorbent assay (ELISA) as described previously (29). Briefly, 96-well plates were coated with 1 μg/ml SIVmac239 gp140 protein (Immune Tech, New York, NY) by overnight incubation at 4°C and then incubated with serial dilutions of plasma for 2 h followed by detection using horseradish peroxidase (HRP)-conjugated goat anti-monkey IgG (AlphaDiagnostic, Owings Mills, MD) (1:10,000 dilution) and 3,3′,5,5′-tetramethylbenzidine (TMB) substrate. A dilution series of a SIVmac239 gp140-specific IgG standard with a known concentration (generously provided by P. Kozlowski, Louisiana State University) was included in parallel.
Nucleotide sequence accession numbers.
The sequences of the SIV env clones described in this study have been submitted to GenBank under the following accession numbers: KF276976 to KF277047.
RESULTS
CD4-independent SIV Env emerges in CD4+ T cell-depleted rhesus macaques.
SIVmac251 infection of animals following antibody-mediated depletion of CD4+ T cells from peripheral blood, lymph node, and bone marrow but not mucosal sites resulted in extensive in vivo macrophage infection, high postpeak viremia, and rapid disease progression (21). We previously showed that Env clones derived from postpeak (day 42) plasma of these CD4-depleted SIV-infected rhesus macaques were able to enter target cells expressing human CCR5 in the absence of CD4, reaching levels approximately 50% of that seen in cells expressing CCR5 together with CD4 (21). Here we asked whether CD4-independent variants were present in plasma at an earlier time point, during peak viremia (day 11), and if CD4 independence was maintained using species-matched rhesus macaque CCR5 and CD4 (6). Single-genome amplification (SGA)-derived Env clones were generated from 4 depleted and 2 control animals and 6 Envs from each animal at each time point (72 total Envs) and were used to generate a panel of Env-pseudotyped reporter viruses. Pseudotyped viruses were tested for their ability to enter 293T cells transfected with CCR5 in the presence or absence of CD4 (Fig. 1).
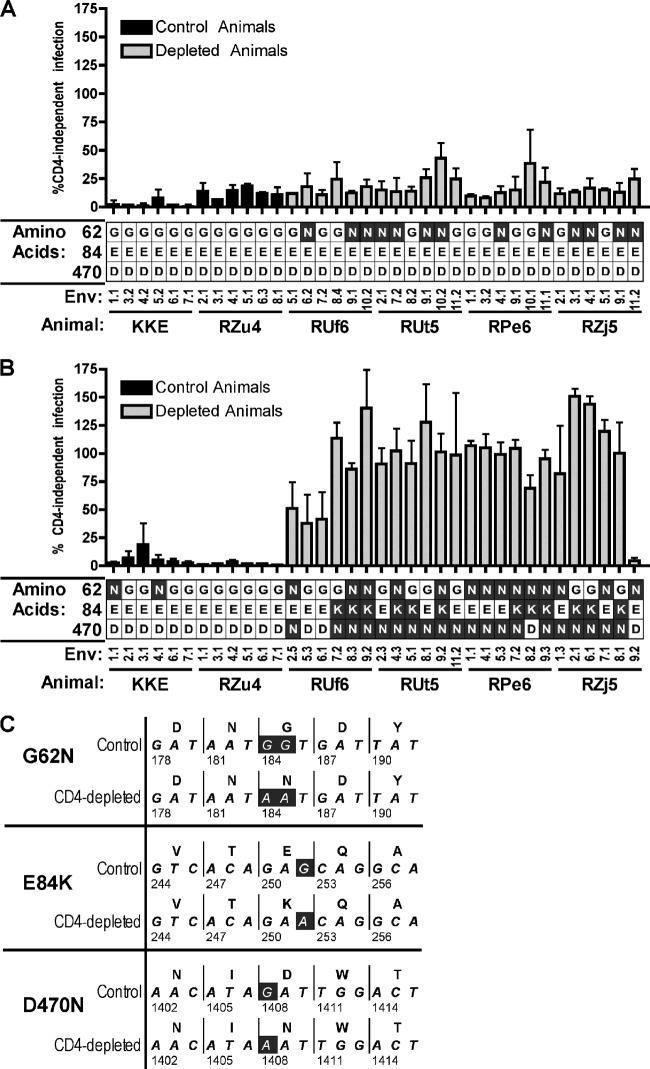
Fig 1.
Envs from CD4+ T cell-depleted macaques mediate CD4-independent entry. (A and B) Env clones obtained by single-genome amplification (SGA) from plasma at day 11 (A) and day 42 (B) after infection from 2 control animals (black bars) and 4 CD4-depleted animals (grey bars) were used to generate HIV virions pseudotyped with SIV Env and containing a luciferase reporter. These virions were used to infect 293T cells transfected with rmCCR5 and with and without rmCD4. Values indicated are expressed as percent entry in the absence versus presence of CD4 on target cells (means ± standard errors of the means [SEM] of the results of triplicate experiments). Predicted amino acids at sites 62, 84, and 470 (based on SIVmac239 numbering) are indicated below the clone number and are also shown in the context of amino acid and nucleotide sequences (C) for a representative Env from a control animal (RZu4.d42.7.1) and from a CD4-depleted animal (RPe6.d42.7.2).
Envs from both the CD4-depleted and control animals required CD4 at day 11 for efficient entry, both with rhesus CCR5 (Fig. 1A) and with human CCR5 (data not shown). A few Envs mediated low-level entry in the absence of CD4, and while this appeared to be slightly greater among CD4-depleted than control day 11 Envs, the level was variable and did not reach statistical significance. In contrast, nearly all day 42 Envs from the CD4-depleted group mediated robust CD4-independent entry (Fig. 1B). In fact, using these rhesus-derived receptors, most clones were essentially indifferent to the presence of CD4 (mean CD4-independent entry was 94% ± 7% compared with CD4-dependent entry). In contrast, day 42 Envs from the control animals all required CD4 for entry through rhesus CCR5. These data indicate that in animals depleted of CD4+ T cells, CD4-independent variants emerged in vivo during the postpeak phase of SIV infection and dominated in plasma during chronic infection.
Emergence of SIV Env D470N and E84K confers CD4 independence.
We next asked what molecular determinants conferred the CD4-independent phenotype that arose in vivo in multiple CD4-depleted animals. We aligned the amino acid sequences of the 72 day 11 and day 42 SGA Envs and found that 3 amino acid changes in gp120, G62N, E84K, and D470N (based on SIVmac239 numbering [24]) were enriched in CD4-depleted RM Envs compared to controls (Fig. 1). E84K and D470N were found exclusively in day 42 CD4-depleted RM Envs, and both were significantly associated with CD4-independent entry (P < 0.001; McNemar test for associations). In contrast, G62N was found in CD4-depleted RM Envs at both the day 11 and day 42 time points, as well as in a few control Envs, and was not significantly associated with CD4-independent entry (P = 0.14). One partially CD4-independent Env clone, RUf6.d42.6.1, carried neither E84K nor D470N but did contain D385N, G383R, and D180N, which had each been previously described in CD4-independent SIV Envs (30). Only one day 42 Env from a CD4-depleted animal did not mediate appreciable CD4-independent entry (RZj5.D42.9.2) and did not carry D470N, E84K, or any previously described polymorphism(s) associated with CD4 independence. G62N, E84K, and D470N have not been explicitly characterized in previous studies, but an analysis of SIVmac sequences revealed that G62N has ∼5% frequency within SIVmac251 viral stocks (31–34), while D470N and E84K are extremely rare and present mainly in in vivo-passaged SIVmac251-related isolates (34–38). Of note, all instances of G62N, E84K, and D470N that arose in these animals were the result of G-to-A nucleotide changes (Fig. 1C).
To determine whether G62N, E84K, and D470N regulate CD4-independent entry, we introduced each of these mutations, as well as E84K plus D470N, into a representative control Env, RZu4.d42.7.1 (Fig. 2A). G62N had negligible effects on RZu4.d42.7.1 entry and did not enable CD4 independence. E84K and D470N each severely attenuated total entry, although the signal remained significantly above the assay background and represented bona fide Env-mediated entry. Despite the overall low entry levels, however, levels of D470N entry were similar whether or not CD4 was present, while E84K remained CD4 dependent. When E84K and D470N were simultaneously introduced into RZu4.d42.7.1, robust entry was restored and the levels of entry were equivalent in the presence and absence of CD4. Thus, D470N appears to play a central role in CD4 independence but results in attenuation, while E84K restores overall efficiency of entry in this Env background.
Fig 2.
D470N and E84K regulate CD4-independent entry. (A and B) Mutations G62N, E84K, D470N, and D470Q were introduced into a representative CD4-dependent Env (RZu4.d42.7.1) (A), while N62G, K84E, N470D, and N470Q were introduced into a representative CD4-independent Env (RUt5.d42.4.3) (B). Columns and error bars indicate the means and standard errors, respectively, of the results from 3 independent experiments. The mutant Envs were tested for their ability to mediate infection of 293T cells transfected with rmCCR5, with and without rmCD4, indicated by relative light units (RLU). (C and D) The same mutations were introduced into an extended panel of CD4-dependent (C) and CD4-independent (D) Envs, and CD4-independent entry is shown as a percentage of entry in the presence of CD4, with each individual Env clone indicated as a point and each horizontal line indicating mean percent entry among the various clones. Mean percent CD4-independent entry values were compared using a pairwise t test. Note that in one CD4-independent Env (RUf6.d42.2.5), E84 was present in the parental form, so only N470D was introduced. (E) The locations of E84 (green) and D470 (blue) are shown based on a core structure of SIVmac32H (24), with the predicted Env-CD4 binding site indicated based on homology with HIV-HxB Env-CD4 direct contact (red) and loss of solvent accessibility (magenta) residues. Peptide residues are colored white, and carbohydrates are colored gray. (F) The potential N-linked glycosylation site introduced by D470N and nonglycosylated mutant D470Q are shown.
We also introduced the reciprocal mutations, including N62G, K84E, N470D, and K84E plus N470D, into a representative CD4-independent Env, RPe6.d42.7.2 (Fig. 2B). Similarly, N62G had little effect. K84E severely attenuated overall entry and also modestly reduced relative CD4-independent entry. N470D also reduced CD4-independent entry, although again only modestly (76% reduction). However, the combination of mutations in K84E plus N470D restored the level of overall entry to that seen with the parental Env and dramatically reduced CD4-independent entry (95% reduction). Thus, in the CD4-independent background, both N470D and E84K contribute to CD4 independence and interact to maintain overall efficiency of entry.
To see if the effects of G62N, E84K, and D470N in these 2 representative Envs were generalizable to other viruses from additional animals, we introduced D470N and E84K plus D470N into a larger panel of CD4-dependent Envs from the animals in this study, including Envs from day 11 control, day 11 CD4-depleted, and day 42 control animals (n = 7). We also tested the reciprocal mutations in an expanded panel of CD4-independent Envs from day 42 CD4+ T cell-depleted animals (n = 4). Similar to the representative Env results, D470N and, more potently, D470N plus E84K regulated CD4-independent entry of multiple Envs, including CD4-independent variants that arose independently in different animals (Fig. 2C and D).
Structural significance of CD4-independent mutations that arise in vivo.
Using Jmol (25), we mapped predicted CD4-binding residues onto the previously described core structure of SIVmac32H gp120 (24) based on homology with known HIV gp120-binding residues (24, 26) and highlighted residues E84 and D470 (Fig. 2E) (the published core structure of SIV gp120 is N-terminally truncated and excludes G62). This analysis indicates that D470 is a predicted CD4 contact residue residing in a cavity flanked by other CD4 contact residues, while E84 is proximal to the predicted gp120 CD4-binding surface, situated on the first alpha helix, and at a distance of 13.3Å to 19.2Å from D470. In addition, owing to its linear position upstream from a threonine residue in SIVmac251 gp120, D470N introduces a potential N-linked glycosylation site (PNLG) within a predicted CD4-binding pocket (Fig. 2F).
To address whether the PNLG introduced into SIV Env by D470N was important in CD4-independent entry (Fig. 2F), we introduced D470Q and N470Q changes into our representative CD4-dependent and CD4-independent Envs (Fig. 2A and B) as well as the extended panel of Envs (Fig. 2C and D). Q was chosen as a conservative change compared to N that would maintain the charge but abrogate glycosylation, as described previously (39). D470Q in control Envs did not substantially increase CD4-independent entry (Fig. 2A and C), and N470Q in CD4-independent Envs reduced CD4 independence similarly to N470D (Fig. 2B and D). These findings suggest that the PNLG in this deep pocket on the CD4-binding surface is an important component of CD4-independent entry.
Constitutive sensitivity of CD4-independent Envs to MAb neutralization.
Since Envs from CD4+ T cell-depleted animals were capable of utilizing CCR5 for entry in the absence of CD4, we performed a neutralization assay using the Env monoclonal antibody (MAb) 7D3 (Fig. 3), which targets the predicted CD4-induced (CD4-i) binding-binding site of SIV Env (27, 40, 41). As shown in Fig. 3A, control Env RZu4d11.2.1 was resistant to neutralization by 7D3 but became sensitive when preincubated with sCD4. This confirmed that 7D3 targets a CD4-i neutralizing epitope. In contrast to the control Env, the CD4-independent Env RUt5d42.4.3 was potently neutralized by 7D3 regardless of sCD4 preincubation, suggesting that this Env constitutively exposes the 7D3 CD4-i neutralization epitope. We then tested the extended panel of day 42 CD4-independent Envs that arose independently in multiple CD4-depleted animals and confirmed 7D3 neutralizing sensitivity for all of these variants. Furthermore, the other CD4-dependent Envs from day 11 or control animals were all resistant but became sensitive following sCD4 exposure (data not shown).
Fig 3.
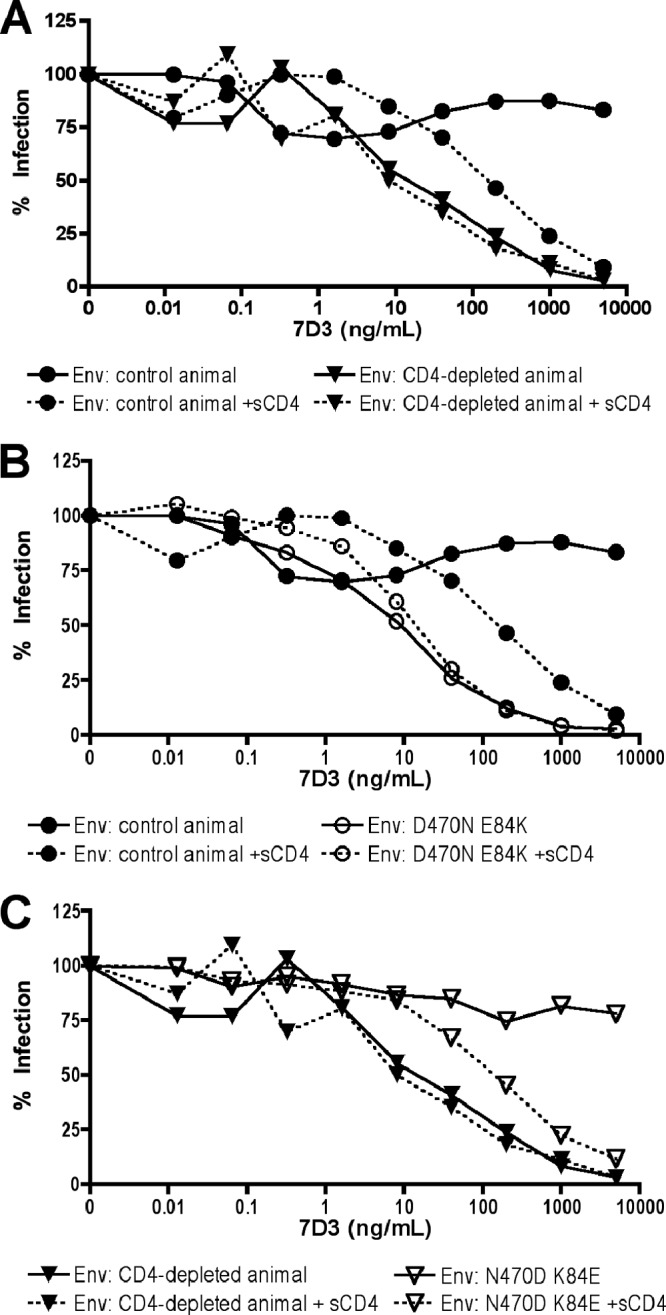
A CD4-inducible epitope monoclonal antibody neutralizes CD4-independent Envs. (A) Pseudotype viruses carrying a representative CD4-dependent Env from a control animal (RZu4.d11.2.1) and CD4-independent Env from a CD4-depleted animal (RPe6.42.7.2) were incubated with or without sCD4 followed by various concentrations of monoclonal antibody 7D3 prior to infection of 293T cells transfected with rmCD4 plus rmCCR5. (B) The parental and E84K-plus-D470N mutant forms of the CD4-dependent Env were similarly tested for neutralization by 7D3. (C) The parental and K84E-plus-N470D mutant forms of the CD4-independent Env were tested for 7D3 neutralization.
We then asked whether the same molecular determinants that regulate CD4-independent entry also determine sensitivity to neutralization by 7D3. As shown in Fig. 3B, introduction of D470N and E84K into control Env RZu4d11.2.1 rendered the virus sensitive to 7D3 neutralization in the absence of sCD4 triggering. When the reciprocal mutations, N470D and K84E, were introduced into the CD4-independent Env RUt5d42.4.3, it rendered this Env resistant to 7D3 neutralization unless first exposed to sCD4 (Fig. 3C). Thus, the combination of E84K plus D470N is principally responsible for sensitivity to neutralization by 7D3.
To determine whether these amino acids were determinants of CD4-inducible antibody neutralization among other Envs from this panel and the role of each residue specifically, we introduced single mutations at positions 84 and 470 into our extended panel of CD4-dependent and -independent Envs. As shown in Fig. 4, introduction of D470N into CD4-dependent Envs conferred sensitivity to 7D3 whereas introduction of E84K alone did not. Furthermore, introduction of D470Q into the CD4-dependent Envs did not confer 7D3 sensitivity, implicating glycosylation at the N470 PNLG site. In contrast, neither N470D nor K84E alone made CD4-independent Envs resistant to 7D3 (with the exception of Ruf6.d42.2.5, which already carries E84 and was rendered resistant to 7D3 by N470D alone; Fig. 1B and data not shown). Introduction of N470D and K84E together, however, led to 7D3 resistance for all Envs tested. Thus, both D470N and E84K contribute to CD4-i antibody neutralization sensitivity.
Fig 4.
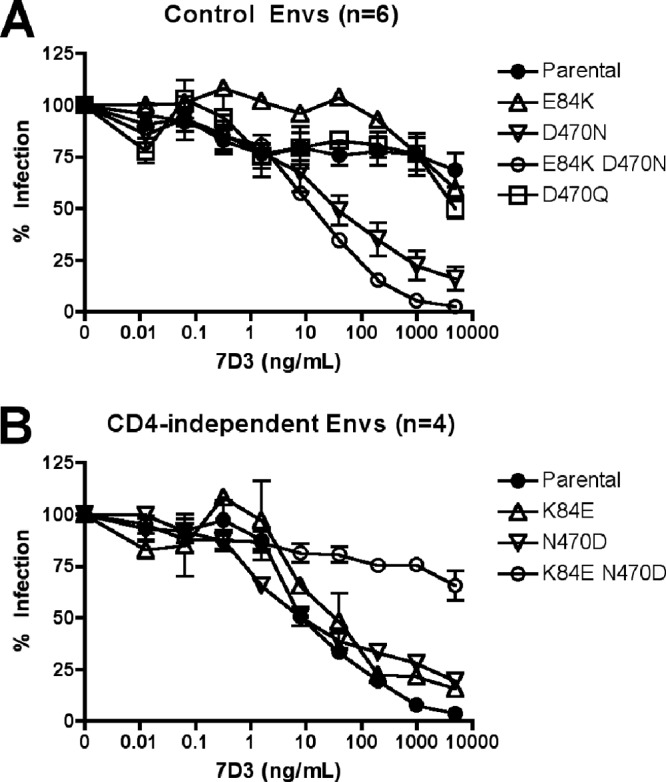
Molecular determinants of 7D3 neutralization are consistent across the extended panel of Envs. (A) CD4-dependent Envs from either control animals or day 11 CD4-depleted animals (n = 6) were mutagenized to introduce E84K or D470N or both or non-PNLG mutant D470Q and then tested for neutralization by monoclonal antibody 7D3. Data show the means ± SEM of the results from all Envs analyzed. (B) Means ± SEM for 7D3 neutralization of CD4-independent Envs from CD4-depleted day 42 animals and Env mutants are shown.
We then tested a panel of additional SIV Env monoclonal antibodies with differing target epitopes (27) for the ability to neutralize control macaque CD4-dependent Envs without and with sCD4 pre-exposure and neutralization of these Envs carrying E84K, D470N, and D470Q mutations (Fig. 5). Both a V3-specific MAb (Fig. 5A and B) and an anti-gp120 conformational epitope MAb (Fig. 5C and D) showed patterns similar to those seen with 7D3, in which Env was neutralized only if pre-exposed to sCD4 or if mutated to carry D470N (with or without E84K) but not D470Q. In contrast, a V1/V2-specific MAb was poorly neutralizing regardless of sCD4 pretreatment but did inefficiently neutralize the mutant Envs (Fig. 5E and F), whereas a gp41 MAb failed to neutralize either the sCD4-treated or mutant Envs (Fig. 5G and H). Thus, both sCD4 pretriggering and the D470N mutation that emerged in CD4-depleted RM render SIV Envs sensitive to antibodies with several distinct, but not all, epitope targets.
Fig 5.
CD4-inducible epitope-targeted monoclonal antibodies neutralize CD4-independent Envs. (A, C, E, and G) Representative CD4-dependent (RZu4.d11.2.1) Env-pseudotyped virus was incubated with or without sCD4 followed by various concentrations of monoclonal antibodies with indicated specificities prior to infection of 293T cells transfected with rmCD4 plus rmCCR5. (B, D, F, and H) The parental and E84K-plus-D470N mutant forms of multiple CD4-dependent Envs were similarly tested for neutralization, and the results are expressed as the mean values for the various clones tested ± SEM (n = 4). 17A11 is believed to target a conformational epitope that overlaps both the CD4- and CCR5-binding sites.
CD4-independent Envs are sensitive to neutralization by CD4-i activity in SIV+ plasma.
Previous studies have shown that commonly used SIV strains, such as mac239, mac251, and SM-E660, are highly resistant to neutralization by most SIV+ plasma (21, 42–44). However, it has also been shown that HIV+ plasma contains antibodies that target broadly conserved CD4-i binding-binding structures and can neutralize viruses if pretriggered with sCD4 (9). Therefore, we asked if SIV+ plasma contains antibodies with similar activities and whether these SIV Envs could be neutralized by pooled plasma from chronically SIVmac251-infected macaques (Fig. 6).
Fig 6.
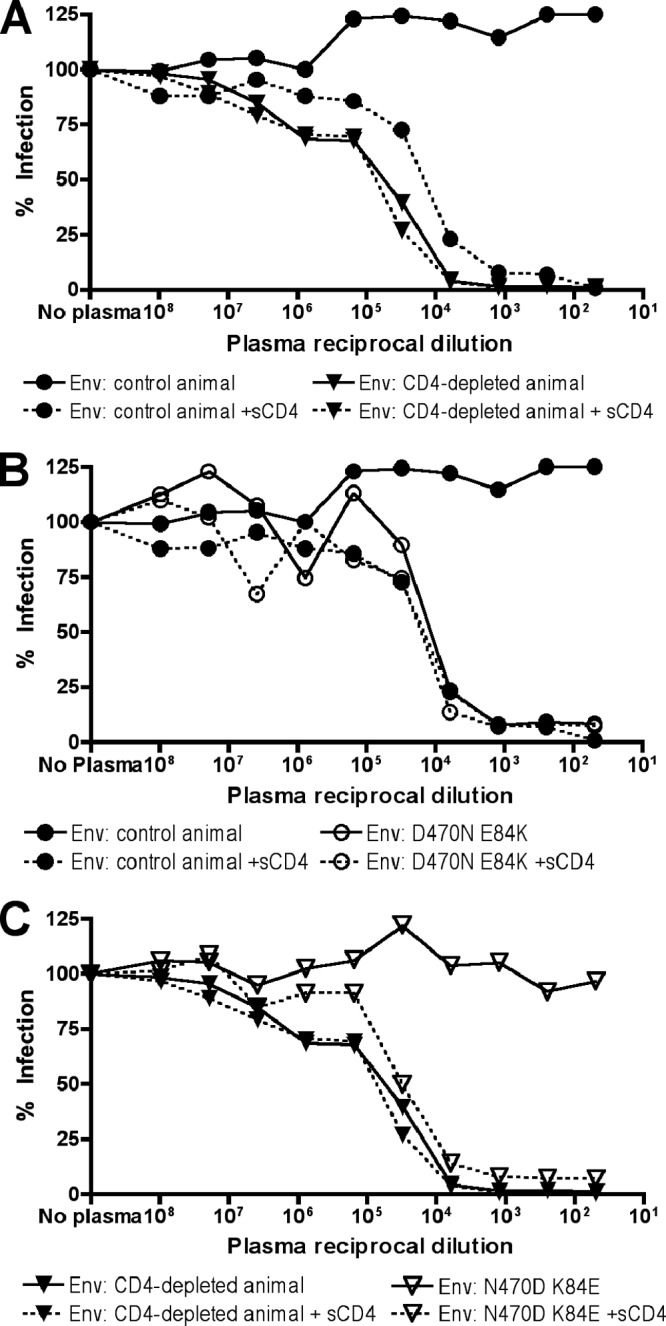
Pooled SIV+ plasma neutralizes CD4-independent Envs and CD4-triggered CD4-dependent Env. (A) Representative CD4-dependent (Rzu4.d11.2.1) and CD4-independent (Rpe6.42.7.2) Env-pseudotyped viruses were incubated with or without 50 μg/ml sCD4 and then incubated with serial dilutions of SIV+ plasma pooled from two chronically SIVmac251-infected macaques (not from this study). Viruses were then used to infect 293T cells transfected with rmCD4 plus rmCCR5. (B) The parental and E84K-plus-D470N mutant forms of a CD4-dependent Env were tested for plasma neutralization. (C) The parental and K84E-plus-N470D mutant forms of a CD4-independent Env were tested for plasma neutralization.
As expected, CD4-dependent control Env RZu4d11.2.1 was resistant to plasma neutralization (Fig. 6A). However, preincubation with sCD4 rendered this Env sensitive to neutralization, confirming that SIV+ plasma contains CD4-i neutralizing activity. In contrast, CD4-independent Env RUt5d42.4.3 from a day 42 CD4-depleted animal was sensitive to neutralization regardless of CD4 pretriggering. The D470N mutation, either alone or in combination with E84K, rendered the control CD4-dependent Env sensitive to neutralization by SIV+ plasma without sCD4 pre-exposure (Fig. 6B and data not shown). Similarly, K84E/N470D rendered the CD4-independent Env resistant to plasma neutralization unless pre-exposed to sCD4 (Fig. 6C), while, like MAb 7D3, neither N470D nor K84E alone rendered CD4-independent Envs plasma neutralization resistant (data not shown).
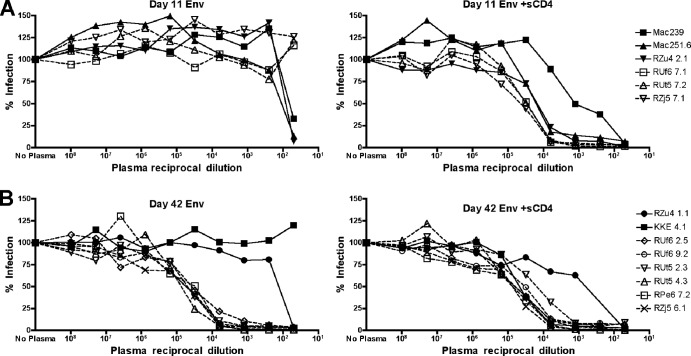
To determine whether this sensitivity to SIV+ plasma neutralization was common among the viruses that arose in CD4-depleted animals, we tested a larger panel of Envs from our study along with SIVmac239 and SIVmac251.6 reference Envs for comparison. As shown in Fig. 7A, day 11 Envs from both control RM (solid lines) and CD4+ T cell-depleted RM (dotted lines) were resistant to pooled plasma neutralization but became sensitive if first exposed to sCD4. SIVmac239 and SIVmac251.6 showed similar patterns of CD4-induced neutralization sensitivity (Fig. 7A). At day 42, all Envs from CD4+ T cell-depleted animals were sensitive to plasma neutralization regardless of sCD4 pretreatment (Fig. 7B). Control animal day 42 Envs were resistant to plasma neutralization but became more sensitive with sCD4 pretreatment (Fig. 7B). Some Envs exhibited a drop in infectivity at the highest concentrations of plasma, but this was also observed with uninfected control plasma (data not shown) and likely due to nonspecific effects of high concentrations of plasma as has been previously described (28).
Fig 7.
SIV+ plasma neutralization of the extended panel of CD4-independent and control Envs and prototype SIV Envs. Pseudotype viruses carrying Envs from day 11 animals (A) and day 42 animals (B) were preincubated without (left) or with (right) sCD4, exposed to various dilutions of pooled SIV+ plasma, and used to infect 293T cells expressing rmCD4 and rmCCR5. Envs from CD4-depleted animals are represented by dotted lines, and Envs from control animals are represented by solid lines. Reference Envs from SIVmac239 and SIVmac251.6 were tested in parallel and are included in panel A.
Thus, the CD4-independent Envs that arose in CD4-depleted macaques are constitutively sensitive to CD4-inducible neutralization by SIV+ plasma, which targets Env conformations regulated by D470N and E84K. These findings also suggested that this antibody activity generated during normal SIV infection could restrict the emergence of CD4-independent variants in vivo.
CD4+ T cell-depleted macaques lack robust plasma CD4-inducible neutralization activity.
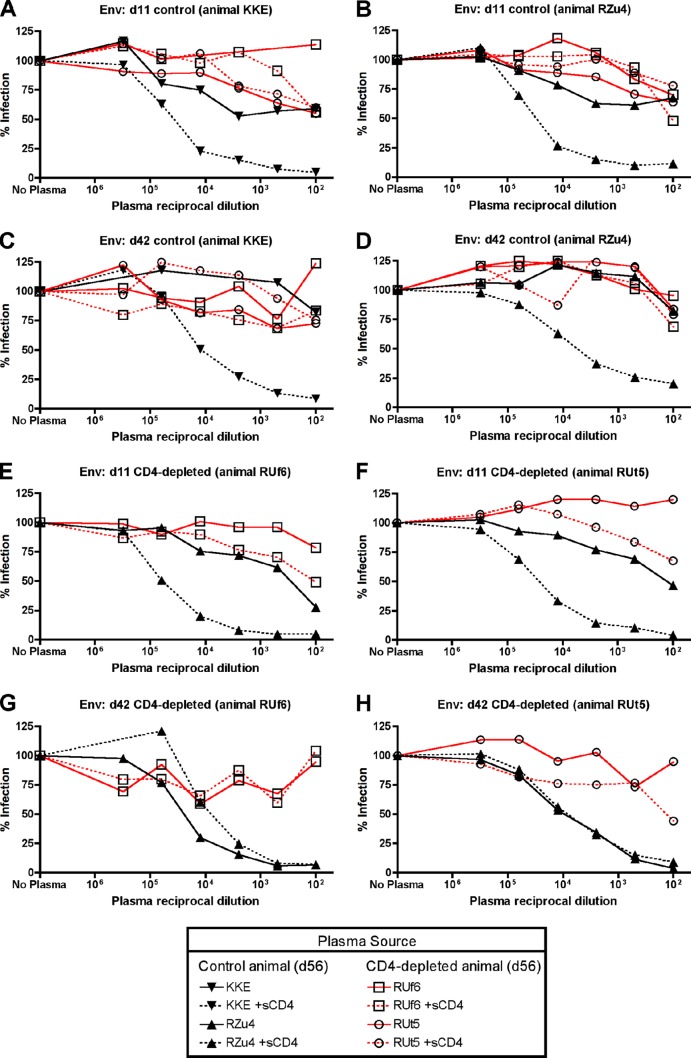
Both the CD4+ T cell-depleted and control animals in this study seroconverted by Western blot analysis and ELISA (21). Having observed the marked sensitivity to neutralization by SIV+ plasma of CD4-independent Envs that emerged in CD4+ T cell-depleted animals, we hypothesized that these animals might lack CD4-i neutralization activity. To address this, we used chronic infection plasma samples (day 56) from the CD4+ T cell-depleted and control animals and tested their ability to neutralize autologous and heterologous Envs (Fig. 8). As expected, plasma from control animals did not neutralize autologous day 11 or day 42 Envs in the absence of sCD4 but did neutralize Envs pre-exposed to sCD4, confirming the presence of CD4-i activity (Fig. 8A to D; black lines). In addition, plasma from control animals also neutralized heterologous CD4-independent Envs from day 42 CD4-depleted animals even in the absence of sCD4 (Fig. 8E to H; black lines). In sharp contrast, plasma from CD4-depleted animals showed a markedly reduced ability to neutralize heterologous CD4-dependent Envs (Fig. 8A to D; red lines), autologous day 11 CD4-dependent Envs (Fig. 8E and G; red lines), or autologous day 42 CD4-independent Envs (Fig. 8F and H; red lines), regardless of sCD4 treatment (although the possibility of the occurrence of very weak neutralization at high plasma concentrations cannot be completely excluded). Thus, plasma from CD4+ T cell-depleted animals in which CD4-independent viruses emerged lacks robust CD4-i neutralizing activity. Plasma from earlier time points, day 14 and day 25, had no detectable CD4-i neutralizing activity (data not shown), indicating that CD4-i neutralizing antibody activity emerges between days 25 and 56 after infection in normal SIV infection.
Fig 8.
Reduced CD4-inducible SIV Env neutralization activity in plasma from CD4+ T cell-depleted animals. Chronic infection plasma (day 56) from control animals (KKE, RZu4; black lines) or CD4-depleted animals (RUf6, RUt5; red lines) was used in neutralization studies with pseudotyped viruses carrying Envs from control animals (A to D) or CD4-depleted animals (E to H), with or without preincubation with sCD4. Black lines in panels A to D represent neutralization by autologous non-CD4-depleted plasma, while red lines represent neutralization by heterologous plasma from CD4-depleted animals. Red lines in panels E to H represent neutralization by autologous CD4-depleted plasma, while black lines represent neutralization by heterologous non-CD4-depleted plasma.
Finally, we used an ELISA against SIVmac230 gp140 to ask if total anti-Env IgG was reduced in the plasma of CD4 T cell-depleted animals. As shown in Fig. 9, Env-binding antibodies were present in all macaques, but titers were significantly lower in the CD4 T cell-depleted animals. Thus, CD4 T cell depletion results in global reduction of antigen-specific antibody production, consistent with the poor CD4-inducible neutralization capacity of CD4 T cell-depleted plasma.
Fig 9.
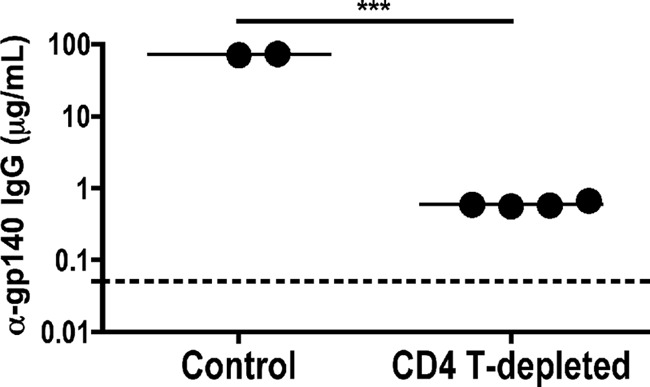
Reduced antigen-specific IgG in CD4 T cell-depleted animals. Total anti-gp140 plasma IgG from control and CD4 T cell-depleted macaques at day 42 postinfection was measured by ELISA. The mean anti-gp140 titers differed significantly between control (n = 2) and CD4-depleted (n = 4) animals by t test. The assay threshold of detection is indicated by a dotted horizontal line.
DISCUSSION
SIV infection of rhesus macaques that were largely depleted of CD4+ T cells, as described here and in reference 21, reveals that CD4-independent viral variants emerge and consistently dominate circulating virus in this setting. Emergence of CD4-independent Env variants corresponded to extensive in vivo infection of macrophages (21), a relationship which has been previously addressed in vitro (6, 45–47) and in the CNS or lung compartments (48–54). Here we demonstrate for the first time the factors governing CD4 dependence and target cell tropism in the systemic compartment in vivo. Animals that were not CD4+ T cell depleted before infection produced a robust antibody response which likely contributes to maintaining strict CD4 dependence and CD4+ T cell tropism. Such antibodies, which are nonneutralizing against control virus unless pretriggered with sCD4, are also abundantly present in chronic HIV-1 infection (9). The absence of such antibodies in the CD4+ T cell-depleted animals was associated with amino acid changes in Env gp120 that confer entry through CCR5 independently of CD4, enabling a shift in tropism to macrophages. These findings highlight the critical role that CD4-inducible neutralizing antibody activity has in vivo in shaping Env function, regulating CD4 use, and defining cell targeting early after infection.
We mapped the molecular determinants of CD4 independence to two amino acids in Env gp120, D470N and E84K. These changes were strongly associated with CD4 independence among Env variants, arose independently in multiple animals, and conferred the phenotype as shown by mutagenesis studies. Several pathways to CD4 independence in HIV and SIV have previously been described (7, 30, 48, 55–57), but our observation suggests that this particular pathway is especially favored in vivo, at least in the context of this SIVmac251 infection. SIVmac251 is a heterogeneous swarm, so it is conceivable that E84K and D470N were present as polymorphisms in the inoculum used to infect the animals in this study. However, our review of database sequences indicates that these amino acids are exceedingly uncommon, and the fact that D470N and E84K were found neither in Envs from control animals nor in those from day 11 CD4-depleted animals demonstrates that these changes are not favored until postpeak infection in the relative absence of CD4+ T cells. In contrast to D470N and E84K, G62N was enriched in CD4-depleted animals at both day 11 and day 42 but was not significantly associated with CD4-independent entry. G62N is present at an overall frequency of 4% to 5% among SIVmac sequences and occasionally represents the dominant sequence among some SIVmac251 swarms (33, 34). Thus, additional selective forces in the CD4-depleted animals other than CD4 independence per se appear to be responsible for selection at this residue. Of note, all occurrences of G62N, E84K, and D470N resulted from G-to-A nucleotide changes, implicating the APOBEC3 family of proteins (58) and suggesting an important role for this mechanism in achieving diversity rapidly in the context of this selective environment in vivo.
While both E84K and D470N contribute to the full CD4-independent phenotype, D470N appears to have a primary role, with E84K reflecting at least in part a compensatory change that reversed the degree of attenuation induced by D470N. Structural mapping of D470N on the unliganded core crystal structure of SIV gp120 (24) places a potential glycosylation site deep within a pocket of CD4-binding residues. In contrast to N at that location, Q470 Envs were not able to efficiently use CCR5 independently of CD4, suggesting that glycosylation rather than simply loss of a negative charge may be responsible for allowing CD4-independent entry. It is possible that placement of a bulky carbohydrate within that pocket could affect gp120 structure in a manner similar to that seen with docking of CD4 itself, causing Env to adopt a CD4-bound conformation and expose the conserved coreceptor-binding site. Whether differences in the structure of an intact SIV gp120 compared to the core structure employed here (as has been suggested for HIV-1 gp120 [40, 41, 59–61]) would affect the position of this residue is unknown. The homologous HIV-1 gp120 amino acid (D457) maps to the CD4-binding region in the HIV-1 gp120 core crystal structure and trimeric cryo-electron microscopy (cryo-EM) models (26, 59, 60), suggesting that the structural significance of D470 modeled here may be generalizable. How E84K might contribute to CD4 independence or to reverse D470N attenuation is unclear. This residue is near the predicted CD4-binding region but not a predicted contact point and lies within an alpha-helix thought to move with CD4 engagement (24, 62, 63). This drastic charge change may have indirect effects that affect the structure or perhaps may stabilize inter- or intramolecular interactions induced by the D470N effects that mimic CD4 engagement.
Although these residues have not been explicitly characterized in previous studies, analysis of database sequences reveals that both E84K and D470N are very rare. E84K is present at a low frequency in some stocks of SIVmacBK28 and SIVmac251 (33, 34, 36), and D470N is present in a few brain and plasma sequences in a model of rapid disease progression (37). E84K is also present in an in vitro-derived CD4-independent variant of SIVmac239 (35, 64), although its specific contribution to CD4 independence was not assessed. In addition, both E84K and D470N were found in Envs from experimentally CD8-depleted, SIVmac251-infected rhesus macaques experiencing brain disease (38). CNS disease is driven largely by macrophage-tropic infection of myeloid cells in the brain, in the overall immune-privileged CNS environment (52). The presence of these mutations in the brain and in highly immunodeficient animals in previous studies is consistent with our observation of macrophage infection in vivo in CD4 T cell-depleted animals (21).
Recently, it has been suggested that the generally greater sensitivity of CD4-independent viruses to antibody neutralization is due to greater “intrinsic reactivity” of these Env variants (10). This property enables them to spontaneously sample a range of conformations in the absence of CD4 binding, leading to both greater efficiency of coreceptor engagement and greater sensitivity to structural perturbation (and thus neutralization) by antibodies with a range of epitope specificities and not just those reflecting the coreceptor-binding site exposed upon binding to CD4. Our CD4-independent variants were sensitive to antibodies targeting several (although not all) SIV gp120 epitopes (Fig. 4 and 5), which is consistent with this model. Thus, the mutations that arose in these Envs may not only expose discrete CD4-i coreceptor-binding-site epitopes on gp120 but may also increase the intrinsic reactivity, rendering them more sensitive to neutralization by multiple epitope antibodies. This is the first report to directly demonstrate in vivo the functional constraints on Env imposed by this mechanism and the consequences for coreceptor use and target cell tropism that are governed by such constraints.
In our model, CD4+ T cells were profoundly depleted from blood and inductive lymphoid compartments, although incompletely from mucosal sites (21). It has been proposed that CD4-i epitope antibodies arise in vivo through B cell recognition of Env complexed with CD4, which exposes the conserved binding-binding site to the B cell receptor (65). Our observation that CD4+ T cell depletion leads to decreased CD4-inducible epitope antibody activity, despite the availability of these epitopes directly on these CD4-independent Envs for potential recognition by B cells, suggests that it is the loss of CD4+ T helper function in these animals that leads to the dysfunctional antibody response.
Along with the in vivo tropism shift toward macrophage target cells, CD4+ T cell-depleted animals exhibited rapid disease progression and an absence of postpeak decline in plasma viremia. While the role of antibody activity in shaping altered tropism in the CD4-depleted animals seems clear, the mechanisms responsible for sustained high-level plasma viremia are less clear. Robust autologous or heterologous neutralizing activity was not detected in the control animals even at the day 56 time point (21) (Fig. 8). In addition, studies differ on whether direct B cell depletion prior to infection of rhesus macaques affects the postpeak viral setpoint (66–68). Similarly, although CD8+ T cells clearly play a central role in postpeak viral decline (69–73), the current paradigms are that CD4+ T cell help is dispensable for primary CD8 T cell responses (74–76), and indeed no differences in CD8+ T cell responses were seen between control and CD4-depleted animals (21). In contrast, a variety of direct HIV- or SIV-specific antiviral activities have been ascribed to CD4+ T cells (77), including a role in postpeak decline following acute infection (21, 78). CD4+ T cells have also been reported to have particular antiviral activity against SIV-infected macrophages (79). Thus, loss of direct CD4+ T cell antiviral mechanisms may be responsible for the sustained viremia. At the same time, loss of strict CD4+ T cell dependence with consequent expansion to macrophages as a second robust target cell population may be an additional factor favoring sustained postpeak viremia, particularly in this setting of limited CD4+ T cells.
Some SIV-infected macaques that progress rapidly to AIDS fail to seroconvert (80, 81), a phenomenon that has been linked to early B cell defects following infection (82). In addition, a few CD4-independent SIV Env variants have previously been reported to arise spontaneously in SIV rapid progressors (30). Brain disease in SIV infection is also far more common in rapid- than normal-progressor animals (83), and macrophage infection within the CNS was also seen in these CD4 T cell-depleted animals (data not shown). Our results suggest that failure to develop CD4-inducible neutralization activity, whether as a consequence of direct virus-induced B cell defects as in spontaneous rapid progressive disease or of experimental abrogation of CD4+ T cell help as shown here, may be a common factor that enables emergence of these macrophage-tropic variants with an expanded host range and a propensity for CNS infection. Furthermore, while most HIV and SIV blood variants exhibit modest macrophage tropism, highly macrophage-tropic variants may emerge in very-late-stage disease when profound CD4+ T cell loss has developed (18–20). Infection with X4-tropic simian-human immunodeficiency virus (SHIV) also results in rapid profound CD4 depletion, with emergence of macrophage-tropic variants (19, 20). Furthermore, brain disease developed in all animals in another recent study that involved the use of CD4-depleted macaques prior to SIV infection using an approach slightly different from ours, although CD4 independence was not assessed (76). Future studies will help elucidate whether variations in antibody activity in different stages of disease may also regulate tropism and cell targeting.
In summary, our findings reveal a critical role for CD4 T cell-dependent production of CD4-inducible neutralizing activity early after infection in vivo in shaping Env function and target cell tropism. These data suggest that, normally, there is a balance in which CD4+ T cells support the production by B cells of antibodies that, paradoxically, enforce strict CD4+ T cell tropism for the virus. In this model, CD4+ T cell depletion results in both a profound reduction of levels of CD4+ T cell targets and impaired production of these antibodies. As a result, strict CD4 dependence is lost and expanded tropism for CD4-low macrophages emerges. In addition to the forces regulating normal infection, these findings have implications as well for infection in the CNS and, possibly, late-stage disease upon severe CD4+ T cell depletion.
ACKNOWLEDGMENT
We thank F. Shaheen and S. Bryan for technical assistance; N. Riddick, A. Yadav, and members of the Collman and Hoxie laboratories for useful discussions; and G. Shaw and R. Doms for valuable suggestions. We thank B. Keele and R. Kohli for assistance in analyzing sequences, J. Sodroski for advice on gp120 structure, and S. Bellamy for assistance with statistical analysis. We thank P. Carnathan and P. Marx for SIV+ plasma and P. Kozlowski for ELISA standards and acknowledge assistance of the Penn CFAR Viral/Molecular, Immunology, Biostatistics, and Non-Human Primate Cores (P30-AI045008).
N.F. and S.T.C.E. were supported by T32-AI007632. This work was supported by NIH grants AI091516 and MH61139 (R.G.C.), AI58706 (C.A.D.), and AI066998 (G.S.). We also acknowledge NIH support to the Yerkes National Primate Research Center (P51-RR000165 and P51-OD011132).
Footnotes
Published ahead of print 3 July 2013
REFERENCES
- 1.Peters PJ, Sullivan WM, Duenas-Decamp MJ, Bhattacharya J, Ankghuambom C, Brown R, Luzuriaga K, Bell J, Simmonds P, Ball J, Clapham PR. 2006. Non-macrophage-tropic human immunodeficiency virus type 1 R5 envelopes predominate in blood, lymph nodes, and semen: implications for transmission and pathogenesis. J. Virol. 80:6324–6332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Collman R, Godfrey B, Cutilli J, Rhodes A, Hassan NF, Sweet R, Douglas SD, Friedman H, Nathanson N, Gonzalez-Scarano F. 1990. Macrophage-tropic strains of human immunodeficiency virus type 1 utilize the CD4 receptor. J. Virol. 64:4468–4476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goodenow MM, Collman RG. 2006. HIV-1 coreceptor preference is distinct from target cell tropism: a dual-parameter nomenclature to define viral phenotypes. J. Leukoc. Biol. 80:965–972 [DOI] [PubMed] [Google Scholar]
- 4.Duncan CJ, Sattentau QJ. 2011. Viral determinants of HIV-1 macrophage tropism. Viruses 3:2255–2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thomas ER, Dunfee RL, Stanton J, Bogdan D, Taylor J, Kunstman K, Bell JE, Wolinsky SM, Gabuzda D. 2007. Macrophage entry mediated by HIV Envs from brain and lymphoid tissues is determined by the capacity to use low CD4 levels and overall efficiency of fusion. Virology 360:105–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Puffer BA, Pohlmann S, Edinger AL, Carlin D, Sanchez MD, Reitter J, Watry DD, Fox HS, Desrosiers RC, Doms RW. 2002. CD4 independence of simian immunodeficiency virus Envs is associated with macrophage tropism, neutralization sensitivity, and attenuated pathogenicity. J. Virol. 76:2595–2605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoffman TL, LaBranche CC, Zhang W, Canziani G, Robinson J, Chaiken I, Hoxie JA, Doms RW. 1999. Stable exposure of the coreceptor-binding site in a CD4-independent HIV-1 envelope protein. Proc. Natl. Acad. Sci. U. S. A. 96:6359–6364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kolchinsky P, Kiprilov E, Sodroski J. 2001. Increased neutralization sensitivity of CD4-independent human immunodeficiency virus variants. J. Virol. 75:2041–2050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Decker JM, Bibollet-Ruche F, Wei X, Wang S, Levy DN, Wang W, Delaporte E, Peeters M, Derdeyn CA, Allen S, Hunter E, Saag MS, Hoxie JA, Hahn BH, Kwong PD, Robinson JE, Shaw GM. 2005. Antigenic conservation and immunogenicity of the HIV coreceptor binding site. J. Exp. Med. 201:1407–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haim H, Strack B, Kassa A, Madani N, Wang L, Courter JR, Princiotto A, McGee K, Pacheco B, Seaman MS, Smith AB, III, Sodroski J. 2011. Contribution of intrinsic reactivity of the HIV-1 envelope glycoproteins to CD4-independent infection and global inhibitor sensitivity. PLoS Pathog. 7:e1002101. 10.1371/journal.ppat.1002101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luciw PA, Shaw KE, Unger RE, Planelles V, Stout MW, Lackner JE, Pratt-Lowe E, Leung NJ, Banapour B, Marthas ML. 1992. Genetic and biological comparisons of pathogenic and nonpathogenic molecular clones of simian immunodeficiency virus (SIVmac). AIDS Res. Hum. Retroviruses 8:395–402 [DOI] [PubMed] [Google Scholar]
- 12.Mankowski JL, Flaherty MT, Spelman JP, Hauer DA, Didier PJ, Amedee AM, Murphey-Corb M, Kirstein LM, Munoz A, Clements JE, Zink MC. 1997. Pathogenesis of simian immunodeficiency virus encephalitis: viral determinants of neurovirulence. J. Virol. 71:6055–6060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koenig S, Gendelman HE, Orenstein JM, Dal Canto MC, Pezeshkpour GH, Yungbluth M, Janotta F, Aksamit A, Martin MA, Fauci AS. 1986. Detection of AIDS virus in macrophages in brain tissue from AIDS patients with encephalopathy. Science 233:1089–1093 [DOI] [PubMed] [Google Scholar]
- 14.Ghorpade A, Nukuna A, Che M, Haggerty S, Persidsky Y, Carter E, Carhart L, Shafer L, Gendelman HE. 1998. Human immunodeficiency virus neurotropism: an analysis of viral replication and cytopathicity for divergent strains in monocytes and microglia. J. Virol. 72:3340–3350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peters PJ, Bhattacharya J, Hibbitts S, Dittmar MT, Simmons G, Bell J, Simmonds P, Clapham PR. 2004. Biological analysis of human immunodeficiency virus type 1 R5 envelopes amplified from brain and lymph node tissues of AIDS patients with neuropathology reveals two distinct tropism phenotypes and identifies envelopes in the brain that confer an enhanced tropism and fusigenicity for macrophages. J. Virol. 78:6915–6926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walter BL, Wehrly K, Swanstrom R, Platt E, Kabat D, Chesebro B. 2005. Role of low CD4 levels in the influence of human immunodeficiency virus type 1 envelope V1 and V2 regions on entry and spread in macrophages. J. Virol. 79:4828–4837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schnell G, Spudich S, Harrington P, Price RW, Swanstrom R. 2009. Compartmentalized human immunodeficiency virus type 1 originates from long-lived cells in some subjects with HIV-1-associated dementia. PLoS Pathog. 5:e1000395. 10.1371/journal.ppat.1000395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li S, Juarez J, Alali M, Dwyer D, Collman R, Cunningham A, Naif HM. 1999. Persistent CCR5 utilization and enhanced macrophage tropism by primary blood human immunodeficiency virus type 1 isolates from advanced stages of disease and comparison to tissue-derived isolates. J. Virol. 73:9741–9755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Igarashi T, Brown CR, Endo Y, Buckler-White A, Plishka R, Bischofberger N, Hirsch V, Martin MA. 2001. Macrophage are the principal reservoir and sustain high virus loads in rhesus macaques after the depletion of CD4+ T cells by a highly pathogenic simian immunodeficiency virus/HIV type 1 chimera (SHIV): implications for HIV-1 infections of humans. Proc. Natl. Acad. Sci. U. S. A. 98:658–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Igarashi T, Imamichi H, Brown CR, Hirsch VM, Martin MA. 2003. The emergence and characterization of macrophage-tropic SIV/HIV chimeric viruses (SHIVs) present in CD4+ T cell-depleted rhesus monkeys. J. Leukoc. Biol. 74:772–780 [DOI] [PubMed] [Google Scholar]
- 21.Ortiz AM, Klatt NR, Li B, Yi Y, Tabb B, Hao XP, Sternberg L, Lawson B, Carnathan PM, Cramer EM, Engram JC, Little DM, Ryzhova E, Gonzalez-Scarano F, Paiardini M, Ansari AA, Ratcliffe S, Else JG, Brenchley JM, Collman RG, Estes JD, Derdeyn CA, Silvestri G. 2011. Depletion of CD4(+) T cells abrogates post-peak decline of viremia in SIV-infected rhesus macaques. J. Clin. Invest. 121:4433–4445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elliott ST, Riddick NE, Francella N, Paiardini M, Vanderford TH, Li B, Apetrei C, Sodora DL, Derdeyn CA, Silvestri G, Collman RG. 2012. Cloning and analysis of sooty mangabey alternative coreceptors that support simian immunodeficiency virus SIVsmm entry independently of CCR5. J. Virol. 86:898–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Riddick NE, Hermann EA, Loftin LM, Elliott ST, Wey WC, Cervasi B, Taaffe J, Engram JC, Li B, Else JG, Li Y, Hahn BH, Derdeyn CA, Sodora DL, Apetrei C, Paiardini M, Silvestri G, Collman RG. 2010. A novel CCR5 mutation common in sooty mangabeys reveals SIVsmm infection of CCR5-null natural hosts and efficient alternative coreceptor use in vivo. PLoS Pathog. 6:e1001064. 10.1371/journal.ppat.1001064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen B, Vogan EM, Gong H, Skehel JJ, Wiley DC, Harrison SC. 2005. Structure of an unliganded simian immunodeficiency virus gp120 core. Nature 433:834–841 [DOI] [PubMed] [Google Scholar]
- 25.Jmol 2012. Jmol: an open-source Java viewer for chemical structures in 3D. http://www.jmol.org
- 26.Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Edinger AL, Ahuja M, Sung T, Baxter KC, Haggarty B, Doms RW, Hoxie JA. 2000. Characterization and epitope mapping of neutralizing monoclonal antibodies produced by immunization with oligomeric simian immunodeficiency virus envelope protein. J. Virol. 74:7922–7935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Montefiori DC. 2005. Evaluating neutralizing antibodies against HIV, SIV, and SHIV in luciferase reporter gene assays. Curr. Protoc. Immunol. 2005:Chapter 12:Unit 12.11 [DOI] [PubMed] [Google Scholar]
- 29.Xiao P, Zhao J, Patterson LJ, Brocca-Cofano E, Venzon D, Kozlowski PA, Hidajat R, Demberg T, Robert-Guroff M. 2010. Multiple vaccine-elicited nonneutralizing antienvelope antibody activities contribute to protective efficacy by reducing both acute and chronic viremia following simian/human immunodeficiency virus SHIV89.6P challenge in rhesus macaques. J. Virol. 84:7161–7173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ryzhova E, Whitbeck JC, Canziani G, Westmoreland SV, Cohen GH, Eisenberg RJ, Lackner A, Gonzalez-Scarano F. 2002. Rapid progression to simian AIDS can be accompanied by selection of CD4-independent gp120 variants with impaired ability to bind CD4. J. Virol. 76:7903–7909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keele BF, Li H, Learn GH, Hraber P, Giorgi EE, Grayson T, Sun C, Chen Y, Yeh WW, Letvin NL, Mascola JR, Nabel GJ, Haynes BF, Bhattacharya T, Perelson AS, Korber BT, Hahn BH, Shaw GM. 2009. Low-dose rectal inoculation of rhesus macaques by SIVsmE660 or SIVmac251 recapitulates human mucosal infection by HIV-1. J. Exp. Med. 206:1117–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stone M, Keele BF, Ma ZM, Bailes E, Dutra J, Hahn BH, Shaw GM, Miller CJ. 2010. A limited number of simian immunodeficiency virus (SIV) env variants are transmitted to rhesus macaques vaginally inoculated with SIVmac251. J. Virol. 84:7083–7095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Strickland SL, Gray RR, Lamers SL, Burdo TH, Huenink E, Nolan DJ, Nowlin B, Alvarez X, Midkiff CC, Goodenow MM, Williams K, Salemi M. 2011. Significant genetic heterogeneity of the SIVmac251 viral swarm derived from different sources. AIDS Res. Hum. Retroviruses 27:1327–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Del Prete GQ, Scarlotta M, Newman L, Reid C, Parodi LM, Roser JD, Oswald K, Marx PA, Miller CJ, Desrosiers RC, Barouch DH, Pal R, Piatak M, Jr, Chertova E, Giavedoni LD, O'Connor DH, Lifson JD, Keele BF. 2013. Comparative characterization of transfection- and infection-derived simian immunodeficiency virus challenge stocks for in vivo nonhuman primate studies. J. Virol. 87:4584–4595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.LaBranche CC, Sauter MM, Haggarty BS, Vance PJ, Romano J, Hart TK, Bugelski PJ, Marsh M, Hoxie JA. 1995. A single amino acid change in the cytoplasmic domain of the simian immunodeficiency virus transmembrane molecule increases envelope glycoprotein expression on infected cells. J. Virol. 69:5217–5227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Edmonson P, Murphey-Corb M, Martin LN, Delahunty C, Heeney J, Kornfeld H, Donahue PR, Learn GH, Hood L, Mullins JI. 1998. Evolution of a simian immunodeficiency virus pathogen. J. Virol. 72:405–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rivera-Amill V, Noel RJ, Jr, Garcia Y, Rivera I, Iszard M, Buch S, Kumar A. 2010. Accelerated evolution of SIV env within the cerebral compartment in the setting of morphine-dependent rapid disease progression. Virology 398:201–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Strickland SL, Gray RR, Lamers SL, Burdo TH, Huenink E, Nolan DJ, Nowlin B, Alvarez X, Midkiff CC, Goodenow MM, Williams K, Salemi M. 2012. Efficient transmission and persistence of low-frequency SIVmac251 variants in CD8-depleted rhesus macaques with different neuropathology. J. Gen. Virol. 93:925–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pikora C, Wittish C, Desrosiers RC. 2005. Identification of two N-linked glycosylation sites within the core of the simian immunodeficiency virus glycoprotein whose removal enhances sensitivity to soluble CD4. J. Virol. 79:12575–12583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.White TA, Bartesaghi A, Borgnia MJ, de la Cruz MJ, Nandwani R, Hoxie JA, Bess JW, Lifson JD, Milne JL, Subramaniam S. 2011. Three-dimensional structures of soluble CD4-bound states of trimeric simian immunodeficiency virus envelope glycoproteins determined by using cryo-electron tomography. J. Virol. 85:12114–12123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.White TA, Bartesaghi A, Borgnia MJ, Meyerson JR, de la Cruz MJ, Bess JW, Nandwani R, Hoxie JA, Lifson JD, Milne JL, Subramaniam S. 2010. Molecular architectures of trimeric SIV and HIV-1 envelope glycoproteins on intact viruses: strain-dependent variation in quaternary structure. PLoS Pathog. 6:e1001249. 10.1371/journal.ppat.1001249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burns DP, Collignon C, Desrosiers RC. 1993. Simian immunodeficiency virus mutants resistant to serum neutralization arise during persistent infection of rhesus monkeys. J. Virol. 67:4104–4113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hirsch V, Adger-Johnson D, Campbell B, Goldstein S, Brown C, Elkins WR, Montefiori DC. 1997. A molecularly cloned, pathogenic, neutralization-resistant simian immunodeficiency virus, SIVsmE543-3. J. Virol. 71:1608–1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cole KS, Rowles JL, Jagerski BA, Murphey-Corb M, Unangst T, Clements JE, Robinson J, Wyand MS, Desrosiers RC, Montelaro RC. 1997. Evolution of envelope-specific antibody responses in monkeys experimentally infected or immunized with simian immunodeficiency virus and its association with the development of protective immunity. J. Virol. 71:5069–5079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Platt EJ, Wehrly K, Kuhmann SE, Chesebro B, Kabat D. 1998. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J. Virol. 72:2855–2864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dunfee RL, Thomas ER, Gorry PR, Wang J, Taylor J, Kunstman K, Wolinsky SM, Gabuzda D. 2006. The HIV Env variant N283 enhances macrophage tropism and is associated with brain infection and dementia. Proc. Natl. Acad. Sci. U. S. A. 103:15160–15165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Duenas-Decamp MJ, Peters PJ, Burton D, Clapham PR. 2009. Determinants flanking the CD4 binding loop modulate macrophage tropism of human immunodeficiency virus type 1 R5 envelopes. J. Virol. 83:2575–2583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Edinger AL, Mankowski JL, Doranz BJ, Margulies BJ, Lee B, Rucker J, Sharron M, Hoffman TL, Berson JF, Zink MC, Hirsch VM, Clements JE, Doms RW. 1997. CD4-independent, CCR5-dependent infection of brain capillary endothelial cells by a neurovirulent simian immunodeficiency virus strain. Proc. Natl. Acad. Sci. U. S. A. 94:14742–14747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Korber BT, Kunstman KJ, Patterson BK, Furtado M, McEvilly MM, Levy R, Wolinsky SM. 1994. Genetic differences between blood- and brain-derived viral sequences from human immunodeficiency virus type 1-infected patients: evidence of conserved elements in the V3 region of the envelope protein of brain-derived sequences. J. Virol. 68:7467–7481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hughes ES, Bell JE, Simmonds P. 1997. Investigation of the dynamics of the spread of human immunodeficiency virus to brain and other tissues by evolutionary analysis of sequences from the p17gag and env genes. J. Virol. 71:1272–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gorry PR, Bristol G, Zack JA, Ritola K, Swanstrom R, Birch CJ, Bell JE, Bannert N, Crawford K, Wang H, Schols D, De Clercq E, Kunstman K, Wolinsky SM, Gabuzda D. 2001. Macrophage tropism of human immunodeficiency virus type 1 isolates from brain and lymphoid tissues predicts neurotropism independent of coreceptor specificity. J. Virol. 75:10073–10089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.González-Scarano F, Martín-García J. 2005. The neuropathogenesis of AIDS. Nat. Rev. Immunol. 5:69–81 [DOI] [PubMed] [Google Scholar]
- 53.Peters PJ, Duenas-Decamp MJ, Sullivan WM, Brown R, Ankghuambom C, Luzuriaga K, Robinson J, Burton DR, Bell J, Simmonds P, Ball J, Clapham PR. 2008. Variation in HIV-1 R5 macrophage-tropism correlates with sensitivity to reagents that block envelope: CD4 interactions but not with sensitivity to other entry inhibitors. Retrovirology 5:5. 10.1186/1742-4690-5-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Means RE, Matthews T, Hoxie JA, Malim MH, Kodama T, Desrosiers RC. 2001. Ability of the V3 loop of simian immunodeficiency virus to serve as a target for antibody-mediated neutralization: correlation of neutralization sensitivity, growth in macrophages, and decreased dependence on CD4. J. Virol. 75:3903–3915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kolchinsky P, Mirzabekov T, Farzan M, Kiprilov E, Cayabyab M, Mooney LJ, Choe H, Sodroski J. 1999. Adaptation of a CCR5-using, primary human immunodeficiency virus type 1 isolate for CD4-independent replication. J. Virol. 73:8120–8126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Clapham PR, McKnight A, Weiss RA. 1992. Human immunodeficiency virus type 2 infection and fusion of CD4-negative human cell lines: induction and enhancement by soluble CD4. J. Virol. 66:3531–3537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dehghani H, Puffer BA, Doms RW, Hirsch VM. 2003. Unique pattern of convergent envelope evolution in simian immunodeficiency virus-infected rapid progressor macaques: association with CD4-independent usage of CCR5. J. Virol. 77:6405–6418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jern P, Russell RA, Pathak VK, Coffin JM. 2009. Likely role of APOBEC3G-mediated G-to-A mutations in HIV-1 evolution and drug resistance. PLoS Pathog. 5:e1000367. 10.1371/journal.ppat.1000367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. 2008. Molecular architecture of native HIV-1 gp120 trimers. Nature 455:109–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kwon YD, Finzi A, Wu X, Dogo-Isonagie C, Lee LK, Moore LR, Schmidt SD, Stuckey J, Yang Y, Zhou T, Zhu J, Vicic DA, Debnath AK, Shapiro L, Bewley CA, Mascola JR, Sodroski JG, Kwong PD. 2012. Unliganded HIV-1 gp120 core structures assume the CD4-bound conformation with regulation by quaternary interactions and variable loops. Proc. Natl. Acad. Sci. U. S. A. 109:5663–5668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mao Y, Wang L, Gu C, Herschhorn A, Xiang SH, Haim H, Yang X, Sodroski J. 2012. Subunit organization of the membrane-bound HIV-1 envelope glycoprotein trimer. Nat. Struct. Mol. Biol. 19:893–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kwong PD, Wyatt R, Majeed S, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. 2000. Structures of HIV-1 gp120 envelope glycoproteins from laboratory-adapted and primary isolates. Structure 8:1329–1339 [DOI] [PubMed] [Google Scholar]
- 63.Huang CC, Venturi M, Majeed S, Moore MJ, Phogat S, Zhang MY, Dimitrov DS, Hendrickson WA, Robinson J, Sodroski J, Wyatt R, Choe H, Farzan M, Kwong PD. 2004. Structural basis of tyrosine sulfation and VH-gene usage in antibodies that recognize the HIV type 1 coreceptor-binding site on gp120. Proc. Natl. Acad. Sci. U. S. A. 101:2706–2711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Edinger AL, Blanpain C, Kunstman KJ, Wolinsky SM, Parmentier M, Doms RW. 1999. Functional dissection of CCR5 coreceptor function through the use of CD4-independent simian immunodeficiency virus strains. J. Virol. 73:4062–4073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Forsell MN, Dey B, Morner A, Svehla K, O'Dell S, Hogerkorp CM, Voss G, Thorstensson R, Shaw GM, Mascola JR, Karlsson Hedestam GB, Wyatt RT. 2008. B cell recognition of the conserved HIV-1 co-receptor binding site is altered by endogenous primate CD4. PLoS Pathog. 4:e1000171. 10.1371/journal.ppat.1000171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schmitz JE, Kuroda MJ, Santra S, Simon MA, Lifton MA, Lin W, Khunkhun R, Piatak M, Lifson JD, Grosschupff G, Gelman RS, Racz P, Tenner-Racz K, Mansfield KA, Letvin NL, Montefiori DC, Reimann KA. 2003. Effect of humoral immune responses on controlling viremia during primary infection of rhesus monkeys with simian immunodeficiency virus. J. Virol. 77:2165–2173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mao H, Lafont BA, Igarashi T, Nishimura Y, Brown C, Hirsch V, Buckler-White A, Sadjadpour R, Martin MA. 2005. CD8+ and CD20+ lymphocytes cooperate to control acute simian immunodeficiency virus/human immunodeficiency virus chimeric virus infections in rhesus monkeys: modulation by major histocompatibility complex genotype. J. Virol. 79:14887–14898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Miller CJ, Genesca M, Abel K, Montefiori D, Forthal D, Bost K, Li J, Favre D, McCune JM. 2007. Antiviral antibodies are necessary for control of simian immunodeficiency virus replication. J. Virol. 81:5024–5035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Koup RA, Safrit JT, Cao Y, Andrews CA, McLeod G, Borkowsky W, Farthing C, Ho DD. 1994. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J. Virol. 68:4650–4655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Borrow P, Lewicki H, Hahn BH, Shaw GM, Oldstone MB. 1994. Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. J. Virol. 68:6103–6110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schmitz JE, Kuroda MJ, Santra S, Sasseville VG, Simon MA, Lifton MA, Racz P, Tenner-Racz K, Dalesandro M, Scallon BJ, Ghrayeb J, Forman MA, Montefiori DC, Rieber EP, Letvin NL, Reimann KA. 1999. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science 283:857–860 [DOI] [PubMed] [Google Scholar]
- 72.Jin X, Bauer DE, Tuttleton SE, Lewin S, Gettie A, Blanchard J, Irwin CE, Safrit JT, Mittler J, Weinberger L, Kostrikis LG, Zhang L, Perelson AS, Ho DD. 1999. Dramatic rise in plasma viremia after CD8(+) T cell depletion in simian immunodeficiency virus-infected macaques. J. Exp. Med. 189:991–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Goulder PJ, Watkins DI. 2008. Impact of MHC class I diversity on immune control of immunodeficiency virus replication. Nat. Rev. Immunol. 8:619–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kalams SA, Buchbinder SP, Rosenberg ES, Billingsley JM, Colbert DS, Jones NG, Shea AK, Trocha AK, Walker BD. 1999. Association between virus-specific cytotoxic T-lymphocyte and helper responses in human immunodeficiency virus type 1 infection. J. Virol. 73:6715–6720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ostrowski MA, Justement SJ, Ehler L, Mizell SB, Lui S, Mican J, Walker BD, Thomas EK, Seder R, Fauci AS. 2000. The role of CD4+ T cell help and CD40 ligand in the in vitro expansion of HIV-1-specific memory cytotoxic CD8+ T cell responses. J. Immunol. 165:6133–6141 [DOI] [PubMed] [Google Scholar]
- 76.Okoye AA, Rohankhedkar M, Abana C, Pattenn A, Reyes M, Pexton C, Lum R, Sylwester A, Planer SL, Legasse A, Park BS, Piatak M, Jr, Lifson JD, Axthelm MK, Picker LJ. 2012. Naive T cells are dispensable for memory CD4+ T cell homeostasis in progressive simian immunodeficiency virus infection. J. Exp. Med. 209:641–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Norris PJ, Sumaroka M, Brander C, Moffett HF, Boswell SL, Nguyen T, Sykulev Y, Walker BD, Rosenberg ES. 2001. Multiple effector functions mediated by human immunodeficiency virus-specific CD4(+) T-cell clones. J. Virol. 75:9771–9779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Soghoian DZ, Jessen H, Flanders M, Sierra-Davidson K, Cutler S, Pertel T, Ranasinghe S, Lindqvist M, Davis I, Lane K, Rychert J, Rosenberg ES, Piechocka-Trocha A, Brass AL, Brenchley JM, Walker BD, Streeck H. 2012. HIV-specific cytolytic CD4 T cell responses during acute HIV infection predict disease outcome. Sci. Transl. Med. 4:123ra125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sacha JB, Giraldo-Vela JP, Buechler MB, Martins MA, Maness NJ, Chung C, Wallace LT, Leon EJ, Friedrich TC, Wilson NA, Hiraoka A, Watkins DI. 2009. Gag- and Nef-specific CD4+ T cells recognize and inhibit SIV replication in infected macrophages early after infection. Proc. Natl. Acad. Sci. U. S. A. 106:9791–9796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dykhuizen M, Mitchen JL, Montefiori DC, Thomson J, Acker L, Lardy H, Pauza CD. 1998. Determinants of disease in the simian immunodeficiency virus-infected rhesus macaque: characterizing animals with low antibody responses and rapid progression. J. Gen. Virol. 79(Pt 10):2461–2467 [DOI] [PubMed] [Google Scholar]
- 81.Hirsch VM, Santra S, Goldstein S, Plishka R, Buckler-White A, Seth A, Ourmanov I, Brown CR, Engle R, Montefiori D, Glowczwskie J, Kunstman K, Wolinsky S, Letvin NL. 2004. Immune failure in the absence of profound CD4+ T-lymphocyte depletion in simian immunodeficiency virus-infected rapid progressor macaques. J. Virol. 78:275–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Titanji K, Velu V, Chennareddi L, Vijay-Kumar M, Gewirtz AT, Freeman GJ, Amara RR. 2010. Acute depletion of activated memory B cells involves the PD-1 pathway in rapidly progressing SIV-infected macaques. J. Clin. Invest. 120:3878–3890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Westmoreland SV, Halpern E, Lackner AA. 1998. Simian immunodeficiency virus encephalitis in rhesus macaques is associated with rapid disease progression. J. Neurovirol. 4:260–268 [DOI] [PubMed] [Google Scholar]