Abstract
Soil microeukaryotes may trophically benefit from plant biopolymers. However, carbon transfer from cellulose into soil microeukaryotes has not been demonstrated so far. Microeukaryotes assimilating cellulose-derived carbon in oxic and anoxic soil slurries were therefore examined by rRNA-based stable-isotope probing. Bacteriovorous flagellates and ciliates and, likely, mixotrophic algae and saprotrophic fungi incorporated carbon from supplemental [U-13C]cellulose under oxic conditions. A previous study using the same soil suggested that cellulolytic Bacteria assimilated 13C of supplemental cellulose. Thus, it can be assumed that ciliates, cercozoa, and chrysophytes assimilated carbon by grazing upon and utilizing metabolic products of Bacteria that hydrolyzed cellulose in the soil slurries.
TEXT
Cellulose is the most abundant organic compound of plant litter (40% of dry weight), is mineralized primarily by soil microorganisms, and contributes significantly to carbon dioxide release of terrestrial ecosystems (1, 2). Bacteria and fungi are generally assumed to be the main consumers of cellulose in terrestrial ecosystems. Ascomycota, Basidiomycota, Zygomycota, and Chytridiomycota are important fungal groups for cellulose degradation, since they comprise saprotrophic species (3, 4). Several protistan functional groups, i.e., bacterivorous, fungivorous, and cytotrophic protists, might also benefit from cellulose-derived carbon mainly by consuming products and microorganisms that have utilized cellulose. Protists have a pivotal role in the microbial loop, which facilitates the transfer of carbon from detrital organic material (e.g., cellulose) to higher eukaryotes that feed on soil microorganisms (5, 6). Many cellulolytic Bacteria are known (2), and recently, the utilization of hydrolysis and fermentations products of cellulose by noncellulolytic soil prokaryotes was resolved using nucleic acid stable-isotope probing (NA-SIP) (7–9). Such studies demonstrated that also noncellulolytic prokaryotes contribute to and benefit from carbon flow from cellulose (7–9).
Prokaryotic taxa of an agricultural soil involved in cellulose degradation under oxic and anoxic conditions were resolved in a previous study using rRNA SIP (9). The objectives of the current study were to test in the same experimental setup whether microeukaryotes incorporate carbon from cellulose and to identify the actively involved eukaryotes in order to shed light on their putative roles in the cellulose-dependent food web.
Experimental setup.
Slurries with samples from the upper 20 cm of an agricultural soil (dystric cambisol) were prepared as previously described (9) and were supplemented with 0.2 g of [U-13C]cellulose and one with [U-12C]cellulose (9). Since substantial labeling of prokaryotes occurred after 35 days (9), samples for analyzing microeukaryotes (i.e., eukaryotes of a higher trophic level) were taken after 35 and 70 days to ensure enough 13C incorporation in rRNA. Tightly closed gas flasks contained 80 ml of slurry in a total volume of 500 ml for anoxic and 1,000 ml for oxic treatments. All flasks were incubated in the dark in an end-over-end shaker (9). Per treatment (i.e., in the absence and presence of oxygen), two replicated flasks were tested. SIP analyses were done from combined RNA extracts at a given time point. In one case (Fig. 1 and 2), SIP gradients from each experimental replicate were conducted separately to evaluate experimental variations. The headspaces were regularly exchanged with sterile air (every 2 days) or dinitrogen (every 4 days). Under oxic incubations, carbon dioxide was the sole carbonaceous product and molecular hydrogen was not detected, suggesting that these treatments were truly oxic (9). Slurries were incubated for 70 days at 15°C in the dark. RNA was extracted from 0.6 g of soil slurry at the start of the experiment and after 35 and 70 days (10). 13C-labeled RNA was separated from nonlabeled RNA by isopycnic centrifugation with a cesium trifluoroacetate-based gradient medium according to previous protocols (9, 11). Fractions with buoyant densities between 1.767 and 1.776 g ml−1 were regarded as containing unlabeled RNA, and fractions with buoyant densities between 1.813 and 1.821 g ml−1 were regarded as containing 13C-labeled RNA. RNA was precipitated, quantified from these fractions (12), and reverse transcribed (9). To minimize analytical efforts, 18S rRNA-based analyses were conducted from pooled RNAs from two light and two heavy fractions per gradient (9).
Fig 1.
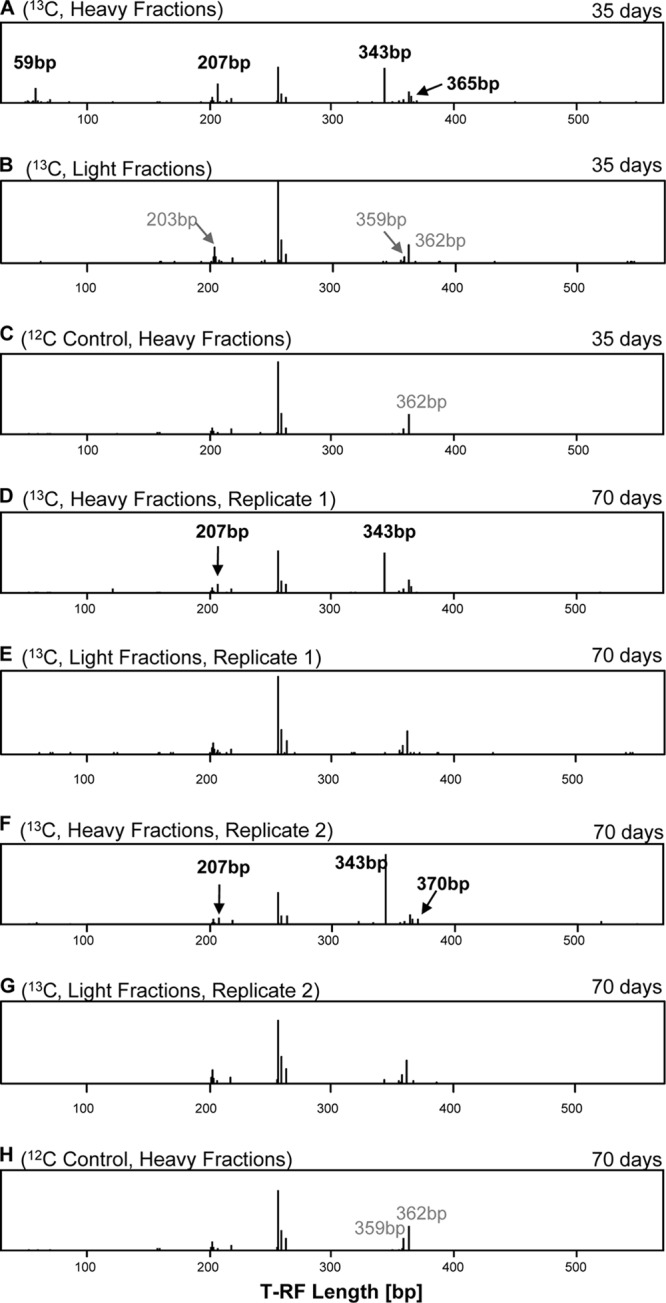
T-RFLP patterns obtained with the eukaryote-targeting primer set from oxic treatments. Anoxic treatments did not reveal any labeling. Patterns are retrieved from heavy fractions of treatments that were supplemented with [U-13C]- and [12C]cellulose. Samples were taken after 35 days (A, B, and C) and 70 days (D, E, F, G, H) of incubation. Black numbers, T-RFs that represented 13C-labeled phylotypes in the heavy fractions. Identification of labeled phylotypes was based on comparison with the T-RFLP patterns of the heavy fractions of the 12C treatment at the same time point and on comparison with the T-RFLP patterns of the light fractions of the same gradient. Gray numbers, nonlabeled T-RFs in the light fractions of the same gradient or in heavy fractions of a 12C control that have a size similar to but different from those of T-RFs that represented labeled phylotypes. The T-RFLP patterns of panels D, E, F, and G are each based on a single gradient from combined RNAs of two experimental replicates.
Fig 2.
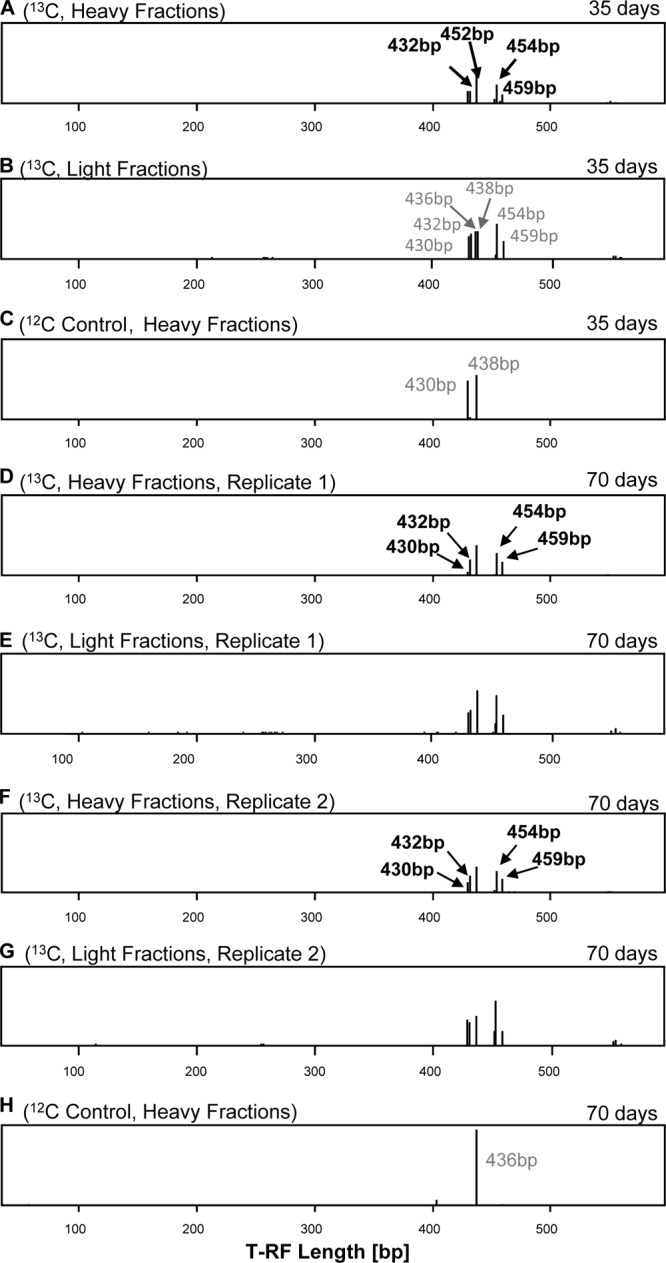
T-RFLP patterns obtained with the Chrysophyceae-targeting primer set from oxic treatments. Anoxic treatments did not reveal any labeling. Patterns are retrieved from heavy fractions of treatments that were supplemented with [U-13C]- and [12C]cellulose. Samples were taken after 35 days (A, B, and C) and 70 days (D, E, F, G, H) of incubation. Black numbers, T-RFs that represented 13C-labeled phylotypes in the heavy fractions. Identification of labeled phylotypes was according to comparison with the T-RFLP patterns of the gradient of the heavy fractions of the 12C treatment at the same oxygen availability and time point and by comparison with the light fractions of the same gradient. Gray numbers, nonlabeled T-RFs in the light fractions of the same gradient or in heavy fractions of a 12C control that have a size similar to but different from those T-RFs that represented labeled phylotypes. The T-RFLP patterns of panels D, E, F, and G are each based on a single gradient from combined RNAs of two experimental replicates.
18S rRNA libraries.
Two 18S rRNA cDNA libraries using the eukaryote-specific primers 20fNS1 and 516r were analyzed to assess shifts in the microeukaryotic community between the start and the end of the experiment (13). RNA was extracted from fractions that represented labeled RNA (“heavy” fractions) and fractions that represented unlabeled RNA (“light” fractions). 18S rRNA libraries from pooled light and heavy fractions at the start were dominated by fungi (Ascomycota), chlorophyta (Chlamydomonadales and Scenedesmaceae), and metazoan phylotypes (Annelida), while most protist phylotypes were affiliated with Alveolata, Amoebozoa, Cercozoa, stramenopiles, and yet-uncultured microeukaryotic taxa (Table 1 and see Table S1 in the supplemental material). Since gene libraries were not set up from total RNA, there might have been a bias toward labeled phylotypes. Nonetheless, if substantial changes (i.e., changes of several percent) in relative abundances occurred, they were regarded as true.
Table 1.
Phylogenetic affiliation and relative abundances of 18S rRNA sequences in clone libraries derived from pooled light and heavy RNAs
| Taxonb | Relative abundance (%) under indicated conditiona at: |
|||
|---|---|---|---|---|
| Start |
70 days |
|||
| Anoxic (n = 252) | Oxic (n = 254) | Anoxic (n = 120) | Oxic (n = 125) | |
| Alveolata (Ciliophora) | 0.8 | 0.0 | 2.5 | 44.8 |
| Alveolata (other) | 1.2 | 3.9 | 5.8 | 0.8 |
| Amoebozoa | 7.1 | 0.0 | 6.7 | 3.2 |
| Apuzoa | 0.0 | 0.4 | 0.0 | 0.0 |
| Ascomycota | 30.6 | 24.0 | 28.3 | 21.6 |
| Basidiomycota | 1.2 | 3.5 | 4.2 | 0.0 |
| Glomeromycota | 0.4 | 0.0 | 0.0 | 0.0 |
| Fungi incertae sedis | 0.8 | 4.7 | 0.0 | 2.4 |
| Fungi (Chytridiomycota) | 0.0 | 0.4 | 0.0 | 0.0 |
| Zygomycota | 1.2 | 0.0 | 0.8 | 0.8 |
| Cercozoa | 3.2 | 5.1 | 6.7 | 2.4 |
| Chlorophyta | 18.3 | 15.0 | 35.0 | 22.4 |
| Heterolobosea | 0.4 | 0.4 | 0.0 | 0.0 |
| Metazoa | 30.6 | 34.3 | 0.0 | 0.0 |
| Stramenopiles | 2.0 | 5.5 | 6.7 | 0.0 |
| Streptophyta | 2.4 | 1.6 | 1.7 | 1.6 |
| Unknown eukaryota | 0.0 | 1.2 | 1.7 | 0.0 |
Values are relative abundances of phylogenetic groups in the respective gene libraries. Incubation conditions, i.e., without (anoxic) and with (oxic) oxygen in the headspace, are noted. Numbers in parentheses are total numbers of analyzed clone insert sequences.
Clone insert sequences had a sequence similarity to known phylotypes of >97% based on BLAST searches in the nucleotide databases of GenBank.
Oxic conditions resulted in an apparent increase of Alveolata (i.e., peritrich ciliates) and amoebozoan phylotypes affiliating with the naked lobose amoeba group Euamoebida. Phylotypes affiliating with the parasitic apicomplexan family Eimeriidae, the Cercozoa, and the Basidiomycota were either reduced in relative abundance or disappeared after 70 days. Unlike in a previous study (14), relative abundances of several stramenopile algae and fungus-like stramenopile plant pathogens (i.e., Pythium and Aphanomyces) (15, 16) that were present at the start of the experiment decreased in response to cellulose supplementation in the oxic slurries. Members of the chlorophytes changed over the incubation period, and after 70 days, Scenedesmaceae were predominant. Anoxic conditions caused shifts within the phylotypes of amoebozoa, alveolata, and chlorophyta (e.g., an enrichment of phylotypes affiliating with the green-algal class Trebouxiophyceae).
Fungal phylotypes at the start of incubations belonged mainly to mitosporic Ascomycota (i.e., genera Hyphozyma and Tetracladium) and to several groups within the subphylum Pezizomycotina, such as the Sordariomycetes. Mitosporic Ascomycota and Pezizomycotina include parasitic fungi and strains that are decomposers and grow on soil, leaf litter, and wood (see Table S2 in the supplemental material) (17). Cellulases have, for instance, been proved to exist in Lecythophora (18). Abundances of phylotypes of mitosporic Ascomycota and several saprophytic fungi, such as those of the genus Acremonium, increased under oxic conditions, while several genera within the Sordariomycetes either decreased in abundance or disappeared after 70 days. The predominant fungi under anoxic conditions were related to the Dothideomycetes, which include many plant pathogens (19), or the Microascales, which are known as plant and insect pathogens or coprophilous (20). The Chaetomiaceae, which are capable of lignocellulose degradation (21), were no longer detected after 70 days.
Identification of 13C-labeled 18S rRNA phylotypes by T-RFLP.
Labeled microeukaryotes were examined by 18S rRNA terminal restriction fragment length polymorphism (T-RFLP) analysis. To reduce the likelihood that 12C RNA would be identified as labeled, T-RF patterns of heavy fractions of [U-12C]cellulose-supplemented slurries were compared to those of [U-13C]cellulose (22, 23). T-RFs that were enriched in heavy fractions of 13C treatments but not in 12C treatments were regarded as 13C labeled, even if those T-RFs occurred in the light fractions of 13C treatments. The eukaryotic primer set, as well as primers targeting two ubiquitously distributed bacterivorous flagellate groups, i.e., Chrysophyceae and Kinetoplastidae, and the ciliates (24, 25) were applied. T-RFLP analysis was conducted as described in a previous study (26) using the restriction enzymes MspI, RsaI, MseI, and Cfr and RsaI for 18S rRNA amplicons of total eukaryotes, ciliates, Kinetoplastidae, and Chrysophyceae, respectively.
Labeled T-RFs were identified only in oxic incubations using the general eukaryote-targeting (i.e., T-RFs of 59, 207, 343, 365, and 370 bp) and the Chrysophyceae-targeting (i.e., T-RFs of 430, 432, 452, 454, and 459 bp) primers (Fig. 1 and 2; see also Fig. S1 and Tables S3 and S4 in the supplemental material). An affiliation of several peaks was possible by comparing the restriction patterns with the T-RFs of cloned sequences originating from the heavy fractions (Table S4). Based on this assignment of T-RFs to phylotypes, the ciliate Opisthonecta minima (343 bp), the cercozoan Proleptomonas faecicola (370 bp), several chrysophytes related to Chrysophyceae sp. strain CCCM41 (430 bp), Mallomonas peroneides (432 bp), and a Leukarachnion strain (454 bp; i.e., an amoeboid sister group to the Chrysophyceae) likely assimilated carbon from supplemental [U-13C]cellulose. A dominant peak at 257 bp belonged to the cellulolytic ascomycete fungus Lecythophora; this peak was present in both the 12C- and 13C-labeled fractions at 35 and 70 days, suggesting that members of this group were active, but carbon assimilation from cellulose cannot unambiguously be proven. Overall, the presence of several protistan sequences in the clone libraries from 70 days and the shift in heavy T-RFLP patterns (Tables 1, S1, and S3) suggested that some flagellates, ciliates, and amoebae incorporated cellulose-derived carbon by bacterivory on cellulose-degrading bacteria, confirming that protists contribute to the biopolymer-degrading food webs in soil by top-down control of the bacterial community. Among the flagellates, several sequences affiliating with cercozoa were detected in the 18S rRNA libraries from both oxic and anoxic incubations. The presence of cercozoa under oxygen-limited conditions has recently been shown also in flooded rice soils, indicating that members of the cercozoa are capable of coping with limited oxygen availability (27). Moreover, soil protists include several additional trophic groups that might be directly or indirectly involved in organic matter degradation and, thus, explain the observed assimilation of carbon of supplemental cellulose (28, 29). For instance, the occurrence of phylotypes closely related to the nonphagotrophic cercozoan Proleptomonas faecicola (Table S3) (30) and the occurrence of algal phylotypes (i.e., stramenopiles and Chlorophyceae) in 18S rRNA libraries of oxic and anoxic slurries suggested that mixotrophic protists contributed to the cellulose-derived carbon flow. The importance of heterotrophy within mixotrophic algal species can vary between species of the same genus and is not related to its taxonomic affiliation (31). Its impact on soil food webs and the distinction between direct degradation of cellulose, degradation of hydrolysis products, or grazing-mediated incorporation of 13C warrants further studies.
Metabolically active phylotypes belonging to the Ascomycota were detected, but unlike in another study (32), 13C labeling was not observed, indicating that the detectable Ascomycota were not primarily involved in the hydrolysis of cellulose. Nonetheless, cellulolytic Ascomycota were detected in heavy-gradient fractions. Intermediate fractions of SIP gradients were not evaluated. Thus, the possibility that Ascomycota assimilated smaller amounts of the 13C cannot be excluded. Basidiomycota did not respond to experimental conditions. The experimental setup included shaking, which most likely restricted the growth of hypha-forming members of the Basidiomycetes (33), and thus, experimental conditions may have selected for microorganisms that were capable of adaptation to the slurry conditions. Whether the detected microeukaryotes are similarly important in undisturbed and dry soil warrants further investigations. Thus, there is a need to improve the analytic accessibility of metabolically active microorganisms in largely intact soil samples.
Conclusions.
Previous studies identified in the same experiment various bacterial taxa that assimilated cellulose-derived carbon under anoxic (e.g., Kineosporaceae, Streptomycetaceae, Clostridiaceae) and under oxic (e.g., Bacteroidetes) conditions and demonstrated differential responses to oxygen availability (23), while members of the Archaea were not labeled (9). Results of both the previous and the current study indicate that Bacteria hydrolyzed primarily cellulose and that ciliates, cercozoa, and chrysophytes assimilated carbon by grazing or utilizing products of cellulolytic bacterial metabolisms in the experiment. The lack of evidence for 13C labeling under anoxic conditions suggests that detectable microeukaryotes played a minor role in carbon flux under oxygen-limited conditions. SIP experiments using 13C-labeled-cellulose-derived hydrolysis and degradation products (9), prey bacteria (34), or litter are promising approaches to reveal further trophic aspects of cellulose degradation with regard to soil microeukaryotes.
Nucleotide sequence accession numbers.
Sequences of clone inserts (RNA extracted from heavy and light fractions) were deposited in the GenBank repository under the accession numbers KF356737 to KF357523.
Supplementary Material
ACKNOWLEDGMENTS
The study was financed by the Helmholtz Association and the Deutsche Forschungsgemeinschaft (grants Ko2912/3-1 and Ko2912/3-2).
We thank Verena Jaschik for technical assistance.
Footnotes
Published ahead of print 12 July 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01598-13.
REFERENCES
- 1.Lal R. 2008. Sequestration of atmospheric CO2 in global carbon pools. Energy Environ. Sci. 1:86–100 [Google Scholar]
- 2.Lynd LR, Weimer PJ, van Zyl WH, Pretorius IS. 2002. Microbial cellulose utilization: fundamentals and biotechnology. Microbiol. Mol. Biol. Rev. 66:506–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kirk PM, Cannon PF, Minter DW, Stalpers JA. (ed). 2008. Dictionary of the fungi, 10th ed. ABI, Wallingford, United Kingdom [Google Scholar]
- 4.Freeman K, Martin A, Karki D, Lynch R, Mitter M, Meyer A, Longcore J, Simmons D, Schmidt S. 2009. Evidence that chytrids dominate fungal communities in high-elevation soils. Proc. Natl. Acad. Sci. U. S. A. 106:18315–18320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pomeroy LR. 1974. The ocean's food web: a changing paradigm. Bioscience 24:409–504 [Google Scholar]
- 6.Coleman DC. 1994. The microbial loop concept as used in terrestrial soil ecology studies. Microb. Ecol. 28:245–250 [DOI] [PubMed] [Google Scholar]
- 7.Haichar FEZ, Achouak W, Christen R, Heulin T, Marol C, Marais MF, Mougel C, Ranjard L, Balesdent J, Berge O. 2007. Identification of cellulolytic bacteria in soil by stable isotope probing. Environ. Microbiol. 9:625–634 [DOI] [PubMed] [Google Scholar]
- 8.Lee CG, Watanabe T, Sato Y, Murase J, Asakawa S, Kimura M. 2011. Bacterial populations assimilating carbon from C-13-labeled plant residue in soil: analysis by a DNA-SIP approach. Soil Biol. Biochem. 43:814–822 [Google Scholar]
- 9.Schellenberger S, Kolb S, Drake HL. 2010. Metabolic responses of novel cellulolytic and saccharolytic agricultural soil Bacteria to oxygen. Environ. Microbiol. 12:845–861 [DOI] [PubMed] [Google Scholar]
- 10.Griffiths RI, Whiteley AS, O'Donnell AG, Bailey MJ. 2000. Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA- and rRNA-based microbial community composition. Appl. Environ. Microbiol. 66:5488–5491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lueders T, Manefield M, Friedrich MW. 2004. Enhanced sensitivity of DNA- and rRNA-based stable isotope probing by fractionation and quantitative analysis of isopycnic centrifugation gradients. Environ. Microbiol. 6:73–78 [DOI] [PubMed] [Google Scholar]
- 12.Degelmann DM, Kolb S, Dumont M, Murrell JC, Drake HL. 2009. Enterobacteriaceae facilitate the anaerobic degradation of glucose by a forest soil. FEMS Microbiol. Ecol. 68:312–319 [DOI] [PubMed] [Google Scholar]
- 13.Amann RI, Binder BJ, Olson RJ, Chisholm SW, Devereux R, Stahl DA. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murase J, Shibata M, Lee CG, Watanabe T, Asakawa S, Kimura M. 2012. Incorporation of plant residue-derived carbon into the microeukaryotic community in a rice field soil revealed by DNA stable-isotope probing. FEMS Microbiol. Ecol. 79:371–379 [DOI] [PubMed] [Google Scholar]
- 15.Thorn RG, Lynch MDJ. 2007. Fungi and eukaryotic algae, p 145–163 In Pau EA. (ed), Soil microbiology, ecology and biochemistry. Academic Press, Oxford, United Kingdom [Google Scholar]
- 16.Gaulin E, Jaquet C, Bottin A, Dumas B. 2007. Root rot disease of legumes caused by Aphanomyces euteiches. Mol. Plant Pathol. 8:539–548 [DOI] [PubMed] [Google Scholar]
- 17.Schoch CL, Sung GH, López-Giráldez F, Townsend JP, Miadlikowska J, Hofstetter V, Robbertse B, Matheny PB, Kauff F, Wang Z, Gueidan C, Andrie RM, Trippe K, Ciufetti LM, Wynns A, Fraker E, Hodkinson BP, Bonito G, Groenewald JZ, Arzanlou M, de Hoog GS, Crous PW, Hewitt D, Pfister DH, Peterson K, Gryzenhout M, Wingfield MJ, Aptroot A, Suh SO, Blackwell M, Hillis DM, Griffith GW, Castlebury LA, Rossman AY, Lumbsch HT, Lücking R, Büdel B, Rauhut A, Diederich P, Ertz D, Geiser DM, Hosaka K, Inderbitzin P, Kohlmeyer J, Volkmann-Kohlmeyer B, Mostert L, O'Donnell K, Sipman H, Rogers JD, Shoemaker RA, Sugiyama J, Summerbell RC, Untereiner W, Johnston PR, Stenroos S, Zuccaro A, Dyer PS, Crittenden PD, Cole MS, Hansen K, Trappe JM, Yahr R, Lutzoni F, Spatafora JW. 2009. The Ascomycota tree of life: a phylum-wide phylogeny clarifies the origin and evolution of fundamental reproductive and ecological traits. Syst. Biol. 58:224–239 [DOI] [PubMed] [Google Scholar]
- 18.Nilsson T. 1973. Studies on wood degradation and cellulolytic activity of microfungi. Stud. For. Suec. 104:2–40 [Google Scholar]
- 19.Ohm RA, Feau N, Henrissat B, Schoch CL, Horwitz BA, Feau N, Henrissat B, Schoch CL, Horwitz BA, Barry KW, Condon BJ, Copeland AC, Dhillon B, Glaser F, Hesse CN, Kosti I, LaButti K, Lindquist EA, Lucas S, Salamov AA, Bradshaw RE, Ciuffetti L, Hamelin RC, Kema GHJ, Lawrence C, Scott JA, Spatafora JW, Turgeon BG, de Wit PJGM, Zhong S, Goodwin SB, Grigoriev IV. 2012. Diverse lifestyles and strategies of plant pathogenesis encoded in the genomes of eighteen Dothideomycetes fungi. PLoS Pathog. 8:e1003037. 10.1371/journal.ppat.1003037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barron GL, Cain RF, Gilman LC. 1961. The genus Microascus. Can. J. Bot. 39:1609–1631 [Google Scholar]
- 21.Song F, Tian X, Fan X, He X. 2010. Decomposing ability of filamentous fungi on litter is involved in a subtropical mixed forest. Mycologia 102:20–26 [DOI] [PubMed] [Google Scholar]
- 22.Manefield M, Whiteley AS, Griffith RI, Bailey M. 2002. RNA stable isotope probing, a novel means of linking microbial community function to phylogeny. Appl. Environ. Microbiol. 68:5367–5373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schellenberger S, Drake HL, Kolb S. 2011. Functionally redundant cellobiose-degrading soil bacteria respond differentially to oxygen. Appl. Environ. Microbiol. 77:6043–6048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lara E, Berney C, Harms H, Chatzinotas A. 2007. Cultivation-independent analysis reveals a shift in ciliate 18S rRNA gene diversity in a polycyclic aromatic hydrocarbon polluted soil. FEMS Microbiol. Ecol. 62:365–373 [DOI] [PubMed] [Google Scholar]
- 25.Jousset E, Lara M, Nikolausz H, Harms A, Chatzinotas A. 2010. Application of the denaturing gradient gel electrophoresis (DGGE) technique as an efficient diagnostic tool for ciliate communities in soil. Sci. Total Environ. 408:1221–1225 [DOI] [PubMed] [Google Scholar]
- 26.Wu QL, Chatzinotas A, Wang J, Boenigk J. 2009. Genetic diversity of eukaryotic plankton assemblages in eastern Tibetan lakes differing by their salinity and altitude. Microb. Ecol. 58:569–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sugano A, Tsuchimoto H, Tun CC, Asakawa S, Kimura M. 2007. Succession and phylogenetic profile of eukaryotic communities in rice-straw incorporated into a rice field: estimation by PCR-DGGE and sequence analysis. Soil Sci. Plant Nutr. 53:585–594 [Google Scholar]
- 28.Adl MS. 2003. The ecology of soil decomposition. CABI Publishing, Wallingford, United Kingdom [Google Scholar]
- 29.Adl MS, Gupta VVSR. 2006. Protists in soil ecology and forest nutrient cycling. Can. J. For. Res. 36:1805–1817 [Google Scholar]
- 30.Vickerman K, Le Ray D, Hoef-Emden K, De Jonckheere J. 2002. The soil flagellate Proleptomonas faecicola: cell organisation and phylogeny suggest that the only described free-living trypanosomatid is not a kinetoplastid but has cercomonad affinities. Protist 153:9–24 [DOI] [PubMed] [Google Scholar]
- 31.Jones HLJ. 1997. A classification of mixotrophic protists based on their behavior. Freshwater Biol. 37:35–43 [Google Scholar]
- 32.Eichorst SA, Kuske CR. 2012. Identification of cellulose-responsive bacterial and fungal communities in geographically and edaphically different soils by using stable isotope probing. Appl. Environ. Microbiol. 78:2316–2327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lindahl BD, de Boer W, Finlay RD. 2010. Disruption of root carbon transport into forest humus stimulates fungal opportunists at the expense of mycorrhizal fungi. ISME J. 4:872–881 [DOI] [PubMed] [Google Scholar]
- 34.Kuppardt S, Chatzinotas A, Kästner M. 2010. Development of a fatty acid and RNA stable isotope probing-based method for tracking protist grazing on bacteria in wastewater. Appl. Environ. Microbiol. 76:8222–8230 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


