Abstract
Viruses are the most abundant and diverse biological entities within soils, yet their ecological impact is largely unknown. Defining how soil viral communities change with perturbation or across environments will contribute to understanding the larger ecological significance of soil viruses. A new approach to examining the composition of soil viral communities based on random PCR amplification of polymorphic DNA (RAPD-PCR) was developed. A key methodological improvement was the use of viral metagenomic sequence data for the design of RAPD-PCR primers. This metagenomically informed approach to primer design enabled the optimization of RAPD-PCR sensitivity for examining changes in soil viral communities. Initial application of RAPD-PCR viral fingerprinting to soil viral communities demonstrated that the composition of autochthonous soil viral assemblages noticeably changed over a distance of meters along a transect of Antarctic soils and across soils subjected to different land uses. For Antarctic soils, viral assemblages segregated upslope from the edge of dry valley lakes. In the case of temperate soils at the Kellogg Biological Station, viral communities clustered according to land use treatment. In both environments, soil viral communities changed along with environmental factors known to shape the composition of bacterial host communities. Overall, this work demonstrates that RAPD-PCR fingerprinting is an inexpensive, high-throughput means for addressing first-order questions of viral community dynamics within environmental samples and thus fills a methodological gap between narrow single-gene approaches and comprehensive shotgun metagenomic sequencing for the analysis of viral community diversity.
INTRODUCTION
Although only ∼30% of our planet is covered by landmass, terrestrial ecosystems are responsible for 50% of global net primary productivity (2). Because soils are the essential basis of all terrestrial plant communities, global productivity ultimately relies on the health and fertility of soils. In turn, the fertility of soils is dictated by the activity of microorganisms, which are the key facilitators of biogeochemical cycles. It is these cycles that maintain the nutrient balance of soils (6–8). Bacterially mediated redox reactions of elements and molecules contribute substantially to nutrient transformations occurring in the biosphere (9–12). Agricultural practices such as application of biofertilizers (e.g., nitrogen-fixing and phosphate-solubilizing bacteria) have leveraged these microbial processes to increase levels of plant-available nutrients in soils (13, 14). In a similar way, bioremediation strategies for environmental cleanup are aimed at empowering specific bacterial populations responsible for the transformation of pollutant compounds to less harmful forms (15–17). Collectively, these services provided by soil bacteria are critical to ecosystem health, yet our understanding of the factors that regulate the magnitude and rates of such services is limited.
Predator-prey and host-parasite interactions are considered key ecological factors regulating the composition and activity of bacterial communities (18–22). Nematodes, protozoa, viruses, and microarthropods form the principle bactivorous fraction of soil microbiota, which help to regulate bacterial abundance and therefore influence bacterial processes within soils. Eloquent ex situ model studies have credited soil viruses with modulating bacterial diversity (23), and observations of phage and bacterial populations indicate viral regulation of bacterial activity in Artic soil ecosystems (24). In addition, the sheer abundance (∼109 viruses g−1) and diversity of soil viruses (25, 26), along with an absence of any metabolic requirements for survival outside the host cell, argue that viruses may be persistent microbial parasites within soils. In support of this idea, investigations into the fate and transport of pathogenic viruses have found that viable virus particles show long-term persistence in soils (27–29).
For aquatic environments, we now appreciate that virioplankton populations have significant influence in the flow of carbon and energy (30). In highly productive marine ecosystems, virioplankton assemblages are dynamic, showing fast turnover rates (31, 32) and high diversity (33). Viruses in aquatic environments are known to regulate the growth of bacterial communities directly through host cell lysis and indirectly through the lytic release of nutrients, which benefit the growth of uninfected populations (34). Through these and other mechanisms, aquatic viruses contribute to ecosystem productivity and the flux of nutrient elements through global biogeochemical cycles. In contrast, the degree to which viral activities influence soil biogeochemical processes remains an open question. Documenting the dynamics of soil viral populations with changes in edaphic and geographic factors is one important step toward understanding the larger role of soil viral communities.
Recently, random PCR amplification of polymorphic DNA (RAPD-PCR) has provided a valuable means for examining fine-scale changes in the composition of virioplankton within aquatic environments (1, 3–5, 35, 36). However, no comparable high-throughput approaches have been developed for examining short-term changes in the composition of soil viral assemblages at high resolution. To address these first-order questions surrounding the dynamics of soil viral assemblages, we developed a RAPD-PCR fingerprinting approach suitable for the particular demands of soil samples. Subsequently, limited proof-of-concept experiments were conducted to test the use of RAPD-PCR for documenting changes in soil viral assemblages across geographic scales and with changes in land use.
MATERIALS AND METHODS
Soil samples.
Antarctic soil samples were collected as ∼1-kg frozen cores of 0 to 10 cm in depth along a transect at the eastern rim of Tom's Pond (ETP) and the southern rim of Obelisk Pond (SOB), Antarctica. Three soil samples were analyzed over each transect. These soils were provided by M. Uhle and M. Howard, University of Tennessee, Knoxville, TN, and were stored at −20°C. Delaware soils were Matapeake silt loam composite samples of 0 to 10 cm in depth collected across a cornfield at the Agricultural Experimental Station, University of Delaware, Newark, DE. The Delaware soil samples were maintained at 12.7% moisture and 4°C until use. Properties of the Antarctic and Delaware soils can be found in a publication from Williamson et al. (37). Soils from Kellogg Biological Station (KBS) Long-Term Ecological Research (LTER) treatment plots were either Kalamazoo fine loamy or Oshtemo coarse loamy. The following soil samples were collected on 19 May 2008 from KBS treatment plots (http://lter.kbs.msu.edu/): T1, corn/soybean/wheat, conventional till and inputs; T4, corn/soybean/wheat, organic (reduced inputs, winter cover); T7, early successional community, annually burned; T8, early successional community, annually mowed, never tilled; and SF2, mid- to late successional forest (40 to 60 years postagriculture).
Preparation of virus and cell concentrates from soil samples.
Viruses were extracted from soils using a potassium citrate extraction procedure described elsewhere (38). Briefly, soils were vortexed with potassium citrate extraction buffer and kept on ice during the sonication treatment. After sonication, the soil-buffer mixture was centrifuged at 3,000 × g for 30 min at 4°C. The resulting supernatant was filtered through a 0.22-μm-pore-size filter (Sterivex; Millipore Corp., Bedford, MA). This viral extract was snap-frozen in liquid nitrogen before storage at −80°C. To increase the concentration of virus particles, extracts were ultracentrifuged at >200,000 × g at 4°C for 2 h (41,000 rpm in an SW 41 Ti rotor; Beckman Coulter Inc., Fullerton, CA). The supernatant was carefully decanted, and viral pellets were resuspended to a total volume of 200 μl of the potassium citrate viral extraction buffer. Dilutions of this viral concentrate were prepared using extraction buffer. Extracted viruses were enumerated using epifluorescence microscopy as described by Williamson et al. (38).
Prior to analysis by RAPD-PCR, viral concentrates were treated with a high concentration of DNase I to remove free, extraviral DNA (39). Five units of DNase I (RQ1 RNase-free DNase; Fisher Scientific, Pittsburg, PA), 2 μl of 10× DNase buffer, and 20 μl of viral concentrate or viral extract were mixed, and the mixture was incubated at 37°C for 30 min. Reactions were terminated by addition of 2 μl of DNase I stop solution, and the reaction mixtures were incubated at 65°C for 10 min.
RAPD-PCR primer design.
The total population and frequency of all possible decamers having ≥70% G+C content were recorded within 22 viral metagenome libraries (see Table S2 in the supplemental material). The PERL script RDP-M (RAPD primer design from metagenomes), used to calculate decamer frequencies, is located at http://rpd-m.sourceforge.net. The decamer frequency of occurrence within different viral metagenomic libraries was normalized through multiplication by use of a normalizing factor unique to each library. Library normalizing factors were calculated as the ratio of the number of base pairs of DNA sequence within each library to the number of base pairs of DNA sequence within the largest library in the collection of 22 viral metagenome libraries. Normalization was done to allow comparison of the decamer frequency of occurrence across libraries, as no two libraries were of similar size in terms of the numbers of base pairs of DNA.
RAPD-PCR analysis.
RAPD-PCR assay mixtures of a 25-μl total volume contained 2.5 μl of 10× Ex Taq buffer (Mg2+ plus), 1.60 μl of a deoxynucleoside triphosphate (dNTP) mixture (0.16 mM each), 2 μl of a 50-pmol μl−1 primer stock (final concentration, 4 μM), 0.50 μl of TaKaRa Ex Taq Hot-Start DNA polymerase (5 units/μl; Hot-Start version; TaKaRa Bio Inc., Otsu, Shiga, Japan), and 1 μl of template (104, 105, 106, or 107 viruses). The primers used in the investigations were HDC-1 (5′-CGCCGCCGCC-3′), HCB-1 (5′-CCAGCAGCAG-3′), LWHS-1 (5′-GTTCGGGTCG-3′), RLWLS-1 (5′-GCGATCCACG-3′), and AAWZS-1 (5′-CACCACCTGC-3′). Single primers served as both forward and reverse primers in the PCR amplification. Thermocycler conditions were as follows: (i) 94°C for 10 min for denaturing the protein capsids and releasing the viral DNA, (ii) primer annealing at the indicated temperature for each primer for 3 min (HDC-1, 55°C; HCB-1, 47°C; LWHS-1, 43°C; RLWLS-1, 44°C; AAWZS-1, 43°C), (iii) primer extension at 72°C for 1 min, (iv) 94°C for 30 s, (v) repeat of steps ii through iv for an additional 29 cycles, (vi) annealing at the temperature for each primer indicated above for 3 min, (vii) final primer extension at 72°C for 10 min, and (viii) hold at 4°C.
RAPD-PCR amplicons were separated on a 1.8% high-resolution agarose gel (Agarose–Hi-Res separation, ≤1,000 bp [Affymetrix, Inc., Santa Clara, CA] or GenePure HiRes agarose [ISC BioExpress, Kaysville, UT]) made in 1× Tris-borate-EDTA (TBE; 10× TBE is 890 mM Tris base, 890 mM boric acid, 20 mM EDTA, pH 8.3). Running buffer consisted of 0.5× TBE. Molecular size markers were loaded into two terminal lanes and one central lane of the gel (100-bp DNA ladder plus; Fermentas Gene Ruler, Burlington, ON, Canada). Electrophoresis was conducted using a 21-cm (distance between electrodes) gel box and a 13.5-cm gel. After electrophoresis, the gel was stained with SYBR gold (Molecular Probes Inc., Eugene, OR), according to the manufacturer's instructions, in 500 ml of 1× Tris-EDTA (TE; 10× TE is 100 mM Tris-Cl, 10 mM EDTA, pH 8) buffer. Stained gels were imaged using an MD Typhoon 6500 variable-mode imager (Amersham Biosciences, Buckinghamshire, United Kingdom).
Banding patterns from gel images were analyzed using GelCompar II software (version 4.5; Applied Maths, Sint-Martens-Latem, Belgium). Bands were initially selected by screening the fingerprints with 5% minimum profiling and 2% gray zone criteria before manually checking for band assignment. Matrices of banding pattern similarity were determined using Dice's binary coefficient (40). Dendrograms of banding pattern similarity were obtained using the unweighted-pair group method using arithmetic averages (UPGMA) (4).
The reproducibility of the RAPD-PCR banding patterns across PCRs and gels was investigated. The experimental design consisted of two individual PCR master mixes (MMs) each aliquoted twice. Each master mix contained diethyl pyrocarbonate-treated water, reaction buffer, dNTPs, primer, and DNA polymerase. A single template of 105 soil viruses that had been DNase I treated was assayed in two aliquots of two MMs for a total of four independent reactions. These independent reactions were each run on two different gels to compare the between and within reproducibility of the RAPD-PCR banding patterns according to master mix and gel.
Statistical and other analyses.
Statistical tests were conducted using JMP software (SAS Institute Inc., Cary, NC). Viral and bacterial quantitative data were plotted with Prism software (GraphPad Software, La Jolla, CA).
RESULTS
Primer design and testing.
A total of 22 viral metagenome libraries (see Table S2 in the supplemental material) were used in designing RAPD-PCR decamer primers. All libraries are publicly available on the Viral Informatics Resource for Metagenome Exploration (VIROME) web portal (virome.dbi.udel.edu) (41). The theoretical maximum numbers of unique decamers was 1,048,576; however, for greater primer specificity and binding strength, only decamers having a ≥70% G+C content were selected (42, 43). High-G+C-content primers also favored amplification of soil viral DNA, as soil viruses are known to have genomes of high G+C content (26, 44). High-G+C-content decamers were obtained through analysis of viral metagenomic DNA sequences, and their frequency of occurrence in viral metagenome libraries from water, soils, extreme environments, and sludges was recorded (see Table S1 in the supplemental material). Occurrence frequency was subsequently normalized according to the size of a given library in terms of the total numbers of base pairs of DNA. Altogether, 190,279 high-G+C-content decamers occurred in at least one of the viral metagenome libraries.
Sixteen decamers were selected for initial tests in RAPD-PCR assays against soil viral extracts from Delaware soil. Decamer HDC-1 had the highest normalized frequency of occurrence in the Matapeake soil viral metagenome library and, similar to decamer HKCHD-1, also occurred relatively frequently in other soil and sludge viral libraries. Primer HDPC-1 had a high average normalized frequency of occurrence in soil and water viral libraries. The frequency of HDPC-1 was highly variable across libraries due to its frequent occurrence in the Wisconsin soil and Chesapeake Bay (CBAY_Bench) viral libraries. Decamer HCCG-1 was the most common in aquatic viral libraries, including extreme aquatic environments. Primer AAWZS-1 had a low frequency of occurrence in soils, and its frequency was zero in Desert, Kansas, and Peru soils (data not included for frequencies in individual libraries). Decamer LWHS-1 had a zero frequency in viral metagenomes from sludge and a low occurrence in water and extreme environments.
For each selected decamer, the optimal PCR annealing temperature was determined by testing a gradient of annealing temperatures; for example, the optimal annealing temperature of primer HCB-1 was found to be 47°C (see Fig. S1 in the supplemental material) rather than the predicted temperature of 37.9°C. Annealing temperatures of 47.1°C and 47.8°C provided the most similar banding patterns among the annealing temperatures tested. This higher annealing temperature was also chosen to avoid nonspecific DNA binding (45) and false-positive banding in no-template control samples (46, 47). Among the group of 16 primers, a subset was selected on the basis of the number of RAPD-PCR bands produced and the banding pattern spread of RAPD-PCR fingerprints across the gel lane. Primer HDC-1, which had the highest frequency of occurrence in the Matapeake soil viral library, produced an excessive number of bands and was not used in RAPD-PCR fingerprint analysis of soil viruses.
Primer frequency and banding patterns.
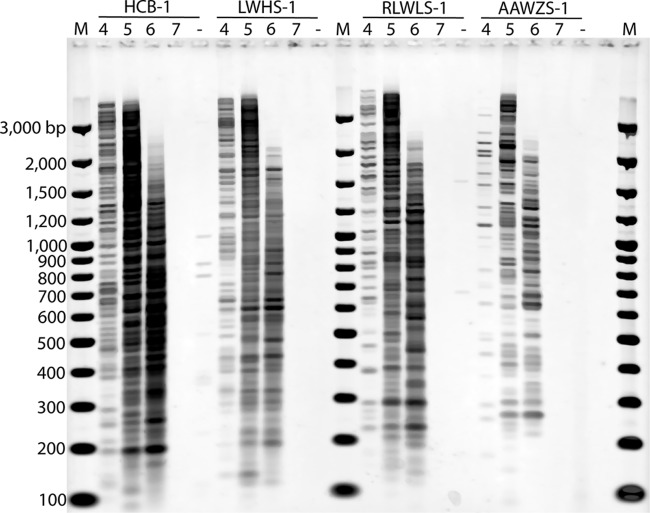
Four primers, HCB-1, LWHS-1, RLWLS-1, and AAWZS-1 (listed according to decreasing frequency within the Matapeake [Delaware] soil viral metagenome library), were used in RAPD-PCR trials against various concentrations of viruses extracted from a single soil sample (Fig. 1). Primer HCB-1 produced more bands at the lower template concentration (104 viruses) than the lower-frequency primers LWHS-1, RLWLS-1, and AAWZS-1 (Fig. 1). Band intensity also increased with increasing template concentration up to 106 viruses. At any given template concentration, primers with a greater frequency of occurrence produced more bands in the RAPD-PCR assay (Fig. 1). As the number of template viral particles in each PCR assay increased, higher-molecular-size bands tended to disappear and the number and intensity of smaller amplification products increased. At template concentrations of 107 viruses per reaction mixture, none of the primers produced amplicons. The absence of RAPD-PCR amplicons at a high template concentration might have been due to the high ratio of sites for priming to primer molecules within the PCR. This high ratio would have scavenged primer molecules within early PCR cycles, preventing effective amplification in later cycles. Alternatively, PCR inhibition at high template concentrations could have been caused by inhibitory substances, such as humic acids, within soil viral extracts (48). Considering the loss of higher-molecular-size bands at template concentrations of 106 viruses per PCR mixture and the complete loss of amplification with 107 viruses per PCR mixture, a template concentration of 105 viruses per 25-μl PCR mixture was determined to be the optimal amount of starting template in the RAPD-PCR assay.
Fig 1.
RAPD-PCR of viral assemblages from a Delaware soil sample using four different decamer primers at four levels of viral template DNA. Lanes 4, 5, 6, and 7 indicate 104, 105, 106, and 107 viruses per PCR, respectively. Lanes M, molecular size marker lanes; lanes −, no-template control. A decrease in decamer frequency in soil viral metagenomes is shown from left to right: HCB-1 > LWHS-1 > RLWLS-1 > AAWZS-1.
Reproducibility.
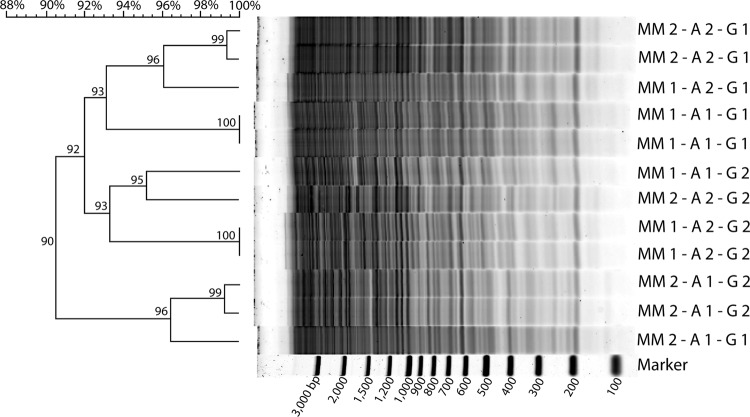
Because the ultimate objective of RAPD-PCR analysis is the comparison of polymorphic DNA templates, the reproducibility of whole fingerprints is of paramount importance to unbiased assessment of similarities and differences between samples. To assess the reproducibility of RAPD-PCR analysis of soil viral assemblages, RAPD-PCR fingerprints obtained using a single sample and a single template concentration were produced from two master mixes, each of which was split into two aliquots. The resulting RAPD-PCR amplicons were resolved on two independent agarose gels.
Fingerprints from the same master mix and aliquot resolved on a single gel were 99 to 100% similar. However, this similarity dropped to 92 to 97% across gels (Fig. 2). Similarity fell to 91 to 95% for RAPD-PCR fingerprints from the same master mix but from different master mix aliquots resolved on a single gel. Across gels, the similarity of these reactions fell to 89 to 92%. Fingerprint similarities were 90 to 96% and 90 to 95% from different master mixes resolved on a single gel or two different gels, respectively. Most fingerprints tended to cluster with fingerprints run on the same gel, irrespective of the master mix or the different aliquots used during PCR. Thus, for amplicons produced from the same master mix and master mix aliquot and resolved on the same gel, a similarity above 99% would be considered identical. For fingerprints produced from different master mixes and run on different gels, those with similarity values above 89% would be considered identical.
Fig 2.
Reproducibility of RAPD-PCR fingerprints using the HCB-1 decamer against whole viral assemblages from a single Delaware soil sample. The scale at the upper top left is the percent similarity of RAPD-PCR fingerprints. Numbers at the nodes of the dendrogram indicate percent similarity. RAPD-PCR fingerprints were produced from two aliquots (A) each from two different PCR master mixes (MMs). Subsequently, a total of 3 lanes per master mix aliquot were run: 2 lanes on one gel (G) and 1 lane on the other gel. The sizes of marker bands are given as numbers of base pairs of DNA.
RAPD-PCR analysis of soil viral communities.
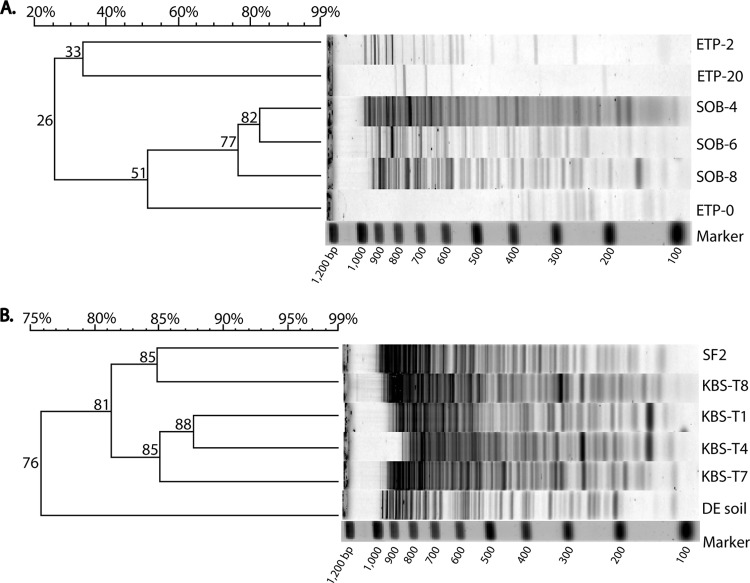
To explore the influence of environmental factors on soil viral communities, primer AAWZS-1 was used to investigate differences in the composition of viral assemblages in soils from Antarctica, the Kellogg Biological Station, and Delaware (Fig. 3). Viral RAPD-PCR fingerprints from Antarctic soils included three samples from a transect near the eastern rim of Tom's Pond (ETP) and three from a transect of the southern rim of Obelisk Pond (SOB) in the dry valleys. RAPD-PCR fingerprints showed these viral assemblages to be highly distinct from one other (Fig. 3A). Viral fingerprints from the SOB transect clustered together at 77% similarity, with two upgradient samples, SOB sample 4 (SOB-4) and SOB-6, clustering most tightly at 82%. The SOB viral assemblages were very different from those in the ETP soils, with the closest similarity between the two sites being 51%. While the SOB viral communities clustered tightly, the RAPD-PCR fingerprints from the ETP site indicated that the soil viral assemblage composition changed dramatically across geographic distances as short as 20 m. The viral communities in ETP soil sample 0 (ETP-0), adjacent to the edge of Tom's Pond, shared only 26% similarity to those in the upslope ETP-2 and ETP-20 sites, and the viral communities in the ETP-2 and ETP-20 sites shared only 33% similarity in RAPD-PCR banding patterns (Fig. 3A). Thus, it appears that the changing intensities of environmental factors along the ETP transect, such as levels of carbon, moisture, and salinity and the number of hours of sunlight exposure, resulted in substantial changes in the composition of soil viral assemblages within this extreme soil environment.
Fig 3.
RAPD-PCR fingerprints of viral communities in soils from Antarctica, Kellogg Biological Station (KBS), and Delaware. (A) RAPD-PCR fingerprints from Antarctic soils (ETP and SOB series). (B) RAPD-PCR fingerprints from KBS soils (T1, T4, T7, T8, and SF2) and Delaware soils (DE soil) under different land use practices. A percent similarity scale is shown at the top left of each panel. Values at the nodes in the dendrogram are percent similarity values for the clusters. Banding patterns with similarity values of ≥89% are considered identical.
The impact of land use practices on soil viral community composition was evaluated through the use of viral RAPD-PCR fingerprints from experimental treatment plots of the Kellogg Biological Station LTER and a Delaware farm soil under conventional corn cultivation. The greatest differences in viral RAPD-PCR fingerprints (76% similarity) occurred between the KBS treatment plots and Delaware agricultural soil, despite the fact that the treatment used in the KBS T1 plot was similar to the cultivation practices used in the Delaware soil (Fig. 3B). Thus, within this sampling design, geographic distance and differences in edaphic properties appeared to be the greatest contributors to the differences in soil viral communities between the two sites. The samples with the highest similarity (88%) were from KBS plots T1 and T4, which had similar cropping practices, albeit with conventional till and inputs and with organic reduced till and winter cover crops, respectively. Viral assemblages within soils from KBS treatment site T7 (with annually burned early successional community treatment) clustered with those within soils from plots T1 and T4 at 85% similarity. Soil viral assemblages within KBS treatment plot T8 (annually mowed, never tilled, early successional community treatment) did not cluster with those within T7 and instead clustered at 85% with those within plot SF2, a mid- to late successional forest treatment (40 to 60 years postagriculture).
DISCUSSION
Comparative analysis of the microbial community composition across environmental samples is critical for revealing important structure-function relationships within microbial communities that may have emergent impacts at the ecosystem scale. An ideal method for measuring microbial community composition would be capable of identifying all populations comprising a community and enumerating all individuals within each population. No existing microbial community composition method fulfills these ideal requirements; however, for bacterial communities, techniques utilizing the 16S rRNA gene have continued to inch toward this goal and have become the de facto standard for comparative analyses. For viruses, no universally shared genetic marker exists; thus, marker gene approaches to analyzing viral communities are incapable of fully capturing viral diversity. Moreover, because the genetic markers used for viral diversity studies have all been protein-encoding genes, it has been notoriously difficult to design degenerate PCR primer pairs capable of detecting all viruses carrying the gene without spurious amplification of nontarget sequences (5, 36). To a point, shotgun metagenomics approaches have come closer to the ideal requirements of a method for measuring viral community composition; however, these approaches are still expensive (despite the falling costs of sequencing) and do not allow the sample throughput necessary for ecological investigations (49). Furthermore, the short read length of current next-generation sequencing platforms is not well suited to unambiguously identifying viral populations (50, 51). The RAPD-PCR approach fills a methodological gap between more narrowly focused marker gene approaches and broadly focused, low-throughput, expensive shotgun metagenomics approaches for semiquantitative estimation of viral diversity in environmental samples.
Several studies have applied RAPD-PCR to examinations of genotypic changes in virioplankton communities over time and by location in marine ecosystems. Offering greater sensitivity to virioplankton community changes than pulsed-field gel electrophoresis methods (36), RAPD-PCR has demonstrated shifts in viral communities across depth zones in the pelagic ocean (1), over seasonal scales in estuarine ecosystems (36), and most recently, in permanently anoxic freshwater sediments (35). This community fingerprinting approach was also important in demonstrating that lysogenic viral populations in deep-sea hydrothermal vent environments are a less diverse subset of the virioplankton communities found at the vents (4). When coupled with direct sequencing of bands, RAPD-PCR has provided a means for limited exploration of the genetic capabilities and taxonomic identity of viruses within environmental samples (3–5). Like shotgun metagenomic sequences, RAPD-PCR sequences have shown a similar distribution of sequences that are known (i.e., showing homology to a sequence in a known organism), unknown, and novel, with known sequences accounting for less than half of the RAPD-PCR amplicons. This study is the first to demonstrate that RAPD-PCR can be used to fingerprint soil viral communities and subsequently compare the composition of these communities across different soil samples.
Advantages of metagenomically informed RAPD-PCR primer design.
Primer design was a key difference between this study and previous RAPD-PCR studies of viral diversity. This study utilized shotgun viral metagenome sequence data to inform the design of RAPD-PCR primers according to their frequency of occurrence in the environment, whereas earlier RAPD-PCR studies used existing RAPD-PCR primers initially tested for use in genotyping plant (52) or bacterial (53) species. Conventionally, RAPD-PCR primers have been randomly selected without previous knowledge of the target sequence (53–55). While random primer selection can avoid bias, it might not be optimal for analyzing a heterogeneous mixture of template DNA (56). Indeed, virioplankton RAPD-PCR assays using plant or bacterial RAPD-PCR primers produced relatively few bands (∼20) compared to the larger number of bands in the more complex patterns resulting from the use of primers designed using viral shotgun metagenome sequence data (Fig. 1) (5, 36). It was encouraging that decamers showing a greater frequency of occurrence in viral metagenome sequence data tended to provide a greater number of bands in the RAPD-PCR fingerprinting assay (Fig. 1). This phenomenon indicates that decamers that are more common within viral metagenome sequence libraries more frequently prime PCR amplification from environmental viral template DNA. Similar observations have been reported for RAPD primers designed using sequence information from the Arabidopsis thaliana genome (57). It is also notable that different decamers showed different frequencies within metagenomes from different environments (see Table S1 in the supplemental material). In future applications, choosing from a range of primers with different frequencies of occurrence in different environments will enable optimization of RAPD-PCR assays to provide greater or lesser sensitivity in detecting differences between viral communities.
It was also encouraging to see that the RAPD-PCR fingerprint patterns obtained using these new, metagenomically informed primers showed a high degree of reproducibility across gels and PCRs. Past virioplankton studies found that identical templates run under identical reaction conditions and resolved on a single agarose gel produced RAPD-PCR fingerprint patterns showing no more than 80% similarity (5, 36). In contrast, the metagenomically designed HCB-1 decamer used in this study showed greater than 90% similarity in banding patterns for identical templates amplified in two independent PCR master mixes and run on separate gels (Fig. 2). This similarity increased to 99% for identical templates analyzed with a single master mix on the same gel. The improved reproducibility in RAPD-PCR fingerprinting is welcomed, although the source of the improved reproducibility is not known. However, we are certain that adherence to a strict set of experimental conditions is critical to reproducibility. As others have noted and we have observed, even slight changes in the template concentration or annealing temperature can alter the observed banding patterns (58–60). Because of the inherent variability in RAPD fingerprint patterns, we established that patterns with ≥89% similarity should be considered identical.
Initial application of RAPD-PCR fingerprinting to soil viral ecology.
It is known that soil biotic and abiotic properties can change at microscales (61) and that temporal and spatial variations in Antarctic soils are critical in influencing the richness and structure of microbial communities (62). RAPD-PCR fingerprints showed that the heterogeneous conditions among Antarctic dry valley soils influenced the composition of soil viral assemblages in this unique and extreme environment. Past work examining the abundance of viruses and bacteria within Antarctic soils found that the ratio of viruses to bacteria increased as water and organic matter content decreased when moving upslope from the edge of ephemeral dry valley lakes (37). The proportion of inducible lysogenic cells also increased along the upgradient transects; however, this trend did not correlate with any measured soil edaphic factors (37). RAPD-PCR fingerprint analyses corroborate these earlier observations by showing that the composition of viral assemblages changes when moving upgradient along the ETP and SOB transects. Whether these changes in soil viral assemblages were related to an increasing predominance of temperate phages is not known; however, future work using sequence data from RAPD-PCR bands (3–5) or shotgun metagenomic surveys could provide data on whether the relative prevalence of temperate phages changes with edaphic factors within dry valley soils.
Differences in land use practices can result in changes in bacterial communities that are likely connected to changes in soil properties. For example, slash-and-burn agricultural practices have been shown to cause shifts in bacterial community structure that are linked with ash nutrient inputs (63). Agricultural land use leads to increases in methane-oxidizing bacteria, sulfate-reducing bacteria, and the diversity of denitrifying bacteria (64, 65). RAPD-PCR analysis of viral communities across different land use treatments at the Kellogg Biological Station indicated that shifts in viral communities also occur with changes in land use practices.
Previous microbial community analysis of KBS agricultural plots T1, T4, and T7 showed that they had remarkably similar bacterial community structures (66). Although crop cultivation within plot T7 (an annually burned, early successional plant community) ceased in 1989, it shared a similar bacterial community structure with the agricultural plots (T1 and T4). Our RAPD-PCR analyses found that the viral assemblages in plots T1, T4, and T7 were the most similar to one another (at least 85% similar), in agreement with earlier observations of the bacterial community structure in these land use plots (Fig. 3B). In contrast, the soil viral communities present in KBS plots SF2 and T8, which support stable successional plant communities, were more distant from the viral communities present in plots T1, T4, and T7. Earlier work in SF2 and T8 found that these plots supported microbial communities different from those seen in T1, T4, and T7 (67). Despite their close geographic proximity, the cultivated (T1, T4, and T7) and uncultivated (SF2 and T8) plots showed differences in soil biogeochemical properties that ultimately appear to influence soil microbial communities (67). It is known that cultivation can perturb common soil properties like pH, C/N ratio, organic matter content, exchangeable cation content, nutrient availability, electrical conductivity, and bulk density, all of which can influence the microbial community structure (64). Once a soil microbial community is disturbed by cultivation (68), it takes many years after cultivation ceases for soil properties and, therefore, microbial communities to return to their original state (69). This scenario likely explains why the viral assemblages seen in plot SF2 (40 to 60 years postagriculture) are similar to those seen in uncultivated plot T8 and more distant from those seen in the T1, T4, and T7 cultivated plots.
Overall, these data indicate that RAPD-PCR fingerprinting can capture changes in the composition of soil viral communities that occur with environmental features known to affect the composition of host microbial communities. Our initial applications also demonstrate the utility of RAPD-PCR fingerprinting as an inexpensive and high-throughput tool for comparative analyses of viral assemblages across soil environments and a diverse range of soil types. More broadly interpreted, these data support the idea that viruses are active within soil ecosystems and capable of influencing the activity and population ecology of coexisting microbial host communities.
Supplementary Material
ACKNOWLEDGMENTS
This project was supported by National Research Initiative competitive grant no. 2005-35107-15214 and 2007-35319-18432 from the USDA Cooperative State Research, Education, and Extension Service. Support for this research was also provided by the NSF Long-Term Ecological Research Program at the Kellogg Biological Station and by the Michigan Agricultural Experiment Station.
Footnotes
Published ahead of print 21 June 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00268-13.
REFERENCES
- 1.De Corte D, Sintes E, Winter C, Yokokawa T, Reinthaler T, Herndl GJ. 2010. Links between viral and prokaryotic communities throughout the water column in the (sub)tropical Atlantic Ocean. ISME J. 4:1431–1442 [DOI] [PubMed] [Google Scholar]
- 2.Field CB, Behrenfeld MJ, Randerson JT, Falkowski P. 1998. Primary production of the biosphere: integrating terrestrial and oceanic components. Science 281:237–240 [DOI] [PubMed] [Google Scholar]
- 3.Helton RR, Wommack KE. 2009. Seasonal dynamics and metagenomic characterization of estuarine viriobenthos assemblages by randomly amplified polymorphic DNA PCR. Appl. Environ. Microbiol. 75:2259–2265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Williamson SJ, Cary SC, Williamson KE, Helton RR, Bench SR, Winget D, Wommack KE. 2008. Lysogenic virus-host interactions predominate at deep-sea diffuse-flow hydrothermal vents. ISME J. 2:1112–1121 [DOI] [PubMed] [Google Scholar]
- 5.Winget DM, Wommack KE. 2008. Randomly amplified polymorphic DNA PCR as a tool for assessment of marine viral richness. Appl. Environ. Microbiol. 74:2612–2618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ceja-Navarro JA, Rivera-Orduna FN, Patino-Zuniga L, Vila-Sanjurjo A, Crossa J, Govaerts B, Dendooven L. 2010. Phylogenetic and multivariate analyses to determine the effects of different tillage and residue management practices on soil bacterial communities. Appl. Environ. Microbiol. 76:3685–3691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Uroz S, Calvaruso C, Turpault M-P, Frey-Klett P. 2009. Mineral weathering by bacteria: ecology, actors and mechanisms. Trends Microbiol. 17:378–387 [DOI] [PubMed] [Google Scholar]
- 8.Williams HTP, Lenton TM. 2008. Environmental regulation in a network of simulated microbial ecosystems. Proc. Natl. Acad. Sci. U. S. A. 105:10432–10437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conrad R. 1996. Soil microorganisms as controllers of atmospheric trace gases (H2, CO, CH4, OCS, N2O, and NO). Microbiol. Rev. 60:609–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Falkowski PG, Fenchel T, Delong EF. 2008. The microbial engines that drive Earth's biogeochemical cycles. Science 320:1034–1039 [DOI] [PubMed] [Google Scholar]
- 11.McKinlay JB, Harwood CS. 2010. Carbon dioxide fixation as a central redox cofactor recycling mechanism in bacteria. Proc. Natl. Acad. Sci. U. S. A. 107:11669–11675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richardson D, Felgate H, Watmough N, Thomson A, Baggs E. 2009. Mitigating release of the potent greenhouse gas N2O from the nitrogen cycle—could enzymic regulation hold the key? Trends Biotechnol. 27:388–397 [DOI] [PubMed] [Google Scholar]
- 13.Elkoca E, Kantar F, Sahin F. 2008. Influence of nitrogen fixing and phosphorus solubilizing bacteria on the nodulation, plant growth, and yield of chickpea. J. Plant Nutr. 31:157–171 [Google Scholar]
- 14.Sahin F, Cakmakci R, Kantar F. 2004. Sugar beet and barley yields in relation to inoculation with N2-fixing and phosphate solubilizing bacteria. Plant Soil 265:123–129 [Google Scholar]
- 15.Alisi C, Musella R, Tasso F, Ubaldi C, Manzo S, Cremisini C, Sprocati AR. 2009. Bioremediation of diesel oil in a co-contaminated soil by bioaugmentation with a microbial formula tailored with native strains selected for heavy metals resistance. Sci. Total Environ. 407:3024–3032 [DOI] [PubMed] [Google Scholar]
- 16.N′Guessan AL, Elifantz H, Nevin KP, Mouser PJ, Methe B, Lwoodard T, Manley K, Williams KH, Wilkins MJ, Larsen JT, Long PE, Lovley DR. 2010. Molecular analysis of phosphate limitation in Geobacteraceae during the bioremediation of a uranium-contaminated aquifer. ISME J. 4:253–266 [DOI] [PubMed] [Google Scholar]
- 17.Vazquez S, Nogales B, Ruberto L, Hernandez E, Christie-Oleza J, Lo Balbo A, Bosch R, Lalucat J, MacCormack W. 2009. Bacterial community dynamics during bioremediation of diesel oil-contaminated Antarctic soil. Microb. Ecol. 57:598–610 [DOI] [PubMed] [Google Scholar]
- 18.Jousset A, Rochat L, Pechy-Tarr M, Keel C, Scheu S, Bonkowski M. 2009. Predators promote defence of rhizosphere bacterial populations by selective feeding on non-toxic cheaters. ISME J. 3:666–674 [DOI] [PubMed] [Google Scholar]
- 19.Lambert C, Chang C-Y, Capeness MJ, Sockett RE. 2010. The first bite—profiling the predatosome in the bacterial pathogen Bdellovibrio. PLoS One 5:e8599. 10.1371/journal.pone.0008599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pearson H. 2008. ‘Virophage' suggests viruses are alive. Nature 454:677. [DOI] [PubMed] [Google Scholar]
- 21.Rosenberg K, Bertaux J, Krome K, Hartmann A, Scheu S, Bonkowski M. 2009. Soil amoebae rapidly change bacterial community composition in the rhizosphere of Arabidopsis thaliana. ISME J. 3:675–684 [DOI] [PubMed] [Google Scholar]
- 22.Vos M, Birkett PJ, Birch E, Griffiths RI, Buckling A. 2009. Local adaptation of bacteriophages to their bacterial hosts in soil. Science 325:833. [DOI] [PubMed] [Google Scholar]
- 23.Buckling A, Rainey PB. 2002. The role of parasites in sympatric and allopatric host diversification. Nature 420:496–499 [DOI] [PubMed] [Google Scholar]
- 24.Allen B, Willner D, Oechel WC, Lipson D. 2010. Top-down control of microbial activity and biomass in an Arctic soil ecosystem. Environ. Microbiol. 12:642–648 [DOI] [PubMed] [Google Scholar]
- 25.Fierer N, Breitbart M, Nulton J, Salamon P, Lozupone C, Jones R, Robeson M, Edwards RA, Felts B, Rayhawk S, Knight R, Rohwer F, Jackson RB. 2007. Metagenomic and small-subunit rRNA analyses reveal the genetic diversity of bacteria, archaea, fungi, and viruses in soil. Appl. Environ. Microbiol. 73:7059–7066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Srinivasiah S, Bhavsar J, Thapar K, Liles M, Schoenfeld T, Wommack KE. 2008. Phages across the biosphere: contrasts of viruses in soil and aquatic environments. Res. Microbiol. 159:349–357 [DOI] [PubMed] [Google Scholar]
- 27.Hurst CJ, Gerba CP, Cech I. 1980. Effects of environmental variables and soil characteristics on virus survival in soil. Appl. Environ. Microbiol. 40:1067–1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sinclair R, Boone SA, Greenberg D, Keim P, Gerba CP. 2008. Persistence of category A select agents in the environment. Appl. Environ. Microbiol. 74:555–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao B, Zhang H, Zhang J, Jin Y. 2008. Virus adsorption and inactivation in soil as influenced by autochthonous microorganisms and water content. Soil Biol. Biochem. 40:649–659 [Google Scholar]
- 30.Suttle CA. 2005. Viruses in the sea. Nature 437:356–361 [DOI] [PubMed] [Google Scholar]
- 31.Winget DM, Helton RR, Williamson KE, Bench SR, Williamson SJ, Wommack KE. 2011. Repeating patterns of virioplankton production within an estuarine ecosystem. Proc. Natl. Acad. Sci. U. S. A. 108:11506–11511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Winget DM, Wommack KE. 2009. Diel and daily fluctuations in virioplankton production in coastal ecosystems. Environ. Microbiol. 11:2904–2914 [DOI] [PubMed] [Google Scholar]
- 33.Bench SR, Hanson TE, Williamson KE, Ghosh D, Radosovich M, Wang K, Wommack KE. 2007. Metagenomic characterization of Chesapeake Bay virioplankton. Appl. Environ. Microbiol. 73:7629–7641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poorvin L, Rinta-Kanto JM, Hutchins DA, Wilhelm SW. 2004. Viral release of iron and its bioavailability to marine plankton. Limnol. Oceanogr. 49:1734–1741 [Google Scholar]
- 35.Borrel GG, Colombet JJ, Robin AA, Lehours A-CA, Prangishvili DD, Sime-Ngando TT. 2012. Unexpected and novel putative viruses in the sediments of a deep-dark permanently anoxic freshwater habitat. ISME J. 6:2119–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jamindar S, Polson SW, Srinivasiah S, Waidner L. 2012. Evaluation of two approaches for assessing the genetic similarity of virioplankton populations as defined by genome size. Appl. Environ. Microbiol. 78:8773–8783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Williamson KE, Radosevich M, Smith DW, Wommack KE. 2007. Incidence of lysogeny within temperate and extreme soil environments. Environ. Microbiol. 9:2563–2574 [DOI] [PubMed] [Google Scholar]
- 38.Williamson KE, Wommack KE, Radosevich M. 2003. Sampling natural viral communities from soil for culture-independent analyses. Appl. Environ. Microbiol. 69:6628–6633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Casas V, Rohwer F. 2007. Phage metagenomics. Methods Enzymol. 421:259–268 [DOI] [PubMed] [Google Scholar]
- 40.Clegg CD, Lovell RDL, Hobbs PJ. 2003. The impact of grassland management regime on the community structure of selected bacterial groups in soils. FEMS Microbiol. Ecol. 43:263–270 [DOI] [PubMed] [Google Scholar]
- 41.Wommack KE, Bhavsar J, Polson SW, Chen J, Dumas M, Srinivasiah S, Furman M, Jamindar S, Nasko DJ. 2012. VIROME: a standard operating procedure for analysis of viral metagenome sequences. Stand. Genomic Sci. 6:427–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kubelik AR, Szabo LJ. 1995. High-GC primers are useful in RAPD analysis of fungi. Curr. Genet. 28:384–389 [DOI] [PubMed] [Google Scholar]
- 43.Lim SH, Phua DCY, Tan HTW. 2000. Primer design and optimization for RAPD analysis of Nepenthes. Biol. Plantarum 43:153–155 [Google Scholar]
- 44.Pope WH, Jacobs-Sera D, Russell DA, Peebles CL, Al-Atrache Z, Alcoser TA, Alexander LM, Alfano MB, Alford ST, Amy NE, Anderson MD, Anderson AG, Ang AAS, Ares M, Barber AJ, Barker LP, Barrett JM, Barshop WD, Bauerle CM, Bayles IM, Belfield KL, Best AA, Borjon A, Bowman CA, Boyer CA, Bradley KW, Bradley VA, Broadway LN, Budwal K, Busby KN, Campbell IW, Campbell AM, Carey A, Caruso SM, Chew RD, Cockburn CL, Cohen LB, Corajod JM, Cresawn SG, Davis KR, Deng L, Denver DR, Dixon BR, Ekram S, Elgin SCR, Engelsen AE, English BEV, Erb ML, Estrada C, Filliger LZ, et al. 2011. Expanding the diversity of mycobacteriophages: insights into genome architecture and evolution. PLoS One 6:e16329. 10.1371/journal.pone.0016329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rychlik W, Spencer WJ, Rhoads RE. 1990. Optimization of the annealing temperature for DNA amplification in vitro. Nucleic Acids Res. 18:6409–6412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Atienzar F, Evenden A, Jha A, Savva D, Depledge M. 2000. Optimized RAPD analysis generates high-quality genomic DNA profiles at high annealing temperature. Biotechniques 28:52–54 [DOI] [PubMed] [Google Scholar]
- 47.Atienzar FA, Jha AN. 2006. The random amplified polymorphic DNA (RAPD) assay and related techniques applied to genotoxicity and carcinogenesis studies: a critical review. Mutat. Res. 613:76–102 [DOI] [PubMed] [Google Scholar]
- 48.Tebbe CC, Vahjen W. 1993. Interference of humic acids and DNA extracted directly from soil in detection and transformation of recombinant DNA from bacteria and a yeast. Appl. Environ. Microbiol. 59:2657–2665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Polson SW, Wilhelm SW, Wommack KE. 2011. Unraveling the viral tapestry (from inside the capsid out). ISME J. 5:165–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wommack KE, Bhavsar J, Ravel J. 2008. Metagenomics: read length matters. Appl. Environ. Microbiol. 74:1453–1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wommack KE, Srinivasiah S, Liles M, Bhavsar J, Bench S, Williamson KE, Polson SW. 2011. Metagenomic contrasts of viruses in soil and aquatic environments, p 25–36 In de BruijnFJ. (ed), Handbook of molecular microbial ecology, vol II. Metagenomics in different habitats. John Wiley & Sons, Inc, New York, NY [Google Scholar]
- 52.Huff DR, Peakall R, Smouse PE. 1993. RAPD variation within and among natural populations of outcrossing buffalograss [Buchloë dactyloides (Nutt.) Engelm.]. Theor. Appl. Genet. 86:927–934 [DOI] [PubMed] [Google Scholar]
- 53.Akopyanz N, Bukanov NO, Westblom TU, Kresovich S, Berg DE. 1992. DNA diversity among clinical isolates of Helicobacter pylori detected by PCR-based RAPD fingerprinting. Nucleic Acids Res. 20:5137–5142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fani R, Damiani G, Diserio C, Gallori E, Grifoni A, Bazzicalupo M. 1993. Use of random amplified polymorphic DNA (RAPD) for generating specific DNA probes for microorganisms. Mol. Ecol. 2:243–250 [DOI] [PubMed] [Google Scholar]
- 55.Tabacchioni S, Visca P, Chiarini L, Bevivino A, Diserio C, Fancelli S, Fani R. 1995. Molecular characterization of rhizosphere and clinical isolates of Burkholderia cepacia. Res. Microbiol. 146:531–542 [DOI] [PubMed] [Google Scholar]
- 56.Di Giovanni GD, Watrud LS, Seidler RJ, Widmer F. 1999. Fingerprinting of mixed bacterial strains and BIOLOG gram-negative (GN) substrate communities by enterobacterial repetitive intergenic consensus sequence-PCR (ERIC-PCR). Curr. Microbiol. 38:217–223 [DOI] [PubMed] [Google Scholar]
- 57.Li JJ, Pei GL, Pang HX, Bilderbeck A, Chen SS, Tao SH. 2006. A new method for RAPD primers selection based on primer bias in nucleotide sequence data. J. Biotechnol. 126:415–423 [DOI] [PubMed] [Google Scholar]
- 58.Cusick SM, O'Sullivan DJ. 2000. Use of a single, triplicate arbitrarily primed-PCR procedure for molecular fingerprinting of lactic acid bacteria. Appl. Environ. Microbiol. 66:2227–2231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Elegado FB, Guerra MA, Macayan RA, Mendoza HA, Lirazan MB. 2004. Spectrum of bacteriocin activity of Lactobacillus plantarum BS and fingerprinting by RAPD-PCR. Int. J. Food Microbiol. 95:11–18 [DOI] [PubMed] [Google Scholar]
- 60.Pérez T, Albornoz J, Domínguez A. 1998. An evaluation of RAPD fragment reproducibility and nature. Mol. Ecol. 7:1347–1357 [DOI] [PubMed] [Google Scholar]
- 61.Ranjard L, Richaume A. 2001. Quantitative and qualitative microscale distribution of bacteria in soil. Res. Microbiol. 152:707–716 [DOI] [PubMed] [Google Scholar]
- 62.Yergeau E, Kowalchuk GA. 2008. Responses of Antarctic soil microbial communities and associated functions to temperature and freeze-thaw cycle frequency. Environ. Microbiol. 10:2223–2235 [DOI] [PubMed] [Google Scholar]
- 63.da C Jesus E, Marsh TL, Tiedje JM, Moreira FMD. 2009. Changes in land use alter the structure of bacterial communities in Western Amazon soils. ISME J. 3:1004–1011 [DOI] [PubMed] [Google Scholar]
- 64.Rahman MH, Okubo A, Sugiyama S, Mayland HF. 2008. Physical, chemical and microbiological properties of an andisol as related to land use and tillage practice. Soil Till. Res. 101:10–19 [Google Scholar]
- 65.Stres B, Mahne I, Avgustin G, Tiedje JM. 2004. Nitrous oxide reductase (nosZ) gene fragments differ between native and cultivated Michigan soils. Appl. Environ. Microbiol. 70:301–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Buckley DH, Schmidt TM. 2001. The structure of microbial communities in soil and the lasting impact of cultivation. Microb. Ecol. 42:11–21 [DOI] [PubMed] [Google Scholar]
- 67.Buckley DH, Schmidt TM. 2003. Diversity and dynamics of microbial communities in soils from agro-ecosystems. Environ. Microbiol. 5:441–452 [DOI] [PubMed] [Google Scholar]
- 68.Cookson WR, Osman M, Marschner P, Abaye DA, Clark I, Murphy DV, Stockdale EA, Watson CA. 2007. Controls on soil nitrogen cycling and microbial community composition across land use and incubation temperature. Soil Biol. Biochem. 39:744–756 [Google Scholar]
- 69.Ndour NYB, Achouak W, Christen R, Heulin T, Brauman A, Chotte J-L. 2008. Characteristics of microbial habitats in a tropical soil subject to different fallow management. Appl. Soil Ecol. 38:51–61 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.