Abstract
The present study was carried out to test the colonic mucosal response of rats to oral supplementation with Lactobacillus fermentum BGHI14 and to correlate the tissue reaction to trinitrobenzenesulfonate (TNBS)-induced colitis with mucosal barrier alterations caused by bacterial ingestion. An immune cell-mediated reaction of healthy colonic tissue was noticed after bacterial feeding. After prolonged bacterial treatment, the observed reaction had retreated to normality, but the mRNA levels of proinflammatory cytokines interleukin-1β (IL-1β) and tumor necrosis factor alpha (TNF-α) remained elevated. These data point to the chronic low-grade inflammation that could be caused by long-term probiotic consumption. Although no detrimental effects of bacterial pretreatment were noticed in colitic rats, at least in the acute state of disease, the results obtained in our study point to the necessity of reassessment of existing data on the safety of probiotic preparations. Additionally, probiotic effects in experimental colitis models might depend on time coordination of disease induction with treatment duration.
INTRODUCTION
Gut microorganisms are considered highly beneficial to the host, and disturbance of gut microflora due to inadequate nutrition or antibiotic treatment has been associated with loss of homeostasis and development of gastrointestinal (GI) pathologies (1, 2). Dietary management of aberrant gut microbiota can be achieved by administration of noncommensal beneficial microbes, commonly named probiotics. Nutritional supplements, usually lactobacilli and bifidobacteria of gastrointestinal origin, have been developed relying on the knowledge of the gut microbiota composition of the healthy host (3–5). The coexistence of a large number of symbionts in close proximity to immunologically “armed” mucosal tissue is explained by luminal sequestration of commensals by the host's barrier mechanisms. Beneficial bacteria interact with pattern recognition receptors (PRR) on epithelial cells and induce innate mucosal responses (6) through direct and indirect stimulation of enterocytes and lamina propria phagocytes. Subsequently, cells initiate mucosal cross talk that is essential for maturation and maintenance of barrier integrity. These interactions are regulated through the release of cytokines, including interleukin-6 (IL-6), transforming growth factor β (TGF-β), and tumor necrosis factor alpha (TNF-α), by epithelial cells and underlying phagocytes. This is how commensals keep the mucosa in an alert state prepared for the eventual pathogen encounter. Although the initial reaction of harmless bacteria would stimulate innate cytokine production by epithelial and phagocytic cells, no disruption of the epithelial barrier occurs (6). Probiotic therapy has often been employed for treatment of intestinal inflammations. Studies using animal models of intestinal inflammation demonstrated that impaired innate immune responses lead to exacerbation of tissue pathology (4, 7). The most commonly studied animal model for bowel inflammation is the trinitrobenzenesulfonate (TNBS)-induced colitis model presented by transmural inflammation of colonic wall after intrarectal TNBS instillation (8). Lactobacillus treatment has proved to be effective in TNBS-induced inflammation (9). Acting as antigenic stimuli, administered lactobacilli can promote a time-varying response in mucosal tissue (10), which should be considered when testing the protective effects of the potential probiotics in established colitis models. The aim of our study was to test the effect of Lactobacillus fermentum BGHI14 on the intensity of the TNBS-invoked mucosal lesion after 14 days of bacterial treatment, which is the most commonly used time point for studying the preventive effects of probiotics in TNBS colitis. To gain comprehensive information on the state of mucosa in the moment of lesion induction, we monitored time changes in healthy mucosa continually exposed to bacteria. Expression of proinflammatory cytokines TNF-α, IL-1β, and IL-17F, as well as that of barrier protective elements, including cytoprotective and intercellular junction proteins, was analyzed.
MATERIALS AND METHODS
Preparation of bacteria.
Lactobacillus fermentum strain BGHI14, isolated from newborn feces of a breast-fed infant 2 weeks after the birth, was used in the study. Bacteria were cultivated in De Man-Rogosa-Sharpe (MRS) medium (Merck GmbH, Darmstadt, Germany) at 37°C anaerobically in an anaerobic jar (Merck) using Anaerocult A (Merck). For the animal treatment, 10 ml of overnight culture (containing in total 2 × 1010 to 3 × 1010 bacteria) was pelleted, washed in saline, and finally resuspended in 1 ml of 11% sterile skimmed milk (AD Mlekara Subotica, Subotica, Serbia), as described by Geier et al. (11).
Animals.
Female Wistar rats of the same litter, 5 to 6 weeks old, were purchased from the Farm of the Military Medical Academy, Belgrade, and for research purposes were housed in the animal facility at the Faculty of Pharmacy, University of Belgrade. After 3 days of adaptation to the experimentation environment, at the onset of the experiment, the animals weighed 140 ± 10 g. Rats were housed in cages, four per room, with a controlled 12-h-light/12-h-dark cycle and had unlimited access to standard rat food and tap water. Cages were maintained clean, and bedding was changed daily. All experimental procedures and protocols conformed to institutional guidelines for the care and use of animals in research no. 2/09 (Ethics Committee of the Faculty of Pharmacy, University of Belgrade). Animal manipulations were approved by the Ethical Committee for Experimentation on Laboratory Animals of the Faculty of Pharmacy, University of Belgrade.
Study design.
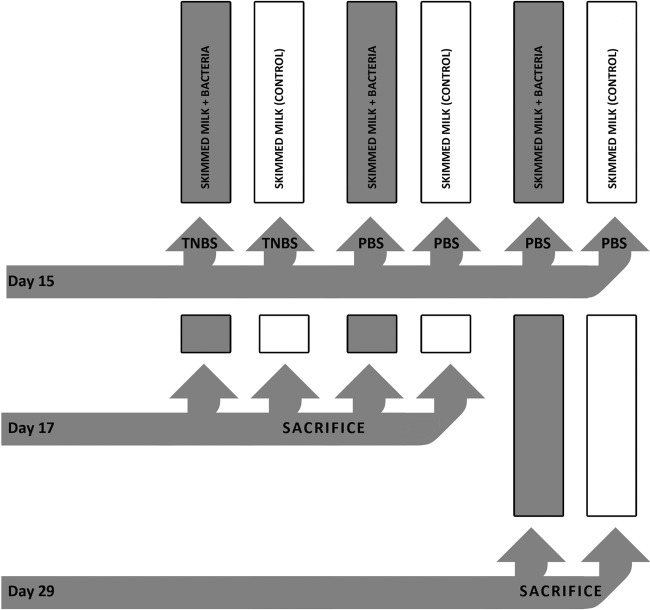
Animals (n = 48) were allocated to six groups (n = 8) (Fig. 1) and gavaged daily using a stainless steel 18-gauge feeding tube (Instech Solomon, Plymouth Meeting, PA). For the first 15 days, rats from three bacterium-treated groups received 1 ml of prepared bacterial suspension in skimmed milk and rats from three control groups received 1 ml of sterile skimmed milk every day. The rat's body weight was recorded on a daily basis throughout the experiment. On day 15, after 36 h of fasting, animals from one bacterium-treated group and one control group were rendered colitic using a 34% solution (vol/vol) of 1 M TNBS (Fluka, Buchs, Switzerland) resuspended in 50% ethanol. Rats were administered 100 ± 10 μl of prepared TNBS solution per 100 g of body weight (12) using a rubber catheter (Ch6; Romed Holland, Wilnis, Netherlands) inserted 6 cm proximally from the anus (13). The other four control groups named as healthy bacterium-treated and healthy control groups, depending on previous treatment, received the same dose of phosphate-buffered saline (PBS) (sham control). The next day (day 16), the rats were treated in the same way. On day 17, 48 h after TNBS or PBS administration, overnight-fasted animals from two colitic groups and from two healthy 16-day groups were sacrificed by increasing the CO2 concentration. The colon from each rat was removed aseptically and opened longitudinally, and the luminal content was washed out with saline. Tissue segments 3 cm proximally from the anus, including all colonic wall layers, were sampled. For histological assessment, tissue was fixed in 10% formalin (14). For mRNA isolation, tissue was immediately frozen at −70°C. Luminal contents from distal intestine of 4 healthy control and 4 healthy bacterium-treated rats were removed and frozen at −70°C for detection of BGHI14 survival in the GI tract. On day 17, for the rats from remaining two groups (the healthy 28-day bacterium-treated group and the healthy 28-day control group), the same treatment continued for next 12 days. On day 29, after overnight fasting, rats were sacrificed, and sampling of colonic tissue was done as described above.
Fig 1.
Study design.
DGGE analysis.
Extraction of bacterial DNA from frozen samples was done using the QIAamp DNA stool minikit (Qiagen, Hilden, Germany). PCR with isolated genomic DNA as a template was set up according to Heilig et al. (15), except for the cycle number, which was decreased to avoid amplification of nonspecific fragments (16). Lactobacillus-specific primer set Lab-0159f and Uni-0515-GCr (Metabion International, Martinsried, Germany) was used. The primer sequences are given in Table 1. Amplification was performed using KAPA Taq DNA polymerase (KAPA Biosystems, Cape Town, South Africa). The reaction was performed in thermal cycler Gene AmpR PCR system 2700 (Applied Biosystems, Foster City, CA). Denaturing gradient gel had been prepared as described by Heilig et al. (15) using glass plates for the denaturing gradient gel electrophoresis (DGGE) apparatus DGGE-2001 (C.B.S. Scientific, San Diego, CA). USAGel manipulation after electrophoresis was performed according to Radojkovic et al. (17).
Table 1.
Primers used in this study
| Primer name | Primer sequence (5′→3′) | Source or reference |
|---|---|---|
| Lab-0159f | GGA AAC AG (A/G) TGC TAA TAC CG | 15 |
| Uni-0515r | ATC GTA TTA CCG CGG CTG CTG GCA | 15 |
| Uni-0515-GCr | ATC GTA TTA CCG CGG CTG CTG GCA CGC CGG GGG CGC GCC CCG GGC GGG GCG GGG GCA CGG GGG G | 15 |
| β-Actin forward | AGC CAT GTA CGT AGC CAT CC | 50 |
| β-Actin reverse | CTC TCA GCT GTG GTGGTG AA | 50 |
| Tjp1 forward | GCA GTG TGA ACA TGG ATT GAA | This work |
| Tjp1 reverse | AGC CAA TGC CTG ACA GTT CT | This work |
| Hsp70 forward | CGC TCC AGG TGT GAT CTA GG | This work |
| Hsp70 reverse | TAC TGG GAA TGC AAA GCA CA | This work |
| IL-17F forward | GGA AAA GCC TCC TTT GAT CC | This work |
| IL-17F reverse | ACG GAG CTT CAA GGA TGT TG | This work |
| IL-1β forward | GGA AGG CAG TGT CAC TCA TTG TG | 51 |
| IL-1β reverse | GGT CCT CAT CCT GGA AGC TCC | 51 |
| TNF-α forward | AAA TGG GCT CCC TCT CAT CAG TTC | 52 |
| TNF-α reverse | TCT GCT TGG TGG TTT GCT ACG AC | 52 |
DGGE fragment sequencing.
Fragments of interest were excised from the gel and macerated, and the suspension was incubated for 10 min at 98°C (18). After incubation, the suspension was centrifuged to pellet gel particles. The supernatant (30 μl) was used in PCR with Lab-0159f and Uni-0515r primers. The obtained PCR products were purified using the QIAquick PCR purification kit (Qiagen) and ligated into the pGEM-T Easy vector system (Promega, Fitchburg, WI) according to the manufacturer's instructions. Ligated constructs were transformed in Ca2+-induced competent DH5α cells (19), and insert-containing transformants were selected as white colonies on Luria agar (LA) plates containing 100 μg ml−1 ampicillin and 20 μg ml−1 X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactoside) as recommended by Promega. For each excised DNA band, one white colony was picked and plasmids were isolated using the QIAprep spin miniprep kit (Qiagen). The sequencing of the isolated insert-containing pGEM-T Easy vectors was done with M13F/R primers at Macrogen Europe Service, Amsterdam, Netherlands (http://dna.macrogen.com/eng/support/seq/seq_uniprimer.jsp).
Histological analysis.
Formalin-fixed colonic tissue was processed for histopathological observation with the protocol adapted from various sources (14, 20, 21). Paraffin blocks were cut on a rotary microtome (RM2125RT, Leica Microsystems, Wetzlar, Germany). Staining of tissue was performed using manually prepared Mayer hematoxylin and eosin (H&E) solutions. Slides were inspected under a Nikon microscope (Nikon Instruments, Inc., Tokyo, Japan). Colon damage was evaluated by pathologist who was blinded to the experimental groups. Scoring was done semiquantitatively on a scale from 0 to 9 modified from Farooq et al. (22), assuming inflammation intensity (score of 0 to 5), crypt architectural changes (score of 0 to 3), and edema presence (score of 0 or 1). Histological sections were photographed using NIS-Elements microscope imaging software 2.3 (Nikon Instruments Inc.).
mRNA isolation.
Extraction of mRNA for quantitative PCR (qPCR) analysis was done according to Chomczynski et al. (23) with slight modifications. Tissue was pulverized in liquid nitrogen and resuspended in denaturing solution (4 M guanidine thiocyanate, 25 mM sodium citrate, 0.1 M β-mercaptoethanol, 0.5% [wt/vol] N-lauroylsarcosinate sodium salt). Two acid phenol (pH 4) extractions were performed. After isopropanol precipitation, the pellet was again resuspended in denaturing solution and the phenol extraction step was repeated. All isolation steps were performed restricted to the precautions for RNA handling. Isolated RNA was stored at −70°C until further manipulations.
Real-time PCR (qPCR).
Reverse transcription was performed according to the protocol provided by the reverse transcriptase manufacturer (Thermo Scientific). Random hexamers (Applied Biosystems) and RiboLock RNase inhibitor (Thermo Scientific) were utilized in the reaction. Controls without reverse transcriptase were included for the genomic DNA contamination check. All reaction steps were performed in the thermal cycler Gene AmpR PCR system 2700 (Applied Biosystems).
Prepared cDNA was amplified in qPCRs using the 7500 real-time PCR system (Applied Biosystems) with the primer sets listed in Table 1. Primers (Invitrogen, Paisley, United Kingdom) were designed utilizing Primer3 software, available online (http://frodo.wi.mit.edu). The reaction and cycling were done as recommended by the manufacturer of the KAPA SYBR Fast universal master mix (KAPA Biosystems). The conditions of the two-step qPCR were set as follows: activation for 3 min at 95°C followed by 40 cycles of 15 s at 95°C and 60 s at 60°C. In relative quantification analysis, a 1/10 dilution of cDNA template was used with sample from the control 16-day treatment group set as a calibrator. Controls without reverse transcriptase were amplified to ensure that DNA contamination in the isolated mRNA is acceptable (24).
Data presentation and statistical analysis.
Results are presented as mean values with standard errors graphically using column bar charts, and differences are reported as statistically significant if P (two tailed) is <0.05. Data normality was evaluated by the Shapiro-Wilk test, and normally distributed data were analyzed using a two-sample Student's t test assuming unequal variances. Nonnormally distributed data were log10 transformed, and if the transformed data were normal, the t test was applied as described above. If the normality could not be achieved by transformation, a nonparametric Mann-Whitney U test was applied on nontransformed results. Statistical analyses were performed using SPSS 12.0 for Windows. Graphs were drawn in Microsoft Office Excel 2007.
RESULTS
Body weight assessment.
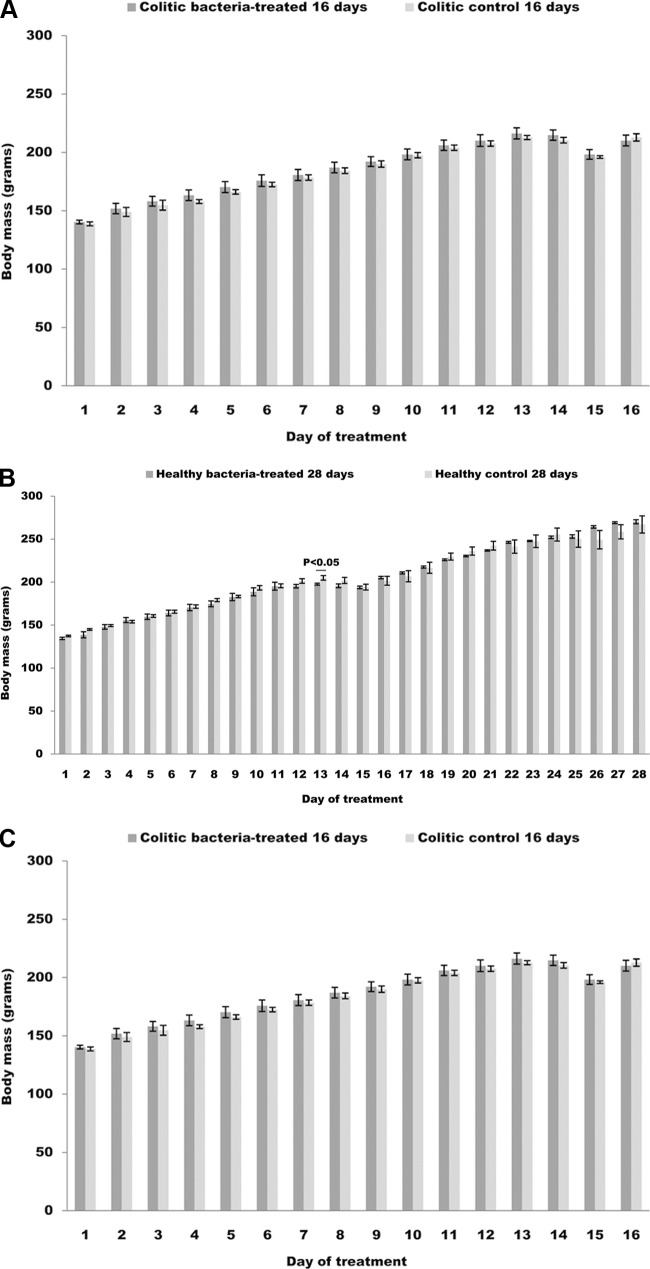
During the treatment, the body weight of rats was assessed on a daily basis. Rats belonging to both 16-day and 28-day healthy control groups showed similar patterns in weight changes during first 16 days of treatment and differed from the patterns obtained for both 16-day and 28-day healthy bacterium-treated rats (Fig. 2A and B). On day 12 from the beginning of the treatment, in the case of healthy 16-day bacterium-treated groups, bacterial treatment resulted in significant decrease (P < 0.05) in the rats' weights compared to those of the controls (Fig. 2A). For rats belonging to the healthy 28-day treatment groups, weight decline due to bacterial treatment reached statistical significance (P < 0.05) on day 12 from the onset of the study (Fig. 2B). No weight differences were detected during the rest of the treatment for all four healthy groups. Rats from the bacterium-treated colitic group didn't present a statistically significant weight difference at any time point compared to colitic control rats (Fig. 2C). According to direct observation of rats in the course of the experiment, bacterial intake led to slight drops in body masses that became most conspicuous ∼2 weeks after the beginning of the treatment.
Fig 2.
Daily body mass changes during the course of the study of bacterium-treated and control rats belonging to (A) healthy 16-day treated groups, (B) healthy 28-day treated groups, and (C) colitic groups. Probability (P) values obtained for group differences at the indicated time points are reported above the charts.
Detection of the BGHI14 bacterial strain.
Ileal content taken from rats after sacrifice was analyzed as an indicator of microflora composition. The DGGE profile of 16S rRNA gene amplicons obtained from pure culture of BGHI14 strain revealed the existence of several copies of the 16S rRNA gene, with one copy staining most intensively on the gel. DGGE analysis of 16S rRNA products obtained from ileal contents of 2 of 4 analyzed bacterium-treated rats revealed the band corresponding to the dominant band obtained with DNA isolated from strain BGHI14 (Fig. 3). Suspected bands as well as the dominant pure culture band were sequenced, and the sequences were queried against the NCBI genome collection. The results confirmed that all eluted bands belonged to Lactobacillus fermentum. No band corresponding to dominant pure BGHI14 culture band was detected in control rats. Along with the demonstration of the ability of BGHI14 to survive in GI tract of the rats, results obtained by DGGE analysis showed no significant shift in the ileal microbial community of the bacterium-treated rats.
Fig 3.
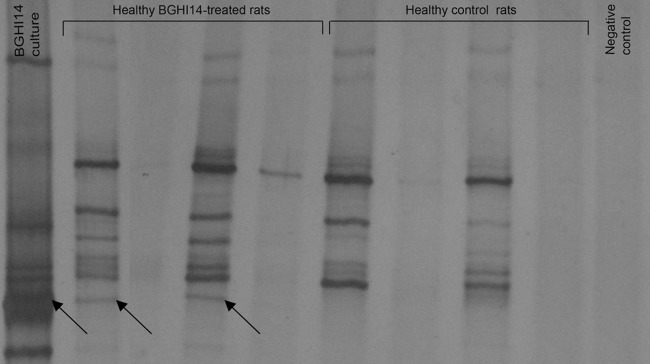
Detection of BGHI14 in GI tract of treated rats. DGGE profiles of lactobacillus-specific 16S rRNA gene amplicons obtained in PCRs with Lab-0159f/Uni-0515r set of primers. Arrows indicate eluted and sequenced bands. Lane 1, BGHI14 pure culture; lanes 2 to 5, ileal contents of four bacterium-treated rats; lanes 6 to 9, ileal contents of four control rats; lane 10, PCR-negative control.
Histological evaluation.
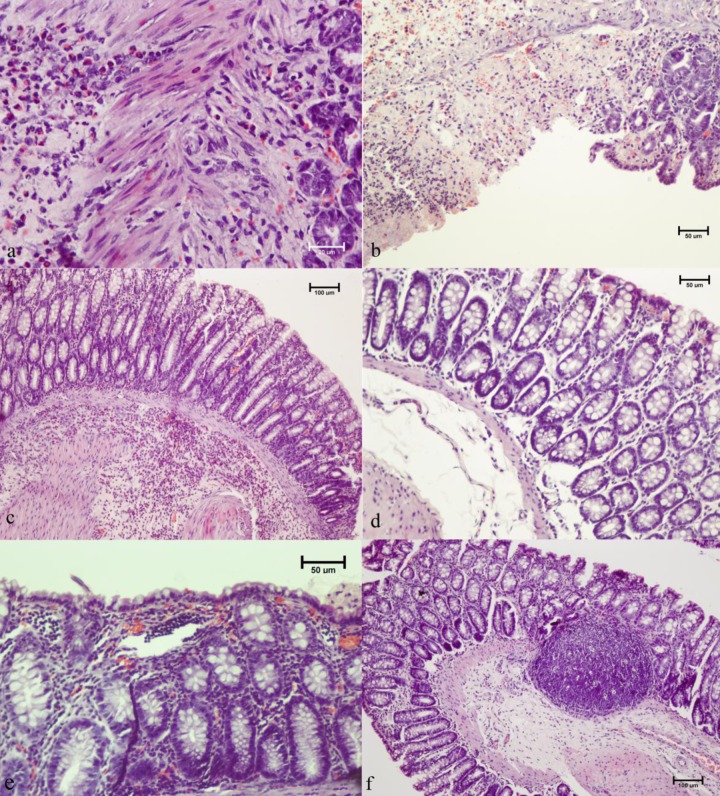
H&E staining was employed for semiquantitative estimation of tissue reaction to bacteria and disease induction. Histological grading of colon tissue, assuming inflammation intensity, crypt distortion, and edema formation (Fig. 4), indicated a significantly higher (P < 0.05) damage score in both colitic bacterium-treated rats and colitic control rats compared to healthy 16-day control rats but not in comparison to healthy 16-day bacterium-treated rats. No differences in total pathohistological scores were observed between the two colitic groups. A significant increase (P < 0.05) in histological score was observed in healthy rats receiving bacteria for 16 days compared to healthy control rats. No edema formation or crypt architectural changes were detected in healthy 16-day bacterium-treated rats (Fig. 5). Although no statistical difference could be reached for healthy 28-day rats relative to 16-day bacterium-treated rats, no difference in pathohistological scores was observed between the healthy 28-day bacterium-treated and control groups. Histological grading pointed to an immune reaction caused by bacteria after the administration in healthy rats but with no deleterious effects of bacterial pretreatment in diseased rats.
Fig 4.
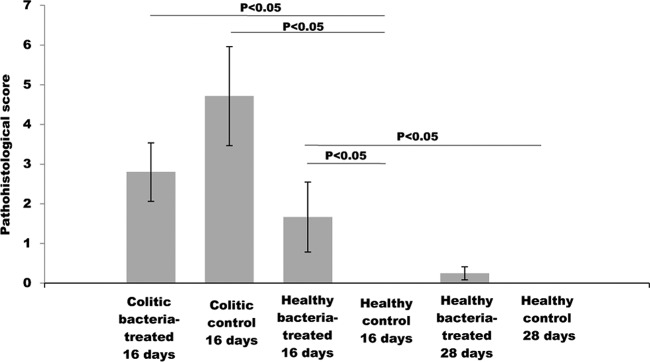
Pathohistological evaluation of the effect of bacterial treatment on colonic tissue morphology after induction of colitis in a defined time point (16 days) and after two different periods (16 and 28 days) of treatment.
Fig 5.
Histopathology of rat colonic tissue. (a) Cross-section of colonic tissue of bacteria-treated colitic rats exhibiting large submucosal infiltrates of polymorphonuclear cells. (b) Colitic control rats presenting a high damage score with crypt distortion and massive neutrophil infiltration. (c) Healthy 16-day bacterium-treated rats presenting diffuse submucosal accumulation of mononuclear cells, eosinophils, and occasional neutrophils. (d) Morphology in healthy 16-day rats. (e) Occasional presence of mucosal and submucosal lymphocyte aggregates was noticed in healthy 28-day bacterium-treated rats. (f) Twenty-eight-day control rats. Magnifications: a, ×600; b, ×200; c, ×100; d, ×200; e, ×200; f, ×100.
Mucosal gene expression.
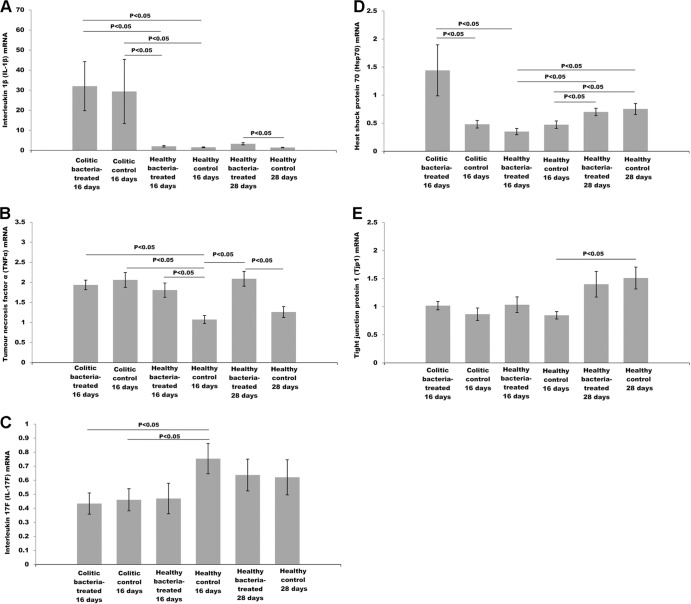
Expression of inflammatory cytokines as well as tissue cytoprotective and barrier maintenance factors was analyzed at the mRNA level. Real-time PCR (qPCR) analysis of IL-1β transcript levels (Fig. 6A) indicated that TNBS administration induced significantly higher IL-1β expression than PBS administration (P < 0.05). There was no difference in levels of IL-1β mRNA induction between bacterium-treated and control colitic rats. Nevertheless, healthy rats treated with bacteria showed different tendencies in IL-1β transcription, depending on the period of bacterial treatment. Sixteen days of bacterial treatment did not have any influence on IL-1β expression in healthy rats. Nevertheless, a 4-week bacterial treatment of healthy rats resulted in a significant increase in IL-1β mRNA synthesis relative to that of the control (P < 0.05).
Fig 6.
Effect of BHGI14 on expression of (A) IL-1β, (B) TNF-α, (C) IL-17F, (D) Hsp70, and (E) Tjp1 mRNA in rats' colonic tissue.
Colitis induction upregulated TNF-α mRNA synthesis in diseased rats relative to the healthy control group (P < 0.05) (Fig. 6B). Bacterial feeding had no influence on TNF-α transcription 48 h after colitis induction. Nevertheless, healthy 16-day as well as 28-day bacterium-treated rats presented significantly higher TNF-α mRNA expression than their control groups (P < 0.05).
TNBS administration significantly decreased IL-17F mRNA expression compared to that in healthy control rats (P < 0.05) (Fig. 6C). No difference was detected in IL-17F mRNA levels between colitic bacterium-treated and colitic control rats. Similarly, BGHI14 did not influence IL-17F mRNA transcription in healthy rats. Bacterial pretreatment resulted in significantly higher heat shock protein 70 (Hsp70) mRNA transcription 48 h after colitis induction compared to that in the colitic control group as well as the healthy 16-day bacterium-treated group (P < 0.5) (Fig. 6D). No such difference was detected relative to the healthy 16-day control group. Also, healthy 28-day bacterium-treated rats as well as 28-day healthy control rats presented significantly higher Hsp70 transcript levels than the 16-day corresponding groups (P < 0.05), but no effect of bacterial treatment in healthy rats was observed.
Colitis induction had no influence on the Tjp1 (tight junction protein 1) mRNA transcription (Fig. 6E). Also, BGHI14 treatment of healthy and colitic rats did not induce Tjp1 mRNA transcription. Rats from the 28-day control group exhibited significantly higher Tjp1 mRNA expression than those from the 16-day control group (P < 0.05).
RNA messenger levels point to the induction of key proinflammatory cytokines in healthy rats exposed to BGHI14, although not to the level detected in diseased rats. Additionally, induction of cytoprotective Hsp70 was noticed in colitic bacterium-treated rats.
DISCUSSION
Enrichment of gut microbiota with nonpathogenic bacteria strengthens the host's GI barrier actively shaping mucosal functions (25). Most commonly tested potential probiotics are lactobacillus strains that possess the plethora of highly desirable properties related to host health. Lactobacillus fermentum has long been studied for its probiotic properties because of its human origin, adherence capacity in GI tract, as well as protection in animal models of bowel inflammation (26, 27). Experimental data also support its role in mucosal barrier fortification (28). Also, clinical trials including L. fermentum have been undertaken with promising results (27).
Due to vigorous conditions along the proximal intestine, the first site of physiologically relevant (29) bacterial colonization is the ileum (30), with lactobacilli being among most abundant residents of this part of the gut (31). It is assumed that bacterial DNA detection by DGGE analysis of ileal luminal content is an indicator of GI passage survival because the DNA of dead bacteria would have been degraded by nucleases released in duodenal lumen (32). Results obtained in our study indicated that L. fermentum BGHI14 successfully survives passage through the proximal part of the digestive tract. Moreover, bacterium-treated rats displayed a microflora profile that was similar to that of control rats. Probiotics should not lead to perturbation of gut microflora of the healthy host, as this could be an indicator of the invasive potential of ingested microbes (33).
The immune reaction detected at a histological level in rats exposed to L. fermentum BGHI14 for 16 days is an indicator of the inflammatory response of the mucosal barrier to ingested bacteria. Similarly, inflammatory cell infiltration has been observed in lactobacillus-free mice inoculated with the closely related species Lactobacillus reuteri (34). The large dose of lactobacillus given to rats (∼1010 bacteria/animal/day) might present an alarming signal to mucosal defense system, which in turn elicits effective innate mechanisms to fight the unknown bacteria. This could be taken as an indicator of the potential risk associated with probiotic consumption in immunodeficient patients (33). According to the rats' weight changes, the peak of immune response to ingested bacteria might have occurred several days earlier. Although the treatment with bacteria was continued for next 12 days, symptoms of chronic inflammation were not observed, as evident from histological data. Moreover, body weights of 28-day bacterium-treated rats reached values similar to those of control rats.
Although the reaction of the healthy rat's mucosa to lactobacilli had retreated in spite of continued bacterial treatment, it can be considered that, potentially, BGHI14 reached into deeper layers of mucosal tissue. The reaction of the healthy host to lactobacillus treatment could be individual, and particular precautions should be taken in the case of the immunocompromised host.
Stimulation of proinflammatory cytokines has been observed after oral administration of lactobacilli, with cytokine production ascribed to innate immune cells, including macrophages and dendritic cells (35). Treatment of healthy animals with BGHI14 increased IL-1β and TNF-α expression in healthy bacterium-treated animals after 28 and 14 days, respectively. Both proinflammatory cytokine levels remained increased, although the initial innate immune cell-mediated reaction had subsided, judging by the tissue sections. Both beneficial and detrimental roles for TNF-α have been described using animal models of inflammation, depending on the dose applied (36). A low TNF-α dose suppressed the infection, while a high TNF-α dose, although lessening the infection, was followed by serious inflammatory reaction. However, chronic low-dose TNF-α stimulation could lead to cancerous cell growth (37). Contrary to TNF-α, the primary regulation of which resides at the transcriptional level, increased IL-1β mRNA levels indicate accumulation of inactive IL-1β precursor, which presents a danger in the case of inflammasome assembly leading potentially to extensive inflammation (38). This issue should be considered in cases of long-term lactobacillus supplementation.
Synthesis of IL-17F-type mRNA has been detected in colonic epithelial cells, indicating its function in mucosal barrier fortification (39). It is possible that IL-17F serves as a first frontier of the mucosal surfaces that is prepared to alarm the recruitment of innate defense army in the case of stress or injury.
Although numerous studies reported TNBS-mediated induction of the IL-17 cytokine (40), the results of our research revealed downregulation of IL-17F mRNA 48 h after TNBS injury. One of the possible explanations was suggested by Ruyssers et al. (41), where IL-17 mRNA transcription as an early response to stress stimuli is counterregulated by the protective host response mounted in later phases of the acute response.
Data available from the literature pointed to the strain-dependent effects of lactobacilli in acute inflammatory phase of TNBS-induced colitis (42). Although the protective role of TNF-α has been demonstrated for this model (43), the observed protective abilities of lactobacilli were attributed to their anti-inflammatory actions in the GI tract (9). There are no published data that would associate the time-varying reaction of colonic tissue to ingested bacteria with the inflammation induced after TNBS damage in a defined time point with respect to the length of bacterial treatment. Chemical damage by TNBS in our study was inflicted in the moment that, according to histological and cytokine-level evaluations, corresponded to the immune reaction of colonic tissue to BGHI14. Since TNBS induces mucosal damage that is perpetuated by superinfection with luminal bacteria (44), it might be expected that the presence of virulent microbes would substantially exacerbate the disease score. However, no adverse effects or amelioration of acute inflammatory pathology by BGHI14 treatment was observed.
The timing of TNBS administration relative to the barrier breaching moment as well as the dose instilled are variables influencing the outcome of lactobacillus pretreatment in examined disease model. Depending on the TNBS dose applied, protection might not be achieved in an early inflammation phase but with resolution in late acute or chronic phases of disease (45). A study was published in which the aggravation of colitis was observed in the first week of postinduction treatment with tested bacteria, and at the later time point, a protective effect of probiotic administration was observed (26). This implies that the inflammatory reaction of the colonic mucosa to ingested lactobacilli might occur in the first few days of treatment, as observed in our study.
Heat shock proteins (HSP) are crucial for cellular resistance to environmental stress, and the expression of Hsp70 is increased at colonic mucosa surfaces due to continuous exposure to luminal stressors. Accordingly, in the case of bowel inflammation, Hsp70 expression is considered to play an important role in GI tract barrier maintenance (46). Induction of heat shock proteins by probiotics, including lactobacilli, in injured GI mucosa has been reported, and these findings were substantiated by in vitro systems using intestinal epithelial cells (47). Nevertheless, decreased Hsp70 expression with a posttranscriptional level of regulation was observed in inflamed bowel tissue (48). In the present study, we demonstrated the upregulation of Hsp70 mRNA in colitic rats treated with BGHI14, which is in agreement with the effects described for nonpathogenic bacteria on cytoprotective protein induction (49). Moreover, our results showed no change in Hsp70 transcription after colitis induction. As mentioned above, posttranscriptional regulation might be involved in the acute phase of TNBS-caused inflammation. Moreover, elevated Hsp70 mRNA synthesis was observed in 28-day healthy control and bacterium-treated groups, which could be attributed to milk's effects on the increase of overall microbial count in the rat's colon, as reported previously (32).
Tight junction proteins (Tjp) are key stabilizers of the epithelial barrier, lactobacilli are well known to influence that function, and most studies described posttranscriptional regulation of Tjp expression and localization in intestinal enterocytes (47). However, in our study, healthy rats from the 28-day control group presented higher levels of Tjp1 mRNA than 16-day treatment control rats. Aside from potential prebiotic milk effects (32), this could also be the result of epithelial “maturation” due to the progression from the preadult to adult developmental stage of the animals. Thus, the effects of lactobacilli could be more pronounced in less “mature” mucosa, which has an implication for age-related probiotic interventions.
In this study, we have shown that the cellular immune response of healthy rat mucosa is evident after 16 days of lactobacillus treatment. Although the observed reaction to bacteria subsided after the prolonged treatment, the obtained results imply that ingested lactobacilli probably came into close contact with the immune cells of the lamina propria. Although bacterial prophylaxis did not lead to exacerbation of colitis pathology in the acute phase, bacterial administration should not be considered innocuous to the healthy host. Having the above facts, if eventual protection of probiotics in experimental colitis models is to be achieved, strategies should be developed to consider probiotic safety and the “vulnerability” of the mucosal barrier at the moment the chemical injury is inflicted.
ACKNOWLEDGMENTS
Jovanka Lukic is grateful to Petar Milosavljevic from the Military Medical Academy, Belgrade, Serbia, for photographing and inspecting the histological sections.
The Ministry of Education, Science and Technological Development of the Republic of Serbia supported this work (grant no. 173019).
The authors of this article declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Published ahead of print 12 July 2013
REFERENCES
- 1.Wlodarska M, Willing B, Keeney KM, Menendez A, Bergstrom KS, Gill N, Russell SL, Vallance BA, Finlay BB. 2011. Antibiotic treatment alters the colonic mucus layer and predisposes the host to exacerbated Citrobacter rodentium-induced colitis. Infect. Immun. 79:1536–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoffmann KM, Deutschmann A, Weitzer C, Joainig M, Zechner E, Högenauer C, Hauer AC. 2010. Antibiotic-associated hemorrhagic colitis caused by cytotoxin-producing Klebsiella oxytoca. Pediatrics 125:e960–e963 [DOI] [PubMed] [Google Scholar]
- 3.Bezkorovainy A. 2001. Probiotics: determinants of survival and growth in the gut. Am. J. Clin. Nutr. 73(Suppl 2):399S–405S [DOI] [PubMed] [Google Scholar]
- 4.Rescigno M. 2011. The intestinal epithelial barrier in the control of homeostasis and immunity. Trends Immunol. 32:256–264 [DOI] [PubMed] [Google Scholar]
- 5.Collado MC, Bäuerl C, Pérez-Martínez G. 2012. Defining microbiota for developing new probiotics. Microb. Ecol. Health Dis. 23:18579 http://dx.doi.org/10.3402/mehd.v23i0.18579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Galdeano CM, De Moreno de LeBlank A, Vinderola G, Bibas Bonet ME, Perdigón G. 2007. Proposed model: mechanisms of immunomodulation induced by probiotic bacteria. Clin. Vaccine Immunol. 14:485–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corridoni D, Pastorelli L, Mattioli B, Locovei S, Ishikawa D, Arseneau KO, Chieppa M, Cominelli F, Pizarro T. 2012. Probiotic bacteria regulate intestinal epithelial permeability in experimental ileitis by a TNF-dependent mechanism. PLoS One 7:e42067. 10.1371/journal.pone.0042067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ardite E, Sans M, Panés J, Romero FJ, Piqué JM, Fernández-Checa JC. 2000. Replenishment of glutathione levels improves mucosal function in experimental acute colitis. Lab. Invest. 80:735–744 [DOI] [PubMed] [Google Scholar]
- 9.Peran L, Camuesco D, Comalada M, Bailon E, Henriksson A, Xaus J, Zarzuelo A, Galvez J. 2007. A comparative study of the preventive effects exerted by three probiotics, Bifidobacterium lactis, Lactobacillus casei and Lactobacillus acidophilus, in the TNBS model of rat colitis. J. Appl. Microbiol. 103:836–844 [DOI] [PubMed] [Google Scholar]
- 10.Khurana I. 2006. Textbook of medical physiology. Elsevier, Noida, India [Google Scholar]
- 11.Geier MS, Butler RN, Giffard PM, Howarth GS. 2007. Lactobacillus fermentum BR11, a potential new probiotic, alleviates symptoms of colitis induced by dextran sulphate sodium (DSS) in rats. Int. J. Food Microbiol. 114:267–274 [DOI] [PubMed] [Google Scholar]
- 12.Yokoyama K, Nakajima N, Ito Y, Iwasaki A, Arakawa Y. 2006. Histoimmunological evaluation for the efficacy of entero nutrient containing n-3 fatty acids in TNBS rat colitis model. J. Clin. Biochem. Nutr. 39:88–97 [Google Scholar]
- 13.Zheng L, Gao ZQ, Wang SX. 2000. A chronic ulcerative colitis model in rats. World J. Gastroenterol. 6:150–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marjit B. 2009. General anatomy, genetics, histology and embryology, 5th ed. Bimar Kumal Dhur of Academic Publishers, Kolkata, India [Google Scholar]
- 15.Heilig HGHJ, Zoetendal EG, Vaughan EE, Marteau P, Akkermans ADL, de Vos WDL. 2002. Molecular diversity of Lactobacillus spp. and other lactic acid bacteria in the human intestine by specific amplification of 16S ribosomal DNA. Appl. Environ. Microbiol. 68:114–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park JW, Crowley DE. 2010. Nested PCR bias: a case study of Pseudomonas spp. in soil microcosms. J. Environ. Monit. 12:985–988 [DOI] [PubMed] [Google Scholar]
- 17.Radojkovic D, Kusic J. 2000. Silver staining of denaturing gradient gel electrophoresis gels. Clin. Chem. 46:883–884 [PubMed] [Google Scholar]
- 18.An C, Takahashi H, Kimura B, Kuda T. 2011. Comparison of PCR-DGGE and PCR-SSCP analysis for microflora of kaburazushi and daikonzushi, traditional fermented foods made from fish and vegetables. J. Food Technol. 9:1–8 [Google Scholar]
- 19.Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557–580 [DOI] [PubMed] [Google Scholar]
- 20.Dietrich D, Krieger H. 2009. Histological analysis of endocrine disruptive effects in small laboratory fish. John Wiley & Sons, Hoboken, NJ [Google Scholar]
- 21.Roberts RJ, Smail DA, Munro ES. 2012. Laboratory methods, p 439–481 In Roberts RJ. (ed), Fish pathology. Wiley-Blackwell, Hoboken, NJ [Google Scholar]
- 22.Farooq SM, Stillie R, Svensson M, Svanborg C, Strieter RM, Stadnyk AW. 2009. Therapeutic effect of blocking CXCR2 on neutrophil recruitment and dextran sodium sulphate-induced colitis. J. Pharmacol. Exp. Ther. 329:123–129 [DOI] [PubMed] [Google Scholar]
- 23.Chomczynski P, Sacchi N. 2006. The single-step method of RNA isolation by acid guanidium thiocyanate-phenol chloroform extraction: twenty-something years on. Nat. Protoc. 1:581–585 [DOI] [PubMed] [Google Scholar]
- 24.Cheeseman JF, Winnebeck EC, Millar CD, Kirkland LS, Sleigh J, Goodwin M, Pawley MDM, Bloch G, Lehmann K, Menzel R, Warman GR. 2012. General anesthesia alters time perception by phase shifting the circadian clock. Proc. Natl. Acad. Sci. U. S. A. 109:7061–7066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ng SC, Hart AL, Kamm MA, Stagg AJ, Knight SC. 2009. Mechanisms of action of probiotics: recent advances. Inflamm. Bowel Dis. 15:300–310 [DOI] [PubMed] [Google Scholar]
- 26.Mañé J, Lorén V, Pedrosa E, Ojanguren I, Xaus J, Cabré E, Doménech E, Gassull MA. 2009. Lactobacillus fermentum CECT 5716 prevents and reverts intestinal damage on TNBS-induced colitis in mice. Inflamm. Bowel Dis. 15:1155–1163 [DOI] [PubMed] [Google Scholar]
- 27.Jiménez E, Langa S, Martín V, Arroyo R, Martín R, Fernández L, Rodríguez JM. 2010. Complete genome sequence of Lactobacillus fermentum CECT 5716, a probiotic strain isolated from human milk. J. Bacteriol. 192:4800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schlee M, Harder J, Köten B, Stange EF, Wehkamp J, Fellermann K. 2008. Probiotic lactobacilli and VSL#3 induce enterocyte β-defensin 2. Clin. Exp. Immunol. 151:528–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kosin B, Rakshit SK. 2006. Microbial and processing criteria for production of probiotics: a review. Food Technol. Biotechnol. 44:371–379 [Google Scholar]
- 30.Isolauri E, Sütas Y, Kankaanpää P, Arvilommi H, Salminen S. 2001. Probiotics: effects on immunity. Am. J. Clin. Nutr. 73(Suppl):444S–450S [DOI] [PubMed] [Google Scholar]
- 31.Hayashu H, Takahashi R, Nishi T, Sakamoto M, Benno Y. 2005. Molecular analysis of jejunal, ileal, caecal and recto-sigmoidal human colonic microbiota using 16S rRNA gene libraries and terminal restriction fragment length polymorphism. J. Med. Microbiol. 54:1093–1101 [DOI] [PubMed] [Google Scholar]
- 32.Bertazzoni-Minelli E, Benini A, Marzotto M, Sbarbati A, Ruzzenente O, Ferrario R, Hendriks H, Dellaglio F. 2004. Assessment of novel probiotic Lactobacillus casei strains for the production of functional dairy foods. Int. Dairy J. 14:723–736 [Google Scholar]
- 33.Boyle RJ, Robins-Browne RM, Tang MLK. 2006. Probiotics use in clinical practice: what are the risks? Am. J. Clin. Nutr. 83:1256–1264 [DOI] [PubMed] [Google Scholar]
- 34.Hoffmann M, Rath E, Hölzwimmer G, Quintanilla-Martinez L, Loach D, Tannock D, Haller D. 2008. Lactobacillus reuteri 100-23 transiently activates intestinal epithelial cells of mice that have a complex microbiota during early stages of colonization. J. Nutr. 138:1684–1691 [DOI] [PubMed] [Google Scholar]
- 35.Galdeano CM, Perdigón G. 2006. The probiotic bacterium Lactobacillus casei induces activation of the gut mucosal immune system through innate immunity. Clin. Vaccine Immunol. 13:219–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bekker LG, Moreira AL, Bergtold A, Freeman S, Ryffel B, Kaplan G. 2000. Immunopathologic effects of tumor necrosis factor alpha in murine mycobacterial infection are dose dependent. Infect. Immun. 68:6954–6961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Onizawa M, Nagaishi T, Kanai T, Nagano K, Oshima S, Nemoto Y, Yoshioka A, Totsuka T, Okamoto R, Nakamura T, Sakamoto N, Tsuchiya K, Aoki K, Ohya K, Yagita H, Watanabe M. 2009. Signaling pathway via TNF-α/NF-κB in intestinal epithelial cells may be directly involved in colitis-associated carcinogenesis. Am. J. Physiol. Gastrointest. Liver Physiol. 296:G850–G859 [DOI] [PubMed] [Google Scholar]
- 38.Sahoo M, Ceballos-Olvera I, del Barrio L, Re F. 2011. Role of inflammasome, IL-1β, and IL-18 in bacterial infections. ScientificWorldJournal 11:2037–2050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Iwakura Y, Ishigame H, Saijo S, Nakae S. 2011. Functional specialization of interleukin-17 family members. Immunity 34:149–162 [DOI] [PubMed] [Google Scholar]
- 40.Zhang Z, Zheng M, Bindas J, Schwarzenberger P, Kolls JK. 2006. Critical role of IL-17 receptor signaling in acute TNBS-induced colitis. Inflamm. Bowel Dis. 12:382–388 [DOI] [PubMed] [Google Scholar]
- 41.Ruyssers NE, De Winter BY, De Man JG, Ruyssers ND, Van Gils AJ, Loukas A, Pearson MS, Weinstock JV, Pelckmans PA, Moreels TG. 2010. Schistosoma mansoni proteins attenuate gastrointestinal motility disturbances during experimental colitis in mice. World J. Gastroenterol. 16:703–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Daniel C, Poiret S, Goudercourt D, Dennin V, Leyer G, Pot B. 2006. Selecting lactic acid bacteria for their safety and functionality by use of a mouse colitis model. Appl. Environ. Microbiol. 72:5799–5805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ebach DR, Newberry R, Stenson WF. 2005. Differential role of tumor necrosis factor receptors in TNBS colitis. Inflamm. Bowel Dis. 11:533–540 [DOI] [PubMed] [Google Scholar]
- 44.Guarner F, Malagelada JR. 2003. Role of bacteria in experimental colitis. Best Pract. Res. Clin. Gastroenterol. 17:793–804 [DOI] [PubMed] [Google Scholar]
- 45.Wan YM, Zhu YQ, Xia B, Luo J. 2011. Treating TNBS-induced colitis in rats with probiotics. Turk. J. Gastroenterol. 22:486–493 [DOI] [PubMed] [Google Scholar]
- 46.Barbatis C, Tsopanomichalou M. 2009. Heat shock proteins in inflammatory bowel disease. Ann. Gastroenterol. 22:244–247 [Google Scholar]
- 47.Madsen KL. 2012. Enhancement of epithelial barrier function by probiotics. J. Epithel. Biol. Pharmacol. 5(Suppl 1-M8):55–59 [Google Scholar]
- 48.Hu S, Wang Y, Lichtenstein L, Tao Y, Musch MW, Jabri B, Antonopoulos D, Claud EC, Chang EB. 2010. Regional differences in colonic mucosa-associated microbiota determine the physiological expression of host heat shock proteins. Am. J. Physiol. Gastrointest. Liver Physiol. 299:G1266–G1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Konturek PC, Sliwowski Z, Koziel J, Ptak-Belowska A, Burnat G, Brzozowski T, Konturek SJ. 2009. Probiotic bacteria Escherichia coli strain Nissle 1917 attenuates acute gastric lesions induced by stress. J. Physiol. Pharmacol. Suppl. 6:41–48 [PubMed] [Google Scholar]
- 50.Mousavi SA, Fønhus MS, Berg T. 2009. Up-regulation of uPARAP/Endo180 during culture activation of rat hepatic stellate cells and its presence in hepatic stellate cell lines from different species. BMC Cell Biol. 10:39. 10.1186/1471-2121-10-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tilleux S, Berger J, Hermans E. 2007. Induction of astrogliosis by activated microglia is associated with a down-regulation of metabotropic glutamate receptor 5. J. Neuroimmunol. 189:23–30 [DOI] [PubMed] [Google Scholar]
- 52.Peinnequin A, Mouret C, Birot O, Alonso A, Mathieu J, Clarençon D, Agay D, Chancerelle Y, Multon E. 2004. Rat pro-inflammatory cytokine and cytokine related mRNA quantification by real-time polymerase chain reaction using SYBR green. BMC Immunol. 5:3. 10.1186/1471-2172-5-3 [DOI] [PMC free article] [PubMed] [Google Scholar]