Abstract
Listeria monocytogenes is a food-borne bacterial pathogen and the causative agent of human and animal listeriosis. Among the three major genetic lineages of L. monocytogenes (i.e., LI, LII, and LIII), LI and LII are predominantly associated with food-borne listeriosis outbreaks, whereas LIII is rarely implicated in human infections. In a previous study, we identified a Crp/Fnr family transcription factor gene, lmo0753, that was highly specific to outbreak-associated LI and LII but absent from LIII. Lmo0753 shares two conserved functional domains, including a DNA binding domain, with the well-characterized master virulence regulator PrfA in L. monocytogenes. In this study, we constructed lmo0753 deletion and complementation mutants in two fully sequenced L. monocytogenes LII strains, 10403S and EGDe, and compared the flagellar motility, phospholipase C production, hemolysis, and intracellular growth of the mutants and their respective wild types. Our results suggested that lmo0753 plays a role in hemolytic activity in both EGDe and 10403S. More interestingly, we found that deletion of lmo0753 led to the loss of l-rhamnose utilization in EGDe, but not in 10403S. RNA-seq analysis of EGDe Δ0753 incubated in phenol red medium containing l-rhamnose as the sole carbon source revealed that 126 (4.5%) and 546 (19.5%) out of 2,798 genes in the EGDe genome were up- and downregulated more than 2-fold, respectively, compared to the wild-type strain. Genes related to biotin biosynthesis, general stress response, and rhamnose metabolism were shown to be differentially regulated. Findings from this study collectively suggested varied functional roles of lmo0753 in different LII L. monocytogenes strain backgrounds associated with human listeriosis outbreaks.
INTRODUCTION
Listeria monocytogenes is a Gram-positive, non-spore-forming, facultative anaerobic bacterium and the causative agent of human and animal listeriosis. Listeriosis is often caused by consumption of contaminated food products, such as raw milk, cheese, and ready-to-eat (RTE) meat products (1, 2). Listeriosis in healthy individuals may cause self-limiting gastroenteritis, whereas the disease in immunocompromised individuals, such as the elderly and pregnant women, may cause more severe complications, such as meningitis and encephalitis (1, 3, 4). Due to the high rates of hospitalizations (90%) and deaths (20%) (4, 5), a zero-tolerance policy has been implemented for the RTE meat and poultry industry in the United States (6). L. monocytogenes is ubiquitous in nature and is capable of adapting to and proliferating in various environmental niches, such as soil, rotting vegetation, and sewage (1, 3), which makes complete eradication of the persistent pathogen from industrial settings particularly challenging.
The species L. monocytogenes can be phylogenetically grouped into at least three major genetic lineages by various genotyping techniques (7). Lineage I (LI) mainly comprises serotypes 1/2b, 3b, 4b, 4d, and 4e; lineage II (LII) contains serotypes 1/2a, 1/2c, 3a, and 3c; and lineage III (LIII) consists of serotypes 4a, some 4b, and 4c (7). Previously reported human listeriosis outbreaks were predominantly caused by LI and LII strains, but rarely by strains in the LIII group (1). In our previous study, we conducted pangenomic analysis of 26 L. monocytogenes strains, including eight from LIII (8). We identified 86 disparately distributed genes (DDGs) and eight noncoding small RNAs that were highly conserved in LI and LII genomes but absent in LIII genomes. The majority of these DDGs were associated with cell wall structure, carbohydrate metabolism, and transcriptional regulation. We speculated that some of these DDGs contribute to the environmental persistence and full virulence of LI and LII strains during infection, which led to major food-borne listeriosis outbreaks.
One interesting DDG identified from our previous study was lmo0753, which encodes a putative Crp/Fnr family transcription factor. Lmo0753 has two functional domains that are specific to Crp/Fnr transcription factors, similar to the well-characterized positive regulatory factor A, or PrfA. The two domains are an N-terminally located cofactor binding domain and a C-terminally located helix-turn-helix Crp-type DNA binding domain (9). Crp/Fnr family transcription factors function as positive regulators in bacteria and are capable of responding to various environmental signals, such as anoxia, temperature, and oxidative stress (9). Due to its conservation in human outbreak-associated lineages, we hypothesized that lmo0753 plays a role in regulating environmental survival or virulence-related mechanisms in L. monocytogenes.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains used in this study are described in Table 1. EGDe (ATCC BAA-679) was obtained from the American Type Culture Collection (ATCC) (Manassas, VA). 10403S was provided by Nancy Freitag (University of Illinois). Stocks of bacterial cultures were maintained at −80°C. Unless otherwise stated, all strains were grown overnight (18 h) at 37°C in brain heart infusion (BHI) broth (Becton, Dickinson and Co., Franklin Lakes, NJ) supplemented with chloramphenicol (10 μg ml−1), kanamycin (30 μg ml−1), or streptomycin (200 μg ml−1) when necessary.
Table 1.
Bacterial strains and plasmids used in this study
Construction of lmo0753 deletion mutants and complements.
L. monocytogenes gene deletions in 10403S and EGDe were constructed using methods described by Alonzo et al. (15). All primers used are listed in Table 2. Briefly, upstream and downstream regions of the wild-type gene lmo0753 were fused via splice overlap extension (SOE) PCR to create a 1,373-bp fragment. The SOE PCR fragment was cloned into the temperature-sensitive shuttle vector pKSV7 using the BamHI and HindIII restriction sites to create plasmid JS-0753. The plasmid was then transformed into 10403S and EGDe as previously described (16), and the Δ0753 mutation was introduced into the chromosome via allelic exchange (17). The mutation was verified by colony PCR and DNA sequencing.
Table 2.
Primers used for construction of deletion mutants and complements
| Primer | Sequence (5′ to 3′) (restriction site)a |
|---|---|
| lmo0753 deletion | |
| 0753 A1 | GGCGGATCCCGACGTGCCTGTTTATATTTT (BamHI) |
| 0753 A2 | ACTAGTTACTCAATAAAAGTAGAATGTTCATTTTGCTTCTCC |
| 0753 B1 | GGAGAAGCAAAATGAACATTCTACTTTTATTGAGTAACTAGT |
| 0753 B2 | GGCAAGCTTAGCTATCCTTTTTAAATTATT (HindIII) |
| lmo0753 cloned into pIMK2 | |
| 0753 compF | GGCGGAATCTCATGAGCCACCAATCAATA (BamHI) |
| 0753 compR | GCGCCCGGGCCCAGTTCCTTGAAACCAG (SmaI) |
Restriction sites are underlined.
Complementation strains were constructed by amplifying the entire open reading frame of lmo0753 by PCR and cloning the fragment into pIMK2 via the BamHI and SmaI restriction sites to generate JS-c0753. The lmo0753 coding sequence in pIMK2 was under the control of the Phelp promoter, resulting in constitutive gene expression. JS-c0753 was transformed into L. monocytogenes Δ0753 strains (16), and the presence of the lmo0753 gene was verified by PCR and DNA sequencing.
Broad-range phospholipase C activity.
Phospholipase C activity was measured using the method of Alonzo et al. (18). Briefly, single colonies of each strain were streaked onto LB agar containing 2.5% chicken egg yolk. The agar plates were incubated for 24 and 48 h at 37°C, and the diameter of the zone of white precipitate surrounding the streaks was measured. Data were normalized against the negative-control strain 10403S ΔprfA to account for bacterial growth. Experiments were repeated at least three times with independent cultures.
Bacterial motility.
The flagellar-swimming motility of each strain was assessed using the soft-agar assay previously described by Shetron-Rama et al. (19) with slight modifications. Briefly, 1 μl of an overnight culture of each strain was dropped onto the surfaces of two BHI soft-agar plates containing 0.3% agar. The agar plates were incubated for 24 h at either 37°C or 25°C, and the diameters of the zones of growth for each strain were measured. Experiments were repeated at least three times with independent cultures.
LLO-associated hemolytic activity.
Hemolytic activity was measured as described previously by Alonzo et al. (18) with slight modifications. Briefly, overnight BHI cultures of each strain were diluted 1:10 in fresh BHI broth and incubated for 5 h at 37°C with shaking. After 5 h, the optical density at 600 nm (OD600) was measured and normalized for each strain. Serial dilutions of the supernatants were made using phosphate-buffered saline (PBS) containing 1 mM dithiothreitol (pH 5.0), followed by the addition of PBS-washed sheep red blood cells (Innovative Research, Inc., Novi, MI). After 30 min of incubation at 37°C, the mixtures were centrifuged, and the OD450 of the supernatant was measured. Hemolytic activity was represented by the OD600 divided by the OD450. Experiments were repeated at least three times with independent cultures.
Intracellular growth.
Caco-2 human intestinal epithelial cells (ATCC HTB-37) and J774 murine macrophage-like cells were maintained as previously described (20), and intracellular assays were performed using the method of Alonzo et al. (18). Monolayers of cells were grown to confluence on sterile glass coverslips. The cells were infected with L. monocytogenes strains at a multiplicity of infection of 100:1 for Caco-2 cells and 0.1:1 for J774 cells. After 1 h, the coverslips were washed three times with 37°C PBS, followed by the addition of fresh 37°C tissue culture medium containing gentamicin (15 μg/ml) to inactivate extracellular bacteria. Thereafter, every 2 h for up to 9 h, the coverslips were removed and eukaryotic cells were lysed by vortexing in 5 ml sterile water. Cell lysates were dilution plated onto LB agar plates to enumerate bacteria. All experiments were repeated at least three times with independent cultures.
Biolog phenotypic microarray.
Phenotypic microarray analysis was performed at Biolog Inc. (Hayward, CA). All mutants and wild-type strains were assayed on 20 96-well microtiter plates specific for bacterial metabolic and chemical sensitivity properties according to the manufacturer's specifications (21). Select gained or lost phenotypes of mutants were verified by individual broth assays using HTM minimal medium (22).
l-Rhamnose utilization.
l-Rhamnose utilization growth curves were constructed in phenol red minimal medium (10 g pancreatic digest of casein, 5 g sodium chloride, 0.018 g phenol red, and 5 g glucose or rhamnose per liter, pH 7.0) using a Bioscreen C automatic growth curve system (Growth Curves, Piscataway, NJ). Bacterial growth was monitored by recording the cell turbidity every 5 min over a period of 48 h. Experiments were performed at least three times with quadruplicate samples and verified by plate counting. pH curves were drawn using the same medium, and pH values were measured at 8, 24, 32, and 48 h. Experiments were performed three times with independent cultures.
Transcriptomic profiling of l-rhamnose utilization.
The L. monocytogenes EGDe wild type and its Δ0753 mutant were grown overnight in BHI broth at 37°C, washed three times with PBS, and subcultured at 1:100 into 100-ml phenol red broth containing either glucose or rhamnose as the sole carbon source. Bacteria were grown at 37°C for 4 h to mid-log phase, and total RNA was extracted using an Ambion RiboPure Bacterial Kit (Life Technologies Corporation, Carlsbad, CA). Total RNA from two biological replicates was prepared from independent cultures on different days to ensure the reproducibility of RNA-seq data. mRNA was purified using the NuGen Prokaryotic RNA-Seq system (NuGen, San Carlos, CA). Deep RNA sequencing was conducted on an Illumina HiSeq 2000 instrument (12.5 million filter-passed 50- plus 50-nucleotide [nt] paired-end reads; Illumina Inc., San Diego, CA). The reads were mapped to the L. monocytogenes 10403S sequence (available at http://www.broadinstitute.org/annotation/genome/listeria_group/GenomesIndex.html using GSNAP3) (23). Gene and transcript expression levels were computed using Cufflinks (24–26). Quality control of transcripts was performed using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) (Table 3).
Table 3.
Quality control summary statistics for each RNA-seq sample generated via FastQC
| Sample | Sample yield (Mb) | % PFa clusters | Mismatch rate (%) | % bases ≥Q30b (PF) | Mean quality score (PF) | Total no. of reads | No. of reads with at least one reported alignment | % R1 reads with at least one reported alignment |
|---|---|---|---|---|---|---|---|---|
| EGDe WTc rhamnose-1 | 2,672 | 93.86 | 0 | 94.66 | 37.27 | 56,932,882 | 43,535,535 | 76.5 |
| EGDe WT rhamnose-2 | 2,297 | 94.15 | 0 | 94.87 | 37.36 | 48,790,452 | 34,139,379 | 70.0 |
| EGDe 0753 rhamnose-1 | 1,998 | 94.30 | 0 | 94.76 | 37.31 | 42,363,954 | 27,339,106 | 64.5 |
| EGDe 0753 rhamnose-2 | 2,614 | 93.98 | 0 | 94.70 | 37.32 | 55,633,416 | 41,570,106 | 74.7 |
PF, purity filtered.
Q30, 99.9% accurate.
WT, wild type.
qRT-PCR.
Quantative real-time transcription-PCR (qRT-PCR) was performed as previously described (27). Briefly, six genes showing significant (P < 0.05) up- or downregulation identified by RNA-seq were selected for verification by qRT-PCR. Primers were designed with Primer3 software to produce amplicons of approximately 150 bp (28) (Table 4). qRT-PCR was performed using a LightCycler 480 (Roche Applied Science, Indianapolis, IN) with cDNA reverse transcribed from 1 μg of purified total RNA using the Transcriptor first-strand cDNA synthesis kit (Roche Applied Science). The relative expression changes were calculated using the method of Pfaffl (29); the 16S rRNA gene was used as an internal reference for data normalization. Average log2 values and standard deviations from quadruplicate samples are reported for each experiment. The experiments were conducted three times for statistical analysis.
Table 4.
Primers used for qRT-PCR analysis
| Gene | Annotation | Primer sequence 5′ to 3′a |
|---|---|---|
| LMRG_00281 | Biotin biosynthesis protein BioY | F: CTATACCACTCGGCCCTATT |
| R: TGTCATTCCTTGGAAAACAG | ||
| LMRG_01174 | Quinolinate synthetase complex A subunit | F: TTTTTGGCGATACGATTTTA |
| R: GAAAATCCGCTCTTTTTGTT | ||
| LMRG_01948 | General stress protein 26 | F: TCCGTTTTAATTGGTTACGA |
| R: ATAACGACAAAGGATGGTGA | ||
| LMRG_00287 | Hypothetical protein | F: ATTTGTCGCTGGTATTGTTG |
| R: TCCTCGAATCAAGTTGAAAA | ||
| LMRG_02489 | YukD protein | F: CACTAATTGGGGAGCAAGTA |
| R: TCGCTTTATTCGTTGTTTTT | ||
| LMRG_00054 | Sec-independent protein translocase TatAy | F: GGACCAGGAAGTATTGCTTT |
| R: TGGTTTCTTCTTTGGAATCA | ||
| LMRG_05501 | 16S rRNA | F: CAGCTAACGCATTAAGCACT |
| R: GTGGTCAAAGGATGTCAAGA |
F, forward primer; R, reverse primer.
Statistical analysis.
Student's t test and analysis of variance (ANOVA) were performed using the GraphPad Prism software package (version 5; GraphPad Software) and Excel software (2010 version; Microsoft).
RNA-seq data accession numbers.
RNA-seq data were deposited in the NCBI database with GEO accession numbers GSM1155399 through GSM1155402.
RESULTS
Virulence-associated mechanisms.
Using LAlign (30), we found the overall percentage of amino acid identity between Lmo0753 and PrfA was 18%. The C-terminal domains shared 27% identity (65% similarity), and the N-terminal domains shared 25% identity (53% similarity). Figure S1 in the supplemental material depicts a sequence alignment highlighting the helix-turn-helix DNA binding domain in each protein. Using BLAST, we determined that the DNA sequences of lmo0753 in 10403S and EGDe were 100% identical. To determine if lmo0753 contributes to virulence-associated mechanisms in L. monocytogenes 10403S and EGDe, we performed in vitro assays to evaluate the phospholipase C production, flagellar motility, hemolytic activity, and intracellular growth of the mutants. For phospholipase C activity (Fig. 1), assays were conducted for both 24 and 48 h. After 24 h, 10403S Δ0753 and EGDe Δ0753 mutants showed no significant difference from their parent strains in phospholipase C activity. After 48 h, 10403S Δ0753 showed a significant (P < 0.05) decrease in phospholipase C activity compared to the parent strain. 10403S prfA*, a constitutively active form of the protein, showed greater phospholipase C activity at both 24 and 48 h (P < 0.0001) than the parent strain. Complementation of 10403S Δ0753 was able to restore the phenotype of the parent strain. This indicated that lmo0753 played a role in phospholipase C activity in 10403S after 48 h, but not in EGDe.
Fig 1.
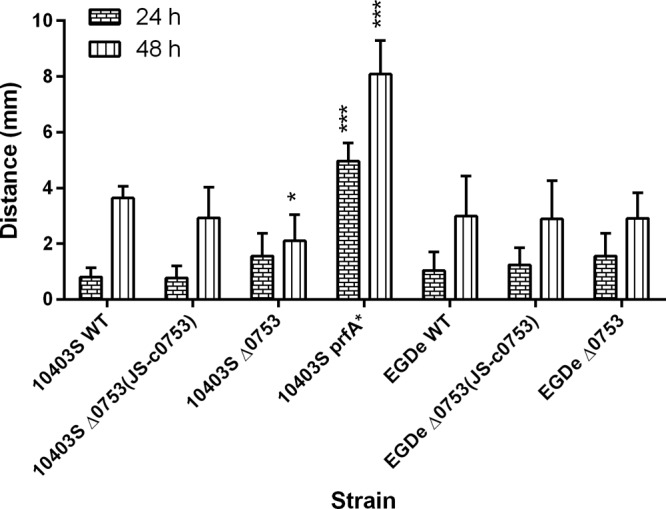
Phospholipase C activity of L. monocytogenes strains at 37°C after 24 and 48 h. The error bars represent standard deviations of three independent experiments. Significant differences in comparison to the parent strains under the same experimental condition are shown as follows: *, P < 0.5; ***, P < 0.0001.
Motility assays were conducted for 48 h at both 37°C and 25°C to determine if lmo0753 is involved in flagellar motility (Fig. 2). Significant motility reduction was observed at 37°C in both strains 10403S Δ0753 (P < 0.05) and 10403S ΔprfA (P < 0.001). At 25°C, slight but insignificant motility reduction was observed for both 10403S Δ0753 and EGDe Δ0753 mutants (P > 0.05). The 10403S complementation mutant was able to restore the motility phenotype to that of the wild-type strain at 37°C. The results indicated that lmo0753 played a minor role in the motility of L. monocytogenes 10403S at 37°C.
Fig 2.
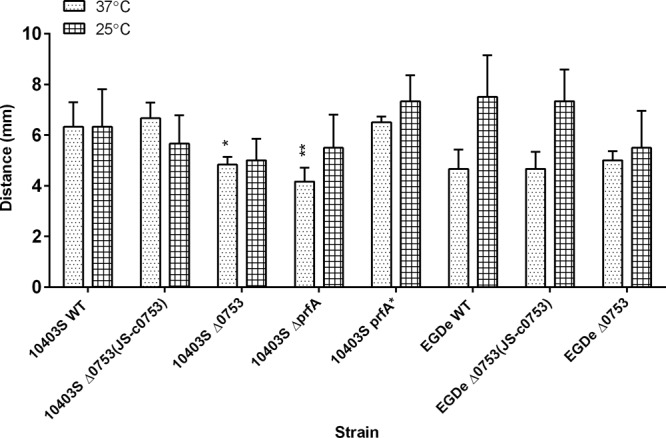
Flagellar-swimming motility of L. monocytogenes strains at 37°C and 25°C after 24 h. The error bars represent standard deviations of three independent experiments. Significant differences in comparison to the parent strains under the same experimental condition are shown as follows: *, P < 0.5; **, P < 0.001.
Hemolytic-activity assays were performed to determine if lmo0753 contributed to the hemolytic activity of L. monocytogenes 10403S and EGDe (Fig. 3A and B, respectively). Various dilutions of bacteria ranging from 1:10 to 1:320 were used to test the hemolytic activity of each strain at each dilution. At dilutions of 1:10, 1:20, 1:40, and 1:80, significant differences (P < 0.0001) were observed for all strains compared to their parent strains. Strains 10403S Δ0753, 10403S ΔprfA, and EGDe Δ0753 showed significantly less hemolytic activity than their respective wild-type strains, whereas 10403S prfA* showed significantly higher hemolytic activity than its wild type (data not shown). Complementation mutants were able to restore the phenotype observed in the wild-type strains. These results suggested that deletion of lmo0753 had a significant impact on the hemolytic activity of both 10403S and EGDe.
Fig 3.
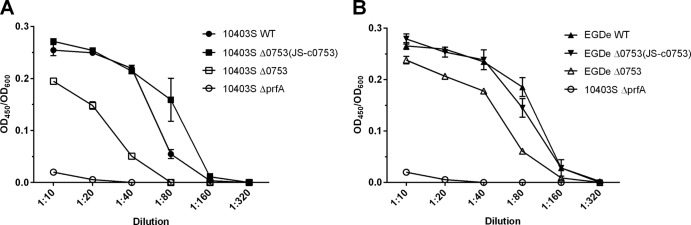
Hemolytic activity of L. monocytogenes strains at 37°C represented as the OD600 divided by the OD450. The error bars represent standard deviations of three independent experiments.
To determine if lmo0753 is involved in intracellular growth, we compared intracellular growth for all L. monocytogenes mutants in both Caco-2 and J774 eukaryotic host cells. 10403S ΔprfA was used as a negative control in these assays. No statistical difference was detected when strains were grown in Caco-2 cells (data not shown), and only slight differences were observed when they were grown in J774 cells (Fig. 4). Intracellular growth of both Δ0753 mutant strains resembled wild-type growth.
Fig 4.
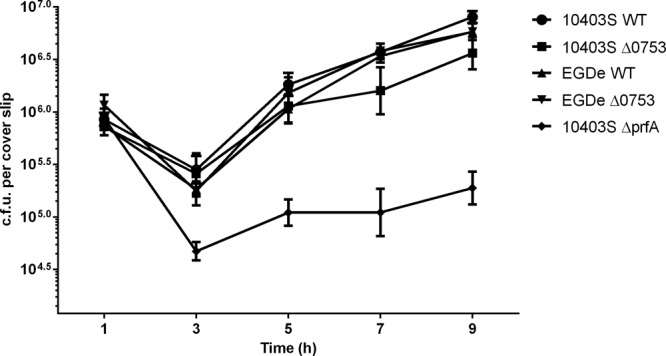
Intracellular growth assays of L. monocytogenes strains in J774 host cells. 10403S ΔprfA was used as a negative control. The error bars represent standard deviations of three independent experiments.
l-Rhamnose utilization.
L. monocytogenes EGDe and 10403S wild types and the respective Δ0753 mutants were subjected to Biolog phenotypic microarray screening to determine if the deletion had an impact on the metabolic utilization or chemical sensitivity of the bacteria. Twenty standard bacterial plates, which tested for metabolic utilization, such as carbon, nitrogen, phosphorus, and sulfur sources; pH and osmolytes; and antibiotic sensitivity, were used. Compared to its parent strain, the metabolic profile of 10403S Δ0753 showed increased metabolism in reactions with phenylmethylsufonylfluoride (PMSF) (a protease inhibitor) and crystal violet and decreased metabolism in reactions with 5,7-dichloro-8-hydroxyquinaldine (a lipophilic chelator). EGDe Δ0753 showed increased metabolism in reactions with domiphen bromide (a cationic detergent) and cesium chloride (a toxic cation) and decreased metabolism in reactions with l-rhamnose, poly-l-lysine (a cationic detergent), and patulin (a microtubulin polymerization inhibitor), in addition to decreased metabolism with nitrogen sources. Select gained/lost phenotypes of mutants were verified by individual broth assays to confirm the Biolog results.
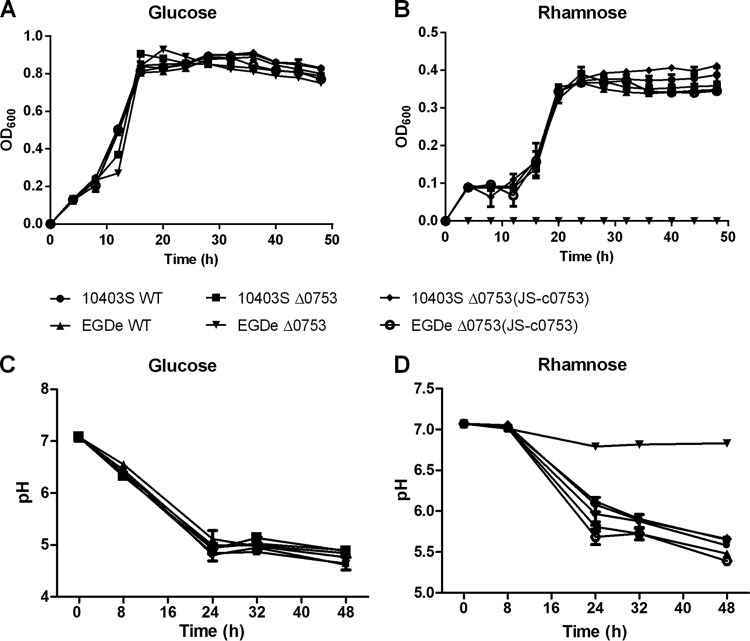
One interesting phenotypic change was the loss of l-rhamnose utilization in EGDe Δ0753. l-Rhamnose is often found as a constituent of pectin in dicotyledonous plant cell walls and is an important carbon source for energy metabolism for many soil bacterial species. To verify this phenotypic change, we constructed Bioscreen C growth curves with l-rhamnose as the sole carbon source in phenol red medium. When grown in phenol red medium with glucose as the sole carbon source (Fig. 5A), 10403S and EGDe showed growth patterns similar to those of their respective Δ0753 mutants. However, when grown in phenol red medium with l-rhamnose as the sole carbon source (Fig. 5B), EGDe Δ0753 did not grow, whereas other strains showed growth patterns similar to those of the wild-type strains. To ensure that the lack of growth was due to the loss of l-rhamnose utilization, pH determinations were performed. When grown in phenol red medium with glucose as the sole carbon source (Fig. 5C), all strains showed similar decreases in pH to 5.0 after 48 h of incubation, indicating the carbon source was metabolized. When grown in phenol red medium with l-rhamnose as the sole carbon source (Fig. 5D), the pH of EGDe Δ0753 remained neutral (pH 6.8) after 48 h (statistically significant; P < 0.0001), whereas the pH for all other strains dropped to approximately 5.5, suggesting that l-rhamnose was not utilized by EGDe Δ0753 for metabolism.
Fig 5.
Phenol red minimal medium growth curves and pH carbohydrate assays of L. monocytogenes strains. The growth curves were constructed using either glucose (A) or l-rhamnose (B) as the sole carbon source. pH carbohydrate assays were conducted with either glucose (C) or l-rhamnose (D) as the sole carbon source. The error bars represent standard deviations of three independent experiments conducted with quadruplicate samples.
Transcriptomic profiling.
To explore the transcriptomic response of L. monocytogenes when grown in phenol red medium with l-rhamnose as the sole carbon source, we performed RNA-seq analysis on the EGDe wild type and its Δ0753 mutant. Purified mRNA samples were sequenced on an Illumina HiSeq 2000 instrument, and all samples had a minimum of 42,000,000 paired-end reads aligned at >65% efficiency with the reference 10403S genome. 10403S was used the reference genome because detailed genome annotation and analytic tools for the strain were available at the Broad Institute website. qRT-PCR was used to validate the RNA-seq data for select genes. As shown in Fig. 6, the fold change detected by qRT-PCR correlated well with the corresponding RNA-seq results.
Fig 6.
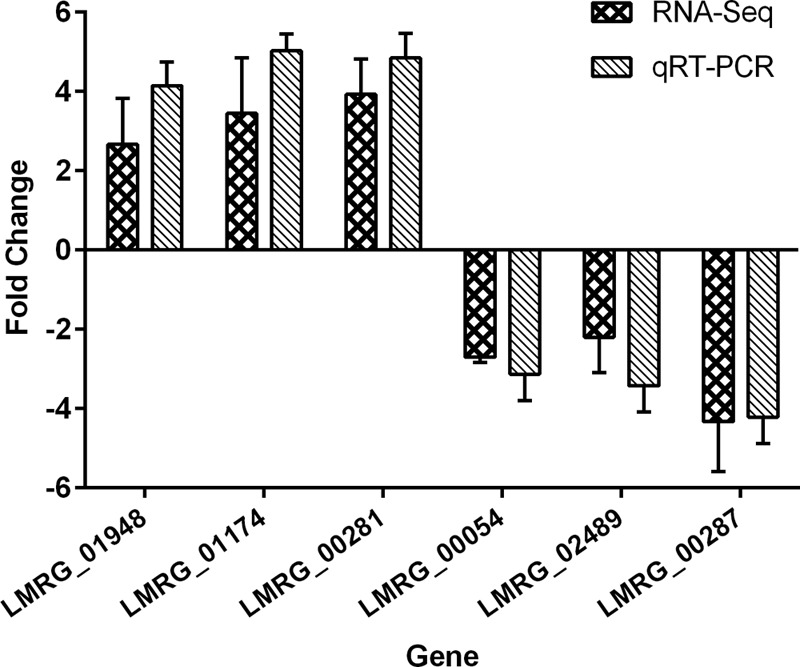
Comparison of gene expression fold changes identified by RNA-seq and qRT-PCR analyses. Six genes (3 upregulated and 3 downregulated) were compared. Fold changes were converted to log2 values for comparison. 16S rRNA (LMRG_05501) was used as an internal reference. The error bars represent standard deviations of three independent experiments conducted with quadruplicate samples.
Table 3 depicts the quality control summary for each sample sequenced. Out of 2,798 genes in the EGDe genome, 126 (4.5%) genes were found to be upregulated more than 2-fold and 43 (1.5%) genes more than 4-fold under the conditions tested (Table 5). The two most upregulated genes in EGDe Δ0753 were those encoding the biotin biosynthesis protein BioY (LMRG_00281; upregulated 23.5-fold) and the quinolinate synthetase complex A subunit (LMRG_01174; upregulated 20.3-fold). Other significantly upregulated transcripts included stress-related proteins, such as universal stress protein (LMRG_00196; upregulated 8.2-fold), general stress protein 26 (LMRG_01948; upregulated 8.1-fold), and a starvation-inducible DNA binding protein (LMRG_02041; upregulated 7.8-fold).
Table 5.
Transcripts with more than a 4-fold increase in EGDe Δ0753 compared to EGDe WT
| Gene | Annotation | Fold change |
|---|---|---|
| LMRG_00281 | Biotin biosynthesis protein BioY | 23.5 |
| LMRG_01174 | Quinolinate synthetase complex A subunit | 20.3 |
| LMRG_00199 | Membrane protein | 16.2 |
| LMRG_01972 | DNA binding 3-demethylubiquinone-9 3-methyltransferase | 15.0 |
| LMRG_01060 | Hypothetical protein | 13.4 |
| LMRG_01479 | Glutamate decarboxylase | 12.6 |
| LMRG_01173 | Nicotinate-nucleotide diphosphorylase | 10.6 |
| LMRG_01172 | l-Aspartate oxidase | 10.5 |
| LMRG_00557 | Cellobiose-specific PTS IIB component | 10.3 |
| LMRG_02808 | Hypothetical protein | 9.6 |
| LMRG_02382 | Hypothetical protein | 9.0 |
| LMRG_02094 | Hypothetical protein | 8.8 |
| LMRG_02304 | Peptidoglycan-bound protein | 8.4 |
| LMRG_00196 | Universal stress protein | 8.2 |
| LMRG_01948 | General stress protein 26 | 8.1 |
| LMRG_02041 | Starvation-inducible DNA binding protein | 7.8 |
| LMRG_00236 | Hypothetical protein | 7.3 |
| LMRG_02013 | Succinate-semialdehyde dehydrogenase | 6.8 |
| LMRG_00280 | Transcription regulator CRP/FNR family protein | 6.5 |
| LMRG_02414 | Sugar uptake protein | 6.2 |
| LMRG_01480 | Glutamate/gamma-aminobutyrate antiporter | 5.9 |
| LMRG_00710 | Hypothetical protein | 5.8 |
| LMRG_00885 | Glutathione reductase | 5.5 |
| LMRG_01794 | Hypothetical protein | 5.5 |
| LMRG_00335 | Magnesium and cobalt transporter CorA | 5.4 |
| LMRG_01912 | Catalase | 5.1 |
| LMRG_02001 | Dihydroxyacetone kinase l subunit | 5.1 |
| LMRG_00293 | Internalin | 5.1 |
| LMRG_00221 | Tagatose 1,6-diphosphate aldolase | 4.9 |
| LMRG_00365 | Flagellar biosynthetic protein FliQ | 4.9 |
| LMRG_00010 | Phosphomethylpyrimidine kinase | 4.9 |
| LMRG_00745 | Host factor I protein | 4.6 |
| LMRG_02602 | Hypothetical protein | 4.6 |
| LMRG_01030 | Chitinase | 4.6 |
| LMRG_01444 | Hypothetical protein | 4.5 |
| LMRG_01763 | PspC domain-containing protein | 4.4 |
| LMRG_01236 | Peptidoglycan binding protein | 4.4 |
| LMRG_00278 | Hypothetical protein | 4.4 |
| LMRG_01602 | Hypothetical protein | 4.2 |
| LMRG_00472 | Mannose-specific PTS IIA component | 4.1 |
| LMRG_01574 | Hypothetical protein | 4.1 |
| LMRG_02611 | Succinyl-diaminopimelate desuccinylase | 4.1 |
| LMRG_01103 | Fur family transcriptional regulator | 4.0 |
| LMRG_01849 | Hypothetical protein | 4.0 |
A total of 546 (19.5%) genes were found to be downregulated more than 2-fold and 22 (0.8%) genes more than 4-fold under the conditions tested (Table 6). The most significantly downregulated genes in EGDe Δ0753 included a hypothetical protein (LMRG_00287; downregulated 33.3-fold), YukD protein (LMRG_02489; downregulated 7.7-fold), and Sec-independent protein translocase TatAy (LMRG_00054; downregulated 7.7-fold). Rhamnose operon genes (31) were also downregulated: l-rhamnose 1-epimerase (LMRG_02417) 2.2-fold, rhamnulose-1-phosphate aldolase (LMRG_02418) 1.9-fold, l-rhamnose isomerase (LMRG_02419) 1.9-fold, and rhamnulokinase (LMRG_02420) 2.1-fold.
Table 6.
Transcripts with more than a 4-fold decrease in EGDe Δ0753 compared to EGDe WT
| Gene | Annotation | Fold change |
|---|---|---|
| LMRG_00287 | Hypothetical protein | 33.3 |
| LMRG_02489 | YukD protein | 7.7 |
| LMRG_00054 | Sec-independent protein translocase TatAy | 7.7 |
| LMRG_02700 | Hypothetical protein | 6.7 |
| LMRG_01268 | Hypothetical protein | 6.7 |
| LMRG_00793 | Competence protein ComGE | 6.3 |
| LMRG_00748 | HTHa-type transcriptional regulator GlnR | 6.3 |
| LMRG_01676 | Hypothetical protein | 5.9 |
| LMRG_01269 | Hypothetical protein | 5.9 |
| LMRG_01809 | Hypothetical protein | 5.9 |
| LMRG_01908 | Hypothetical protein | 5.3 |
| LMRG_02074 | Hypothetical protein | 5.3 |
| LMRG_01761 | Hypothetical protein | 5.3 |
| LMRG_00606 | Ethanolamine and carbon dioxide metabolism | 5.0 |
| LMRG_01655 | Hypothetical protein | 4.8 |
| LMRG_01915 | Cellobiose-specific PTS IIB component | 4.8 |
| LMRG_02072 | Membrane protein | 4.4 |
| LMRG_00868 | Hypothetical protein | 4.4 |
| LMRG_02225 | K+-transporting ATPase C subunit | 4.2 |
| LMRG_02073 | d-Alanine-poly(phosphoribitol) ligase subunit 1 | 4.0 |
| LMRG_01886 | Hypothetical protein | 4.0 |
| LMRG_00177 | Hypothetical protein | 4.0 |
HTH, helix-turn-helix.
DISCUSSION
Lmo0753 is a putative Crp/Fnr transcriptional regulator that shares two similar functional domains with PrfA, the master virulence regulator in L. monocytogenes (8). The shared functional domains are an N-terminally located cofactor binding domain and a C-terminally located helix-turn-helix domain consisting of two α-helices that are capable of resting in the major groove in DNA (9). Crp/Fnr family transcription factors are a very large and diverse family of proteins, spanning functions such as regulation of virulence, metabolic pathways, and stress response (9). One distinct function of PrfA is its ability to sense environmental changes and switch on various types of virulence factors. When environmental conditions, such as temperature, pH, and access to nutrients, change, PrfA is capable of positively regulating a number of virulence factors and thus aids in pathogenicity (3, 32–34). Because lmo0753 is highly conserved only in the human outbreak-associated lineages of L. monocytogenes (8), we hypothesized that this putative transcription factor likely played a role in environmental-signal sensing and pathogenicity.
It is well known that PrfA regulates L. monocytogenes virulence by switching on genes in the PrfA regulon (35). These genes are essential for bacterial invasion of host cells, cell-to-cell spread of the pathogen, and intracellular growth. Broad-range phospholipase C activity, encoded by plcB, is important during intracellular growth, mainly for phagosomal-vacuole escape (36). Deletion of lmo0753 modestly reduced phospholipase C activity in 10403S, but not in EGDe. This indicated that Lmo0753 had different impacts on plcB-associated phospholipase C activity in different strain backgrounds. It should be noted that deletion of prfA from 10403S also led to less production of phospholipase C and reduced invasive capability of the pathogen; conversely, constitutive activation of prfA led to greater phospholipase C activity and thus greater virulence (37). 10403S and EGDe remain motile at body temperature (37°C) (38). Deletion of lmo0753 resulted in less flagellar-associated motility in 10403S at 37°C, but not in EGDe. Decreased flagellar motility is generally associated with decreased virulence, although this is not always the case (38–40). Our results showed that 10403S ΔprfA had decreased flagellar motility compared to the parent strain, which was contradictory to other reports (41). This discrepancy could be explained by the different methods used in different studies for evaluating bacterial motility, such as stab inoculation versus drop inoculation on agar plates. Our study also showed that 10403S prfA* led to a slight decrease in motility compared with its parent strain. This was consistent with previous reports, although not to the same extent (41).
Virulent strains of L. monocytogenes can effectively escape from the phagosome and enter the cytosol during host cell infection because of the PrfA-dependent gene hly (3, 37, 42). Strains lacking hly are essentially avirulent (43). We demonstrated that both 10403S Δ0753 and EGDe Δ0753 showed reduced hemolytic activity; however, we did not detect a deficiency of either mutant in intracellular growth in Caco-2 cells and only insignificant differences in intracellular growth in J774 cells.
One criterion to differentiate L. monocytogenes from other, nonpathogenic Listeria species is that L. monocytogenes is able to utilize l-rhamnose as a carbon source. Certain atypical LIII strains that are deficient in rhamnose fermentation (44) display attenuated virulence (45), as well as reduced resistance to temperature shifts (46). Rhamnose pathway genes have been shown to be highly upregulated during intracellular growth (47). In this study, EGDe Δ0753 displayed a marked phenotypic loss of l-rhamnose utilization in the Biolog phenotypic assays. To verify the l-rhamnose deficiency in EGDe Δ0753, we constructed growth curves and performed pH assays using l-rhamnose as the sole carbon source. EGDe Δ0753 was found to be deficient in both growth and utilization of this carbon source for metabolism. Interestingly, unlike EGDe Δ0753, 10403S Δ0753 did not display deficiency in l-rhamnose utilization, suggesting that Lmo0753 functions differently in different strain backgrounds.
Transcriptomic profiling via RNA-seq was performed to detect differentially regulated genes when EGDe Δ0753 was incubated in medium containing l-rhamnose as the sole carbon source. A vast upregulation (23.5-fold) was seen in the biotin biosynthesis protein BioY. Biotin is an essential vitamin that functions as a cofactor in carboxylation and decarboxylation reactions. A recent study suggested roles for the vitamin in cell signaling and gene expression (48). Bacteria typically acquire biotin from exogenous sources (49) and transport it using an energy-coupling-factor transporter that consists of three main components: an S component that provides substrate specificity, a membrane component, and an ATPase A component (50–52). Such an increase in BioY may be due to an increase in stress resulting from starvation of the cells during incubation with l-rhamnose, as seen by the upregulation of three stress-specific genes, including a universal stress protein, general stress protein 26, and a starvation-inducible DNA binding protein. The upregulation of these transcripts was likely due to starvation-associated stress induction and not necessarily l-rhamnose itself.
Two other significantly upregulated genes in EGDe Δ0753 were those encoding the two components of the quinolinate synthetase complex: quinolinate synthetase complex A subunit (upregulated 20.3-fold) and l-aspartate oxidase (B subunit) (upregulated 10.5-fold). Quinolinate synthetase is an oxygen-sensitive complex required for NADP (NADP+) synthesis (53, 54). Quinolinate synthetase complexes are found in all sequenced genomes of bacilli and other pathogenic microorganisms, including Streptococcus and Enterococcus, and are necessary for the survival of Bacillus subtilis (55). Upregulation of these transcripts may be attributed to the cellular need to increase the de novo biosynthetic pathway for metabolic purposes.
YukD protein, a ubiquitin-like transporter (56), was significantly downregulated (7.7-fold) in EGDe Δ0753 during incubation in medium with l-rhamnose. YukD is an essential part of the type VII WXG100 protein secretory pathway found in Listeria and other Gram-positive bacteria (57, 58). WXG100 proteins are relatively small, approximately 100 amino acids in length; have a conserved WXG amino acid motif in the center of the protein; and lack canonical signal peptides (59). The proteins are thought to be secretory effectors that perform important functions once pathogens are inside host cells (58). This secretion system is deemed critical for cell-to-cell spread in host cells for Mycobacterium marinum and is also involved in mechanisms of dissemination and colonization in Staphylococcus aureus (60). However, it was reported that this type VII secretion system is not required for L. monocytogenes virulence (60). Two additional downregulated transporters were TatAy, a Sec-independent protein translocase (61) (downregulated 7.7-fold) and cellobiose-specific phosphotransferase system (PTS) IIB component, a major carbohydrate active-transport system (62) (downregulated 4.8-fold). Downregulation of protein and carbohydrate secretion systems may be attributed to a stress response mechanism.
In summary, the results from this study clearly demonstrated that lmo0753 plays a major role in l-rhamnose metabolism in L. monocytogenes strain EGDe. The absence of lmo0753 in L. monocytogenes LIII genomes potentially leads to an impaired ability of the bacteria to utilize l-rhamnose as an environmental carbon source, which partially explains the relative infrequency of the lineage in human food-borne disease outbreaks. Although 10403S and EGDe share highly similar genomic contents and belong to the same genetic lineage and serovar of L. monocytogenes, we observed marked functional differences of lmo0753 in the two strains. This observation highlights the need for a thorough examination of gene functions using different strain backgrounds in future studies.
Supplementary Material
ACKNOWLEDGMENTS
We thank Bobbi Xayarath for helpful discussions.
This work was supported by U.S. Food and Drug Administration research funds to the Illinois Institute of Technology. The work was also partly supported by a Chu Tian Lecturing Professorship from the Department of Education of Hubei Province to the Wuhan Polytechnic University.
The sponsors had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Published ahead of print 8 July 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01812-13.
REFERENCES
- 1.Kathariou S. 2002. Listeria monocytogenes virulence and pathogenicity, a food safety perspective. J. Food Prot. 65:1811–1829 [DOI] [PubMed] [Google Scholar]
- 2.Farber JM, Peterkin PI. 1991. Listeria monocytogenes, a food-borne pathogen. Microbiol. Rev. 55:476–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Freitag NE, Port GC, Miner MD. 2009. Listeria monocytogenes: from saprophyte to intracellular pathogen. Nat. Rev. Microbiol. 7:623–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Swaminathan B, Gerner-Smidt P. 2007. The epidemiology of human listeriosis. Microbes Infect. 9:1236–1243 [DOI] [PubMed] [Google Scholar]
- 5.Mead PS, Slutsker L, Dietz V, McCaig LF, Bresee JS, Shapiro C, Griffin PM, Tauxe RV. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.CDC 2011. Vital signs: incidence and trends of infection with pathogens transmitted commonly through food—foodborne diseases active surveillance network, 10 U.S. sites, 1996–2010. MMWR Morb. Mortal. Wkly. Rep. 60:749–755 [PubMed] [Google Scholar]
- 7.Ragon M, Wirth T, Hollandt F, Lavenir R, Lecuit M, Le Monnier A, Brisse S. 2008. A new perspective on Listeria monocytogenes evolution. PLoS Pathog. 4:e1000146. 10.1371/journal.ppat.1000146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deng X, Phillippy AM, Li Z, Salzberg SL, Zhang W. 2010. Probing the pan-genome of Listeria monocytogenes: new insights into intraspecific niche expansion and genomic diversification. BMC Genomics 11:500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Körner H, Sofia HJ, Zumft WG. 2003. Phylogeny of the bacterial superfamily of Crp-Fnr transcription regulators: exploiting the metabolic spectrum by controlling alternative gene programs. FEMS Microbiol. Rev. 27:559–592 [DOI] [PubMed] [Google Scholar]
- 10.Wong KK, Freitag NE. 2004. A novel mutation within the central Listeria monocytogenes regulator PrfA that results in constitutive expression of virulence gene products. J. Bacteriol. 186:6265–6276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miner MD, Port GC, Freitag NE. 2008. Functional impact of mutational activation on the Listeria monocytogenes central virulence regulator PrfA. Microbiology 154:3579–3589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ripio MT, Domínguez-Bernal G, Lara M, Suárez M, Vazquez-Boland JA. 1997. A Gly145Ser substitution in the transcriptional activator PrfA causes constitutive overexpression of virulence factors in Listeria monocytogenes. J. Bacteriol. 179:1533–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith K, Youngman P. 1992. Use of a new integrational vector to investigate compartment-specific expression of the Bacillus subtilis spoIIM gene. Biochimie 74:705–711 [DOI] [PubMed] [Google Scholar]
- 14.Lauer P, Chow MY, Loessner MJ, Portnoy DA, Calendar R. 2002. Construction, characterization, and use of two Listeria monocytogenes site-specific phage integration vectors. J. Bacteriol. 184:4177–4186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alonzo F, McMullen PD, Freitag NE. 2011. Actin polymerization drives septation of Listeria monocytogenes namA hydrolase mutants, demonstrating host correction of a bacterial defect. Infect. Immun. 79:1458–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park SF, Stewart GS. 1990. High-efficiency transformation of Listeria monocytogenes by electroporation of penicillin-treated cells. Gene 94:129–132 [DOI] [PubMed] [Google Scholar]
- 17.Camilli A, Tilney LG, Portnoy DA. 1993. Dual roles of plcA in Listeria monocytogenes pathogenesis. Mol. Microbiol. 8:143–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alonzo F, Port GC, Cao M, Freitag NE. 2009. The posttranslocation chaperone PrsA2 contributes to multiple facets of Listeria monocytogenes pathogenesis. Infect. Immun. 77:2612–2623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shetron-Rama LM, Marquis H, Bouwer HG, Freitag NE. 2002. Intracellular induction of Listeria monocytogenes actA expression. Infect. Immun. 70:1087–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brundage RA, Smith GA, Camilli A, Theriot JA, Portnoy DA. 1993. Expression and phosphorylation of the Listeria monocytogenes ActA protein in mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 90:11890–11894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bochner BR, Gadzinski P, Panomitros E. 2001. Phenotype microarrays for high-throughput phenotypic testing and assay of gene function. Genome Res. 11:1246–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsai HN, Hodgson DA. 2003. Development of a synthetic minimal medium for Listeria monocytogenes. Appl. Environ. Microbiol. 69:6943–6945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu TD, Nacu S. 2010. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 26:873–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28:511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roberts A, Pimentel H, Trapnell C, Pachter L. 2011. Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinformatics 27:2325–2329 [DOI] [PubMed] [Google Scholar]
- 26.Roberts A, Trapnell C, Donaghey J, Rinn JL, Pachter L. 2011. Improving RNA-Seq expression estimates by correcting for fragment bias. Genome Biol. 12:R22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang S, Deng K, Zaremba S, Deng X, Lin C, Wang Q, Tortorello ML, Zhang W. 2009. Transcriptomic response of Escherichia coli O157:H7 to oxidative stress. Appl. Environ. Microbiol. 75:6110–6123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rozen S, Skaletsky H. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132:365–386 [DOI] [PubMed] [Google Scholar]
- 29.Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang X, Miller W. 1991. LAlign (v 36.3.6) finds non-overlapping local alignments. Adv. Appl. Math. 12:373–381 [Google Scholar]
- 31.Fieseler L, Schmitter S, Teiserskas J, Loessner MJ. 2012. Rhamnose-inducible gene expression in Listeria monocytogenes. PLoS One 7:e43444. 10.1371/journal.pone.0043444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Piveteau P, Depret G, Pivato B, Garmyn D, Hartmann A. 2011. Changes in gene expression during adaptation of Listeria monocytogenes to the soil environment. PLoS One 6:e24881. 10.1371/journal.pone.0024881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sheehan B, Klarsfeld A, Msadek T, Cossart P. 1995. Differential activation of virulence gene expression by PrfA, the Listeria monocytogenes virulence regulator. J. Bacteriol. 177:6469–6476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ivy RA, Chan YC, Bowen BM, Boor KJ, Wiedmann M. 2010. Growth temperature-dependent contributions of response regulators, σB, PrfA, and motility factors to Listeria monocytogenes invasion of Caco-2 cells. Foodborne Pathog. Dis. 7:1337–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scortti M, Monzó HJ, Lacharme-Lora L, Lewis DA, Vázquez-Boland JA. 2007. The PrfA virulence regulon. Microbes Infect. 9:1196–1207 [DOI] [PubMed] [Google Scholar]
- 36.Mengaud J, Braun-Breton C, Cossart P. 1991. Identification of phosphatidylinositol-specific phospholipase C activity in Listeria monocytogenes: a novel type of virulence factor? Mol. Microbiol. 5:367–372 [DOI] [PubMed] [Google Scholar]
- 37.Mueller KJ, Freitag NE. 2005. Pleiotropic enhancement of bacterial pathogenesis resulting from the constitutive activation of the Listeria monocytogenes regulatory factor PrfA. Infect. Immun. 73:1917–1926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Way SS, Thompson LJ, Lopes JE, Hajjar AM, Kollmann TR, Freitag NE, Wilson CB. 2004. Characterization of flagellin expression and its role in Listeria monocytogenes infection and immunity. Cell. Microbiol. 6:235–242 [DOI] [PubMed] [Google Scholar]
- 39.Gorski L, Duhé JM, Flaherty D. 2009. The use of flagella and motility for plant colonization and fitness by different strains of the foodborne pathogen Listeria monocytogenes. PLoS One 4:e5142. 10.1371/journal.pone.0005142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O'Neil HS, Marquis H. 2006. Listeria monocytogenes flagella are used for motility, not as adhesins, to increase host cell invasion. Infect. Immun. 74:6675–6681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Port GC, Freitag NE. 2007. Identification of novel Listeria monocytogenes secreted virulence factors following mutational activation of the central virulence regulator, PrfA. Infect. Immun. 75:5886–5897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hamon MA, Ribet D, Stavru F, Cossart P. 2012. Listeriolysin O: the Swiss army knife of Listeria. Trends Microbiol. 20:360–368 [DOI] [PubMed] [Google Scholar]
- 43.Dramsi S, Cossart P. 2002. Listeriolysin O: a genuine cytolysin optimized for an intracellular parasite. J. Cell Biol. 156:943–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roberts A, Nightingale K, Jeffers G, Fortes E, Kongo JM, Wiedmann M. 2006. Genetic and phenotypic characterization of Listeria monocytogenes lineage III. Microbiology 152:685–693 [DOI] [PubMed] [Google Scholar]
- 45.Liu D, Lawrence ML, Wiedmann M, Gorski L, Mandrell RE, Ainsworth AJ, Austin FW. 2006. Listeria monocytogenes subgroups IIIA, IIIB, and IIIC delineate genetically distinct populations with varied pathogenic potential. J. Clin. Microbiol. 44:4229–4233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.De Jesús AJ, Whiting RC. 2003. Thermal inactivation, growth, and survival studies of Listeria monocytogenes strains belonging to three distinct genotypic lineages. J. Food Prot. 66:1611–1617 [DOI] [PubMed] [Google Scholar]
- 47.Lobel L, Sigal N, Borovok I, Ruppin E, Herskovits AA. 2012. Integrative genomic analysis identifies isoleucine and CodY as regulators of Listeria monocytogenes virulence. PLoS Genet. 8:e1002887. 10.1371/journal.pgen.1002887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zempleni J, Wijeratne SS, Hassan YI. 2009. Biotin. Biofactors. 35:36–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin S, Cronan JE. 2011. Closing in on complete pathways of biotin biosynthesis. Mol. Biosyst. 7:1811–1821 [DOI] [PubMed] [Google Scholar]
- 50.Streit WR, Entcheva P. 2003. Biotin in microbes, the genes involved in its biosynthesis, its biochemical role and perspectives for biotechnological production. Appl. Microbiol. Biotechnol. 61:21–31 [DOI] [PubMed] [Google Scholar]
- 51.Prakash O, Eisenberg MA. 1974. Active transport of biotin in Escherichia coli K-12. J. Bacteriol. 120:785–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hebbeln P, Rodionov DA, Alfandega A, Eitinger T. 2007. Biotin uptake in prokaryotes by solute transporters with an optional ATP-binding cassette-containing module. Proc. Natl. Acad. Sci. U. S. A. 104:2909–2914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gardner PR, Fridovich I. 1991. Quinolinate synthetase: the oxygen-sensitive site of de novo NAD(P)+ biosynthesis. Arch. Biochem. Biophys. 284:106–111 [DOI] [PubMed] [Google Scholar]
- 54.Ceciliani F, Caramori T, Ronchi S, Tedeschi G, Mortarino M, Galizzi A. 2000. Cloning, overexpression, and purification of Escherichia coli quinolinate synthetase. Protein Expr. Purif. 18:64–70 [DOI] [PubMed] [Google Scholar]
- 55.Rossolillo P, Marinoni I, Galli E, Colosimo A, Albertini AM. 2005. YrxA is the transcriptional regulator that represses de novo NAD biosynthesis in Bacillus subtilis. J. Bacteriol. 187:7155–7160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van den Ent F, Löwe J. 2005. Crystal structure of the ubiquitin-like protein YukD from Bacillus subtilis. FEBS Lett. 579:3837–3841 [DOI] [PubMed] [Google Scholar]
- 57.Desvaux M, Hébraud M. 2006. The protein secretion systems in Listeria: inside out bacterial virulence. FEMS Microbiol. Rev. 30:774–805 [DOI] [PubMed] [Google Scholar]
- 58.Simeone R, Bottai D, Brosch R. 2009. ESX/type VII secretion systems and their role in host-pathogen interaction. Curr. Opin. Microbiol. 12:4–10 [DOI] [PubMed] [Google Scholar]
- 59.Pallen MJ. 2002. The ESAT-6/WXG100 superfamily—and a new Gram-positive secretion system? Trends Microbiol. 10:209–212 [DOI] [PubMed] [Google Scholar]
- 60.Abdallah AM, Gey van Pittius NC, Champion PA, Cox J, Luirink J, Vandenbroucke-Grauls CM, Appelmelk BJ, Bitter W. 2007. Type VII secretion—mycobacteria show the way. Nat. Rev. Microbiol. 5:883–891 [DOI] [PubMed] [Google Scholar]
- 61.Barnett JP, van der Ploeg R, Eijlander RT, Nenninger A, Mendel S, Rozeboom R, Kuipers OP, van Dijl JM, Robinson C. 2009. The twin-arginine translocation (Tat) systems from Bacillus subtilis display a conserved mode of complex organization and similar substrate recognition requirements. FEBS J. 276:232–243 [DOI] [PubMed] [Google Scholar]
- 62.Lai X, Ingram LO. 1993. Cloning and sequencing of a cellobiose phosphotransferase system operon from Bacillus stearothermophilus XL-65-6 and functional expression in Escherichia coli. J. Bacteriol. 175:6441–6450 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.