Abstract
It has previously been shown that the tomato pathogen Clavibacter michiganensis subsp. michiganensis secretes a 14-kDa protein, C. michiganensis subsp. michiganensis AMP-I (CmmAMP-I), that inhibits growth of Clavibacter michiganensis subsp. sepedonicus, the causal agent of bacterial ring rot of potato. Using sequences obtained from tryptic fragments, we have identified the gene encoding CmmAMP-I and we have recombinantly produced the protein with an N-terminal intein tag. The gene sequence showed that CmmAMP-I contains a typical N-terminal signal peptide for Sec-dependent secretion. The recombinant protein was highly active, with 50% growth inhibition (IC50) of approximately 10 pmol, but was not toxic to potato leaves or tubers. CmmAMP-I does not resemble any known protein and thus represents a completely new type of bacteriocin. Due to its high antimicrobial activity and its very narrow inhibitory spectrum, CmmAMP-1 may be of interest in combating potato ring rot disease.
INTRODUCTION
Many bacteria produce antimicrobial compounds called bacteriocins. These are ribosomally synthesized peptides or proteins that most often act on species that are closely related to the producer strain. Bacteriocins are thought to participate in the competition between similar species occupying the same ecological niche (1–3). Bacteriocins produced by phytopathogens are of great interest because they may contribute to determining the composition of microbial ecosystems in, e.g., the rhizosphere, which again may affect the emergence and severity of plant disease outbreaks. Furthermore, such compounds are of interest because they may provide new tools for combating plant pathogens. In this respect, it is helpful that bacteriocins tend to have a narrow target cell spectrum and lack a “general” antibiotic activity that could hamper their potential application. Despite their obvious importance and potential, relatively little is known about bacteriocins from plant pathogens (4).
We have previously shown that the Gram-positive tomato pathogen Clavibacter michiganensis subsp. michiganensis, an actinomycete, produces two proteinaceous antimicrobial compounds: a heat-stable 2,145-Da posttranslationally modified peptide called michiganin A (5) and a heat-sensitive antimicrobial protein with a molecular mass of approximately 14 kDa called C. michiganensis subsp. michiganensis AMP-I (CmmAMP-I) (6). Michiganin A was studied in detail and turned out to be a globular posttranslationally modified peptide bacteriocin belonging to the so-called type B lantibiotics. The CmmAMP-I protein was purified to apparent homogeneity, but Edman degradation for protein sequencing did not yield sufficiently clear results for protein identification. The antimicrobial activity of CmmAMP-I was confirmed in microtiter plate-based bacteriocin assays. Furthermore, upon overlaying renatured SDS-PAGE gels with indicator cells, the lane containing purified CmmAMP-I showed a clear halo at approximately 14 kDa (6). Most interestingly, so far, the only identified organism sensitive to both Michiganin A and AMP-I is C. michiganensis subsp. sepedonicus, the causal agent of bacterial ring rot in potatoes. Since potato ring rot is a regulated quarantine disease causing economic losses in North America, Asia, and Europe, natural compounds that inhibit C. michiganensis subsp. sepedonicus may be of value from a biocontrol perspective.
Michiganin A belongs to a class of bacteriocins that require a variety of posttranslational modifications and dedicated secretion machinery (7, 8). Thus, exploiting michiganin A is not straightforward, as both industrial-scale synthesis of the peptide and expression of the peptide in, e.g., transgenic plants would require large and complex genetic manipulations. On the other hand, CmmAMP-I could in principle be a normal secreted protein that would be easier to produce and exploit.
Here we describe the identification of the structural gene for CmmAMP-I via mass spectrometry (MS)-based sequence analysis of tryptic fragments of the purified protein. The gene encoding CmmAMP-I was identified in the genome of C. michiganensis subsp. michiganensis (9) and successfully expressed using an Impact system in Escherichia coli. Using purified heterologously expressed CmmAMP-I, we showed that the protein kills C. michiganensis subsp. sepedonicus at low concentrations and thus presents an interesting potential tool for developing new control strategies for bacterial ring rot in potatoes. Interestingly, CmmAMP-I does not resemble any known protein and thus represents a completely new type of bacteriocin.
MATERIALS AND METHODS
Bacterial strains and media.
C. michiganensis subsp. michiganensis NCPPB 1468 and C. michiganensis subsp. sepedonicus NCPPB 2136 (National Collection of Plant Pathogenic Bacteria, Harpenden, United Kingdom) and Bioforsk strains 1977-2, 1979-9, 1982-4, 1982-11, and 2006-A55 (all from the Norwegian county of Akershus; the year of collection is indicated by the first part of the strain designation) were grown in NBY medium (10) with shaking at 100 rpm and at 20°C. Escherichia coli TOP10 (Invitrogen), used as the host cell for plasmid construction, and E. coli strain C3016 (New England BioLabs), used as the host cell for protein expression, were grown in 2× TY medium with ampicillin (16 g/liter tryptone, 10 g/liter yeast extract, 5 g/liter NaCl, 150 mg/liter ampicillin) at 37°C with shaking. For plates, the media were solidified with 15 g/liter agar.
Strains tested for AMP-I sensitivity included the following: Bacillus subtilis BAB-1, Enterococcus faecalis V583, E. coli Top10, E. coli C3016, Lactobacillus plantarum NC8, Lactobacillus sakei Lb790, Listeria innocua 4202, Pediococcus pentosaceus LMGT 2001, Serratia marcescens BJL200, Streptococcus faecalis NCDO581, and 6 strains of Streptomyces spp. from a local culture collection numbered 1-A, 1-B, 5-A, 5-B, 6-A, and 10-A. L. plantarum, L. sakei, S. faecalis, and P. pentosaceus were grown in MRS medium (Oxoid Ltd., Hampshire, England) at 30°C. L. innocua was grown in BHI (brain heart infusion) medium (Oxoid Ltd., Hampshire, England) at 30°C. E. faecalis was grown in BHI medium at 37°C. S. marcescens and E. coli were grown in LB medium (10 g/liter Bacto–tryptone, 5 g/liter Bacto–yeast extract, 10 g/liter NaCl) at 30°C. Streptomyces spp. were grown in GYM Streptomyces medium (4 g/liter glucose, 4 g/liter yeast extract, 10 g/liter malt extract, 2 g/liter CaCO3, pH 7.2) at 30°C. The media were solidified by adding 15 g/liter agar.
Tryptic mapping and sequence analysis of CmmAMP-I.
CmmAMP-I was produced and purified from a culture supernatant of C. michiganensis subsp. michiganensis NCPPB 1468 as described previously (6). Purified protein was dried in a vacuum and redissolved in 50 μl 100 mM (NH4)HCO3. For reduction and alkylation of cysteine residues, 2.5 μl 200 mM dithiothreitol (DTT)–100 mM (NH4)HCO3 was added and the solution was incubated at 56°C for 30 min. After cooling to room temperature, 5 μl 0.55 M iodoacetamide–100 mM (NH4)HCO3 was added, and the solution was incubated at room temperature in the dark for 30 min. Finally, 3 μl 1 M DTT–100 mM (NH4)HCO3 was added, and the solution was incubated for an additional 15 min.
Tryptic digestion was performed overnight (16 h) by adding sequencing-grade modified trypsin (Promega, Madison, WI) to reach a final concentration of 5 ng/μl (giving molar trypsin/protein ratios of 1:50 to 1:100). The reaction was terminated by adding trifluoroacetic acid to reach a final concentration of 0.5% (vol/vol), and the tryptic peptides were purified using a STAGE column (11).
Tryptic fragments were analyzed using a matrix-assisted laser desorption ionization–tandem time of flight (MALDI-TOF/TOF) instrument (Ultraflex; Bruker Daltonics) as described previously (12). For de novo sequencing, an Ettan chemically assisted fragmentation-MALDI (CAF-MALDI) sequencing kit (GE Healthcare, Little Chalfont, United Kingdom) was employed. The MS/MS spectra recorded for derivatized peptides were manually annotated using flexAnalysis software (v.3.3; Bruker Daltonics).
Cloning, expression, and purification of CmmAMP-I.
The gene encoding CmmAMP-I (NC_009480.1; 1899661-to-1900137 complement; locus tag CMM_1675), excluding the signal sequence, was amplified by PCR from genomic DNA of C. michiganensis subsp. michiganensis NCPPB 1468, which was purified by phenol-chloroform extraction and ethanol precipitation after disruption of the cells with glass beads in a beadbeater (Biospec). The primers used were as follows: AMP-1 fw (5′-GGTGGTCATATGGCCACGATCTCCAGCCCCGAT-3′; NdeI site italicized) and AMP-1 rev (5′-GGTGGTTGCTCTTCCGCAGTTGCTGAATCCACCCAGCGGAAC-3′; SapI site italicized). The PCR product was purified by standard agarose gel electrophoresis (1% gel), digested with NdeI and SapI, and ligated into NdeI-SapI-digested vector pTYB1, thus coupling the gene to an N-terminal intein tag (Impact System; New England BioLabs, Beverly, MA). After transformation of the ligation mix into chemically competent E. coli TOP10 cells, plasmids were isolated using a NucleoSpin plasmid miniprep kit (Macherey-Nagel, Düren, Germany) and the presence of a correct insertion was confirmed by sequencing. A correct plasmid was then transformed into chemically competent E. coli C3016 cells for protein expression. A positive transformant was grown in 2× TY medium containing ampicillin at 37°C. When the A600 reached 0.5, gene expression was induced by addition of IPTG (isopropyl-β-d-1-thiogalactopyranoside) to reach a final concentration of 0.4 mM. After overnight growth at 25°C, cells collected by centrifugation were resuspended in 20 mM Tris-HCl–500 mM NaCl, pH 8.5. The cells were lysed by sonication at 33% amplitude with 30 5-s pulses (with a 5-s delay between pulses) on ice, with a Vibra cell ultrasonic processor, converter model CV33, equipped with a 3-mm probe (Sonics, Newtown, CT). The sonicated material was centrifuged at 23,700 × g for 30 min at 4°C to obtain a cell-free protein extract.
Recombinant CmmAMP-I was enriched with a chitin bead column, according to the manufacturer's instructions (Impact kit; New England BioLabs, Beverly, MA). As part of this procedure, the intein tag was removed while on the column by application of 50 mM DTT. The detagged eluted protein was further purified by standard gel filtration chromatography using a prepacked resin column (HiLoad 16/60; GE Healthcare) operated in TN buffer (20 mM Tris [pH 8.0], 20 mM NaCl), analyzed by SDS-polyacrylamide gel electrophoresis, and stored at −20°C in TN buffer containing 50% (vol/vol) glycerol. Protein concentrations were determined using a Bio-Rad protein assay kit (Bio-Rad), with bovine serum albumin as a standard. Immediately before use, glycerol was removed from the CmmAMP-I stock solutions using a centrifugal filter (Millipore) (molecular weight [MW], 3,000) for buffer exchange to TN buffer.
Agar plate assays for screening of antimicrobial activity.
To test AMP-I activity, different strains were plated on solidified media in 100-mm-diameter petri dishes. Immediately after spreading the cells, holes punched into the agar using a cut pipet tip were filled with 25 μl of 5 μM CmmAMP-I in TN buffer or with TN buffer (negative control). Zones of inhibition were assessed after incubating the plates at appropriate temperatures for 2 to 3 days or as soon as a confluent layer of bacteria was visible.
A similar plate assay was used to analyze whether C. michiganensis subsp. michiganensis NCPPB 1468 and C. michiganensis subsp. sepedonicus NCPPB 2136 could inhibit each other. Supernatants were derived from 2-day-old liquid cultures of C. michiganensis subsp. michiganensis and C. michiganensis subsp. sepedonicus in NBY medium and sterilized by filtration using a 0.22-μm-pore-size filter. C. michiganensis subsp. michiganensis or C. michiganensis subsp. sepedonicus was plated on NBY plates, and holes punched in the plate with a cut pipet tip were filled with 50 μl of culture supernatant. The plates were incubated for 3 days at 20°C, after which the occurrence of inhibition zones was assessed.
Growth inhibition in liquid cultures and determination of IC50.
Growth inhibition was further analyzed by microtiter plate dilution assays and quantified by determining the CmmAMP-I concentration required for 50% growth inhibition (IC50), essentially as described in reference 13. The indicator strain (C. michiganensis subsp. sepedonicus) was grown in liquid NBY medium until the A600 reached 0.5 and then diluted 200 times with medium. Liquid NBY medium (50 μl) was added to each well of a 96-well microtiter plate. A 50-μl volume of a 0.5 mg/ml CmmAMP-I solution in TN buffer was added to the first well. Then a serial dilution (1:2) was made by stepwise transferral of 50 μl to subsequent wells. Serially diluted buffer was used as a negative control. After preparing the serial dilutions, 150 μl of the diluted indicator strain was added to each well (this yields a final AMP-I concentration of 4.8 μM in the first well). After incubation of the microtiter plate at 20°C and 100 rpm for 40 h, A600 values were measured using a microplate reader. The IC50 was estimated by calculating the protein concentration in the wells with an A600 value equal to approximately 50% of the average A600 value in wells without AMP-I (i.e., noninhibited growth).
To analyze whether CmmAMP-I kills or only inhibits C. michiganensis subsp. sepedonicus, survival of the indicator strain was analyzed. Cells were collected from appropriate microtiter plate wells, washed with NBY medium, and grown on NBY plates for colony counting.
Phytoxicity tests.
Potato (Solanum tuberosum) cultivar Bintje and tomato (Solanum lycopersicum) cultivar Espero were cultivated in the greenhouse under conditions of natural day light supplemented with artificial light (16-h photoperiod) at 16 to 18°C. Leaflets and fruits were harvested at the start of the experiments, whereas tubers were from cold storage. Fruits and tubers were surface disinfected using 0.5% sodium hypochlorite and rinsed in autoclaved Milli-Q water before the start of the experiment. The experiment was performed in large humid chambers (trays lined with moist filter paper under an aluminum grid and covered with plastic). Detached leaflets were placed directly on the metal grid, while tubers and fruits were placed in 150-mm-diameter petri dishes. Drops (5 μl) of AMP-I solution (55 ng/μl in TN buffer) were placed onto each of 10 leaflets, 5 tuber slices, and 3 tubers of potato and 10 leaflets, 5 fruit slices, and 3 whole fruits of tomato. Controls (same numbers) were treated with double-distilled water (dH2O), TN buffer, or the nonselective contact herbicide Reglone (active ingredient dikvat-dibromid; Syngenta). The plant samples were incubated at 16°C for 7 days, leaflets under conditions of a 16-h photoperiod, and tubers and fruits in the dark.
RESULTS
Identification of the gene encoding CmmAMP-I.
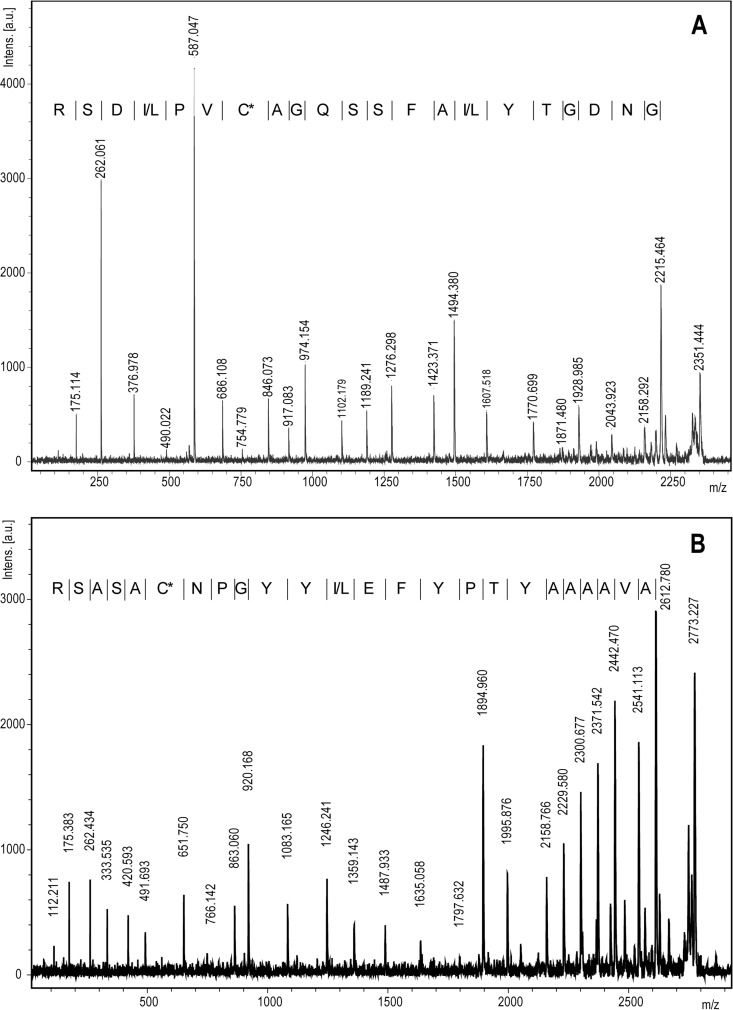
Analysis of tryptic fragments obtained from CmmAMP-I yielded good-quality sequence information for two peptides: AVAAAAYTPYFE(I/L)YYGPNCASASR and GNDGTY(I/L)AFSSQGACVP(I/L)DSR (Fig. 1), which led to the identification of a locus called CMM_1675 as the gene encoding the protein (NC_009480.1; 1899661-to-1900137 complement). This gene encodes a 158-residue protein of which the first 37 amino acids (aa) are predicted to be a Sec-type signal peptide according to SignalP (14). The remaining 121-residue protein has a predicted mass of 13,012 Da, which is in good agreement which the results from SDS-PAGE (6). The theoretical pI (15) of the protein is 8.69.
Fig 1.
De novo sequencing of tryptic peptides from AMP-I. MALDI MS/MS spectra of the two tryptic peptides GNDGTY(I/L)AFSSQGACVP(I/L)DSR (A) and AVAAAAYTPYFE(I/L)YYGPNCASASR (B). The derivatization methods used (CAF; see Materials and Methods) lead to a predominance of y-ions in the MS/MS spectra, thereby significantly simplifying interpretation and annotation. C* denotes a carbamidomethylated cysteine (see Materials and Methods). Intens. [a.u.], intensity [arbitrary units].
BLAST searches (16) with the CmmAMP-I protein sequence against the translated nucleotide database yielded only two significant hits. One of these hits was a protein with unknown function from C. michiganensis subsp. sepedonicus (CMS_0380) annotated as “putatively secreted.” This protein has the same number of residues as CmmAMP-I and shows 68% sequence identity (Fig. 2). The only other significant hit was to part of a postulated insertion sequence (IS) element in the related bacterium Leifsonia xyli subsp. cynodontis (previously Clavibacter xyli subsp. cynodontis), the causal agent of ratoon stunting disease in sugarcane (17). In this case, a 41% sequence identity was observed for a stretch of 105 residues.
Fig 2.

Sequence alignment of AMP-I from C. michiganensis subsp. michiganensis (CmmAMP-I) with a homologous protein from C. michiganensis subsp. sepedonicus (CmsAMP). Peptide sequences that led to the identification of the AMP-I open reading frame (ORF) (see Fig. 1) are printed in bold face and underlined. According to SignalP predictions, both proteins contain a signal peptidase I cleavage site for Sec-driven secretion. The signal peptides are printed in italics, and the first amino acid of the mature secreted protein is marked with a solid triangle. The gene encoding CmsAMP (locus CMS0380) seems to be incorrectly annotated in the publicly available genome sequence of C. michiganensis subsp. sepedonicus. In the gene bank entry, the ORF is covering nucleotides 417755 to 418165, whereas an ORF spanning nucleotides 417689 to 418165 seems more correct. Here, the corrected ORF has been used.
The CMM_1675 gene in C. michiganensis subsp. michiganensis is encoded on the reverse strand, and no obvious operon structure can be observed. CMM_1674 encodes a putative endo-1,4-beta-xylanase A and is located as much as 966 bp upstream on the forward strand. CMM_1676, located 108 bp downstream on the forward strand, encodes a putative protein of 162 aa with no similarity to any other proteins (meaning that this protein is absent in C. michiganensis subsp. sepedonicus).
Expression of CmmAMP-I.
AMP-I was produced with an N-terminal intein tag that permitted purification with a chitin column and precise on-column removal of the tag, as described in Materials and Methods. The fusion protein was expressed in soluble form in E. coli C3016. After chitin affinity chromatography with on-column tag removal, the protein preparation was subjected to one-step size exclusion chromatography to obtain maximal purity. Figure 3 shows that this procedure led to the production of reasonably pure material. The typical yield of this procedure was 0.5 mg of pure protein from 1 liter of culture of E. coli C3016. An agar plate assay showed that the recombinantly produced protein was active (Fig. 4).
Fig 3.
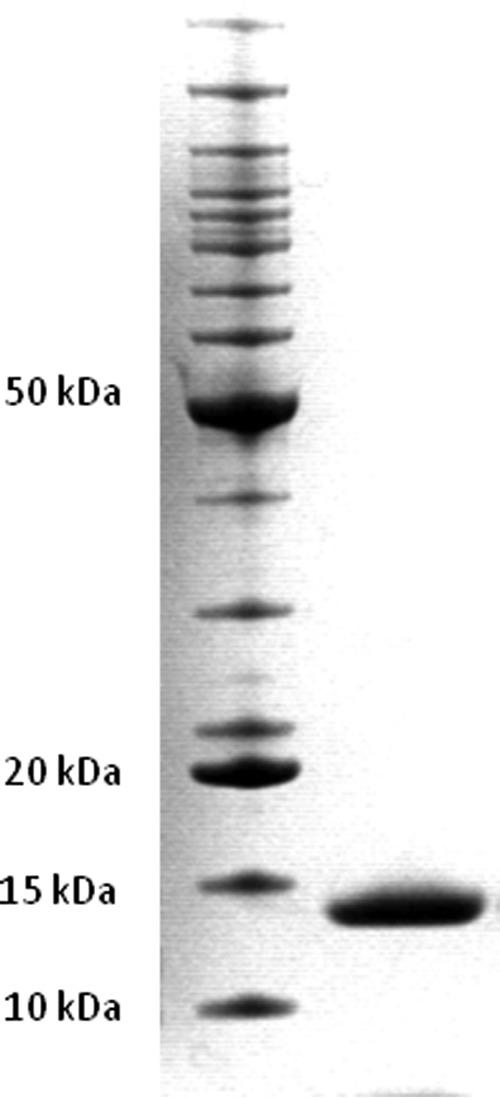
SDS-PAGE analysis of purified recombinant CmmAMP-I. Right lane, purified CmmAMP-I; left lane, BenchMark protein ladder (Life Technologies) with 15 proteins ranging from 10 to 220 kDa.
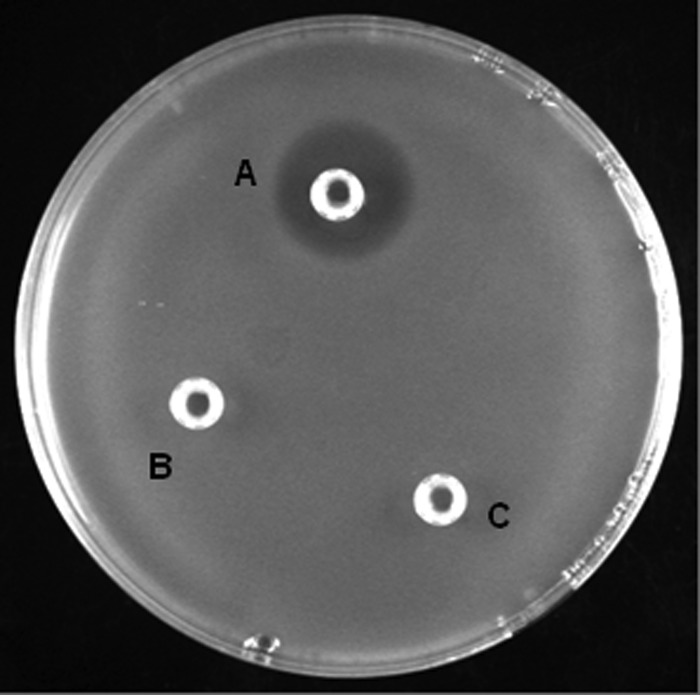
Fig 4.
Inhibition of C. michiganensis subsp. sepedonicus by recombinant CmmAMP-I. An NBY plate with a confluent layer of bacterial C. michiganensis subsp. sepedonicus cells, except for a halo around the well labeled “A,” is shown. The following samples were added to the wells: A, 25 μl of 5 μM CmmAMP-I in TN buffer; B and C, 25 μl TN buffer.
Antimicrobial activity.
The antimicrobial activity of purified CmmAMP-I was tested using plate assays with a total of 23 indicator strains: C. michiganensis subsp. michiganensis NCPPB1468 (the producer strain), C. michiganensis subsp. sepedonicus NCPPB2136, 1977-2, 1979-9, 1982-4, 1982-11, and 2006-A55, L. plantarum NC8, L. sakei Lb790, L. innocua 4202, E. faecalis V583, S. marcesens BJL200, S. faecalis NCDO581, P. pentosaceus LMGT2001, E. coli Top10, E. coli C3016, B. subtilis BAB-1, and 6 strains of Streptomyces spp. called 1-A, 1-B, 5-A, 5-B, 6-A, and 10-A. Using the setup and the concentrations shown in Fig. 4, no inhibitory activity was detected, except for the clear inhibitory activity on C. michiganensis subsp. sepedonicus illustrated by Fig. 4. Five of the six tested C. michiganensis subsp. sepedonicus strains were sensitive to CmmAMP-I, showing approximately similar halo sizes, whereas one, strain 1982-11, was not sensitive. These results show that CmmAMP-I has a very narrow spectrum of target organisms.
The IC50 for CmmAMP-I acting on C. michiganensis subsp. sepedonicus in liquid culture was estimated using microtiter plate dilution assays, which consistently yielded an IC50 of approximately 10 pM (i.e., CmmAMP-I was diluted 219 times relative to the concentration in well 1). Plating of cells recovered from the microtiter plate wells, and washed with NBY medium before plating, showed that all indicator cells were killed in wells containing CmmAMP-I at concentrations of approximately 40 pM and higher.
Despite several attempts, we did not manage to recombinantly produce the putative homologous AMP from C. michiganensis subsp. sepedonicus (Fig. 2). The same intein-tag-based approach that was successful for CmmAMP-I consistently yielded inclusion bodies under all conditions tested (various promoters, various expression temperatures). Attempts to refold the protein in the inclusion bodies failed. We also tested the possible presence of a C. michiganensis subsp. michiganensis strain inhibiting activity in the supernatant from C. michiganensis subsp. sepedonicus and did not observe any such inhibition.
Phytotoxicity.
For application in bacterial ring rot control, CmmAMP-I should have no toxic effects on potato plants. To test the effect of CmmAMP-I, 275 ng (55 ng/μl) of the bacteriocin was spotted on potato leaflets and tubers as well as on slices of tubers. Because of its origin in a tomato pathogen, phytotoxicity was also tested on leaflets and on whole and sliced fruits of tomatoes. No visible effects could be observed on any of the plant parts treated with CmmAMP-I after 1 week, while the herbicide-treated leaflet controls, in particular, the potato leaflets, showed extensive necrosis.
DISCUSSION
The present data show that CmmAMP-I is a novel type of protein bacteriocin which currently does not have any reported characterized homologues. This novel bacteriocin kills its target bacteria at concentrations in the low picomolar range, corresponding to 0.1 to 0.5 ng/ml. Thus, CmmAMP-I is at least as powerful as the most powerful of peptide bacteriocins (18). Most known protein bacteriocins from plant pathogens are from Gram-negative species, as these are the most dominant among plant-pathogenic bacteria. Known protein bacteriocins from the plant-pathogenic bacteria Pseudomonas syringae, Pectobacterium carotovorum, Xylella fastidiosa, and Xanthomonas oryzae are related to the high-molecular-mass (>50-kDa) colicins (19–22). Another group of high-molecular-mass protein bacteriocins from Gram-negative plant-pathogenic bacteria comprises proteins that affect the target cell membrane by self-assembly into particles resembling the tails of bacteriophages. The best-studied of the latter bacteriocins is probably carotovoricin (23). There are several examples of plant pathogens producing other protein bacteriocins that do not resemble the “standard” types of antimicrobial proteins mentioned above and/or that have not been sufficiently characterized to allow some sort of classification (4).
In general, very little is known about protein bacteriocins from Gram-positive bacteria and evidently even less about protein bacteriocins from Gram-positive plant pathogens. Protein bacteriocins from Gram-positive species show large variations in structure, and their activities seem to involve a wide range of mechanisms. Several protein bacteriocins from Gram-positive species are in fact cell-wall hydrolyzing (murolytic) enzymes, such as lysostaphin (24). Others, with largely unknown mechanisms, include the 37-kDa helveticin J (25), the 17-kDa streptococcin A-M57 (26), the 21.5-kDa dysgalacticin (27), and the similar 17-kDa proteins SA-M57, enterococcin V583, and corynicin JK (28). The three latter proteins have been studied in some detail, and it has been shown that the basic C-terminal regions of these otherwise acidic proteins are responsible for their killing activity. The best-studied protein bacteriocin from plant-pathogenic Gram-positive species is ipomicin, a heat-sensitive 10-kDa protein produced by the sweet-potato pathogen Streptomyces ipomoea (29). The antagonistic activity of ipomicin seems limited to closely related strains, i.e., primarily other strains of S. ipomoea. It is known that ipomicin works by lysing the target cells, but the exact mechanism remains to be elucidated (29). CmmAMP-I did not show any features that suggest a functional relationship with any of these known protein bacteriocins.
Interestingly, one of the two homologous sequences found in the database is that of a secreted protein of unknown function in C. michiganensis subsp. sepedonicus. It is tempting to speculate that, by analogy to the function of CmmAMP-I, this C. michiganensis subsp. sepedonicus protein could be a selective inhibitor of C. michiganensis subsp. michiganensis. Despite quite massive attempts, as described above, we have not been able to demonstrate such an activity. Recombinant production of the protein failed, whereas we could not detect activity against C. michiganensis subsp. michiganensis in culture supernatants of C. michiganensis subsp. sepedonicus. The present results show that more work on the potential C. michiganensis subsp. sepedonicus AMP (CmsAMP) is warranted. It is quite likely that this protein will be produced and secreted under certain conditions and that it will have an antimicrobial activity, albeit perhaps not against C. michiganensis subsp. michiganensis.
CmmAMP-I has a strong antibacterial activity in vitro, specifically inhibiting the growth of the ring rot pathogen by killing it. Studies of phytoxicity showed no apparent toxic effect on potato (or tomato) plants when droplets of CmmAMP-I were applied with concentrations approximately 5 orders of magnitude higher that the IC50. Hence, in vitro data suggest that CmmAMP is a good candidate for potential management of potato ring rot. Since there currently are no direct tools available to combat potato ring rot, the commercial potential of a new, environmentally friendly combat strategy is significant. Studies on other systems (30) indicate that one could consider using the producing organism itself as a biocontrol agent. This, however, is not possible because C. michiganensis subsp. michiganensis has the status of a serious plant pathogen itself. This leaves two potential application routes for CmmAMP-I. One is to overproduce the bacteriocin and apply it directly to plants and seed potatoes, a strategy that has worked well for other bacterial plant diseases (31). The other is to develop transgenic potatoes that express CmsAMP-I and hence are likely to be resistant to C. michiganensis subsp. sepedonicus. The latter strategy is well established in potato (32).
ACKNOWLEDGMENTS
This research was supported by two grants from The Research Council of Norway (grants 148024 and 199718). P.M. thanks the China Scholarship Council for a visiting scholarship.
We thank Vinh Hong Le for performing the phytotoxicity test.
Footnotes
Published ahead of print 12 July 2013
REFERENCES
- 1.Eijsink VG, Axelsson L, Diep DB, Håvarstein LS, Holo H, Nes IF. 2002. Production of class II bacteriocins by lactic acid bacteria; an example of biological warfare and communication. Antonie Van Leeuwenhoek 81:639–654 [DOI] [PubMed] [Google Scholar]
- 2.Riley MA, Gordon DM. 1999. The ecological role of bacteriocins in bacterial competition. Trends Microbiol. 7:129–133 [DOI] [PubMed] [Google Scholar]
- 3.Vidaver AK. 1983. Bacteriocins: the lure and the reality. Plant Dis. 67:471–475 [Google Scholar]
- 4.Holtsmark I, Eijsink VG, Brurberg MB. 2008. Bacteriocins from plant pathogenic bacteria. FEMS Microbiol. Lett. 280:1–7 [DOI] [PubMed] [Google Scholar]
- 5.Holtsmark I, Mantzilas D, Eijsink VG, Brurberg MB. 2006. Purification, characterization, and gene sequence of michiganin A, an actagardine-like lantibiotic produced by the tomato pathogen Clavibacter michiganensis subsp. michiganensis. Appl. Environ. Microbiol. 72:5814–5821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holtsmark I, Mantzilas D, Eijsink VG, Brurberg MB. 2007. The tomato pathogen Clavibacter michiganensis ssp. michiganensis: producer of several antimicrobial substances. J. Appl. Microbiol. 102:416–423 [DOI] [PubMed] [Google Scholar]
- 7.Chatterjee C, Paul M, Xie L, van der Donk WA. 2005. Biosynthesis and mode of action of lantibiotics. Chem. Rev. 105:633–684 [DOI] [PubMed] [Google Scholar]
- 8.Bierbaum G, Sahl HG. 2009. Lantibiotics: mode of action, biosynthesis and bioengineering. Curr. Pharm. Biotechnol. 10:2–18 [DOI] [PubMed] [Google Scholar]
- 9.Gartemann KH, Abt B, Bekel T, Burger A, Engemann J, Flügel M, Gaigalat L, Goesmann A, Gräfen I, Kalinowski J, Kaup O, Kirchner O, Krause L, Linke B, McHardy A, Meyer F, Pohle S, Rückert C, Schneiker S, Zellermann EM, Pühler A, Eichenlaub R, Kaiser O, Bartels D. 2008. The genome sequence of the tomato-pathogenic actinomycete Clavibacter michiganensis subsp. michiganensis NCPPB382 reveals a large island involved in pathogenicity. J. Bacteriol. 190:2138–2149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schaad NW. 1980. Laboratory guide for identification of plant pathogenic bacteria. American Phytopathological Society, St; Paul, MN [Google Scholar]
- 11.Rappsilber J, Ishihama Y, Mann M. 2003. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 75:663–670 [DOI] [PubMed] [Google Scholar]
- 12.Diep DB, Skaugen M, Salehian Z, Holo H, Nes IF. 2007. Common mechanisms of target cell recognition and immunity for class II bacteriocins. Proc. Natl. Acad. Sci. U. S. A. 104:2384–2389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geis A, Singh J, Teuber M. 1983. Potential of lactic streptococci to produce bacteriocin. Appl. Environ. Microbiol. 45:205–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8:785–786 [DOI] [PubMed] [Google Scholar]
- 15.Gasteiger EHC, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. 2005. Protein identification and analysis tools on the ExPASy Server, p 571–607 In Walker JM. (ed), The proteomics protocols handbook. Humana Press, Totowa, NJ [Google Scholar]
- 16.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 17.Zerillo MM, Van Sluys MA, Camargo LE, Monteiro-Vitorello CB. 2008. Characterization of new IS elements and studies of their dispersion in two subspecies of Leifsonia xyli. BMC Microbiol. 8:127. 10.1186/1471-2180-8-127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eijsink VG, Skeie M, Middelhoven PH, Brurberg MB, Nes IF. 1998. Comparative studies of class IIa bacteriocins of lactic acid bacteria. Appl. Environ. Microbiol. 64:3275–3281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feil H, Feil WS, Chain P, Larimer F, DiBartolo G, Copeland A, Lykidis A, Trong S, Nolan M, Goltsman E, Thiel J, Malfatti S, Loper JE, Lapidus A, Detter JC, Land M, Richardson PM, Kyrpides NC, Ivanova N, Lindow SE. 2005. Comparison of the complete genome sequences of Pseudomonas syringae pv. syringae B728a and pv. tomato DC3000. Proc. Natl. Acad. Sci. U. S. A. 102:11064–11069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chuang DY, Chien YC, Wu HP. 2007. Cloning and expression of the Erwinia carotovora subsp. carotovora gene encoding the low-molecular-weight bacteriocin carocin S1. J. Bacteriol. 189:620–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van Sluys MA, de Oliveira MC, Monteiro-Vitorello CB, Miyaki CY, Furlan LR, Camargo LEA, da Silva ACR, Moon DH, Takita MA, Lemos EGM, Machado MA, Ferro MIT, da Silva FR, Goldman MHS, Goldman GH, Lemos MVF, El-Dorry H, Tsai SM, Carrer H, Carraro DM, de Oliveira RC, Nunes LR, Siqueira WJ, Coutinho LL, Kimura ET, Ferro ES, Harakava R, Kuramae EE, Marino CL, Giglioti E, Abreu IL, Alves LMC, do Amaral AM, Baia GS, Blanco SR, Brito MS, Cannavan FS, Celestino AV, da Cunha AF, Fenille RC, Ferro JA, Formighieri EF, Kishi LT, Leoni SG, Oliveira AR, Rosa VE, Sassaki FT, Sena JAD, de Souza AA, Truffi D, Tsukumo F, Yanai GM, Zaros LG, Civerolo EL, Simpson AJG, Almeida NF, Setubal JC, Kitajima JP. 2003. Comparative analyses of the complete genome sequences of Pierce's disease and citrus variegated chlorosis strains of Xylella fastidiosa. J. Bacteriol. 185:1018–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ochiai H, Inoue V, Takeya M, Sasaki A, Kaku H. 2005. Genome sequence of Xanthomonas oryzae pv. oryzae suggests contribution of large numbers of effector genes and insertion sequences to its race diversity. Jpn. Agric. Res. Q. 39:275–287 [Google Scholar]
- 23.Yamada K, Hirota M, Niimi Y, Nguyen HA, Takahara Y, Kamio Y, Kaneko J. 2006. Nucleotide sequences and organization of the genes for carotovoricin (Ctv) from Erwinia carotovora indicate that Ctv evolved from the same ancestor as Salmonella typhi prophage. Biosci. Biotechnol. Biochem. 70:2236–2247 [DOI] [PubMed] [Google Scholar]
- 24.Bastos M, Coutinho B, Coelho M. 2010. Lysostaphin: a staphylococcal bacteriolysin with potential clinical applications. Pharmaceuticals 3:1139–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Joerger MC, Klaenhammer TR. 1990. Cloning, expression, and nucleotide sequence of the Lactobacillus helveticus 481 gene encoding the bacteriocin helveticin J. J. Bacteriol. 172:6339–6347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heng NC, Burtenshaw GA, Jack RW, Tagg JR. 2004. Sequence analysis of pDN571, a plasmid encoding novel bacteriocin production in M-type 57 Streptococcus pyogenes. Plasmid 52:225–229 [DOI] [PubMed] [Google Scholar]
- 27.Heng NC, Ragland NL, Swe PM, Baird HJ, Inglis MA, Tagg JR, Jack RW. 2006. Dysgalacticin: a novel, plasmid-encoded antimicrobial protein (bacteriocin) produced by Streptococcus dysgalactiae subsp. equisimilis. Microbiology 152:1991–2001 [DOI] [PubMed] [Google Scholar]
- 28.Swe PM, Heng NC, Ting YT, Baird HJ, Carne A, Tauch A, Tagg JR, Jack RW. 2007. ef1097 and ypkK encode enterococcin V583 and corynicin JK, members of a new family of antimicrobial proteins (bacteriocins) with modular structure from Gram-positive bacteria. Microbiology 153:3218–3227 [DOI] [PubMed] [Google Scholar]
- 29.Zhang X, Clark CA, Pettis GS. 2003. Interstrain inhibition in the sweet potato pathogen Streptomyces ipomoeae: purification and characterization of a highly specific bacteriocin and cloning of its structural gene. Appl. Environ. Microbiol. 69:2201–2208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hert AP, Marutani M, Momol MT, Roberts PD, Olson SM, Jones JB. 2009. Suppression of the bacterial spot pathogen Xanthomonas euvesicatoria on tomato leaves by an attenuated mutant of Xanthomonas perforans. Appl. Environ. Microbiol. 75:3323–3330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lavermicocca P, Lonigro SL, Valerio F, Evidente A, Visconti A. 2002. Reduction of olive knot disease by a bacteriocin from Pseudomonas syringae pv. ciccaronei. Appl. Environ. Microbiol. 68:1403–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rivero M, Furman N, Mencacci N, Picca P, Toum L, Lentz E, Bravo-Almonacid F, Mentaberry A. 2012. Stacking of antimicrobial genes in potato transgenic plants confers increased resistance to bacterial and fungal pathogens. J. Biotechnol. 157:334–343 [DOI] [PubMed] [Google Scholar]