Abstract
Stenotrophomonas maltophilia is an important global opportunistic pathogen for which limited therapeutics are available because of the emergence of multidrug-resistant strains. A novel bacteriocin, maltocin P28, which is produced by S. maltophilia strain P28, may be the first identified phage tail-like bacteriocin from S. maltophilia. Maltocin P28 resembles a contractile but nonflexible phage tail structure based on electron microscopy, and it is sensitive to trypsin, proteinase K, and heat. SDS-PAGE analysis of maltocin P28 revealed two major protein bands of approximately 43 and 20 kDa. The N-terminal amino acid residues of these two major subunits were sequenced, and the maltocin P28 gene cluster was located on the S. maltophilia P28 chromosome. Our sequence analysis results indicate that this maltocin gene cluster consists of 23 open reading frames (ORFs), and that its gene organization is similar to that of the P2 phage genome and R2 pyocin gene cluster. ORF17 and ORF18 encode the two major structural proteins, which correspond to gpFI (tail sheath) and gpFII (tail tube) of P2 phage, respectively. We found that maltocin P28 had bactericidal activity against 38 of 81 tested S. maltophilia strains. Therefore, maltocin P28 is a promising therapeutic substitute for antibiotics for S. maltophilia infections.
INTRODUCTION
Stenotrophomonas maltophilia is an aerobic, nonfermentative, Gram-negative bacillus found in a variety of aquatic, soil, and plant rhizosphere environments (1). Over the last 2 decades, it has emerged as a global opportunistic pathogen that causes considerable morbidity and mortality in immunosuppressed patients (2, 3). S. maltophilia can cause serious infections, including pneumonia, bloodstream infections, wound/soft-tissue infections, and urinary tract infections (2, 4, 5). Recently, fatal infections in humans have been documented (6). S. maltophilia is also associated with equine, canine, and feline respiratory infections and bovine mastitis (7–9). Furthermore, S. maltophilia is an environmental multidrug-resistant organism that exhibits high-level intrinsic resistance to most currently available broad-spectrum antibiotic agents (10, 11). Similar to other pathogenic strains, S. maltophilia can acquire antibiotic resistance during therapy (12, 13). Thus, managing S. maltophilia infections is difficult because of the lack of effective antimicrobials, and novel treatment strategies are required (2, 3).
Bacteriocins are a promising alternative to antibiotics for treating bacterial infections. They are ribosomally synthesized proteinaceous compounds that only have bactericidal activity against bacteria that are closely related to the producing strain (14–16). Based on their molecular weight, these bacteriocins can be divided into two distinct groups: high-molecular-weight (HMW) and low-molecular-weight bacteriocins. HMW bacteriocins are easily isolated and visible using electron microscopy (17, 18). The most intensively studied HMW bacteriocins are the colicins produced by Escherichia coli (19), as well as the pyocins from Pseudomonas aeruginosa (20). Although colicins are plasmid encoded, pyocin gene clusters are exclusively located on the chromosome (20). There are three types of pyocins: S-, R-, and F-type. S-type pyocins are colicin-like proteins, whereas R- and F-type pyocins are phage tail-like particles. R-type pyocin resembles a nonflexible but contractile phage tail structure, while F-type pyocin is similar to the flexible but noncontractile tails (20). R- and F-type pyocins are related to P2 phage and lambda phage of E. coli, respectively (21).
Bacteriocins have been described in many species of Gram-negative and Gram-positive bacteria. It is believed that the majority of bacteria can produce at least one bacteriocin (18, 22, 23), but some bacteria simultaneously produce various types of bacteriocins. For example, P. aeruginosa strain PAO1 can synthesize all three types of pyocins (20). However, no bacteriocin from S. maltophilia has previously been identified. By evaluating several S. maltophilia isolates, we found that strain P28 produces a novel phage tail-like bacteriocin, which was designated maltocin P28 according to the typical bacteriocin nomenclature (17). In this context, maltocin P28 was purified and characterized, and the maltocin P28 gene cluster location was determined.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The strains and plasmids used in this work are listed in Table 1, and the oligonucleotides used are provided in Table 2. All S. maltophilia strains were isolated from either soil (36 strains) or clinical (44 strains) samples. S. maltophilia P28, the producer of maltocin P28, is an environmental isolate. The clinical S. maltophilia strain c6 was used as the sensitive indicator strain for most experiments. Other strains (besides S. maltophilia) used to determine the inhibitory activities of maltocin are the following: Aeromonas media AB208100, Bacillus subtilis AB93017, Enterobacter aerogenes AB91102, Escherichia coli ATCC 47076, Proteus vulgaris AB91103, Pseudomonas aeruginosa ATCC 15692, DSM19882, Pseudomonas fluorescens AB92001, Pseudomonas putida ATCC 12633, Staphylococcus aureus AB91053, and Xanthomonas campestris AB96030. Strain names beginning with “AB” were obtained from the China Center for Type Culture Collection (CCTCC). All strains were cultured in Luria-Bertani (LB) medium at 30°C, except for E. coli, which was cultured at 37°C. The medium was supplemented with ampicillin (100 μg/ml) and chloramphenicol (34 μg/ml) when required.
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| E. coli S17-1λpir | hsdR pro recA; RP4-2 in chromosome; Km::Tn7 (Tc::Mu) | 32 |
| S. maltophilia P28 | Maltocin P28 producer, Ampr | Laboratory collection |
| S. maltophilia P28 Δorf17 | S. maltophilia P28 orf17 deletion mutant | This study |
| S. maltophilia P28 Δorf17/pBBR1MCS | S. maltophilia P28 Δorf17 complemented with pBBR1MCS | This study |
| S. maltophilia P28 Δorf17/pBBR1MCS-orf17 | S. maltophilia P28 Δorf17 complemented with pBBR1MCS-orf17 | This study |
| S. maltophilia P28 Δorf17/pBBR1MCSPlac-orf17 | S. maltophilia P28 Δorf17 complemented with pBBR1MCSPlac-orf17 | This study |
| Plasmids | ||
| pBBR1MCS | Mobilizable broad-host-range cloning vector, Cmr | 33 |
| pBBR1MCS-orf17 | pBBR1MCS with a complete orf17 gene | This study |
| pBBR1MCSPlac-orf17 | pBBR1MCS-orf17 with an rrn terminator and lacZ promoter | This study |
| pDM4 | pir dependent with sacAB genes; oriR6K; Cmr | 32 |
| pDM4-17 | pDM4 containing orf17 fragment for construction of P28 Δorf17 strain | This study |
Table 2.
Primers used in this study
| Primer | Sequencea (5′→3′) | Use |
|---|---|---|
| RepF | GGGTCTTGAACTGGATGAAACGG | Detection of ϕSHP2 |
| RepR | TTCCTTGCGATTGGAGTTGCG | Detection of ϕSHP2 |
| 1038R | AAGAAGGAGAACTGGTCGTTCGGCAA | Smlt1039-1044 amplicon |
| 1045F | GGACTTCGACTTCACTCCGACCTACCC | Smlt1039-1044 amplicon |
| 1045R | CGCACCACGCACAGGAACTGCCACTT | Smlt1046-1053 amplicon |
| 1054F | CAGAAGGAGCACGCACTGGAAACACT | Smlt1046-1053 amplicon |
| 1054R | TCGGAATGTCGGTGCCTTCGGATAC | Smlt1055-1063 amplicon |
| 1064F | CTCCCATCACATCCTGACCACACTGC | Smlt1055-1063 amplicon |
| 1055F | CAGATCGCCCAGCAGAG | Smlt1054 amplicon |
| 1053R | TCGATGTCCCGTTGCAG | Smlt1054 amplicon |
| 1046F | GTCCGCCAGCTACGAAT | Smlt1044-1045 amplicon |
| 1043R | GGTGTCGGTGGGGGT | Smlt1044-1045 amplicon |
| Sheath LA-F | AAACTCGAGTCTCGATCCTGATGTATG | P28Δorf17 |
| Sheath LA-R | AAATCTAGATCGGGGTTCTTCCTCGG | P28Δorf17 |
| Sheath RA-F | AAATCTAGAGCATGACGCGCAAGATCCG | P28Δorf17 |
| Sheath RA-R | AAAGAGCTCGGGCGATCATGTTGAC | P28Δorf17 |
| Knock verify-F | AGGGCTACTCGGCAAGCAAT | P28Δorf17 |
| Knock verify-R | CATCGCTGCCCTTGTCGT | P28Δorf17 |
| Sheath-F | AAACTCGAGCGGCACACACCCACAC | Complementation of strain P28Δorf17 |
| Sheath-R | AAAGAGCTCTTGCGCGTCATGCTTAGA | Complementation of strain P28Δorf17 |
| Plac-F | AAACTCGAGACCGCGGAAGCTTAGC | Complementation of strain P28Δorf17 |
| Plac-R | AAAGGGCCCATTATTGCATGCTGCC | Complementation of strain P28Δorf17 |
Added restriction enzyme recognition sites in primers are underlined.
Production and purification of maltocin P28.
S. maltophilia strain P28 was grown in 200 ml of LB medium with shaking (200 rpm) at 30°C. The culture was harvested during early exponential growth (A600 = 0.5) and divided in two. Half was treated with mitomycin C (0.5 μg/ml). After incubation overnight at 30°C, both the induced and noninduced cultures were centrifuged at 10,000 × g for 10 min at 4°C. The supernatants were precipitated with 4% (wt/vol) polyethylene glycol (PEG) 8000 and 3% (wt/vol) NaCl at 4°C for 24 h. Pellets were collected by centrifugation at 14,000 × g for 30 min at 4°C and resuspended in 1 ml of TE buffer (10 mM Tris-Cl and 1.0 mM EDTA, pH 8.0). After storage at 4°C overnight, the suspensions were centrifuged at 10,000 × g for 10 min at 4°C to remove impurities. Afterwards, the resulting supernatants were passed through a 0.45-μm filter.
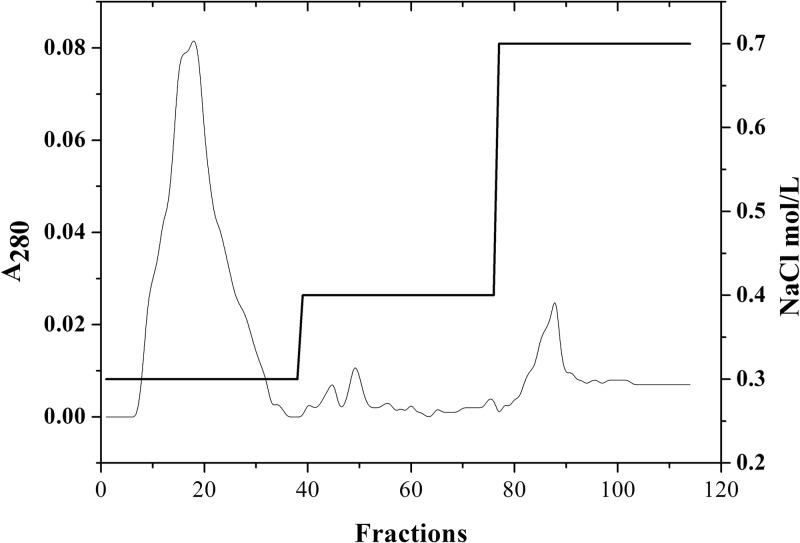
For maltocin purification, the partial preparation was chromatographed on a DEAE-cellulose (DEAE) column (10 by 175 mm) with a nonlinear gradient of NaCl (0, 0.3, 0.4, and 0.7 M) in 0.01 M phosphate buffer (pH 6.8). All fractions absorbing at 280 nm were tested for the presence of maltocin P28 or phage ϕSHP2 based on bactericidal activity against the sensitive strain, followed by PCR analysis using the primer pair (Table 2) designed according to the sequence of ϕSHP2 genomic DNA.
Transmission electron microscopy.
Samples were dropped onto a 300- by 300-mesh copper grid. After 3 min, particles were stained with 2% phosphotungstic acid for 2 min and then examined using a Tecnai G2 transmission electron microscope (FEI Co., Hillsboro, OR) at an operating voltage of 200 kV.
Quantification of antimicrobial activity.
Maltocin antimicrobial activity was determined by following a protocol described for other bacteriocins (24, 25). A total of 200 μl of each exponentially growing indicator strain was added to 5 ml of LB soft agar (0.7% agar) at 50°C, mixed, and overlaid on LB plates. Serial (2-fold) dilutions of the maltocin preparations were made in TE buffer, and 5 μl of each dilution was spotted onto a lawn of each indicator strain. The plates were incubated at 30°C overnight. The reciprocal of the highest dilution that formed a clear zone was defined as the relative activity (in activity units [AU]) of maltocin.
Inhibitory action of maltocin P28.
To analyze the killing action of maltocin P28, CFU counts of S. maltophilia c6 were determined upon addition of various concentrations of maltocin as follows. A culture of 50 ml was grown at 30°C to an optical density at 600 nm (OD600) of 0.50 and divided into 10-ml aliquots. Maltocin was added to final concentrations of 2.6 × 104, 2.6 × 103, 2.6 × 102, and 0 AU ml−1, and portions of the treated cultures were removed after 2, 5, 10, 15, 30, 60, and 120 min. The bacteria were immediately washed once with 0.9% (wt/vol) NaCl, and CFU were determined on LB plates.
Sensitivity of maltocin to heat and enzyme treatment.
For thermostability testing of maltocin P28, purified maltocin was heated for 10 min at 30, 40, 45, 50, 55, and 60°C. Bacteriocin enzymatic stability was investigated following treatment with trypsin (Gibco, Carlsbad, CA) (0.5 mg/ml) and proteinase K (Merck, Darmstadt, Germany) (0.2 mg/ml) in TE buffer at 37°C for 1 h. Residual bacteriocin activity following each treatment was determined as described above.
Protein analysis.
SDS-PAGE analysis of purified maltocin and an N-terminal sequence analysis of its major structural subunits were performed as described previously (26). Two major protein bands (approximately 43 and 20 kDa) from maltocin P28 particles separated by SDS-PAGE were electroblotted onto a polyvinylidene difluoride membrane and stained with Coomassie brilliant blue. Membrane strips containing the major protein bands were excised and subjected to Edman degradation to determine the N-terminal sequences using an ABI ProciseTM492cLC (GC320078) protein sequencer.
Location and analysis of the maltocin P28 locus.
The primers used to locate the maltocin P28 locus are listed in Table 2. To identify the gene cluster encoding maltocin proteins, the sequences of five N-terminal amino acid residues of both major subunits (approximately 43 and 20 kDa) were searched against known protein databases of sequenced S. maltophilia strains (27, 28). The two amino acid sequences were separately located in two putative phage-related proteins encoded by conjoined genes (Smlt1045 and Smlt1044) on the S. maltophilia K279a chromosome. A sequence analysis showed that the genes between Smlt1064 and Smlt1038 in strain K279a may be associated with phage genes. To clone the maltocin genes, primers 1046F and 1043R were designed according to sequences of the flanking genes Smlt1046 and Smlt1043 in S. maltophilia K279a. Chromosomal DNA from strain P28 was used as the template. The amplified DNA fragment showed 99% sequence identity to that of the K279a genome. Four primer pairs (1038R and 1045F, 1045R and 1054F, 1054R and 1064F, and 1053R and 1055F) were synthesized according to the genomic sequence of K279a to amplify the complete locus. Using genomic DNA from S. maltophilia P28 as the template, four fragments (5.3, 7.6, 5.5, and 0.8 kb) were amplified and sequenced. The obtained sequences were assembled using DNAMAN software (Lynnon Biosoft, Quebec, Canada) and compared to the genomic sequences of S. maltophilia K279a and R551-3.
In our analyses, open reading frames (ORFs) were defined as those containing either AUG (methionine), GUG (valine), or UUG (leucine) as a start codon and were composed of at least 50 amino acids based on ORF-Finder and BLAST from NCBI (29) (http://www.ncbi.nlm.nih.gov/gorf/gorf.html).
Identifying similarities between predicted maltocin proteins and known proteins in the databases was performed using the internet versions of PSI-BLAST and PHI-BLAST (http://www.ncbi.nlm.nih.gov/BLAST/) (30). Putative transmembrane helices in the proteins were predicted using the ExPASy server (31).
Construction of the ORF17 deletion strain.
The primers used to construct the ORF17 deletion strain are also listed in Table 2. ORF17 was predicted to encode the major tail sheath protein of S. maltophilia P28. The amplified 609- and 529-bp fragments were located upstream and downstream, respectively, of ORF17. Both were subcloned into the suicide vector pDM4 (32). Subsequently, the recombinant plasmid pDM4-17 was transformed into E. coli S17-1 (λpir) and transferred to S. maltophilia P28 by conjugation. S. maltophilia exconjugants containing the first allelic exchange were screened on LB agar supplemented with ampicillin-chloramphenicol. Colonies were added to 5 ml of liquid LB medium. After growth for 12 h, 100 μl of 103-fold-diluted culture was plated on sucrose LB agar (5% sucrose) and incubated at 30°C for 2 days. The ORF17 deletion mutant P28Δorf17 was then screened by PCR.
Complementation of strain P28Δorf17.
A 1,266-bp fragment containing ORF17 and its upstream 30 bp was amplified by PCR and cloned into pBBR1MCS (33) using XhoI and SacI. The resulting plasmid, pBBR1MCS-orf17, was transferred to strain P28Δorf17 by conjugation. Unfortunately, maltocin P28 production could not be restored by the recombinant strains carrying pBBR1MCS-orf17. We then cloned a fragment (including an rrn terminator and lacZ promoter) directly upstream of ORF17 in pBBR1MCS-orf17 using XhoI and ApaI. The recombinant plasmid, pBBR1MCSPlac-orf17, was transferred to strain P28Δorf17, and exconjugants were selected as described above.
Nucleotide sequence accession numbers.
The nucleotide sequences of maltocin P28 and phage ϕSHP2 have been registered in GenBank under accession numbers KC787694 and NC_015586, respectively.
RESULTS
Purification and morphology of maltocin P28 particles.
The complete nucleotide sequence of the high-copy-number plasmid pSH2 from S. maltophilia P28 was determined. Informatics analysis showed that both the gene organization and protein identities of five ORFs were similar to those of ϕSHP1 and ϕSMA9 (26, 34). The putative products of ORF1, ORF2, ORF7, ORF8, and ORF9 contained the Rep_trans superfamily, Phage_DNA_bind superfamily, Zot superfamily, DUF3653 superfamily, and HTH_XRE superfamily, respectively. The potential replication initiation factor (Rep) encoded by ORF1 shared 76.9% identity with Rep of ϕSHP1 and 24.4% identity with RstA of ϕSMA9. Similar to Rep protein, the putative single-stranded DNA binding protein (SSB) encoded by ORF2 shared high identity (77.3%) with SSB of ϕSHP1 and low identity (25.0%) with the ORF5 product of ϕSMA9. The identity of the ORF7 product to the predicted Zot of ϕSHP1 and ϕSMA9 is 27.6 and 20.8%, respectively. The product of ORF8 had the same identity (57.5%) to both the ORF9 product of ϕSHP1 and the ORF2 product of ϕSMA9. However, the ORF9 protein had slightly lower identity to the related proteins, the ORF10 product of ϕSHP1 (19.0%) and ORF1 protein of ϕSMA9 (22.8%). Thus, we propose that plasmid pSH2 is the replication form of a novel filamentous phage, named ϕSHP2 (see Fig. S1 and Table S1 in the supplemental material). The prepared samples contained single-stranded DNA, and its sequence coincided with that of plasmid ϕSH2, which was deposited in GenBank under accession number NC_015586.
Electron micrographs of the partial preparation revealed a mixture of phage tails and filamentous phages (Fig. 1A). The sample suspension was separated using DEAE-cellulose column chromatography, which generated a major eluted fraction at 0.3 M NaCl and a minor fraction at 0.7 M NaCl (Fig. 2). Electron micrographs taken from the peak fractions contained phage tail-like particles (120 by 20 nm) in the major eluted fraction (Fig. 1B) and filamentous particles (800 by 10 nm) in the minor eluted fraction (Fig. 1C). These two fractions were also analyzed for the presence of DNA and bactericidal activity against S. maltophilia c6. We found that the minor eluted fraction had no antimicrobial activity and contained single-stranded DNA, corresponding to ϕSHP2 genomic DNA based on both enzymatic digestion and primer extension (data not shown). However, the major eluted fraction had antimicrobial activity but contained no genetic material. Taken together, these results indicate that a novel filamentous phage, ϕSHP2, and a novel phage tail-like bacteriocin, designated maltocin P28, were simultaneously produced by S. maltophilia strain P28. In this study, only maltocin P28 was characterized.
Fig 1.
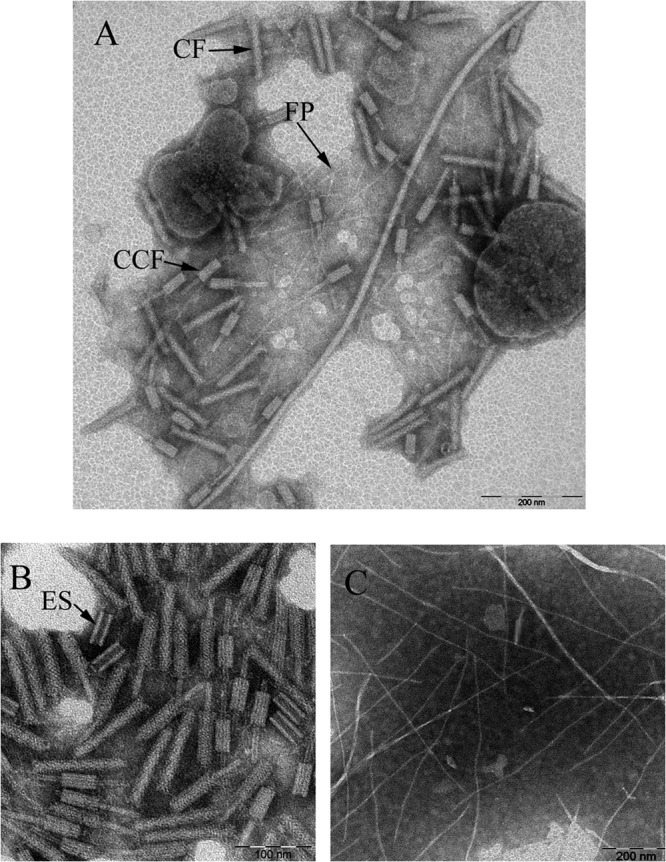
Electron micrographs of the partially purified sample (A), the major DEAE-eluted fraction at 0.3 M NaCl (B), and the minor DEAE-eluted fraction at 0.7 M NaCl (C). The maltocin P28 particles with different structural elements, namely, empty sheath, complete form, and complete contracted form, are marked ES, CF, and CCF, respectively. The filamentous phage is marked FP.
Fig 2.
DEAE chromatography of the partially purified maltocin P28. The partial preparation was chromatographed on a DEAE-cellulose (DEAE) column (10 by 175 mm) with a nonlinear gradient of NaCl (0, 0.3, 0.4, and 0.7 M) (thick line) in 0.01 M phosphate buffer (pH 6.8). The elution profile of the maltocin preparation was measured at an absorbance of 280 nm (thin line).
Antimicrobial activity spectrum of maltocin.
Purified maltocin P28 was tested against various strains collected in our laboratory. Among the 92 strains examined, no other bacteria, excluding 38 S. maltophilia strains (19 environmental strains and 19 clinical strains), showed growth sensitivity to maltocin P28. The most sensitive strain was c6; it was selected as the indicator strain. In a culture of strain P28, maximum spontaneous maltocin production (above 2.5 × 104 AU/ml) occurred after 20 h at 30°C with heavy shaking, and the yield could be increased to 4 × 105 AU/ml upon induction with mitomycin C (0.5 μg/ml).
Inhibitory action of maltocin P28.
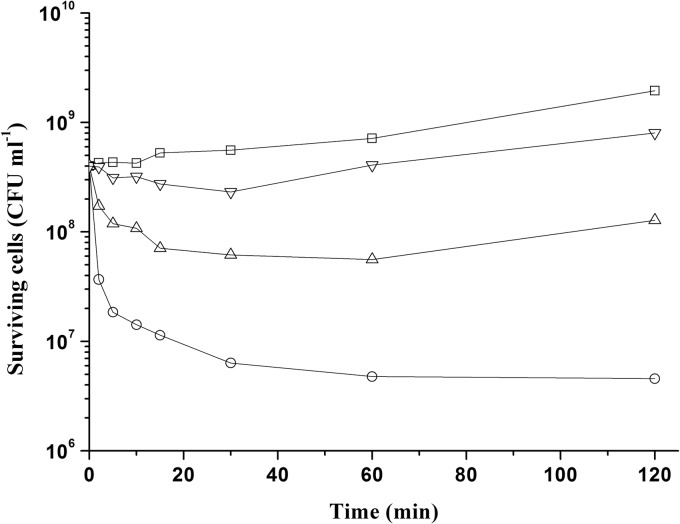
The lysis of the susceptible strain S. maltophilia c6 was determined by viable cell count upon addition of various concentrations of maltocin to an early exponential culture. As shown in Fig. 3, after treatment with a final maltocin concentration of 2.6 × 104 AU ml−1 for 120 min, the viable cell numbers could be reduced by about 98.9%. The other two lower concentrations (2.6 × 103 and 2.6 × 102 AU ml−1) showed weaker bactericidal activity and could delay the growth of the cultures for 60 and 30 min, respectively.
Fig 3.
Killing of S. maltophilia c6 after addition of maltocin P28 at final concentrations of 2.6 × 104 (○), 2.6 × 103 (Δ), 2.6 × 102 (▽), and 0 (□) AU ml−1. CFU were determined in triplicate, and the mean value at each time point was used to make the curve. The experiment was repeated twice with similar results.
Sensitivity of maltocin P28 to heat and enzyme treatment.
Purified maltocin P28 could be stored at 4°C for several months without loss of activity (data not shown). We also analyzed the sensitivity of the maltocin preparation to enzyme hydrolysis and heat treatment (Table 3). When maltocin P28 preparations were incubated for 10 min at different temperatures, no loss of bactericidal activity was detected up to 40°C, but more than 99% of the activity was lost at 45°C, and no activity was retained after incubation at 50°C. Incubation in the presence of trypsin for 1 h reduced maltocin activity to less than 10%; however, proteinase K treatment completely abrogated bactericidal activity.
Table 3.
Sensitivity of maltocin to heat and enzyme treatment
| Sensitivity toa: | |||||||
|---|---|---|---|---|---|---|---|
| Heat (°C) |
Enzyme |
||||||
| 30 | 40 | 45 | 50 | 55 | 60 | Trypsin (0.5 mg/ml) | Proteinase K (0.2 mg/ml) |
| 211 | 211 | 24 | 0 | 0 | 0 | 27 | 0 |
Residual antimicrobial activity (×200 AU/ml) was determined following each treatment. Heat treatment was for 10 min. Enzyme treatment was for 1 h at 37°C.
SDS-PAGE and N-terminal amino acid sequence of the maltocin P28 major subunits.
An analysis of maltocin P28 by SDS-PAGE displayed two major protein bands of about 43 and 20 kDa, as well as several minor bands (Fig. 4). The sequences of five N-terminal amino acid residues of the 43- and 20-kDa subunits were TEFLH and TRKIR, respectively. Homology searches of these two amino acid sequences using known protein databases of S. maltophilia revealed that they are identical to the N-terminal sequences of the Smlt1044 and Smlt1045 products, respectively, of S. maltophilia K279a.
Fig 4.
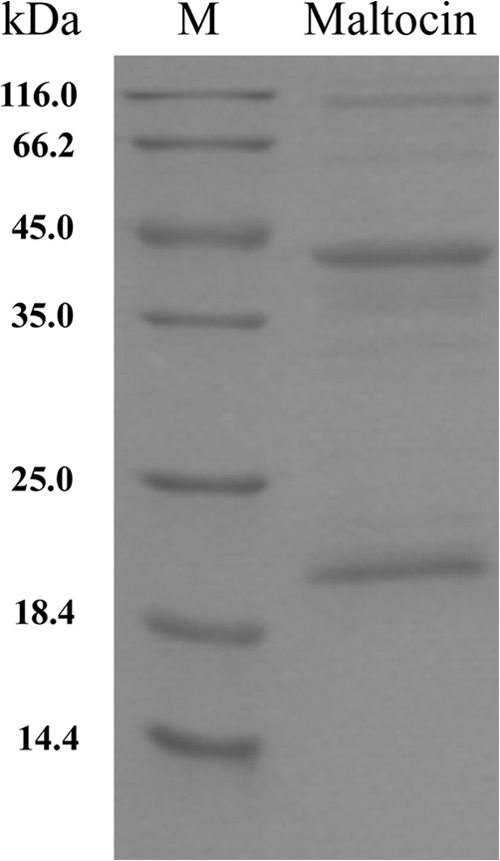
SDS-PAGE of the DEAE-purified maltocin P28 particles.
Genetic organization of the maltocin P28 gene locus.
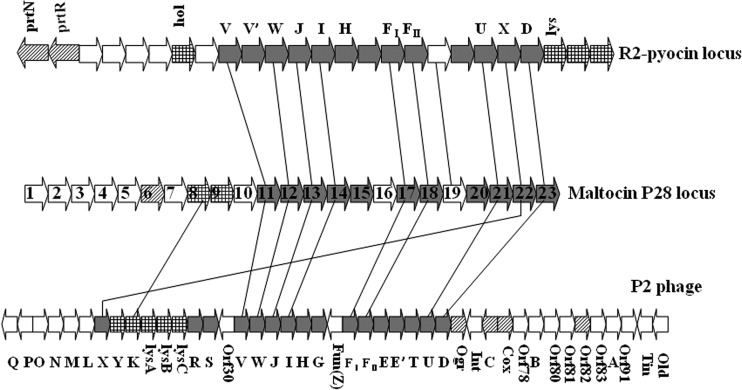
A 19.9-kb fragment was cloned from the S. maltophilia P28 genome. The %G+C was 66.2%, in agreement with the sequenced S. maltophilia chromosomes. A total of 25 ORFs were predicted after comparison to the nonredundant peptide sequence database using the BLAST program (BLAST algorithm) and ORF Finder software. Twenty-three ORFs were proposed to be maltocin P28-related genes (Table 4), all of which were transcribed in the same direction (opposite the directions of the 1st and 25th ORFs in the set). The genetic organization of maltocin P28 was similar to that of the R2 pyocin gene cluster and P2 phage genome (Fig. 5).
Table 4.
List of ORFs of genes in the maltocin P28 cluster
| ORF | Position (5′–3′) | Length of protein (aa) | Identity (%) throughout the whole length to: |
Conserved domain(s) | Predicted function | |
|---|---|---|---|---|---|---|
| P2 | Pyocin R2 | |||||
| 1 | 712–3048 | 778 | TOPRIM_primases superfamily, virulence-associated protein E superfamily | Unknown | ||
| 2 | 3049–3324 | 91 | Unknown | |||
| 3 | 3399–3626 | 75 | CE4-SF superfamily | Unknown | ||
| 4 | 3668–4306 | 212 | 1 superfamily (deoxynucleotide monophosphate kinase) | Unknown | ||
| 5 | 4459–4842 | 127 | Unknown | |||
| 6 | 5177–5626 | 149 | HTH superfamily, Mor superfamily | Transcriptional regulator | ||
| 7 | 5623–5973 | 116 | Unknown | |||
| 8 | 5970–6455 | 161 | gpK (48.8) | lysozyme_like superfamily | Endolysin | |
| 9 | 6452–6829 | 125 | Holin | |||
| 10 | 7182–7736 | 184 | Unknown | |||
| 11 | 7733–8323 | 196 | gpV (32.7) | VR2 (29.5) | Phage_base_V superfamily | Tail spike |
| 12 | 8376–8714 | 112 | gpW (33.6) | WR2 (41.1) | GPW_gp25 superfamily | Baseplate-assembly |
| 13 | 8717–9613 | 298 | gpJ (45.3) | JR2 (50.5) | Baseplate_J superfamily | Baseplate-assembly |
| 14 | 9606–10385 | 259 | gpI (22.3) | IR2 (20.5) | Tail formation | |
| 15 | 10393–12360 | 655 | Tail fiber | |||
| 16 | 12362–13021 | 219 | Unknown | |||
| 17 | 13128–14330 | 400 | gpFI (34.1) | FIR2 (43.5) | Phage_sheath_1 superfamily | Major tail sheath |
| 18 | 14333–14836 | 167 | gpFII (29.1) | FIIR2 (37.5) | Phage_tube superfamily | Major tail tube |
| 19 | 14917–15207 | 96 | PRF19 (24.4) | FluMu_gp41 superfamily | Unknown | |
| 20 | 15328–17775 | 815 | Tape_meas_TP901 family | Tail tape measure | ||
| 21 | 17778–18245 | 155 | gpU (24.3) | UR2 (16.9) | Phage_P2_GpU superfamily | Tail formation |
| 22 | 18229–18444 | 71 | gpX (39.4) | XR2 (39.4) | Phage_tail_X superfamily | Tail formation |
| 23 | 18501–19508 | 335 | gpD (24.2) | DR2 (26.9) | Phage_GPD superfamily | Tail formation |
Fig 5.
Comparison of genetic organization among the gene locus for the maltocin P28, pyocin R2, and P2 phage. Genes (indicated as large arrows) are not drawn to scale for simplicity. Homologous genes are connected by lines. Open reading frames encoding phage proteins involved in the tail phage synthesis are indicated by gray arrows. Open reading frames encoding phage regulators and lytic proteins are indicated by diagonal and gridding arrows separately. Open reading frames encoding proteins with unknown functions are indicated in white.
The predicted proteins encoded by ORF11 to ORF23 were related to phage tail proteins, nine of which contained conserved domains and showed significant homology to the tail proteins of P2 phage and R2 pyocin (Table 4). The arrangement of these genes was the same as that of R2 pyocin. ORF11 to ORF14 were homologous to V, W, J, and I, respectively, of P2 phage. Although ORF15 and ORF16 showed no homology to P2 family phages and R2 pyocin genes, their locations and product sizes were consistent with those of the P2 H (tail fiber) gene and G (tail fiber assembly) gene, respectively.
In addition, the predicted molecular masses were 41.7 and 18.2 kDa for ORF17 and ORF18, respectively, which coincided with the two most abundant structural components (43 and 20 kDa) of maltocin P28 as determined using SDS-PAGE analysis. The sequences of five N-terminal amino acid residues of the 43- and 20-kDa subunits were consistent with the predicted amino acid sequences of ORF17 and ORF18, excluding removal of the N-terminal methionine. The protein product of ORF17 was in the phage_sheath_1 superfamily and was similar to the phage tail sheath protein gpFI from P2 phage. The protein product of ORF18 was in the phage_tube superfamily and corresponded to the phage tail tube protein gpFII of P2 phage.
The products of ORF19 and ORF20 were in the FluMu_gp41 superfamily and Tape_meas_TP901 family, respectively. Neither protein showed homology to P2 phage proteins, while the ORF19 product showed significant homology to the predicted protein PRF19 in R2 pyocin (24.4% identity throughout the length), which is thought to correspond to the P2 E (essential tail protein) gene. As a corresponding ortholog of gpT of P2 phage, the ORF20 product was proposed to be the tail length determinator protein. Similar to the XR2 gene of R2 pyocin, ORF22 (a homolog of X from P2) was also located at the U-D intergenic region in P2 phage.
The termination and initiation codons overlapped between ORF10 and ORF11. PRF10 of R2 pyocin is thought to replace the P2 R or S genes responsible for tail completion. No homology of P2 R or S genes was found in the maltocin gene cluster. Similar to PRF10 of R2 pyocin, ORF10 was located upstream of structural genes. This suggests that the ORF10 product is involved in maltocin tail assembly, like PRF10 of R2 pyocin.
ORF6 to ORF9 should be included in one transcription unit, since the termination and initiation codons overlapped between the former ORF and the next ORF. The protein encoded by ORF6 had two conserved domains; namely, the helix-turn-helix superfamily and Mor superfamily, suggesting that ORF6 encodes a regulator. A sequence analysis indicated that ORF8 and ORF9 are related to lysis. The ORF8 protein contained a lysozyme_like superfamily conserved domain and exhibited significant homology to P2 gpK (48.8% identity throughout the length), which is an endolysin for disrupting the host cell wall. The protein encoded by ORF9 possessed the distinctive features of holins (N-terminal hydrophobic transmembrane and C-terminal hydrophilic domains), which could play a role in endolysin translocation (35). However, ORF7 did not have a homolog in P2 phage or other phages. Thus, determining whether ORF7 belongs to the lytic system or functions as a regulator requires further analysis.
The remaining ORFs (ORF1 to ORF5) on the maltocin P28 gene locus encoded five unknown proteins. The product of ORF1 had TOPRIM_primases superfamily and VirE superfamily domains on its N and C termini, respectively. ORF3 and ORF4 also had conserved domains. The ORF2 protein showed significant homology to a hypothetical protein of Escherichia phage vB_EcoM_ECO1230-10.
To confirm that the cloned gene cluster was responsible for maltocin P28 production, we constructed a mutant strain in which ORF17, coding for the tail sheath protein, was deleted. Bactericidal activity and the two major structural subunits of maltocin were not detected in the ORF17 deletion mutant preparation (Fig. 6). Furthermore, in trans complementation with ORF17 cloned into pBBR1MCS could restore bactericidal activity (Fig. 6). Thus, the maltocin P28 gene locus was identified.
Fig 6.
SDS-PAGE (A) and bactericidal activity (B) of partially purified maltocin sample from S. maltophilia P28 (lane 2), S. maltophilia P28 Δorf17 (lane 3), S. maltophilia P28 Δorf17/pBBR1MCSPlac-orf17 (lane 4), and S. maltophilia P28 Δorf17/pBBR1MCS (lane 5). The partially purified maltocin sample from MMC-induced S. maltophilia P28 (lane 1) is used as a maker. The tail sheath and tail tube proteins are indicated with arrows.
DISCUSSION
As for many phage tail-like bacteriocins (20, 24, 25, 36), maltocin P28 was mitomycin C inducible, thermolabile, and sensitive to proteinase K. Electron microscopy revealed that the structure of maltocin P28 was the same as that of R-type pyocins and resembled the inflexible and contractile tails of P2-related phages. Most predicted maltocin genes have homologs in the P2 phage genome and R2 pyocin gene cluster. Moreover, the arrangement of maltocin P28 genes was similar to that of the R2 pyocin cluster. To the best of our knowledge, maltocin P28 is the first identified bacteriocin produced by S. maltophilia. These results show that maltocin P28 is a novel phage tail-like bacteriocin.
In the P2 phage family, the C-terminal regions of the tail fiber proteins are responsible for binding to host receptors (37, 38). However, no similarity was detected in the C-terminal region of putative fiber proteins encoded by ORF15, corresponding to the P2 H (tail fiber) gene. These results implied that the bactericidal activity spectrum of maltocin differs from that of other reported phage tail-like bacteriocins. Until now, maltocin P28 was only known to have antimicrobial activity against S. maltophilia strains.
A lytic system composed of a lytic enzyme (endolysin), a holin, and their modulators is required to break down bacterial peptidoglycan at the terminal stage of the phage or phage tail-like bacteriocin (21, 35). Phage lytic system genes are commonly organized on a gene cassette (35), while the lytic genes of P. aeruginosa strain PAO1 are distributed on both sides of the R2 pyocin locus (20, 21). Two homologous lysis genes (ORF8 and ORF9), coding for an endolysin and holin, were adjacent and located upstream of structural genes in the maltocin P28 gene locus. Otherwise, we could not identify any modulatory genes in the lytic system.
The predicted ORF6 protein contained conserved HTH superfamily and Mor superfamily domains. This suggests that the ORF6 product is a regulator of maltocin gene expression. Indeed, maltocin P28 could be induced by mitomycin C, as shown for many bacteriocins. The pyocin gene cluster has two regulatory genes (prtN and prtR), which are transcribed in a direction opposite that of all other ORFs (39). The only predicted regulatory gene (ORF6) in the maltocin locus was transcribed as being in the same direction as the remaining ORFs, and it showed no homology with other known regulatory proteins. Other regulators (excluding ORF6) were not identified. Thus, the regulation of maltocin likely differs from that of R2 pyocin or P2 phage.
More than 90% of P. aeruginosa strains can produce R- and F-type pyocins (20). However, the maltocin P28 gene-like clusters are also present in four out of the five chromosomes of S. maltophilia strains whose full genomic sequences are available (NC_010943, NC_011071, NC_017671, NC_015947, and NZ_ACDV00000000). This implies that phage tail-like bacteriocins are widely distributed in S. maltophilia strains, similar to pyocins in P. aeruginosa. To examine the bacteriocins in S. maltophilia, a total of 20 S. maltophilia strains (10 clinical and 10 environmental strains) were selected at random. Ten preparations of S. maltophilia strains displayed maltocin P28-like protein bands by SDS-PAGE. Six of these samples were further examined using electron microscopy to visualize the phage tail-like particles (data not shown).
S. maltophilia is the only species of Stenotrophomonas known to cause human disease (40). S. maltophilia has significant resistance to broad-spectrum antibiotics, resulting in limited treatment options (2, 3, 6). Currently, the bacteriostatic compound trimethoprim-sulfamethoxazole (TMP-SMX) is the preferred treatment for S. maltophilia infections. However, resistance is increasing due to the spread of acquired mobile resistance determinants, and in many patients TMP-SMX therapy is contraindicated (41). In our analysis, maltocin P28 exhibited antimicrobial activity against 19 out of 44 clinical S. maltophilia strains. This indicates that maltocin P28 is a novel therapeutic agent against some S. maltophilia infections.
Supplementary Material
ACKNOWLEDGMENTS
We thank Zhenrong Peng for kindly providing the environmental strains of Stenotrophomonas maltophilia.
This work was supported by the National Natural Science Foundation of China (no. 30970070).
Footnotes
Published ahead of print 8 July 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01648-13.
REFERENCES
- 1.Ryan RP, Monchy S, Cardinale M, Taghavi S, Crossman L, Avison MB, Berg G, Lelie D, Dow JM. 2009. The versatility and adaptation of bacteria from the genus Stenotrophomonas. Nat. Rev. Microbiol. 7:514–525 [DOI] [PubMed] [Google Scholar]
- 2.Brooke JS. 2012. Stenotrophomonas maltophilia: an emerging global opportunistic pathogen. Clin. Microbiol. Rev. 25:2–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Looney WJ, Narita M, Mühlemann K. 2009. Stenotrophomonas maltophilia: an emerging opportunist human pathogen. Lancet Infect. Dis. 9:312–323 [DOI] [PubMed] [Google Scholar]
- 4.Bin Abdulhak AA, Zimmerman V, Al Beirouti BT, Baddour LM, Tleyjeh IM. 2009. Stenotrophomonas maltophilia infections of intact skin: a systematic review of the literature. Diagn. Microbiol. Infect. Dis. 63:330–333 [DOI] [PubMed] [Google Scholar]
- 5.Paez JI, Tengan FM, Barone AA, Levin AS, Costa SF. 2008. Factors associated with mortality in patients with bloodstream infection and pneumonia due to Stenotrophomonas maltophilia. Eur. J. Clin. Microbiol. Infect. Dis. 27:901–906 [DOI] [PubMed] [Google Scholar]
- 6.Fihman V, Le Monnier A, Corvec S, Jaureguy F, Tankovic J, Jacquier H, Carbonnelle E, Bille E, Illiaquer M, Cattoir V, Zahar JR. 2012. Stenotrophomonas maltophilia—the most worrisome threat among unusual non-fermentative gram-negative bacilli from hospitalized patients: a prospective multicenter study. J. Infect. 64:391–398 [DOI] [PubMed] [Google Scholar]
- 7.Albini S, Abril C, Franchini M, Hussy D, Filioussis G. 2009. Stenotrophomonas maltophilia isolated from the airways of animals with chronic respiratory disease. Schweizer Arch. Tierheilkd. 151:323–328 [DOI] [PubMed] [Google Scholar]
- 8.Winther L, Andersen RM, Baptiste KE, Aalbǽk B, Guardabassi L. 2010. Association of Stenotrophomonas maltophilia infection with lower airway disease in the horse: a retrospective case series. Vet. J. 186:358–363 [DOI] [PubMed] [Google Scholar]
- 9.Ohnishi M, Sawada T, Marumo K, Harada K, Hirose K, Shimizu A, Hayashimoto M, Sato R, Uchida N, Kato H. 2012. Antimicrobial susceptibility and genetic relatedness of bovine Stenotrophomonas maltophilia isolates from a mastitis outbreak. Lett. Appl. Microbiol. 54:572–576 [DOI] [PubMed] [Google Scholar]
- 10.Alonso A, Martínez JL. 1997. Multiple antibiotic resistance in Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 41:1140–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang L, Li XZ, Poole K. 2000. Multiple antibiotic resistance in Stenotrophomonas maltophilia: involvement of a multidrug efflux system. Antimicrob. Agents Chemother. 44:287–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fedler KA, Biedenbach DJ, Jones RN. 2006. Assessment of pathogen frequency and resistance patterns among pediatric patient isolates: report from the 2004 SENTRY Antimicrobial Surveillance Program on 3 continents. Diagn. Microbiol. Infect. Dis. 56:427–436 [DOI] [PubMed] [Google Scholar]
- 13.Tan CK, Liaw SJ, Yu CJ, Teng LJ, Hsueh PR. 2008. Extensively drug-resistant Stenotrophomonas maltophilia in a tertiary care hospital in Taiwan: microbiologic characteristics, clinical features, and outcomes. Diagn. Microbiol. Infect. Dis. 60:205–210 [DOI] [PubMed] [Google Scholar]
- 14.Cotter PD, Ross RP, Hill C. 2013. Bacteriocins—a viable alternative to antibiotics? Nat. Rev. Microbiol. 11:95–105 [DOI] [PubMed] [Google Scholar]
- 15.Gillor O, Nigro LM, Riley MA. 2005. Genetically engineered bacteriocins and their potential as the next generation of antimicrobials. Curr. Pharm. Des. 11:1067–1075 [DOI] [PubMed] [Google Scholar]
- 16.Sang Y, Blecha F. 2008. Antimicrobial peptides and bacteriocins: alternatives to traditional antibiotics. Anim. Health Res. Rev. 9:227–235 [DOI] [PubMed] [Google Scholar]
- 17.Dykes GA. 1995. Bacteriocins: ecological and evolutionary significance. Trends Ecol. Evol. 10:186–189 [DOI] [PubMed] [Google Scholar]
- 18.Daw MA, Falkiner FR. 1996. Bacteriocins: nature, function and structure. Micron 27:467–479 [DOI] [PubMed] [Google Scholar]
- 19.Cascales E, Buchanan SK, Duché D, Kleanthous C, Lloubès R, Postle K, Riley M, Slatin S, Cavard D. 2007. Colicin biology. Microbiol. Mol. Biol. Rev. 71:158–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Michel-Briand Y, Baysse C. 2002. The pyocins of Pseudomonas aeruginosa. Biochimie 84:449–510 [DOI] [PubMed] [Google Scholar]
- 21.Nakayama K, Takashima K, Ishihara H, Shinomiya T, Kageyama M, Kanaya S, Ohnishi M, Murata T, Mori H, Hayashi T. 2000. The R-type pyocin of Pseudomonas aeruginosa is related to P2 phage, and the F-type is related to lambda phage. Mol. Microbiol. 38:213–231 [DOI] [PubMed] [Google Scholar]
- 22.Nes IF, Diep DB, Holo H. 2007. Bacteriocin diversity in Streptococcus and Enterococcus. J. Bacteriol. 189:1189–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abriouel H, Franz CM, Ben Omar N, Gálvez A. 2011. Diversity and applications of Bacillus bacteriocins. FEMS Microbiol. Rev. 35:201–232 [DOI] [PubMed] [Google Scholar]
- 24.Jabrane A, Sabri A, Compere A, Jacques P, Vandenberghe I, Beeumen JV, Thonart P. 2002. Characterization of serracin P, a phage-tail-like bacteriocin, and its activity against Erwinia amylovora, the fire blight pathogen. Appl. Environ. Microbiol. 68:5704–5710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Strauch E, Kaspar H, Schaudinn C, Dersch P, Madela K, Gewinner C, Hertwig S, Wecke J, Appel B. 2001. Characterization of enterocoliticin, a phage tail-like bacteriocin, and its effect on pathogenic Yersinia enterocolitica strains. Appl. Environ. Microbiol. 61:5634–5642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu J, Liu Q, Shen P, Huang YP. 2012. Isolation and characterization of a novel filamentous phage from Stenotrophomonas maltophilia. Arch. Virol. 157:1643–1650 [DOI] [PubMed] [Google Scholar]
- 27.Crossman LC, Gould VC, Dow JM, Vernikos GS, Okazaki A, Sebaihia M, Saunders D, Arrowsmith C, Carver T, Peters N, Adlem E, Kerhornou A, Lord A, Murphy L, Seeger K, Squares R, Rutter S, Quail MA, Rajandream MA, Harris D, Churcher C, Bentley SD, Parkhill J, Thomson NR, Avison MB. 2008. The complete genome, comparative and functional analysis of Stenotrophomonas maltophilia reveals an organism heavily shielded by drug resistance determinants. Genome Biol. 9:R74. 10.1186/gb-2008-9-4-r74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lira F, Hernández A, Belda E, Sánchez MB, Moya A, Silva FJ, Martíneza JL. 2012. Whole genome sequence of Stenotrophomonas maltophilia D457, a clinical isolate and a model strain. J. Bacteriol. 194:3563–3564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 30.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. 2005. Protein identification and analysis tools on the ExPASy server, p 571–607 In Walker JM. (ed), The proteomics protocols handbook. Humana Press, Clifton, NJ [Google Scholar]
- 32.Milton DL, O'Toole R, Horstedt P, Wolf-Watz H. 1996. Flagellin A is essential for the virulence of Vibrio anguillarum. J. Bacteriol. 178:1310–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kovach ME, Phillips RW, Elzer PH, Roop RM, Jr, Peterson KM. 1994. pBBR1MCS: a broad-host-range cloning vector. Biotechniques 16:800–802 [PubMed] [Google Scholar]
- 34.Hagemann M, Hasse D, Berg G. 2006. Detection of a phage genome carrying a zonula occludens like toxin gene (zot) in clinical isolates of Stenotrophomonas maltophilia. Arch. Microbiol. 185:449–458 [DOI] [PubMed] [Google Scholar]
- 35.Young R. 1992. Bacteriophage lysis: mechanism and regulation. Microbiol. Rev. 56:430–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smarda J, Benada O. 2005. Phage tail-like (high-molecular-weight) bacteriocins of Budvicia aquatica and Pragia fontium (Enterobacteriaceae). Appl. Environ. Microbiol. 71:8970–8973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haggard-Ljungquist E, Halling C, Calendar R. 1992. DNA sequences of the tail fiber genes of bacteriophage P2: evidence for horizontal transfer of tail fiber genes among unrelated bacteriophages. J. Bacteriol. 174:1462–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sandmeier H. 1994. Acquisition and rearrangement of sequence motifs in the evolution of bacteriophage tail fibres. Mol. Microbiol. 12:343–350 [DOI] [PubMed] [Google Scholar]
- 39.Matsui H, Sano Y, Ishihara H, Shinomiya T. 1993. Regulation of pyocin genes in Pseudomonas aeruginosa by positive (prtN) and negative (prtR) regulatory genes. J. Bacteriol. 175:1257–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Coenye T, Vanlaere E, Falsen E, Vandamme P. 2004. Stenotrophomonas africana Drancourt et al. 1997 is a later synonym of Stenotrophomonas maltophilia (Hugh 1981) Palleroni and Bradbury 1993. Int. J. Syst. Evol. Microbiol. 54:1235–1237 [DOI] [PubMed] [Google Scholar]
- 41.Hu LF, Chang X, Ye Y, Wang ZX, Shao YB, Shi W, Li X, Li JB. 2011. Stenotrophomonas maltophilia resistance to trimethoprim/sulfamethoxazole mediated by acquisition of sul and dfrA genes in a plasmid-mediated class 1 integron. Int. J. Antimicrob. Agents 37:230–234 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.