Abstract
Chicken-pathogenic Escherichia coli is severely endangering the poultry industry in China and worldwide, and antibiotic therapy is facing an increasing problem of antibiotic resistance. Bacteriophages can kill bacteria with no known activity in human or animal cells, making them an attractive alternative to antibiotics. In this study, we present the characteristics of a novel virulent bacteriophage, Bp7, specifically infecting pathogenic multidrug-resistant E. coli. Phage Bp7 was isolated from chicken feces. Bp7 belongs to the family Myoviridae, possessing an elongated icosahedral head and contractile sheathed tail. It has a 168-kb double-stranded DNA genome. For larger yields, its optimal multiplicity of infection (MOI) to infect E. coli was about 0.001. The latent period was 10 to 15 min, and the burst size was 90 PFU/infected cell. It was stable both at pH 5.0 to 10.0 and at 40°C or 50°C for at least 1 h. Bp7 could infect 46% of pathogenic clinical E. coli strains. Bp7 harbored 791 open reading frames (ORFs) and 263 possible genes. Among the 263 genes, 199 possessed amino acid sequence identities with ORFs of phage T4, 62 had identities with other T4-like phages, and only one lacked any database match. The genome of Bp7 manifested obvious division and rearrangement compared to phages T4, JS98, and IME08. Bp7 is a new member of the “T4-like” genus, family Myoviridae. Its wide host range, strong cell-killing activity, and high stability to pH make it an alternative to antimicrobials for controlling drug-resistant E. coli in chickens.
INTRODUCTION
Chicken colibacillosis is one of the main bacterial diseases and severely endangers the poultry industry in China and worldwide. Escherichia coli has been identified as a major pathogen (1). Antibiotics are widely used to control chicken colibacillosis, but it is very common for E. coli to be resistant to antibiotics (2, 3). In recent years, nearly 80% of E. coli isolates from diseased animals have manifested severe resistance to antimicrobial drugs (4, 5), so antibiotic therapy is facing an increasing problem of antibiotic resistance. Bacteriophages are now considered a good alternative to antibiotics (6, 7).
However, there are many problems with phage therapy, and not every phage strain is appropriate for such therapy. Based upon their replication methods, phages are classified as either virulent or lysogenic. Virulent phages replicate in their bacterial hosts and destroy them in the process, but lysogenic phages insert their genomes into their hosts' genomes (8). As it has turned out, both lysogenic and virulent bacteriophages are actively involved in the evolution of bacteria, including pathogens (9). A troubling possibility is that there are virulence genes in some phages and these genes can change the pathogenicity of their host bacteria. Lysogenic phages transfer genes that express toxin proteins or pathogenic factors among bacterial species (8, 10). For safety reasons, lysogenic phages are not allowed to be used in phage therapy, and if a phage is permitted to be an alternative to antibiotics, clear genome information is needed. Phages are reported to be the most abundant and diverse forms of life on earth and are ubiquitous in nature (8), but few of them have been sequenced completely. There is still a lot of work to do in the process of phage therapy.
Here, we present an analysis of the biological characterization and genome of phage Bp7, a T4-like phage with a wide host range (4) isolated from chicken feces and related to phages JS98 (NC-010105) and IME08 (NC-014260). The analysis of its genome indicated that it was a new member of the “T4-like virus” genus.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains used in this study are listed in Table 1. All the strains were cultivated in Luria-Bertani (LB) broth (Biocorp) at 37°C and stored in 30% glycerol in a freezer at −80°C.
Table 1.
E. coli strains used for determination of the Bp7 host range
| Strain no.a | Strain | Serotype | Infectivityb |
|---|---|---|---|
| 1 | HL041025A | O24 | C |
| 2 | CH041025A | O15 | N |
| 3 | ZC041025A | O78 | N |
| 4 | H041027A | O88 | C |
| 5 | C041027A | X | C |
| 6 | EC041029A | O78 | C |
| 7 | EH041029B | O78 | C |
| 8 | C041029A | O78 | C |
| 9 | H041029 | O78 | C |
| 10 | L041030A | O78 | N |
| 11 | Z041030B | O1 | N |
| 12 | EZ041122A | O73 | C |
| 13 | EZ041030A | O35 | N |
| 14 | EZ041030B | O78 | N |
| 15 | L041030B | O78 | C |
| 16 | L041030C | O78 | N |
| 17 | C041101A | O35 | N |
| 18 | EC041101A | O35 | N |
| 19 | H041103A | O23 | C |
| 20 | H041103D | O88 | C |
| 21 | H041103B | O78 | N |
| 22 | H041104A | O24 | N |
| 23 | CK041105 | O78 | N |
| 24 | H041108C | O76 | N |
| 25 | H041108B | O78 | N |
| 26 | Z041108D | O5 | N |
| 27 | Z041108A | O78 | N |
| 28 | EH041110B | O76 | N |
| 29 | EH041110D | X | C |
| 30 | ER041118B | X | C |
| 31 | G041126A | O93 | N |
| 32 | EHJJ041116A | O78 | N |
| 33 | ER041118C | X | C |
| 34 | ER041118B | X | C |
| 35 | ER041118A | O157 | C |
| 36 | BL21 | C | |
| 37 | JM109 | C | |
| 38 | JM110 | C | |
| 39 | DH5α | C |
Strains 1 to 35 were isolated from diseased chicken organs and identified in 2004 and are multiresistant to antibiotics at different levels. Strains 36 to 39 are laboratory E. coli strains used widely in gene engineering.
C, clear lysis; N, no lysis.
Isolation and identification of E. coli bacteriophages.
E. coli bacteriophages were isolated from chicken feces in Shandong Province, China. Feces samples were homogenized in saline and filtered through a sieve. The supernatant from the feces was added to a mixture of 35 clinical E. coli strains (Table 1) and incubated overnight at 250 rpm and 37°C. The supernatant was clarified by centrifugation (10,000 rpm; 20 min) and then passed through a 0.22-μm bacterial filter (Millipore). A double-layer agar method was used to examine whether the filtrate contained phage (11). Filtrate samples (50 μl) were mixed with 0.1 ml of an overnight culture of E. coli and 2.5 ml LB broth with 0.7% agar and then plated onto LB agar plates and incubated overnight at 37°C. The 35 E. coli strains were used for phage isolation on double-layer agar plates. A single phage plaque was picked out with a sterile Pasteur pipette into 1 ml LB broth and stored at 4°C. Successive single-plaque isolations were performed at least three times. Bacteriophage Bp7 was propagated on E. coli in LB broth.
Electron microscopy of phage Bp7.
Phage Bp7 and culture liquid from E. coli (EC041029A, serotype O78 [Table 1]) were mixed in fresh LB broth with shaking at 250 rpm and 37°C. Aliquots were taken, and 2.5% glutaric dialdehyde was added at different times in order to stop the reproduction of the phage. The cell pellet was harvested by centrifugation at 4,000 rpm for 10 min; 2.5% glutaric dialdehyde (20 times the volume of the aliquot) was added for fixation for 4 h at 4°C. The specimens were postfixed for 1 h at 4°C in 1% osmium tetroxide in 0.1 M cacodylate buffer and 4.8% uranyl acetate for 24 h at 4°C. The samples were dehydrated in graded ethanol (70% to 100%) and embedded in Epon 812 (12). Ultrathin sections were cut with glass knives and stained with uranyl acetate and lead citrate, and the reproduction process of phage Bp7 was examined by transmission electron microscopy (TEM) (JEM-1200EX; Japan Electronics and Optics Laboratory [JEOL], Tokyo, Japan) at an accelerating voltage of 80 kV.
After the coculture of phage Bp7 and E. coli (EC041029A, serotype O78) was clear, the supernatant was clarified by centrifugation at 10,000 rpm for 10 min. Two times the volume of 2.5% glutaric dialdehyde was added to it. A phage sample was negatively stained with 2% (wt/vol) aqueous uranyl acetate (pH 4.0) on a carbon-coated grid and examined with a JEM-1200EX microscope (JEOL).
Host range analysis.
The host range of phage Bp7 for 35 clinical E. coli strains and 4 laboratory E. coli strains was determined by spotting 105 PFU of Bp7 onto freshly seeded lawns of bacteria on agar plates (13). Each test was repeated three times.
Optimal MOI of phage Bp7.
The optimal multiplicity of infection (MOI) of Bp7 was determined in a coculture of E. coli (EC041029A, serotype O78) in early exponential phase (5 × 107 CFU/ml) and phage in 10 ml of LB broth at different MOIs (0.001, 0.01, 0.1, 1, and 10). A culture of bacteria without Bp7 was used as a control. The preparations were incubated with shaking at 250 rpm and 37°C. Aliquots were taken at 0, 1, 2, 3, and 4 h postinfection (p.i.) for determination of the optical density at 600 nm (OD600). The supernatant of the coculture was collected at 4 h p.i. by centrifugation (10,000 rpm/10 min). Phage titers were determined by the double-layer agar method. The MOI resulting in the highest phage titer within 4 h was considered the optimal MOI and used in subsequent large-scale phage production.
One-step growth of Bp7 phage.
The test of the one-step growth curve was performed as described previously with some modifications (14, 15). Briefly, 30 ml of an early-exponential-phase culture of E. coli (EC041029A, serotype O78) was harvested by centrifugation (10,000 rpm; 30 s; 4°C) and resuspended in one-fifth of the initial volume of LB broth. Phage Bp7 was added at the optimal MOI and allowed to adsorb for 10 min at 37°C with shaking at 250 rpm. The suspension was then centrifuged at 10,000 rpm for 30 s and resuspended in 30 ml of LB broth. Serial dilutions of the suspension were made and incubated at 37°C. Aliquots were taken at 5-min intervals, and the titers in the aliquots were immediately determined by the double-layer agar method. The assay was performed in triplicate. The latent period was defined as the time interval between adsorption (not including 10 min of preincubation) and the beginning of the initial rise in the phage titer. The burst size was calculated as the ratio of the final count of liberated phage particles to the initial count of infected bacterial cells during the latent period (11).
Thermal/pH stability of phage Bp7.
For the thermal-stability assay, phage suspensions (4 × 109 PFU/ml) were incubated at 40°C, 50°C, 60°C, 70°C, and 80°C, and aliquots were taken after 20, 40, and 60 min of incubation. For the pH stability assay, a phage suspension (3 × 106 PFU/ml) was inoculated in a series of tubes containing fresh LB broth at pH 2.0, 3.0, 4.0, 5.0, 6.0, 7.0, 8.0, 9.0, 10.0, 11.0, 12.0, and 13.0 and incubated at 37°C; aliquots were taken at 1, 2, and 3 h p.i. Phage titers were determined with E. coli (EC041029A, serotype O78) as host cells by the double-layer agar method. All assays were performed in triplicate.
Sequencing and genomic analysis of Bp7 phage.
CsCl density gradient-purified phage particles were subjected to phenol-chloroform extraction and ethanol precipitation. DNA for sequencing was extracted from phage pellet suspensions using a High Pure PCR Template Preparation Kit (Roche) and sent to Beijing Sunbiotech Co., Ltd., for commercial sequencing. A random library of Bp7 phage was constructed by the shotgun library method and sequenced with an ABI Prism 3100-Avant genetic analyzer (Applied Biosystems, Foster City, CA). Sequences were assembled using the Phred-Phrap program (DNA Sequencing Facility, University of Cambridge, Cambridge, United Kingdom) to form the complete genome of phage Bp7. The nucleotide sequences were compared to the NCBI GenBank database. The potential open reading frames (ORFs) were predicted using Microbial Genome Annotation Tools (http://www.ncbi.nlm.nih.gov/genomes/MICROBES/glimmer_3.cgi). The basic prerequisites for an ORF were ATG, TTG, or GTG as a possible start codon and a minimum size of 30 codons. The putative functions of the ORFs were analyzed by BLASTP search at NCBI and with the DNA Master program (http://cobamide2.bio.pitt.edu/computer.htm). The genome map was drawn using the VectorNTI program. Searching for tRNA genes was done with the tRNAscan-SE program, version 1.21 (16). The genome of Bp7 was pairwise analyzed using the Artemis Comparison Tool (ACT) with T4 and the closest homologs, JS98 and IME08 (17).
Nucleotide sequence accession number.
The sequence data were deposited at GenBank under accession number HQ829472.
RESULTS
Isolation and morphology of phage Bp7.
Bacteriophage Bp7 was isolated from chicken feces after continuous cultivation and plaque purification. Its plaques averaged 1 to 2 mm in diameter with a clear lytic zone. Electron microscopy confirmed that Bp7 belongs to the T4-like virus genus, order Caudovirales, family Myoviridae (elongated icosahedral head and contractile sheathed tails) according to the classification of Ackermann (18). Phage Bp7 had a hexagonal head ca. 93 nm in length and ca. 73 nm in diameter, a contractile tail of ca. 106 nm, and a tail sheath of ca. 22 nm (Fig. 1d). Different stages of phage reproduction are shown in Fig. 1a, b, and c. The adsorption, maturity, and release phases clearly existed, which was similar to phage T4.
Fig 1.
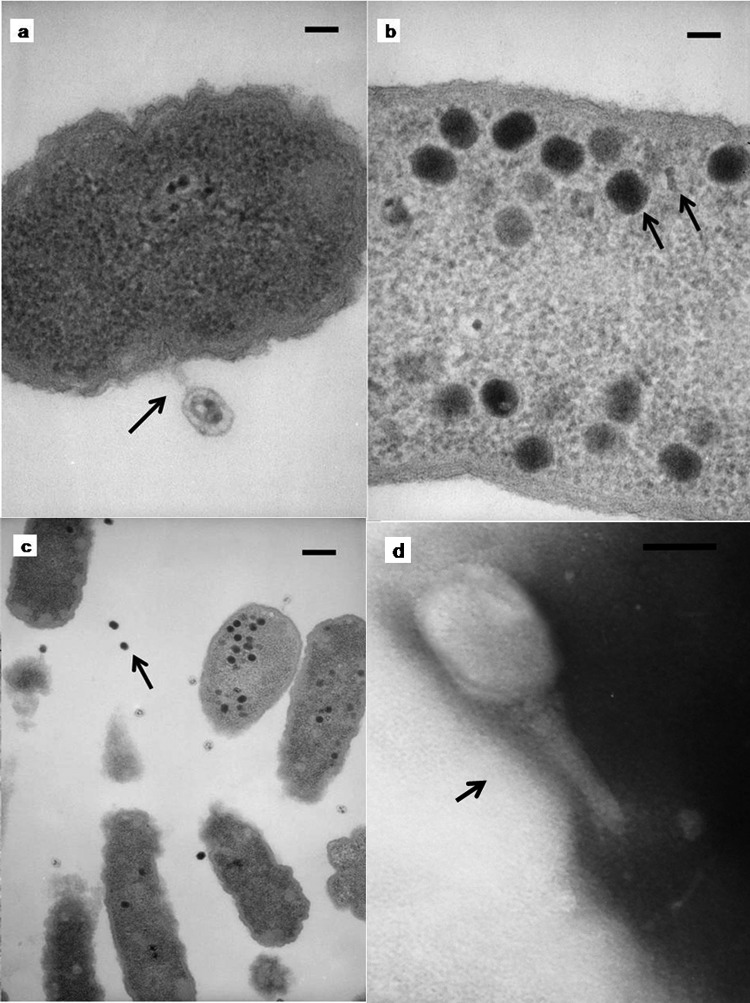
Morphology and reproduction process of phage Bp7 examined with a JEM-1200EX microscope (JEOL). (a to c) Different stages of phage reproduction. (a) E. coli cell and phage Bp7 in the adsorption phase (bar = 50 nm; the arrow indicates the adsorption of phage Bp7). (b) Phage Bp7 in the maturity phase (the arrows indicate the elongated head and tail; bar = 50 nm). (c) Phage Bp7 in the release period (the arrow indicates phage released by host cells; bar = 200 nm). (d) Transmission electron micrograph of a phage Bp7 virion negatively stained with 2% uranyl acetate (bar = 50 nm; the arrow indicates the phage with the elongated head and tail).
Host range of phage Bp7.
The host range of phage Bp7 was determined by the double-layer agar method with 35 clinical strains of E. coli we isolated and 4 laboratory E. coli strains as host cells. Overall, 16 clinical strains and all 4 laboratory strains were sensitive to phage Bp7. Among the 16 clinical strains, there were 6 known serotypes (O23, 1 strain; O24, 1 strain; O73, 1 strain; O78, 5 strains; O88, 2 strains; and O157, 1 strain), while the serotypes of the other 5 strains remained unknown (Table 1). The results showed phage Bp7 had broad infectivity against pathogenic E. coli strains (46%) and laboratory E. coli strains (100%); among the 11 phage-sensitive clinical strains with known serotypes, 45% were serotype O78, which meant that O78 was the dominant serotype as the host of Bp7.
Optimal MOI of phage Bp7.
The curve of OD600 versus time showed that the bacterial number increased continuously within 4 h of incubation, when there were no phage in the medium; phage Bp7 caused reduction of the bacterial number (see Fig. SA1 in the supplemental material). At MOIs of 0.001 and 0.01, the growth curve increased slowly until 2 h p.i., which showed retarded bacterial growth compared to the control group, and afterwards decreased gradually. At MOIs of 0.1 and 1, the growth curve decreased from the beginning and reached 0 at 3 h p.i. For larger yields, the optimal MOI of phage Bp7 was about 0.001 (data not shown).
One-step growth and thermal/pH stability of phage Bp7.
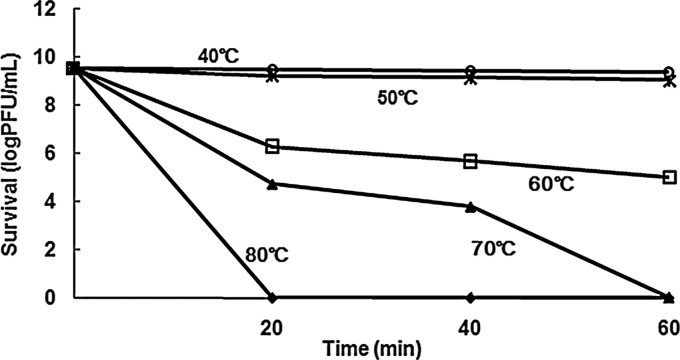
The growth curve of Bp7 is shown in Fig. SA2 in the supplemental material. The latent period was estimated to be approximately 10 to 15 min, and the average burst size of Bp7 was about 90 PFU/infected cell. The stability of Bp7 was tested by recording phage survival rates under different pH and temperature conditions. A survival curve of the phage at different temperatures is shown in Fig. 2. Bp7 could basically keep its infectivity for at least 1 h at 40°C or 50°C. With increased temperature and longer time, phage titers decreased. Phage were almost inactivated after 1 h of incubation at 70°C or 80°C. In the range of pH 2.0 to 13.0, significant inactivation appeared at lower or higher pH values (Fig. 3). Little to almost no reduction in PFU was observed in 1-, 2-, and 3-h incubation assays at pH 7.0 or in 1 h between pH 5.0 and 10.0. Phage titers decreased below the detection limit (20 PFU/ml) after 1 h of incubation at pH 2.0 or pH 13.0 and after 2 h of incubation at pH 3.0 and pH 13.0 but could be detected between pH 4.0 and pH 11.0 after 3 h of incubation. The results suggested that extreme pH conditions affected the stability of phage Bp7.
Fig 2.
Stability of Bp7 under different temperature conditions. Bp7 could keep its basic infectivity at 40°C or 50°C. With the increased temperature and longer time, phage titers decreased.
Fig 3.
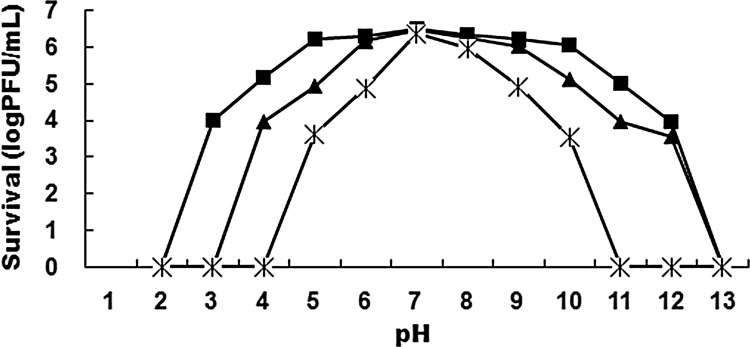
Stability of Bp7 under different pH conditions. In the range of pH 2.0 to 13.0, approximately neutral conditions had little or no effect on phage Bp7, while extreme pH conditions affected the stability of phage Bp7.
Characterization of the Bp7 genome sequence.
A random library of Bp7 phage was constructed by a shotgun library method and sequenced (10-fold coverage). According to the morphology, phage Bp7 is a member of the T4-like virus genus. Bp7 has a linear double-stranded DNA (dsDNA) of 168,066 bp, similar to those of phages T4 (168,903 bp; NC_000866), JS98 (170,523 bp; NC_010105), and IME08 (172,253 bp; NC_014260). Phages T4, JS98, and IME08 belonged to the T4-like virus genus (19). The G+C content of Bp7 was 39.49%, which is slightly higher than that of T4 (34.50%) but similar to those of JS98 (39.51%) and IME08 (39.59%). The highest G+C content in the Bp7 genome was 57%, located in the gp38 tail fiber gene (positions 77,158 to 77,949). The genome sequence of Bp7 had more than 90% homology with those of JS98 and IME08 but displayed lower query coverage with T4 (4).
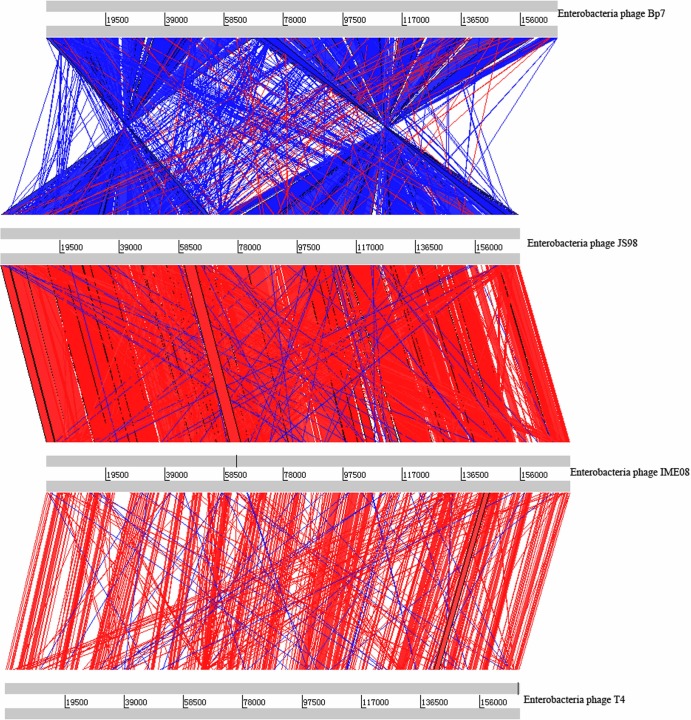
Phage Bp7 harbored 791 ORFs, and 263 possible genes were predicted by homology analysis in the Protein Data Bank (see Table SA1 in the supplemental material). The shortest ORF contained only 36 codons. Most of the predicated ORFs had an ATG initiation codon, 7 began with GTG, and 5 began with TTG (data not shown). To determine the number of T4-like phages that had a homologous ORF with Bp7, the predicated ORFs were analyzed with the DNA Master program, and the protein sequence of each ORF was searched by BLAST against the NCBI database. In the phage Bp7 genome, there were 199 ORFs that shared protein sequence identities with phage T4; the average identity was only 76.56%, but the average identities with JS98, JS10, and IME08 were more than 94%. There were 64 ORFs in phage Bp7 with no match in phage T4. Significantly, 50 of the 64 ORFs in phage Bp7 shared protein sequence identity with JS98, JS10 (NC_012741), or IME08; 13 of them shared protein sequence identity with phage vB_EcoM-VR7 (NC_014792), SP18 (NC_014595), or STML-198 (AFU64017.1) (all of these phages belong to the T4-like phages); and 1 ORF (ORF101) shared protein sequence identity (47%) with a hypothetical protein of Nectria haematococca mpVI 77-13-4 (NZ_ACJF00000000.1). Only one ORF (ORF56) lacked any match in the GenBank database (see Table SA1 in the supplemental material). Moreover, the Bp7 genome showed a gene arrangement different from those of T4, JS98, and IME08. Based on the ACT comparison result, the genome of Bp7 could be divided into two parts (ORF1 to ORF136 and ORF137 to ORF263), which were reversed (a large-scale genome translocation) from the ORF arrangement of phages T4, JS98, and IME08 (Fig. 4). ORF136 (rIIA) and ORF137 (rIIB) were predicted to be protective against prophage-induced early lysis and were adjacent to each other in Bp7 (Fig. 5) but were separated and located at the two ends of the T4, JS98, and IME08 genomes (Fig. 4). This indicated that phage Bp7 was a new member of the T4-like virus genus.
Fig 4.
Comparison of the Bp7 genome with those of JS98, IME08, and T4. Regions with similarity are highlighted by connecting red or blue lines between the genomes; red lines indicate homologous blocks of sequence, and blue lines indicate inversions. The gray bars represent the forward and reverse strands. Gene arrangements and orientations were highly conserved among phages JS98, IME08, and T4, but the genome of Bp7 showed a large-scale genome translocation and can be divided into two parts that are the reverse of the arrangement of phages JS98, IME08, and T4. As seen by the large red blocks connecting both genomes, there is a smaller degree of similarity between Bp7 and T4.
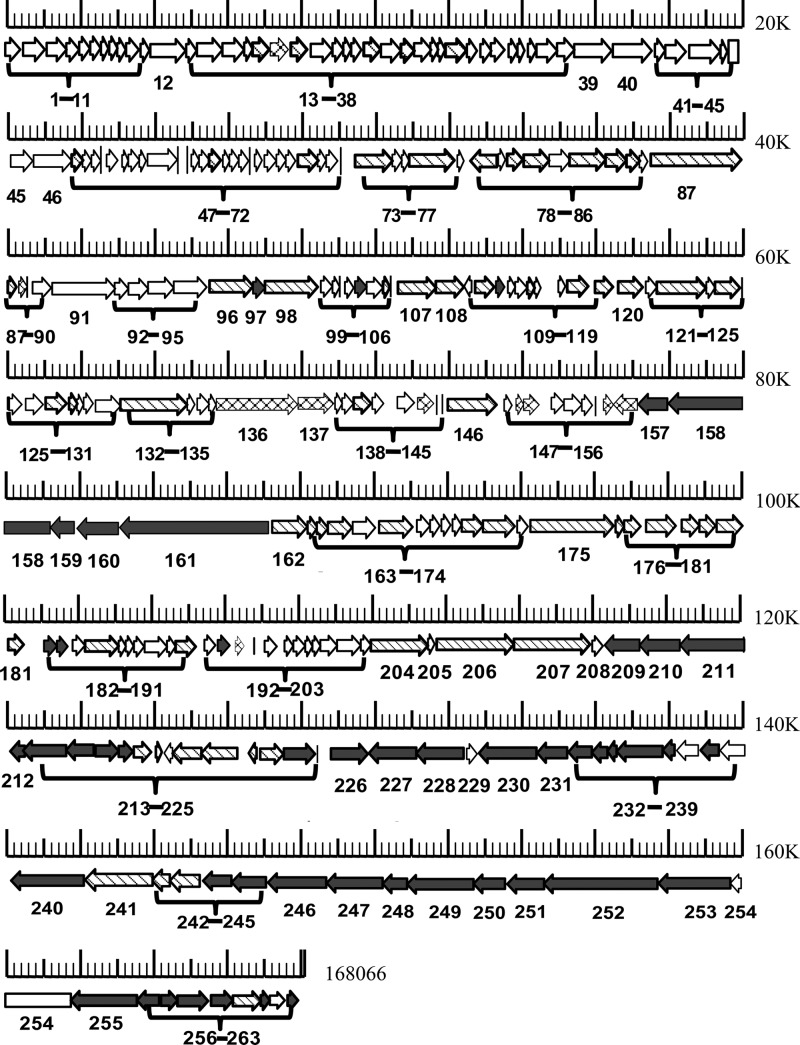
Fig 5.
Annotated genome map of bacteriophage Bp7. The genome can be read from left to right and from top to bottom. ORFs with right arrows or left arrows are genes on the positive or negative strand, respectively. The predicted genes were classified into three groups based on their different functions, as shown by different patterns: solid, phage structure and packaging gene; hatched, DNA replication-, transcription-, and translation-related gene; crosshatched, host lysis gene; open, genes with unknown functions. The tRNA coding region was not found. The numbers (1 to 263) show predicted ORFs listed in Table SA1 in the supplemental material.
The annotated genes were clustered into 3 functional groups, as we described previously (4). Notably, the major structural genes were mainly located in a tight genome cluster downstream and on the negative strand. In contrast, the DNA replication genes were randomly located in all genomes and mainly located on the positive strand (Fig. 5). Interestingly, no tRNA was detected. Concrete gene information, such as the position, size, molecular weight, and putative function of each ORF of Bp7, is shown in Table SA1 in the supplemental material.
DISCUSSION
Colibacillosis can cause high mortality in chickens and bring huge economic loss (1) and is very common in chickens, especially broilers, in China. Nearly 2 billion chickens are slaughtered each year in Shandong Province, located in east China, accounting for more than 50% of the chicken market of China. In 2004, 35 E. coli strains were isolated from chickens with clinical symptoms in the province and subjected to antimicrobial susceptibility testing (Table 1). Most strains were multidrug resistant. For example, 97% of them were resistant to erythromycin, 80% to ciprofloxacin, 71% to norfloxacin, 57% to streptomycin, and 29% to gentamicin (detailed data not shown). Zhang et al. (5) surveyed the resistance of avian E. coli in north China and found that the highest resistance rates were 84.76% to tetracycline and 70.12% to doxycycline; the lowest resistance rate was 4.88% to minocycline. Multiresistance is a worldwide problem and endangers the health of humans and animals. Despite great advances in antimicrobial therapy, the problem of multiresistance is still very serious. Phages are viruses that kill bacteria. They are plentiful in nature, with no known activity in human or animal cells, making them an attractive alternative to antibiotics. Phage therapy is attracting more and more attention, and it seems to be highlighted in the headlines of research subjects recently (20, 21). The process of phage therapy for pathogen control is largely dependent upon broad-range virulent phages with rapid adsorptive properties and strong cell-killing activity; furthermore, the phages must be capable of being mass produced and stable during preparation and storage; third, the phages must be safe and effective upon application (22).
Phage Bp7 was isolated from chicken feces and showed a wide host range among pathogenic E. coli strains (46%). All 35 E. coli strains used in the host range assay were isolated from different chicken farms at different times and manifested different drug resistance properties. Bp7 could infect several serotypes of E. coli, such as O23, O24, O73, O78, O88, and O157 (Table 1). O78 is a main serotype of E. coli prevalent in chickens in China. Yang et al. (23) verified that O78 was the most common serogroup identified (63%) among the chicken E. coli isolates. Our study showed that O78 was the dominant serotype as the host of Bp7, because 45% of the serotypes of the phage-sensitive clinical strains were O78, so phage Bp7 should be effective in controlling chicken colibacillosis in China. Phage Bp7 was identified as virulent for E. coli strains because of the lytic effect at an MOI of 0.001. Bp7 exhibited rapid adsorption (>99% adsorbed within 5 min [data not shown]), a large burst size (90 PFU per cell), high stability at a wide range of pH values, and lytic activity toward a broad range of E. coli strains with different serotypes. Moreover, in our former research work, 28 E. coli strains were isolated from normal chicken intestines and were tested for phage infectivity, and the results showed that all but 1 strain revealed resistance to phage Bp7 (data not shown). Since the receptors of phages on the bacterial cell wall are mainly virulence factors (8), a phage-resistant bacterium would be less virulent. It seemed that phage Bp7 could distinguish pathogenic E. coli from nonpathogenic E. coli and had little influence on the microbial flora of the animal body. Overall, phage Bp7 was remarkable enough to be developed as an alternative to antimicrobials to control the occurrence and spread of chicken colibacillosis.
In this study, the classification of Bp7 was analyzed. The classification of enterobacterial phages (especially T4-like viruses) was first suggested by morphological similarity and later confirmed by genomic comparison (24, 25). Phages with tails belong to the order Caudovirales and are classified into three families: Myoviridae, Siphoviridae, and Podoviridae (26). The T4-like viruses have been morphologically described (25). Bp7 has an elongated icosahedral head and contractile sheathed tails, which indicate that it is a T4-like phage and is classified in the order Caudovirales and family Myoviridae (19). The process of phage Bp7 host invasion was observed by TEM, which showed an infecting mechanism similar to that of T4 (Fig. 1). A genomic comparison was also used in phage classification. About 40 T4-like phages have been sequenced and annotated in the last several years. These phages were grouped into 23 different types by the ORF composition of the T4 genome (27). Lavigne et al. (25) identified four distinct subtypes with >70% protein similarity among T4-like viruses, i.e., T4-type, 44Rr-type, RB43-type, and RB49-type viruses. The genome of Bp7 is about 168 kb, which is similar to those of other T4-like phages in size (25). Homology analysis of the Bp7 genome showed the highest similarity to phage IME08, followed by JS98 and JS10, which all belong to the T4-like phages (28, 29). Genome analysis of Bp7 at the amino acid level displayed a high degree of identity to genes encoding structural function, DNA replication, and metabolism from T4-like phages. Of 263 predicted genes of phage Bp7, a total of 199 had amino acid identity with those of T4 (see Table SA1 in the supplemental material). Based on these results, Bp7 was closely related to phage T4 by genetic, as well as morphological, characteristics and might have similar phage DNA metabolism and morphogenesis systems.
We also tried to find the reason why Bp7 had such a wide host range by genome analysis. The host interaction-associated genes of Bp7 were analyzed. A BLAST search for lysis-associated genes of Bp7 determined that the e, rIII, sp, and t (rIIA and rIIB) genes were similar to those of T4 (73% to 93.9%), but the rI gene was not found in Bp7. Bp7 contained three sets of T4-like fiber structures with distinct functions: wac (whisker antigen control protein), tail fiber (long tail fiber and short tail fiber), and base plate. These structural genes indicated that Bp7 was closely related to the T4 phage (30). Phage-host interaction genes of T4 have been reported. In these reports, the ac, gt, dam, pin, stp, and gol genes appeared in the T4 genome but were not found in Bp7 (see Table SA1 in the supplemental material). Since the Myoviridae are mainly influenced by vertical evolution rather than by horizontal gene transfer (25), how phage Bp7 evolved into a virus with a broad host range remains unknown.
The genome of Bp7 was compared with those of JS98, IME08, and T4 in this study. Interestingly, phage Bp7 showed genome division and rearrangement compared to phages T4, JS98, and IME08. There was an obvious division located at position 67880. The rIIA and rIIB genes were adjacent to each other in Bp7 (Fig. 5) but were separated and located the two ends of the T4, JS98, and IME08 genomes (Fig. 4). According to the Bp7 sequence, a pair of primers (forward, 5′-TTA TGC TGA AGC CTT AGA CG; reverse, 5′-GCC AAT AGT AAC TTC AAC GG) were designed to test and verify the division. A 581-bp fragment including position 67880 was amplified, which strongly verified that there was no error in our sequencing results. The reason the Bp7 genome is arranged in this way is unknown, but the arrangement of the genome makes it functional. There was a report about a similar division and rearrangement phenomenon in the T4-like bacteriophage phiAS5 (31). The Bp7 genome analysis showed that it is a new member of the T4-like virus genus, family Myoviridae.
Phages of the family Myoviridae have been successfully used in phage therapy study (8, 32). The wide host range of phage Bp7 suggests its polyvalent nature and signifies its importance as an alternative to antimicrobials for controlling drug-resistant E. coli in chickens.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by a grant from the Nature Science Foundation of Shandong Province, China (ZR2009DM009).
Footnotes
Published ahead of print 8 July 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01505-13.
REFERENCES
- 1.Olsen RH, Frantzen C, Christensen H, Bisgaard M. 2012. An investigation on first-week mortality in layers. Avian Dis. 56:51–57 [DOI] [PubMed] [Google Scholar]
- 2.Dupont HL, Jiang ZD, Belkind-Gerson J, Okhuysen PC, Ericsson CD, Ke S, Huang DB, Dupont MW, Adachi JA, De La Cabada FJ, Taylor DN, Jaini S, Martinez Sandoval F. 2007. Treatment of travelers' diarrhea: randomized trial comparing rifaximin, rifaximin plus loperamide, and loperamide alone. Clin. Gastroenterol. Hepatol. 5:451–456 [DOI] [PubMed] [Google Scholar]
- 3.Merson MH, Sack RB, Islam S, Saklayen G, Huda N, Huq I, Zulich AW, Yolken RH, Kapikian AZ. 1980. Disease due to enterotoxigenic Escherichia coli in Bangladeshi adults: clinical aspects and a controlled trial of tetracycline. J. Infect. Dis. 141:702–711 [DOI] [PubMed] [Google Scholar]
- 4.Zhang C, Liu WH, Ren HY. 2012. Complete genome sequence of Bp7, an Escherichia coli bacteriophage with wide host range. J. Virol. 86:13832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang T, Wang CG, Lv JC, Wang RS, Zhong XH. 2012. Survey on tetracycline resistance and antibiotic-resistant genotype of avian Escherichia coli in North China. Poult. Sci. 91:2774–2777 [DOI] [PubMed] [Google Scholar]
- 6.Li H, Ma ML, Xie HJ, Kong J. 2012. Biosafety evaluation of bacteriophages for treatment of diarrhea due to intestinal pathogen Escherichia coli 3-2 infection of chickens. World J. Microbiol. Biotechnol. 28:1–6 [DOI] [PubMed] [Google Scholar]
- 7.Burrowes B, Harper DR, Anderson J, McConville M, Enright MC. 2011. Bacteriophage therapy: potential uses in the control of antibiotic-resistant pathogens. Expert Rev. Anti Infect. Ther. 9:775–785 [DOI] [PubMed] [Google Scholar]
- 8.Sabouri Ghannad M, Mohammadi A. 2012. Bacteriophage: time to re-evaluate the potential of phage therapy as a promising agent to control multidrug-resistant bacteria. Iran J. Basic Med. Sci. 15:693–701 [PMC free article] [PubMed] [Google Scholar]
- 9.Krylov V, Shaburova O, Krylov S, Pleteneva E. 2013. A genetic approach to the development of new therapeutic phages to fight Pseudomonas aeruginosa in wound infections. Viruses 5:15–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Joerger RD. 2003. Alternatives to antibiotics: bactericins, antimicrobial peptides and bacteriophages. Poult. Sci. 82:640–647 [DOI] [PubMed] [Google Scholar]
- 11.Adams MH. 1959. Bacteriophage. Interscience Publishers, New York, NY [Google Scholar]
- 12.Vuković I, Lacković V, Todorivić V, Kanjuh V, Ilić S. 2004. Cytohistologic and immunohistochemical characteristics of the aortic intima and media in coarctation of the aorta of the adult type. Srp. Arh. Celok. Lek. 132:66–71. (In Serbian.) [DOI] [PubMed] [Google Scholar]
- 13.Kropinski AM, Waddell T, Meng J, Franklin K, Ackermann HW, Ahmed R, Mazzocco A, Yates J, Lingohr EJ, Johnson RP. 2013. The host-range, genomics and proteomics of Escherichia coli O157:H7 bacteriophage rV5. Virol. J. 10:76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun WJ, Liu CF, Yu L, Cui FJ, Zhou Q, Yu SL, Sun L. 2012. A novel bacteriophage KSL-1 of 2-keto-gluconic acid producer Pseudomonas fluorescens K1005: isolation, characterization and its remedial action. BMC Microbiol. 12:127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pajunen M, Kiljunen S, Skurnik M. 2000. Bacteriophage ϕYeO3-12, specific for Yersinia enterocolitica serotype O:3, is related to coliphages T3 and T7. J. Bacteriol. 182:5114–5120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carver T, Rutherford KM, Berriman M, Rajandream M, Barrell BG, Parkhill J. 2005. ACT: the Artemis comparison tool. Bioinfomatics 21:3422–3423 [DOI] [PubMed] [Google Scholar]
- 18.Ackermann HW. 2007. Salmonella phages examined in the electron microscope. Methods Mol. Biol. 394:213–234 [DOI] [PubMed] [Google Scholar]
- 19.Miller ES, Kutter E, Mosig G, Arisaka F, Kunisawa T, Rüger W. 2003. Bacteriophage T4 genome. Microbiol. Mol. Biol. Rev. 67:86–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanlon GW. 2007. Bacteriophages: an appraisal of their role in the treatment of bacterial infections. Int. J. Antimicrob. Agents 30:118–128 [DOI] [PubMed] [Google Scholar]
- 21.Skurnik M, Strauch E. 2006. Phage therapy: facts and fiction. Int. J. Med. Microbiol. 296:5–14 [DOI] [PubMed] [Google Scholar]
- 22.Kropinski AM, Lingohr EJ, Moyles DM, Ojha S, Mazzocco A, She YM, Bach SJ, Rozema EA, Stanford K, McAllister TA, Johnson RP. 2012. Endemic bacteriophages: a cautionary tale for evaluation of bacteriophage therapy and other interventions for infection control in animals. Virol. J. 17:207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang H, Chen S, White DG, Zhao S, McDermott P, Walker R, Meng J. 2004. Characterization of multiple-antimicrobial-resistant Escherichia coli isolates from diseased chickens and swine in China. J. Clin. Microbiol. 42:3483–3489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Comeau AM, Bertrand C, Letarov A, Tétart F, Krisch HM. 2007. Modular architecture of the T4 phage superfamily: a conserved core genome and a plastic periphery. Virology 362:384–396 [DOI] [PubMed] [Google Scholar]
- 25.Lavigne R, Darius P, Summer EJ, Seto D, Mahadevan P, Nilsson AS, Ackermann HW, Kropinski AM. 2009. Classification of Myoviridae bacteriophages using protein sequence similarity. BMC Microbiol. 9:224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball A. 2005. Virus taxonomy, p 35–85, In VIIIth report of the International Committee on Taxonomy of Viruses Elsevier Academic Press, New York, NY [Google Scholar]
- 27.Petrov VM, Ratnayaka S, Nolan JM, Miller ES, Karam JD. 2010. Genomes of the T4-related bacteriophages as windows on microbial genome evolution. Virol. J. 7:292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang H, Jiang X, Wang S, Li C, Chen B, An X, Mi Z, Chen J, Tong Y. 2011. The complete genome sequence of a novel T4-like bacteriophage, IME08. Arch. Virol. 156:1489–1492 [DOI] [PubMed] [Google Scholar]
- 29.Zuber S, Ngom-Bru C, Barretto C, Bruttin A, Brüssow H, Denou E. 2007. Genome analysis of phage JS98 defines a fourth major subgroup of T4-like phages in Escherichia coli. J. Bacteriol. 189:8206–8214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Letarov A, Manival X, Desplats C, Krisch HM. 2005. gpwac of the T4-type bacteriophages: structure, function, and evolution of a segmented coiled-coil protein that controls viral infectivity. J. Bacteriol. 187:1055–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim JH, Son JS, Choi YJ, Choresca CH, Jr, Shin SP, Han JE, Jun JW, Park SC. 2012. Complete genome sequence and characterization of a broad-host range T4-like bacteriophage phiAS5 infecting Aeromonas salmonicida subsp. salmonicida. Vet. Microbiol. 157:164–171 [DOI] [PubMed] [Google Scholar]
- 32.Huff WE, Huff GR, Rath NC, Donoghue AM. 2013. Method of administration affects the ability of bacteriophage to prevent colibacillosis in 1-day-old broiler chickens. Poult. Sci. 92:930–934 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.