Abstract
Intestinal microbial dysbiosis contributes to the dysmetabolism of luminal factors, including steroid hormones (sterones) that affect the development of chronic gastrointestinal inflammation and the incidence of sterone-responsive cancers of the breast, prostate, and colon. Little is known, however, about the role of specific host sterone nucleoreceptors, including estrogen receptor β (ERβ), in microbiota maintenance. Herein, we test the hypothesis that ERβ status affects microbiota composition and determine if such compositionally distinct microbiota respond differently to changes in diet complexity that favor Proteobacteria enrichment. To this end, conventionally raised female ERβ+/+ and ERβ−/− C57BL/6J mice (mean age of 27 weeks) were initially reared on 8604, a complex diet containing estrogenic isoflavones, and then fed AIN-76, an isoflavone-free semisynthetic diet, for 2 weeks. 16S rRNA gene surveys revealed that the fecal microbiota of 8604-fed mice and AIN-76-fed mice differed, as expected. The relative diversity of Proteobacteria, especially the Alphaproteobacteria and Gammaproteobacteria, increased significantly following the transition to AIN-76. Distinct patterns for beneficial Lactobacillales were exclusive to and highly abundant among 8604-fed mice, whereas several Proteobacteria were exclusive to AIN-76-fed mice. Interestingly, representative orders of the phyla Proteobacteria, Bacteroidetes, and Firmicutes, including the Lactobacillales, also differed as a function of murine ERβ status. Overall, these interactions suggest that sterone nucleoreceptor status and diet complexity may play important roles in microbiota maintenance. Furthermore, we envision that this model for gastrointestinal dysbiosis may be used to identify novel probiotics, prebiotics, nutritional strategies, and pharmaceuticals for the prevention and resolution of Proteobacteria-rich dysbiosis.
INTRODUCTION
The phenotypic manifestation (phenome) of the hydrolytic capacity of the gastrointestinal microbiome vastly expands the efficiency by which the host assimilates dietary nutrients and energy, especially from the otherwise indigestible dietary components, including select fibers and prebiotics (1, 2). Cooperative metabolism (cometabolism) of low-molecular-weight compounds by the host and luminal microbiota also plays a part in these energy-salvaging activities (3). Together, these hydrolytic and cometabolic activities have undoubtedly played a vital role in the natural selection of species, especially when supplies of safe, nutrient-dense food were scarce (4).
Today, cometabolism of steroid hormones (i.e., sterones), including endogenous sterones (e.g., estradiol) and low-molecular-weight dietary compounds with hormone-like activities (e.g., phytoestrogenic isoflavones), continues to play an important role in maintaining healthy colonic tissues (5). In soy-based products, for example, the isoflavone genistein is typically consumed as genistin, an isoflavone glucoside (3). Genistein (aglycone) exhibits strong estrogenic activities that may reduce the incidence of chronic low-grade gastrointestinal inflammation (6). Many of the biological activities associated with phytoestrogen consumption may arise from direct absorption of isoflavones by mammalian cells; however, the intestinal microbiota clearly acts to coregulate these bioactivities (7). Indeed, β-glucosidase enzymes produced by intestinal bacteria liberate genistein, which may be cometabolized into biochemically and functionally distinct metabolites by a variety of intestinal microorganisms (8). These sterone metabolites mediate their bioactive actions through their association with host cell-encoded transcriptional regulators, principally estrogen receptor α (ERα) and estrogen receptor β (ERβ) (6).
ERβ is the most abundant estrogen receptor in the colon, where it regulates the permeability of colonic epithelia (9). Interestingly, ERβ-null mice exhibit a number of prepathogenic phenotypes, including abnormal colonic architecture and disrupted cell-to-cell tight junctions (9). These structural abnormalities facilitate invasion of host tissues by luminal bacteria, which leads to localized infection and increased levels of colonic inflammation (10). Unfortunately, little is known about the role of host ERβ status on the structure of the gastrointestinal microbiota and their downstream effects on host physiology. Nevertheless, other studies illustrate that variation of even a single host gene can significantly alter host-driven selective pressures that help to shape the structure and function of the commensal gastrointestinal microbiota (11, 12).
While eubiotic microbiota may transition between distinct ecosystem optima, they can also degenerate into dysbiosis in response to dietary and physiological changes. Such abnormal structures and injurious functions of autochthonous gastrointestinal microbiota also contribute to the development of chronic low-grade gastrointestinal inflammation and its related comorbidities, including obesity, dysmetabolic syndrome, diabetes mellitus (type II), atherosclerosis, inflammatory bowel diseases, and certain cancers (13, 14). Once a dysbiotic microbiota has been established, regardless of its etiology, emerging evidence shows a striking correlation between Proteobacteria-rich dysbiotic microbiota and chronic inflammatory bowel diseases, including Crohn's disease and ulcerative colitis (15). These findings are relevant because many Proteobacteria are known to elicit strong, proinflammatory immune responses due to the presence of lipopolysaccharide (LPS) in the outer leaflet of their outer membrane (16).
In this study, we developed a murine model to test the hypothesis that a dysbiotic intestinal microbiota characterized by a relatively high abundance of Proteobacteria develops in response to large-scale changes in diet complexity, specifically in response to a transition from a biochemically complex diet to one that is highly derivatized and biochemically simple. Since microbiota composition may contribute to the dysmetabolism of luminal compounds, including sterones, we also use this model system to test the hypothesis that ERβ plays an important role in the selection of intestinal microbiota.
MATERIALS AND METHODS
Animal study design.
The compositions of the diets used in this study are listed in Table 1, while their analyzed constituents are in Table S1 in the supplemental material. All animal procedures were performed under a protocol approved by the Texas A&M University Institutional Animal Care and Use Committee. Mice (Mus musculus Linnaeus) were housed individually at the Texas A&M University Laboratory Animal Resources and Research Facility, where they were provided food and water ad libitum. Wild-type C57BL/6J mice and their ERβ-null (Esr2−/−) derivatives (17) (Jackson Laboratory, Bar Harbor, ME) were crossed to produce a Mendelian distribution of wild-type C57BL/6 (Esr2+/−), ERβ+/+ (Esr2+/+), and ERβ−/− (Esr2−/−) progeny, which were verified by tail snip and PCR as described previously (5). Mice were maintained on a complex diet, Tekland rodent diet 8604 (Harlan Laboratories, Madison, WI), prior to the initiation of the study; from birth until the start of the study, the mean age [μage] of the mice was 27 weeks. Freshly voided feces were collected from 8604-fed female ERβ+/+ mice (n = 21) and their ERβ−/− littermates (n = 21) at the start of the study (day 0) and then again after the mice had consumed a simple semisynthetic diet, American Institute of Nutrition rodent diet 76 (AIN-76) (Lab Supply, Highland Village, TX), for a 2-week period (day 14). Fecal pellets collected over the course of the study were weighed and frozen (−80°C) for downstream DNA extraction and analyses.
Table 1.
Formulation of the animal diets used in this study
| Fraction | 8604 |
AIN-76 |
||||
|---|---|---|---|---|---|---|
| Ingredient(s) | kcal g−1 | Energy (%) | Ingredient(s) | kcal g−1 | Energy (%) | |
| Carbohydrate | Flaked corn, ground corn, wheat middling, cane molasses, ground wheat | 1.608 | 54 | Sucrose, dextrin, cellulose | 2.656 | 69.2 |
| Protein | Dehulled soybean meal, fish meal, dried whey | 0.972 | 32 | Casein, dl-methionine | 0.728 | 19 |
| Lipid | Soybean oil | 0.423 | 14 | Corn oil | 0.45 | 11.7 |
| Micronutrients | Brewers dried yeast, dicalcium phosphate, calcium carbonate, iodized salt, choline chloride, kaolin, magnesium oxide, ferrous sulfate, vitamin E acetate, menadione sodium bisulfite, manganous oxide, zinc oxide, niacin, thiamine mononitrate, vitamin A acetate, vitamin D3, calcium pantothenate, pyridoxine hydrochloride, riboflavin, vitamin B12, calcium iodate, folic acid, biotin, cobalt carbonate | AIN-76 mineral mix, AIN-76 vitamin mix, choline bitartrate | ||||
| Total energy | 3.003 | 100 | 3.84 | 99.9 | ||
Extraction of total DNA from fecal samples.
Unless indicated otherwise, reagents were of analytical grade or higher and obtained from Sigma-Aldrich Chemical Company (St. Louis, MO). In brief, fecal samples from day 0 and day 14 were thawed on ice and homogenized in Tris buffer (pH 8) for 2 min at 4 m s−1 using a FastPrep-24 instrument (MP Biomedicals, Solon, OH). Total DNA was extracted from the resultant murine fecal sample homogenates (500 μl) by using the Fast DNA spin kit for soil (MP Biomedicals). Purified DNA was resuspended in sterile deionized water, analyzed by spectrophotometry (NanoDrop 1000; Thermo Scientific, Wilmington, DE), and frozen (−80°C) for downstream terminal restriction fragment length polymorphism (TRFLP) analysis.
TRFLP analysis.
The HotStar HiFidelity polymerase kit (Qiagen, Valencia, CA) was used to amplify the 16S rRNA genes from fecal DNA. DNA (100 ng) served as the template for each 100-μl PCR mixture. Master mixes consisted of 10 μl of 5× reaction buffer, 28.5 μl of molecular biology-grade deionized water, 0.25 μl of 5′-hexachlorofluorescein (HEX)-labeled forward primer 8F-HEX (5′-AGAGTTTGATCMTGGCTCAG-3′, where M is A or C) at 100 μM, 0.25 μl of reverse primer 1492R (5′-GGTTACCTTGTTACGACTT-3′) (18) at 100 μM, and 1 μl (2.5 U) of polymerase. After an initial DNA denaturation step (5 min at 95°C), samples underwent 25 cycles of denaturation (1 min at 95°C), primer annealing (1 min at 50°C), and primer extension (2 min at 72°C), followed by a final extension (10 min at 74°C). In order to minimize the impact of PCR variability on downstream TRFLP analysis, DNA from each fecal sample was amplified by PCR in quadruplicate, and all four PCRs from each fecal sample were pooled prior to cleanup using the QIAquick PCR purification kit (Qiagen).
In order to survey the species dominance and species richness of the 16S rRNA genes amplified from each DNA sample (19), TRFLP analysis was performed at the University of North Carolina Microbiome Core Facility (Chapel Hill, NC). In brief, purified PCR amplicons were treated (separately) with three restriction endonucleases: RsaI, HhaI, and MspI (New England BioLabs, Ipswich, MA). Restriction digests were composed of 10 μl of 10× reaction buffer (buffer 4; New England BioLabs), 1 μl of 100× bovine serum albumin (HhaI only), 1 μl of each enzyme, 30 μl of purified amplicon, and molecular biology-grade water that was added to a final volume of 100 μl. Restriction digests were incubated overnight at 37°C. Following incubation, digested DNA was purified using the QIAquick nucleotide removal kit (Qiagen) according to the manufacturer's instructions, with minor modifications. Following cleanup, 4.5 μl of Hi-Di formamide (Applied Biosystems, Carlsbad, CA) and 0.5 μl of MM-1000 ROX size standard (BioVentures, Murfreesboro, TN) were added to 5 μl of each digestion reaction. Following a brief denaturing step (94°C for 3 min), samples for fragment detection were loaded into an ABI 3130xl capillary sequencer (Applied Biosystems).
qPCR-based measurement of phylotype-specific 16S rRNA gene abundance.
In order to estimate the total number of bacteria, Bacteroidetes, Gammaproteobacteria, and lactobacilli present in the feces of AIN-76-fed mice, BR-SYBR green-based quantitative real-time PCR (qPCR) was performed according to specifications provided by the manufacturer (Quanta Biosciences, Gaithersburg, MD). Purified genomic DNA from Escherichia coli MC1061 (20), Bacteroides thetaiotaomicron E50 (American Type Culture Collection, Manassas, VA, USA), and Lactobacillus acidophilus NCFM (21) was decimally diluted and used as the templates to generate standard curves that allowed for the quantification of fecal bacteria and Gammaproteobacteria, Bacteroidetes, and lactobacilli, respectively. Each reaction (25 μl) included oligonucleotide primers (Invitrogen Corporation, Carlsbad, CA) designed to target group-specific 16S rRNA gene sequences. For the enumeration of bacteria (50 pg per reaction), primer Bact515R (5′-TTACCGCGGCKGCTGGCAC-3′, where K is G or T) was paired with primers 8FM (5′-AGAGTTTGATCMTGGCTCAG-3′, where M is A or C) and 8FB (5′-AGGGTTCGATTCTGGCTCAG-3′), as described elsewhere (22). For the enumeration of Bacteroidetes (10 ng per reaction), primer Bac32F (5′-AACGCTAGCTACAGGCTT-3′) was paired with primer Bac303R (5′-CCAATGTGGGGGACCTTC-3′), as described elsewhere (23). For the enumeration of Gammaproteobacteria (10 ng per reaction), primer γ395f (24) (5′-CMATRCCGCGTGTRTGAA-3′, where M is A or C and R is A or G) was paired with primer γ871r (5′-ACTCCCCAGGCGGTCDACTTA-3′, where D is A, G, or T), as described elsewhere (25). For the enumeration of lactobacilli (5 ng per reaction), primer Lac1 (5′-AGCAGTAGGGAATCTTCCA-3′) was paired with primer Lac2 without the GC clamp (5′-ATTYCACCGCTACACATG-3′, where Y is C or T) (21). All qPCRs were incubated in an iCycler (Bio-Rad Laboratories, Hercules, CA) equipped with an iQ5 multicolor real-time PCR detection system (Bio-Rad Laboratories) using thermal cycling conditions described elsewhere (22). Results are expressed as means ± standard deviations.
Bioinformatics.
Peaks for TRFLP were identified using GeneMapper 4.0 (Applied Biosystems) using the default detection parameters and a minimum peak height of 50 relative fluorescence units (RFU). Following peak detection, peaks that fell outside the size standard (50 to 1,000 bp) were removed, and only terminal restriction fragments (TRFs) with a relative peak area ratio of ≥1% were considered for further analysis (26). The cleaned fragment files were then uploaded to the Web-based TRFLP Phylogenetic Assignment Tool (https://secure.limnology.wisc.edu/trflp/) (27), and each TRF profile was tentatively identified using a custom pattern database created from both an in silico digest of the Web-based Ribosomal Database Program (RDP) database (http://rdp.cme.msu.edu/) (28) and unique clone sequences from the University of North Carolina Microbiome Core Facility sequence bank. The RDP Classifier (29) was used to assign the putative hierarchical taxonomy for each TRF pattern using 16S rRNA gene sequences derived from the National Center for Biotechnology Information (NCBI) nucleotide database.
Biostatistics.
Prior to statistical analysis, TRFLP Phylogenetic Assignment Tool data were compressed to binary (presence/absence) data using custom database macros written for Access and Excel (Microsoft, Redmond, WA). The relative abundance for each taxon (phylum, class, and order) was subsequently calculated as described elsewhere (30). Unless otherwise indicated, all statistical analyses were performed using Paleontological Statistics Software Package (PAST) version 2.12 (31) or R (www.r-project.org) (R Development Core Team, 2008). The Shannon (H′) (32) and Simpson (D) (33) parametric diversity indices were calculated using PAST version 2.12, whereas the nonparametric diversity score (SChao1) was calculated using a Web-based application (http://www.aslo.org/cgi-bin/largeenough.cgi) (34, 35). A diversity t test was performed to test for statistically significant differences in pattern richness between the groups (36).
Cluster analysis with presence/absence binary data using Ward's group linkage method was used to construct a hierarchical tree (37). A nonparametric multidimensional scaling (NMS) analysis based on Bray-Curtis distance (38) was performed to estimate similarities between the bacterial communities as a function of the categorical variables examined in this study: host ERβ status (ERβ+/+ versus ERβ−/−) and diet (8604 versus AIN-76). Analysis of similarities (ANOSIM) was used to test for global differences in bacterial community composition (38). Unlike NMS, ANOSIM tests are not compromised by the approximations necessary to view a two-dimensional ordination pattern, since they utilize the full high-dimensional space of the (rank) dissimilarity matrices (39). The permutation-based, nonparametric statistic R was used to test the null hypothesis (H0) that within-group and between-group distances are the same on average (39). H0 was rejected when distances between samples were more dissimilar between different groups than between samples from the same group (R = −1 < r ≤ 1). If statistically significant (p < 0.05) differences were detected by ANOSIM, the conservation of discrete operational taxonomic units between groups was detected using similarity percentage analysis (SIMPER) using the Bray-Curtis similarity measure (38, 40).
Discrete differences in the bacterial taxa were analyzed via regression models that accounted for host genotype, diet, and their interaction. The repeated measurements for each mouse were accommodated by allowing the regression model residuals to be correlated (41). Likelihood ratio statistics were used to carry out hypothesis tests for the genotype and diet effects on gastrointestinal microbiota (42). Effects were called statistically significant if they were assigned a q-value less than 0.05; as a result, we expected no more than 5% of the effects called significant to be false discoveries (43).
RESULTS
Specific dietary components and diet complexity contribute to the enrichment of microorganisms that are important for gastrointestinal function.
8604 and AIN-76 were both produced from undefined ingredients (Table 1). As a result, the exact molecular composition of these diets is unknown. Nevertheless, the biochemically complex 8604 diet is comprised of eight complex food ingredients (e.g., soybean meal), two semidefined food ingredients (e.g., soybean oil), and 24 defined chemical supplements (e.g., riboflavin). The AIN-76 diet, on the other hand, is a biochemically simple semisynthetic diet, as it is comprised of only highly refined ingredients: four semidefined ingredients (e.g., casein, corn oil) and five defined chemical supplements (e.g., AIN-76 vitamin mix). Based on analytical information provided by their manufacturers (see Table S1 in the supplemental material), the soy-derived isoflavones (daidzein and genistein) were exclusive to the complex 8604 diet, as was ash (74 g kg−1 of diet), low levels of cholesterol (0.05 g kg−1), unsaturated fatty acids (7 g kg−1 palmitic acid, 1 g kg−1 stearic acid), and the monounsaturated fatty acid oleic acid (9 g kg−1). In contrast, chromium was exclusive to the semisynthetic AIN-76 diet. The AIN-76 diet had a relatively higher concentration of vitamin B12 (8604 = 0.00005 g kg−1; AIN-76 = 0.01 g kg−1) but lower fiber content (8604 = 164 g kg−1; AIN-76 = 50 g kg−1) than the 8604 diet.
Large-scale structural changes in the autochthonous gastrointestinal microbiota occurred in response to dietary changes.
TRFLP of 16S rRNA genes was used to examine the fecal microbiota of conventionally raised ERβ+/+ mice and their ERβ−/− littermates following the habitual consumption of the complex 8604 diet (day 0) and again after consuming the AIN-76 semisynthetic diet for a 2-week period (day 14). Of the three enzymes tested during TRFLP, HhaI provided the best discrimination between samples in the different groups (data not shown). As a result, the HhaI-derived terminal restriction fragment (TRF) data set was analyzed further. NMS was used to analyze the similarity of HhaI-digested TRFs associated with each sample (Fig. 1). Within the two diet-specific superclusters (Fig. 1), the fecal communities of individual mice within the AIN-76-fed ERβ−/− groups formed tight subclusters. In contrast, the fecal communities from the 8604-fed ERβ+/+, 8604-fed ERβ−/−, and AIN-76-fed ERβ+/+ mice formed relatively diffuse subclusters and, thus, exhibited a greater degree of compositional variability in their TRFLP patterns. Stress in the ordination was likely high due to variability in the data and the approximations required to view the data in two-dimensional space (NMS, stress = 0.21).
Fig 1.
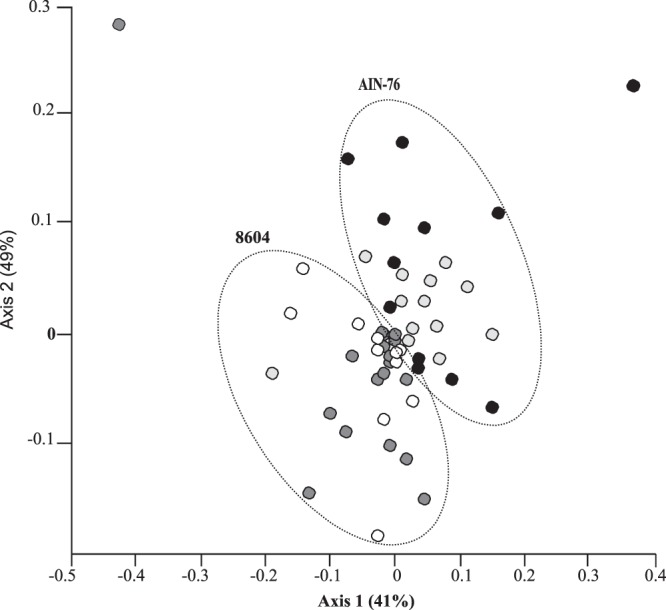
Nonmetric multidimensional scaling analysis of bacterial community TRFLP profiles. ERβ+/+ mice (n = 21) and their ERβ−/− littermates (n = 21) were initially maintained on a complex 8604 diet (day 0). Mice were then transitioned to the AIN-76 semidefined diet for a 2-week period (day 14). Ninety-five percent of the total variance in TRFLP data is represented in the two-dimensional space (stress = 0.21). ○, 8604-fed ERβ+/+ mice;  , 8604-fed ERβ−/− mice; ●, AIN-76-fed ERβ+/+ mice;
, 8604-fed ERβ−/− mice; ●, AIN-76-fed ERβ+/+ mice;  , AIN-76-fed ERβ−/− mice. Dotted circles were manually inserted in order to highlight the clustering of communities according to diet.
, AIN-76-fed ERβ−/− mice. Dotted circles were manually inserted in order to highlight the clustering of communities according to diet.
Estrogen receptor β status may contribute to diet-directed modulation of the autochthonous gastrointestinal microbiota.
Given the group-specific differences in variability that were revealed by NMS, ANOSIM was used to test the hypothesis that host ERβ status (genotype) contributed to group-specific variation in microbial community structure. The fecal microbiota of ERβ+/+ and ERβ−/− mice that consumed the 8604 complex diet were significantly different at the start of the study (ANOSIM, R = 0.237, p < 0.0001). Consumption of the AIN-76 semisynthetic diet for a 2-week period correlated with significant changes in the microbiota of ERβ+/+ mice (p < 0.0001) and their ERβ−/− littermates (p < 0.0001). Interestingly, however, the microbiota from ERβ+/+ and ERβ−/− mice did not differ statistically at the end of the study (p = 0.2078).
ERβ status and diet complexity differentially affect the species richness of abundant gastrointestinal microorganisms.
The species diversity within each group was estimated using two parametric tests (H′ and D) and one nonparametric test (SChao1) (Table 2). Since many of the samples contained rare abundance TRFs (singletons and doubletons), the nonparametric test SChao1 afforded the highest level of discrimination between the categorical variables that were examined in this study (host genotype and diet). Interestingly, a significant difference was found in the species richness due to differences in diet and genotype (diversity t test, p < 0.01). Indeed, ERβ+/+ mice (SChao1 = 1,234.27) that consumed the 8604 diet (day 0) exhibited lower species diversity than their 8604-fed ERβ−/− littermates (SChao1 = 1,717.25). In contrast, following the consumption of the AIN-76 diet, the species richness increased dramatically for ERβ+/+ mice (SChao1 = 1,909.55) but decreased for ERβ−/− mice (SChao1 = 1,649.52).
Table 2.
Microbial pattern diversity indexes
| Genotype | Parametric |
Nonparametric SChao1 |
||||
|---|---|---|---|---|---|---|
|
D |
H′ |
|||||
| 8604 | AIN-76 | 8604 | AIN-76 | 8604 | AIN-76 | |
| ERβ+/+ | 0.96 | 0.97 | 3.50 | 3.90 | 1,234.27 | 1,909.55 |
| ERβ−/− | 0.97 | 0.92 | 3.90 | 3.40 | 1,717.20 | 1,649.50 |
The initial composition of the microbiota determines, in part, its response to dietary change.
To explore the relationship of individual bacterial communities between the samples, a hierarchical tree based on Ward's group linkage method was constructed from a binary (presence/absence) index of the species-level TRF data (Fig. 2). Similar to the results from NMS, microbiota were sorted largely based on diet into two superclusters: I and II. Regardless of host genotype, the fecal microbiota from mice fed the complex 8604 diet sorted almost exclusively into supercluster I (95% from ERβ+/+ mice; 100% from ERβ−/− mice). The proportion of microbiota identified in 8604-fed ERβ+/+ mice (62%) was significantly higher in subcluster IB than in any other subcluster (χ2, p = 0.0003). Similarly, the proportion of microbiota isolated from 8604-fed ERβ−/− mice was higher in subcluster IA than in any other subcluster (48%). Interestingly, the composition of subcluster IC was indeterminate, as it was comprised almost equally of the microbiota from ERβ+/+ and ERβ−/− mice on both diets.
Fig 2.
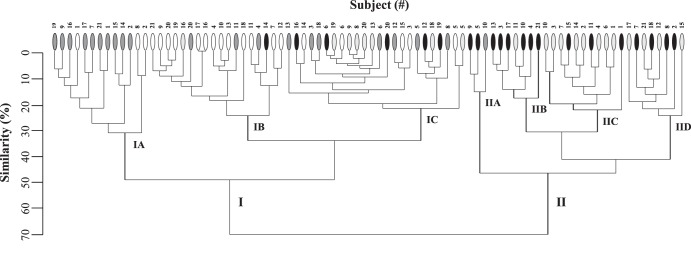
Cluster dendrogram illustrating the degree of relatedness between microbiota isolated from individual subjects. Individual subjects (x axis) were assigned a numerical designation (#) as a function of genotype (for both genotypes, n = 21 mice). The similarity (%) (y axis) of the microbiota between individual subjects is based on an index of TRF presence/absence binary data. Individual mice are represented as follows: ○, 8604-fed ERβ+/+ mice;  , 8604-fed ERβ−/− mice; ●, AIN-76-fed ERβ+/+ mice;
, 8604-fed ERβ−/− mice; ●, AIN-76-fed ERβ+/+ mice;  , AIN-76-fed ERβ−/− mice.
, AIN-76-fed ERβ−/− mice.
In contrast, the microbiota from mice fed the AIN-76 diet were found in both superclusters; however, most sorted into supercluster II (71% from ERβ+/+ mice; 61% from ERβ−/− mice). The microbiota from 8604-fed mice in subcluster IB transitioned into subclusters IID and IIC more frequently than into any other subcluster (46% from ERβ+/+ mice and 33% from ERβ−/− mice). Similarly, microbiota from 8604-fed mice in subcluster IA transitioned into subclusters IIB and IIA more frequently than they transferred into any other subcluster (60% from ERβ+/+ mice and 100% from ERβ−/− mice).
The relative abundance of prominent phylotypes within the murine autochthonous gastrointestinal microbiota differed in their response to diet, ERβ status, and interaction.
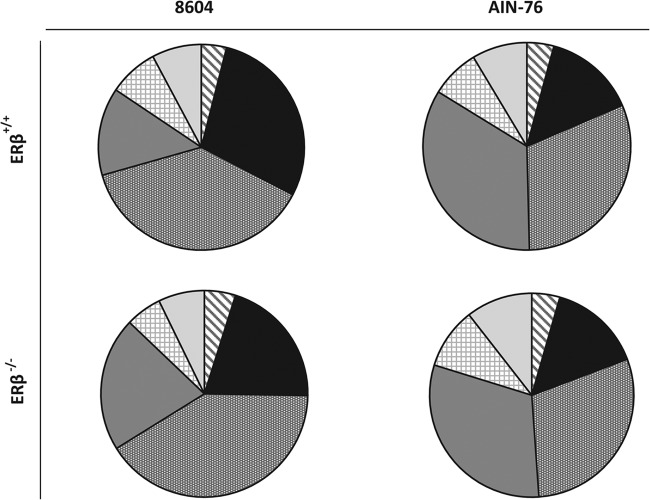
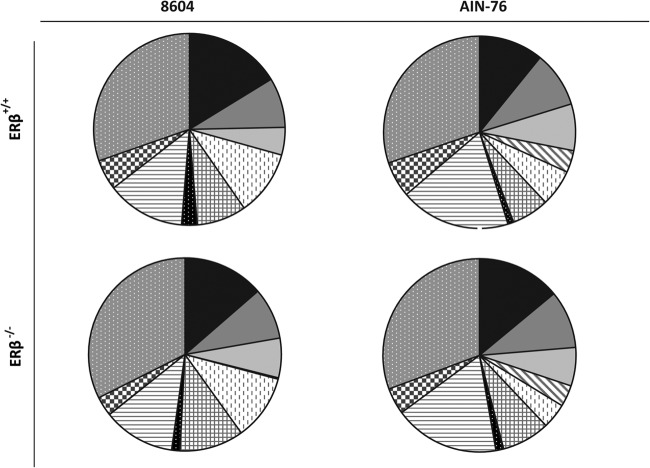
Representatives from 19 phyla consisting of 35 classes and 66 orders were tentatively identified by TRFLP. The relative abundance of these taxa was determined as a function of diet and genotype at the levels of phylum (Fig. 3) and class (Fig. 4). Irrespective of the diet consumed or host genotype, Bacteroidetes, Firmicutes, and Proteobacteria were the most abundant phylotypes in the gastrointestinal tracts of ERβ+/+ and ERβ−/− mice, although lower levels (<5%) of Acidobacteria and Chloroflexi were also detected. In mice fed the complex 8604 diet (day 0), the Firmicutes were the most abundant phylotype detected at the start of the study regardless of host genotype (ERβ+/+ = 38.0%; ERβ−/− = 40.9%) (Fig. 3). The Bacteroidetes levels were relatively higher in ERβ+/+ mice (28.7%) than in their ERβ−/− littermates (20.4%), whereas the Proteobacteria levels were relatively lower in ERβ+/+ mice (13.8%) than in ERβ−/− mice (21.0%). Most of the Firmicutes were Clostridia in 8604-fed mice, regardless of host genotype (ERβ+/+ = 85.2%; ERβ−/− = 90.4%); however, Bacilli levels were relatively higher in ERβ+/+ mice (14.4%) than in ERβ−/− mice (9.2%) (Fig. 4). Within the Proteobacteria, the Epsilonproteobacteria levels were relatively higher in ERβ+/+ mice (7.4%) than in ERβ−/− mice (3.9%), while the Deltaproteobacteria levels were relatively lower in ERβ+/+ mice (23.5%) than in ERβ−/− mice (30.0%).
Fig 3.
Relative abundance (%) of phylum-level taxa within Bacteria. The proportion of each taxonomic group is described by the measure of its central angle, which is relative to the total abundance of all phylotypes (100%).  , Acidobacteria;
, Acidobacteria;  , Bacteroidetes;
, Bacteroidetes;  , Firmicutes;
, Firmicutes;  , Proteobacteria;
, Proteobacteria;  , unclassified bacteria;
, unclassified bacteria;  , other phyla, including taxa from the Actinobacteria, Aquificae, Chlamydiae, Chlorobi, Cyanobacteria, Deferribacteres, Deinococcus-Thermus, Euryarchaeota, Fusobacteria, Gemmatimonadetes, OD1, Planctomycetes, Tenericutes, and Verrucomicrobia.
, other phyla, including taxa from the Actinobacteria, Aquificae, Chlamydiae, Chlorobi, Cyanobacteria, Deferribacteres, Deinococcus-Thermus, Euryarchaeota, Fusobacteria, Gemmatimonadetes, OD1, Planctomycetes, Tenericutes, and Verrucomicrobia.
Fig 4.
Relative abundance (%) of class-level taxa within the phyla Bacteroidetes, Proteobacteria, and Firmicutes. The proportion of each taxonomic group is described by the measure of its central angle. Taxa within the phylum Bacteroidetes:  , Bacteroidia;
, Bacteroidia;  , Flavobacteria;
, Flavobacteria;  , Sphingobacteria. Taxa within the phylum Proteobacteria:
, Sphingobacteria. Taxa within the phylum Proteobacteria:  , Alphaproteobacteria;
, Alphaproteobacteria;  , Betaproteobacteria;
, Betaproteobacteria;  , Deltaproteobacteria;
, Deltaproteobacteria;  , Epsilonproteobacteria;
, Epsilonproteobacteria;  , Gammaproteobacteria. Taxa within the phylum Firmicutes:
, Gammaproteobacteria. Taxa within the phylum Firmicutes:  , Bacilli;
, Bacilli;  , Clostridia.
, Clostridia.
At the end of the study (day 14), there was no significant difference in the relative abundance of the dominant phyla within the murine fecal microbiota, regardless of host ERβ status (for Firmicutes, ERβ+/+ = 31.1%, ERβ−/− = 29.4%; for Proteobacteria, ERβ+/+ = 34.2%, ERβ−/− = 30.9%; for Bacteroidetes, ERβ+/+ = 14.3%, ERβ−/− = 14.8%). In addition to becoming more dominant in AIN-76-fed mice, the relative proportions of phylotypes within the Proteobacteria also changed. Within the Proteobacteria, Gammaproteobacteria were enriched in the feces of AIN-76-fed mice (ERβ+/+ = 51.1%, ERβ−/− = 49.8%), while Betaproteobacteria levels were lower at the end of the study (ERβ+/+ = 17.0%, ERβ−/− = 11.9%), although their relative decrease differed by ERβ status (ERβ+/+ = −19%, ERβ−/− = −14%). Interestingly, while negligible levels (<1%) of Alphaproteobacteria were initially detected at the start of the study, they were significantly enriched following the consumption of the AIN-76 diet, regardless of genotype (ERβ+/+ = 10.2%, ERβ−/− = 10.1%).
Regression analysis was used to determine the statistical significance of order-level relative differences as a function of genotype. Within the Firmicutes, the relative abundance of the orders Clostridiales (q < 0.0001) and Lactobacillales (q = 0.002) differed significantly as a function of ERβ status. Indeed, the Lachnospiraceae and Lactobacillales levels were both relatively higher in 8604-fed ERβ+/+ mice than in their ERβ−/−-null littermates and decreased following the consumption of the AIN-76 diet. These analyses also indicated that the Anaerolineales (Chloroflexi, q = 0.002), Burkholderiales (Betaproteobacteria, q = 0.043), Deinococcales (Deinococcus-Thermus, q = 0.036), Desulfovibrionales (Deltaproteobacteria, q < 0.0001), Sphingobacteriales (Bacteroidetes, q = 0.027), and unclassified bacteria (q < 0.0001) also differed as a function of genotype.
Select taxonomic biomarkers may be diagnostic of ERβ status and diet-induced dysbiosis.
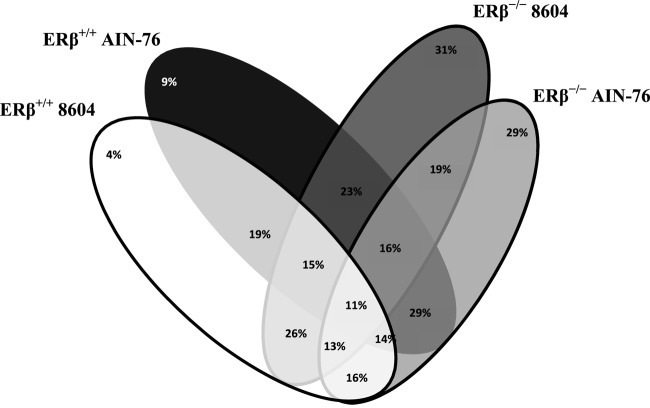
Approximately 1,000 taxa were tentatively identified in this study. SIMPER was used to identify taxonomic biomarkers that were diagnostic as a function of host ERβ status and diet. The similarity (intersection) and dissimilarity (exclusivity) percentages between the groups were determined (Fig. 5). As shown in Fig. 5, only 11% of the taxa were common in C57BL/6 mice regardless of host ERβ status (genotype) or diet consumed (i.e., were common to 8604-fed ERβ+/+ and ERβ−/− mice as well as AIN-76-fed ERβ+/+ and ERβ−/− mice). Of the 697 taxa that were found in the feces of ERβ+/+ mice, the vast majority (81%) were tentatively identified in both diet groups, while only 4% of the taxa were exclusive to the 8604 diet (day 0) and 9% of the taxa were unique to the AIN-76 diet (day 14). A similar number of taxa (783) was found in the feces of ERβ−/− mice. Of these taxa, 31% were exclusive to the 8604 diet (day 0), while 29% were unique to the AIN-76 diet (day 14); far fewer (19%) of these taxa were identified in both diet groups.
Fig 5.
Venn diagram showing similarity and dissimilarity between the taxa identified in this study. The number at the top of each oval indicates the unique species percentage within that group.
The highly discriminant taxa within the ERβ+/+ and ERβ−/− mice were determined as a function of diet. Taxa were classified as highly discriminant if they were exclusively present in 6 or more of the 21 mice. Twenty four highly discriminate TRF patterns were exclusive to mice fed the complex 8604 diet (Table 3). Five of these TRFs were exclusive to ERβ+/+ mice, whereas the remaining 19 TRFs were exclusive to ERβ−/− mice. Interestingly, all five of the TRF patterns exclusive to ERβ+/+ mice were tentatively assigned to a single family, Lachnospiraceae (Firmicutes), whereas the 19 TRFs from ERβ−/− mice were more heterogeneous and were tentatively assigned to five different phyla (Firmicutes, Chloroflexi, Bacteroidetes, Proteobacteria, and Aquificae). In contrast, 25 highly discriminate TRF patterns were exclusive to AIN-76-fed mice (Table 4). Twelve of these TRFs were exclusive to ERβ+/+ mice, whereas the remaining 13 TRFs were exclusive to ERβ−/− mice. The 12 TRFs exclusive to ERβ+/+ mice tentatively belonged to five phyla (Proteobacteria, Firmicutes, Chloroflexi, Bacteroidetes, and Acidobacteria), whereas the 13 TRFs exclusive to ERβ−/− mice belonged to six phyla (Proteobacteria, Acidobacteria, Bacteroidetes, Chloroflexi, Chlorobi, and Verrucomicrobia).
Table 3.
Highly discriminant bacterial patterns exclusive to 8604-fed mice
| Genotype | Bacterial pattern | Samplesa | Classification (phylum; class; order; family) |
|---|---|---|---|
| ERβ+/+ | Uncultured bacterium A19 | 12 | Firmicutes; Clostridia; Clostridiales; Lachnospiraceae |
| Uncultured rumen bacterium BRC159 | 8 | Firmicutes; Clostridia; Clostridiales; Lachnospiraceae | |
| Uncultured rumen bacterium 3C3d-17 | 7 | Firmicutes; Clostridia; Clostridiales; Lachnospiraceae | |
| Uncultured bacterium A11 | 6 | Firmicutes; Clostridia; Clostridiales; Lachnospiraceae | |
| Uncultured bacterium TuCc26 | 6 | Firmicutes; Clostridia; Clostridiales; Lachnospiraceae | |
| ERβ−/− | Lactobacillus fermentum KLB 261 | 12 | Firmicutes; Bacilli; Lactobacillales; Lactobacillaceae |
| Uncultured Bacilli bacterium ATB-LH-6349 | 12 | Firmicutes; Bacilli; Lactobacillales; Leuconostocaceae | |
| Uncultured bacterium TTMF57 | 12 | Unclassified bacteria | |
| Uncultured bacterium SJTUF0143 | 11 | Firmicutes; Clostridia; Clostridiales | |
| Uncultured bacterium CI35cm.2.05a | 10 | Chloroflexi; Anaerolineae; Anaerolineales; Anaerolineaceae | |
| Uncultured bacterium L-121(3) | 10 | Firmicutes; Clostridia; Clostridiales; Lachnospiraceae | |
| Uncultured rumen bacterium BRC11 | 10 | Firmicutes; Clostridia; Clostridiales | |
| Ruminofilibacter xylanolyticum S1 | 9 | Bacteroidetes; Bacteroidia; Bacteroidales; Marinilabiaceae | |
| Uncultured rumen bacterium F24-F06 | 9 | Bacteroidetes; Bacteroidia; Bacteroidales | |
| Uncultured rumen bacterium 3C3d-17 | 8 | Firmicutes; Clostridia; Clostridiales; Lachnospiraceae | |
| Cellulophaga tyrosinoxydans EM41 | 7 | Bacteroidetes; Flavobacteria; Flavobacteriales; Flavobacteriaceae | |
| Desulfovibrio vulgaris I5 | 6 | Proteobacteria; Deltaproteobacteria; Desulfovibrionales; Desulfovibrionaceae | |
| Uncultured bacterium BacB038 | 6 | Firmicutes; Clostridia; Clostridiales; Clostridiaceae | |
| Uncultured bacterium IIB-27 | 6 | Bacteroidetes; Bacteroidia; Bacteroidales | |
| Uncultured bacterium L-154 | 6 | Firmicutes; Clostridia; Clostridiales; Lachnospiraceae | |
| Uncultured bacterium SJTUD0150 | 6 | Firmicutes; Clostridia; Clostridiales | |
| Uncultured bacterium Y04 | 6 | Aquificae; Aquificae; Aquificales; Aquificaceae | |
| Uncultured Bacteroidetes bacterium VHS-B5-15 | 6 | Bacteroidetes | |
| Uncultured Shuttleworthia sp. 301E01 (oral) | 6 | Firmicutes; Clostridia; Clostridiales; Lachnospiraceae |
Number of samples in which the species pattern was found among the n = 21 samples tested.
Table 4.
Highly discriminant bacterial patterns exclusive to AIN-76-fed mice
| Genotype | Bacterial pattern | Samplesa | Classification (phylum; class; order; family) |
|---|---|---|---|
| ERβ+/+ | Uncultured bacterium A6 | 10 | Proteobacteria; Alphaproteobacteria; Sphingomonadales; Erythrobacteraceae |
| Uncultured bacterium NiASF28 | 9 | Acidobacteria | |
| Uncultured bacterium FGL12B72 | 8 | Unclassified | |
| Uncultured bacterium S26-35 | 8 | Proteobacteria; Alphaproteobacteria; Rhodospirillales; Rhodospirillaceae | |
| Uncultured bacterium SJTUE0255 | 8 | Firmicutes; Clostridia; Clostridiales; Ruminococcaceae | |
| Uncultured organism MAT-CR-P4-C09 | 8 | Chloroflexi; Anaerolineae; Anaerolineales; Anaerolineaceae | |
| Uncultured bacterium D13S-44 | 7 | Proteobacteria; Gammaproteobacteria | |
| Uncultured Cytophaga sp. BD1-15 | 7 | Bacteroidetes; Sphingobacteria; Sphingobacteriales; Rhodothermaceae | |
| Alcanivorax sp. Nag1 | 6 | Proteobacteria; Gammaproteobacteria; Oceanospirillales; Alcanivoracaceae | |
| Granulosicoccus sp. ZS4-22 | 6 | Proteobacteria; Gammaproteobacteria; Chromatiales; Granulosicoccaceae | |
| Pseudomonas cichorii AMP03 | 6 | Proteobacteria; Gammaproteobacteria; Pseudomonadales; Pseudomonadaceae | |
| Uncultured bacterium JH-WHS137 | 6 | Acidobacteria | |
| ERβ−/− | Uncultured bacterium A6 | 14 | Proteobacteria; Alphaproteobacteria; Sphingomonadales; Erythrobacteraceae |
| Uncultured bacterium S26-35 | 12 | Proteobacteria; Alphaproteobacteria; Rhodospirillales; Rhodospirillaceae | |
| Uncultured bacterium FGL12B72 | 11 | Unclassified | |
| Uncultured Acidobacterium sp. 16L4 | 10 | Acidobacteria | |
| Uncultured bacterium Kas162B | 9 | Bacteroidetes | |
| Uncultured Caldilinea sp. B01-03F | 9 | Chloroflexi; Caldilineae; Caldilineales; Caldilineaceae | |
| Uncultured deltaproteobacterium Bac48Flocs | 9 | Proteobacteria | |
| Chlorobium limicola UdG 6045 | 6 | Chlorobi; Chlorobia; Chlorobiales; Chlorobiaceae | |
| Clostridium cellulovorans DSM 3052 | 6 | Firmicutes; Clostridia; Clostridiales | |
| Halomonas sp. P40 | 6 | Proteobacteria; Gammaproteobacteria; Oceanospirillales; Halomonadaceae | |
| Uncultured bacterium B8S-114 | 6 | Proteobacteria; Alphaproteobacteria; Rhodospirillales; Rhodospirillaceae | |
| Uncultured bacterium CCM6b | 6 | Chloroflexi | |
| Uncultured bacterium E12 | 6 | Verrucomicrobia |
Number of samples in which the species pattern was found among the 21 samples examined.
Validation of quantitative differences in select microbial populations.
At the end of the study (day 14), fecal samples from AIN-76-fed ERβ+/+ and ERβ−/− mice were examined for quantitative differences in select bacterial groups. Decimally diluted purified genomic DNAs from Escherichia coli MC1061, Bacteroides thetaiotaomicron E50, and Lactobacillus acidophilus NCFM were used as the templates to generate standard curves to quantify bacteria and Gammaproteobacteria, Bacteroidetes, and the lactobacilli, respectively. The coefficient for determination (r2) for the resultant standard curves indicated strong linearity (r2 > 0.977). The mean numbers of fecal bacteria from ERβ+/+ mice (8.67 ± 0.58 log10 16S rRNA gene copies g−1) and ERβ−/− mice (8.61 ± 0.77 log10 16S rRNA gene copies g−1) were similar (Mann-Whitney, p = 0.8). In contrast, however, the mean numbers of Bacteroidetes from ERβ+/+ mice (9.25 ± 1.61 log10 16S rRNA gene copies g−1) and ERβ−/− mice (8.16 ± 0.33 log10 16S rRNA gene copies g−1) were significantly different (Mann-Whitney, p = 0.004). Similarly, the mean numbers of Gammaproteobacteria from ERβ+/+ mice (5.92 ± 0.68 log10 16S rRNA gene copies g−1) and ERβ−/− mice (6.75 ± 1.10 log10 16S rRNA gene copies g−1) were different (Mann-Whitney, p = 0.001).
Mean numbers of fecal lactobacilli from ERβ+/+ mice (5.66 ± 1.29 log10 16S rRNA gene copies g−1) and ERβ−/− mice (5.73 ± 0.79 log10 16S rRNA gene copies g−1) were not significantly different (Mann-Whitney, p = 0.4). Melt curve analysis of the L. acidophilus NCFM-derived 16S rRNA gene amplicon showed a single melting temperature (Tm) maximum (85°C); however, two distinct Tm maxima (85°C and 86.5°C) were detected among the experimental fecal samples (data not shown). While most fecal samples generated PCR products with both maxima, there was no obvious relationship between the distinct Tm maxima and ERβ status or diet complexity.
DISCUSSION
In this pilot preclinical study, we used a diet-based murine model to test the hypothesis that ERβ status affects the composition of the autochthonous gastrointestinal microbiota of female mice and that microbiota enriched from differential ERβ expression will respond differently to changes in diet complexity. The following major conclusions were derived as a result of this study. First, the consumption of a biochemically complex diet rich in isoflavones and fiber resulted in a nondysbiotic and likely mutualistic (i.e., eubiotic) microbiota. Relatively low levels of Proteobacteria and a relatively high abundance of Bacteroidetes defined these eubiotic microbiota. Second, microbiota characterized by relatively abundant Proteobacteria and low levels of Bacteroidetes occurred in response to the consumption of a compositionally simple, sucrose-based diet that was low in fiber and devoid of isoflavones. Third, we showed that ERβ status affects the diet-directed community structure of the gastrointestinal microbiota. Lastly, we identified taxonomic biomarkers that were differentially enriched as a function of ERβ status and diet complexity. The following experimental evidences support these conclusions.
At the start of the study, regardless of host genotype, the dominant phylotype isolated from the feces of 8604-fed eubiotic mice was Firmicutes (ERβ+/+ = 38.0% and ERβ−/− = 40.9%). Low levels of Bacteroidetes and Proteobacteria were also found; however, the relative abundance of Bacteroidetes (ERβ+/+ = 28.7%, ERβ−/− = 20.4%) and Proteobacteria (ERβ+/+ = 13.8%, ERβ−/− = 21.0%) differed as a function of ERβ status. The relative abundance of fecal Bacteroidetes, Firmicutes, and Proteobacteria from 8604-fed mice agreed with findings from similar studies performed previously, as expected. Indeed, Proteobacteria usually comprise a very small proportion of the eubiotic microbiota in healthy mice (1 to 15%), whereas the Firmicutes (30 to 70%) and Bacteroidetes (10 to 40%) are typically more abundant by comparison (11, 44). These findings are important for two reasons. First, they suggest that the methodology used in this study (TRFLP) was robust, since it produced values that approximated those that were previously determined by pyrosequencing (11, 44, 45). Second, they suggest that the microbiota of 8604-fed mice were likely eubiotic, as their microbiota composition was similar to other healthy mice that consumed complex rodent chows (45, 46). It is important to note, however, that the proportion of Proteobacteria in 8604-fed ERβ−/− mice (21.0%) was slightly higher than both their ERβ+/+ littermates (13.8%) and the upper limit (15%) that is typically reported in eubiotic microbiota derived from other healthy mice (44).
In 8604-fed female ERβ+/+ mice, all of the highly discriminant bacterial patterns were tentatively assigned to a single family within the Firmicutes: Lachnospiraceae (Clostridia). Interestingly, previous studies have shown that many Lachnospiraceae are capable of producing butyrate from the hydrolysis of dietary fiber (47). As a result, the Lachnospiraceae are largely believed to be health-promoting species that are important to help maintain healthy colonic tissues (48). In contrast, the highly discriminant bacterial patterns from 8604-fed ERβ−/− mice were more diverse than those for their ERβ+/+ littermates. Distinct Lachnospiraceae patterns were seen in both host backgrounds, however. In addition, distinct Lactobacillaceae and Leuconostocaceae from the order Lactobacillales were highly abundant in 8604-fed ERβ−/− mice. This finding might be important, since many Lactobacillales, especially members of the well-studied genus Lactobacillus, have been shown to exhibit a wide range of health-promoting functionalities in vivo (4, 12, 49).
TRFLP analysis revealed that the fecal communities from mice that had habitually consumed a complex diet that was rich in isoflavones and fiber (8604) for approximately 27 weeks were readily distinguishable from those that had consumed a biochemically simple diet (AIN-76) that was rich in simple sugars for a 2-week period. While the relative abundance of the three major phyla (i.e., Firmicutes, Bacteroidetes, and Proteobacteria) differed between 8604-fed ERβ+/+ and ERβ−/− mice, differences in the phyla of AIN-76-fed mice were not significant as a function of ERβ status. The numbers of fecal bacteria in ERβ+/+ and ERβ−/− mice were also similar, as expected. Together, these results suggest that the transition from the 8604 complex diet to the AIN-76 semisynthetic diet by ERβ+/+ and ERβ−/− mice correlated with a convergence of their initially distinct and likely eubiotic microbiota. This convergence serves as another reminder that diet is a powerful tool that can be used to manage the composition of the intestinal microbiota, including constituent members that arise in response to differences in host genotype and, likely, host gene expression.
Compared to the eubiotic microbiota profiles seen in 8604-fed mice, those of AIN-76-fed mice were clearly distinct. The Firmicutes levels decreased marginally in AIN-76-fed mice regardless of their ERβ status, while the relative abundance of Bacteroidetes also decreased by approximately half. More interestingly, the Proteobacteria became more dominant members of the microbiota, although this relative increase in Proteobacteria was significantly greater for ERβ+/+ mice than for their ERβ−/− littermates. A closer examination of the expanded Proteobacteria revealed that their community structure also reorganized following the consumption of the AIN-76 diet. Gammaproteobacteria and Alphaproteobacteria levels increased in AIN-76-fed mice, regardless of the genotype. The Betaproteobacteria levels decreased in AIN-76-fed mice; however, this decrease was more pronounced in ERβ−/− mice than in ERβ+/+ mice. Based on these observations, we conclude that the consumption of fiber-poor, biochemically simple diets might promote intestinal dysbiosis by the gradual and preferential enrichment of Proteobacteria at the expense of the Bacteroidetes.
Since Lactobacillales bacterial patterns were initially abundant in ERβ−/− mice but not ERβ+/+ mice, we used qPCR and group-specific primers (i.e., Lac1 and Lac2) to quantitatively assess the response of the Lactobacillus group to the AIN-76 diet. At the end of the study, however, the Lactobacillus group taxa were not significantly different between ERβ+/+ and ERβ−/− mice. Interestingly, melt curve data analysis revealed that the amplicons were heterogeneous and defined by two distinct Tm maxima, although neither of these maxima were ERβ status or diet dependent. Since the qPCR primer Lac2 has a 2-fold degeneracy at its fourth nucleotide position, the commercial primer suspension is a mix of two primers in equal molar abundance, which we call Lac2_4C (5′-ATTCCACCGCTACACATG-3′) and Lac2_4T (5′-ATTTCACCGCTACACATG-3′). TRFLP illustrated that the fecal samples contained a heterogeneous mixture of bacteria that included distinct subpopulations within the Lactobacillus group. As a result, amplicons with different G+C content and distinct Tm maxima may have resulted from the differential enrichment by Lac2_4C or Lac2_4T from the distinct subpopulations within the phylogenetic supergroup that the Lac2 primer was originally designed to target, which includes species of Lactobacillus, Pediococcus, Leuconostoc, and Weissella (21). Alternatively, given the small difference in the observed Tm maxima (85°C versus 86.5°C), these primers might have promiscuously bound to and amplified 16S rRNA gene sequences from other, more GC-rich species. Additional research is required to explore this possibility, however.
ERβ+/+ mice showed a lower diversity while on the complex, isoflavone-containing 8604 diet than on the AIN-76 diet. This finding is consistent with the negative relationship that has been shown to exist between species diversity and the presence of estrogenic compounds in wastewater (50). However, the TRFLP analysis used in this study provides a superficial examination of the microbiota. As a result, the diversity shown here may represent only the most highly abundant species present in the murine fecal samples we examined. As a result, additional experimentation based on pyrosequencing is required in order to provide a better understanding of the overall diversity and to obtain a higher level of confidence in the role of specific bacterial taxonomic markers.
Both of the commercial diets used in this study were formulated by their manufacturers to provide similar levels of energy from carbohydrates, proteins, and lipids and meet the minimum macronutrient and micronutrient requirements for rodents; however, the effects of diet on the structure and function of the murine intestinal microbiota was likely not considered during diet formulation. Nevertheless, the compositions of the two diets and, perhaps more importantly, their complexities differed substantially (Table 1; see also Table S1 in the supplemental material). We speculate that the compositional simplicity of the semisynthetic diet, and not just its composition or the overall level of ingredient refinement per se, might have contributed to the enrichment of Proteobacteria (Fig. 3). If this supposition is supported, then biochemically simple diets may not be ideal for the long-term care of rodents, as they might result in intestinal dysbiosis that might affect the health, husbandry, and genetic integrity of the line. Furthermore, if these findings were translatable to humans, then overconsumption of biochemically simple foods might also act to gradually enrich for Proteobacteria, establish intestinal dysbiosis, alter the luminal environment, and ultimately favor the development of dysbiosis-related pathologies.
Most of the probiotic and prebiotic research that has been performed to date has been conducted using complex diets and domesticated rodent strains. Since domesticated rodents and their microbiota are now well adapted to these diets, these results might help to explain, in part, some of the variability and sometimes poor efficacy that has been observed in many probiotic and prebiotic studies (51, 52). Differences in diet variety and complexity might also explain why fecal Proteobacteria are typically more abundant in chow-fed domesticated mice (15%) than in humans (3.7%), since humans consume a variety of whole and processed foods (44). Furthermore, since long-term consumption of AIN-76A, a formulary derivative of AIN-76, has been shown to accelerate the development of heart failure in rats with spontaneously hypersensitive heart failure (53), it is intriguing to consider the possibility that the functional properties of the microbiota might have contributed to these negative study events.
In conclusion, we show that ERβ status affects the composition of the intestinal microbiota in female mice and that these microbiota respond differently to changes in diet complexity. These findings may prove to be important since the expression of ERβ and serum concentrations of steroidal hormones, especially estradiol, is known to change throughout the life cycle. Furthermore, we are currently using this diet-induced model for gastrointestinal dysbiosis to both study the role of host genotype in dysbiosis development and to identify novel probiotics, prebiotics, nutritional strategies, and pharmaceuticals for dysbiosis prevention and resolution.
Supplementary Material
ACKNOWLEDGMENTS
This study was funded by NIH/NCI Biostatistics Training Program grant CA090301-11 (J.M.S. and C.D.A.), NIH grant P30 DK34987 (Microbiome Core Support), American Institute for Cancer Research grant 07B080 (C.D.A.), and Texas A&M AgriLife Research (J.M.S.).
The sponsors had no role in the study design, the data collection or analysis, the production of the submitted manuscript, or the decision to submit the manuscript for publication.
We thank Autumn Billimek, Kimberly Allred, and the University of North Carolina Microbiome Core for technical assistance. In addition, we thank the anonymous reviewers for their valuable comments.
Footnotes
Published ahead of print 19 July 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01182-13.
REFERENCES
- 1.Bochner BR. 2011. Metagenomics and complementary approaches, p 533–540 In de BruijnFJ. (ed), Handbook of molecular microbiology and ecology. John Wiley & Sons, Hoboken, NJ [Google Scholar]
- 2.Sturino J, Zorych I, Mallick B, Pokusaeva K, Chang YY, Carroll RJ, Bliznuyk N. 2010. Statistical methods for comparative phenomics using high-throughput phenotype microarrays. Int. J. Biostat. 6:1–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Setchell K. 1998. Phytoestrogens: the biochemistry, physiology, and implications for human health of soy isoflavones. Am. J. Clin. Nutr. 68:1333S–1346S [DOI] [PubMed] [Google Scholar]
- 4.Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. 2005. Host-bacterial mutualism in the human intestine. Science 307:1915–1920 [DOI] [PubMed] [Google Scholar]
- 5.Weige CC, Allred KF, Allred CD. 2009. Estradiol alters cell growth in nonmalignant colonocytes and reduces the formation of preneoplastic lesions in the colon. Cancer Res. 69:9118–9124 [DOI] [PubMed] [Google Scholar]
- 6.Arai N, Strom A, Rafter JR, Gustafsson JA. 2000. Estrogen receptor β mRNA in colon cancer cells: growth effects of estrogen and genistein. Biochem. Biophys. Res. Commun. 270:425–431 [DOI] [PubMed] [Google Scholar]
- 7.Atkinson C, Frankenfeld CL, Lampe JW. 2005. Gut bacterial metabolism of the soy isoflavone daidzein: exploring the relevance to human health. Exp. Biol. Med. 230:155–170 [DOI] [PubMed] [Google Scholar]
- 8.Yuan JP, Wang JH, Liu X. 2007. Metabolism of dietary soy isoflavones to equol by human intestinal microflora—implications for health. Mol. Nutr. Food Res. 51:765–781 [DOI] [PubMed] [Google Scholar]
- 9.Looijer-van Langen M, Hotte N, Dieleman LA, Albert E, Mulder C, Madsen KL. 2011. Estrogen receptor signaling modulates epithelial barrier function. Am. J. Physiol. Gastroenterol. Liver Physiol. 300:G621–G626 [DOI] [PubMed] [Google Scholar]
- 10.Wada-Hiraike O, Imamov O, Hiraike H, Hultenby K, Schwend T, Omoto Y, Warner M, Gustafsson J-Å. 2006. Role of estrogen receptor β in colonic epithelium. Proc. Natl. Acad. Sci. U. S. A. 103:2959–2964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benson AK, Kelly SA, Legge R, Ma F, Low SJ, Kim J, Zhang M, Oh PL, Nehrenberg D, Hua K, Kachman SD, Moriyama EN, Walter J, Peterson DA, Pomp D. 2010. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc. Natl. Acad. Sci. U. S. A. 107:18933–18938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walter J, Ley R. 2011. The human gut microbiome: ecology and recent evolutionary changes. Annu. Rev. Microbiol. 65:411–429 [DOI] [PubMed] [Google Scholar]
- 13.Guo X, Li H, Xu H, Halim V, Thomas LN, Woo SL, Huo Y, Chen YE, Sturino JM, Wu C. 2013. Disruption of inducible 6-phosphofructo-2-kinase impairs the suppressive effect of PPARγ activation on diet-induced intestine inflammatory response. J. Nutr. Biochem. 24:770–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hooper LV, Littman DR, Macpherson AJ. 2012. Interactions between the microbiota and the immune system. Science 336:1268–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sartor RB. 2008. Microbial influences in inflammatory bowel diseases. Gastroenterology 134:577–594 [DOI] [PubMed] [Google Scholar]
- 16.Carvalho FA, Koren O, Goodrich JK, Johansson MEV, Nalbantoglu I, Aitken JD, Su Y, Chassaing B, Walters WA, González A. 2012. Transient inability to manage proteobacteria promotes chronic gut inflammation in TLR5-deficient mice. Cell Host Microbe 12:139–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krege JH, Hodgin JB, Couse JF, Enmark E, Warner M, Mahler JF, Sar M, Korach KS, Gustafsson JÅ, Smithies O. 1998. Generation and reproductive phenotypes of mice lacking estrogen receptor β. Proc. Natl. Acad. Sci. U. S. A. 95:15677–15682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson MC, Devine AA, Ellis JC, Grunden AM, Fellner V. 2009. Effects of antibiotics and oil on microbial profiles and fermentation in mixed cultures of ruminal microorganisms. J. Dairy Sci. 92:4467–4480 [DOI] [PubMed] [Google Scholar]
- 19.Engelbrektson AL, Korzenik JR, Sanders ME, Clement BG, Leyer G, Klaenhammer TR, Kitts CL. 2006. Analysis of treatment effects on the microbial ecology of the human intestine. FEMS Microbiol. Ecol. 57:239–250 [DOI] [PubMed] [Google Scholar]
- 20.Huynh TV, Young RA, Davis RW. 1985. Constructing and screening cDNA libraries in λgt10 and λgt11, p 49–78 In Glover DM. (ed), DNA cloning: a practical approach, vol 1 IRL Press, Oxford, United Kingdom [Google Scholar]
- 21.Walter J, Hertel C, Tannock GW, Lis CM, Munro K, Hammes WP. 2001. Detection of Lactobacillus, Pediococcus, Leuconostoc, and Weissella species in human feces by using group-specific PCR primers and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 67:2578–2585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. 2007. Development of the human infant intestinal microbiota. PLoS Biol. 5:e177. 10.1371/journal.pbio.0050177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frank DN, Amand ALS, Feldman RA, Boedeker EC, Harpaz N, Pace NR. 2007. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. U. S. A. 104:13780–13785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thomas MC, Thomas DK, Selinger LB, Inglis GD. 2011. SPYDER, a new method for in silico design and assessment of 16S rRNA gene primers for molecular microbial ecology. FEMS Microbiol. Let. 320:152–159 [DOI] [PubMed] [Google Scholar]
- 25.Mühling M, Woolven-Allen J, Murrell JC, Joint I. 2008. Improved group-specific PCR primers for denaturing gradient gel electrophoresis analysis of the genetic diversity of complex microbial communities. ISME J. 2:379–392 [DOI] [PubMed] [Google Scholar]
- 26.Li F, Hullar MA, Lampe JW. 2007. Optimization of terminal restriction fragment polymorphism (TRFLP) analysis of human gut microbiota. J. Microbiol. Methods 68:303–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kent AD, Smith DJ, Benson BJ, Triplett EW. 2003. Web-based phylogenetic assignment tool for analysis of terminal restriction fragment length polymorphism profiles of microbial communities. Appl. Environ. Microbiol. 69:6768–6776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cole J, Wang Q, Cardenas E, Fish J, Chai B, Farris R, Kulam-Syed-Mohideen A, McGarrell D, Marsh T, Garrity G. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37:D141–D145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261–5267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lukow T, Dunfield PF, Liesack W. 2000. Use of the T RFLP technique to assess spatial and temporal changes in the bacterial community structure within an agricultural soil planted with transgenic and non-transgenic potato plants. FEMS Microbiol. Ecol. 32:241–247 [DOI] [PubMed] [Google Scholar]
- 31.Hammer Ø, Harper D, Ryan P. 2009. PAST: paleontological statistics software package for education and data analysis. Palaeontol. Electronica 4:1–9 [Google Scholar]
- 32.Shannon CE, Weaver W. 1949. The mathematical theory of communication. University of Illinois Press, Urbana, IL [Google Scholar]
- 33.Magurran AE. 1988. Ecological diversity and its measurement. Princeton University Press, Princeton, NJ [Google Scholar]
- 34.Kemp PF, Aller JY. 2004. Estimating prokaryotic diversity: when are 16S rDNA libraries large enough? Limnol. Oceanogr. Methods 2:114–125 [Google Scholar]
- 35.Chao A. 1987. Estimating the population size for capture-recapture data with unequal catchability. Biometrics 43:783–791 [PubMed] [Google Scholar]
- 36.Poole RW. 1974. An introduction to quantitative ecology. McGraw-Hill, New York, NY [Google Scholar]
- 37.Blackwood CB, Marsh T, Kim SH, Paul EA. 2003. Terminal restriction fragment length polymorphism data analysis for quantitative comparison of microbial communities. Appl. Environ. Microbiol. 69:926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clarke K. 1993. Non-parametric multivariate analyses of changes in community structure. Aust. J. Ecol. 18:117–143 [Google Scholar]
- 39.Clarke K, Somerfield P, Chapman M. 2006. On resemblance measures for ecological studies, including taxonomic dissimilarities and a zero-adjusted Bray-Curtis coefficient for denuded assemblages. J. Exp. Mar. Biol. Ecol. 330:55–80 [Google Scholar]
- 40.Rees GN, Baldwin DS, Watson GO, Perryman S, Nielsen DL. 2004. Ordination and significance testing of microbial community composition derived from terminal restriction fragment length polymorphisms: application of multivariate statistics. Antonie Van Leeuwenhoek 86:339–347 [DOI] [PubMed] [Google Scholar]
- 41.Diggle P, Heagerty P, Liang KY, Zeger S. 2002. Analysis of longitudinal data, 2nd ed. Oxford University Press, New York, NY [Google Scholar]
- 42.Seber GAF, Lee AJ. 2012. Linear regression analysis, 2nd ed. Wiley, New York, NY [Google Scholar]
- 43.Storey JD, Tibshirani R. 2003. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. U. S. A. 100:9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carroll IM, Threadgill DW, Threadgill DS. 2009. The gastrointestinal microbiome: a malleable, third genome of mammals. Mamm. Genome 20:395–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang C, Zhang M, Pang X, Zhao Y, Wang L, Zhao L. 2012. Structural resilience of the gut microbiota in adult mice under high-fat dietary perturbations. ISME J. 6:1848–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang C, Zhang M, Wang S, Han R, Cao Y, Hua W, Mao Y, Zhang X, Pang X, Wei C. 2009. Interactions between gut microbiota, host genetics and diet relevant to development of metabolic syndromes in mice. ISME J. 4:232–241 [DOI] [PubMed] [Google Scholar]
- 47.Van den Abbeele P, Belzer C, Goossens M, Kleerebezem M, De Vos WM, Thas O, De Weirdt R, Kerckhof FM, Van de Wiele T. 2012. Butyrate-producing Clostridium cluster XIVa species specifically colonize mucins in an in vitro gut model. ISME J. 7:949–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reeves AE, Koenigsknecht MJ, Bergin IL, Young VB. 2012. Suppression of Clostridium difficile in the gastrointestinal tract of germ-free mice inoculated with a murine Lachnospiraceae isolate. Infect. Immun. 80:3786–3794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Altermann E, Russell WM, Azcarate-Peril MA, Barrangou R, Buck BL, McAuliffe O, Souther N, Dobson A, Duong T, Callanan M. 2005. Complete genome sequence of the probiotic lactic acid bacterium Lactobacillus acidophilus NCFM. Proc. Natl. Acad. Sci. U. S. A. 102:3906–3912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu CP, Ahuja R, Sayler G, Chu KH. 2005. Quantitative molecular assay for fingerprinting microbial communities of wastewater and estrogen-degrading consortia. Appl. Environ. Microbiol. 71:1433–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shanahan F. 2003. Probiotics: a perspective on problems and pitfalls. Scand. J. Gastroenterol. 38:34–36 [DOI] [PubMed] [Google Scholar]
- 52.Klaenhammer TR, Kleerebezem M, Kopp MV, Rescigno M. 2012. The impact of probiotics and prebiotics on the immune system. Nat. Rev. Immunol. 12:728–734 [DOI] [PubMed] [Google Scholar]
- 53.Rees M, Gioscia-Ryan R, McCune S, Browder J, Zachman D, Chicco A, Johnson C, Murphy R, Moore R, Sparagna G. 9 January 2013. The AIN-76A defined rodent diet accelerates the development of heart failure in SHHF rats: a cautionary note on its use in cardiac studies. J. Anim. Physiol. Anim. Nutr. 10.1111/jpn.12031 [Epub ahead of print.] [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.