Abstract
The total cellular lipids of Porphyromas gingivalis, a known periodontal pathogen, were previously shown to promote dendritic cell activation and inhibition of osteoblasts through engagement of Toll-like receptor 2 (TLR2). The purpose of the present investigation was to fractionate all lipids of P. gingivalis and define which lipid classes account for the TLR2 engagement, based on both in vitro human cell assays and in vivo studies in mice. Specific serine-containing lipids of P. gingivalis, called lipid 654 and lipid 430, were identified in specific high-performance liquid chromatography fractions as the TLR2-activating lipids. The structures of these lipids were defined using tandem mass spectrometry and nuclear magnetic resonance methods. In vitro, both lipid 654 and lipid 430 activated TLR2-expressing HEK cells, and this activation was inhibited by anti-TLR2 antibody. In contrast, TLR4-expressing HEK cells failed to be activated by either lipid 654 or lipid 430. Wild-type (WT) or TLR2-deficient (TLR2−/−) mice were injected with either lipid 654 or lipid 430, and the effects on serum levels of the chemokine CCL2 were measured 4 h later. Administration of either lipid 654 or lipid 430 to WT mice resulted in a significant increase in serum CCL2 levels; in contrast, the administration of lipid 654 or lipid 430 to TLR2−/− mice resulted in no increase in serum CCL2. These results thus identify a new class of TLR2 ligands that are produced by P. gingivalis that likely play a significant role in mediating inflammatory responses both at periodontal sites and, potentially, in other tissues where these lipids might accumulate.
INTRODUCTION
Toll-like receptors (TLRs) represent a diverse family of molecules that play a critical role in activating the innate immune system in response to pathogens (1, 2). Toll-like receptor 2 (TLR2) recognizes diverse molecular structures of microbial cell wall origin, including lipoteichoic acid, lipoproteins, peptidoglycan from Gram-positive bacteria, lipoarabinomannan from mycobacteria, and zymosan from yeast cell walls. TLR2 is reported to be activated by many other microbial products, including phenol-soluble modulins (3) and Porphyromonas gingivalis lipoprotein (4), lipopolysaccharide (LPS) (5–7), and fimbriae (8–10). However, two recent reports have questioned the extent to which lipoprotein, LPS, or fimbriae mediate TLR2 engagement by P. gingivalis (11, 12).
We previously reported that the total lipid extract of P. gingivalis promotes activation of mouse dendritic cells and inhibits osteoblast-mediated bone deposition through engagement of TLR2 (13, 14). These effects were attributed to the dominant phosphorylated dihydroceramide lipids of P. gingivalis, in particular, phosphoethanolamine dihydroceramides. These studies reported engagement of TLR2 only in vitro in mouse cells. Recent reports have demonstrated TLR2-dependent periodontal bone loss in mice following oral infection with P. gingivalis (15, 16). Most recently, cell adhesion mediated through the expression of fimbriae by P. gingivalis has been implicated in promoting of TLR2-dependent oral bone loss (17). In contrast, two recent reports indicated that the capacity of fimbriae to engage TLR2 is dependent on the presence of a contaminating factor that is susceptible to hydrolysis by lipoprotein lipase (11, 18).
In addition to effects on mouse cells, the phosphorylated dihydroceramide lipids of P. gingivalis have been shown to promote proinflammatory responses in human fibroblasts and to cause disruption of human fibroblast adherence/vitality in culture (19). However, it is not clear whether these effects require engagement of TLR2. Since the total lipid extract of P. gingivalis has been shown to activate TLR2 in mice and in mouse cells, the primary purpose of this investigation was to further identify and characterize the specific lipid classes of P. gingivalis that are responsible for engagement of TLR2 and, specifically, to determine whether similar effects are observed in cells expressing human TLR2.
MATERIALS AND METHODS
Reagents.
BBL Biosate peptone, Trypticase peptone, yeast extract, and brain heart infusion (BHI) broth were obtained from Fisher Scientific. Neutralizing human and mouse anti-TLR2 antibodies, anti-TLR6 antibodies, and anti-TLR1 antibodies were obtained from InvivoGen, San Diego, CA. CCL2 enzyme-linked immunosorbent assay (ELISA) kits were obtained from R&D Systems, Minneapolis, MN. Lipoteichoic acid was obtained from InvivoGen, San Diego, CA. MMP is a synthetic bacterial lipoprotein and TLR2 ligand [palmitoyl-Cys (R,S)-2,3-di(palmitoyloxy)-propyl)-Ser-Ser-Asn-Ala-OH(pam3-Cys-Ser-Ser Asn-Ala-OH; Bachem H-9460] (20). Deuterated solvents (CCl3D, D3COD, and D3COH) and [1-13C]sodium acetate were obtained from Cambridge Isotope Laboratories, Andover, MA. Nuclear magnetic resonance (NMR) tubes were obtained from Norrell, Landiville, NJ. Gas chromatography-mass spectrometry (GC-MS) derivatizing agents were obtained from Pierce, Rockford, IL.
Bacterial growth.
Bacteria were grown in broth culture as previously described. P. gingivalis (ATCC 33277, type strain) was inoculated into basal medium (peptone, Trypticase, and yeast extract) supplemented with hemin and menadione (Sigma, St. Louis, MO) and brain heart infusion (BHI) broth (19). Culture purity was verified by lack of growth in aerobic culture and formation of uniform colonies when inoculated on brain heart infusion agar plates and grown under anaerobic conditions. The suspension cultures were incubated for 4 days in an anaerobic chamber flushed with N2 (80%), CO2 (10%), and H2 (10%) at 37°C, and the bacteria were harvested by centrifugation (3,000 × g for 20 min).
Lipid extraction, fractionation, and characterization.
Lipids were extracted from lyophilized bacterial pellets. Generally, 2 to 4 g of bacterial pellet was extracted for each semipreparative fractionation. The bacterial samples were weighed and dissolved in chloroform-methanol-water (1.33:2.67:1 [vol/vol/vol]; 2 g of bacterial pellet in a total of 16 ml of solvent). The mixture was vortexed at 15-min intervals for 2 h, and the mixture was supplemented with 6 ml of chloroform and 6 ml of a combination of 2 N KCl and 0.5 N K2HPO4. The mixture was vortexed and centrifuged at 20°C for 45 min. The lower organic phase was removed and dried under nitrogen. The dried extract was reconstituted in high-performace liquid chromatography (HPLC) solvent (hexane-isopropanol-water [6:8:0.75, vol/vol/vol; 18-ml total volume) and vortexed. The sample was centrifuged at 2,500 × g for 10 min, and the supernatant was removed for HPLC analysis. Semipreparative HPLC fractionation was accomplished by using a Shimadzu HPLC system equipped with dual pumps (LC-10ADvp), an automated controller (SCL-10Avp), and an in-line UV detector (SPD-10Avp). Lipids were fractionated by using normal phase separation (AscentisSi; 25 cm by 10 mm by 5 μm; Supelco Analytical) with a solvent flow of 1.8 ml/min and 1-min fractions. The effluent was monitored at 205 nm. Replicate fractionations were pooled and dried under nitrogen. The dried samples were reconstituted in HPLC solvent for MS analysis as described below. Based on the MS profiles, selected fractions were weighed and aliquoted for biological testing as described below.
HPLC fractionations also included analytical normal-phase HPLC using an AscentisSi column (25 cm by 4.6 mm by 5 mm; Supelco Analytical), as described below in Results. This column was used with a flow of 0.5 ml/min, and effluent was monitored as described above. Lipid samples to be analyzed by NMR were first repurified by this method before dissolving them in deuterated NMR solvent.
Mass spectrometry.
HPLC fractions derived either from semipreparative purification or analytical column enrichment were infused at a low flow rate (0.1 ml/min) into an ABSciex 4000 Qtrap instrument. Lipid samples were dissolved in the HPLC solvent described above. For mass spectrometry analyses, a short normal-phase column (AscentisSi; 3 cm by 2.1 mm by 5 μm; Supelco Analytical) was used for separation of the injected lipids fractions. HPLC solvent was delivered under isocratic conditions with a Shimadzu LC-10ADvp pump. Total ion chromatograms were acquired using a mass range of 100 to 1,800 atomic mass units (amu), and tandem MS (MS/MS) acquisitions used parameters optimized for the specific lipid products under analysis. Collision energies for negative ion products were typically between −30 and −55 V, depending on the precursor ion under investigation.
Fatty acid analysis of P. gingivalis lipids included transesterification or base-catalyzed hydrolysis, using either sodium methoxide (0.5 ml of 0.5 N in dry methanol; 40°C for 20 min) or potassium hydroxide (0.5 ml of 4 N, 100°C for 2 h), respectively. Fatty acid methyl esters were recovered by extraction into hexane (three times; 1 ml) after the addition of 1.0 ml of water to the sodium methoxide hydrolysis solution. The hexane extracts were then dried, reconstituted with N,O-bis(trimethylsilyl)trifluoroacetamide, and allowed to stand overnight before analysis. The potassium hydroxide hydrolysis reaction was stopped with the addition of 0.15 ml of concentrated HCl and 1 ml of water. The hydrolysate was extracted in triplicate with chloroform, and the combined extracts were dried under nitrogen. The dry extract was then treated to form pentafluorobenzyl ester, trimethylsilyl (TMS) ether derivatives as previously described (21).
Serine was hydrolyzed from the target lipids by adding 0.1 ml of 6 N HCl and heating the sample for 4 min in a microwave oven (22). The residue was dried and prepared to form methyl ester-pentafluoropropyl ether/amide derivatives for analysis according to the method of Fuchs et al. (23). The dried samples were first treated with acetyl chloride-methanol (1:4 [vol/vol[; 100 μl; 70°C for 45 min) and dried. The samples were then treated with chloroform-pentofluoropropionic anhydride (4:1 [vol/vol]; 500 μl; 100°C for 20 min) and dried. The residues were dissolved in chloroform and analyzed by GC-MS.
Fatty acid and serine analyses.
Experiments were performed on an Agilent 5975C GC-MS apparatus. Fatty acid samples were run on an Agilent HP-5 M column with a helium flow rate of 1 ml/min for both positive and negative ionization conditions. The column was generally heated from 100 to 290°C, and the injection block and transfer line were maintained at 280° and 290°C, respectively. Methyl ester-TMS products were run in the electron impact mode, and pentafluorobenzyl-TMS derivatives were run in the negative chemical ionization mode. Fatty acid quantification was accomplished by electronic integration of selected ion chromatograms. Serine quantification was accomplished using a chiral column (CP-Chirasil-l-Val; 25-m by 0.25-mm by 0.25-μm column; Agilent). The serine derivatives (pentafluoropropyl ether/amide-methyl ester derivatives) of each sample were run from 80°C to 150°C with the injection block and transfer line both held at 150°C. d- and l-serine standards were prepared in parallel to determine the epimeric form of serine recovered in the serine lipids of P. gingivalis.
NMR spectroscopy.
All NMR experiments were performed on Agilent VNMRS spectrometers equipped with cryogenically cooled HCN triple resonance probes at 18.8 T (1H and 13C enhanced) and 11.7 T (1H enhanced). All NMR experiments were performed at the natural abundance of 13C and 15N with a lipid concentration of approximately 1.5 mM at 25°C with a sample volume of 600 μl in a 5-mm sample tube. The lipid sample was dissolved in deuterated solvent (CD3Cl-D3COD, 2:1 [vol/vol]), which gave narrow line widths, and the following experimental data were collected; one-dimensional (1D) 1H, 1D 13C (24), 2D TOCSY (25), 2D DQF-COSY (26), 2D 1H-13C HSQC (27), 2D 1H-13C HMBC (28), and 2D 1H-13C H2BC (29). For evaluation of proton substitution of nitrogen, the lipid sample was dissolved in CD3Cl-D3COH (2:1 [vol/vol]) and analyzed as a 2D 1H-15N HSQC spectrum and two 1D 1H-15N HSQC spectra (1H detected) run in a mode that only observed primary or secondary amines, respectively. The 1H-13C HMBC spectrum was collected as four different experiments, each enhanced for a different 1H-13C multiple-bond coupling (3 Hz, 5 Hz, 8 Hz, and 10 Hz) and added together after processing the individual spectra. The 1D 13C spectrum was collected at 18.8 T by using a spin-echo sequence, which gave perfectly flat baselines, along with chirp pulses to obtain uniform excitation over a 52,000-Hz sweep width (pulse sequence provided by Agilent). All NMR data were processed and analyzed using either MestReNova or NMRPipe software. The structure was reconciled through correlations in the HMBC, H2BC, DQF COSY, and TOCSY spectra along with 1H-1H splittings, 1H integrations, and 13C chemical shifts.
Mice.
Female C57BL/6 (WT) mice were purchased from Jackson Laboratory (Bar Harbor, ME). TLR2−/− mice, bred onto a C57BL/6 background, were a generous gift of S. Akira (Osaka University, Japan) (30). All mice were between 6 and 12 weeks old when used, and all mice were maintained and bred in accordance with University of Connecticut Center for Laboratory Animal Care regulations.
Cell lines and assays.
Human embryonic kidney cells (HEK293 cells), either nontransfected or transfected with human TLR2 or human TLR4 and stably expressing MD-2 and CD14, were purchased from InvivoGen. Cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco) containing 4.5 g/liter l-glucose and 10% fetal bovine serum (FBS). The activities of specific TLR agonists were measured through a colorimetric assay for the secretory embryonic alkaline phosphatase (SEAP), a reporter gene that is linked to NF-κB activation. Measurement of SEAP activity using the Quanti-blue substrate (InvivoGen) after TLR agonist treatment was carried out in test medium (DMEM, 10% FBS) without antibiotics according to the manufacturer's instructions. NF-κB activation was expressed as a response ratio for each stimulus relative to SEAP activity in unstimulated (vehicle control) cells. For in vitro testing, all lipid preparations tested were solubilized in a 50% mixture of dimethyl sulfoxide (DMSO)-water (approximately 1.11% DMSO in the final culture medium).
In vivo assays.
Wild-type (WT) C57BL/6 mice or TLR2−/− mice were injected intraperitoneally (i.p.) or intravenously (i.v.) with vehicle control or specific lipids. Four hours later, blood samples were obtained from mice, and the mice were euthanized in accordance with University of Connecticut Center for Laboratory Animal Care regulations. Serum was separated from the blood samples and frozen at −80°C until analysis. Serum samples were analyzed for levels of CCL2 by ELISA (R&D, Minneapolis, MN).
Assessment of lipid 654 contamination of P. gingivalis LPS.
Crude LPS of P. gingivalis was prepared using the TRI reagent method of Yi and Hackett (31), and the crude LPS was precipitated with cold magnesium chloride in 95% ethanol (31). After three additional precipitations with 95% ethanol followed by precipitation with 100% ethanol (31), the LPS preparation was dried. Aliquots of LPS (20 μg) were dispensed into glass vials to which known amounts of serine lipid internal standard was added. The serine lipid internal standard was prepared by culturing P. gingivalis in BHI broth (described above) supplemented with 1-13C-labeled sodium acetate (0.5 g/liter). Bacteria were harvested by centrifugation, and total lipids were extracted and fractionated as described above. A lipid fraction with the retention time of lipid 654 was shown by electrospray ionization (ESI)-MS to have a peak mass of m/z 660. The background m/z 654 lipid in this internal standard lipid preparation was only 1.5% of the abundance of the m/z 660 species. This lipid fraction was used as an internal standard for quantifying lipid 654 contamination of P. gingivalis LPS.
The LPS:internal standard mixtures (prepared in a 0.5-ml volume in water) were supplemented with 2 ml of chloroform-methanol (1:2 [vol/vol]) and vortexed repeatedly over 1 h. The samples were supplemented with chloroform (0.75 ml) and 0.75 ml of 2 N KCl plus 0.5 N K2HPO4. After vortexing, the lower chloroform phase was removed and dried. These samples were subjected to multiple reaction monitoring (MRM)-MS as previously described (32) with instrument parameters optimized for the m/z 654-to-m/z 381 transition (lipid 654 product) and the m/z 660-to-m/z 385 transition (internal standard). The ratio of the electronically integrated peaks for these two transitions was then used to determine the amount of lipid 654 present in 20 μg of P. gingivalis LPS.
Statistical analysis.
Data are expressed as the means ± standard errors. Statistical testing included an analysis of variance (ANOVA)with pairwise comparisons using the Fisher least significant difference (LSD) test or the Student t test for simple group mean comparisons.
RESULTS
Activation of human TLR2 by P. gingivalis lipids.
To determine the ability of P. gingivalis lipids to activate human cells via TLRs, we fractionated the total lipids of P. gingivalis by HPLC, and an aliquot of each fraction was dried and dissolved in 50% DMSO in water. Each HPLC fraction was then tested for cell activation by using HEK293 cells stably transfected with human TLR2, CD14, MD-2, and SEAP genes. In addition, the HEK293 cells naturally express variable levels of TLRs 1, 3, 5, 6, 7, and 9. The SEAP reporter gene is under the control of the beta interferon minimal promoter fused to five NF-κB and AP-1 binding sites. Stimulation with a TLR2 ligand activates NF-κB and AP-1, which induces the production of SEAP. SEAP is then quantitated via a colorimetric change in medium samples following the addition of a suitable enzyme substrate. Using this cell screen for human TLR2 engagement, each HPLC lipid fraction was screened for cell activation. We observed HEK cell activation, depicted as the response ratio relative to the vehicle control culture, only in HPLC fractions 33 through 40, as shown in Fig. 1A. Using mass spectrometry, we then examined these HPLC fractions for lipid ions that correlated with the observed HEK cell activation. HEK-TLR2 cell activation directly correlated with levels of lipids that produced negative ions of m/z 654, 640, and 626 (here termed lipid 654) or negative ions of m/z 430, 416, and 402 (here termed lipid 430). The ion abundances of the m/z 654 and 430 negative ions are depicted for HPLC fractions 33 through 40 in Fig. 1B and C, and these negative ions represent the most abundant lipid species within lipid 654 and lipid 430 classes. Although these TLR2-activating HPLC fractions contained small amounts of previously characterized phosphatidylethanolamine and phosphoethanolamine dihydroceramide lipids as determined by mass spectrometric analysis (19), the minimal levels of these contaminating lipids did not correlate with TLR2 cell activation (see below).
Fig 1.
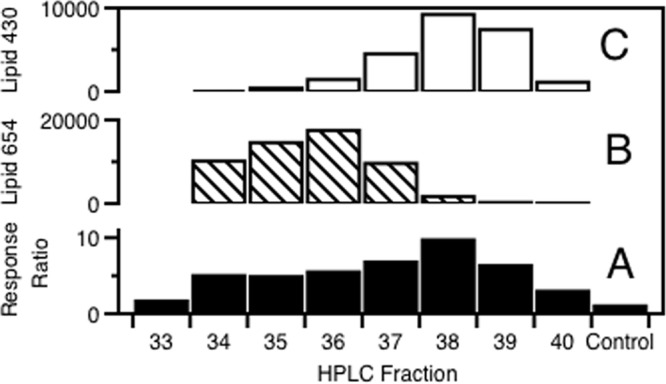
HEK293 activation by lipids recovered in HPLC fractions of P. gingivalis total lipids. HEK293 cells, transfected with the human TLR2, MD-2, CD14, and SEAP genes, were used to assay the functions of P. gingivalis lipid fractions in vitro. A defined volume of each HPLC fraction was dried and reconstituted in 50% DMSO in water. The final concentration of DMSO achieved in culture medium was 1.11%. HEK293 cells were stimulated for 24 h with a defined amount of each lipid fraction. Results are expressed as the stimulated/nonstimulated (DMSO control) response ratio of HEK293 cells (graph A). Graphs B and C show the ion abundances within each HPLC fraction of lipids that produced negative ions of m/z 654 and 430, respectively.
MS and NMR analysis of lipid 654.
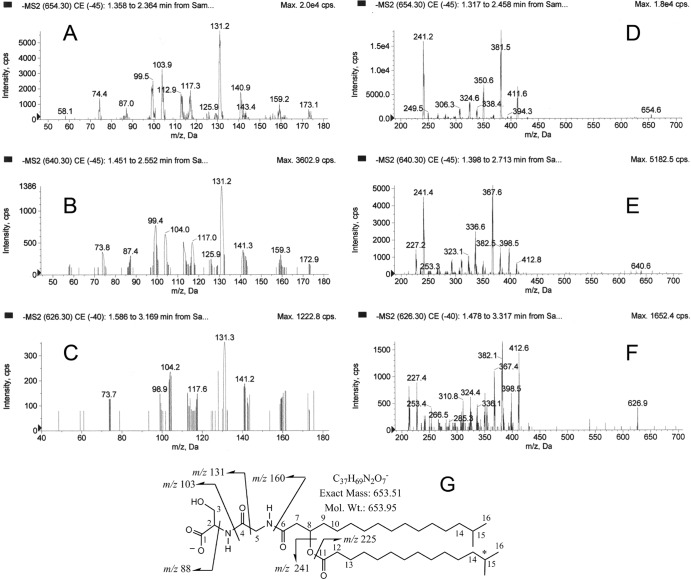
The negative ion MS/MS analysis of the m/z 654, 640, and 626 precursor ions revealed the MS/MS spectra shown in Fig. 2A to F. The proposed lipid structure in Fig. 2G shows the most abundant species of the lipid 654 class. The structure depicts two fatty acids linked by a β-carbon ester, and the hydroxyl fatty acid is held in amide linkage to a dipeptide composed of glycine and a terminal serine. Other fatty acids can be substituted into this lipid class as described below, and these alternate fatty acid substitutions account for the m/z 654, 640, and 626 parent molecules. Lipid extracts from the broth medium used to culture P. gingivalis showed no m/z 654, 640, or 626 ions (data not shown). Reconciliation of the ion fragments for the lipid 654 structure is provided in the legend for Fig. 2. Positive ion mass spectra revealed molecular ion masses of m/z 656, 643, and 628, indicating at least one elemental nitrogen atom in the component molecular species of the lipid 654 class (data not shown).
Fig 2.
MS/MS profiles of 654, 640, and 626 lipid species. A highly enriched preparation of lipid 654 was subjected to MS/MS analysis as described in Materials and Methods. The partial mass spectra are depicted for the m/z 654 lipid species (A and D), the m/z 640 lipid species (B and E), and the m/z 626 lipid species (C and F). The proposed structure of the most abundant species in the lipid 654 class is shown in graph G. The numbers adjacent to specific carbons refer to the NMR assignments shown in Table 1. The major low-mass product ions (<200 amu) detected with the three parent lipid species were essentially identical (A, B, and C), and these product ions represent cleavage products of the serine-glycine head group as depicted in the proposed chemical structure (G). Loss of the -CH2-OH from the m/z 103 fragment and addition of two protons yields an m/z 74 product ion. Loss of a proton from the m/z 88 fragment yields an m/z 87 product ion. Loss of the -CH2-OH from the m/z 131 fragment and loss of a proton yield an m/z 99 product ion. Loss of the -CH2-OH from the m/z 160 fragment and loss of a proton yield an m/z 129 product ion. Cleavage between carbons 7 and 8 and loss of the -CH2-OH group plus addition of two protons yield the m/z 173 ion product. The m/z 160 fragment forms an imine with loss of a proton to yield an m/z 159 product ion. The m/z 412 fragment results from loss of the C15:0 fatty acid (241 amu) from the 654 precursor ion. The m/z 381 fragment results from loss of C15:0 fatty acid and -CH2-OH from the 654 precursor ion. Fragmentation of the serine-glycine dipeptide together with loss of the C15:0 fatty acid results in the other ion fragments (m/z 250 to 420) depicted in graph D. Additional fatty acids described elsewhere in Results account for the 640 and 626 lipid species.
The multiple-bond 1H-13C correlations from the HMBC, 1H-1H correlations from the DQF-COSY and TOCSY experiments, 13C chemical shifts, 1H integrations, and 1H-1H coupling constants were sufficient to map the structure of the lipid for all atoms except the central CH2 groups in the fatty acid aliphatic chains, due to significant overlap in the NMR spectra. Information used in the structure determination is shown in Table 1, with carbon numbers corresponding to those listed in the chemical structure in Fig. 2G. Integration of the large overlapped peak in the 1H 1D NMR spectrum corresponding to 17 CH2 groups yielded a value of 35.9, slightly higher than the expected 34 but well within the expected error and consistent with the length of fatty acid aliphatic chains. Coupling patterns and integrations confirmed that approximately 85% of the fatty acids are isobranched, with 15% being anteisobranched. The 1H-15N HSQC confirmed that there were two protonated nitrogens, and both were shown to be secondary amines. The 1D 13C NMR spectrum confirmed the presence of four carbonyl carbons, although the signal for carbonyl 1 was weak due to a longer T1 relaxation time (a 3-s recycle delay was used). The four carbonyls were also observed by long-range couplings in the 1H-13C HMBC. The three methylene groups at atoms C-3, C-5, and C-7 gave unique chemical shifts for the two protons, demonstrating a lack of bond rotation. From these proton and carbon assignments, listed in Table 1, together with the mass spectrometric results, we confirmed that this lipid class represents the previously reported lipid called flavolipin (33–36). However, as discussed below, lipid 654 is clearly distinct from flavolipin, both in its biological activity and in the range of bacteria from which it can be derived.
Table 1.
NMR analysis of the lipid 654 classa
| Carbon no. or amino acid | Group | Integration | 1H δ (ppm) | 13C/15N δ (ppm) | 1H-1H J (Hz) | DQF-COSY correlation(s) | TOCSY correlations | HMBC correlation(s) |
|---|---|---|---|---|---|---|---|---|
| 1 | C=O | 174.877 | 2, Ser Ni | |||||
| 2 | CH | 0.94 | 4.465 | 57.571 | 3.76 (3, 3′) | 3, 3′, 17 | 3, 3′, 17 | 1, 3, 4 |
| 3 | CH2b | 2.16c | 3.873, 3.794 | 64.582 | 11.58 (gemj), [3,47, 4.19] | 2, 3, 3′ | 2, 3, 3′, 17 | 2i |
| 4 | C=O | 172.033 | 2, Ser N, 5 | |||||
| 5 | CH2b | 1.77c | 3.843, 3.801 | 45.209 | 16.76 (gemj) | 5, 5′ | 5, 5′ | 4, 6 |
| 6 | C=O | 174.142 | 5, Gly N, 7, 8 | |||||
| 7 | CH2b | 1.97 | 2.460, 2.412 | 43.506 | 14.53 (gemj), [7.58, 5.07] | 8 | 8, 9, 10, FA | 6, 8, 9 |
| 8 | CH | 1.0 | 5.122 | 73.82 | —g | 7, 7′, 9 | 7, 7′, 9, 10, FA | 6, 7, 9, 10 |
| 9 | CH2 | 4.28d, e | 1.533 | 36.711 | —g | 8, 10 | 7, 7′, 8, 10, FA | 7, 8, 10, FA |
| 10 | CH2 | —e | 1.225 | 27.668 | —g | 9, FAh | 7, 7′, 8, 9, FA | 9, FA |
| 11 | C=O | 176.732 | 8, 12, 13 | |||||
| 12 | CH2 | 2.00 | 2.222 | 36.984 | 7.5, 2.97, 2.66 | 13 | 13, FA | 11, 13, FA |
| 13 | CH2 | 4.28d, e | 1.509 | 27.508 | —g | 12, FA | 12, FA | 11, 12, FA |
| 14 | (CH2)2 | 3.91 | 1.051 | 41.545 | —g | FA, 15 | FA, 15, 16 | FA, 15, 16 |
| 15 | (CH)2 | 1.69e | 1.430 | 30.437 | 6.68 | 14, 16 | FA, 14, 16 | FA, 14, 16 |
| 16 | (CH3)4 | 11.80 | 0.778 | 29.947 | 6.68 | 15 | FA, 14, 15 | 14, 15 |
| Ser | NH-Ser | 1.0 | 7.504 | 112.19 | 2 | 2, 3, 3′ | 2, 1, 4 | |
| Gly | NH-Gly | 1.0 | 7.756 | 110.82 | 5, 5′ | 5, 5′ | 5, 6 |
For NMR evaluation of lipid 654, a highly purified lipid sample was processed as described in Materials and Methods. For the NMR proton assignments, all integrations were normalized to the proton on C-8 (integrated to 1.0). The carbon assignments shown in Fig. 1G correspond to the carbon numbers listed in the first column.
Methylene groups for C-3, C-5, and C-7 gave unique proton chemical shifts, indicating a lack of proton rotation.
Proton resonances for C-3 and C-5 overlap, causing the individual integrations to be slightly deviated, but taken together they integrated to 4.04, a result very close to the predicted value of 4.0.
Proton resonances for C-9 and C-13 overlap and were integrated together.
The integration of the peak for C-15 yielded 1.69, 15% lower than the predicted 2.0, indicating that approximately 85% of the fatty acids are isobranched. Integration of peaks C-9 and C-13 yielded 4.28, which is slightly higher than the predicted 4.0 due to an extra signal from the approximately 15% of anteisobranched fatty acid.
Peak overlaps, with the intense peak from the fatty acid. The total integration of the intense peak was 35.92, slightly higher than the predicted 34.
We could not determine couplings due to overlap.
FA, fatty acid.
Coupling between C-1 and C-3 was not observed due to overlap between the proton chemical shifts of C-3 and C-5.
gem, geminal protons.
MS analysis of lipid 430.
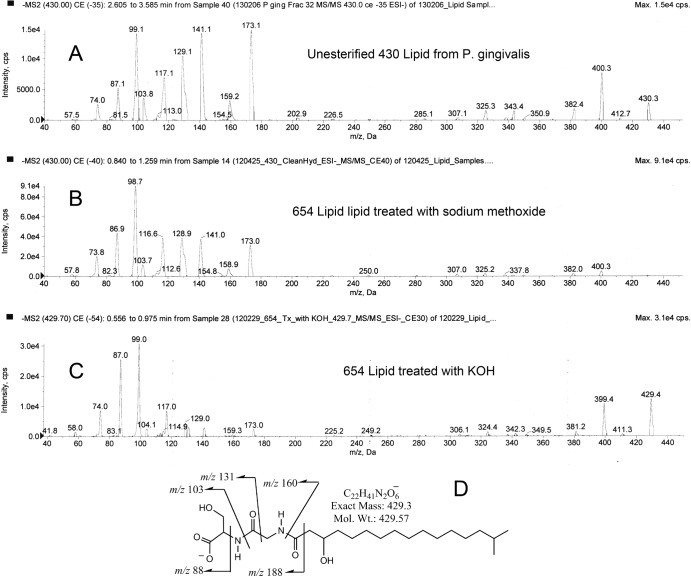
The molecular weights of the three major lipid species within the lipid 430 class could be consistent with the loss of an esterified fatty acid from the respective constituent lipid species of the lipid 654 class. To analyze this possibility, a sample of HPLC lipid fraction 35 containing highly enriched lipid 654 was subjected to base-catalyzed hydrolysis with either sodium methoxide or KOH in order to release ester-linked fatty acids. Both hydrolysis methods yielded low levels of lipids that produced negative ions of m/z 430, 416, and very small amounts of 402 as measured by ESI-MS (Fig. 3). The nonesterified 430 lipid class recovered in HPLC fraction 39 of the total lipid extract of P. gingivalis (Fig. 3A) showed an MS/MS spectrum similar to the m/z 430 lipids recovered after sodium methoxide treatment of lipid 654 (Fig. 3B) or KOH treatment of lipid 654 (Fig. 3C). The m/z 430 and 416 negative ions of the nonesterified lipid 430 were evaluated by MS/MS and revealed low-mass product ions (<200 amu), similar to those produced from m/z 654, 640, and 626 lipid species (Fig. 2). By increasing the collision energy for gas-phase fragmentation of precursor ions, the low-mass ion fragments (<200 amu) of the m/z 654 precursor increased in abundance to that shown for the lipid 430 fragmentations shown in Fig. 3 (precursor data not shown). As with the lipid 654 class, lipid extracts from the broth medium used to culture P. gingivalis showed no m/z 430, 416, or 402 ions (data not shown). These results demonstrate that the lipid 430 class represents the deesterified or nonesterified lipid 654 class (Fig. 3D, lipid 430 structure) and that the three lipid species contain the same amino acids within their respective head groups. However, the base-catalyzed hydrolysis of either the lipid 654 or lipid 430 classes eliminated their ability to activate TLR2-expressing HEK293 cells due to substantial breakdown of the lipid 654/430 products, as verified by thin-layer chromatography.
Fig 3.
MS/MS profiles of 430 lipid species. Samples of the highly enriched preparation of lipid 654 were subjected to either KOH or NaOCH3 treatment as described in Materials and Methods. The highly enriched lipid 430 (Fig. 1, fraction 39) prepared from the total lipids of P. gingivalis was compared with the KOH- or NaOCH3-hydrolyzed lipid 654 samples by using MS/MS analysis as described in Materials and Methods. The partial mass spectra are depicted for the m/z 430 lipid species recovered from HPLC-fractionated total lipids of P. gingivalis (A), NaOCH3-treated lipid 654 (B), or KOH-treated lipid 654 (C). The proposed structure shown in panel D represents the deesterified form of the 654 lipid species shown in Fig. 2G. See the Fig. 2 legend for reconciliation of the low-mass product ions (<200 amu).
Fatty acid and serine constituents in the lipid 654 class.
Hexane extraction of the sodium methoxide-treated lipid 654 followed by GC-MS analysis yielded fatty acid methyl esters, including branched CH3-C15:0 with lesser amounts of CH3-C14:0 (2.3%) and CH3-isobranched C13:0 (0.44%). Hexane extracts of the KOH-treated lipid 654 were processed to form pentafluorobenyl ester, TMS ether derivatives and were prepared in parallel with synthetic standards of anteisobranched and isobranched C15:0 and 3-OH fatty acid standards. Negative-ion GC-MS revealed that the C15:0 is approximately 88% isobranched, with the remainder anteisobranched C15:0. Straight-chain C15:0 was not observed. Negative-ion GC-MS of the 3-OH fatty acids revealed 69.8% as 3-OH iso-C17:0, 25.5% as 3-OH C16:0, and 4.7% as 3-OH iso-C15:0. By comparison, the average distribution of lipid species within the lipid 654 class was as follows: m/z 654 (61.8%), m/z 640 (31%), and m/z 626 (7.2%) ions. Therefore, the distribution of the hydroxy fatty acids, rather than the ester-linked fatty acids, in the lipid 654 class appears to account for the distribution of its three characteristic lipid species. The epimeric configuration of serine in the 654 lipid class was determined by chiral GC-MS analysis. This analysis demonstrated that the 654 lipid class contains only l-serine (data not shown). The stereochemistry of C-8 (Fig. 1G) has not been determined for lipid 654, nor has the stereochemistry of C-14 for the anteiso-C15:0 fatty acid been determined.
Lipid 654 and lipid 430 effects in vitro: dose responses and biological activities relative to other major lipid classes of P. gingivalis.
We next evaluated the biological activity dose-response characteristics of lipid 654 and lipid 430 and compared these responses with well-characterized TLR2 agonists as well as other prevalent lipid classes of P. gingivalis. The HPLC fractions containing either highly enriched lipid 654 (fraction 35) or lipid 430 (fraction 39) were evaluated for their abilities to activate TLR2-expressing HEK293 cells compared with the substituted phosphoglycerol dihydroceramides (subPG-DHC), unsubstituted phosphoglycerol dihydroceramide lipids (unPG-DHC), phosphoethanolamine dihydroceramide lipids (PE-DHC), and phosphatidylethanolamine (PEA) lipids of P. gingivalis (Fig. 4). Compared with the known TLR2 ligand positive controls MMP (20) and lipoteichoic acid (LTA), lipid 654 and lipid 430 promoted significant HEK cell activation over the control cells (DMSO-treated cells). MMP (molecular weight of 1,269.82) was used at a concentration of 0.2 μg/ml, or 0.158 μM. Using the molecular weights and distributions of the three lipid species within each lipid class, lipid 654 and lipid 430 at a concentration of 0.69 μg/ml represented doses of 1.066 μM and 1.621 μM, respectively. Lipid 654 and lipid 430 used at a concentration of 0.17 μg/ml represented 0.259 μM and 0.395 μM, respectively. The molecular weight of LTA was not provided by the supplier, and the molar concentration could not be calculated. Note that all other major lipid classes of P. gingivalis, previously isolated to very high purity (19, 37), showed little capacity to activate TLR2 in HEK cells. Therefore, we concluded that the phosphorylated dihydroceramide lipids of P. gingivalis do not account for the HEK cell activation observed in the total lipid extract of P. gingivalis. Instead, the HPLC fractions containing lipid 654 and lipid 430 accounted for the majority of the HEK cell activation observed with the total lipid extract. Figure 4 also shows the dose-response characteristics of lipid 654 and lipid 430 classes and confirms that these lipid classes are capable of activating HEK cells at low concentrations.
Fig 4.
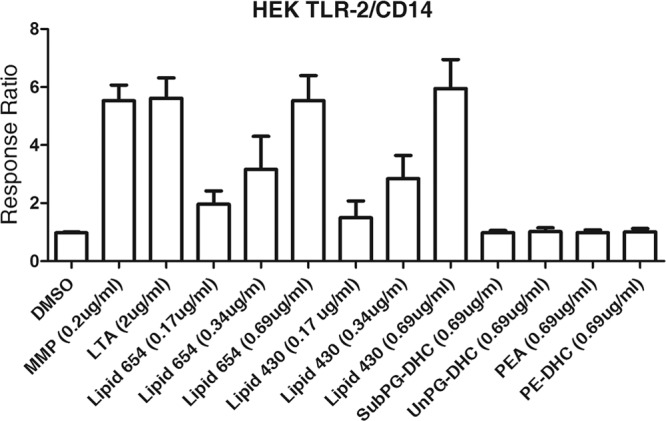
HEK cell activation by lipid classes derived from P. gingivalis. HEK293 cells, transfected with the human TLR2, MD-2, CD14, and SEAP genes, were used to assay the function of P. gingivalis lipid classes in vitro. HEK293 cells were stimulated for 24 h with the following treatments: DMSO (vehicle; 50% mixture of DMSO-water, approximately 1.11% DMSO in the final culture medium; n = 23), the known TLR2 ligand MMP (0.2 μg/ml; n = 34), LTA (2 μg/ml; n = 17), lipid 654 (0.17 μg/ml [n = 5]; 0.34 μg/ml [n = 2]; 0.69 μg/ml [n = 22]), lipid 430 (0.17 μg/ml [n = 2]; 0.34 μg/ml [n = 3]; 0.69 μg/ml [n = 18]), subPG-DHC (0.69 μg/ml; n = 4), unPG-DHC (0.69 μg/ml; n = 4), PE-DHC (0.69 μg/ml; n = 4), and PEA (0.69 μg/ml; n = 9). The phospholipid preparations were prepared from P. gingivalis total lipids as previously described (19, 37). Responses were assessed after 24 h, and results are expressed as the ratio of stimulated versus nonstimulated (DMSO) responses. By one-way ANOVA and Fisher LSD pairwise comparisons, HEK cell activation levels by MMP, LTA, lipid 654, and lipid 430 (both at 0.69 μg/ml) were significantly elevated over the DMSO vehicle (P < 0.05).
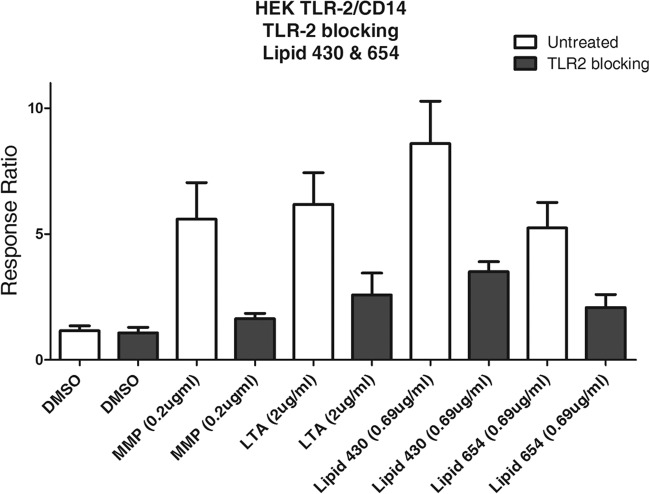
Lipid 654 and lipid 430 in vitro: TLR2 dependence of biological activities.
As shown in Fig. 5, MMP and LTA demonstrated TLR2 cell activation that was inhibited by pretreatment with anti-human TLR2 antibody. HEK-TLR2 cell responses to lipid 654 or lipid 430 preparations of P. gingivalis were also significantly inhibited by pretreatment with anti-human TLR2 antibody (Fig. 5). These results showed that lipid 654 and lipid 430 are ligands for human TLR2.
Fig 5.
TLR2-mediated stimulation levels by lipid 654 and lipid 430. HEK293 cells, transfected with the human TLR2, MD-2, CD14, and SEAP genes, were used to assay the function of lipid 654 and lipid 430 in vitro. For antibody blocking, cells were preincubated for 1 h with neutralizing anti-human TLR2 antibody (10 μg/ml; InvivoGen). HEK293 cells were stimulated for 24 h with DMSO (vehicle; 50% mixture of DMSO-water; approximately 1.11% DMSO in final culture medium; n = 4), the known TLR2 ligand MMP (0.1 μg/ml; n = 6), the known TLR2 ligand LTA (1 μg/ml; n = 6), lipid 430 (0.69 μg/ml; n = 2), and lipid 654 (0.69 μg/ml; n = 7). The sample sizes (n) for each treatment refer to the number of both untreated and TLR2-blocked samples. Responses were assessed 24 h after the addition of TLR2 ligands, and results are expressed as the ratio of stimulated to nonstimulated (DMSO) responses.
Lack of TLR4 activation by lipid 654 and lipid 430.
A previous report demonstrated that flavolipin induced innate immune activation by acting as an agonist for TLR4 (35). To test whether lipid 654 and lipid 430 can function as ligands for TLR4, we next utilized HEK293 cells transfected with the human TLR4, CD14, and MD-2 genes but not expressing TLR2. As shown in Fig. 6, the known TLR4 agonist LPS (derived either from Salmonella enterica or P. gingivalis) demonstrated the ability to stimulate the TLR4-expressing HEK293 cells. Consistent with a previous report (38), LPS from P. gingivalis was considerably weaker than enterobacterial LPS in stimulating HEK cells. In contrast, the TLR2 agonists MMP and LTA showed no activity. Most importantly, lipid 654 and lipid 430 showed no capacity to activate the TLR4-expressing cell line. These results indicate that lipid 654 and lipid 430, in contrast to flavolipin, can activate via TLR2 but are unable to function as TLR4 ligands. In additional studies, we demonstrated that the HEK null cells (HEK cells with the SEAP reporter gene but without transfected TLRs) also do not respond to either the TLR4 or TLR2 agonists (data not shown).
Fig 6.
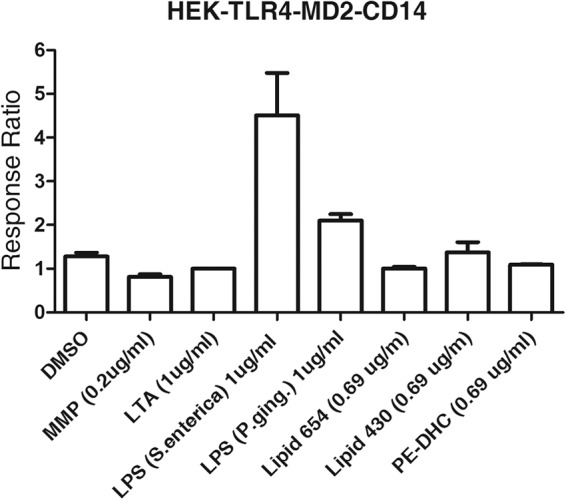
Lipid 654 and lipid 430 are not agonists for TLR4. HEK293 cells, transfected with the human TLR4, CD14, MD-2, and SEAP genes, were used to assay the function of lipid 654 and lipid 430 in vitro. HEK293 cells were stimulated for 24 h with DMSO (vehicle; 50% DMSO in water), the known TLR2 ligands MMP and LTA, lipid 654 (0.69 μg/ml), lipid 430 (0.69 μg/ml), and two different preparations of known TLR4 agonists, LPS derived from Salmonella enterica or P. gingivalis (1 μg/ml). Responses were assessed at 24 h, and results are expressed as the ratio of stimulated to nonstimulated (DMSO) responses (n = 2).
Bioactivities of lipid 654 and lipid 430 in vivo.
We next sought to confirm the in vitro functional effects of lipid 654 and lipid 430 by using in vivo approaches. We injected mice with lipid 654 or lipid 430 and analyzed the effect on serum levels of the chemokine CCL2 (also known as monocyte chemoattractant protein 1). CCL2 plays a major role in mediating the migration of inflammatory macrophages into tissue sites of inflammation, and it has been previously documented that administration of TLR agonists to mice can result in expression of serum CCL2 (39). Furthermore, this chemokine has been suggested to be important in the pathogenesis of autoimmune diseases and has been shown to be critical for the development of experimental autoimmune encephalomyelitis (EAE), the murine model of multiple sclerosis, and is also believed to be critical in the pathogenesis of human multiple sclerosis (40–43).
In our studies, lipid 654 was injected i.p. in 50% DMSO and lipid 430 was injected i.v. in phosphate-buffered saline (PBS). We injected WT female C57BL/6 mice and TLR2−/− female mice with either vehicle, lipid 654, or lipid 430. Four hours later, serum was recovered from these mice and analyzed for levels of CCL2. As seen in Fig. 7, lipid 654 induced a significant increase in serum levels of CCL2 in WT mice but failed to do so when injected into TLR2−/− mice (Fig. 7A). The same was true for lipid 430. Lipid 430 induced a significant increase in serum levels of CCL2 in WT mice but failed to do so when injected into TLR2−/− mice (Fig. 7B). These results demonstrated that both lipid 654 and lipid 430 have proinflammatory effects in vivo and, further, that these effects are dependent on TLR2.
Fig 7.
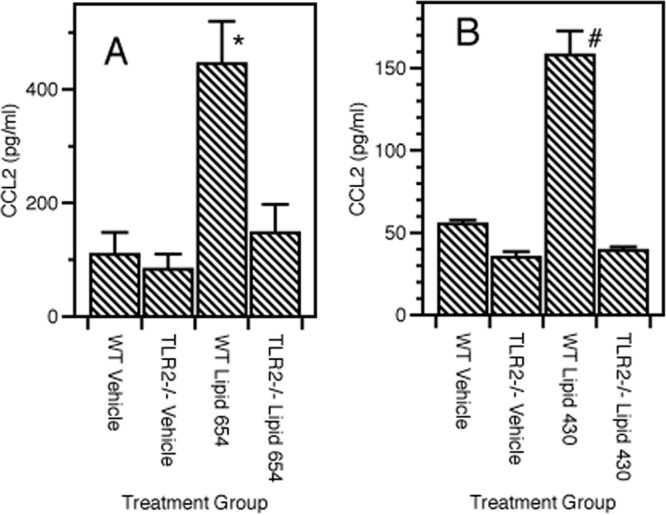
In vivo administration of lipid 654 or lipid 430 to wild-type and TLR2−/− mice. (A) Lipid 654 (1 μg) or vehicle (50% mixture of DMSO-water) was injected i.p. into either WT or TLR2−/− mice. Four hours later, the mice were bled and the sera assayed for levels of CCL2 by ELISA. Histogram bars represent the mean ± standard error of the mean (SEM) for 3 or 4 trials. (B) Lipid 430 (2.5 μg) or vehicle (PBS) was injected i.v. into either WT or TLR2−/− mice. Four hours later, the mice were bled and the sera assayed for levels of CCL2 by ELISA. Histogram bars represent the mean ± SEM for 3 trials. Statistical significance was assessed using the Student t test, with symbols indicating the following significance levels: *, P = 0.0164; #, P = 0.0012 for WT versus TLR−/− lipid responses.
Lipid 654 contamination of P. gingivalis LPS.
The LPS extract of P. gingivalis prepared by the method of Yi and Hackett (31) was shown to contain 3.82% ± 0.18% lipid 654 (mean ± standard deviation; n = 3). This LPS preparation, at a concentration of 0.69 μg/ml, produced a 1.8-fold increase in TLR2 activation in HEK cells, whereas MMP (0.2 μg/ml) produced a 3.7-fold increase. The final concentration of lipid 654 in this LPS assay preparation was calculated to be 0.039 μM, whereas the concentration of MMP used in the assay was 0.158 μM.
DISCUSSION
By using HPLC together with mass spectrometric/nuclear magnetic resonance spectroscopy, we identified two new lipid classes of P. gingivalis, lipid 654 and lipid 430, that act as ligands for human and mouse TLR2. While lipid 654 shares most structural characteristics with the previously described lipid class that includes flavolipin (33–36) and topostin (44), we found significant differences between lipid 654 and flavolipin in their respective biological activities. Flavolipin was named after the organisms of the Flavobacterium genus, from which it was originally isolated. According to previous reports, flavolipin represents approximately 20% of the total cellular lipids of Flavobacterium meningosepticum and is capable of acting as a TLR4 ligand (35, 36). In contrast, the results of our study demonstrated that neither lipid 654, nor lipid 430, activate via TLR4 but instead function as ligands for TLR2. Since both lipid 654 and lipid 430 act as TLR2 ligands but do not appear to activate through engagement of TLR4, it is possible that a lipid contaminant in the flavolipin preparations accounted for the previously reported TLR4 engagement (35).
An additional important distinction between flavolipin and lipid 654 is that, while only Flavobacterium species were originally reported to produce flavolipin (33, 35), we now report that lipid 654 is produced by more than Flavobacterium species. In addition to P. gingivalis, analysis of lipid extracts from Prevotella intermedia, Tannerella forsythia, Capnocytophaga ochracea, Capnocytophaga gingivalis, and Capnocytophaga sputigena revealed that each of these oral Bacteroidetes produce the lipid 654 class (data not shown). Selected intestinal Bacteroidetes, including Prevotella copri, Parabacteroides merdea, Parabacteroides distasonis, Bacteroides fragilis, Bacteroides stercoris, Bacteroides uniformis, and Bacteroides vulgatis, also produce the lipid 654 class (data not shown). A complete characterization of commensal organisms capable of producing lipid 654 and lipid 430 will be the focus of future studies. Nevertheless, our evidence thus far indicates that lipid 654 is produced by a wide variety of Bacteroidetes species and that, unlike flavolipin, it is not produced exclusively by Flavobacterium species (35, 36).
Using HEK293 cells stably transfected with TLR2 along with other relevant receptors, we found that lipid 654 and lipid 430 of P. gingivalis activate human cells via TLR2. Lipid 430, which is structurally related to lipid 654, is unique as a TLR2 agonist in that it contains only one fatty acid linked to a glycine-serine head group. In contrast, lipid 654 contains two fatty acids held in an ester linkage. Furthermore, the lipid 430 class is soluble in aqueous solvents of either neutral or basic pH, but this lipid class becomes considerably less soluble in acidic aqueous solvent. In contrast, the lipid 654 class will not dissolve in aqueous solvent unless it is sonicated to form liposome preparations. These differences in lipid structure and solubility will be the subjects of future investigations into the structural requirements for TLR2 engagement. The synthesis of lipid 430 by organisms other than P. gingivalis is likely, but this remains to be determined.
Activation of TLR2-expressing HEK293 cells was not observed to be mediated by the phosphorylated dihydroceramides or phospholipid preparations of P. gingivalis. With normal-phase HPLC fractionation, lipid 654 of P. gingivalis elutes slightly earlier than the PE DHC lipids. With the use of state-of-the-art HPLC equipment, which was not available at the time of our earlier work, the lipid 654 class can now be isolated largely free of PE DHC lipids. Since the purified PE DHC lipids demonstrate no capability to activate human cells through TLR2, we conclude that the previously reported activity of PE DHC lipids of P. gingivalis to engage TLR2 in mouse dendritic cells (13) was likely related to contamination of PE DHC lipids with the lipid 654 class.
Pretreatment of HEK-TLR2 cells with anti-human TLR2 antibody inhibited the effects of both lipid 654 and lipid 430 classes on TLR2-expressing HEK cell activation. The in vitro effects of anti-TLR2 antibody were consistent with the in vivo effects of lipid 654 and lipid 430 on serum CCL2 levels in wild-type and TLR2−/− mice. We therefore conclude from this evidence that lipid 654 and lipid 430 are structurally unique TLR2 ligands derived from P. gingivalis. In attempting to clarify the role of TLR2 coreceptors in the engagement of lipid 654 or lipid 430, we observed that neutralizing antibody against either TLR1 or TLR6 partially inhibited HEK cell activation (data not shown), but not to the extent that anti-TLR2 antibody inhibited HEK cell responses. In addition, the pattern of inhibition with the coreceptor antibodies appeared to differ between lipid 654 and lipid 430. Further work is needed to confirm these observations.
In unpublished studies, we recently evaluated lipid extracts of human impacted third molars using multiple reaction monitoring MS, specifically quantifying the lipid 654 class. We found that these lipid extracts contained minimal amounts of lipid 654 and essentially no lipid 430 (data not shown). These results suggest that in human tissues not directly exposed to bacteria, these lipids are not recovered in appreciable levels. In contrast, our recent studies revealed that lipid extracts from diseased human gingival tissue, carotid atheroma, and serum demonstrate lipid 654 (data not shown) in levels far exceeding the phosphorylated dihydroceramide lipids of P. gingivalis (32). Therefore, future studies will focus on the recovery of 654 and 430 lipids in healthy and diseased human tissues.
Although we have previously shown that LPS and lipid A of P. gingivalis are significantly contaminated with the phosphorylated dihydroceramide lipids (12), we have now determined that LPS of P. gingivalis prepared according to the methods of Yi and Hacket and Caroff (31, 45) contains an average 3.81% by weight of lipid 654. LPS contamination with lipid 654 supports the conclusion that the TLR2 activity previously attributed to be LPS/lipid A of P. gingivalis (46) is due in part to the presence of the 654 lipid class. However, other factors could be involved, as described below. A more recent report (11) indicated that the same phospholipid extraction method used in our laboratory (47, 48) yields an aqueous soluble factor from P. gingivalis that engages TLR2/TLR1 to a greater extent than the total lipids recovered in the organic solvent. We recently confirmed that lipid 430 and lipid 654 can be recovered from the aqueous extract of P. gingivalis following acidification of the aqueous phase with acetic acid and extraction of lipids into chloroform. The lipid 430 present in the aqueous phase alone could have produced the TLR2 effects observed by Jain et al. (11). Jain et al. also concluded that an aqueous soluble TLR2/TLR1 agonist copurifies with LPS and is released when the LPS is hydrolyzed to produce lipid A. This LPS-associated factor is attenuated by lipoprotein lipase treatment (11). Lipoprotein lipase will hydrolyze ester-linked fatty acids from triglycerides of lipoproteins. While lipid 430 does not contain an ester-linked fatty acid, the lipid 654 class does contain an ester-linked fatty acid. Although highly purified lipid 654 is not soluble in aqueous solvents, LPS, which is soluble in aqueous solutions and is contaminated with lipid 654 and lesser amounts of lipid 430 (data not shown), likely acts as a carrier for lipid 654 in aqueous extracts of P. gingivalis. It is also possible that another TLR2 ligand or a lipoprotein that has not yet been identified is present in the aqueous extracts of P. gingivalis. However, the previously reported lipopeptide of P. gingivalis (4, 49), which is soluble in aqueous solutions, was not the primary TLR2/TLR1 ligand present in the P. gingivalis aqueous extract. Until the putative water-soluble factor of P. gingivalis described by Jain et al. (11) is isolated and structurally characterized, lipid 430 and lipid 654 constitute the primary TLR2 ligands that are recovered in the aqueous extract of P. gingivalis. Future work will focus on the water-soluble TLR2 factors produced by P. gingivalis.
Although many virulence factors of P. gingivalis have been proposed to contribute to periodontal bone and tissue destruction (16, 17, 50), we have reported that various complex lipids of P. gingivalis are readily detected in periodontal disease sites without concurrent bacterial invasion or LPS recovery (21, 51). We have now determined that specific lipid classes that are relatively minor constituents of this organism are responsible for activation of TLR2 and that dihydroceramide lipids are not responsible for these TLR2-mediated biological responses. Our findings are consistent with those of other investigators who have reported that P. gingivalis colonization of teeth in experimental animals mediates bone loss through engagement of TLR2 (17, 52, 53). We believe that the lipid 654 and lipid 430 classes produced by P. gingivalis and other oral Bacteroidetes play a critical role in the development of destructive periodontal disease by promoting bone loss and inhibition of bone formation, as we have previously reported (14). We further suggest that lipid 654 and lipid 430 may contribute to the development of other chronic inflammatory diseases of humans, where these bacterial lipids accumulate to substantial levels due to contributions from microbes of the oral cavity, gastrointestinal tract, and other anatomical sites.
In summary, this investigation identified two serine lipid classes produced by P. gingivalis that act as newly defined ligands for TLR2 both in vitro and in vivo. These lipid classes are comprised of a group of lipid species with similar base structures as depicted for lipid 430. Addition of an esterified fatty acid to these base structures yields the three constituent lipid species of lipid 654. However, it is unclear at this time if the lipid species of lipid 430 represent a precursor or breakdown products (or both) for the constituent lipid species of lipid 654, and this will be a focus of future investigations. It is notable that the lipid 654 and lipid 430 classes differ substantially in their solubility characteristics in aqueous solvents. We believe the different solubility characteristics and the enzyme susceptibilities of these lipids could provide a novel framework for different modes of local delivery and innate immune system activation by these lipids. Thus, the two classes of lipids working together could potentially result in the both bone and soft tissue destruction that is associated with chronic periodontitis in humans. Further research will clarify these questions.
ACKNOWLEDGMENTS
We thank Julie Downes for preparing intestinal bacterial isolates for lipid evaluation.
This work was supported by NIH 1 R01 DE021055-01A1 (F.C.N.), National MS Society RG4070-A-6 (R.B.C.), and NIAID AI090166 (J.C.S.).
Footnotes
Published ahead of print 8 July 2013
REFERENCES
- 1.Medzhitov R. 2001. Toll-like receptors and innate immunity. Nat. Rev. Immunol. 1:135–145 [DOI] [PubMed] [Google Scholar]
- 2.Medzhitov R, Preston-Hurlburt P, Janeway CA., Jr 1997. A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature 388:394–397 [DOI] [PubMed] [Google Scholar]
- 3.Zahringer U, Lindner B, Inamura S, Heine H, Alexander C. 2008. TLR2: promiscuous or specific? A critical re-evaluation of a receptor expressing apparent broad specificity. Immunobiology 213:205–224 [DOI] [PubMed] [Google Scholar]
- 4.Hashimoto M, Asai Y, Ogawa T. 2004. Separation and structural analysis of lipoprotein in a lipopolysaccharide preparation from Porphyromonas gingivalis. Int. Immunol. 16:1431–1437 [DOI] [PubMed] [Google Scholar]
- 5.Bainbridge BW, Coats SR, Darveau RP. 2002. Porphyromonas gingivalis lipopolysaccharide displays functionally diverse interactions with the innate host defense system. Ann. Periodontol. 7:29–37 [DOI] [PubMed] [Google Scholar]
- 6.Bainbridge BW, Darveau RP. 2001. Porphyromonas gingivalis lipopolysaccharide: an unusual pattern recognition receptor ligand for the innate host defense system. Acta Odontol. Scand. 59:131–138 [DOI] [PubMed] [Google Scholar]
- 7.Triantafilou M, Gamper FG, Lepper PM, Mouratis MA, Schumann C, Harokopakis E, Schifferle RE, Hajishengallis G, Triantafilou K. 2007. Lipopolysaccharides from atherosclerosis-associated bacteria antagonize TLR4, induce formation of TLR2/1/CD36 complexes in lipid rafts and trigger TLR2-induced inflammatory responses in human vascular endothelial cells. Cell, Microbiol. 9:2030–2039 [DOI] [PubMed] [Google Scholar]
- 8.Davey M, Liu X, Ukai T, Jain V, Gudino C, Gibson FC, III, Golenbock D, Visintin A, Genco CA. 2008. Bacterial fimbriae stimulate proinflammatory activation in the endothelium through distinct TLRs. J. Immunol. 180:2187–2195 [DOI] [PubMed] [Google Scholar]
- 9.Hajishengallis G, Tapping RI, Harokopakis E, Nishiyama S, Ratti P, Schifferle RE, Lyle EA, Triantafilou M, Triantafilou K, Yoshimura F. 2006. Differential interactions of fimbriae and lipopolysaccharide from Porphyromonas gingivalis with the Toll-like receptor 2-centred pattern recognition apparatus. Cell, Microbiol. 8:1557–1570 [DOI] [PubMed] [Google Scholar]
- 10.Ogawa T, Asai Y, Hashimoto M, Uchida H. 2002. Bacterial fimbriae activate human peripheral blood monocytes utilizing TLR2, CD14 and CD11a/CD18 as cellular receptors. Eur. J. Immunol. 32:2543–2550 [DOI] [PubMed] [Google Scholar]
- 11.Jain S, Coats SR, Chang AM, Darveau RP. 2013. A novel class of lipoprotein lipase-sensitive molecules mediates Toll-like receptor 2 activation by Porphyromonas gingivalis. Infect. Immun. 81:1277–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nichols FC, Bajrami B, Clark RB, Housley W, Yao X. 2012. Free lipid A isolated from Porphyromonas gingivalis lipopolysaccharide is contaminated with phosphorylated dihydroceramide lipids: recovery in diseased dental samples. Infect. Immun. 80:860–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nichols FC, Housley WJ, O'Conor CA, Manning T, Wu S, Clark RB. 2009. Unique lipids from a common human bacterium represent a new class of Toll-like receptor 2 ligands capable of enhancing autoimmunity. Am. J. Pathol. 175:2430–2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang YH, Jiang J, Zhu Q, Alanezi AZ, Clark RB, Jiang X, Rowe DW, Nichols FC. 2010. Porphyromonas gingivalis lipids inhibit osteoblastic differentiation and function. Infect. Immun. 78:3726–3735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gibson FC, III, Genco CA. 2007. Porphyromonas gingivalis mediated periodontal disease and atherosclerosis: disparate diseases with commonalities in pathogenesis through TLRs. Curr. Pharm. Des. 13:3665–3675 [DOI] [PubMed] [Google Scholar]
- 16.Ukai T, Yumoto H, Gibson FC, III, Genco CA. 2008. Macrophage-elicited osteoclastogenesis in response to bacterial stimulation requires Toll-like receptor 2-dependent tumor necrosis factor-alpha production. Infect. Immun. 76:812–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Papadopoulos G, Weinberg EO, Massari P, Gibson FC, III, Wetzler LM, Morgan EF, Genco CA. 2013. Macrophage-specific TLR2 signaling mediates pathogen-induced TNF-dependent inflammatory oral bone loss. J. Immunol. 190:1148–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aoki Y, Tabeta K, Murakami Y, Yoshimura F, Yamazaki K. 2010. Analysis of immunostimulatory activity of Porphyromonas gingivalis fimbriae conferred by Toll-like receptor 2. Biochem. Biophys. Res. Commun. 398:86–91 [DOI] [PubMed] [Google Scholar]
- 19.Nichols FC, Riep B, Mun J, Morton MD, Bojarski MT, Dewhirst FE, Smith MB. 2004. Structures and biological activity of phosphorylated dihydroceramides of Porphyromonas gingivalis. J. Lipid Res. 45:2317–2330 [DOI] [PubMed] [Google Scholar]
- 20.Aliprantis AO, Yang RB, Mark MR, Suggett S, Devaux B, Radolf JD, Klimpel GR, Godowski P, Zychlinsky A. 1999. Cell activation and apoptosis by bacterial lipoproteins through toll-like receptor-2. Science 285:736–739 [DOI] [PubMed] [Google Scholar]
- 21.Nichols FC. 1994. Distribution of 3-hydroxy iC17:0 in subgingival plaque and gingival tissue samples: relationship to adult periodontitis. Infect. Immun. 62:3753–3760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chiou SH, Wang KT. 1989. Peptide and protein hydrolysis by microwave irradiation. J. Chromatogr. 491:424–431 [DOI] [PubMed] [Google Scholar]
- 23.Fuchs SA, de Sain-van der Velden MG, de Barse MM, Roeleveld MW, Hendriks M, Dorland L, Klomp LW, Berger R, de Koning TJ. 2008. Two mass-spectrometric techniques for quantifying serine enantiomers and glycine in cerebrospinal fluid: potential confounders and age-dependent ranges. Clin. Chem. 54:1443–1450 [DOI] [PubMed] [Google Scholar]
- 24.Xia Y, Moran S, Nikonowitz EP. 2008. Z-restored spin-echo 13C 1D spectrum of straight baseline free of hump, dip and roll. Magn. Reson. Chem. 46:432–435 [DOI] [PubMed] [Google Scholar]
- 25.Fulton DB, Hrabal R, Ni F. 1996. Gradient-enhanced TOCSY experiments with improved sensitivity and solvent suppression. J. Biomol. NMR 8:213–218 [DOI] [PubMed] [Google Scholar]
- 26.Piantini U, Sorenson OW, Ernst RR. 1982. Multiple quantum filters for elucidating NMR coupling networks. J. Am. Chem. Soc. 104:6800–6801 [Google Scholar]
- 27.Bodenhausen G, Ruben DJ. 1980. Natural abundance nitrogen-15 NMR by enhanced heteronuclear spectroscopy. Chem. Phys. Lett. 69:185–189 [Google Scholar]
- 28.Bax A, Summers M. 1986. 1H and 13C assignments form sensitivity-enhanced detection of heteronuclear multiple-bond connectivity by 2D multiple quantum NMR. J. Am. Chem. Soc. 108:2093–2094 [Google Scholar]
- 29.Nyberg NT, Duus JO, Sorenson OW. 2005. Heteronuclear two-bond correlation: suppressing heteronuclear three-bond or higher NMR correlations while enhancing two-bond correlations even for vanishing 2J(CH). J. Am. Chem. Soc. 127:6154–6155 [DOI] [PubMed] [Google Scholar]
- 30.Takeuchi O, Hoshino K, Kawai T, Sanjo H, Takada H, Ogawa T, Takeda K, Akira S. 1999. Differential roles of TLR2 and TLR4 in recognition of gram-negative and gram-positive bacterial cell wall components. Immunity 11:443–451 [DOI] [PubMed] [Google Scholar]
- 31.Yi EC, Hackett M. 2000. Rapid isolation method for lipopolysaccharide and lipid A from gram-negative bacteria. Analyst 125:651–656 [DOI] [PubMed] [Google Scholar]
- 32.Nichols FC, Yao X, Bajrami B, Downes J, Finegold SM, Knee E, Gallagher JJ, Housley WJ, Clark RB. 2011. Phosphorylated dihydroceramides from common human bacteria are recovered in human tissues. PLoS One 6(2):e16771. 10.1371/journal.pone.0016771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kawai Y, Yano I, Kaneda K. 1988. Various kinds of lipoamino acids including a novel serine-containing lipid in an opportunistic pathogen Flavobacterium. Their structures and biological activities on erythrocytes. Eur. J. Biochem. 171:73–80 [DOI] [PubMed] [Google Scholar]
- 34.Shiozaki M, Degucki N, Mochizuki T, Wakabayashi T, Ishikawa T, Haruyama H, Kawai Y, Nishijima M. 1998. Revised structure of flavolipin and synthesis of its isomers. Tetrahedron Lett. 39:4497–4500 [Google Scholar]
- 35.Gomi K, Kawasaki K, Kawai Y, Shiozaki M, Nishijima M. 2002. Toll-like receptor 4-MD-2 complex mediates the signal transduction induced by flavolipin, an amino acid-containing lipid unique to Flavobacterium meningosepticum. J. Immunol. 168:2939–2943 [DOI] [PubMed] [Google Scholar]
- 36.Kawasaki K, Gomi K, Kawai Y, Shiozaki M, Nishijima M. 2003. Molecular basis for lipopolysaccharide mimetic action of Taxol and flavolipin. J. Endotoxin Res. 9:301–307 [DOI] [PubMed] [Google Scholar]
- 37.Nichols FC, Riep B, Mun J, Morton MD, Kawai T, Dewhirst FE, Smith MB. 2006. Structures and biological activities of novel phosphatidylethanolamine lipids of Porphyromonas gingivalis. J. Lipid Res. 47:844–853 [DOI] [PubMed] [Google Scholar]
- 38.Garrison SW, Holt SC, Nichols FC. 1988. Lipopolysaccharide-stimulated PGE2 release from human monocytes. Comparison of lipopolysaccharides prepared from suspected periodontal pathogens. J. Periodontol. 59:684–687 [DOI] [PubMed] [Google Scholar]
- 39.Shi C, Jia T, Mendez-Ferrer S, Hohl TM, Serbina NV, Lipuma L, Leiner I, Li MO, Frenette PS, Pamer EG. 2011. Bone marrow mesenchymal stem and progenitor cells induce monocyte emigration in response to circulating toll-like receptor ligands. Immunity 34:590–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fife BT, Huffnagle GB, Kuziel WA, Karpus WJ. 2000. CC chemokine receptor 2 is critical for induction of experimental autoimmune encephalomyelitis. J. Exp. Med. 192:899–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Izikson L, Klein RS, Charo IF, Weiner HL, Luster AD. 2000. Resistance to experimental autoimmune encephalomyelitis in mice lacking the CC chemokine receptor (CCR)2. J. Exp. Med. 192:1075–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simpson J, Rezaie P, Newcombe J, Cuzner ML, Male D, Woodroofe MN. 2000. Expression of the beta-chemokine receptors CCR2, CCR3 and CCR5 in multiple sclerosis central nervous system tissue. J. Neuroimmunol. 108:192–200 [DOI] [PubMed] [Google Scholar]
- 43.Sorensen TL, Ransohoff RM, Strieter RM, Sellebjerg F. 2004. Chemokine CCL2 and chemokine receptor CCR2 in early active multiple sclerosis. Eur. J. Neurol. 11:445–449 [DOI] [PubMed] [Google Scholar]
- 44.Shiori T, Terao Y, Irako N, Aoyama T. 1998. Synthesis of topostins B567 and D654 (WB-3559D, flavolipin), DNA toposimerase I inhibitors of bacterial origin. Tetrahedron 54:15701–15710 [Google Scholar]
- 45.Cann-Moisan C, Caroff J, Girin E. 1988. Determination of flavin adenine dinucleotide in biological tissues by high-performance liquid chromatography with electrochemical detection. J. Chromatogr. 442:441–443 [DOI] [PubMed] [Google Scholar]
- 46.Darveau RP, Pham TT, Lemley K, Reife RA, Bainbridge BW, Coats SR, Howald WN, Way SS, Hajjar AM. 2004. Porphyromonas gingivalis lipopolysaccharide contains multiple lipid A species that functionally interact with both toll-like receptors 2 and 4. Infect. Immun. 72:5041–5051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bligh EG, Dyer WJ. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37:911–917 [DOI] [PubMed] [Google Scholar]
- 48.Garbus J, DeLuca HF, Loomas ME, Strong FM. 1963. The rapid incorporation of phosphate into mitochondrial lipids. J. Biol. Chem. 238:59–63 [PubMed] [Google Scholar]
- 49.Makimura Y, Asai Y, Taiji Y, Sugiyama A, Tamai R, Ogawa T. 2006. Correlation between chemical structure and biological activities of Porphyromonas gingivalis synthetic lipopeptide derivatives. Clin. Exp. Immunol. 146:159–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gibson FC, III, Hong C, Chou HH, Yumoto H, Chen J, Lien E, Wong J, Genco CA. 2004. Innate immune recognition of invasive bacteria accelerates atherosclerosis in apolipoprotein E-deficient mice. Circulation 109:2801–2806 [DOI] [PubMed] [Google Scholar]
- 51.Nichols FC, Rojanasomsith K. 2006. Porphyromonas gingivalis lipids and diseased dental tissues. Oral Microbiol. Immunol. 21:84–92 [DOI] [PubMed] [Google Scholar]
- 52.Gibson FC, III, Ukai T, Genco CA. 2008. Engagement of specific innate immune signaling pathways during Porphyromonas gingivalis induced chronic inflammation and atherosclerosis. Front. Biosci. 13:2041–2059 [DOI] [PubMed] [Google Scholar]
- 53.Liang S, Krauss JL, Domon H, McIntosh ML, Hosur KB, Qu H, Li F, Tzekou A, Lambris JD, Hajishengallis G. 2011. The C5a receptor impairs IL-12-dependent clearance of Porphyromonas gingivalis and is required for induction of periodontal bone loss. J. Immunol. 186:869–877 [DOI] [PMC free article] [PubMed] [Google Scholar]