Abstract
Sepsis is an infection-induced systemic inflammatory response syndrome. Upstream recognition molecules, like CD14, play key roles in the pathogenesis. The aim of the present study was to investigate the effect of systemic CD14 inhibition on local inflammatory responses in organs from septic pigs. Pigs (n = 34) receiving Escherichia coli-bacteria or E. coli-lipopolysaccharide (LPS) were treated with an anti-CD14 monoclonal antibody or an isotype-matched control. Lungs, liver, spleen, and kidneys were examined for bacteria and inflammatory biomarkers. E. coli and LPS were found in large amounts in the lungs compared to the liver, spleen, and kidneys. Notably, the bacterial load did not predict the respective organ inflammatory response. There was a marked variation in biomarker induction in the organs and in the effect of anti-CD14. Generally, the spleen produced the most cytokines per weight unit, whereas the liver contributed the most to the total load. All cytokines were significantly inhibited in the spleen. Interleukin-6 (IL-6) was significantly inhibited in all organs, IL-1β and IP-10 were significantly inhibited in liver, spleen, and kidneys, and tumor necrosis factor, IL-8, and PAI-1 were inhibited only in the spleen. ICAM-1 and VCAM-1 was significantly inhibited in the kidneys. Systemic CD14-inhibition efficiently, though organ dependent, attenuated local inflammatory responses. Detailed knowledge on how the different organs respond to systemic inflammation in vivo, beyond the information gained by blood examination, is important for our understanding of the nature of systemic inflammation and is required for future mediator-directed therapy in sepsis. Inhibition of CD14 seems to be a good candidate for such treatment.
INTRODUCTION
Sepsis is a serious and life-threatening systemic inflammatory response syndrome caused by infection (1). The inflammatory response is vast and complex. The innate immune system plays a key role in the onset and maintenance of the inflammation (2, 3). The main trigger of inflammation is the recognition of microbes and damaged tissue by receptors of the innate immune system (4, 5). Recognition molecules include both soluble and cell-bound molecules, e.g., the Toll-like receptors (TLRs) (6). Soluble recognition molecules extra- or intracellularly play in concert with the cell-bound receptors in initiating the inflammatory response (7). These receptors are targeting conserved features of microbes and thereby detect a wide range of microbial diversity (8, 9). The inflammation in sepsis becomes a threat to the host when it spreads out of control (10).
The TLRs are a broad group of recognition molecules in the innate immune system (6). They recognize a range of molecules (11). TLR4, MD-2, and CD14 form the TLR4 complex, which recognizes the lipopolysaccharides (LPS) of Gram-negative bacteria (12, 13). Interestingly, CD14 is also a comolecule for several other TLRs (14), showing its broad functionality and promiscuity as an innate immune receptor. It has been shown that CD14 plays a part in inflammation in organs, as in LPS-induced lung inflammation (15) and in the recruitment of neutrophils in liver microcirculation (16). A recent publication showed that CD14 controls the endocytosis of TLR4 upon the recognition of LPS and subsequent intracellular signaling (17). These findings point to CD14 as a key upstream receptor of innate immunity.
Studies of human and animal sepsis models have examined the inflammatory response in blood (2, 18, 19). The effects of sepsis on single organs, such as the heart or liver (20, 21), or on the endothelium have also been studied clinically and molecularly (22). However, the inflammatory response to sepsis in organs in vivo is less well known. Thus, the aim of the present study was to investigate the inflammatory responses in various organs to systemic E. coli whole bacteria or LPS-induced inflammation and the effects of inhibiting CD14 on these responses. For this purpose, we used a porcine model which, in light of a recent publication, perhaps represents a more relevant model for mimicking human disease and in particular inflammation compared to murine models (23).
MATERIALS AND METHODS
Animals and experimental groups.
The animals and experimental design used here have been described in detail previously (24). Briefly, ∼7-week-old pigs (Sus scrofa domesticus, outbred stock), weighing 15 kg (range, 14.5 to 17 kg), were used. Clinically healthy pigs were allocated into one of two groups. The E. coli group consisted of subgroups receiving E. coli and anti-CD14 (n = 7), E. coli and isotype-matched control antibody (n = 6), E. coli only (n = 6), or bacterial culture medium (n = 6). The LPS group consisted of subgroups receiving LPS and anti-CD14 (n = 6) or LPS and isotype-matched control antibody (positive control group) (n = 3). The results from the LPS group are presented in the supplemental material. Exclusion criteria included unhealthy animals prior to the experiments, hemoglobin at <5 g/dl during the stabilization phase, or SaO2 < 90% with FiO2 = 0.30 in the stabilization phase. No animals were excluded. Animals were anesthetized and surgery was conducted according to procedures described in detail previously (25). Mouse anti-porcine CD14 monoclonal antibody clone MIL-2, isotype IgG2b, was purchased from AbD Serotec (Oxford, United Kingdom). A mouse anti-human IgG2b (clone BH1, product no. 2070) was purchased from Diatec monoclonal antibodies AS (Oslo, Norway) and used as an isotype-matched control. Live E. coli (strain LE392; ATCC no. 33572) was obtained from American Type Culture Collection (Manassas, VA). E. coli was infused intravenously at an increasing rate: 3.75 × 107 bacteria/h from 0 to 60 min, 1.5 × 108 bacteria/h from 60 to 90 min, and 6.0 × 108 bacteria/h from 90 to 240 min. Infusions of bacteria started immediately after anti-CD14 or control treatment was given. A total amount of 1.075 × 108 bacteria/kg, corresponding to 1.1 × 106 bacteria/ml of blood, was infused into each animal in the E. coli group. Ultra Pure E. coli LPS (strain O111:B4) was purchased from InvivoGen (San Diego, CA). The LPS was dissolved in sterile water and thereafter in sterile saline water. A total amount of 0.03 mg of LPS/kg was infused intravenously over 30 min, immediately after anti-CD14 or isotype-matched control antibody was given.
Preparation of tissues and homogenization.
Tissue samples from the right lung, liver, spleen, and kidney were harvested immediately after euthanasia, put on ice, cut, and transferred to Nunc tubes (Roskilde, Denmark) on dry ice, before storage at −70°C. All tissue samples were cut, weighed, and transferred to gentle MACS M tubes (Thermo Fischer Scientific, Gothenburg, Sweden) for homogenization, during which the samples, and all equipment used were kept on dry ice.
For the cytokine analysis, 50 mg of tissue sample was added to a mix of 495 μl of CytoBuster protein extraction reagent (Novagen, San Diego, CA) and 5 μl of protease inhibitor cocktail set 1 (Calbiochem, Darmstadt, Germany), followed by homogenization with Xiril Dispomix (Xiril AG, Hombrechtikon, Switzerland). After completion, the samples were incubated for 5 min on ice and thereafter centrifuged at 2,500 × g for 20 min at 4°C. The supernatants were transferred to Nunc tubes and stored at −70°C until analysis.
For the E. coli analysis, 50 mg of tissue sample was added to 400 μl of MagNa Pure DNA tissue lysis buffer (Roche Diagnostics GmbH, Indianapolis, IN), followed by homogenization with Xiril Dispomix. After completion, the samples were incubated for 30 min at room temperature for better lysis. The samples were transferred to Safe-Lock tubes, PCR Clean (Eppendorf AG, Hamburg, Germany) was added, and the samples were stored at −70°C until analysis.
For the LPS analysis, 50 mg of tissue sample was added to 500 μl of LAL reagent water (Associates of Cape Cod), followed by homogenization with Xiril Dispomix. After completion, the samples were transferred to Nunc tubes and stored at −70°C until analysis.
For the RNA analysis, 20 mg of tissue sample was added to 800 μl of TRIzol reagent (Invitrogen, Carlsbad, CA), followed by homogenization with Xiril Dispomix. After completion, the samples were incubated for 5 min at room temperature, transferred to Safe-Lock tubes, and stored at −70°C until analysis. Precautions were taken to avoid RNase and DNA contamination. All conditions of homogenization were optimized and validated before analysis.
Total protein measurement.
The total protein amount in all tissue cytokine homogenates was measured by using a DC protein assay (Bio-Rad, Hercules, CA), using 30% albumin solution from bovine serum (Sigma-Aldrich, St. Louis, MO) diluted with CytoBuster protein extraction reagent for the standard curve. Conditions were optimized and validated before analysis.
E. coli DNA analysis.
E. coli DNA from 80 μl of homogenized sample was isolated by robotic DNA purification using a MagNA Pure LC DNA isolation kit II (tissue variant) on a MagNA Pure LC instrument (Roche Applied Science, Mannheim, Germany). Quantification of E. coli DNA was performed by detection of the seqA gene as described earlier (24) using real-time PCR (AB 7500 Fast; Applied Biosystems, Warrington, United Kingdom). The PCR amplification was performed in a total volume of 10 μl, using 2 μl of DNA, 5 μl of 2× TaqMan Universal FAST Master Mix (Applied Biosystems), 0.5 μl of gene-specific 20× gene expression assay mix (Applied Biosystems), and 2.5 μl of RNase/DNase-free water. The cycling conditions were as follows: 20 s of polymerase activation at 95°C was followed by 40 cycles of 95°C for 3 s and 60°C for 30 s. All samples were amplified in triplicates.
LPS quantification.
Endotoxin levels were quantified in a Pyrochrome Limulus amebocyte lysate (LAL) assay by an endpoint chromogenic method using a diazo-coupling assay kit (Associates of Cape Cod). Homogenates were diluted in depyrogenated Pyrotube-D tubes with LAL reagent water (both from Associates of Cape Cod). The diluted samples were heat treated at 75°C for 10 min, mixed with pyrochrome solved in Glucashield a β-glucan inhibiting buffer, and incubated in a 96-well Pyroplate (both from Associates of Cape Cod) at 37°C on a dry block incubator. Normal human serum was used as negative control. After incubation, the procedure was followed according to the instructions from the manufacturer.
Protein quantification of inflammatory mediators.
Cytokines were analyzed using commercially available kits. Quantikine porcine immunoassay kits from R&D Systems (Minneapolis, MN) were used for analyses of tumor necrosis factor (TNF), interleukin-1β (IL-1β), IL-6, and IL-8. IL-10 was analyzed by swine IL-10 enzyme-linked immunosorbent assay kit from BioSource Invitrogen (Camarillo, CA).
qPCR analyses. (i) RNA preparation.
Total RNA was isolated from the homogenates using chloroform extraction with an RNeasy MinElute cleanup kit (Qiagen, Hilden, Germany). Sufficient RNA quantity and quality was assessed by a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA) and by an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). Contaminating DNA was removed from the samples using DNA-free DNase treatment and removal reagent (Ambion, Inc., Austin, TX) as recommended by the manufacturer. DNase-treated RNA samples were then stored at −70°C.
(ii) cDNA synthesis.
cDNA was produced from 500 ng of each RNA sample by a one-vial reverse transcriptase PCR using the high-capacity cDNA reverse transcriptase kit (Applied Biosystems, Foster City, CA). PCR conditions were set to 120 min of reverse transcription at 37°C and 5 s at 85°C to stop the reaction.
(iii) Candidate genes.
FAM-MGB dye-labeled TaqMan qPCR primers for E-selectin (Ss03394520_m1), ICAM-1 (Ss03392384_m1), VCAM-1 (Ss03390909_m1), PAI-1 (Ss03392656_u1), IL-1β (Ss03393804_m1), IP-10 (Ss03391846_m1), IL-10 (Ss03382372_u1), CD14 (Ss03818718_s1), TLR-4 (Ss03389780_m1), Caspase-1 (Ss03394227_m1), and C5aR1 (Ss03375530_u1) were obtained from Applied Biosystems. GAPDH (glyceraldehyde-3-phosphate dehydrogenase; Ss03374854_g1) was used as an endogenous control since it was confirmed stably expressed in all groups and organs in control experiments.
(iv) qPCRs.
Three qPCRs per sample were performed for each candidate gene. A 15-μl reaction volume consisting of 5 ng of cDNA, 0.75 μl of 20× TaqMan gene expression assay mix (Applied Biosystems), 7.5 μl of TaqMan Universal PCR Master Mix (Roche, Branchburg, NJ), and 6.25 μl of nuclease-free water (Sigma-Aldrich) was added to 96-well MicroAmp optical reaction plates (Applied Biosystems) for analysis. PCR amplification was performed in a StepOnePlus machine (Applied Biosystems) with the following cycling conditions: 50°C for 2 min and 95°C for 10 min, followed by 40 two-segment cycles (95°C for 15 s and 60°C for 1 min). Fluorescence was automatically measured during each two-segment cycle.
Data presentation and statistical analysis.
All data are presented per mg of tissue. The two positive control groups in the E. coli group (i.e., the E. coli and isotype-matched control antibody group and the E. coli-only group) showed identical results and were therefore merged into one group for the LPS analyses, E. coli DNA analyses, and the cytokine analyses. (For an outline of the groups, please see reference 24.) Differences in organ LPS load in the control group of the E. coli group was analyzed using one-way analysis of variance (ANOVA) with Tukey's multiple comparisons post hoc test. Differences in organ LPS load between the treatment group and the positive control group were analyzed using a two-sample t test. Differences in organ cytokine load in the E. coli control group were analyzed using one-way ANOVA with Tukey's multiple-comparison post hoc test. Differences in organ cytokine load between the treatment group and the positive control group were analyzed using a Mann-Whitney U test, as normal distribution of the data could not be assumed. The mRNA analyses were only performed on the E. coli group. The group that only received E. coli was used as a positive control group in the mRNA analyses. qPCR results were quantified by calculating the fold changes using the 2−ΔΔCT method and normalized to the negative control group (culture medium group). Differences between the groups were calculated using one-way ANOVA with the Bonferroni post hoc test corrected for multiple pairwise comparisons.
The results for the LPS group are presented without statistical analysis, since the positive control group was limited to three animals. For all of the statistical analyses, a P value of <0.05 was considered statistical significant. The results were analyzed using GraphPad Prism (La Jolla, CA) and PASW Statistics 18 (IBM, Chicago, IL).
Ethics.
The study was approved by the Norwegian Animal Research Authority, and animals were treated according to the Norwegian Laboratory Animal Regulations.
RESULTS
Total protein amount.
The total protein amount was measured in all of the samples and correlated to the weight of the tissue. No differences were found between the homogenates (data not shown).
E. coli group. (i) E. coli DNA.
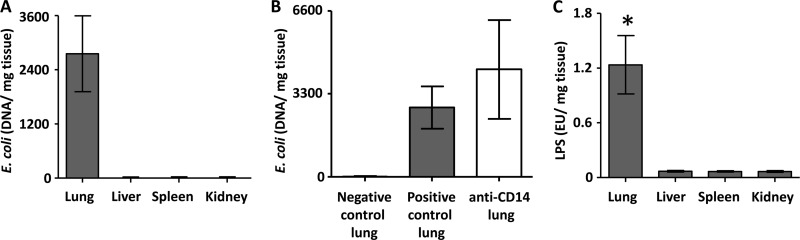
E. coli DNA was only detected in the homogenates from the lungs (Fig. 1A). Since the threshold for detection was 7,000 copies per ml of homogenate, the presence of lower amounts of E. coli in the other organs could not be detected. There was a nonsignificant trend (P = 0.086) toward a higher count of E. coli DNA in the anti-CD14 group compared to the positive control group in the lungs (Fig. 1B). No E. coli DNA was found in the negative control group.
Fig 1.
E. coli DNA and LPS load in the organs and effect of anti-CD14. E. coli DNA levels in the organs in the positive control group in the E. coli group (A) and E. coli DNA levels in the lung in the anti-CD14 group compared to the positive control group are shown (B). (C) LPS loads in organs in the positive control group in the E. coli group. The data are presented as means and 95% confidence intervals. *, P < 0.05.
(ii) LPS load.
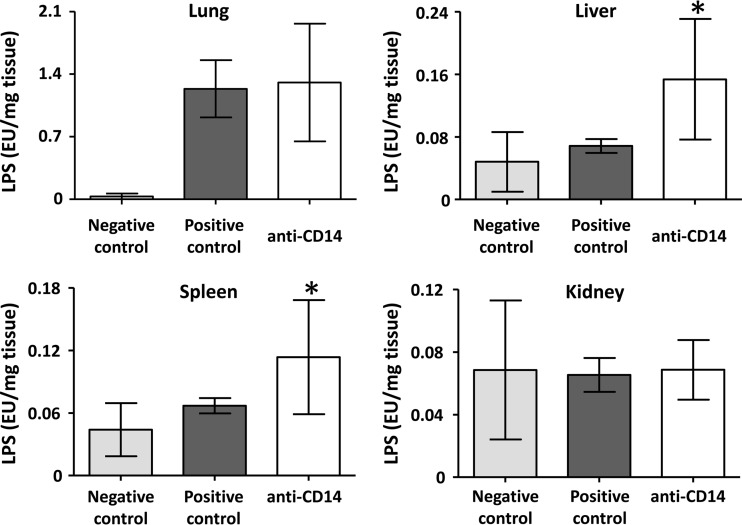
There was a substantially and significantly higher amount of LPS in the lung samples compared to the other organs (P < 0.0001), and no difference between the other organs (Fig. 1C). In the liver and spleen, there was a significantly higher LPS amount in the anti-CD14 group compared to the positive control group (P = 0.0003 and 0.0141, respectively), and no difference in lungs and kidneys (Fig. 2).
Fig 2.
Effect of anti-CD14 on LPS load in the organs. The LPS loads in the organs in the negative control group, in the positive control group, and in the anti-CD14 group are shown for various organs. The anti-CD14 group is statistically compared to the positive control group. The data are presented as means and 95% confidence intervals. *, P < 0.05.
Inflammatory biomarkers. (i) Cytokines in the positive control group measured by enzyme immunoassays.
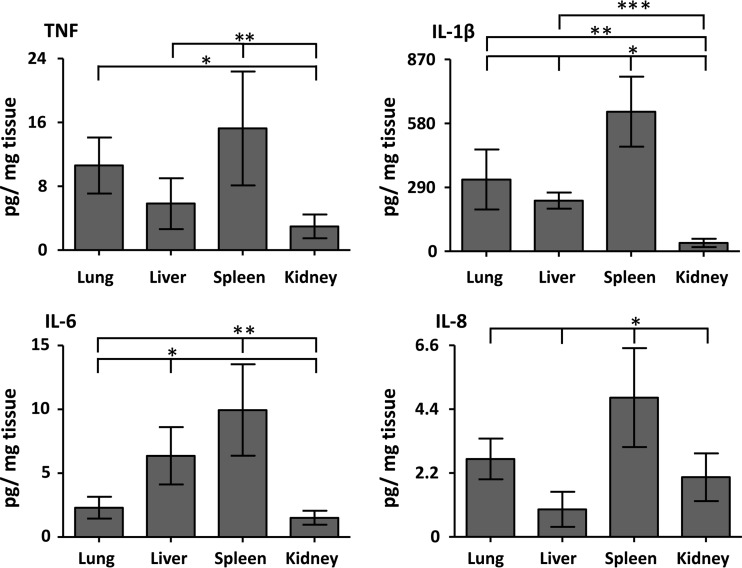
TNF, IL-1β, IL-6, and IL-8 were increased in all of the organs in the positive control group (Fig. 3). There was an overall highly significant difference between the loads of TNF (P = 0.0004), IL-1β, IL-6, and IL-8 (P < 0.0001) in the organs. TNF was significantly higher in the spleen compared to the liver and kidney and in the lung compared to the kidney (P < 0.05). IL-1β was significantly higher in the spleen compared to the lung, liver, and kidney, in the lung compared to the kidney, and in the liver compared to the kidney (P < 0.05). IL-6 was significantly higher in the spleen compared to the lung and kidney and in the liver compared to the lung and kidney (P < 0.05). IL-8 was significantly higher in the spleen compared to the lung, liver, and kidney (P < 0.05).
Fig 3.
Cytokine load in the organs. The organ load of the proinflammatory cytokines TNF, IL-1β, IL-6, and IL-8 in the organs in the positive control group is shown in the E. coli group. The data are presented as means and 95% confidence intervals. TNF: *, lung versus kidney; **, spleen versus liver and kidney. IL-1β: *, spleen versus lung, liver, and kidney; **, lung versus kidney; ***, liver versus kidney. IL-6: *, liver versus lung and kidney; **, spleen versus lung and kidney. IL-8: *, spleen versus lung, liver, and kidney. All, P < 0.05.
IL-10 was detected only in the liver and kidney, but no increase was found in the positive control group compared to the negative control group (data not shown). None of the cytokines, except for IL-10 in the liver and kidney, was detected in the negative control group (data not shown).
(ii) Effect of anti-CD14 on the cytokine load.
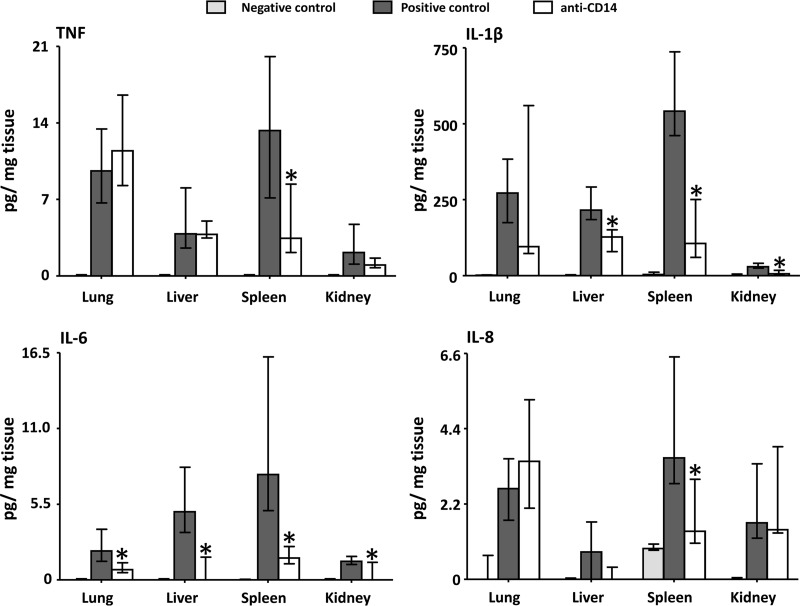
In the lung, IL-6 was significantly reduced (P = 0.024) in the anti-CD14 group compared to the positive control group (Fig. 4). TNF, IL-1β, and IL-8 were not significantly affected by anti-CD14. In the liver, IL-1β (P = 0.0023) and IL-6 (P = 0.0022) were significantly reduced in the anti-CD14 group compared to the positive control group (Fig. 4). TNF and IL-8 were not significantly affected by anti-CD14. In the spleen, all of the measured cytokines—TNF (P = 0.017), IL-1β (P = 0.0076), IL-6 (P = 0.0009), and IL-8 (P = 0.01)—were significantly reduced in the anti-CD14 group compared to the positive control group (Fig. 4). In the kidney, IL-1β (P = 0.013) and IL-6 (P = 0.033) were significantly reduced in the anti-CD14 group compared to the positive control group (Fig. 4). TNF and IL-8 were not significantly affected by anti-CD14.
Fig 4.
Effect of anti-CD14 on the proinflammatory cytokine load in the organs. The loads of the proinflammatory cytokines TNF, IL-1β, IL-6, and IL-8 in the negative control group, the positive control group, and the anti-CD14 group were determined. The anti-CD14 group is statistically compared to the positive control group. The data are presented as medians and interquartile ranges. *, P < 0.05.
(iii) Contribution of the different organs to the total cytokine load and the effect of anti-CD14.
The data reported thus far were all calculated per mg of tissue. The total contribution from each of the four organs was then examined according to their weight (Table 1). The total load, the percent contribution of each of the organs to total cytokine load, and the percent change in cytokine load by anti-CD14 in each organ are presented. The liver contributed by >50% to the load of TNF, IL-1β, and in particular IL-6 with 79% of the total load. The lungs contributed substantially by ca. 30% to the load of TNF, IL-1β, and IL-8. The spleen and kidney contributed with 2 to 15% of the total cytokine load. Anti-CD14 substantially reduced the total cytokine load in the organs, with the exception of TNF, IL-1β, and IL-8 in the lungs and IL-8 in the kidneys. The load of IL-6 was reduced by 65 to 87% by anti-CD14 in all of the organs. The cytokines in the spleen, which was the most efficient organ in producing cytokines per weight unit, were overall most extensively reduced by anti-CD14, with a reduction from 63 to 82% for all four cytokines.
Table 1.
Contribution of organs to cytokine load and effect of anti-CD14a
| Cytokine | Organ | Cytokine load (pg/mg of tissue) | Total cytokine loadb (ng) | % contribution of each organ to the total loadc | % change in load per organ by anti-CD14 |
|---|---|---|---|---|---|
| TNF | Lungs | 10 | 1,733 | 33 | 19 |
| Liver | 5.8 | 2,761 | 53 | –23 | |
| Spleen | 15 | 435 | 8.4 | –69 | |
| Kidneys | 3.0 | 245 | 4.7 | –58 | |
| IL-1β | Lungs | 325 | 53,187 | 29 | 8.8 |
| Liver | 230 | 109,067 | 59 | –48 | |
| Spleen | 633 | 18,046 | 9.8 | –73 | |
| Kidneys | 37 | 3,121 | 1.7 | –70 | |
| IL-6 | Lungs | 2.3 | 376 | 9.9 | –65 |
| Liver | 6.4 | 3,014 | 79 | –87 | |
| Spleen | 10 | 284 | 7.5 | –82 | |
| Kidneys | 1.5 | 124 | 3.3 | –70 | |
| IL-8 | Lungs | 2.7 | 439 | 36 | 30 |
| Liver | 1.0 | 450 | 37 | –75 | |
| Spleen | 4.8 | 137 | 11 | –62 | |
| Kidneys | 2.1 | 170 | 14 | 27 |
Four organs were included (lungs, liver, spleen, and kidneys).
Organ size was calculated from the mean weight of organs in 15-kg pigs (50).
That is, of the four organs.
Adhesion molecules measured by qPCR.
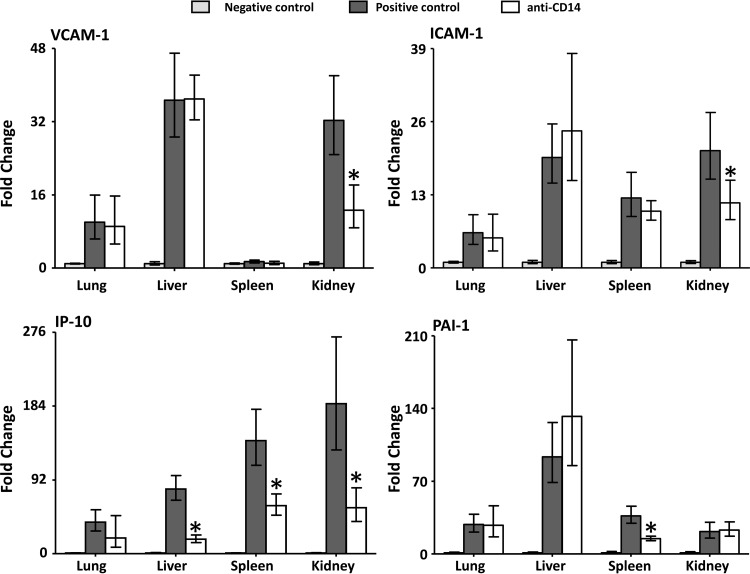
VCAM-1 mRNA was upregulated in all organs except for the spleen and ICAM-1 in all organs (Fig. 5). The fold change for both was highest in the liver and kidney. Anti-CD14 significantly (P < 0.05) reduced the upregulation for both VCAM-1 and ICAM-1 in the kidney, in contrast to the other organs, where no effect was seen. E-selectin was substantially upregulated in all organs (data not shown) with no significant effect of anti-CD14.
Fig 5.
Effect of anti-CD14 on the mRNA induction of the adhesion molecules IP-10 and PAI-1. Fold change in the mRNA expression of the adhesion molecules VCAM-1 and ICAM-1, the chemokine IP-10, and the inhibitor of fibrinolysis, PAI-1, in the negative control group, the positive control group, and the anti-CD14 group. The anti-CD14 group is statistically compared to the positive control group. The data are presented as means and 95% confidence intervals. *, P < 0.05.
Selected cytokines measured by qPCR.
IP-10 mRNA was upregulated in all organs (Fig. 5). The fold change in upregulation was highest in the spleen and kidney. Anti-CD14 significantly (P < 0.05) reduced the upregulation in the liver, spleen, and kidney, in contrast to the lung, where the difference was not significant. IL-1β mRNA was upregulated in all organs (data not shown). Anti-CD14 had no significant effect on this upregulation. IL-10 mRNA was upregulated in the spleen and largely unchanged in the other organs (data not shown). Anti-CD14 reduced the upregulation in the spleen nonsignificantly.
Hemostatic marker (PAI-1) measured by qPCR.
PAI-1 mRNA was upregulated in all organs (Fig. 5). The fold change was highest in the liver. Anti-CD14 significantly reduced (P < 0.05) the upregulation only in the spleen.
DISCUSSION
This study documents the organ distribution of bacteria and bacterial LPS and the subsequent differential local inflammatory responses in a pig model of E. coli sepsis. This inflammatory response was inhibited in an organ-dependent manner by anti-CD14. The data indicate an organ-specific accumulation of the bacteria and LPS to the lungs, with very low levels in other organs. However, the inflammatory response in the organs, measured as the cytokine load, did not reflect the skewed E. coli distribution. Detailed knowledge on how the different organs respond to systemic inflammation in vivo, beyond the information gained by blood examination, is important for our understanding of the nature of systemic inflammation and is required for future mediator-directed therapy in sepsis. Importantly, large-animal models, such as the one described here, are needed to better mimic the situation in humans.
In order to compare the E. coli whole-bacterium group, which we consider a clinically more relevant sepsis model than the frequently used endotoxin (LPS) model, we included an LPS group for comparison. The LPS group showed by far the same trends as the E. coli group. We used a laboratory bacterial strain. Although an isolate of a human E. coli strain from a sepsis patient would have been clinically more relevant, the currently used strain works well to induce septic shock in the pigs. Since Gram-negative sepsis with whole bacteria was the main focus here, our discussion below focuses on the E. coli group.
LPS from the E. coli bacteria accumulated in the lungs of the pigs and was detected only in very low quantities in other organs. E. coli DNA was only detected in the lungs. Compared to the organ biopsy specimens, including the lungs, high detectable levels of LPS and moderate levels of E. coli DNA were found in blood in these animals (24). The lungs have a special position in the immune system in pigs, in particular since they harbor intravascular macrophages similar to liver Kupffer cells in humans (26). It seems that the bacteria were trapped in the lungs throughout the experiment since the tissue samples were obtained after a 4-h observation time. Interestingly, treatment with anti-CD14 did not have any significant effect on the E. coli or LPS levels in the lungs. This finding is in accordance with a previous study showing no effect of the TLR4 pathway in the elimination of bacteria in E. coli-induced pneumonia in mice (27). No effect of anti-CD14 inhibition was seen on E. coli or the LPS levels in the same animals in blood (24). Of interest, anti-CD14 significantly increased the LPS levels in the liver and spleen. One might speculate that the saturation of CD14 molecules by anti-CD14 leads to increased levels of free, and thus measurable, LPS in these organs. However, the LPS levels in the spleen and liver were very low, so the importance of this finding is uncertain.
The cytokine load showed striking differences between the organs. Some of the differences observed between control and treatment groups, although statistically significant, were small, and these should thus be interpreted with caution regarding biological significance. The huge LPS load in the lungs did not lead to an overwhelming increase in proinflammatory cytokines per mg of tissue compared to the other organs. Two studies have shown that already stimulated alveolar macrophages are hyporesponsive to further LPS stimuli in terms of cytokine production (28, 29). Thus, an overwhelming LPS load from the bacteria, as seen in our study, could thus be an explanation for the moderate cytokine production. Of the other organs, the spleen, in particular, had significantly higher levels of IL-1β, IL-6, and IL-8 per mg of tissue compared to the lungs. Even in the kidneys where LPS could not be detected, IL-6 and IL-8 reached approximately the same levels as in the lungs per mg of tissue. The spleen showed the highest levels of all measured cytokines compared to the other organs. Interestingly, the load of all cytokines was reduced by 60 to 80% with anti-CD14 in the spleen, which overall was by far the most efficient inhibition. In a pig model study wherein E. coli LPS was infused in an experimental period from 60 to 240 min, the authors also found a differential cytokine pattern in various organs (30). The spleen was not found to have the same prominent cytokine content as in our study. There could be several reasons for this in terms of the experimental design, the sizes of the pigs, the types of LPS, and the fact that we used a more clinically relevant whole-bacterium model and not only LPS as an inductor of inflammation.
The concept of “compartmentalization,” referring to different pathophysiological events in different organs (compartments) of the inflammatory response in sepsis, is well established (31). Gene profiles in sepsis, for instance, may vary substantially between organs, as shown in a CLP model in rats (32). In the CLP-rat model, some gene clusters were upregulated in the liver and downregulated in the spleen and vice versa. There are also indications that downstream intracellular signaling of common inflammatory pathways may differ from organ to organ, e.g., NF-κB activation by E. coli LPS was dependent on TNF and IL-1β receptors in the liver but not in the lungs in a mouse model (33). These findings support our data showing that the cytokine load varies substantially between the organs in sepsis. Basic knowledge of these inflammatory differences between the organs leads to a better understanding of the inflammatory process and potentially to a more selective, organ-targeted therapy in sepsis.
IL-1β, which seems to be more CD14 dependent in pigs than in humans (34, 35), was significantly inhibited in all organs except for the lungs. IL-1β is one of the principle products of caspase-1 of the inflammasome (36). It is known that the CD14/TLR4-complex “primes” inflammasomes upon activation (36), but it has also been shown that caspase-1 is downregulated and inert to inhibition in Gram-negative sepsis (37). We found a profound inhibition of IL-1β on the protein level by anti-CD14, but no inhibition of the mRNA upregulation. These data suggest that the inhibition of IL-1β by anti-CD14 is due to inhibition of the processing of pro-IL-1β to its active form by the inflammasome and not the pretranslational mRNA upregulation. It seemed that the lungs were in a special position versus the other organs in regard to the missing effect of anti-CD14 inhibition. The overwhelming inflammatory response caused by rapid accumulation of the bacteria might have blurred an effect of the treatment and thereby visible inhibition.
IL-6 was significantly inhibited in all four organs by anti-CD14. This is consistent with our previous finding in blood (24). Induction of IL-6 in Gram-negative sepsis seems to be closely tied to CD14 in several species, including primates (38). Reduction of induction is seen both when the receptor is inhibited by, for instance, an antibody (39), or by short interfering RNA (siRNA) targeting the CD14 gene (40). Human whole blood challenged in vitro with Gram-negative bacteria revealed IL-6 as one of the most CD14-dependent proinflammatory cytokines (35), indicating a species similarity between human and pigs with respect to CD14-dependent IL-6 production.
IL-8 is in general more dependent on complement, the other main inflammation-inducing system of innate immunity, in vitro both in humans and in pigs (34, 35). However, in vivo in pigs anti-CD14 abolished the IL-8 response in blood (24). We found in the present study a significant reduction of IL-8 in the spleen but not in the other organs. It seems that the inhibition of CD14 in vivo has a good effect on blood cells (39), which are indeed abundantly present in the spleen. The same pattern of inhibition was found for TNF and PAI-1. Thus, it seems that these mediators first and foremost are inhibited where blood cells are abundant.
Interestingly, IP-10 was virtually abolished by anti-CD14 on the mRNA level in all of the organs except the lungs. IP-10 is a multifunctional chemokine and is produced by a range of cells, including endothelial cells and T cells. It has various biological functions, such as the chemoattraction of T cells, the promotion of T-cell adhesion to endothelial cells, angiogenesis, and antitumor effects (41). We have previously shown that IP-10 was inhibited significantly by anti-CD14 in vitro in whole blood in humans (39) and show here for the first time that the same striking effect is seen in vivo in organs in pigs. IP-10 is also important in sterile inflammation, i.e., the rejection of liver grafts (42, 43). Importantly, because of its profound effect on T cells, IP-10 seems to be a link in the inflammatory network between innate and adaptive immunity.
Adhesion molecules are found on the endothelium and on white blood cells. These molecules direct the immune cells to the site of inflammation by the process of rolling, activation, adhesion, and transmigration (44). The selectins are essential in the initial rolling (45). We found that E-selectin, which is known to be upregulated on inflamed endothelium, was upregulated in all of the organs examined. The upregulation was not inhibited by anti-CD14. In an ex vivo xenotransplantation model, upregulation of E-selectin was found to be highly and almost completely complement dependent (46). Furthermore, in an E. coli sepsis model in baboons, endothelial cell integrity was preserved by inhibition of complement (47). Consequently, a combined treatment with both CD14 and complement could be rational in sepsis to attenuate the inflammatory responses initiated by innate immune danger signaling, as we have proposed previously (48). VCAM-1 and ICAM-1 are also expressed on inflamed endothelium and contribute to rolling and firm adhesion of the white cells. We found that VCAM-1 and ICAM-1 were highly upregulated in the liver and kidneys, only moderately upregulated in the lungs and, with regard to VCAM-1, not at all upregulated in the spleen. It is known that there are some differences between the organs in the way the leukocyte adhesion cascade is organized. For instance, the leukocyte adhesion cascade differs in particular in respect to the role of the selectins in liver sinusoids compared to other tissues (49). Tissue differences such as these probably also explain the difference in effect of anti-CD14 inhibition, which efficiently inhibited both VCAM-1 and ICAM-1 in the kidneys, but not in the liver, despite similarly increased expression of the molecules in these organs.
In conclusion, we have demonstrated here how the systemic inflammatory response makes its footprint in the organs in sepsis and how anti-CD14 has an inhibitory effect on these responses. Detailed knowledge on how the different organs respond to systemic inflammation in vivo, beyond the information gained by blood examination, is important for our understanding of the nature of systemic inflammation and is required for future mediator-directed therapy in sepsis. The inhibition of CD14 seems to be a good candidate for such a treatment regimen.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by Helse Sør-Øst, by Vestre Viken HF, by the Research Council of Norway, and by The Odd Fellow Foundation.
We thank Kjersti Wendt, Hilde Nilsen, and Carmen Louwerens for excellent technical assistance and Dorte Christiansen for growing and preparing the bacteria.
Footnotes
Published ahead of print 17 June 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.00390-13.
REFERENCES
- 1.Dellinger RP, Levy MM, Carlet JM, Bion J, Parker MM, Jaeschke R, Reinhart K, Angus DC, Brun-Buisson C, Beale R, Calandra T, Dhainaut JF, Gerlach H, Harvey M, Marini JJ, Marshall J, Ranieri M, Ramsay G, Sevransky J, Thompson BT, Townsend S, Vender JS, Zimmerman JL, Vincent JL. 2008. Surviving sepsis campaign: international guidelines for management of severe sepsis and septic shock: 2008. Crit. Care Med. 36:296–327 [DOI] [PubMed] [Google Scholar]
- 2.Sriskandan S, Altmann DM. 2008. The immunology of sepsis. J. Pathol. 214:211–223 [DOI] [PubMed] [Google Scholar]
- 3.Barton GM. 2008. A calculated response: control of inflammation by the innate immune system. J. Clin. Invest. 118:413–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Medzhitov R. 2007. Recognition of microorganisms and activation of the immune response. Nature 449:819–826 [DOI] [PubMed] [Google Scholar]
- 5.Chen GY, Nunez G. 2010. Sterile inflammation: sensing and reacting to damage. Nat. Rev. Immunol. 10:826–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kumar H, Kawai T, Akira S. 2009. Toll-like receptors and innate immunity. Biochem. Biophys. Res. Commun. 388:621–625 [DOI] [PubMed] [Google Scholar]
- 7.de Jong HK, van der Poll T, Wiersinga WJ. 2010. The systemic proinflammatory response in sepsis. J. Innate Immun. 2:422–430 [DOI] [PubMed] [Google Scholar]
- 8.Janeway CA, Jr, Medzhitov R. 2002. Innate immune recognition. Annu. Rev. Immunol. 20:197–216 [DOI] [PubMed] [Google Scholar]
- 9.Medzhitov R. 2009. Approaching the asymptote: 20 years later. Immunity 30:766–775 [DOI] [PubMed] [Google Scholar]
- 10.Castellheim A, Brekke OL, Espevik T, Harboe M, Mollnes TE. 2009. Innate immune responses to danger signals in systemic inflammatory response syndrome and sepsis. Scand. J. Immunol. 69:479–491 [DOI] [PubMed] [Google Scholar]
- 11.Takeda K, Akira S. 2007. Toll-like receptors. Curr. Protoc. Immunol. Chapter 14:14.12.1–14.12.13 [DOI] [PubMed] [Google Scholar]
- 12.Jiang Z, Georgel P, Du X, Shamel L, Sovath S, Mudd S, Huber M, Kalis C, Keck S, Galanos C, Freudenberg M, Beutler B. 2005. CD14 is required for MyD88-independent LPS signaling. Nat. Immunol. 6:565–570 [DOI] [PubMed] [Google Scholar]
- 13.Shimazu R, Akashi S, Ogata H, Nagai Y, Fukudome K, Miyake K, Kimoto M. 1999. MD-2, a molecule that confers lipopolysaccharide responsiveness on Toll-like receptor 4. J. Exp. Med. 189:1777–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee CC, Avalos AM, Ploegh HL. 2012. Accessory molecules for Toll-like receptors and their function. Nat. Rev. Immunol. 12:168–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anas AA, Hovius JWR, der Poll T, de Vos AF. 2010. Role of CD14 in a mouse model of acute lung inflammation induced by different lipopolysaccharide chemotypes. PLoS One 5:e10183. 10.1371/journal.pone.0010183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McAvoy EF, McDonald B, Parsons SA, Wong CH, Landmann R, Kubes P. 2011. The role of CD14 in neutrophil recruitment within the liver microcirculation during endotoxemia. J. Immunol. 186:2592–2601 [DOI] [PubMed] [Google Scholar]
- 17.Zanoni I, Ostuni R, Marek LR, Barresi S, Barbalat R, Barton GM, Granucci F, Kagan JC. 2011. CD14 controls the LPS-induced endocytosis of Toll-like receptor 4. Cell 147:868–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ulloa L, Tracey KJ. 2005. The “cytokine profile”: a code for sepsis. Trends Mol. Med. 11:56–63 [DOI] [PubMed] [Google Scholar]
- 19.Adib-Conquy M, Cavaillon JM. 2007. Stress molecules in sepsis and systemic inflammatory response syndrome. FEBS Lett. 581:3723–3733 [DOI] [PubMed] [Google Scholar]
- 20.Hunter JD, Doddi M. 2010. Sepsis and the heart. Br. J. Anaesth. 104:3–11 [DOI] [PubMed] [Google Scholar]
- 21.Kortgen A, Paxian M, Werth M, Recknagel P, Rauchfuss F, Lupp A, Krenn CG, Muller D, Claus RA, Reinhart K, Settmacher U, Bauer M. 2009. Prospective assessment of hepatic function and mechanisms of dysfunction in the critically ill. Shock 32:358–365 [DOI] [PubMed] [Google Scholar]
- 22.Schouten M, Wiersinga WJ, Levi M, van der Poll T. 2008. Inflammation, endothelium, and coagulation in sepsis. J. Leukoc. Biol. 83:536–545 [DOI] [PubMed] [Google Scholar]
- 23.Seok J, Warren HS, Cuenca AG, Mindrinos MN, Baker HV, Xu W, Richards DR, Donald-Smith GP, Gao H, Hennessy L, Finnerty CC, Lopez CM, Honari S, Moore EE, Minei JP, Cuschieri J, Bankey PE, Johnson JL, Sperry J, Nathens AB, Billiar TR, West MA, Jeschke MG, Klein MB, Gamelli RL, Gibran NS, Brownstein BH, Miller-Graziano C, Calvano SE, Mason PH, Cobb JP, Rahme LG, Lowry SF, Maier RV, Moldawer LL, Herndon DN, Davis RW, Xiao W, Tompkins RG. 2013. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc. Natl. Acad. Sci. U. S. A. 110:3507–3512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thorgersen EB, Hellerud BC, Nielsen EW, Barratt-Due A, Fure H, Lindstad JK, Pharo A, Fosse E, Tonnessen TI, Johansen HT, Castellheim A, Mollnes TE. 2010. CD14 inhibition efficiently attenuates early inflammatory and hemostatic responses in Escherichia coli sepsis in pigs. FASEB J. 24:712–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Castellheim A, Thorgersen EB, Hellerud BC, Pharo A, Johansen HT, Brosstad F, Gaustad P, Brun H, Fosse E, Tonnessen TI, Nielsen EW, Mollnes TE. 2008. New biomarkers in an acute model of live Escherichia coli-induced sepsis in pigs. Scand. J. Immunol. 68:75–84 [DOI] [PubMed] [Google Scholar]
- 26.Winkler GC. 1988. Pulmonary intravascular macrophages in domestic animal species: review of structural and functional properties. Am. J. Anat. 181:217–234 [DOI] [PubMed] [Google Scholar]
- 27.Lee JS, Frevert CW, Matute-Bello G, Wurfel MM, Wong VA, Lin SM, Ruzinski J, Mongovin S, Goodman RB, Martin TR. 2005. TLR-4 pathway mediates the inflammatory response but not bacterial elimination in Escherichia coli pneumonia. Am. J. Physiol. Lung Cell. Mol. Physiol. 289:L731–L738 [DOI] [PubMed] [Google Scholar]
- 28.Ayala A, Perrin MM, Kisala JM, Ertel W, Chaudry IH. 1992. Polymicrobial sepsis selectively activates peritoneal but not alveolar macrophages to release inflammatory mediators (interleukins-1 and -6 and tumor necrosis factor). Circ. Shock 36:191–199 [PubMed] [Google Scholar]
- 29.Dehoux MS, Boutten A, Ostinelli J, Seta N, Dombret MC, Crestani B, Deschenes M, Trouillet JL, Aubier M. 1994. Compartmentalized cytokine production within the human lung in unilateral pneumonia. Am. J. Respir. Crit. Care Med. 150:710–716 [DOI] [PubMed] [Google Scholar]
- 30.Brix-Christensen V, Gjedsted J, Andersen SK, Vestergaard C, Nielsen J, Rix T, Nyboe R, Andersen NT, Larsson A, Schmitz O, Tonnesen E. 2005. Inflammatory response during hyperglycemia and hyperinsulinemia in a porcine endotoxemic model: the contribution of essential organs. Acta Anaesthesiol. Scand. 49:991–998 [DOI] [PubMed] [Google Scholar]
- 31.Cavaillon JM, Annane D. 2006. Compartmentalization of the inflammatory response in sepsis and SIRS. J. Endotoxin Res. 12:151–170 [DOI] [PubMed] [Google Scholar]
- 32.Chinnaiyan AM, Huber-Lang M, Kumar-Sinha C, Barrette TR, Shankar-Sinha S, Sarma VJ, Padgaonkar VA, Ward PA. 2001. Molecular signatures of sepsis: multiorgan gene expression profiles of systemic inflammation. Am. J. Pathol. 159:1199–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koay MA, Christman JW, Wudel LJ, Allos T, Cheng DS, Chapman WC, Blackwell TS. 2002. Modulation of endotoxin-induced NF-κB activation in lung and liver through TNF type 1 and IL-1 receptors. Am. J. Physiol. Lung Cell. Mol. Physiol. 283:L1247–L1254 [DOI] [PubMed] [Google Scholar]
- 34.Thorgersen EB, Pharo A, Haverson K, Axelsen AK, Gaustad P, Kotwal GJ, Sfyroera G, Mollnes TE. 2008. Complement- and CD14-inhibition attenuate Escherichia coli-induced inflammatory response in porcine whole blood. Infect. Immun. 77:725–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lappegard KT, Christiansen D, Pharo A, Thorgersen EB, Hellerud BC, Lindstad J, Nielsen EW, Bergseth G, Fadnes D, Abrahamsen TG, Hoiby EA, Schejbel L, Garred P, Lambris JD, Harboe M, Mollnes TE. 2009. Human genetic deficiencies reveal the roles of complement in the inflammatory network: lessons from nature. Proc. Natl. Acad. Sci. U. S. A. 106:15861–15866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cinel I, Opal SM. 2009. Molecular biology of inflammation and sepsis: a primer. Crit. Care Med. 37:291–304 [DOI] [PubMed] [Google Scholar]
- 37.Giamarellos-Bourboulis EJ, van d, Mouktaroudi VM, Raftogiannis M, Antonopoulou A, Joosten LA, Pickkers P, Savva A, Georgitsi M, van der Meer JW, Netea MG. 2011. Inhibition of caspase-1 activation in Gram-negative sepsis and experimental endotoxemia. Crit. Care 15:R27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leturcq DJ, Moriarty AM, Talbott G, Winn RK, Martin TR, Ulevitch RJ. 1996. Antibodies against CD14 protect primates from endotoxin-induced shock. J. Clin. Invest. 98:1533–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brekke OL, Christiansen D, Fure H, Pharo A, Fung M, Riesenfeld J, Mollnes TE. 2008. Combined inhibition of complement and CD14 abolish Escherichia coli-induced cytokine-, chemokine-, and growth factor-synthesis in human whole blood. Mol. Immunol. 45:3804–3813 [DOI] [PubMed] [Google Scholar]
- 40.Lei M, Jiao H, Liu T, Du L, Cheng Y, Zhang D, Hao Y, Man C, Wang F. 2011. siRNA targeting mCD14 inhibits TNF-α, MIP-2, and IL-6 secretion and NO production from LPS-induced RAW264.7 cells. Appl. Microbiol. Biotechnol. 92:115–124 [DOI] [PubMed] [Google Scholar]
- 41.Angiolillo AL, Sgadari C, Taub DD, Liao F, Farber JM, Maheshwari S, Kleinman HK, Reaman GH, Tosato G. 1995. Human interferon-inducible protein 10 is a potent inhibitor of angiogenesis in vivo. J. Exp. Med. 182:155–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Waelgaard L, Thorgersen EB, Line PD, Foss A, Mollnes TE, Tonnessen TI. 2008. Microdialysis monitoring of liver grafts by metabolic parameters, cytokine production, and complement activation. Transplantation 86:1096–1103 [DOI] [PubMed] [Google Scholar]
- 43.Haugaa H, Thorgersen EB, Pharo A, Boberg KM, Foss A, Line PD, Sanengen T, Almaas R, Grindheim G, Waelgaard L, Pischke SE, Mollnes TE, Tonnessen TI. 2012. Inflammatory markers sampled by microdialysis catheters discriminate rejection from ischemia in liver grafts. Liver Transpl. 18:1421–1429 [DOI] [PubMed] [Google Scholar]
- 44.Ley K, Laudanna C, Cybulsky MI, Nourshargh S. 2007. Getting to the site of inflammation: the leukocyte adhesion cascade updated. Nat. Rev. Immunol. 7:678–689 [DOI] [PubMed] [Google Scholar]
- 45.Kansas GS. 1996. Selectins and their ligands: current concepts and controversies. Blood 88:3259–3287 [PubMed] [Google Scholar]
- 46.Saethre M, Schneider MK, Lambris JD, Magotti P, Haraldsen G, Seebach JD, Mollnes TE. 2008. Cytokine secretion depends on Galα(1,3)Gal expression in a pig-to-human whole blood model. J. Immunol. 180:6346–6353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Silasi-Mansat R, Zhu H, Popescu NI, Peer G, Sfyroera G, Magotti P, Ivanciu L, Lupu C, Mollnes TE, Taylor FB, Kinasewitz G, Lambris JD, Lupu F. 2010. Complement inhibition decreases the procoagulant response and confers organ protection in a baboon model of Escherichia coli sepsis. Blood 116:1002–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mollnes TE, Christiansen D, Brekke OL, Espevik T. 2008. Hypothesis: combined inhibition of complement and CD14 as treatment regimen to attenuate the inflammatory response. Adv. Exp. Med. Biol. 632:253–263 [PubMed] [Google Scholar]
- 49.Liu L, Kubes P. 2003. Molecular mechanisms of leukocyte recruitment: organ-specific mechanisms of action. Thromb. Haemost. 89:213–220 [PubMed] [Google Scholar]
- 50.Swindle MM. 2007. Swine in the laboratory: surgery, anesthesia, imaging, and experimental techniques. CRC Press/Taylor & Francis Group, Boca Raton, FL [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.