Abstract
Moraxella catarrhalis is a human respiratory tract pathogen that causes otitis media in children and lower respiratory tract infections in adults with chronic obstructive pulmonary disease. We have identified and characterized a zinc uptake ABC transporter that is present in all strains of M. catarrhalis tested. A mutant in which the znu gene cluster is knocked out shows markedly impaired growth compared to the wild type in medium that contains trace zinc; growth is restored to wild-type levels by supplementing medium with zinc but not with other divalent cations. Thermal-shift assays showed that the purified recombinant substrate binding protein ZnuA binds zinc but does not bind other divalent cations. Invasion assays with human respiratory epithelial cells demonstrated that the zinc ABC transporter of M. catarrhalis is critical for invasion of respiratory epithelial cells, an observation that is especially relevant because an intracellular reservoir of M. catarrhalis is present in the human respiratory tract and this reservoir is important for persistence. The znu knockout mutant showed marked impairment in its capacity to persist in the respiratory tract compared to the wild type in a mouse pulmonary clearance model. We conclude that the zinc uptake ABC transporter mediates uptake of zinc in environments with very low zinc concentrations and is critical for full virulence of M. catarrhalis in the respiratory tract in facilitating intracellular invasion of epithelial cells and persistence in the respiratory tract.
INTRODUCTION
Moraxella catarrhalis is a Gram-negative diplococcus that is an exclusively human pathogen whose ecological niche is the human respiratory tract. In addition to being a common commensal in the upper respiratory tract of children, M. catarrhalis is the third-most-common cause of otitis media after nontypeable Haemophilus influenzae and Streptococcus pneumoniae (1). Otitis media is the most common reason for children to receive antibiotics, with 70% experiencing at least one episode by age 3 (2–6). M. catarrhalis is associated with up to 25% of cases of acute otitis media as a copathogen or sole pathogen and with 46.4% of chronic middle ear effusions as determined by PCR (1, 7). The prevalence of nasopharyngeal colonization by M. catarrhalis in children is high (up to 75%), and the frequency of colonization is associated with the development of otitis media (8–11). Furthermore, the rate of colonization may be increasing in regions where pneumococcal conjugate vaccines are in widespread use (12).
M. catarrhalis causes approximately 10% of exacerbations of chronic obstructive pulmonary disease (COPD), accounting for 2 million to 4 million episodes annually in the United States (13, 14). COPD is a chronic debilitating disease with a huge global burden and is the third-most-common cause of death in the United States (15, 16). The course of the disease is characterized by intermittent exacerbations, most caused by infection, which are associated with enormous morbidity.
Given the importance of M. catarrhalis as a human pathogen, there is strong interest in understanding mechanisms of pathogenesis, which can lead to the development of interventions to better treat and prevent infections both in the setting of otitis media in children and in adults with COPD (17–20). During the course of work on a genome mining approach to identify candidate vaccine antigens (21–23), we identified a gene cluster in the M. catarrhalis genome with characteristics of an ATP binding cassette (ABC) transporter that has homology to zinc uptake systems. Zinc is essential for all living cells, playing catalytic or structural roles in many enzymes. Indeed, pathways that mediate efficient uptake of zinc are important for bacteria to survive and multiply in the host where zinc is not freely available (24–30).
In this study, we demonstrated that the znu gene cluster (zinc ABC transporter) is critical for growth of M. catarrhalis when zinc is present in trace concentrations. We further demonstrated that the zinc ABC transporter plays a key role in intracellular survival of M. catarrhalis in respiratory epithelial cells and in persistence in the murine respiratory tract. These observations are consistent with the concept that the zinc ABC transporter is a “nutritional” virulence factor for M. catarrhalis infection of the respiratory tract.
MATERIALS AND METHODS
Bacterial strains and plasmids.
M. catarrhalis strain O35E, provided by Eric Hansen, is a prototype otitis media strain that was isolated from middle ear fluid from a child with otitis media in Dallas, TX. M. catarrhalis strains 135, 238, 5488, 7169, and 9483, provided by Diane Dryja and Howard Faden, were isolated from middle ear fluid of children with otitis media in Buffalo, NY. M. catarrhalis strains 0601040VIR, 0701057VIL, 0701064V3L, 0702076SV4R, and 0801070VIL, provided by Diana Adlowitz and Michael Pichichero, were isolated from middle ear fluid of children with otitis media in Rochester, NY. M. catarrhalis strains 1P18B1, 6P29B1, 10P66B1, 19P54B1, and 47P31B1 were isolated from the sputum of adults with COPD experiencing an exacerbation as part of a prospective study in Buffalo (14, 31). M. catarrhalis strains M2, M3, M4, M5, and M6, provided by Daniel Musher, were isolated from the sputum of adults with COPD experiencing an exacerbation. Escherichia coli strains TOP10 and BL21(DE3) were purchased from Invitrogen.
Plasmid pET101D-TOPO was purchased from Invitrogen. Plasmid pWW115 was a gift of Wei Wang and Eric Hansen (32). Plasmid pUCK18K was a gift of Anthony Campagnari.
A549 cells (CC685) were purchased from the American Type Culture Collection.
Construction of mutants.
Two mutants were constructed in M. catarrhalis strain O35E in which the znuA gene was knocked out and in which the entire znu gene cluster was knocked out (see Fig. 1) by using overlap extension PCR (33) and homologous recombination as described previously (23, 33). Briefly, the transforming DNA for the znuA mutant was composed of 3 overlapping fragments that included 1 kb upstream of znuA (fragment 1), the nonpolar kanamycin resistance cassette amplified from plasmid pUCK18K (34) (fragment 2), and 1 kb downstream of znuA (fragment 3) using the oligonucleotide primers listed in Table 1. The znuA mutant was constructed by transformation of strain O35E with a fragment composed of fragments 1, 2, and 3 and selection on brain heart infusion (BHI) plates containing 40 μg/ml of kanamycin. Mutant colonies were examined by PCR as described in Results.
Fig 1.
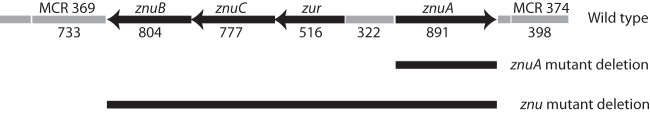
Diagram of the znu gene cluster in the wild-type strain and the deleted segments in znuA and znu knockout mutants. Numbers denote base pairs. Black arrows show open reading frames of the znu gene cluster. The predicted open reading frames surrounding the znu gene cluster are shown.
Table 1.
Oligonucleotide primer sequences
| Primer | Gene or operon | Expt | Direction | Sequence |
|---|---|---|---|---|
| ZnuAfrag1-5′ | znuA | Mutant construction | Forward | TAATAAGCCCAAGATAAGC |
| ZnuA frag1-3′ | znuA | Mutant construction | Reverse | TAGTTAGTCATGATAAGCACCCTGATGTCT |
| ZnuA frag2-5′ | znuA | Mutant construction | Forward | GTGCTTATCATGACTAACTAGGAGGAATA |
| ZnuA frag2 3′ | znuA | Mutant construction | Reverse | GAAGCAAGAACATTATTCCCTCCAGGTACT |
| ZnuA frag3-5′ | znuA | Mutant construction | Forward | GGGAATAATGTTCTTGCTTCGTGTCAGATG |
| ZnuA frag3-3′ | znuA | Mutant construction | Reverse | AGAAAAATTGGTGATAAACG |
| ZnuA F1 | znuA | PCR | Forward | ATGACAACATTTGCATTACG |
| ZnuA R1 | znuA | PCR | Reverse | TTATTTGATGCCAGCACAT |
| Zinc frag1-5′ | znu operon | Mutant construction | Forward | ATATAGTCTTCGCCTTCAG |
| Zinc frag1-3′ | znu operon | Mutant construction | Reverse | TAGTTAGTCACATCACCAACAACCATACCT |
| Zinc frag2-5′ | znu operon | Mutant construction | Forward | GTTGGTGATGTGACTAACTAGGAGGAATAA |
| Zinc frag2-3′ | znu operon | Mutant construction | Reverse | GAAGCAAGAACATTATTCCCTCCAGGTACT |
| Zinc frag3-5′ | znu operon | Mutant construction | Forward | GGGAATAATGTTCTTGCTTCGTGTCAGATG |
| Zinc frag3-3′ | znu operon | Mutant construction | Reverse | AGAAAAATTGGTGATAAACG |
| Zinc F1 | znu operon | PCR | Forward | TTAGGCATATTTGCTTAGGA |
| Zinc R1 | znu operon | PCR | Reverse | TATTTGATGCCAGCACAT |
| mature znuA F1 | znuA | Cloning | Forward | CACCGGCATGGTCAGCGTTAGTAA |
| mature znuA R1 | znuA | Cloning | Reverse | TTATTTGATGCCAGCACAT |
A mutant in which the entire 3,282-bp znu gene cluster was knocked out was constructed by the same method using the primers noted in Table 1. The transforming DNA was composed of 3 overlapping fragments that included 1 kb upstream of znuB, the nonpolar kanamycin resistance cassette amplified from plasmid pUCK18K (34), and 1 kb downstream of znuA. The insert and surrounding sequences of both the znuA and znu knockout mutants were confirmed by sequence analysis.
Construction of znu revertant.
In an effort to complement the znu mutant in trans, the znu gene fragment was cloned into plasmid pWW115 (32) and the znu mutant was transformed and electroporated with the plasmid using a variety of conditions. No transformants were recovered following multiple attempts, likely due to the large size of the plasmid that has an ∼4.4-kb insert.
Therefore, we constructed a revertant strain by introducing the wild-type znu gene cluster into the znu mutant. The znu gene cluster and flanking sequences were amplified by PCR using primers Zinc frag1-5′ and Zinc frag3-3′ (Table 1), the same primers used to construct the mutant. The znu mutant was transformed with the PCR product, and transformants were selected on chocolate agar on which the znu mutant demonstrates a growth defect (see Results). Kanamycin-susceptible revertants were recovered.
Analysis by PCR with specific primers demonstrated that the znu sequence was absent from the mutant and that the kanamycin cassette sequence was present. As expected, using the same primers, PCR confirmed that the znu gene cluster was restored in the znu revertant and the kanamycin cassette was absent. The znu revertant was susceptible to kanamycin, consistent with replacement of the kanamycin cassette. These constructs were confirmed by sequence analysis.
Cloning of the znuA gene.
To express recombinant ZnuA, the 784-bp gene encoding the mature ZnuA protein was amplified by PCR from genomic DNA of strain O35E using primers noted in Table 1 and ligated into plasmid pET 101 D-TOPO (Invitrogen). The ligation mixture was transformed into the chemically competent E. coli strain Top10 and grown on BHI plates containing 50 μg/ml kanamycin. The expression plasmid was named pZnuA.
Expression and purification of recombinant ZnuA.
Plasmid pZnuA was transformed into E. coli strain BL21(DE3) to express ZnuA with a six-histidine amino-terminal tag. A volume of 100 ml of LB broth containing 300 μg/ml carbenicillin was inoculated with 5 ml overnight culture of bacteria containing the expression vector. Following growth to an optical density at 600 nm (OD600) of O.6 to 0.8, ZnuA expression was induced with 4 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 4 h at 37°C. The bacteria were then harvested by centrifugation at 4,000 × g for 15 min at 4°C. The pellet was suspended in 10 ml of lysis buffer (20 mM sodium phosphate, 500 mM NaCl, 1 mg/ml lysozyme, 1× Protease Arrest [G Biosciences] [pH 7.4]) and mixed by nutation for 30 min at 4°C. The suspension was then sonicated with a Branson Sonifier 450 at setting 5, using an 80% pulsed cycle of four 30-s bursts with 2-min pauses. The sonicated bacterial lysate was centrifuged 10,000 × g for 20 min at 4°C. The pellet, which contained ZnuA, was suspended in binding buffer (20 mM sodium phosphate, 500 mM NaCl, 6 M guanidinium chloride, pH 7.4).
Recombinant ZnuA was purified with Talon metal affinity resin (BD Biosciences, Palo Alto, CA) according to the manufacturer's instructions. Two milliliters of a 50% suspension of Talon resin was centrifuged at 2,400 × g for 5 min, and the resin storage buffer was removed with a pipette. The beads were equilibrated with binding buffer and were then incubated with 10 ml of the bacterial lysate pellet suspension for 20 min at room temperature with nutation. The resin with bound protein was centrifuged as described above and washed 2 times with 20 ml of binding buffer, followed by a single wash with phosphate-buffered saline (PBS). Protein was eluted by incubating the resin with 1 ml of elution buffer (PBS plus 150 mM imidazole, pH 7.4) for 10 min with nutation, followed by centrifugation. This elution step was repeated once, and the two eluates were pooled. The concentration of the purified protein was determined using the Lowry assay (Sigma).
Development of antiserum to recombinant ZnuA.
Purified recombinant ZnuA was sent to Covance (Denver, PA) for antibody production in New Zealand White rabbits using a 59-day protocol. Briefly, 250 μg purified ZnuA was emulsified 1:1 in complete Freund's adjuvant for initial subcutaneous immunization. Subsequent immunization followed a 3-week cycle of boosts with 125 μg ZnuA emulsified 1:1 in incomplete Freund's adjuvant. Serum was collected 2 weeks after the second boost.
To remove background antibodies, the antiserum was adsorbed with the ZnuA mutant as follows. A 50-ml late-logarithmic-phase culture of the ZnuA mutant was harvested by centrifugation, washed in PBS, and suspended in 1 ml of a 1:100 dilution of serum. The serum was incubated at 4°C with nutation for 30 min, and bacterial cells were removed by centrifugation at 4,000 × g for 15 min at 4°C. This adsorption was performed twice. The absorbed serum was then filter sterilized using a 0.45-μm filter.
Thermal-shift assay.
Thermal-shift assays were performed using the Stratagene Mx3005P real-time PCR instrument (Stratagene, La Jolla, CA) as previously described (35). Purified ZnuA in PBS was studied at a concentration of 10 μg in a 30-μl volume to which selected divalent cation salts were added to a final concentration of 200 μmol. SYPRO orange was added as a fluorescence reporter at a 1,000-fold dilution from its stock solution. The change in fluorescence was monitored using the Cy3 filter, with excitation-and-emission wavelengths of 545 nM and 568 nM, respectively. The temperature was raised from 25°C to 98°C in 0.5°C intervals over the course of 45 min, with fluorescence readings taken at each interval. The fluorescence data were plotted and normalized, and the first derivative of the curve calculated to provide the melting temperatures (Tm) using the software program GraphPad Prism 5.0 as previously described (35, 36). Results are reported as the averages ± standard deviations of 6 values which were obtained from 3 independent experiments performed in duplicate.
Assessment of bacterial growth.
Growth curves were performed using the Bioscreen C automated growth curve analysis system (Oy Growth Curves AB Ltd., Helsinki, Finland). M. catarrhalis strains were grown in broth overnight with shaking. Overnight cultures were diluted (1:500 in the case of BHI and 1:100 in the case of chemically defined medium [CDM]) and were used to inoculate cultures to perform growth curves in 300-μl volumes with 5 replicate wells for each growth condition in each experiment. Each experiment was performed at least twice. Optical density measurements were taken at 600 nM at 30 min intervals with the Bioscreen C system at 37°C with constant shaking (machine settings: fast speed, high amplitude).
Adherence-and-invasion assays with respiratory epithelial cells.
Quantitative adherence and invasion assays were performed with A549 cells (human type II alveolar lung epithelium; ATCC CCL85) grown in F-12K medium (Gibco) plus 10% fetal bovine serum. Briefly, 2 × 105 A549 cells were seeded into each well of a 24-well tissue culture plate and incubated for ∼48 h when cells showed confluent growth. Cells were inoculated with broth-grown log-phase bacteria (multiplicity of infection = 1), and the plates were centrifuged at 170 × g for 5 min at room temperature to facilitate contact between bacteria and A549 cells. Plates were incubated for 3 h at 37°C. Nonadherent cells were removed by gently washing the wells 3 times with PBS. To quantify adherent cells, 200 μl of trypsin (0.25%) was added to each well and plates were incubated at 37°C for 10 min to remove adherent cells. A 300-μl volume of 1% saponin was applied to each well, and contents were pipetted into microcentrifuge tubes and after vigorous vortexing were plated in duplicate to perform bacterial cell counts. Adherence is measured as CFU per ml. Results of assays with mutants (CFU/ml) are expressed as a percentage of the result with the wild type (CFU/ml) that was determined simultaneously. Each experiment was repeated 3 times.
To measure invasion, gentamicin (100 μg/ml) was added to wells after 3 h of incubation of A549 cells with bacteria. Nonadherent cells were removed by washing, and wells were incubated with gentamicin for 1 h at 37°C. Cells were removed with trypsin and lysed with saponin as described above and then plated in duplicate. Invasion is measured as CFU per ml. Results of assays with mutants (CFU/ml) are expressed as a percentage of the result with the wild type (CFU/ml) that was determined simultaneously. Each experiment was repeated 3 times.
Pulmonary clearance model.
All procedures were performed in compliance with Veterans Affairs Western New York Healthcare System IACUC guidelines. BALB/c mice were challenged simultaneously with the wild type and the znu mutant, and clearance of the strains was assessed. The model has been described previously (22). Briefly, 1 ml (each) of overnight cultures of wild-type M. catarrhalis O35E and the znu mutant were used to inoculate 50 ml of BHI broth cultures which were grown to log phase (OD600 of ∼0.3 to 0.4 or ∼108 CFU/ml). The cultures were harvested by centrifugation, and each was resuspended in 5 ml of phosphate-buffered saline with gelatin, calcium, and magnesium (PBSG) (137 mM NaCl, 2.7 mM KCl, 4.3 mM NaHPO4, 1.4 mM KH2PO4, 0.125 mM CaCl2, 0.5 mM MgCl2, and 0.1% gelatin, pH 7.3). A volume of 5 ml of each culture suspension (total, ∼109 CFU) was placed in the nebulizer of an inhalational exposure system, model 099C A4212 (Glas-Col, Terre Haute, IN). Aliquots of the cultures were serially diluted and plated to determine the starting number of bacteria. The equipment settings were as follows: 10 min of preheating, 40 min of nebulization, 30 min of cloud decay, 10 min of decontamination, vacuum flow meter at 60 cubic feet/h, and compressed air flow meter at 10 cubic feet/h. BALB/c mice (n = 10 per group) were placed in the chamber during this time.
Three hours postchallenge, the mice were euthanized by inhalation of isoflurane. Lungs were harvested, placed in 5 ml PBSG, and homogenized on ice using a tissue homogenizer. Following homogenization, 50 μl of each lung homogenate was plated on BHI agar, and a second aliquot was plated on BHI agar containing 50 μg/ml of kanamycin and incubated at 35°C with 5% CO2 overnight. Colonies were counted the following day to determine the concentration of bacteria in the lungs 3 h after aerosol challenge. The number of colonies on the kanamycin plates was used to calculate the concentration of the znu mutant. The number of colonies on the kanamycin plate was subtracted from the number of colonies on plates with no antibiotic to calculate the concentration of wild-type bacteria in lungs. Statistical significance was determined by performing a two-tailed t test. A P value of ≤0.05 was considered significant.
RESULTS
Identification of the znu gene cluster.
As part of a genome mining approach to screen for genes that encode surface antigens of M. catarrhalis (21), we identified a gene cluster with homology to ABC zinc transporters (Fig. 1). The gene cluster includes znuB, which is predicted to encode a permease protein, znuC, which is predicted to encode an ATPase subunit, a predicted zinc uptake regulator (zur), and znuA, which is predicted to encode a periplasmic substrate binding protein. This predicted zinc ABC transporter belongs to superfamily 3.A.1.15 in the Transporter Classification system (tcdb.org). An open reading frame downstream of znuA (MCR 374) is predicted to encode a hypothetical protein, and an open reading frame downstream of znuB (MCR 369) is predicted to encode a calcineurin-like phosphoesterase of unknown function (Fig. 1).
To assess the presence of the znu gene cluster in clinical isolates of M. catarrhalis, DNA purified from 40 clinical isolates was used as a template in a PCR with primers (Table 1) that were designed to amplify the znu gene cluster. The clinical isolates included 20 middle ear fluid isolates obtained by tympanocentesis from children with acute otitis media and 20 sputum isolates from adults who were experiencing exacerbations of COPD. A band of ∼3,200 bp with a size identical to that of strain O35E was detected in all 40 strains. A negative control in which DNA template was replaced with water showed no band. We conclude that the znu gene cluster is present in the genomes of all clinical isolates of M. catarrhalis tested.
Characterization of recombinant ZnuA and antiserum to ZnuA.
The znuA gene encodes a predicted protein of 259 amino acids with a 37-amino-acid signal peptide at the N terminus. Recombinant ZnuA corresponding to the mature protein was expressed with an N-terminal hexahistidine tag and purified by affinity chromatography. An SDS-PAGE gel, subjected to Coomassie blue and silver staining, showed that purified recombinant ZnuA is a single band of ∼34 kDa as predicted (Fig. 2).
Fig 2.
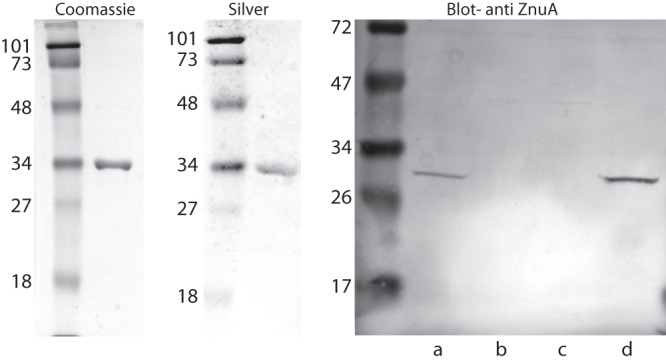
(Left) Coomassie blue-stained sodium dodecyl sulfate-polyacrylamide gel of 10 μg of purified recombinant ZnuA. (Center) Silver-stained gel of 10 μg of purified recombinant ZnuA. (Right) Immunoblot assay probed with adsorbed rabbit antiserum raised to purified recombinant ZnuA (1:1,000). Lanes contain whole bacterial cell lysates of strain O35E: a, wild type; b, znu knockout mutant; c, znuA knockout mutant; d, znu revertant. Molecular mass markers are noted on the left of each panel in kilodaltons (kDa).
Immunoblot assays with absorbed rabbit antiserum to recombinant ZnuA detected purified ZnuA and also detected a band of ∼30 kDa in immunoblot assays with a whole-cell lysate of the wild-type strain O35E (Fig. 2). The band was absent from a whole-cell lysate of the znu and znuA knockout mutants (Fig. 2), confirming that the antiserum recognized epitopes on native ZnuA. The difference in apparent molecular masses between recombinant and native ZnuA (∼34 kDa versus ∼30 kDa) is due to the histidine tag and associated sequences on the amino terminus of the recombinant protein. An immunoblot assay of bacterial whole-cell lysates of 40 clinical isolates of M. catarrhalis with antiserum to ZnuA revealed an ∼30-kDa band in all 40 strains (not shown).
Binding of zinc and ZnuA.
Thermal-shift assays were used to assess the binding of zinc and other divalent cations to recombinant purified ZnuA. In the absence of zinc, the protein showed a bimodal unfolding with melting temperatures (Tm) of 47.5 ± 0.8°C and 59.0 ± 3.0°C, suggesting that the protein has an unstable domain (Fig. 3). When ZnuA was in buffer with no added zinc, addition of zinc chloride (200 μM) resulted in a sharp Tm of 67°C, indicating that the unstable domain was stabilized by zinc binding. A similar result was observed with zinc sulfate and zinc acetate. The presence of zinc is associated with an upward shift of the Tm to 68 ± 0.2°C (Table 2). In contrast, incubation with other divalent cations, including manganese, nickel, cobalt, and magnesium, did not cause an upward shift in Tm, indicating that ZnuA binds zinc specifically. In the presence of magnesium and manganese, the ZnuA protein showed the same bimodal pattern that was observed in the absence of zinc. The addition of nickel and cobalt resulted in a single peak, but the Tm was not shifted upward as was observed with zinc. Thus, nickel and cobalt appeared to stabilize an unstable domain of the protein but did not bind to the same extent as zinc, which causes a significant shift in Tm (Fig. 3 and Table 2).
Fig 3.

Results of thermal-shift assays assessing the effect of divalent cations as noted on the thermal stability of purified recombinant ZnuA. Each curve represents the average of 6 values. The x axis is the melting temperature (Tm) in °C, and the y axis is the normalized first derivative of the data.
Table 2.
Results of thermal-shift assays with recombinant purified ZnuA
| Divalent cation | Tma (°C) ± SD | Tma (°C) ± SD, secondary peak (if present) |
|---|---|---|
| None | 45.7 ± 0.8 | 59.0 ± 3.0 |
| Zinc chloride | 68.0 ± 0.2 | None |
| Zinc acetate | 68.0 ± 0.2 | None |
| Nickel chloride | 55.8 ± 0.2 | None |
| Cobalt chloride | 58.8 ± 0.1 | None |
| Magnesium acetate | 45.7 ± 0.8 | 57.9 ± 2.4 |
| Manganese chloride | 47.7 ± 0.6 | 58.6 ± 4.1 |
| Nickel sulfate | 55.9 | None |
Average of 6 readings (3 independent experiments, each performed in duplicate). Tm, melting temperature.
Characterization of mutants and revertant.
The znu mutant was constructed by replacing the entire 3,282-bp gene cluster with a nonpolar kanamycin cassette via homologous recombination (Fig. 1). The znuA mutant was constructed by replacing the 891-bp znuA gene. Analysis by immunoblot assay with absorbed rabbit antiserum to recombinant ZnuA showed that ZnuA was not expressed in the znu mutant or in the znuA mutant. As expected, ZnuA is expressed in wild-type O35E and the znu revertant (Fig. 2).
Role of znu in bacterial growth.
Growth of the znu knockout mutant is impaired in vitro. Colonies are smaller on BHI plates and chocolate agar plates than those of the wild type. The znu mutant colonies grow to a larger size on BHI plates than colonies on chocolate agar, a characteristic that we used to select the znu revertant (see Methods).
In BHI broth, a rich laboratory medium, the znu mutant shows growth impairment compared to the wild type, showing a lower growth rate and a reduced density in stationary phase (Fig. 4). The znuA mutant shows a growth rate intermediate between those of the wild type and the znu mutant but reaches a final culture density similar to that of the wild type. The znu revertant shows growth characteristics identical to those of the wild type, confirming that the growth impairments observed in the mutants are due to znu genes (Fig. 4).
Fig 4.
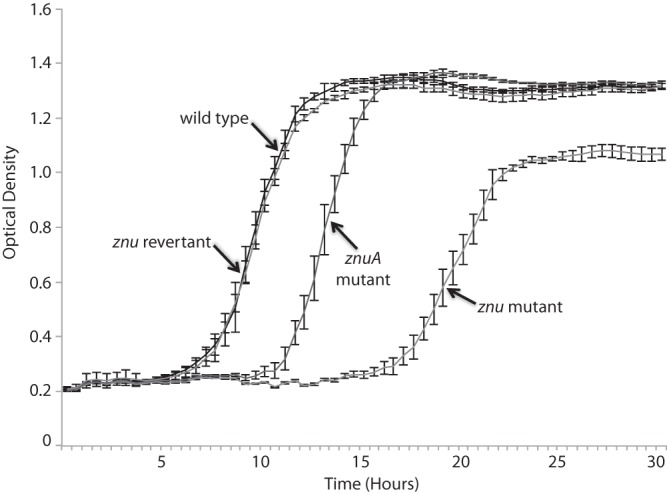
Growth curves in brain heart infusion broth of the M. catarrhalis O35E wild type, znu mutant, znuA mutant, and znu revertant as indicated. The x axis is time in hours, and the y axis is the optical density at 600 nm. Each point is the average for 5 duplicate wells. Error bars are standard deviations.
Wild-type M. catarrhalis grows in chemically defined medium (CDM) to a culture density approximately one-third the density in BHI (Fig. 5, bottom panel). The znu mutant shows minimal or no growth in CDM, which does not have added zinc but contains trace zinc. Addition of 20 μM ZnSO4 to CDM restored growth of the znu mutant to wild-type levels (Fig. 5, top panel). Addition of other divalent cations, including MnCl2, NiCl2, CoCl2, and CuSO4, had no effect on growth of the znu mutant. Addition of ZnSO4 (and other divalent cations) had no effect on growth of the wild type. We conclude that genes in the znu gene cluster (in particular znuB, znuC, and zur) are required for growth of M. catarrhalis under conditions of trace zinc and that addition of free zinc salt allows uptake of zinc by a mechanism independent of that of the znu genes. The observation that other divalent cations do not restore growth of the znu mutant indicates specificity of the znu gene cluster for uptake of zinc.
Fig 5.

Results of growth of the znu mutant (top) and wild-type strain O35E (bottom) in chemically defined medium (CDM) supplemented with divalent cation salts as noted. The x axis is time in hours, and the y axis is the optical density at 600 nm. Each point is the average for 5 duplicate wells. Error bars (CDM + ZnSO4) are standard deviations. For clarity, error bars are not shown for the other curves as they are quite narrow and obscure the curves.
Role of znu genes in adherence and invasion of human respiratory epithelial cells.
Wild-type, znu mutant, and znuA mutant strains were assayed for their capacity to adhere to and invade human respiratory epithelial cell line A549 (type 2 pneumocytes). The level of adherence to cells after 3 h of incubation (1.35 × 107 CFU/ml) was the same for the wild type and the 2 mutants. In contrast, the znu knockout mutant showed a ∼10-fold reduction in invasion of epithelial cells compared to that of the wild type (2.14 × 103 CFU/ml, compared to 2.48 × 102 CFU/ml) (P < 0.001, two-tailed t test) (Fig. 6). The znuA knockout mutant retained its capacity to invade cells. We conclude that the gene products encoded by znuB, znuC, and zur play an important role in invasion of respiratory epithelial cells by M. catarrhalis.
Fig 6.
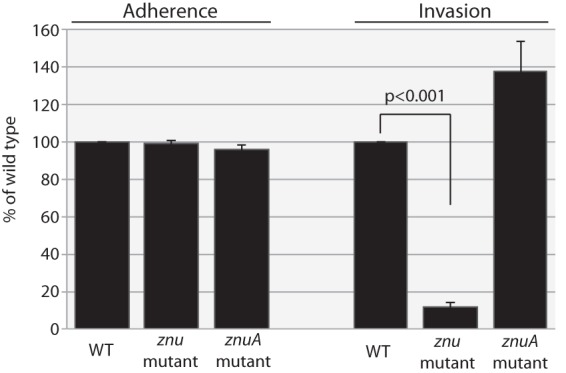
Results of adherence (left) and invasion (right) assays with the A549 respiratory epithelial cell line with the M. catarrhalis wild-type strain O35E (WT), znu mutant, and znuA mutant, as noted at the bottom. The y axis shows results relative to those for the wild type. Results are calculated from CFU/ml and are averages of 3 independent assays; error bars show standard deviations.
Role of znu genes in persistence in the murine respiratory tract.
To assess the role of the znu gene cluster in persistence in the murine respiratory tract following aerosol challenge, groups of BALB/c mice (n = 10 per group) were subjected to aerosol challenge with an equal inoculum of wild-type and znu mutant strains simultaneously. An ∼10-fold reduction in the concentration of the znu mutant in lungs 3 h after challenge compared to that of the wild type (P < 0.001, two-tailed t test) was observed (Fig. 7). Thus, the znu mutant showed marked impairment in its capacity to persist in the murine respiratory tract compared to wild-type M. catarrhalis.
Fig 7.
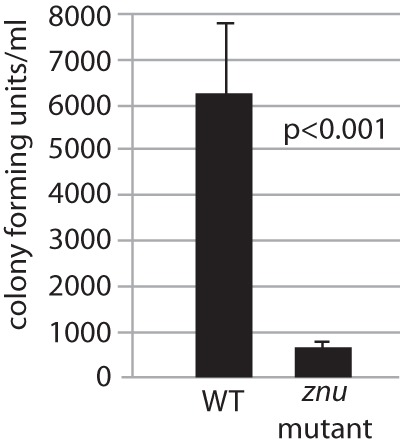
Results of pulmonary clearance in mice following simultaneous aerosol challenge by equal amounts of the M. catarrhalis strain O35E wild type (WT) and znu knockout mutant strain (109 CFU total). The y axis shows the number of CFU per ml in homogenized lung tissue 3 h following challenge. Results are averages for 10 animals per group; error bars show standard deviations.
DISCUSSION
This study contributes to an understanding of mechanisms by which M. catarrhalis colonizes and infects the respiratory tract through the following observations. (i) A newly identified ABC transporter is critical for growth of M. catarrhalis when zinc is present in trace amounts. (ii) The substrate binding protein, ZnuA, specifically binds zinc based on results of thermal-shift assays. (iii) The predicted zinc ABC transporter of M. catarrhalis is critical for invasion of respiratory epithelial cells, an observation that is especially relevant because an intracellular reservoir of M. catarrhalis is present in the human respiratory tract and this reservoir is important for persistence (37, 38). And (iv) the predicted zinc ABC transporter facilitates persistence of M. catarrhalis in the respiratory tract in a murine model.
Zinc is a trace element that plays an essential role as a structural or catalytic cofactor in a large number of proteins (39, 40). We show that M. catarrhalis has a predicted high-affinity zinc transporter, ZnuABC, consisting of a permease (ZnuB), an ATP binding protein (ZnuC), and a substrate binding protein (ZnuA), similar to other bacterial species. A mutant in which the predicted ZnuABC transporter of M. catarrhalis has been knocked out shows minimal or no growth compared to the wild type in medium that contains trace zinc. Thus, the transporter is important when bacteria grow in environments characterized by very low zinc availability. Supplementation of the medium with zinc restores growth to wild-type levels, indicating that other mechanisms of zinc import operate at higher zinc concentrations. The observation that addition of other divalent cations to the medium fails to restore growth indicates that the predicted ZnuABC transporter is specific for zinc. This conclusion is further supported by the result that ZnuA binds zinc but does not bind other divalent cations in thermal-shift assays. Overall, these results indicate that a functional ZnuABC transporter is crucial for efficient uptake of zinc and growth in environments where zinc is scarce. The predicted Znu ABC transporter of M. catarrhalis shares characteristics with similar transporters in other bacterial species (41–44) and also has characteristics that are unique, particularly related to mechanisms of respiratory tract persistence (see below).
The observation that growth of the mutant in which the substrate binding protein ZnuA alone is knocked out reaches wild-type levels (Fig. 4) suggests that other periplasmic proteins may have redundant function with ZnuA, mediating binding of zinc. This characteristic differs from that of E. coli and Salmonella enterica, in which znuA knockout mutants show a marked growth defect (24, 26).
Some bacterial species express a protein called ZinT (formerly known as YodA), which is a periplasmic protein that is involved in zinc recruitment (26, 45, 46). For example, in Salmonella enterica serovar Typhimurium, ZinT participates in zinc uptake mediated by ZnuABC through a mechanism involving direct interaction with ZnuA (25). We investigated whether the M. catarrhalis genome contains a zinT gene with BLAST searches of the genomes in GenBank and found no genes with homology to zinT. Furthermore, the M. catarrhalis genomes that have been annotated contain no genes annotated as zinT or yodA (47, 48). Thus, based on these searches, it does not appear that M. catarrhalis expresses a ZinT protein. While this conclusion is limited by the fact that it is based solely on the absence of genes with high homology to zinT, the conclusion is consistent with the observation that knocking out the ZnuABC transporter in M. catarrhalis results in essentially absent growth in trace zinc, suggesting the absence of compensatory zinc uptake mechanisms.
The results of thermal-shift assays with purified recombinant ZnuA are interesting. In the absence of zinc, the protein showed a bimodal unfolding with Tms of ∼46°C and ∼59°C, indicating that the protein has a partially unfolded conformation (Fig. 3). Addition of zinc resulted in a sharp Tm of 68°C, indicating that zinc stabilized the unstable domain and caused a significant upward shift in Tm. The bimodal curve shifted to a single sharp peak with the addition of nickel and cobalt as well but did not show an upward shift of Tm with these divalent cations (Fig. 3 and Table 2). The recombinant ZnuA has a hexahistidine tag which has specificity for nickel and cobalt; therefore, this transition from a bimodal curve to a single peak is likely due to binding of the hexahistidine tag. The absence of an upward shift of Tm with divalent cations other than zinc indicates that the ZnuA protein itself shows specific binding of zinc.
M. catarrhalis colonizes the nasopharynx of infants and children beginning in the first year of life. Careful studies of tonsils and adenoids show that in addition to colonizing the surface of the upper human respiratory tract mucosa, M. catarrhalis resides in intracellular niches (37). These intracellular bacteria act as a reservoir to persist in the host. The observation that the ZnuABC transporter is critical for intracellular survival is important in understanding mechanisms by which M. catarrhalis persists in the human respiratory tract. This observation, coupled with the marked defect of the znu knockout mutant in persisting in the murine pulmonary clearance model (Fig. 7), indicates that the Znu ABC transporter is required for full virulence by M. catarrhalis. Future studies may be designed to develop interventions (topical agents being an example) to exploit this virulence factor which could reduce intracellular survival and thus interrupt colonization by M. catarrhalis.
In summary, this study has identified and characterized a Znu ABC transporter in M. catarrhalis that plays a role in the uptake of zinc in environments with very low zinc concentrations. The ZnuA periplasmic substrate binding protein binds zinc specifically. The Znu ABC transporter is critical for virulence of M. catarrhalis by facilitating intracellular invasion and persistence in the respiratory tract.
ACKNOWLEDGMENTS
This work was supported by NIH grant DC 012200 to T.F.M. and grant GM094611 to M.G.M.
We thank Howard Faden, Diane Dryja, Diana Adlowitz, Michael Pichichero, and Daniel Musher for bacterial strains. We thank Wei Wang, Eric Hansen, and Anthony Campagnari for plasmids. Anthony Campagnari provided helpful advice.
Footnotes
Published ahead of print 1 July 2013
REFERENCES
- 1.Murphy TF, Parameswaran GI. 2009. Moraxella catarrhalis, a human respiratory tract pathogen. Clin. Infect. Dis. 49:124–131 [DOI] [PubMed] [Google Scholar]
- 2.Vergison A, Dagan R, Arguedas A, Bonhoeffer J, Cohen R, Dhooge I, Hoberman A, Liese J, Marchisio P, Palmu AA, Ray GT, Sanders EA, Simoes EA, Uhari M, van Eldere J, Pelton SI. 2010. Otitis media and its consequences: beyond the earache. Lancet Infect. Dis. 10:195–203 [DOI] [PubMed] [Google Scholar]
- 3.Paradise JL, Rockette HE, Colborn DK, Bernard BS, Smith CG, Kurs-Lasky M, Janosky JE. 1997. Otitis media in 2253 Pittsburgh-area infants: prevalence and risk factors during the first two years of life. Pediatrics 99:318–333 [DOI] [PubMed] [Google Scholar]
- 4.Teele DW, Klein JD, Rosner B, The Greater Boston Ottis Media Study Group 1989. Epidemiology of otitis media during the first seven years of life in children in greater Boston: a prospective, cohort study. J. Infect. Dis. 160:83–94 [DOI] [PubMed] [Google Scholar]
- 5.Gonzales R, Malone DC, Maselli JH, Sande MA. 2001. Excessive antibiotic use for acute respiratory infections in the United States. Clin. Infect. Dis. 33:757–762 [DOI] [PubMed] [Google Scholar]
- 6.Plasschaert AI, Rovers MM, Schilder AG, Verheij TJ, Hak E. 2006. Trends in doctor consultations, antibiotic prescription, and specialist referrals for otitis media in children: 1995–2003. Pediatrics 117:1879–1886 [DOI] [PubMed] [Google Scholar]
- 7.Post JC, Preston RA, Aul JJ, Larkins-Pettigrew M, Rydquist-White J, Anderson KW, Wadowsky RM, Reagan DR, Walker ES, Kingsley LA, Magit AE, Ehrlich GD. 1995. Molecular analysis of bacterial pathogens in otitis media with effusion. JAMA 273:1598–1604 [PubMed] [Google Scholar]
- 8.Faden H, Harabuchi Y, Hong JJ, Pediatrics TW. 1994. Epidemiology of Moraxella catarrhalis in children during the first 2 years of life: relationship to otitis media. J. Infect. Dis. 169:1312–1317 [DOI] [PubMed] [Google Scholar]
- 9.Revai K, Mamidi D, Chonmaitree T. 2008. Association of nasopharyngeal bacterial colonization during upper respiratory tract infection and the development of acute otitis media. Clin. Infect. Dis. 46:e34–e37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Faden H, Duffy L, Wasielewski R, Wolf J, Krystofik D, Tung Y, Tonawanda Williamsville Pediatrics 1997. Relationship between nasopharyngeal colonization and the development of otitis media in children. J. Infect. Dis. 175:1440–1445 [DOI] [PubMed] [Google Scholar]
- 11.Leach AJ, Boswell JB, Asche V, Nienhuys TG, Mathews JD. 1994. Bacterial colonization of the nasopharynx predicts very early onset and persistence of otitis media in Australian Aboriginal infants. Pediatr. Infect. Dis. J. 13:983–989 [DOI] [PubMed] [Google Scholar]
- 12.Revai K, McCormick DP, Patel J, Grady JJ, Saeed K, Chonmaitree T. 2006. Effect of pneumococcal conjugate vaccine on nasopharyngeal bacterial colonization during acute otitis media. Pediatrics 117:1823–1829 [DOI] [PubMed] [Google Scholar]
- 13.Sethi S, Murphy TF. 2008. Infection in the pathogenesis and course of chronic obstructive pulmonary disease. N. Engl. J. Med. 359:2355–2365 [DOI] [PubMed] [Google Scholar]
- 14.Murphy TF, Brauer AL, Grant BJ, Sethi S. 2005. Moraxella catarrhalis in chronic obstructive pulmonary disease. Burden of disease and immune response. Am. J. Respir. Crit. Care Med. 172:195–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mannino DM, Buist AS. 2007. Global burden of COPD: risk factors, prevalence, and future trends. Lancet 370:765–773 [DOI] [PubMed] [Google Scholar]
- 16.Lopez AD, Shibuya K, Rao C, Mathers CD, Hansell AL, Held LS, Schmid V, Buist S. 2006. Chronic obstructive pulmonary disease: current burden and future projections. Eur. Respir. J. 27:397–412 [DOI] [PubMed] [Google Scholar]
- 17.Murphy TF. 2009. Vaccine development for Moraxella catarrhalis: rationale, approaches and challenges. Expert Rev. Vaccines 8:655–658 [DOI] [PubMed] [Google Scholar]
- 18.Mawas F, Ho MM, Corbel MJ. 2009. Current progress with Moraxella catarrhalis antigens as vaccine candidates. Expert Rev. Vaccines 8:77–90 [DOI] [PubMed] [Google Scholar]
- 19.Tan TT, Riesbeck K. 2007. Current progress of adhesins as vaccine candidates for Moraxella catarrhalis. Expert Rev. Vaccines 6:949–956 [DOI] [PubMed] [Google Scholar]
- 20.de Vries SP, Bootsma HJ, Hays JP, Hermans PW. 2009. Molecular aspects of Moraxella catarrhalis pathogenesis. Microbiol. Mol. Biol. Rev. 73:389–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruckdeschel EA, Kirkham C, Lesse AJ, Hu Z, Murphy TF. 2008. Mining the Moraxella catarrhalis genome: identification of potential vaccine antigens expressed during human infection. Infect. Immun. 76:1599–1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ruckdeschel EA, Brauer AL, Johnson A, Murphy TF. 2009. Characterization of proteins Msp22 and Msp75 as vaccine antigens of Moraxella catarrhalis. Vaccine 27:7065–7072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang M, Johnson A, Murphy TF. 2011. Characterization and evaluation of the Moraxella catarrhalis oligopeptide permease A as a mucosal vaccine antigen. Infect. Immun. 79:846–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ammendola S, Pasquali P, Pistoia C, Petrucci P, Petrarca P, Rotilio G, Battistoni A. 2007. High-affinity Zn2+ uptake system ZnuABC is required for bacterial zinc homeostasis in intracellular environments and contributes to the virulence of Salmonella enterica. Infect. Immun. 75:5867–5876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petrarca P, Ammendola S, Pasquali P, Battistoni A. 2010. The Zur-regulated ZinT protein is an auxiliary component of the high-affinity ZnuABC zinc transporter that facilitates metal recruitment during severe zinc shortage. J. Bacteriol. 192:1553–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gabbianelli R, Scotti R, Ammendola S, Petrarca P, Nicolini L, Battistoni A. 2011. Role of ZnuABC and ZinT in Escherichia coli O157:H7 zinc acquisition and interaction with epithelial cells. BMC Microbiol. 11:36. 10.1186/1471-2180-11-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang X, Becker T, Walters N, Pascual DW. 2006. Deletion of znuA virulence factor attenuates Brucella abortus and confers protection against wild-type challenge. Infect. Immun. 74:3874–3879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Campoy S, Jara M, Busquets N, Perez De Rozas AM, Badiola I, Barbe J. 2002. Role of the high-affinity zinc uptake znuABC system in Salmonella enterica serovar typhimurium virulence. Infect. Immun. 70:4721–4725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gunasekera TS, Herre AH, Crowder MW. 2009. Absence of ZnuABC-mediated zinc uptake affects virulence-associated phenotypes of uropathogenic Escherichia coli CFT073 under Zn(II)-depleted conditions. FEMS Microbiol. Lett. 300:36–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weston BF, Brenot A, Caparon MG. 2009. The metal homeostasis protein, Lsp, of Streptococcus pyogenes is necessary for acquisition of zinc and virulence. Infect. Immun. 77:2840–2848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sethi S, Evans N, Grant BJB, Murphy TF. 2002. New strains of bacteria and exacerbations of chronic obstructive pulmonary disease. N. Engl. J. Med. 347:465–471 [DOI] [PubMed] [Google Scholar]
- 32.Wang W, Hansen EJ. 2006. Plasmid pWW115, a cloning vector for use with Moraxella catarrhalis. Plasmid 56:133–137 [DOI] [PubMed] [Google Scholar]
- 33.Shevchuk NA, Bryksin AV, Nusinovich YA, Cabello FC, Sutherland M, Ladisch S. 2004. Construction of long DNA molecules using long PCR-based fusion of several fragments simultaneously. Nucleic Acids Res. 32:e19. 10.1093/nar/gnh014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Menard R, Sansonetti PJ, Parsot C. 1993. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J. Bacteriol. 175:5899–5906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koszelak-Rosenblum M, Krol AC, Simmons DM, Goulah CC, Wroblewski L, Malkowski MG. 2008. His-311 and Arg-559 are key residues involved in fatty acid oxygenation in pathogen-inducible oxygenase. J. Biol. Chem. 283:24962–24971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yeh AP, McMillan A, Stowell MH. 2006. Rapid and simple protein-stability screens: application to membrane proteins. Acta Crystallogr. D Biol. Crystallogr. 62:451–457 [DOI] [PubMed] [Google Scholar]
- 37.Heiniger N, Spaniol V, Troller R, Vischer M, Aebi C. 2007. A reservoir of Moraxella catarrhalis in human pharyngeal lymphoid tissue. J. Infect. Dis. 196:1080–1087 [DOI] [PubMed] [Google Scholar]
- 38.Spaniol V, Heiniger N, Troller R, Aebi C. 2008. Outer membrane protein UspA1 and lipooligosaccharide are involved in invasion of human epithelial cells by Moraxella catarrhalis. Microbes Infect. 10:3–11 [DOI] [PubMed] [Google Scholar]
- 39.Waldron KJ, Rutherford JC, Ford D, Robinson NJ. 2009. Metalloproteins and metal sensing. Nature 460:823–830 [DOI] [PubMed] [Google Scholar]
- 40.Hantke K. 2005. Bacterial zinc uptake and regulators. Curr. Opin. Microbiol. 8:196–202 [DOI] [PubMed] [Google Scholar]
- 41.Patzer SI, Hantke K. 1998. The ZnuABC high-affinity zinc uptake system and its regulator Zur in Escherichia coli. Mol. Microbiol. 28:1199–1210 [DOI] [PubMed] [Google Scholar]
- 42.Lewis DA, Klesney-Tait J, Lumbley SR, Ward CK, Latimer JL, Ison CA, Hansen EJ. 1999. Identification of the znuA-encoded periplasmic zinc transport protein of Haemophilus ducreyi. Infect. Immun. 67:5060–5068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen CY, Morse SA. 2001. Identification and characterization of a high-affinity zinc uptake system in Neisseria gonorrhoeae. FEMS Microbiol. Lett. 202(1):67–71 [DOI] [PubMed] [Google Scholar]
- 44.Garrido ME, Bosch M, Medina R, Llagostera M, Perez de Rozas AM, Badiola I, Barbe J. 2003. The high-affinity zinc-uptake system znuACB is under control of the iron-uptake regulator (fur) gene in the animal pathogen Pasteurella multocida. FEMS Microbiol. Lett. 221:31–37 [DOI] [PubMed] [Google Scholar]
- 45.Graham AI, Hunt S, Stokes SL, Bramall N, Bunch J, Cox AG, McLeod CW, Poole RK. 2009. Severe zinc depletion of Escherichia coli: roles for high affinity zinc binding by ZinT, zinc transport and zinc-independent proteins. J. Biol. Chem. 284:18377–18389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kershaw CJ, Brown NL, Hobman JL. 2007. Zinc dependence of zinT (yodA) mutants and binding of zinc, cadmium and mercury by ZinT. Biochem. Biophys. Res. Commun. 364:66–71 [DOI] [PubMed] [Google Scholar]
- 47.de Vries SP, van Hijum SA, Schueler W, Riesbeck K, Hays JP, Hermans PW, Bootsma HJ. 2010. Genome analysis of Moraxella catarrhalis strain RH4, a human respiratory tract pathogen. J. Bacteriol. 192:3574–3583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Davie JJ, Earl J, de Vries SP, Ahmed A, Hu FZ, Bootsma HJ, Stol K, Hermans PW, Wadowsky RM, Ehrlich GD, Hays JP, Campagnari AA. 2011. Comparative analysis and supragenome modeling of twelve Moraxella catarrhalis clinical isolates. BMC Genomics 12:70. 10.1186/1471-2164-12-70 [DOI] [PMC free article] [PubMed] [Google Scholar]


