Abstract
Investigating the innate immune response mediators released in milk has manifold implications, spanning from elucidation of the role played by mammary epithelial cells (MECs) in fighting microbial infections to the discovery of novel diagnostic markers for monitoring udder health in dairy animals. Here, we investigated the mammary gland response following a two-step experimental infection of lactating sheep with the mastitis-associated bacterium Streptococcus uberis. The establishment of infection was confirmed both clinically and by molecular methods, including PCR and fluorescent in situ hybridization of mammary tissues. Proteomic investigation of the milk fat globule (MFG), a complex vesicle released by lactating MECs, enabled detection of enrichment of several proteins involved in inflammation, chemotaxis of immune cells, and antimicrobial defense, including cathelicidins and calprotectin (S100A8/S100A9), in infected animals, suggesting the consistent involvement of MECs in the innate immune response to pathogens. The ability of MECs to produce and release antimicrobial and immune defense proteins was then demonstrated by immunohistochemistry and confocal immunomicroscopy of cathelicidin and the calprotectin subunit S100A9 on mammary tissues. The time course of their release in milk was also assessed by Western immunoblotting along the course of the experimental infection, revealing the rapid increase of these proteins in the MFG fraction in response to the presence of bacteria. Our results support an active role of MECs in the innate immune response of the mammary gland and provide new potential for the development of novel and more sensitive tools for monitoring mastitis in dairy animals.
INTRODUCTION
Sheep mastitis is most frequently due to Gram-positive environmental pathogens, including Staphylococcus aureus, Staphylococcus epidermidis, and Streptococcus uberis (1–3), together with bacteria belonging to the class Mollicutes (including Mycoplasma agalactiae, the agent of contagious agalactia). The typical infection route is the teat canal, where microorganisms encounter the first line of host defense (4, 5). The milking machine, in particular, is considered to be detrimental, in that it induces teat end erosion and impairs skin conditions, resulting in a higher level of colonization with environmental pathogens (6). During milking, the sphincter muscles at the teat end open to let out the milk, and about 2 h is needed for the canal to be completely reclosed (7). During this time, mammary tissue is more exposed to invasion and colonization by microbial pathogens. Such colonization can result in infection (mild and acute) and, followed by inflammation of the mammary gland, develops into clinical disease. Clinical mastitis is usually associated with an increase in breast temperature and with functional disorders of the mammary gland, ranging from abnormalities in milk to complete agalactia (2). Microbial invasion of the host can last for long periods of time, requiring prolonged antibiotic therapy. Many microorganisms, including Streptococcus uberis, can also cause subclinical infections that do not lead to clinically evident manifestations, such as fever or swelling of the mammary gland, or to detectable milk abnormalities. These subclinical infections represent a serious problem for breeders, since subclinically infected animals can go undetected and act as pathogen reservoirs. Currently, a convenient and effective procedure to identify animals with subclinical mastitis is not available. In addition to more time-consuming and cumbersome procedures, such as the microbiological culture of milk, the presence of a mammary gland infection can be quickly evaluated by using the milk somatic cell count (SCC), since this parameter is commonly associated with inflammation (8–10). However, these tests are known to possess only moderate sensitivity and specificity (9, 11), since the increase in somatic cells is not exclusively dependent on bacterial infections but can also be influenced by the presence of viral infections, animal age, lactation stage, level of milk production, as well as various sources of stress, such as those caused by vaccinations and other treatments (8, 12–17). Moreover, SCC tests performed without nuclear staining have demonstrated a fair level of reliability in cattle but are subjected to a higher level of error when applied to small ruminants (sheep and goat). In fact, in small ruminants, milk secretion leads to the release of abundant amounts of cell debris in milk, and this debris is enumerated as somatic cells. In addition, the small ruminant milk production cycle is seasonal, and a physiological increase in the number of somatic cells is commonly observed at the end of lactation. There is therefore a need to investigate and elucidate the molecular mechanisms occurring during the establishment of a microbial infection in the lactating mammary gland, in order to identify new potential biomarkers suitable for the sensitive and specific detection of subclinical infections in small ruminants, which might also have potential for translation to other dairy animals.
Recently, we investigated milk fat globules (MFGs) to study the biology of the lactating cell and to evaluate the alterations occurring in sheep naturally infected by the bacterial pathogen Mycoplasma agalactiae (18). MFGs represent a suitable experimental system with which to evaluate the dynamic changes occurring in mammary epithelial cells (MECs) during infection. Indeed, MFGs are a natural product that may be used to sample the assortments of molecular networks that are activated within the lactating cell in vivo. Using the MFG approach, we found that, upon exposure to a bacterial pathogen, sheep lactating MECs release an assortment of proteins and peptides involved in the innate immune defense against pathogens, such as cathelicidin, S100 proteins, serum amyloid A3, and lactoferrin, accompanied by a marked reduction in proteins linked to the physiological pathways of lactation. Notably, alterations in the innate immune defense proteins were also clearly evident in subclinically infected animals.
In general, analysis of mediators of innate immunity might represent the ideal tool for detecting a subclinical infection, since these come into play at early stages and are usually pathogen independent. In addition, their production from infected epithelial cells and locally recruited immune cells can also take place at later stages of infection. Therefore, identifying early immune mediators can generate useful information on potential biomarkers for the development of mastitis detection strategies. While there is a scarcity of molecular studies on sheep mastitis, recent reports on bovine and caprine mastitis have provided a wealth of data that might serve as a model for innate immunity against environmental pathogens in ewes (19–21). According to these findings, once a Gram-positive pathogen enters into the mammary gland, its pathogen-associated molecular patterns (PAMPs; i.e., its lipoteichoic acid) are recognized through activation of pattern recognition receptors (PRRs; i.e., Toll-like receptor 2), triggering an innate immune response in mammary tissues (5, 22, 23). As a result, effector molecules that include antimicrobial peptides (AMPs) and acute-phase proteins (APPs) are produced and released. In this respect, MECs are expected to play a pivotal role from the early stages of innate immunity (24–29).
Efforts are required to elucidate the molecular and cellular mechanisms that influence innate immunity in sheep and to clarify the role played by the secreting mammary epithelium. Indeed, data on how innate immunity is orchestrated in sheep mammary gland tissues can contribute to the development of effective and appropriate treatment protocols aimed at limiting colonization and infections and to great decreases in the impact of mastitis in the dairy sheep industry.
The aim of this study was to gain a deeper knowledge of the events taking place in the infected mammary tissue of sheep and to clarify the role played by MECs by applying MFG proteomics and a combination of molecular and immunological techniques. In addition, the ability of S. uberis to establish a mammary infection in sheep was demonstrated. Protein expression profiles of MECs were assessed by means of two-dimensional (2D) difference-in-gel electrophoresis (DIGE) and SDS-PAGE separation, followed by liquid chromatography-tandem mass spectrometry (GeLC-MS/MS) of milk fat globule-associated proteins, revealing the upregulation in infected animals of a number of proteins involved in the innate immune response against pathogens. Two of these, S100A9 and cathelicidin, were then assessed by immunological methods for their cellular origin and kinetics of release in milk. Useful insights into the contribution of lactating MECs to fighting bacterial infections were obtained, as was an indication of several molecules with the potential to be candidates for use in the implementation of novel strategies for mastitis detection.
MATERIALS AND METHODS
Animal infection and sample collection.
Five Sarda sheep in midlactation with no history of S. uberis infection were chosen for inclusion in the study. Experimental infections were carried out at the Istituto Zooprofilattico Sperimentale della Sardegna (IZS). Here, sheep were confined separately from each other and subsequently tested to assess their suitability (fitness) for the experimental procedures, as described previously (3). All animal-related procedures used in this study were performed in accordance with the policies of IZS. Experimental infection of sheep was performed in the context of a research project entitled “I geni di resistenza e il ruolo dei mediatori dell'infiammazione nelle mastiti ovine,” identification (ID) number IZS SA 002/07, funding program Ricerca Corrente 2007. The project was approved (including ethical approval), financed, and authorized by the Italian Ministry of Health and the IZS. The S. uberis strain used for experimental infection was isolated in Sardinia, Italy, from a sheep with clinical mastitis. Before inoculation, the teat ends were cleaned with disinfectant. Four clinically healthy sheep were inoculated twice and kept in separate, contiguous sheds; a control animal was not inoculated, and it was maintained in a contiguous shed during the infection experiment. The two S. uberis inoculations were performed a week apart, and the inocula were administered into the teat cistern of the left half of the udders of four sheep with a syringe. The right half of the udders was inoculated with sterile phosphate-buffered saline (PBS) as a control. During experimental infection, milk was collected daily from both teats of each sheep. Milk samples were subjected to bacteriological culture and PCR analysis for the detection of S. uberis. Six days after the second inoculation, all animals were sacrificed and subjected to necropsy. Mammary tissue samples were collected during necropsy and immediately frozen at −80°C.
Histopathological grading.
The degree of tissue injury was assessed on hematoxylin-eosin-stained slides and was based on a semiquantitative grading system (30). Histopathological grading was carried out on four animals (three experimentally infected animals and one control animal), since one experimentally infected animal developed agalactia within 24 h after the second bacterial inoculation. Both infected and uninfected half udders were graded for each subject. For each half udder, 5 random microscopy fields were examined at ×20 magnification, and lesions were graded with a scoring grid based on estimation of epithelial desquamation and leukocytic infiltrates (neutrophils, macrophages, and lymphocytes), with scores indicating the presence or absence of lesions, the abundance of the different cell types, and the extent of the lesions, as follows: 1, absent; 2, rare; 3, moderate; 4, severe. Fibrosis in particular was useful to assess if the experimental animals had previously been exposed to mastitis.
Extraction of MFGPs.
MFG proteins (MFGPs) were extracted from raw milk as described previously (30–32). Briefly, milk samples were centrifuged to separate the cream fraction containing MFGs from the remaining protein fraction of milk. In order to eliminate highly abundant milk proteins, the cream was washed twice in phosphate-buffered saline and once in triple-distilled water. Subsequently, the MFGs were subjected to crystallization, to mechanical homogenization, and to heating in order to separate the protein fraction from the lipid fraction. The protein concentration was determined with a 2D Quant kit (GE Healthcare, Uppsala, Sweden), and the quality of the extracts was checked by resuspension in Laemmli buffer (33), separation using 10% acrylamide gels, and staining with SimplyBlue SafeStain (Invitrogen, Carlsbad, CA). The samples were stored at −20°C until analysis.
SDS-PAGE and 2D DIGE.
Proteins extracted from MFGs (MFGPs) were analyzed by 2D DIGE, as described previously (18). Sixty micrograms of proteins was labeled with 400 pmol of N-hydroxysuccinimidyl ester of cyanine dyes Cy3 (preinfection) and Cy5 (postinfection) (GE Healthcare, Uppsala, Sweden). Equal amounts of all experimental samples were properly combined to create the internal pooled standard, which was labeled with the Cy2 cyanine dye. The labeled protein samples were conveniently mixed and brought to the final rehydration volume with immobilized pH gradient (IPG) buffer (GE Healthcare) and Destreak rehydration solution (GE Healthcare, Uppsala, Sweden). The samples were applied to 24-cm IPG strips (pH 3 to 11, nonlinear; GE Healthcare, Uppsala, Sweden) by passive rehydration overnight at room temperature. After rehydration, IPG strips were run together on an Ettan IPGphor 3 apparatus (GE Healthcare, Uppsala, Sweden) using a gradient voltage increase for a total of about 90,000 V h. Afterwards, the strips were reduced and alkylated through the process of equilibration, subjected to second-dimension SDS-PAGE, and digitized with a Typhoon 9400 scanner (GE Healthcare, Uppsala, Sweden) as described previously (18). All images were analyzed with a DeCyder batch processor and differential in-gel analysis (DIA) modules (GE Healthcare, Uppsala, Sweden) for the detection and matching of spots, while statistical analysis of protein-level changes was performed with the DeCyder biological variation analysis (BVA; v.6.5) module. The results related to preinfection and postinfection samples were compared by calculation of fold changes and statistically evaluated by one-way analysis of variance (ANOVA) with the DeCyder BVA module. To minimize the number of false-positive results, the false discovery rate (FDR) was applied to the analysis. Protein spots were selected as differentially expressed if they showed a change in expression of ≥2-fold with a statistically significant variation (P ≤ 0.05). The DeCyder extended data analysis (EDA) module was used for performing cluster analysis by principal component analysis (PCA).
Tandem mass spectrometry.
The proteins differentially expressed according to the 2D DIGE analysis were subjected to identification using preparative 2D polyacrylamide gels. For optimum matching with the 2D DIGE maps, 24-cm IPG strips with a pH gradient (3-11NL; GE Healthcare, Uppsala, Sweden) were used. All strips were rehydrated with 300 μg of protein, focused, and subjected to the second dimension of electrophoresis. After electrophoresis, all gels were stained with SimplyBlue SafeStain (Invitrogen, Carlsbad, CA), and images were acquired by a ImageScanner III apparatus (GE Healthcare, Uppsala, Sweden). Matching between 2D DIGE and 2D polyacrylamide gels was performed using Image Master 2D Platinum (v.6.0.1) software (GE Healthcare, Uppsala, Sweden). Differentially expressed spots detected by 2D DIGE analysis were excised, destained, and subjected to tryptic digestion. Peptide mixtures were acidified, dried, and then resuspended in formic acid. Protein identification was performed on a XCT Ultra 6340 ion trap equipped with a 1200 high-pressure liquid chromatography system and a chip cube (Agilent Technologies, Palo Alto, CA), as described previously (34).
MGF files obtained by LC-MS/MS analysis were analyzed by Proteome Discoverer (v.1.3; Thermo Scientific), which used an in-house Mascot server (v.2.3; Matrix Science) for identification of proteins in the updated UniProt database (release 2012_01), the Mammalia taxonomy, and the following search parameters: precursor mass tolerance, 300 ppm; fragment mass tolerance, 0.6 Da; charge states, +2, +3, and + 4; trypsin enzyme; two missed cleavages; cysteine carbamidomethylation as the static modification; and N-terminal glutamine conversion to pyroglutamic acid and methionine oxidation as dynamic modifications.
Label-free quantification.
This analysis was performed on two preinfection samples (samples obtained on day 0 [D0]) and on two postinfection samples (samples obtained on the third day after the second infection [3DIIi]) as described previously (18). For each sample, 30 μg of protein was separated by SDS-PAGE using 4 to 20% acrylamide gradient gels and stained with SimplyBlue SafeStain (Invitrogen, Carlsbad, CA), and 25 slices were cut from each lane. All gel slices were subjected to bleaching, reduction, alkylation, digestion with trypsin, and recovery of the peptides from the gel before LC-MS/MS analysis, as described previously (30).
Identification of peptides was performed on an LTQ-Orbitrap Velos mass spectrometer (Thermo Scientific, San Jose, CA) interfaced with an UltiMate 3000 RSLCnano LC system (Dionex [now part of Thermo Scientific], Sunnyvale, CA), as described previously (35). Briefly, after loading, the peptide mixtures were concentrated and desalted on a trapping precolumn and separated on a 75-μm (inner diameter) by 25-cm C18 column (Acclaim PepMap rapid separation liquid chromatography [RSLC] C18 column [75-μm by 15-cm nanoViper system; particle size, 2 μm; 100 Å]; Dionex). Peptide mixtures from gel slices were subjected to 60-min runs. The LTQ-Orbitrap Velos mass spectrometer was set up in a data-dependent MS/MS mode under direct control of Xcalibur software (v.1.0.2.65 SP2), where a full-scan spectrum was followed by tandem mass spectra (MS/MS). Peptide ions were selected as the 10 most intense peaks (top 10) of the previous scan. Higher-energy collisional dissociation (HCD) was chosen for fragmentation with nitrogen as the collision gas. The raw files generated by the LTQ-Orbitrap mass spectrometer were analyzed on a Proteome Discoverer platform (v.1.4; Thermo Scientific, Bremen, Germany) to obtain protein identifications. All peak lists were processed against the UniProt database (release 2013_05) with the following search parameters: Mammalia as the taxonomy; peptide tolerance, 10 ppm; MS/MS tolerance, 0.02 Da; charge states, +2, +3, and + 4; cysteine carbamidomethylation as the static modification; N-terminal glutamine conversion to pyroglutammic acid and methionine oxidation as dynamic modifications; trypsin enzyme; and allowing up to two missed cleavages. To evaluate peptide validation, the percolator algorithm was used. For label-free quantification, all peptides with a q value of ≤0.01 and a peptide rank of 1 were included (36). Trypsin and skin keratins were excluded from the final protein list obtained by Proteome Discoverer.
Data analysis.
Label-free quantification based on spectral count (SpC) values was used as a semiquantitative measure to evaluate protein abundance and to compare the expression of the same protein among different samples, as described previously (18, 31). Only the proteins with the highest number of unique peptides and SpCs were selected between different homologue proteins present in each sample. The normalized spectral abundance factor (NSAF) and the SpC log ratio (RSC) were used to express protein abundance and the fold change between different conditions, respectively. NSAF and RSC were calculated according to the methods described by Old et al. (37) and Zybailov et al. (38), respectively. NSAF values, subdivided for cellular component and biological process categories, were used to compare the expression of diverse proteins among samples. A two-tailed t test with a 95% confidence level was used to evaluate the statistical significance of abundance categories for cellular component and biological processes between different conditions, while the beta-binomial test was performed to identify differentially expressed proteins according to the method described by Pham et al. (39). P values were corrected by the FDR.
Western immunoblotting.
After extraction of MFGPs, samples were resuspended in lysis buffer, loaded in 18% acrylamide gels, and subjected to electrophoresis for separation of proteins. Afterwards, proteins were transferred to nitrocellulose membranes (GE Healthcare, Uppsala, Sweden) at 100 V for 90 min using a Trans-Blot cell apparatus (Bio-Rad, Hercules, CA). Western immunoblotting was performed using a SNAP i.d. protein detection system (EMD Millipore) following the instructions recommended by the manufacturer. Primary antibodies were rabbit anti-human S100A9 (Sigma-Aldrich, St. Louis, MO) and rabbit anti-human cathelicidin AMP (CAMP; Sigma-Aldrich, St. Louis, MO). An anti-rabbit IgG (whole molecule)-peroxidase produced in goat (Sigma-Aldrich, St. Louis, MO) was used as the secondary antibody. For the detection of the signal, the chemiluminescent peroxidase substrate (Sigma-Aldrich, St. Louis, MO) was used, and blot images were acquired with a VersaDoc MP 4000 system (Bio-Rad, Hercules, CA).
PCR and probe design.
DNA extraction was carried out from milk and mammary tissue using a DNeasy blood and tissue kit (catalog no. 69506; Qiagen) according to the manufacturer's instructions. PCR was performed using primers Ub F and Ub R, designed on two variable regions of the S. uberis 23S rRNA with the following sequences: 5′-GCGAAGTGGGACATAAAGTTA-3′ (Ub probeF) and 5′-GCGGCTGTCATCGCTGA-3′ (Ub probeR). PCRs were performed in a GeneAmp PCR system 9700 (Applied Biosystems) as follows: an initial denaturation step (95°C for 2 min), followed by 30 cycles of denaturation (95°C for 1 min), annealing (56°C for 1 min), and extension (72°C for 1 min). A final extension step at 72°C for 10 min was included. PCR products of 1,446 bp were observed upon agarose gel electrophoresis according to standard procedures. For hybridization, two S. uberis-specific biotinylated oligonucleotide probes were designed on two variable regions of the 23S rRNA, using primers Ub probeF (5′-biotin-GCGAAGTGGGACATAAAGTTA-3′) and Ub probeR (5′-biotin-GCGGCTGTCATCGCTGA-3′). Two new sets of universal primers external to the probes were also designed: set 1, Ub-F-probeF (5′-GGCGAGCGAAACGGCAGGAG-3′) and Ub-R-probeF (5′-TTTCGCGTGTCTCGCCGTACT-3′) (product size, 182 bp) and set2, Ub-F-probeR (5′-TGAGCTGTGATGGGGAGCGAAA-3′) and Ub-R-probeR (5′-GGGCAGGCGTCACCCCCTAT-3′) (product size, 300 bp). Probes and primers were designed by using the nucleotide sequences of the 23S rRNA of several bacterial strains collected from the GenBank database and aligned using ClustalW software. To assess the specificities of the probes, PCR-dot blot assays were performed on the DNAs isolated from the following microorganisms: Streptococcus dysgalactiae 4065, Streptococcus agalactiae 4066, Enterococcus faecium 3885, Enterococcus faecalis 4063, Streptococcus bovis 1167, Streptococcus uberis 4064, Staphylococcus simulans, Staphylococcus aureus, Staphylococcus chromogenes, Staphylococcus epidermidis, Staphylococcus aureus Oxford, Enterococcus faecalis (wild strain), Streptococcus uberis (strain used in the experimental infection). Genomic DNA extraction was carried out from 24-h cultures of each bacterial species using a DNeasy blood and tissue kit (catalog no. 69506; Qiagen) according to the manufacturer's instructions. The DNAs were amplified using the two new sets of universal primers. The PCRs were performed in a GeneAmp PCR system 9700 (Applied Biosystems) with an initial denaturation step (94°C for 5 min), followed by 40 cycles of denaturation (94°C for 1 min), annealing (60°C for 1 min), and extension (72°C for 1 min). A final extension step at 72°C for 10 min was included. Amplicons of the expected sizes were purified from a 2% agarose gel using a QIAquick gel extraction kit (catalog no. 28704; Qiagen). Dot blot hybridization assays using specific Ub probeF or Ub probeR probes were used in combination with PCRs for validation of the probe specificity.
In situ hybridization.
Serial sections of sheep mammary glands were probed for in situ detection of Streptococcus uberis using the biotin-conjugated specific probes described above, and the signal was revealed with streptavidin-Alexa Fluor 555 conjugate (Invitrogen). Three-micrometer sections were mounted on SuperFrost slides, deparaffinized, and rehydrated according to standard histologic methods. A pepsin digestion (0.8% in 0.2 N HCl solution) was performed at 37°C for 30 min in Tris-buffered saline (TBS; 20 mM Tris-HCl, 150 mM NaCl, pH 7.5). The sections were subjected to a postfixation step by incubation in an alcohol ascending scale (from 50% to 98%) and air dried. The sections were incubated at 55°C for 2 h with 200 μl of prehybridization solution (50% of Hybridization Solution II [43% deionized formamide, 7% nuclease-free water]; Fluka). The prehybridization solution was replaced with 200 μl of hybridization solution containing 1 nM Ub probeF, and the slides were incubated overnight at 55°C. Stringent washes were then performed using 2×, 1×, and 0.5× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]) at room temperature for 5 min each time and 0.1× SSPE at 50°C for 30 min. The sections were washed with TBS-Tween (TBS-T; 20 mM Tris-HCl, 150 mM NaCl, pH 7.5, 0.05% Tween 20), and the blocking of nonspecific sites was performed by incubation with 2% bovine serum albumin in TBS-T for 1 h at 37°C. The signal was revealed by incubating for 45 min in the dark at room temperature with streptavidin-Alexa Fluor 555 conjugate (Invitrogen), and the nuclei were counterstained with Hoechst (Sigma) for 5 min. The slides were washed with Milli-Q water, mounted with an aqueous medium, and then examined in a confocal microscope (Leica TCS SP5). Images were captured and processed with LAS AF Lite application software.
Immunohistochemistry.
After euthanasia, mammary tissue samples were collected from both inoculated and uninoculated half udders. A portion of each sample was fixed in 10% paraformaldehyde in PBS, processed through graded concentrations of alcohol and xylene, and embedded in paraffin wax. Tissues were dewaxed and rehydrated, and endogenous peroxidase activity was blocked by incubation with 0.3% hydrogen peroxide in PBS for 30 min. As primary antibodies, rabbit anti-S100A9 (Sigma-Aldrich, St. Louis, MO), rabbit anti-CAMP (anti-cathelicidin; Sigma-Aldrich, St. Louis, MO), and antipancytokeratin (Dako) antibodies were applied as described by the manufacturers. To reveal antigens that reacted with primary antibodies, a Histostain-Plus kit (diaminobenzidine, broad spectrum; Invitrogen, Carlsbad, CA) was used. The sections were then incubated with 3,3′-diaminobenzidine tetrahydrochloride (Invitrogen, Carlsbad, CA) and lightly counterstained with hematoxylin. Images of all sample tissues were visualized and obtained with a Nikon Eclipse 80i microscope with a Nikon DS-Fi1 camera (Nikon Instruments Inc., Melville, NY).
Immune colocalization.
For immune colocalization analysis, the following antibodies were used: fluorescein isothiocyanate (FITC)-labeled anti-cytokeratin 7 (EMD Millipore), FITC-labeled antineutrophil (7/4; (Abcam, Cambridge, MA), FITC-labeled anti-F4/80 (BM8; Abcam, Cambridge, MA), anti-S100A9 (Sigma-Aldrich, St. Louis, MO), and anti-CAMP (anti-cathelicidin; Sigma-Aldrich, St. Louis, MO). In these experiments, Alexa Fluor-conjugated secondary antibodies (Invitrogen, Carlsbad, CA) were used for the detection of S100A9 and CAMP. Images were acquired using a Leica TCS SP 5 confocal microscope (Leica Microsystems, Germany) and processed using LAS AF Lite application software developed by Leica Microsystems CMS GmbH for contrast and brightness adjustment. Negative controls prepared by omission of primary antibodies did not show any FITC fluorescence under the conditions described above.
RESULTS
Experimental infection of sheep with S. uberis.
The experimental infection was carried out on four animals, while a fifth animal was not inoculated and served as a negative control. Figure 1 illustrates the experimental infection timeline. Preinfection milk samples were collected from all five animals (D0) as a reference to enable evaluation of the changes induced upon infection with S. uberis. Following administration of 1 × 106 CFU of S. uberis to a single half udder (left half udder) and of the same volume of physiological saline into the right half udder of four ewes, none of the inoculated animals showed clinical symptoms of mastitis. The animals were apparently healthy, and there were no evident alterations in milk appearance or quantity. After 6 days, the same half udders were inoculated with 2 × 107 CFU of S. uberis in order to induce acute mastitis. At day 2 after the second infection (2DIIi), udder swelling, rubor, calor, dolor, and reduction of milk volumes were observed in the infected half udder, while the contralateral half udders infused with saline did not show any visible alteration. One of the ewes developed agalactia in the infected half udder, while the neighboring half did not show clinical signs. Bacteriological analysis of milk revealed the presence of S. uberis by the second day of the second infection (2DIIi) only in the infected half udder of all animals, and S. uberis was then detected over the whole course of infection. The control animal remained clinically healthy for the whole experimental period.
Fig 1.
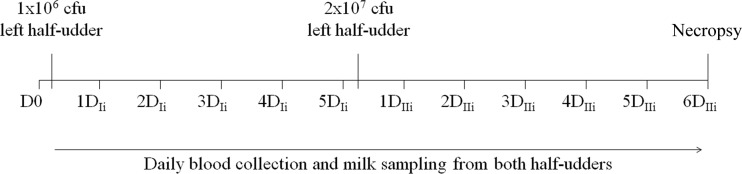
Timeline of experimental infection: sample collection, S. uberis inoculation, and animal sacrifice and necropsy.
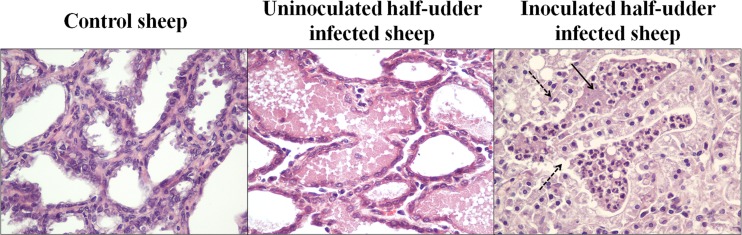
Macroscopic examination of half udders from all animals was performed at necropsy 12 days after the first inoculation. This analysis showed lesions due to acute purulent mastitis in the infected half udders, while the uninfected halves showed no obvious signs of inflammation. All infected animals showed an increase in the volume of the supramammary lymph nodes of the inoculated half udder compared to the volume of the supramammary lymph nodes of the uninoculated one. The control animal was found to be completely free of lesions in all observed sections. Microscopic examination and histopathological grading of mammary tissues highlighted a clear difference between infected and uninfected udder halves. In fact, the latter appeared to be structurally similar to the control udder, showing an intact epithelium of an organ in full lactation. The alveolar lumens were free of exfoliated epithelial cells or immune cells. The intra-alveolar wall displayed a normal thickness, without edema, cellular exudates, or signs of infection. On the contrary, samples from the infected half udders manifested clear indications of acute flogosis, showing traits of degeneration from medium to severe, with necrosis and desquamation affecting both alveolar tubular epithelia. A cellular exudate was present within alveoli and mainly constituted neutrophilic granulocytes, and macrophages were found in the lower portion, indicating an acute innate immune response to the presence of bacteria (Fig. 2). Occasionally, minor foci of lymphoplasmacellular mastitis were observed, but these were completely irrelevant in the observed picture of acute purulent mastitis. These observations were confirmed by the histopathological grading and the final score obtained, revealing a clear picture of mastitis in the infected half udders and a clear picture of normality in the uninfected ones (Table 1).
Fig 2.
Histopathological grading. Representative hematoxylin-eosin-stained sections of tissue from control and experimentally infected sheep are shown. The tissues of the control sheep and uninoculated half udders of infected sheep are histologically normal. Inoculated half udders show intra-alveolar neutrophils (solid arrow) and signs of degeneration of the secretory epithelium (dashed arrows). Magnifications, ×200.
Table 1.
Histopathological grading
| Group and sheep | Histopathological gradea |
||||
|---|---|---|---|---|---|
| Epithelial desquamation | Neutrophilic infiltrate | Macrophagic infiltrate | Lymphocitic infiltrate | Fibrosis | |
| Control, sheep A | 1 | 1 | 1 | 1 | 3 |
| Infected udder halves | |||||
| Sheep B | 1 | 1 | 1 | 1 | 2 |
| Sheep C | 1 | 1 | 1 | 1 | 2 |
| Sheep D | 1 | 2 | 1 | 1 | 2 |
| Mean | 1 | 1 | 1 | 1 | 2 |
| Uninfected udder halves | |||||
| Sheep B | 4 | 3 | 3 | 1 | 2 |
| Sheep C | 4 | 3 | 3 | 1 | 2 |
| Sheep D | 3 | 3 | 3 | 2 | 2 |
| Mean | 4 | 3 | 3 | 1 | 2 |
The scores attributed upon microscopic examination of mammary tissues are reported as follows: 1, absent; 2, rare; 3, moderate; 4, severe.
Alterations induced in the MFGP profile by acute bacterial infection.
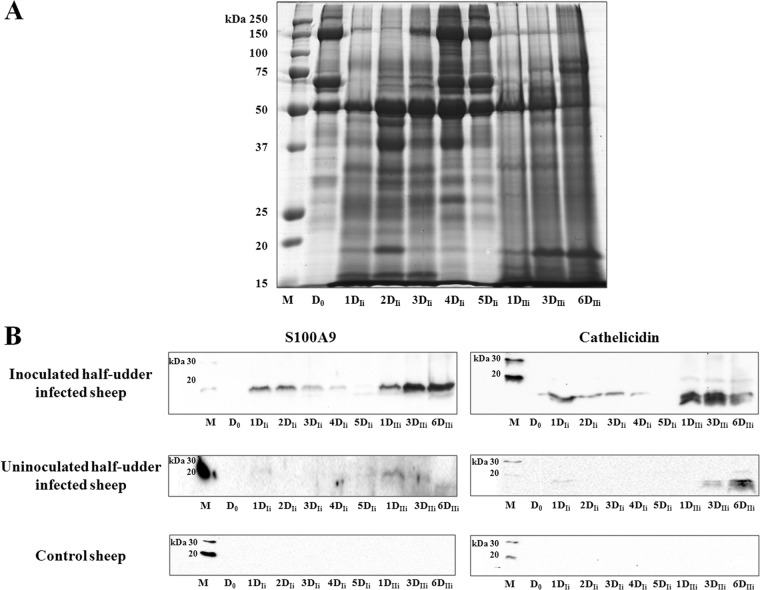
MFG proteins (MFGPs) were investigated as an indicator of the alterations occurring in MECs upon S. uberis infection. Only three infected animals were studied, since an animal developed agalactia within 24 h after the second bacterial inoculation. With the aim of reducing individual variations to a minimum and of maximizing detection of alterations specifically induced by S. uberis, MFGP expression was evaluated by comparing milk samples obtained at D0 with milk from the same animals and half udders obtained at 3DIIi (Fig. 3). Visible alterations in the total protein profile were clearly induced by infection. These were similar for the three infected animals, with numerous overabundant proteins detected at different molecular masses, while the MFGPs of the control animal remained unaffected.
Fig 3.
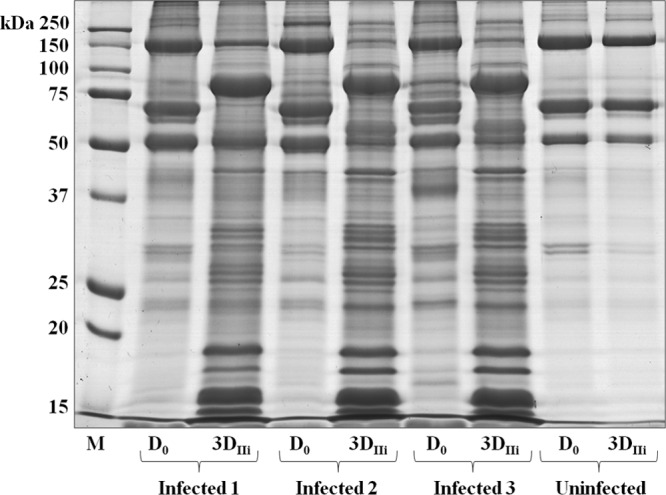
SDS-PAGE of sheep MFGPs before and after infection with S. uberis. The image illustrates a representative gel of all proteins extracted from MFGs isolated from the milk of all animals before infection (D0) and 3 days after the second inoculation (3DIIi). Lane M, molecular mass markers.
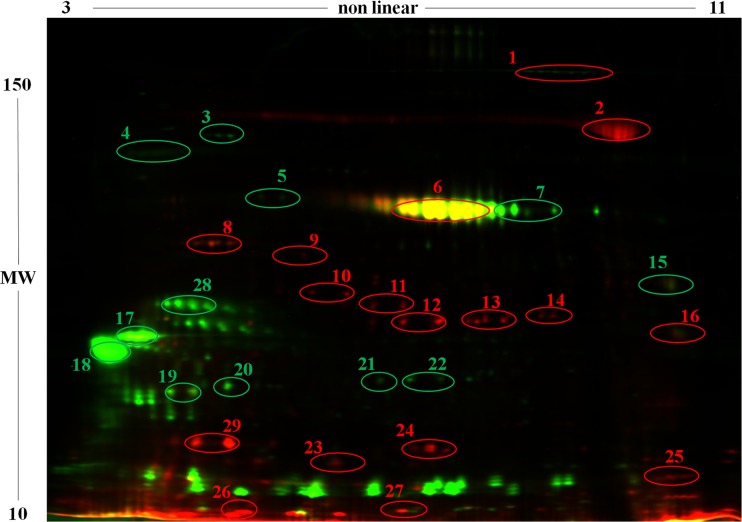
To investigate the nature and intensity of these alterations, the same samples were subjected to 2D DIGE analysis. Figure 4 reports a representative 2D DIGE map. Statistically significant differences in expression were detected for numerous proteins, which were subjected to identification by LC-MS/MS. In total, 28 differential proteins were identified between samples collected before and after infection; of these, 16 proteins were overexpressed and 15 were underexpressed in the presence of mastitis (Fig. 4; Table 2).
Fig 4.
Representative 2D DIGE profile of MFGPs from sheep before (green) and after (red) infection with S. uberis. An overlay image of representative MFGP profiles obtained before (green) and after (red) infection with S. uberis is shown. Yellow spots result from the superimposition of red and green signals and indicate similar levels of protein expression. Spots showing statistically significant differences are circled, and identities according to MS analysis are reported in Table 2. MW, molecular weight (molecular weights are given in thousands).
Table 2.
MFGPs showing statistically significant differences in abundance after S. uberis infection according to 2D DIGE
| Protein function and Ng | Identified protein | Accession no.a | Species | Scoreb | QMc | % coveraged | Avg ratioe | ANOVAf |
|---|---|---|---|---|---|---|---|---|
| Inflammation and host defense | ||||||||
| 2 | Lactotransferrin | Q5MJE8 | Ovis aries | 1,798 | 115 | 47 | 62.89 | 0.0015 |
| 25 | Myeloid antimicrobial peptide (cathelicidin family) | P79360 | Ovis aries | 432 | 19 | 47 | 25.42 | 0.024 |
| 24 | Protein S100A9 | P28783 | Bos taurus | 177 | 5 | 6 | 9.33 | 0.010 |
| 23 | Cathelicidin 1 | P54230 | Ovis aries | 378 | 22 | 50 | 13.15 | 0.016 |
| 27 | Protein S100A11 | P28783 | Bos taurus | 125 | 3 | 5 | 8.82 | 0.029 |
| Cytoskeletal proteins | ||||||||
| 10 | Annexin A3 | Q3SWX7 | Bos taurus | 968 | 52 | 37 | 98.80 | 0.0097 |
| 14 | Annexin AI | P46193 | Bos taurus | 1328 | 35 | 60 | 82.84 | 0.015 |
| 13 | Annexin AI | P46193 | Bos taurus | 1438 | 46 | 59 | 54.96 | 0.010 |
| 12 | Annexin AI | P46193 | Bos taurus | 1467 | 43 | 58 | 43.07 | 0.021 |
| 11 | Annexin A3 | Q3SWX7 | Bos taurus | 1120 | 56 | 39 | 18.08 | 0.022 |
| 9 | Actin, cytoplasmic 2 | P63260 | Mus musculus | 1242 | 83 | 62 | 16.65 | 0.0087 |
| 8 | Actin | P60712 | Bos taurus | 496 | 38 | 27 | 4.93 | 0.029 |
| Proteins involved in lipid metabolism and MFG secretion | ||||||||
| 26 | Fatty acid-binding protein | Q6S4N9 | Capra hircus | 691 | 62 | 66 | 17.04 | 0.0047 |
| 28 | Butyrophilin subfamily 1 member A1 | A3EY52 | Capra hircus | 399 | 10 | 15 | −9.33 | 0.002 |
| 4 | Butyrophilin subfamily 1 member A1 | A3EY52 | Capra hircus | 165 | 4 | 5 | −5.73 | 0.040 |
| 19 | Butyrophilin subfamily 1 member A1 | A3EY52 | Capra hircus | 409 | 12 | 17 | −5.67 | 0.039 |
| 7 | Adipose differentiation-related protein | A6ZE99 | Ovis aries | 364 | 11 | 20 | −5.01 | 0.024 |
| 5 | Adipose differentiation-related protein | A6ZE99 | Ovis aries | 623 | 44 | 27 | −4.74 | 0.024 |
| 6 | Lactadherin | Q95114 | Bos taurus | 496 | 25 | 17 | −4.64 | 0.035 |
| 1 | Xanthine dehydrogenase/oxidase | P80457 | Bos taurus | 819 | 29 | 11 | −3.58 | 0.0046 |
| Enzymes | ||||||||
| 22 | Triosephosphate isomerase | Q5E956 | Bos taurus | 347 | 5 | 15 | −8.36 | 0.017 |
| 15 | Dehydrogenase/reductase (SDR family) member 1 | Q2KIS4 | Bos taurus | 792 | 61 | 41 | −6.40 | 0.017 |
| 21 | Peroxiredoxin 6 | O77834 | Bos taurus | 226 | 7 | 26 | −5.90 | 0.024 |
| 16 | Cell death-inducing DFFA-like effector a | A4FUX1 | Bos taurus | 161 | 5 | 13 | −4.15 | 0.028 |
| Proteins involved in folding, 3 | 70-kDa heat shock protein | Q53GZ6 | Bos taurus | 125 | 3 | 5 | −6.37 | 0.020 |
| Milk proteins | ||||||||
| 29 | Lactoglobulin beta | P02754 | Bos taurus | 157 | 7 | 56 | 8.66 | 0.021 |
| 18 | Alpha-S1-casein | P04653 | Ovis aries | 717 | 40 | 49 | −18.90 | 0.036 |
| 17 | Beta-casein | P33048 | Capra hircus | 301 | 20 | 27 | −15.54 | 0.001 |
| 20 | Kappa-casein | P02669 | Ovis aries | 268 | 12 | 28 | −6.65 | 0.017 |
Accession number in the Uniprot database.
The score represents the probability that the observed match between the experimental data and mass values calculated from a candidate peptide sequence is a random event.
QM, queries matched, indicating the number of matched peptides in the database search.
Percent coverage of the matched peptide in relation to the full-length sequence.
Average ratio between 3DIIi and D0 samples.
By one-way ANOVA (P ≤ 0.05).
N, spot number in Fig. 4.
The proteins overexpressed in milk samples collected from mastitic half udders were mainly involved in (i) immune function, inflammation, and host defense, such as S100 proteins (S100A9 and S100A11), cathelicidin, antimicrobial peptides, and lactotransferrin; (ii) vesicular trafficking, such as annexin and actin; and (iii) lipid metabolism, such as fatty acid-binding proteins. Conversely, underexpressed proteins were mostly involved in the physiological functions of the MFG, such as butyrophilin, lactadherin, adipophilin, and xanthine dehydrogenase/oxidase. Other underexpressed proteins included various enzymes, such as triosephosphate isomerase, dehydrogenase/reductase member 1, peroxiredoxin 6, and cell death-inducing DNA fragmentation factor alpha (DFFA)-like effector A, and several milk proteins, such as alpha-S1-casein, beta-casein, and kappa-casein. PCA of all differentially expressed spots produced a separate clustering of the samples collected at D0 and at 3DIIi, demonstrating their statistically significant divergence.
To integrate the 2D DIGE approach and to compensate for its limitations in analysis of liposoluble and low-abundance proteins, a GeLC-MS/MS analysis was also carried out. In total, 849 unique proteins were identified and assessed for their relative abundance by a label-free quantitative approach (37). The detailed list of all protein identifications is reported in File S1 in the supplemental material. A total of 389 proteins showed a statistically significant (P < 0.05) difference in abundance of at least 1.5 RSCs after S. uberis infection. Among these, 106 underwent a statistically significant increase and are listed in Table 3.
Table 3.
MFGPs showing a statistically significant increase in abundance after S. uberis infectiona
| Accession no.b | Description | RSC |
|---|---|---|
| Q6ECI6 | Integrin beta-2 | 5.85 |
| P05164 | Myeloperoxidase | 5.65 |
| Q3SWX7 | Annexin A3 | 5.49 |
| P02544 | Vimentin | 5.23 |
| P21333 | Filamin A | 5.18 |
| Q92176 | Coronin 1A | 5.07 |
| Q258K2 | Myosin 9 | 4.88 |
| P28783 | Protein S100A9 | 4.84 |
| Q1JPB0 | Leukocyte elastase inhibitor | 4.79 |
| P20000 | Aldehyde dehydrogenase, mitochondrial | 4.33 |
| P20700 | Lamin B | 4.14 |
| Q8SPQ0 | Chitinase-3-like protein 1 | 4.10 |
| P27214 | Annexin A11 | 4.09 |
| O46522 | Cytochrome b245 heavy chain | 4.02 |
| P47843 | Solute carrier family 2, facilitated glucose transport 3 | 3.91 |
| Q29477 | Lactotransferrin | 3.85 |
| Q2UVX4 | Complement C3 | 3.81 |
| P11979 | Pyruvate kinase isozyme M1/M2 | 3.67 |
| O75367 | Core histone macro-H2A.1 | 3.58 |
| B6E141 | Haptoglobin | 3.58 |
| Q8VEK3 | Heterogeneous nuclear ribonucleoprotein U | 3.58 |
| Q9XSJ4 | Alpha-enolase | 3.54 |
| A0A1F3 | l-Lactate dehydrogenase A chain | 3.51 |
| Q32LG3 | Malate dehydrogenase, mitochondrial | 3.43 |
| O46521 | Cytochrome b245 light chain | 3.40 |
| Q58CQ2 | Actin-related protein 2/3 complex subunit 1B | 3.32 |
| P61157 | Actin-related protein 3 | 3.23 |
| Q3SYV4 | Adenylyl cyclase-associated protein 1 | 3.19 |
| P46193 | Annexin A1 | 3.16 |
| P00829 | ATP synthase subunit beta, mitochondrial | 3.13 |
| Q3T0E5 | Adipocyte plasma membrane-associated protein | 3.10 |
| Q5E9B1 | l-Lactate dehydrogenase B chain | 3.05 |
| Q0VCG9 | Pentraxin-related protein PTX3 | 3.05 |
| Q28125 | Intercellular adhesion molecule 3 | 3.00 |
| O95498 | Vascular noninflammatory molecule 2 | 2.95 |
| Q9UM07 | Protein-arginine deiminase type 4 | 2.95 |
| Q32LP0 | Fermitin family homolog 3 | 2.89 |
| P19483 | ATP synthase subunit alpha, mitochondrial | 2.88 |
| P11678 | Eosinophil peroxidase | 2.84 |
| Q3UP87 | Neutrophil elastase | 2.78 |
| Q13349 | Integrin alpha-D | 2.78 |
| P02313 | Nonhistone chromosomal protein HMG-17 | 2.78 |
| P10096 | Glyceraldehyde-3-phosphate dehydrogenase | 2.73 |
| P05044 | Sorcin | 2.72 |
| Q9XT27 | Ceruloplasmin | 2.72 |
| Q63610 | Tropomyosin alpha-3 chain | 2.66 |
| Q99N16 | Leukotriene-B4 omega-hydroxylase 2 | 2.60 |
| P52272 | Heterogeneous nuclear ribonucleoprotein M | 2.52 |
| O00160 | Unconventional myosin If | 2.52 |
| P40673 | High-mobility-group protein B2 | 2.52 |
| Q0VCW4 | l-Serine dehydratase/l-threonine deaminase | 2.45 |
| O77774 | Neutrophil cytosol factor 1 | 2.45 |
| O77775 | Neutrophil cytosol factor 2 | 2.45 |
| A7VJC2 | Heterogeneous nuclear ribonucleoproteins A2/B1 | 2.45 |
| P19134 | Serotransferrin | 2.38 |
| Q14739 | Lamin B receptor | 2.37 |
| Q8IZW8 | Tensin 4 | 2.37 |
| P35246 | Pulmonary surfactant-associated protein D | 2.37 |
| Q5R7W2 | Phosphate carrier protein, mitochondrial | 2.37 |
| O15144 | Actin-related protein 2/3 complex subunit 2 | 2.33 |
| Q8BJS4 | SUN domain-containing protein 2 | 2.29 |
| A7MB62 | Actin-related protein 2 | 2.28 |
| P04157 | Receptor-type tyrosine-protein phosphatase C | 2.21 |
| Q96KK5 | Histone H2A type 1-H | 2.20 |
| P21796 | Voltage-dependent anion-selective channel protein 1 | 2.17 |
| O35737 | Heterogeneous nuclear ribonucleoprotein H | 2.12 |
| Q3SYX9 | Actin-related protein 2/3 complex subunit 5 | 2.12 |
| Q3TRM8 | Hexokinase 3 | 2.12 |
| P32007 | ADP/ATP translocase 3 | 2.10 |
| P29350 | Tyrosine-protein phosphatase non-receptor type 6 | 2.03 |
| Q8VC88 | Grancalcin | 2.03 |
| P12725 | Alpha-1-antiproteinase | 2.02 |
| Q9GL30 | Phospholipase B-like 1 | 2.02 |
| Q68CQ1 | Maestro heat-like repeat-containing protein family member 7 | 2.02 |
| Q13838 | Spliceosome RNA helicase DDX39B | 2.02 |
| P54230 | Cathelicidin 1 | 1.97 |
| P02584 | Profilin 1 | 1.96 |
| O75131 | Copine 3 | 1.92 |
| Q56JZ9 | Glia maturation factor gamma | 1.92 |
| P85521 | Scavenger receptor cysteine-rich type 1 protein M130 | 1.92 |
| Q2KJD0 | Tubulin beta-5 chain | 1.92 |
| P28782 | Protein S100A8 | 1.86 |
| P02253 | Histone H1.2 | 1.82 |
| P30358 | Arachidonate 5-lipoxygenase-activating protein | 1.81 |
| Q5MIB5 | Glycogen phosphorylase, liver form | 1.81 |
| O02849 | Protein-arginine deiminase type 3 | 1.81 |
| D3ZZL9 | GRIP and coiled-coil domain-containing protein 2 | 1.81 |
| Q3T149 | Heat shock protein beta-1 | 1.78 |
| P13796 | Plastin 2 | 1.76 |
| Q3T035 | Actin-related protein 2/3 complex subunit 3 | 1.75 |
| Q95218 | Deleted in malignant brain tumor 1 protein | 1.73 |
| P41976 | Superoxide dismutase [Mn], mitochondrial | 1.72 |
| P48975 | Actin, cytoplasmic 1 | 1.69 |
| Q5E9J1 | Heterogeneous nuclear ribonucleoprotein F | 1.69 |
| A4IF97 | Myosin regulatory light chain 12B | 1.69 |
| P50415 | Cathelicidin 3 | 1.67 |
| P81947 | Tubulin alpha-1B chain | 1.65 |
| Q3ZBV8 | Threonine-tRNA ligase, cytoplasmic | 1.63 |
| Q8BFZ3 | Beta-actin-like protein 2 | 1.60 |
| P62803 | Histone H4 | 1.59 |
| P09769 | Tyrosine-protein kinase Fgr | 1.58 |
| P17453 | Bactericidal permeability-increasing protein | 1.56 |
| P17697 | Clusterin | 1.55 |
| Q148J6 | Actin-related protein 2/3 complex subunit 4 | 1.55 |
| P47791 | Glutathione reductase, mitochondrial | 1.53 |
| A7E3Q8 | Plastin 3 | 1.52 |
Significant (P ≤ 0.05, beta-binomial test) increase in abundance (RSC > 1.5) after S. uberis infection (third day after the second inoculation) according to LTQ-Orbitrap GeLC-MS/MS.
Accession number in the Uniprot database.
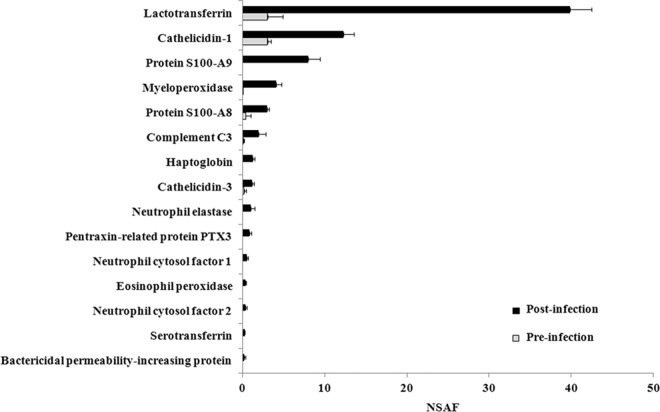
In agreement with the DIGE results, a number of proteins that increased upon infection were involved in innate immune response processes and had direct or indirect antimicrobial activity. These included lactotransferrin, cathelicidins, the calprotectin subunits S100A9 and S100A8, myeloperoxidase, complement C3, haptoglobin, immune cell-related proteases, pentraxin-related protein PTX3, neutrophil cytosolic factors, serotransferrin, and bactericidal permeability-increasing protein. Figure 5 illustrates their normalized spectral abundance factors before and after infection. Several other proteins had roles in inflammatory or immune response processes. Significantly increased upon infection were also the cytoskeletal proteins vimentin, myosins, actins, actin-related proteins, tropomyosins, tubulins, annexins, profilins, coronins, and tensins, many of which are involved in vesicular trafficking (Table 3; see File S1 in the supplemental material).
Fig 5.
Normalized spectral abundance factors (NSAFs) of selected antimicrobial proteins. Bars indicate the abundance and standard deviation abundance of antimicrobial proteins in the milk fat fraction before and after infection. Proteins having direct antimicrobial functions according to the Uniprot Knowledgebase and showing statistically significant changes in abundance (RSC) between the two conditions (Table 3) are reported.
Localization of S100A9 and cathelicidin in mammary tissues.
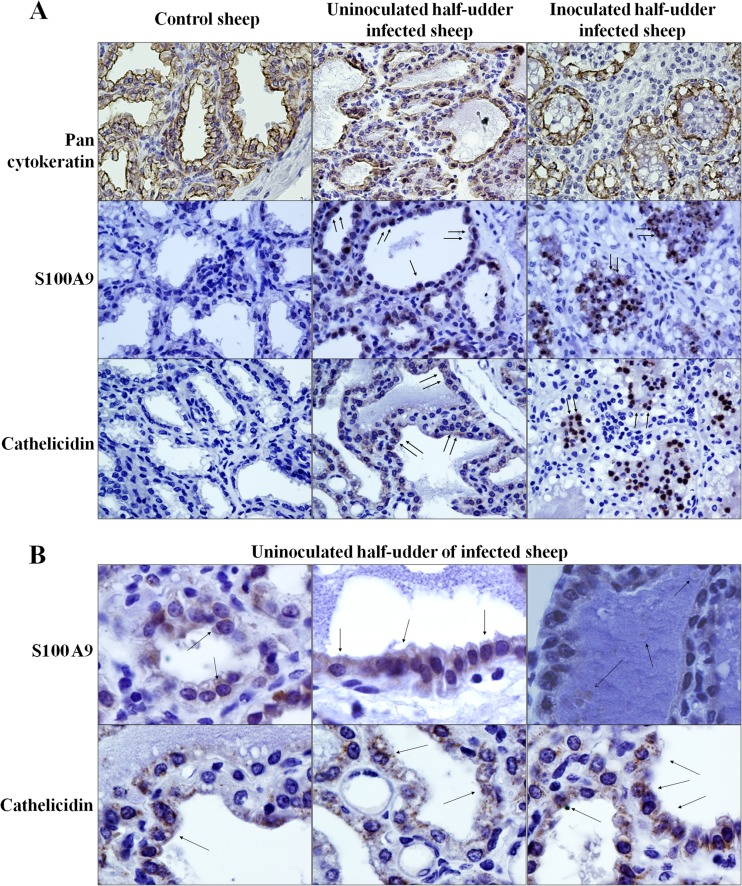
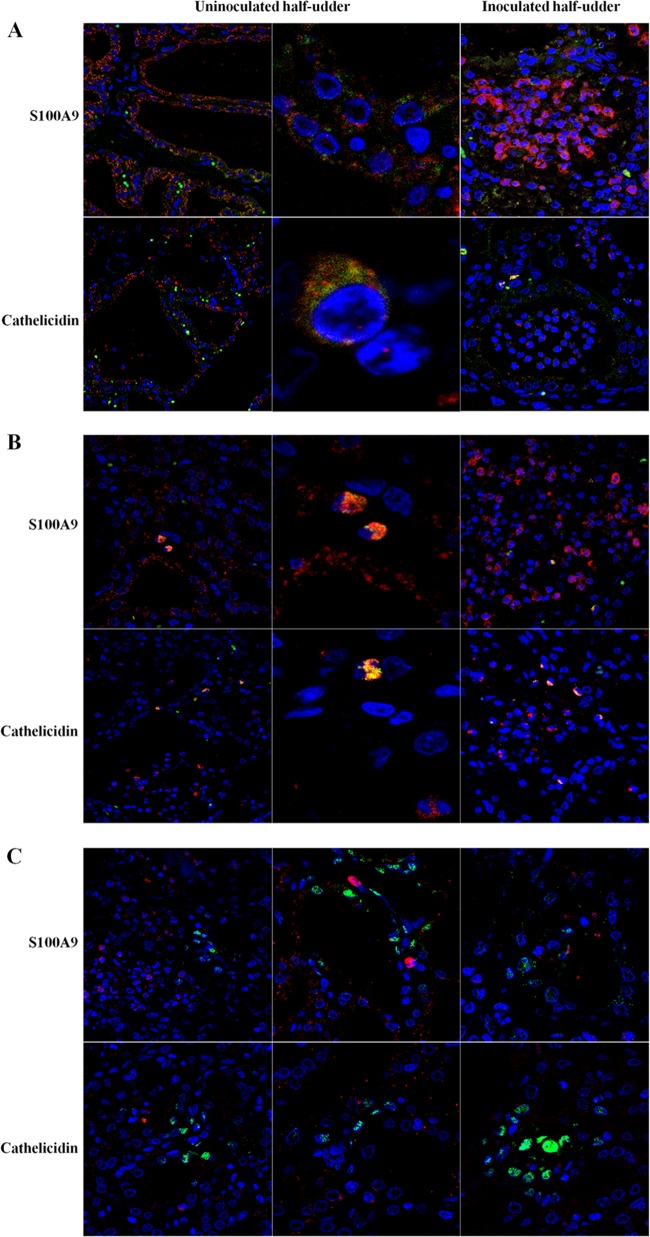
According to the results obtained in this work and the observations that emerged in a previous study on natural infection by Mycoplasma agalactiae (18), cathelicidins and the calprotectin subunits S100A9 and S100A8 are among the proteins undergoing the highest increase upon bacterial infection of mammary tissues. In order to investigate their cellular origin and to gain insights into the validity of the MFG proteomic model to assess the contribution of MECs to innate immune defense responses, mammary tissues collected from all animals at necropsy were evaluated by immunohistochemistry (IHC). Figure 6 reports the results obtained when mammary tissues were probed with antibodies against the calprotectin subunit S100A9 and cathelicidin. Antipancytokeratin antibodies were used to highlight epithelial cells and to obtain specific MEC staining.
Fig 6.
Immunohistochemical analysis of mammary tissues. (A) Reactivity of mammary tissues from the indicated animals for pancytokeratin, S100A9, and cathelicidin. (B) Reactivity of mammary tissues from the uninoculated half udders of infected animals for S100A9 and cathelicidin. All animals included in the experiment were tested in replicate experiments. Arrows, focal points of reactivity.
As expected, the control animal showed no reactivity for either S100A9 or cathelicidin (Fig. 6A, left), while mammary tissue from inoculated half udders showed a strong and specific reactivity for both S100A9 and cathelicidin, localized mainly within the milk duct lumens (Fig. 6A, right). The most interesting information emerged when uninoculated half udders of infected animals were tested (Fig. 6A, middle, and B). Here, a distinctly positive signal was observed in cells lining the milk ducts, suggesting that epithelial cells are likely producing both S100A9 and cathelicidin. In addition, the two proteins showed different patterns of reactivity: S100A9 was more diffusely distributed within the cytoplasm, while cathelicidin displayed a granular staining. In several occurrences, positive staining was observed in the duct lumen inside globular formations within milk, possibly corresponding to proteins localized within MFGs (Fig. 6B, top right).
In order to further investigate the cellular origin of S100A9 and cathelicidin, a colocalization experiment was carried out by confocal microscopy with antibodies against the two proteins and against epithelial cell, neutrophil, and macrophage markers. Figure 7 reports the reactivity of S100A9 and cathelicidin in MECs (Fig. 7A), in neutrophils (Fig. 7B), and in macrophages (Fig. 7C). Cell nuclei are colored blue (Hoechst staining), S100A9 and cathelicidin are colored red, and cell markers are colored green.
Fig 7.
Immune colocalization analysis of mammary tissues. The reactivities of mammary tissues to antibodies against S100A9 and cathelicidin (green signals), together with cell-type-specific antibodies (red signals), are shown. (A) Epithelial cells; (B) neutrophils; (C) macrophages.
As illustrated in Fig. 7A (left and middle), S100A9 and cathelicidin were clearly expressed in MECs of neighboring, uninoculated half udders, with a positive signal (red) located in pancytokeratin-positive cells (green). Notably, both signals were located in cells lining the lactiferous ducts. On the other hand, when inoculated half udders were tested, the S100A9 and cathelicidin signals were mainly located in the milk duct lumens (Fig. 7A, right). This is most likely a consequence of acute mastitis, marked by protein secretion, tissue disintegration, and massive neutrophil recruitment. Accordingly, in neutrophils, a positive signal for the two proteins was observed in both the uninoculated and inoculated halves, mainly outside and inside alveoli, respectively (Fig. 7B). Neither S100A9 nor cathelicidin was detected in macrophages (Fig. 7C).
Detection of S. uberis in milk and mammary tissues.
The detection of specific immunological staining for S100A9 and cathelicidin in uninoculated half udders suggested dissemination of the S. uberis infection from the neighboring inoculated half udder, an occurrence reported in other experimental infections with environmental pathogens (40). To test this hypothesis, all milk and mammary tissue samples collected during the study were tested by PCR for S. uberis. Results are reported in Table 4. All animals, including the uninfected control, were found to be negative preinoculation (D0) and 5 days after the first inoculation (5DIi). Five days after the second inoculation (5DIIi), all inoculated halves were found to be positive for S. uberis (Table 4). The results of milk PCR were in accordance with those of bacterial cultures, since positivity for S. uberis was detected only after the second inoculation. Mammary tissue PCR was positive for S. uberis in both the inoculated and uninoculated halves, supporting transmission of the pathogen from the inoculated half udder to the neighboring one. In the control animal, milk and tissue PCR remained negative throughout the infection experiment.
Table 4.
Results obtained by milk and tissue PCR for detection of S. uberis
| Sample | Time of sample collection | Half udder tested | PCR result |
|---|---|---|---|
| Milk sample_1 | D0 | Uninoculated | − |
| D0 | Inoculated | − | |
| Milk sample_2 | D0 | Uninoculated | − |
| D0 | Inoculated | − | |
| Milk sample_3 | D0 | Uninoculated | − |
| D0 | Inoculated | − | |
| Milk sample_K | D0 | Uninoculated | − |
| Milk sample_1 | 5DIi | Uninoculated | − |
| 5DIi | Inoculated | − | |
| Milk sample_2 | 5DIi | Uninoculated | − |
| 5DIi | Inoculated | − | |
| Milk sample_3 | 5DIi | Uninoculated | − |
| 5DIi | Inoculated | − | |
| Milk sample_K | 5DIi | Uninoculated | − |
| Milk sample_1 | 3DIIi | Uninoculated | − |
| 3DIIi | Inoculated | + | |
| Milk sample_2 | 3DIIi | Uninoculated | − |
| 3DIIi | Inoculated | + | |
| Milk sample_3 | 3DIIi | Uninoculated | − |
| 3DIIi | Inoculated | + | |
| Milk sample_K | 3DIIi | Uninoculated | − |
| Breast tissue sample_1 | Postmortem | Uninoculated | + |
| Postmortem | Inoculated | + | |
| Breast tissue sample_2 | Postmortem | Uninoculated | + |
| Postmortem | Inoculated | + | |
| Breast tissue sample_3 | Postmortem | Uninoculated | + |
| Postmortem | Inoculated | + | |
| Breast tissue sample_K | Postmortem | Uninoculated | − |
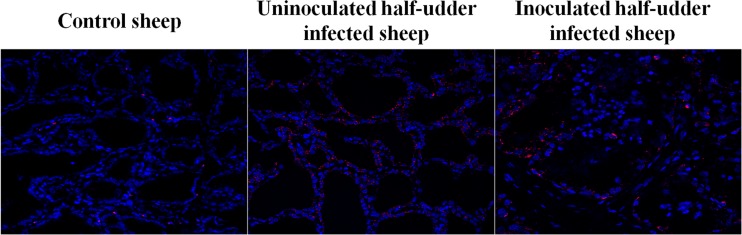
The transmission of S. uberis to the uninoculated half udder of infected animals was also confirmed by fluorescent in situ hybridization (FISH). In fact, positive signals corresponding to S. uberis were detected in both halves, as shown in Fig. 8. In the inoculated half udder, bacteria were mainly localized in macrophages within the milk duct lumens, while in the neighboring half, they were mainly located in the proximity of MECs. In the control animal, mammary tissue from both udder halves was also almost completely negative for S. uberis by FISH, excepted for few scattered spots of reactivity.
Fig 8.
Results of FISH for S. uberis. The reactivity obtained with an S. uberis-specific probe is shown in the mammary tissues of the control sheep, the uninoculated half udders of infected animals, and the inoculated half udders of infected animals.
Time course of S100A9 and cathelicidin release by MECs inferred from MFG proteomics.
In order to validate the results obtained with the proteomics analysis and the immunological and molecular approaches and to assess the kinetics of S100A9 and cathelicidin expression in MECs, a Western immunoblotting experiment was carried out on milk samples collected over the course of the experimental infection. Figure 9 reports the total protein profiles of MFGPs (Fig. 9A) and the reactivity for S100A9 and cathelicidin (Fig. 9B) observed in inoculated and uninoculated half udders, as well as in the negative-control animal. Samples collected preinoculation (D0), from the 1st to the 5th days after the first inoculation (1DIi to 5DIi), and on the 1st, 3rd, and 6th days after the second inoculation (1DIIi, 3DIIi, and 6DIIi) were tested.
Fig 9.
Time course of the MFGP profile and of S100A9 and cathelicidin reactivity over the course of infection. (A). Total MFGP profile over the course of infection, starting with preinoculation (D0) and continuing to the last day of the second inoculation with S. uberis (6DIIi). (B) Reactivity of S100A9 and cathelicidin in inoculated and uninoculated half udders of infected animals and in control animals. Lanes M, molecular mass markers.
Significant alterations in the total protein pattern were already evident 1 day after the first inoculation (1DIi), increasing in intensity and peaking at 2DIi, and then slowly reverting to the initial condition at day 5DIi. Following the second inoculation, the protein profile underwent an immediate and dramatic alteration, which was maintained throughout the 6 days of infection (1DIIi to 6DIIi) The same behavior in the reactivity of S100A9 and cathelicidin was observed by Western immunoblotting (Fig. 9B); an increase in the abundance of both proteins started at day 1 of the first infection (1DIi), decreased after day 3 (3DIi), and then rose again at day 1 after the second inoculation (3DIIi). Remarkably, the same pattern of reactivity, although with a weaker intensity, was observed when the neighboring, uninoculated half udders were tested, indicating the rapid increase of these proteins following an inflammatory stimulus. Both proteins were always undetectable in milk collected from the control animal.
DISCUSSION
In this study, we investigated the events taking place in the mammary gland following experimental infection with S. uberis, a common etiologic agent of mastitis in sheep. S. uberis is considered an environmental pathogen since it is commonly isolated from the environment, milking machines, milkers' hands, and the skin of the teats. Mastitis may therefore originate when the mammary gland is exposed to higher loads of this bacterial species. Inflammation triggered by S. uberis may occur without clinical signs and spontaneously resolve, or it may become chronic. In order to investigate these heterogeneous balances between infecting bacteria and host innate immune response, we have conceived a two-step infection in one udder half, where each individual received two different bacterial inocula, with a 1-log10-unit increase in the bacterial load between the two inocula, 1 week apart. As expected, the first, lower dose of 1 × 106 CFU did not cause any clinical sign of mastitis, nor could S. uberis be isolated in culture or detected in milk by PCR. On the other hand, a second inoculum of 2 × 107 CFU led to the manifestation of clinical mastitis and pathogen detection in milk. This is in agreement with the observations of Lasagno et al. (41). These authors reported that inoculation with S. uberis at between 8.0 × 102 CFU and 2.4 × 106 CFU was not sufficient to induce clinical mastitis in dairy goats; only higher loads of S. uberis (1.7 × 108 CFU) enabled the induction of mild clinical manifestations (41). This model of infection therefore allowed us to evaluate the host immune defense in a framework of infection control and quick bacterial eradication and under a subsequent condition of clinical mastitis with bacterial survival and persistence. Furthermore, in this model of infection, bacterial persistence and replication might easily allow the transmission of infection to the contralateral udder half. Previous studies have reported the frequent occurrence of pathogen dissemination from infected udder halves to contralateral uninfected ones, where infection is established without evidence of clinical signs (40, 42). Indeed, this event occurred during our experimental infection of sheep with S. uberis, probably at the time of the second bacterial infection, with the inoculated udder half modeling for clinical mastitis and the contralateral, uninoculated udder half modeling for a subclinical infection, as demonstrated by PCR, fluorescent in situ hybridization, and changes in the proteomic profile of MFGs. This favorable occurrence enabled us to investigate the phenomena that take place during subclinical mastitis: (i) the role played by MECs as the first line of defense against microbes entering the mammary gland, (ii) the response of the mammary gland to small bacterial loads, and (iii) the mediators released in milk when animals are carrying the bacteria but are not showing any evident clinical signs.
To assess the changes occurring in the actively secreting mammary epithelium and to specifically probe the contents of lactating cells, we used MFGs as a means to investigate protein expression levels within MECs. The use of MFGs to investigate the changes occurring in MECs upon exposure to bacterial pathogens has proven successful in a previous study on natural infection of sheep by Mycoplasma agalactiae (18). The usefulness of the MFG as a surrogate of the lactating cell is also corroborated by the work of Brenaut et al. (43), who exploited MFGs as a main source of mammary RNAs to evaluate the dynamics of the global transcriptional response in MECs during bacterial infection. In fact, MFGs originate from the MEC as vesicles released from the apical part of the polarized secretory epithelium and, during this process, carry portions of the lactating cell cytoplasm with them (44). This phenomenon presents with various intensities in different animal species, and it is believed to be more prominent in small ruminants than in bovines (31, 32, 45, 46).
In the work presented here, the MFG proteomics approach provided results in line with those obtained with M. agalactiae. Upon infection with S. uberis, the fat fraction was strongly enriched in, among other proteins, lactotransferrin, cathelicidins, and calprotectin, according to both DIGE and GeLC-MS/MS, together with several other acute-phase reactants, including haptoglobin, myeloperoxidase, and leukocyte elastase inhibitor, identified by means of the latter approach (Table 3; Fig. 5).
These proteins are mainly released by immune cells as part of the innate response against pathogens (4); however, the role of epithelial cells as the first line of defense, especially in secretory epithelia, is gaining importance. Lactotransferrin is an iron-binding protein mainly synthesized by MECs and, to a lesser extent, by neutrophils (47), having antibacterial, immune-modulating, anti-inflammatory, and antioxidant activity (48, 49). Haptoglobin is found to be increased in plasma and in milk in clinical mastitis and in chronic subclinical mastitis, together with amyloid protein A (24), and several studies indicated that one of the main sources of these two proteins in milk might be MECs (50–52). S100A9 and S100A8 are mainly expressed in the cytoplasm of neutrophils and monocytes but also in activated endothelial and epithelial cells (53–59). As a consequence, S100 proteins are also being proposed to be markers for several inflammatory diseases. Alone or in association as calprotectin (54), S100A8 and S100A9 exert strong proinflammatory and chemotactic activities (60) by promoting leukocyte recruitment (61, 62). In addition, the calprotectin heterodimer exerts direct antimicrobial functions by sequestering zinc and manganese (59, 63–66). The concerted increase in abundance of the two monomers S100A9 and S100A8 observed in MFGs upon infection with S. uberis (Table 3 and Fig. 5) and M. agalactiae (18), together with the detection of S100A9 in MECs by IHC, is strongly suggestive of their ability to produce and release the calprotectin heterodimer.
Cathelicidins are a family of antimicrobial peptides (AMPs) that act as multifunctional effectors in innate immunity (4, 25, 67, 68). Cathelicidins are synthesized as preproproteins and are cleaved by proteases (such as serine proteases, proteinase 3, elastase, and kallikrein) to generate the cathelin domain and a C-terminal AMP (LL-37 in humans) (69, 70). These domains have antimicrobial properties, with the cathelin domain possessing chemotactic and proinflammatory activity and the AMP exploiting a direct lytic activity against bacteria (71, 72). These molecules are the primary constituents of the neutrophil secondary granule, and very high expression levels have been reported under different inflammatory conditions (73). This is in agreement with the strongly positive signal observed for cathelicidin in neutrophils of the inoculated udder halves by both IHC and immune colocalization (Fig. 6A and Fig. 7).
In recent years, cathelicidins have increasingly been detected in other cell types (epithelial cells, mast cells, lymphocytes, and keratinocytes), as well as in a wide variety of tissues (oral cavity, skin, intestine, lungs, cervix, etc.) and body fluids (plasma, breast milk, saliva, gastric juice, semen, sweat, and bronchoalveolar fluid) (74). Concerning MECs, the production of cathelicidins and other proinflammatory/chemotactic mediators has been investigated almost exclusively by means of gene expression approaches either during the course of bovine mastitis or through the use of cultured mammary cells, providing contrasting results (75, 76). However, a study evaluating the production dynamics of cathelicidin release by mammary epithelial cells in humans demonstrated that cathelicidin mRNA is already present in cells and exposure to a bacterial pathogen triggers translation and secretion, accompanied by a net decrease in cathelicidin mRNA levels due to its concomitant degradation (67); this might explain the contrasting results obtained by some investigators attempting to assess the ability of MECs to produce cathelicidin and other mediators of innate immunity. Therefore, the choice of a proteomic approach can help to circumvent this problem by directly assessing the protein levels and enabling their localization within the cells of an infected mammary gland.
In our previous work on the proteomics of milk secretions collected from sheep naturally infected by M. agalactiae, the massive increase of antimicrobial and immune defense proteins within MFGs was strongly suggestive of their MEC origin (18). Here, the ability of MECs to produce cathelicidin and calprotectin in vivo was further demonstrated with the same indirect approach, provided by the proteomic analysis of MFGs, but it was further reinforced by means of a direct approach, that is, IHC and immune colocalization in infected mammary tissues. In addition, this again supports the use of MFGs as a noninvasive model to study what is happening within MECs in vivo, as they are able to provide information about the changes that occur in these cells upon exposure to pathogens. In fact, it can be speculated that these cells also have the potential to produce other immune and antimicrobial proteins, as the proteomic results obtained on the MFG fraction might suggest.
Here, IHC and immune colocalization (Fig. 6 and 7) clearly demonstrated that both S100A9 and cathelicidin are present within the MEC cytoplasm, as well as in neutrophils, as expected. In addition, the staining pattern was compatible with their cellular distribution, since it appeared to be finer and more disperse for S100A9, which is associated with microtubules in the cytoplasm, and more granular and compartmentalized for cathelicidin, which is located within secretory granules. In addition, although the epithelial layer is compromised, both proteins can also be seen in association with epithelial cells by immune colocalization in the acutely mastitic, inoculated half udders. Here, however, the significant positivity of neutrophils and their massive recruitment to the infected site could also be seen, as expected from the known ability of these cells to produce and release AMPs and acute-phase-response proteins, as well as from the strong chemoattractant activity exerted by these proteins at the site of infection (77). Conversely, macrophages remained negative in all samples and under all conditions examined.
The most interesting results about the ability of MECs to produce these innate immunity mediators emerged from analysis of tissue and secretions from the uninoculated half udder. In fact, as mentioned above, PCR and FISH confirmed that transfer of the bacterial infection from the inoculated half udder to the neighboring one occurred, creating conditions that were similar to those occurring in a natural subclinical infection. This leads to speculation on the role that these two proteins may have in early detection and in prediction of infection; in fact, it is now known that both represent valuable markers of inflammation, as their presence has been detected in a number of inflammatory conditions.
The kinetic assessment of cathelicidin and S100A9 release in milk from inoculated udder halves showed that these proteins are produced immediately upon exposure to the pathogen. This trend is consistent with that previously described by Smolenski et al. (78), who observed an immediate increase of cathelicidin in milk after intramammary inoculation of different pathogens into lactating cows. In both cases, strong and specific signals were observed 24 h after the first inoculation. The signals peaked after 48 h and then reverted to the initial conditions when infection was not established or peaked and were maintained at elevated levels when infection occurred. Although milk PCR and bacterial culture remained negative after the first low-dose bacterial inoculation, as well as in the cross-infected, uninoculated half udder, the presence of proinflammatory and antimicrobial proteins was clearly detectable. It is likely that MECs are immediately reacting to bacterial cells by producing and releasing proinflammatory and antimicrobial proteins that can be detected in milk as an indicator of inflammation. In addition, positivity for these markers reverts within a few days if infection is not established, while it persists at high levels when infection occurs. These results support a potential significant role of cathelicidin and the calprotectin subunit S100A9 in predicting the presence of a pathogen in the mammary epithelium regardless of clinical symptoms, suggesting their usefulness as tools for the detection of various disease states, including subclinical mastitis.
Supplementary Material
ACKNOWLEDGMENTS
This work was financed by Sardegna Ricerche with the program Progetto Strategico Biotecnologie and by the Italian Ministry of Health and the Istituto Zooprofilattico della Sardegna with the grant “I geni di resistenza e il ruolo dei mediatori dell'infiammazione nelle mastiti ovine,” ID number IZS SA 002/07.
Footnotes
Published ahead of print 17 June 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.00291-13.
REFERENCES
- 1.Zadoks RN, Middleton JR, McDougall S, Katholm J, Schukken YH. 2011. Molecular epidemiology of mastitis pathogens of dairy cattle and comparative relevance to humans. J. Mammary Gland Biol. Neoplasia 16:357–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Vliegher S, Fox LK, Piepers S, McDougall S, Barkema HW. 2012. Mastitis in dairy heifers: nature of the disease, potential impact, prevention, and control. J. Dairy Sci. 95:1025–1040 [DOI] [PubMed] [Google Scholar]
- 3.Marogna G, Rolesu S, Lollai S, Tola S, Leori S. 2010. Clinical findings in sheep farms affected by recurrent bacterial mastitis. Small Rumin. Res. 88:119–125 [Google Scholar]
- 4.Rainard P, Riollet C. 2006. Innate immunity of the bovine mammary gland. Vet. Res. 37:369–400 [DOI] [PubMed] [Google Scholar]
- 5.Beutler B. 2004. Innate immunity: an overview. Mol. Immunol. 40:845–859 [DOI] [PubMed] [Google Scholar]
- 6.Zecconi A, Hamann J, Bronzo V, Moroni P, Giovannini G, Piccinini R. 2000. Relationship between teat tissue immune defences and intramammary infections, Adv. Exp. Med. Biol. 480:287–293 [DOI] [PubMed] [Google Scholar]
- 7.Schultze WD, Bright SC. 1983. Changes in penetrability of bovine papillary duct to endotoxin after milking. Am. J. Vet. Res. 44:2373–2375 [PubMed] [Google Scholar]
- 8.McDougall S, Lopez-Villalobos N, Prosser CG. 2011. Relationship between estimated breeding value for somatic cell count and prevalence of intramammary infection in dairy goats. N. Z. Vet. J. 59:300–304 [DOI] [PubMed] [Google Scholar]
- 9.Middleton JR, Hardin D, Steevens B, Randle R, Tyler JW. 2004. Use of somatic cell counts and California mastitis test results from individual quarter milk samples to detect subclinical intramammary infection in dairy cattle from a herd with a high bulk tank somatic cell count. J. Am. Vet. Med. Assoc. 224:419–423 [DOI] [PubMed] [Google Scholar]
- 10.De Haas Y, Veerkamp RF, Barkema HW, Gröhn YT, Schukken YH. 2004. Associations between pathogen-specific cases of clinical mastitis and somatic cell count patterns. J. Dairy Sci. 87:95–105 [DOI] [PubMed] [Google Scholar]
- 11.McDougall S, Murdough P, Pankey W, Delaney C, Barlow J, Scruton D. 2001. Relationships among somatic cell count, California mastitis test, impedance and bacteriological status of milk in goats and sheep in early lactation. Small Rumin. Res. 40:245–254 [DOI] [PubMed] [Google Scholar]
- 12.Ryan DP, Greenwood PL, Nicholls PJ. 1993. Effect of caprine arthritis-encephalitis virus infection on milk cell count and N-acetyl-beta-glucosaminidase activity in dairy goats. J. Dairy Res. 60:299–306 [DOI] [PubMed] [Google Scholar]
- 13.Pekelder JJ, Veenink GJ, Akkermans JP, van Eldik P, Elving L, Houwers DJ. 1994. Ovine lentivirus induced indurative lymphocytic mastitis and its effect on the growth of lambs. Vet. Rec. 134:348–350 [DOI] [PubMed] [Google Scholar]
- 14.Lerondelle C, Richard Y, Issartial J. 1992. Factors affecting somatic cell counts in goat milk. Small Rumin. Res. 8:129–139 [Google Scholar]
- 15.Gonzalo C, Carriedo JA, Baro JA, san Primitivo F. 1994. Factors influencing variation of test day milk yield, somatic cell count, fat, and protein in dairy sheep. J. Dairy Sci. 77:1537–1542 [DOI] [PubMed] [Google Scholar]
- 16.Gonzalo C, Carriedo JA, García-Jimeno MC, Pérez-Bilbao M, de la Fuente LF. 2010. Factors influencing variation of bulk milk antibiotic residue occurrence, somatic cell count, and total bacterial count in dairy sheep flocks. J. Dairy Sci. 93:1587–1595 [DOI] [PubMed] [Google Scholar]
- 17.Zeng SS, Escobar EN, Popham T. 1997. Daily variations in somatic cell count, composition, and production of alpine goat milk. Small Rumin. Res. 26:253–260 [Google Scholar]
- 18.Addis MF, Pisanu S, Ghisaura S, Pagnozzi D, Marogna G, Tanca A, Biosa G, Cacciotto C, Alberti A, Pittau M, Roggio T, Uzzau S. 2011. Proteomics and pathway analyses of the milk fat globule in sheep naturally infected by Mycoplasma agalactiae provide indications of the in vivo response of the mammary epithelium to bacterial infection. Infect. Immun. 79:3833–3845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Almeida RA, Gillespie BE, Lewis MJ, Headrick SJ, Oliver SP. 2004. Development of an experimental Streptococcus uberis intramammary infection model, p 282–283 In Annual meeting—National Mastitis Council Incorporated. Proceedings of the 43rd Annual Meeting of the National Mastitis Council [Google Scholar]
- 20.Pedersen L, Aalbaek B, Rontved C, Ingvarstsen K, Sorensen N, Heegaard P, Jensen H. 2003. Early pathogenesis and inflammatory response in experimental bovine mastitis due to Streptococcus uberis. J. Comp. Pathol. 128:156–164 [DOI] [PubMed] [Google Scholar]
- 21.Rambeaud M, Almeida R, Pighetti G, Oliver S. 2003. Dynamics of leukocytes and cytokines during experimentally induced Streptococcus uberis mastitis. Vet. Immunol. Immunopathol. 96:193–205 [DOI] [PubMed] [Google Scholar]
- 22.Schnare M, Barton GM, Holt AC, Takeda K, Akira S, Medzhitov R. 2001. Toll-like receptors control activation of adaptive immune responses. Nat. Immunol. 2:947–950 [DOI] [PubMed] [Google Scholar]
- 23.Akira S, Uematsu S, Takeuchi O. 2006. Pathogen recognition and innate immunity. Cell 124:783–801 [DOI] [PubMed] [Google Scholar]
- 24.Schukken YH, Günther J, Fitzpatrick J, Fontaine MC, Goetze L, Holst O, Leigh J, Petzl W, Schuberth HJ, Sipka A, Smith DG, Quesnell R, Watts J, Yancey R, Zerbe H, Gurjar A, Zadoks RN, Seyfert HM. 2011. Host-response patterns of intramammary infections in dairy cows. Vet. Immunol. Immunopathol. 144:270–289 [DOI] [PubMed] [Google Scholar]
- 25.Van Wetering S, Tjabringa GS, Hiemstra PS. 2005. Interactions between neutrophil-derived antimicrobial peptides and airway epithelial cells. J. Leukoc. Biol. 77:444–450 [DOI] [PubMed] [Google Scholar]
- 26.Bowdish DM, Davidson DJ, Lau YE, Lee K, Scott MG, Hancock RE. 2005. Impact of LL-37 on anti-infective immunity. J. Leukoc. Biol. 77:451–459 [DOI] [PubMed] [Google Scholar]
- 27.Akerstedt M, Waller KP, Sternesjö A. 2009. Haptoglobin and serum amyloid A in bulk tank milk in relation to raw milk quality. J. Dairy Res. 76:483–489 [DOI] [PubMed] [Google Scholar]
- 28.Eckersall PD. 2004. The time is right for acute phase proteins assay. Vet. J. 168:3–5 [DOI] [PubMed] [Google Scholar]
- 29.Eckersall PD, Young FJ, Nolan AM, Knight CH, McComb C, Waterston MM, Hogarth CJ, Scott EM, Fitzpatrick JL. 2006. Acute phase proteins in bovine milk in an experimental model of Staphylococcus aureus subclinical mastitis. J. Dairy Sci. 89:1488–1501 [DOI] [PubMed] [Google Scholar]
- 30.Downing TE, Sporn TA, Bollinger RR, Davis RD, Parker W, Lin SS. 2008. Pulmonary histopathology in an experimental model of chronic aspiration is independent of acidity. Exp. Biol. Med. (Maywood) 233:1202–1212 [DOI] [PubMed] [Google Scholar]
- 31.Pisanu S, Ghisaura S, Pagnozzi D, Biosa G, Tanca A, Roggio T, Uzzau S, Addis MF. 2011. The sheep milk fat globule membrane proteome. J. Proteomics 74:350–358 [DOI] [PubMed] [Google Scholar]
- 32.Pisanu S, Ghisaura S, Pagnozzi D, Falchi G, Biosa G, Tanca A, Roggio T, Uzzau S, Addis MF. 2012. Characterization of sheep milk fat globule proteins by two-dimensional polyacrylamide gel electrophoresis/mass spectrometry and generation of a reference map. Int. Dairy J. 24:78–86 [Google Scholar]
- 33.Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- 34.Terova G, Addis MF, Preziosa E, Pisanu S, Pagnozzi D, Biosa G, Gornati R, Bernardini G, Roggio T, Saroglia M. 2011. Effects of postmortem storage temperature on sea bass (Dicentrarchus labrax) muscle protein degradation: analysis by 2-D DIGE and MS. Proteomics 11:2901–2910 [DOI] [PubMed] [Google Scholar]
- 35.Tanca A, Biosa G, Pagnozzi D, Addis MF, Uzzau S. 2013. Comparison of detergent-based sample preparation workflows for LTQ-Orbitrap analysis of the Escherichia coli proteome. Proteomics 10.1002/pmic.201200478 [DOI] [PubMed] [Google Scholar]
- 36.Spivak M, Weston J, Bottou L, Kall L, Noble WS. 2009. Improvements to the percolator algorithm for peptide identification from shotgun proteomics data sets. J. Proteome Res. 8:3737–3745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Old WM, Meyer-Arendt K, Aveline-Wolf L, Pierce KG, Mendoza A, Sevinsky JR, Resing KA, Ahn NG. 2005. Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Mol. Cell. Proteomics 4:1487–1502 [DOI] [PubMed] [Google Scholar]
- 38.Zybailov B, Mosley AL, Sardiu ME, Coleman MK, Florens L, Washburn MP. 2006. Statistical analysis of membrane proteome expression changes in Saccharomyces cerevisiae. J. Proteome Res. 5:2339–2347 [DOI] [PubMed] [Google Scholar]
- 39.Pham TV, Piersma SR, Warmoes M, Jimenez CR. 2010. On the beta-binomial model for analysis of spectral count data in label-free tandem mass spectrometry based proteomics. Bioinformatics 26:363–369 [DOI] [PubMed] [Google Scholar]
- 40.Mitterhuemer S, Petzl W, Krebs S, Mehne D, Klanner A, Wolf E, Zerbe H, Blum H. 2010. Escherichia coli infection induces distinct local and systemic transcriptome responses in the mammary gland. BMC Genomics 11:138. 10.1186/1471-2164-11-138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lasagno MC, Vissio C, Reinoso EB, Raspanti C, Yaciuk R, Larriestra AJ, Odierno LM. 2012. Development of an experimentally induced Streptococcus uberis subclinical mastitis in goats. Vet. Microbiol. 154:376–383 [DOI] [PubMed] [Google Scholar]
- 42.Dopfer D, Barkema HW, Lam TJ, Schukken YH, Gaastra W. 1999. Recurrent clinical mastitis caused by Escherichia coli in dairy cows. J. Dairy Sci. 82:80–85 [DOI] [PubMed] [Google Scholar]
- 43.Brenaut P, Bangera R, Bevilacqua C, Rebours E, Cebo C, Martin P. 2012. Validation of RNA isolated from milk fat globules to profile mammary epithelial cell expression during lactation and transcriptional response to a bacterial infection. J. Dairy Sci. 95:6130–6144 [DOI] [PubMed] [Google Scholar]
- 44.Heid HW, Keenan TW. 2005. Intracellular origin and secretion of milk fat globules. Eur. J. Cell Biol. 84:245–258 [DOI] [PubMed] [Google Scholar]
- 45.Pisanu S, Marogna G, Pagnozzi D, Piccinini M, Leo G, Tanca A, Roggio AM, Roggio T, Uzzau S, Addis MF. 2012. Characterization of size and composition of milk fat globules from Sarda and Saanen dairy goats. Small Rumin. Res. 109:141–151 [Google Scholar]
- 46.Cebo C, Caillat H, Bouvier P, Martin P. 2010. Major proteins of the goat milk fat globule membrane. J. Dairy Sci. 93:868–876 [DOI] [PubMed] [Google Scholar]
- 47.Harmon RJ, Newbould FHS. 1980. Neutrophil leukocyte as a source of lactoferrin in bovine milk. Am. J. Vet. Res. 41:1603–1606 [Google Scholar]
- 48.Legrand D, Elass E, Pierce A, Mazurier J. 2004. Lactoferrin and host defence: an overview of its immuno-modulating and anti-inflammatory properties. Biometals 17:225–229 [DOI] [PubMed] [Google Scholar]
- 49.Brock J. 1995. Lactoferrin: a multifunctional immunoregulatory protein? Immunol. Today 16:417–419 [DOI] [PubMed] [Google Scholar]
- 50.Petersen HH, Nielsen JP, Heegaard PM. 2004. Application of acute phase protein measurements in veterinary clinical chemistry. Vet. Res. 35:163–187 [DOI] [PubMed] [Google Scholar]
- 51.Wellnitz O, Kerr DE. 2004. Cryopreserved bovine mammary cells to model epithelial response to infection. Vet. Immunol. Immunopathol. 101:191–202 [DOI] [PubMed] [Google Scholar]
- 52.Lai IH, Tsao JH, Lu YP, Lee JW, Zhao X, Chien FL, Mao SJ. 2009. Neutrophils as one of the major haptoglobin sources in mastitis affected milk. Vet. Res. 40:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bargagli E, Olivieri C, Prasse A, Bianchi N, Magi B, Cianti R, Bini L, Rottoli P. 2008. Calgranulin B (S100A9) levels in bronchoalveolar lavage fluid of patients with interstitial lung diseases. Inflammation 31:351–354 [DOI] [PubMed] [Google Scholar]
- 54.Striz I, Trebichavsky I. 2004. Calprotectin: a pleiotropic molecule in acute and chronic inflammation. Physiol. Res. 53:245–253 [PubMed] [Google Scholar]
- 55.Barthe C, Figarella C, Carrere J, Guy-Crotte O. 1991. Identification of “cystic fibrosis protein” as a complex of two calcium-binding proteins present in human cells of myeloid origin. Biochim. Biophys. Acta 1096:175–177 [DOI] [PubMed] [Google Scholar]
- 56.Berntzen HB, Olmez U, Fagerhol MK, Munthe E. 1991. The leukocyte protein L1 in plasma and synovial fluid from patients with rheumatoid arthritis and osteoarthritis. Scand. J. Rheumatol. 20:74–82 [DOI] [PubMed] [Google Scholar]
- 57.Hu SP, Harrison C, Xu K, Cornish CJ, Geczy CL. 1996. Induction of the chemotactic S100 protein, CP-10, in monocyte/macrophages by lipopolysaccharide. Blood 87:3919–3928 [PubMed] [Google Scholar]
- 58.Lusitani D, Malawista SE, Montgomery RR. 2003. Calprotectin, an abundant cytosolic protein from human polymorphonuclear leukocytes, inhibits the growth of Borrelia burgdorferi. Infect. Immun. 71:4711–4716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu JZ, Jellbauer S, Poe AJ, Ton V, Pesciaroli M, Kehl-Fie TE, Restrepo NA, Hosking MP, Edwards RA, Battistoni A, Pasquali P, Lane TE, Chazin WJ, Vogl T, Roth J, Skaar EP, Raffatellu M. 2012. Zinc sequestration by the neutrophil protein calprotectin enhances Salmonella growth in the inflamed gut. Cell Host Microbe 11:227–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Raquil MA, Anceriz N, Rouleau P, Tessier PA. 2008. Blockade of antimicrobial proteins S100A8 and S100A9 inhibits phagocyte migration to the alveoli in streptococcal pneumonia. J. Immunol. 180:3366–3374 [DOI] [PubMed] [Google Scholar]
- 61.Ryckman C, Vandal K, Rouleau P, Talbot M, Tessier PA. 2003. Proinflammatory activities of S100: proteins S100A8, S100A9, and S100A8/A9 induce neutrophil chemotaxis and adhesion. J. Immunol. 170:3233–3242 [DOI] [PubMed] [Google Scholar]
- 62.Vandal K, Rouleau P, Boivin A, Ryckman C, Talbot M, Tessier PA. 2003. Blockade of S100A8 and S100A9 suppresses neutrophil migration in response to lipopolysaccharide. J. Immunol. 171:2602–2609 [DOI] [PubMed] [Google Scholar]
- 63.Corbin BD, Seeley EH, Raab A, Feldmann J, Miller MR, Torres VJ, Anderson KL, Dattilo BM, Dunman PM, Gerads R, Caprioli RM, Nacken W, Chazin WJ, Skaar EP. 2008. Metal chelation and inhibition of bacterial growth in tissue abscesses. Science 319:962–965 [DOI] [PubMed] [Google Scholar]
- 64.Damo SM, Kehl-Fie TE, Sugitani N, Holt ME, Rathi S, Murphy WJ, Zhang Y, Betz C, Hench L, Fritz G, Skaar EP, Chazin WJ. 2013. Molecular basis for manganese sequestration by calprotectin and roles in the innate immune response to invading bacterial pathogens. Proc. Natl. Acad. Sci. U. S. A. 110:3841–3846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hayden JA, Brophy MB, Cunden LS, Nolan EM. 2013. High-affinity manganese coordination by human calprotectin is calcium-dependent and requires the histidine-rich site formed at the dimer interface. J. Am. Chem. Soc. 135:775–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Urban CF, Ermert D, Schmid M, Abu-Abed U, Goosmann C, Nacken W, Brinkmann V, Jungblut PR, Zychlinsky A. 2009. Neutrophil extracellular traps contain calprotectin, a cytosolic protein complex involved in host defense against Candida albicans. PLoS Pathog. 5:e1000639. 10.1371/journal.ppat.1000639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chromek M, Slamová Z, Bergman P, Kovács L, Podracká L, Ehrén I, Hökfelt T, Gudmundsson GH, Gallo RL, Agerberth B, Brauner A. 2006. The antimicrobial peptide cathelicidin protects the urinary tract against invasive bacterial infection. Nat. Med. 12:636–641 [DOI] [PubMed] [Google Scholar]
- 68.Wiesner J, Vilcinskas A. 2010. Antimicrobial peptides: the ancient arm of the human immune system. Virulence 1:440–464 [DOI] [PubMed] [Google Scholar]
- 69.Zanetti M. 2004. Cathelicidins, multifunctional peptides of the innate immunity. J. Leukoc. Biol. 75:39–48 [DOI] [PubMed] [Google Scholar]
- 70.Boman HG. 2003. Antibacterial peptides: basic facts and emerging concepts. J. Intern. Med. 254:197–215 [DOI] [PubMed] [Google Scholar]
- 71.Zaiou M, Nizet V, Gallo RL. 2003. Antimicrobial and protease inhibitory functions of the human cathelicidin (hCAP18/LL-37) prosequence. J. Investig. Dermatol. 120:810–816 [DOI] [PubMed] [Google Scholar]
- 72.Sørensen OE, Follin P, Johnsen AH, Calafat J, Tjabringa GS, Hiemstra PS, Borregaard N. 2001. Human cathelicidin, hCAP-18, is processed to the antimicrobial peptide LL-37 by extracellular cleavage with proteinase 3. Blood 97:3951–3959 [DOI] [PubMed] [Google Scholar]
- 73.Zanetti M. 2005. The role of cathelicidins in the innate host defenses of mammals. Curr. Issues Mol. Biol. 7:179–196 [PubMed] [Google Scholar]
- 74.Nijnik A, Hancock RE. 2009. The roles of cathelicidin LL-37 in immune defences and novel clinical applications. Curr. Opin. Hematol. 16:41–47 [DOI] [PubMed] [Google Scholar]
- 75.Tomasinsig L, De Conti G, Skerlavaj B, Piccinini R, Mazzilli M, D'Este F, Tossi A, Zanetti M. 2010. Broad-spectrum activity against bacterial mastitis pathogens and activation of mammary epithelial cells support a protective role of neutrophil cathelicidins in bovine mastitis. Infect. Immun. 78:1781–1788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ibeagha-Awemu EM, Ibeagha AE, Messier S, Zhao X. 2010. Proteomics, genomics, and pathway analyses of Escherichia coli and Staphylococcus aureus infected milk whey reveal molecular pathways and networks involved in mastitis. J. Proteome Res. 9:4604–4619 [DOI] [PubMed] [Google Scholar]
- 77.Reinhardt TA, Sacco RE, Nonnecke BJ, Lippolis JD. 2013. Bovine milk proteome: quantitative changes in normal milk exosomes, milk fat globule membranes and whey proteomes resulting from Staphylococcus aureus mastitis. J. Proteomics 82:141–154 [DOI] [PubMed] [Google Scholar]
- 78.Smolenski GA, Wieliczko RJ, Pryor SM, Broadhurst MK, Wheeler TT, Haigh BJ. 2011. The abundance of milk cathelicidin proteins during bovine mastitis. Vet. Immunol. Immunopathol. 143:125–130 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.