Abstract
The predominant players in membrane fusion events are the soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) family of proteins. We hypothesize that SNARE proteins mediate fusion events at the chlamydial inclusion and are important for chlamydial lipid acquisition. We have previously demonstrated that trans-Golgi SNARE syntaxin 6 localizes to the chlamydial inclusion. To investigate the role of syntaxin 6 at the chlamydial inclusion, we examined the localization and function of another trans-Golgi SNARE and syntaxin 6-binding partner, vesicle-associated membrane protein 4 (VAMP4), at the chlamydial inclusion. In this study, we demonstrate that syntaxin 6 and VAMP4 colocalize to the chlamydial inclusion and interact at the chlamydial inclusion. Furthermore, in the absence of VAMP4, syntaxin 6 is not retained at the chlamydial inclusion. Small interfering RNA (siRNA) knockdown of VAMP4 inhibited chlamydial sphingomyelin acquisition, correlating with a log decrease in infectious progeny. VAMP4 retention at the inclusion was shown to be dependent on de novo chlamydial protein synthesis, but unlike syntaxin 6, VAMP4 recruitment is observed in a species-dependent manner. Notably, VAMP4 knockdown inhibits sphingomyelin trafficking only to inclusions in which it localizes. These data support the hypothesis that VAMP proteins play a central role in mediating eukaryotic vesicular interactions at the chlamydial inclusion and, thus, support chlamydial lipid acquisition and chlamydial development.
INTRODUCTION
Chlamydia trachomatis is the chlamydial organism most commonly associated with human disease and one of the more-common human pathogens. C. trachomatis serovars A, B, and C cause blinding trachoma (1), serovars D to K cause the most common bacterial sexually transmitted disease (STD), and serovars L1 to L3 cause lymphogranuloma venereum (LGV) (2). It is estimated that 50 to 70% of chlamydial infections are asymptomatic, potentiating their spread and complicating disease progression. Primary chlamydial infections, if left untreated, can result in sequelae such as pelvic inflammatory disease, ectopic pregnancy, and infertility. A hallmark of chlamydial infection is the ability of the pathogen to thrive within the host while limiting a protective immunological response (3).
Chlamydiae have evolved a unique biphasic developmental cycle. To initiate an infection, the infectious metabolically dormant form, termed the elementary body (EB), is endocytosed by the host cell and remains within a vesicle termed the inclusion, where it differentiates into a metabolically active, noninfectious reticulate body (RB). The RBs continue to grow and divide within the inclusion, which expands to accommodate the growing number of organisms. Importantly, the inclusion membrane provides protection for the organisms from the host and a platform which allows the organisms to parasitize nutrients from the host cell (4–6). Upon infection, the nascent inclusion membrane surrounding the infectious EB is plasma membrane derived, but within a few hours, chlamydial type III secreted proteins modify the inclusion membrane (7). These modifications result in the inclusion trafficking to the microtubule organizing center (8, 9) and separation of the inclusion from the classical endosomal/lysosomal pathway (10–15). C. trachomatis expresses distinct inclusion membrane proteins which colocalize to the microtubule organizing center (MTOC) and host centrosomes (16). The exact composition of the inclusion membrane has yet to be fully defined, but it is both bacterial and eukaryotic in nature, likely mimicking trans-Golgi membranes (17).
Chlamydial recruitment of host lipids is not only important to support the growing inclusion membrane, but also, sphingolipids are critical for chlamydial survival (18, 19). An indication that lipid acquisition is at least partly vesicular in nature is that chlamydial acquisition of sphingomyelin and cholesterol is time, temperature, and energy dependent, as well as brefeldin A (BFA) sensitive (20–22). Chlamydial organisms take on the lipid composition of their host cell (6); however, not all host lipids are found within the chlamydial cell wall, indicating specificity as to which lipids are incorporated by the organisms. For example, the chlamydial inclusion intercepts a subset of Golgi apparatus-derived exocytic vesicles containing sphingomyelin and cholesterol, and both are incorporated into chlamydial cell walls (20, 21, 23); however, Golgi apparatus-derived vesicles containing glucosylceramide are excluded from the inclusion and glucosylceramide is not incorporated by the organisms (24). Using a polarized cell model of chlamydial infection, we demonstrated that the sphingomyelin retained by Chlamydia is derived predominately from the basolateral trafficking pathway (24). Therefore, to define eukaryotic and chlamydial inclusion membrane fusion events, we examined proteins that govern membrane fusion along basolateral trafficking pathways. We demonstrated that soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) protein syntaxin 6, a trans-Golgi SNARE protein, localizes to the chlamydial inclusion (17).
SNARE proteins are classified as either R- or Q-SNAREs, based on an arginine (R) or a glutamine (Q) positioned with the alpha helix of the SNARE domain. As the SNARE complexes form, these amino acids are positioned at the “zero ionic layer” at the core of the alpha helix bundle within the complex (25). Q-SNARE proteins are further classified as Qa, Qb, or Qc based on homology with the synaptic SNARE proteins syntaxin 1, N-terminal synaptosomal-associated protein 25 (SNAP25) SNARE, and C-terminal SNAP25 SNARE, respectively (26, 27). To date, all R-SNARE-containing proteins are also all identified as vesicle-associated membrane proteins (VAMPs) (26, 27). SNARE complexes that form in the Golgi apparatus are formed by 4 separate proteins each containing a SNARE motif from a different subfamily, Qabc and R, respectively (28, 29). Specifically, syntaxin 6, a Qc SNARE, mainly localizes to the trans-Golgi network and is involved in both endosomal and trans-Golgi fusion events (30, 31).
Our studies focus on characterizing SNARE complexes that govern the fusion of eukaryotic vesicles with the chlamydial inclusion. As syntaxin 6 recruitment to the inclusion is conserved across chlamydial species and requires chlamydial protein synthesis (17), we hypothesize that syntaxin 6 functions in SNARE complexes at the chlamydial inclusion. Hence, we are examining syntaxin 6 binding partners and their localization to the chlamydial inclusion and ability to interact with syntaxin 6 at this unique microbial subcellular compartment. In this initial study, we examine the nature of syntaxin 6 and VAMP4 interactions in Chlamydia-infected cells. VAMP4, a known syntaxin 6 binding partner (30, 32), has a broad subcellular localization and has been implicated in endosomal and trans-Golgi network vesicular trafficking, as well as regulated exocytosis (33, 34). In this study, we examined VAMP4 localization to the chlamydial inclusion and demonstrated that VAMP4 and syntaxin 6 are binding partners at the chlamydial inclusion. We also identified VAMP4 as an important eukaryotic protein which supports chlamydial lipid acquisition and development. These studies expand our current understanding of how SNARE proteins function at the chlamydial inclusion and will lead to important discoveries about the dynamic nature of membrane fusion events at the chlamydial inclusion.
MATERIALS AND METHODS
Organisms and cell culture.
HeLa 229 cells (CCL-2.1; American Type Culture Collection [ATCC], Manassas, VA) were cultured at 37°C under 5% CO2 in RPMI 1640 (HyClone; Thermo Scientific, Logan, UT) supplemented with 10% fetal bovine serum (FBS) (HyClone) and 10 μg/ml gentamicin (Gibco-BRL; Life Technologies, Grand Island, NY). C2BBe1 cells (ATCC) were cultured at 37°C in 7.5% CO2 in Dulbecco's modified Eagle's medium (DMEM) (HyClone) supplemented with 10% FBS (HyClone), 4 mM l-glutamine (Life Technologies), 10 μg/ml human transferrin (Life Technologies), and 10 μg/ml gentamicin (Life Technologies). HeLa cells were used to propagate Chlamydia trachomatis serovar L2 (LGV 434) for purification using established protocols (35, 36). Chlamydial titers were determined using conventional protocols to establish multiplicities of infection (MOI), which were based on inclusion forming units (IFU) and determined in HeLa cells (36, 37) using a polyclonal rabbit or guinea pig anti-C. trachomatis serovar L2 EB antibody and secondary antibodies conjugated to DyLight fluors (Jackson ImmunoResearch Laboratories, West Grove, PA).
siRNA knock down of syntaxin 6 and VAMP4.
Silencer select small interfering RNAs (siRNAs) against syntaxin 6 (s19959) and VAMP4 (s16525) and a nontargeting control (NT siRNA) (4390843) were used at final concentrations of 10 nM, 5 nM, and 10 nM, respectively (Life Technologies). Reverse transfection using Lipofectamine RNAiMax (Life Technologies) was performed as described by the manufacturer. Briefly, siRNA and lipid were diluted in Opti-MEM (Life Technologies) medium and incubated for 15 min in the culture vessel. HeLa or C2BBe1 cells were diluted in antibiotic-free growth medium and plated directly into the culture vessel containing the siRNA and lipid mixture. The medium was changed after 18 h of incubation at 37°C in 5% CO2 (HeLa) or 37°C in 7.5% CO2 (C2BBe1). After 24 to 48 h of knockdown, the monolayers were infected with C. trachomatis serovar L2 (MOI of 1 to 2). The efficiency of the knockdown was confirmed by either indirect immunofluorescence assay (surveying an entire coverslip) or Western blot assay, and only samples achieving 80% knockdown or more were used in subsequent analyses.
Indirect immunofluorescence and fluorescence microscopy.
To determine the localization of endogenous VAMP4 to the chlamydial inclusion, cells were fixed for 10 min in 4% paraformaldehyde and permeabilized for 5 min with 0.1% saponin. To collapse the Golgi apparatus, cells were treated with 1 μg/ml brefeldin A (BFA) for 2 h prior to fixation (38). The coverslips were processed for indirect immunofluorescence assay using guinea pig anti-C. trachomatis serovar L2 EB antibody, mouse anti-syntaxin 6 antibody (BD Biosciences, San Jose, CA), rabbit anti-VAMP4 antibody (Sigma, St. Louis, MO), or mouse anti-p230 antibody (BD Biosciences), followed by incubation with the appropriate secondary antibodies conjugated to DyLight fluors (Jackson ImmunoResearch Laboratories). The coverslips were mounted onto slides using Prolong Gold antifade mounting medium (Life Technologies). The slides were visualized with an Olympus BX60 fluorescent scope (60× magnification), and images taken with a Nikon DS-Qi1Mc camera.
3×FLAG-syntaxin 6 immunoprecipitation.
C2BBe1 cells were seeded into a 6-well plate and allowed to grow overnight. The cells were transfected with the 3×FLAG vector or 3×FLAG-syntaxin 6 construct (17). Briefly, 1.75 μg DNA was diluted in Opti-MEM (Life Technologies), Plus, and Lipofectamine LTX reagents (Life Technologies) according to the manufacturer's specifications. Then, cells were infected with C. trachomatis serovar L2 (MOI of 3) for 18 h. To preserve SNARE complexes, cells were fixed in 2% paraformaldehyde for 10 min on ice with gentle agitation and the reaction was quenched with 1.25 M glycine. The monolayers were washed three times with phosphate-buffered saline (PBS), suspended in CelLytic M (Sigma) containing protease inhibitor cocktail (Sigma), and lysed by sonication. Cell debris was removed by centrifugation at 12,000 × g for 10 min at 4°C, and the lysate was rotated overnight at 4°C with M2 agarose (Sigma). The beads were washed twice with both wash buffer A (50 mM Tris-HCl, pH 7.4, and 150 mM NaCl) and wash buffer B (50 mM Tris-HCl, pH 7.4, and 250 mM NaCl). Bound proteins were eluted using 3×FLAG peptide in wash buffer A. Samples were analyzed by Western blotting with the primary antibodies mouse anti-M2 FLAG (Sigma) (to detect 3×FLAG-syntaxin 6) and rabbit anti-VAMP4 (Sigma) and secondary antibodies goat anti-rabbit IRDye 800CW- and goat anti-mouse IRDye 680LT-conjugated antibodies (LiCor Biosciences, Lincoln, NE). Images were taken using the Odyssey CLx and processed using Image Studio version 2.0 (LiCor Biosciences).
Duolink PLA.
HeLa cells were seeded and infected with C. trachomatis serovar L2 (MOI of 1) for 18 h as described above. The cells were fixed in 4% paraformaldehyde for 10 min on ice, permeabilized with 0.1% saponin for 5 min at room temperature, and blocked in 3% bovine serum albumin (BSA) for 30 min. Next, the coverslips were incubated with primary antibodies mouse anti-syntaxin 6 antibody (BD Biosciences) and rabbit anti-VAMP4 antibody (Sigma). Following incubation with the primary antibody, the coverslips were removed from the 24-well plate and a PAP Pen (Scientific Device Laboratory, Des Plaines, IL) was used to trace the edges of the coverslip. Proximity ligation assays (PLAs) were carried out using the Duolink II detection kit (Olink Bioscience, Uppsala, Sweden) as described by the manufacturer. All reagents were mixed for a 40-μl reaction mixture volume, and all washes were performed in a 24-well plate using 2 ml of the appropriate wash buffer. Negative controls consisted of samples incubated with secondary antibodies in the absence of primary antibodies. Coverslips were mounted using mounting medium with 4′,6′-diamidino-2-phenylindole (DAPI) (Olink Bioscience), and the edges were sealed with clear nail polish. Imaging was performed essentially as described above.
Infectious progeny.
HeLa cells were reverse transfected with VAMP4, syntaxin 6, or NT siRNA and infected with C. trachomatis serovar L2 (MOI of 2). C2BBe1 cells were reverse transfected with syntaxin 6 and NT siRNA and infected (MOI of 4) as described above for HeLa cells. After 44 h of infection, the infected monolayers were briefly washed with 1 ml of H2O and the cells were lysed in 1 ml of H2O. Serial dilutions of lysate were used to infect a fresh monolayer of HeLa cells on coverslips, in duplicate, and incubated for 30 h at 37°C in 5% CO2. The coverslips were processed for indirect immunofluorescence assay and imaged as described above. VAMP4 data were combined from 2 separate experiments performed in duplicate. Syntaxin 6 data were combined from 4 separate experiments performed in triplicate. The data displayed are the means and standard errors of the means and were calculated using GraphPad Prism 5 software (GraphPad Software, La Jolla, CA) as described below.
Live-cell imaging.
To visualize the retention of fluorescent lipid by the chlamydial inclusion, HeLa cells were reverse transfected with either NT, VAMP4, or syntaxin 6 siRNA and incubated for 48 h prior to infection with C. trachomatis serovar L2 (MOI of 2). Following 18 h of infection, the cells were labeled with 5 μM fluorescent lipid 6-{[N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino]hexanoyl}sphingosine (C6-NBD-ceramide [also called NBD-lipid here]) (Life Technologies) as described previously (21, 39) (the C6-NBD-ceramide metabolites NBD-glucosylceramide and NBD-sphingomyelin are described in Results). Briefly, infected monolayers were incubated for 15 min at 12°C and then labeled with 5 μM C6-NBD-ceramide in cold Eagle's modified essential medium (EMEM) plus 0.035% defatted BSA (Sigma) for 30 min at 12°C. The cells were shifted to 37°C for 5 min prior to the addition of HeLa growth medium (described above) to remove NBD-lipid (defined as NBD-ceramide and its metabolites, which include NBD-glucosylceramide, NBD-galactosylceramide, and NBD-sphingomyelin) not incorporated into the chlamydial inclusion. Phase-contrast and fluorescent live-cell images were taken at 1.5, 6, and 24 h after back-exchange using a 40× phase objective with the Axiovert 200 M Imager and the AxioCam HRm camera (Ziess). The efficiency of the knockdown was determined by indirect immunofluorescence assay as described above.
Thin-layer chromatography (TLC).
To determine the amount of NBD-sphingomyelin retained by the chlamydial organisms recovered from syntaxin 6 or VAMP4 siRNA-treated cells, EBs were isolated and the lipids were extracted (39). Briefly, cells were reverse transfected with siRNA and infected with C. trachomatis serovar L2 (MOI of 3) as described above. After 24 h of infection, cells were labeled with 5 μM C6-NBD-ceramide (Life Technologies), and unincorporated NBD-lipid was back-exchanged for 20 h as described above. The EBs were purified after 40 to 44 h of infection by using a Renografin (Mallinckrodt, Inc., St. Louis, MO) gradient as previously described (39). To control for lipid extraction efficiency, 0.06 μg/ml of NBD-lactosylceramide (Matreya, Pleasant Gap, PA) was added to each sample and then lipids were extracted by a modified Bligh and Dyer chloroform-methanol extraction (24, 39, 40). The lipids were resuspended in a 2:1 chloroform-methanol solution, spotted onto TLC plates precoated with 60Å-pore-size silica gel (Whatman/GE Healthcare, Piscataway, NJ), and resolved using a 40:11.5:1.5 chloroform-methanol-distilled water mixture. The plates were air dried and imaged using the Typhoon 9410 variable-mode imager (GE Healthcare), and densitometry was performed using Image J software (NIH, Bethesda, MD). The densitometry for sphingomyelin was determined and normalized to the amount of EBs loaded as determined by Western blotting and the amount of NBD-lactosylceramide in the extracts.
Statistics and image production.
All quantification and statistical analysis of data were performed with GraphPad Prism 6 software (GraphPad Software, La Jolla, CA). Specifically, statistical significance was determined using two-way analysis of variance (ANOVA) with Bonferroni posttests, calculated by using GraphPad Prism software. All figures were constructed using Adobe Photoshop CS5 (Adobe Systems Incorporated, San Jose, CA). Modifications to images include adjustment to color balance in fluorescent images, applied equally to all images in a single figure. Brightness and contrast were adjusted in scanned images of TLC plates and in Western blot images. All graphed data originated in GraphPad Prism 6.
RESULTS
VAMP4 localization to the chlamydial inclusion.
A recent study demonstrated that syntaxin 6 localizes to the chlamydial inclusion in a manner that is conserved across chlamydial species and requires chlamydial protein synthesis (17). These data suggest that syntaxin 6 localization to the chlamydial inclusion is important; however, the role of syntaxin 6 at the chlamydial inclusion is currently ambiguous. To clarify the function of syntaxin 6 at the chlamydial inclusion, we are examining known syntaxin 6 binding partners and focusing on these interactions in the context of Chlamydia-infected cells in order to characterize relevant interactions at the chlamydial inclusion. Syntaxin 6 has been classified as a promiscuous SNARE protein, having a broad repertoire of binding partners (31). In the current study, we examine one of syntaxin 6's binding partners, VAMP4, and the localization of VAMP4 to the chlamydial inclusion.
Using indirect immunofluorescence microscopy, we examined the localization of endogenous VAMP4 to the chlamydial inclusion. As demonstrated in Fig. 1A, VAMP4 appears to be clustered around the inclusion in close association with the Golgi apparatus. To clarify the localization of VAMP4 relative to the inclusion, we collapsed the surrounding Golgi structures with the fungal metabolite BFA for 2 h prior to fixation in paraformaldehyde and processing for indirect immunofluorescence assay. Evident in these images is that VAMP4 remains associated with the inclusion, while the surrounding Golgi apparatus structure is collapsed (Fig. 1A). Furthermore, we demonstrate that both VAMP4 and syntaxin 6 colocalize and remain stably associated with the chlamydial inclusion in the presence of BFA (Fig. 1B). The localization of VAMP4 to the chlamydial inclusion appears to be more vesicular in nature, as opposed to the rim-like staining pattern previously described for syntaxin 6 (17), albeit treatment with BFA causes inclusion-associated syntaxin 6 to appear less rim-like and more vesicular in nature, as well (Fig. 1B). In our studies, we were able to detect both endogenous syntaxin 6 and VAMP4 in HeLa cells (Fig. 1) and C2BBe1 cells (data not shown), and the localization of both proteins with the chlamydial inclusion was observed in both cell lines.
Fig 1.
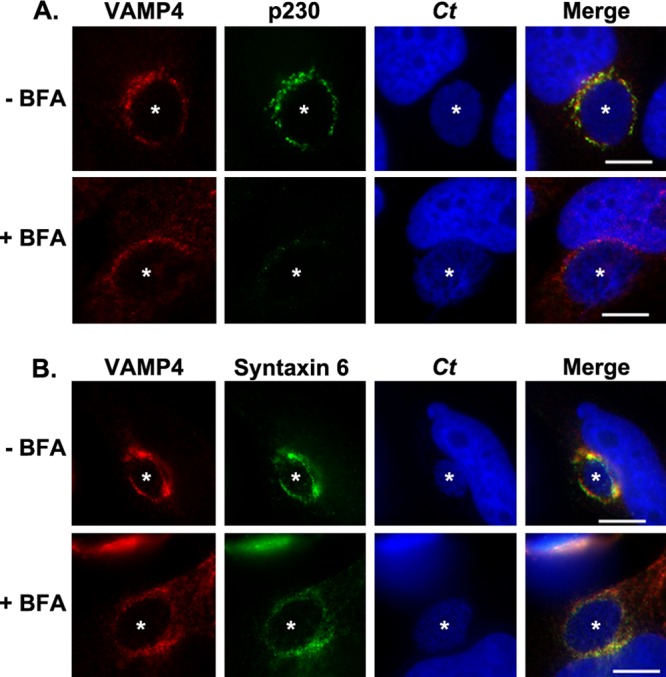
Examination of syntaxin 6 and VAMP4 colocalization at the chlamydial inclusion. HeLa cells were infected with C. trachomatis (Ct) for 16 h and then either mock treated or treated with brefeldin A for an additional 2 h. Then, cells were fixed and processed for indirect immunofluorescence assay as described in Materials and Methods to detect VAMP4 and the Golgi apparatus marker p230 (A) or VAMP4 and syntaxin 6 (B). White stars indicate chlamydial inclusions. The results shown are representative of at least 3 independent experiments. Bars = 10 μm. VAMP4 and syntaxin 6 colocalize with the chlamydial inclusion.
Given that syntaxin 6 and VAMP4 are binding partners (30–32) and colocalize to the chlamydial inclusion, we asked whether siRNA knockdown of either syntaxin 6 or VAMP4 would affect the localization of the other protein to the chlamydial inclusion. For these studies, syntaxin 6 or VAMP4 expression was decreased with siRNA transfection, and then knockdown cells were infected with C. trachomatis for 16 to 18 h. Subsequently, cells were fixed and processed for indirect immunofluorescence assay. In syntaxin 6 knockdown cells, VAMP4 localized to the Golgi apparatus surrounding the inclusion and diffusely around the inclusion itself (Fig. 2). In VAMP4 knockdown cells, syntaxin 6 was found diffusely throughout the cell and around the chlamydial inclusion, but the tight association with the inclusion noted in the control cells was absent (Fig. 2). These data suggested that VAMP4 is involved in syntaxin 6 localization or retention at the chlamydial inclusion. In contrast, the loss of syntaxin 6 had a moderate impact on the localization of VAMP4 with the chlamydial inclusion.
Fig 2.
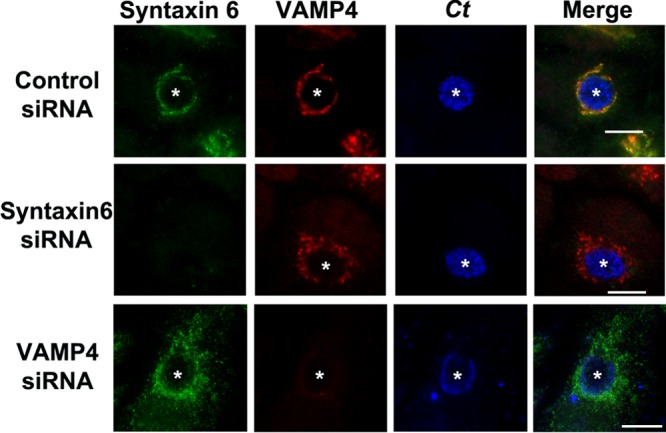
Impact of siRNA knockdown on the localization of syntaxin 6 or VAMP4 to the chlamydial inclusion. HeLa cells were treated with control (nontargeting), syntaxin 6, or VAMP4 siRNA and infected with C. trachomatis (Ct) for 18 h. Then, cells were fixed and processed for indirect immunofluorescence assay to detect chlamydial inclusions, endogenous syntaxin 6, and VAMP4. White stars indicate chlamydial inclusions. The results shown are representative of at least 3 independent experiments. Bars = 10 μm. Knockdown of syntaxin 6 moderately disrupts VAMP4 localization to the inclusion; knockdown of VAMP4 disrupts colocalization of syntaxin 6 with the chlamydial inclusion.
Characterization of syntaxin 6-VAMP4 interactions at the chlamydial inclusion.
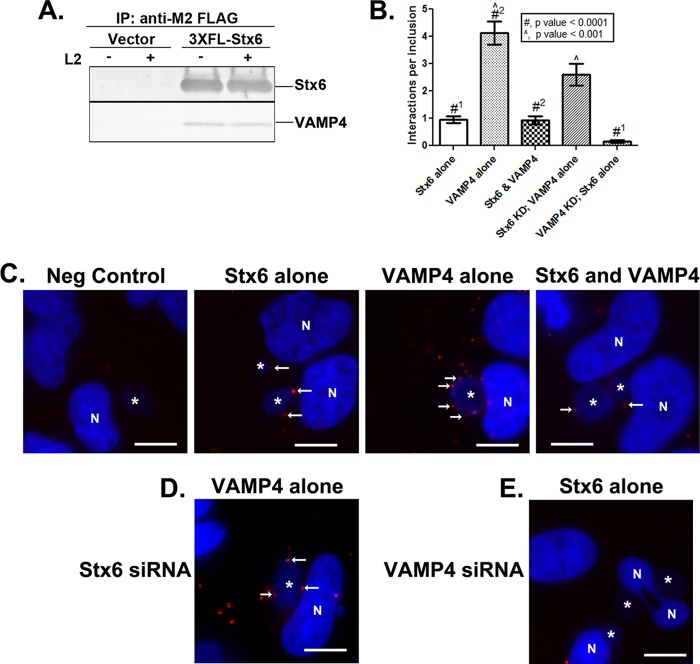
Biochemically, we confirmed previous reports identifying syntaxin 6 and VAMP4 as binding partners (Fig. 3A) (30, 32). To coimmunoprecipitate these proteins, we used a previously described construct, 3×FLAG-syntaxin 6 (17), and transfected C2BBe1 cells (Fig. 3A) or HeLa cells (data not shown) prior to infection with C. trachomatis. Immunoprecipitation of the 3×FLAG-syntaxin 6 constructs demonstrated that syntaxin 6 binds to VAMP4 equally well in mock-infected and Chlamydia-infected cells.
Fig 3.
Characterization of syntaxin 6 and VAMP4 interactions in Chlamydia-infected cells. (A) Coimmunoprecipitation of syntaxin 6 and VAMP4 in Chlamydia-infected cells. C2BBe1 cells were transfected with 3×FLAG-syntaxin 6 (3×FL-Stx6) or 3×FLAG vector only and then mock infected (-) or infected (+) with C. trachomatis serovar L2. 3×FLAG constructs were immunoprecipitated (IP) and resolved by SDS-PAGE, followed by Western blotting against syntaxin 6 or VAMP4. The results shown are representative of at least 3 independent experiments. (B to E) Detection of syntaxin 6 and VAMP4 interactions at the chlamydial inclusion. To detect specific syntaxin 6-VAMP4 interactions at the chlamydial inclusion, samples were processed with Duolink in situ red detection reagents (Olink Biosciences) (see Materials and Methods for specific details) and mounted with Duolink in situ mounting medium with DAPI (detecting both nuclei and chlamydial inclusions). Red punctate dots represent foci of concentrated protein (Stx6 alone and VAMP4 alone) or discrete protein-protein interactions (Stx6 and VAMP4). Negative controls (Neg control) are samples processed with secondary antibodies only (see Materials and Methods for specific details). (B) The foci of protein/interactions around 45 individual inclusions are enumerated. Means and standard deviations are shown. Statistics are one-way ANOVA comparisons performed with GraphPad Prism 5 software (GraphPad Software, Inc.). Statistical significance was determined between Stx6 alone in control and VAMP4 knockdown (KD) cells (#1), VAMP4 alone and Stx6 and VAMP4 interactions (#2), and VAMP4 alone in control and Stx6 KD cells (^). Images representative of those quantified are shown in panels C to E. (C) Examination of endogenous protein around the chlamydial inclusion. (D and E) Examination of endogenous VAMP4 (D) and Stx6 (E) after knockdown with indicated siRNA. White stars indicate chlamydial inclusions, white arrows denote interactions at the periphery of the chlamydial inclusion, and “N”s denote nuclei. Bars = 10 μm. Images are representative of at least 2 independent experiments. Syntaxin 6 and VAMP4 are binding partners in Chlamydia-infected cells and specifically bind in discrete foci around the chlamydial inclusion. Furthermore, knockdown of VAMP4 severely limits syntaxin 6 localization to the chlamydial inclusion.
The limitation of a coimmunoprecipitation assay is that it does not examine interactions that occur at the chlamydial inclusion. The Duolink proximity-dependent ligase assay was utilized to directly examine VAMP4 and syntaxin 6 interactions at the chlamydial inclusion. This technology detects interactions between proteins that are within 40 nm of each other and has been used to confirm protein-protein interactions or to amplify the immunofluorescent signal of a single protein within distinct subcellular locations (41). Positive Duolink reactions result in distinct punctate dots where the signal has been amplified and, hence, will not recapitulate subcellular structures detected by classical indirect immunofluorescence techniques. To determine if syntaxin 6 and VAMP4 were interacting at the chlamydial inclusion, each protein was detected individually and jointly using the Duolink assay (Fig. 3C). In this assay, HeLa cells were infected with C. trachomatis for 16 to 18 h, fixed in 4% paraformaldehyde, and processed as described in Materials and Methods. On average, more VAMP4 molecules (4.13 ± 0.43 [mean ± standard deviations]) than syntaxin 6 molecules (0.957 ± 0.12) were detected around the chlamydial inclusion (Fig. 3B and C). Additionally, similar amounts of syntaxin 6-VAMP4 interactions (0.935 ± 0.15) and syntaxin 6 molecules alone (Fig. 3B and C) were detected, indicating that when syntaxin 6 is found at the chlamydial inclusion, it is found in proximity to VAMP4. Conversely, the data demonstrated that VAMP4 localization to the chlamydial inclusion is not necessarily associated with syntaxin 6 (Fig. 3B and C). The difference between quantified complexes of VAMP4 alone and syntaxin 6-VAMP4 complexes which localize to the inclusion was statistically significant (Fig. 3B, #2) (P < 0.0001).
The Duolink assay was also performed in syntaxin 6 or VAMP4 siRNA-treated cells (Fig. 3D and E). Consistent with the results shown in Fig. 2, syntaxin 6 knockdown results in a moderately reduced, albeit statistically significant amount of VAMP4 molecules which localize to the chlamydial inclusion (2.61 ± 0.40) (Fig. 2B and D). In contrast, VAMP4 knockdown results in a statistically significant deficiency of syntaxin 6 molecules which localize to the chlamydial inclusion (0.152 ± 0.054) (Fig. 3B and E). The data demonstrated that syntaxin 6 localization to the chlamydial inclusion relies, in part, on VAMP4 but VAMP4 can localize independently to the inclusion in the absence of syntaxin 6.
Role of syntaxin 6 and VAMP4 in sphingomyelin trafficking to the chlamydial inclusion.
During the time of rapid multiplication, Chlamydia cells acquire nutrients to support their growth and to compensate for the increasing size of the chlamydial inclusion. Our hypothesis is that SNARE proteins facilitate the trafficking of required nutrients, such as lipids, to the chlamydial inclusion. Therefore, the absence of syntaxin 6 or VAMP4 would hinder chlamydial nutrient acquisition and, thus, development, resulting in a decrease of infectious progeny. A well-established technique to examine Golgi apparatus-derived lipid trafficking to the chlamydial inclusion utilizes the fluorescent lipid, C6-NBD-ceramide (21, 39). C6-NBD-ceramide is a vital stain for the Golgi apparatus and, within the Golgi apparatus, is metabolized into two major metabolites: NBD-glucosylceramide and NBD-sphingomyelin (42–44). The NBD moiety is fluorescent and also allows the lipids to be extracted from the cells by a lipid acceptor in the tissue culture medium in a process termed back-exchange (21, 39, 43). Previous studies utilizing C6-NBD-ceramide established that the chlamydial inclusion intercepts fluorescent lipid, which is incorporated into Chlamydia organisms, and that this lipid is sphingomyelin (21, 23).
The role of syntaxin 6 and VAMP4 in chlamydial lipid acquisition was initially characterized with live-cell imaging (Fig. 4). HeLa cells transfected with control (nontargeting), VAMP4, or syntaxin 6 siRNA were infected with C. trachomatis serovar L2 for 20 h and then labeled with C6-NBD-ceramide as previously described (23, 39). Monolayers were then treated with back-exchange medium for the times indicated below to remove NBD-lipid that was not incorporated into chlamydial organisms from the host cells. At the end of the experiment, knockdown was confirmed by fixing cells and processing the samples for indirect immunofluorescence assay as seen in Fig. 1 (data not shown). At 1.5 h after back-exchange, the majority of the fluorescent lipid in control- or syntaxin 6 siRNA-treated cells was associated with the chlamydial inclusion. By 6 h after back-exchange, all fluorescent lipid was associated with the chlamydial inclusion in these cells. Furthermore, the inclusions propagated in control- or syntaxin 6 siRNA-treated cells remained labeled 24 h after the addition of back-exchange medium, consistent with the notion that lipid that is trafficked to the inclusion does not cycle back to the host cell because it is incorporated into the chlamydial organisms (21, 23, 24, 39).
Fig 4.
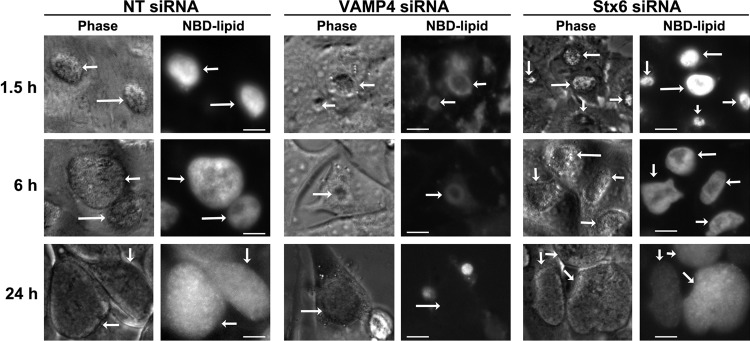
Live-cell imaging of NBD-lipid trafficking to the chlamydial inclusion in VAMP4 and syntaxin 6 knockdown cells. Nontargeting (NT; control), VAMP4, or syntaxin 6 (Stx6) siRNA-treated cells were infected with C. trachomatis serovar L2 for 20 h. Cells were labeled with fluorescent C6-NBD-ceramide (NBD-lipid) and then back-exchanged for the indicated times. The images shown are representative of 3 independent experiments. Phase, phase-contrast microscopy image. White arrows indicate chlamydial inclusions. Bars = 10 μm. Chlamydial inclusions grown in nontargeting or syntaxin 6 knockdown cells acquired fluorescent lipid beginning at 1.5 h after the addition of back-exchange medium. In VAMP4 knockdown cells, chlamydial inclusions did not acquire fluorescent lipid even after 24 h of back-exchange.
In contrast, in VAMP4 knockdown cells, at 1.5 h after the addition of back-exchange medium, the fluorescent lipid remained in Golgi apparatus structures and appeared to accumulate or pool outside the chlamydial inclusion. At this early time point, no fluorescent lipid was associated with chlamydial organisms. These data indicated that VAMP4 may play a role in vesicular trafficking of C6-NBD-ceramide metabolites from the Golgi apparatus to the plasma membrane, consistent with VAMP4's role in regulated exocytosis (33). By 6 h after the addition of back-exchange medium, the only fluorescent lipid remaining in VAMP4 siRNA-treated cells was pooled outside the chlamydial inclusion, but the organisms remained unlabeled. At 24 h after back-exchange in VAMP4 knockdown cells, no fluorescent lipid remained in the cell and no fluorescent lipid was associated with the chlamydial inclusion/organisms. Additionally, the inclusions that developed in VAMP4 knockdown cells were notably smaller at later time points postinfection (20 to 52 h). This is in contrast to the apparent inclusion sizes evident at 16 to 18 h postinfection, which appear comparable to the sizes of inclusions grown in control cells. Importantly, the overall numbers of inclusions formed were similar between cells treated with nontargeting, syntaxin 6, or VAMP4 siRNA (data not shown). Combined, these data suggest that VAMP4 is involved in trafficking sphingomyelin to the chlamydial inclusion and may play a role in chlamydial inclusion expansion.
A more-sensitive technique to quantify sphingomyelin retention by Chlamydia is to purify the organisms from host cell monolayers and perform a modified Bligh and Dyer lipid extraction, followed by examination of the extracts by thin-layer chromatography (TLC) (21, 39, 40). In these studies, C2BBe1 or HeLa cells were treated with syntaxin 6 or VAMP4 siRNA, respectively, or nontargeting (control) siRNA. After 48 h of knockdown, cells were inoculated with C. trachomatis serovar L2 and then labeled with C6-NBD-ceramide, and nonincorporated lipid was back-exchanged overnight. At 40 to 44 h postinfection, chlamydial organisms were purified from the host cells and lipids were extracted using a modified Bligh and Dyer lipid extraction protocol (24, 39, 40). Efficient knockdown of syntaxin 6 or VAMP4 was determined by Western blotting on cell extracts taken at the time of chlamydial purification (Fig. 5B). The lipid extract volumes of equal numbers of EBs were resolved by TLC, and TLC plates were scanned using a Typhoon imager (Fig. 5A). The densitometry results for the NBD-sphingomyelin bands were normalized to chlamydial major outer membrane protein (MOMP) levels and the lipid loading control level (Fig. 5B). Consistent with the live-cell imaging data, there was no inhibition of NBD-sphingomyelin incorporated into Chlamydia grown in syntaxin 6 knockdown cells. Densitometry analysis revealed that there was an increase of about 30% of NBD-sphingomyelin in organisms grown in cells lacking syntaxin 6. In contrast, densitometry analysis of Chlamydia organisms grown in VAMP4 knockdown cells revealed a 27% decrease in NBD-sphingomyelin incorporated into these organisms. The decrease is not as severe as that illustrated with live-cell imaging of Chlamydia organisms grown in VAMP4 knockdown cells. However, the knockdown of VAMP4 is not 100% efficient (Fig. 5B), resulting in some organisms acquiring normal amounts of lipid. Importantly, these data were consistent with the notion that VAMP4 is required for efficient sphingomyelin trafficking to the chlamydial inclusion.
Fig 5.
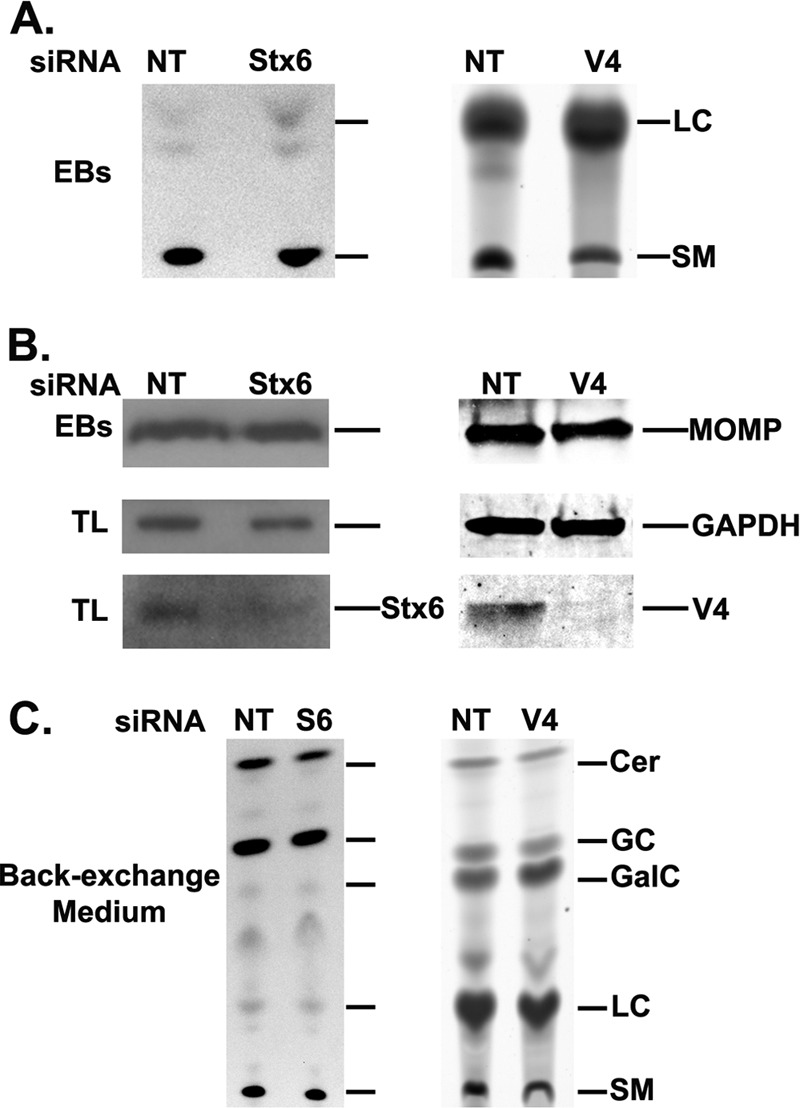
Thin-layer chromatography analysis of VAMP4 and syntaxin 6 in NBD-sphingomyelin trafficking to the chlamydial inclusion. Nontargeting (NT; control), VAMP4 (V4), or syntaxin 6 (Stx6) siRNA-treated cells were infected with C. trachomatis, labeled with fluorescent C6-NBD-ceramide, and back-exchanged as described in Materials and Methods. (A) Lipid extracts of purified organisms were resolved by thin-layer chromatography. (B) Western blot analysis of purified EBs to show equal loading of organisms on thin-layer chromatography plates and total lysates to show efficiency of syntaxin 6 or VAMP4 knockdown. (C) Lipid extracts of back-exchange medium aliquots resolved by thin-layer chromatography. Data shown are representative of at least 2 separate experiments. EBs, elementary bodies; LC, lipid extraction efficiency control (lactosylceramide); SM, sphingomyelin; TL, total lysate; Cer, ceramide; GC, glucosylceramide; GalC, galactosylceramide; GAPDH, glyceraldehyde-3-phosphate dehydrogenase. Consistent with live-cell imaging data, knockdown of VAMP4 decreases the amount of NBD-sphingomyelin trafficked to the chlamydial inclusion, while syntaxin 6 knockdown has little effect on sphingomyelin trafficking to the chlamydial inclusion.
To confirm that C6-NBD-ceramide was being metabolized appropriately in syntaxin 6 or VAMP4 knockdown cells, aliquots of the back-exchange medium were subjected to lipid extraction and resolved by TLC. These data demonstrate that knockdown of syntaxin 6 or VAMP4 did not alter the metabolism of C6-NBD-ceramide into NBD-sphingomyelin, NBD-glucosylceramide, and NBD-galactosylceramide (Fig. 5C). These data demonstrate that the changes observed in chlamydial lipid composition are not due to the effects of syntaxin 6 or VAMP4 knockdown on host cell sphingolipid metabolism.
Effect of siRNA knockdown of VAMP4 and syntaxin 6 on chlamydial infectious progeny.
As we noted no differences in the numbers of inclusions formed (specifically, no abrogation of initial entry and infection) in the absence of VAMP4 or syntaxin 6 compared to the numbers of inclusions in controls (data not shown), we wished to assess the impact of these proteins on chlamydial development by examining infectious progeny. To determine infectious progeny, control (nontargeting), VAMP4, or syntaxin 6 siRNA-treated HeLa cells (VAMP4) or C2BBe1 cells (syntaxin 6) were infected with C. trachomatis serovar L2 for 40 to 42 h and lysed, and titers of lysates containing infectious Chlamydia organisms were used to inoculate a fresh monolayer of HeLa cells as previously described (37, 45). At 30 h postinoculation, the secondary infections were fixed and processed for indirect immunofluorescence assay. Inclusions were enumerated with the premise that a single EB results in a single inclusion (Fig. 6). Knockdown was confirmed by indirect immunofluorescence assay (VAMP4) or Western blot (syntaxin 6) assay as specified in Materials and Methods.
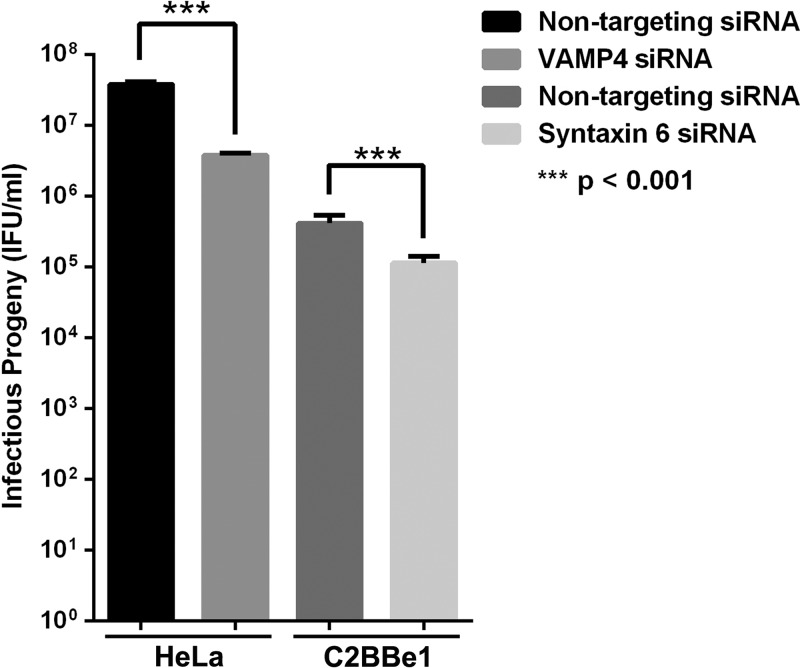
Fig 6.
Effect of syntaxin 6 and VAMP4 on chlamydial infectious progeny. HeLa cells (VAMP4) or C2BBe1 cells (syntaxin 6) were seeded in 24-well plates and treated with the indicated siRNA. Infectious progeny were determined essentially as described in Materials and Methods, and the results are given as inclusion forming units (IFU) per ml. Means and standard errors of the means of 3 independent experiments are shown. Statistics are one-way ANOVA comparisons performed with GraphPad Prism 5 software (GraphPad Software, Inc.). VAMP4 knockdown results in a log decrease in infectious progeny, compared to a nominal decrease in infectious progeny in the absence of syntaxin 6.
Consistent with previous data (24), when comparing progeny obtained from HeLa or C2BBe1 cells treated with control siRNA, more progeny were recovered from HeLa cells (3.8 × 107 ± 3.0 × 106 IFU/ml) than from C2BBe1 cells (4.14 × 105 ± 1.3 × 105 IFU/ml). Knockdown of VAMP4 resulted in a statistically significant log decrease in infectious progeny (3.8 × 106 ± 2.8 × 105 IFU/ml), while knockdown of syntaxin 6 resulted in a modest, albeit statistically significant decrease in infectious progeny (1.1 × 105 ± 2.9 × 104). To eliminate the potential for cell line differences to account for a less-profound effect of syntaxin 6 knockdown on chlamydial development, these studies were repeated in HeLa cells, with similar results. Knockdown of syntaxin 6 resulted in 1.13 × 108 ± 4.0 × 106 IFU/ml, compared to 1.25 × 108 ± 6.0 × 106 IFU/ml obtained in control cells. Overall, these data reveal that VAMP4 is important to the development of C. trachomatis serovar L2 infectious progeny.
Chlamydial protein synthesis and species requirements for localization of VAMP4 to the chlamydial inclusion.
Syntaxin 6 recruitment to the chlamydial inclusion requires chlamydial protein synthesis and is conserved across chlamydial species (17). To determine if VAMP4 localized to the chlamydial inclusion under similar conditions, we first examined whether VAMP4 recruitment to the inclusion is dependent on de novo chlamydial protein synthesis. For these studies, HeLa cells were infected with C. trachomatis serovar L2 for 18 h and then either fixed in 4% paraformaldehyde or treated for an additional 24 h with 200 μg/ml chloramphenicol prior to fixation. Samples were processed for indirect immunofluorescence assay to detect endogenous VAMP4 (Fig. 7). As a control, the localization of syntaxin 6 was also visualized. In cells fixed at 18 h postinfection, both VAMP4 and syntaxin 6 surround and localize the chlamydial inclusion. In chloramphenicol-treated cells, VAMP4 and syntaxin 6 appear to remain localized in the Golgi apparatus and are not retained at the chlamydial inclusion. Therefore, retention of VAMP4 at the chlamydial inclusion is dependent on de novo chlamydial protein synthesis.
Fig 7.
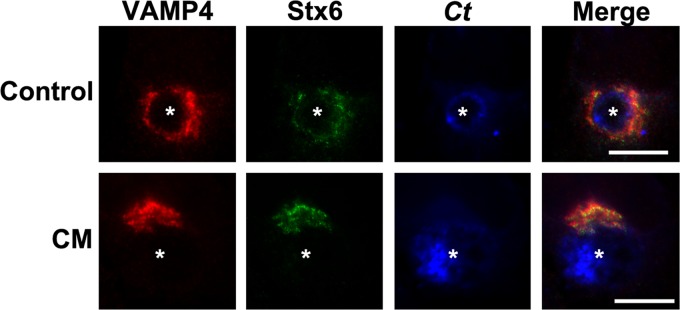
VAMP4 localization to chlamydial inclusion in the absence of chlamydial protein synthesis. HeLa cells were seeded onto glass coverslips in 24-well plates 24 h prior to infection with C. trachomatis serovar L2 (Ct). After 16 h, cells were either fixed in 4% paraformaldehyde (control) or treated with 200 μg/ml chloramphenicol (CM) for an additional 24 h and then fixed and processed for indirect immunofluorescence assay as described in Materials and Methods to detect VAMP4 (red), syntaxin 6 (green), and C. trachomatis (blue). The results shown are representative of at least 2 independent experiments. White stars denote chlamydial inclusions. Bars = 10 μm. Chlamydial protein synthesis is required for VAMP4 retention at the chlamydial inclusion.
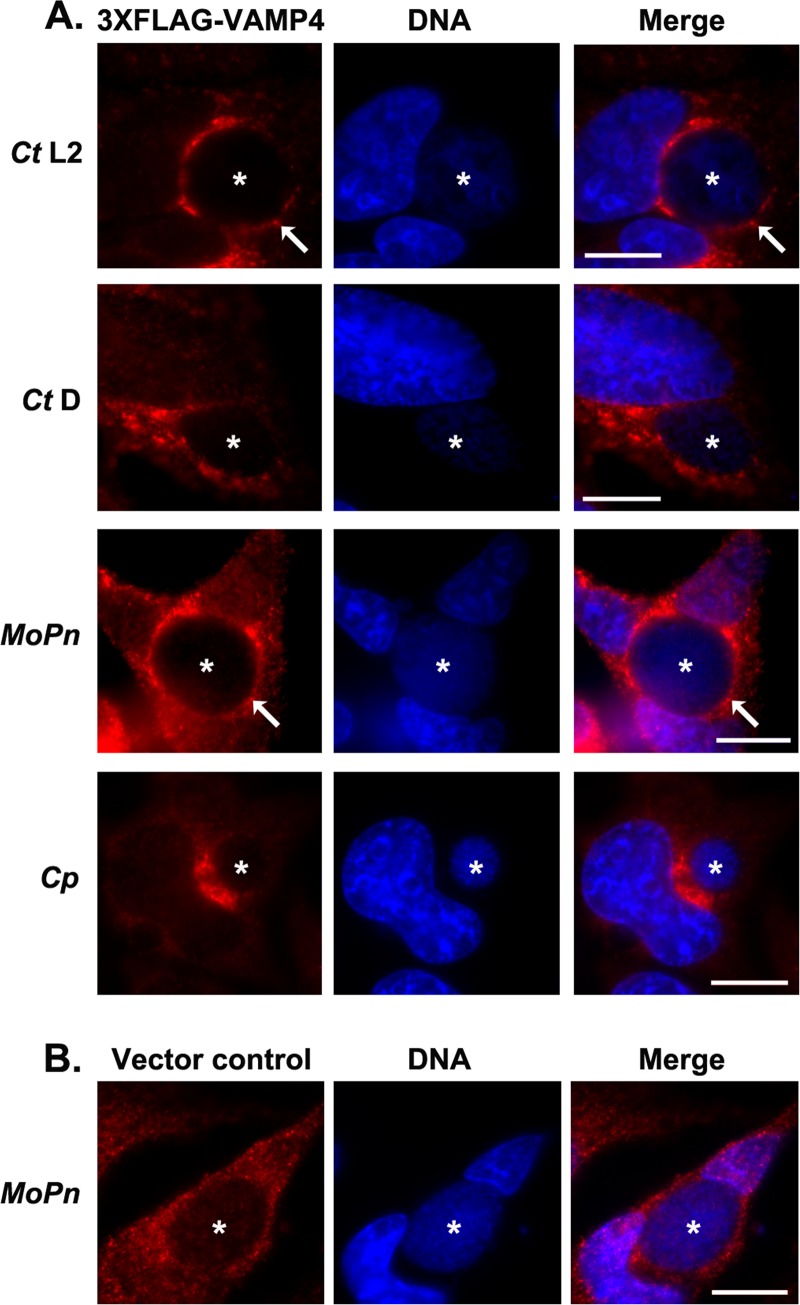
Second, we asked whether VAMP4 localization is conserved among chlamydial species. We have demonstrated that VAMP4 localizes to C. trachomatis serovar L2 inclusions and, furthermore, tested the localization of 3×FLAG-VAMP4 to inclusions containing C. trachomatis serovar D, Chlamydia muridarum, Chlamydia caviae, and Chlamydia pneumoniae (Fig. 8A). These results demonstrated that 3×FLAG-VAMP4 localizes to inclusions containing C. trachomatis serovar L2 and C. muridarum, but 3×FLAG-VAMP4 did not localize to inclusions containing C. trachomatis serovar D or C. pneumoniae. These results were confirmed by examining the localization of endogenous VAMP4 to these chlamydial inclusions (data not shown). As a negative control, cells were transfected with 3×FLAG vector only and infected as indicated above, and the results are illustrated by the results for C. muridarum (Fig. 8B). Notably, 3×FLAG-syntaxin 6 expressed in HeLa cells localized to all chlamydial species examined in the experiment whose results are shown in Fig. 8 (also data not shown), indicating that the differences noted in 3×FLAG-VAMP4 localization are not due to differences between HeLa cells and C2BBe1 cells. The lack of VAMP4 localization to C. trachomatis serovar D chlamydial inclusions is consistent with previous studies which examined the localization of green fluorescent protein-labeled VAMP4 (GFP-VAMP4) constructs to the chlamydial inclusion and demonstrated that GFP-VAMP4 does not localize to the C. trachomatis serovar D inclusions (46, 47). Therefore, unlike syntaxin 6, VAMP4 is recruited to inclusions of specific chlamydial species.
Fig 8.
3×FLAG-VAMP4 localization to different chlamydial species. HeLa cells were transfected with 3×FLAG-VAMP4 (A) or 3×FLAG vector only (B) and infected with C. trachomatis serovar L2 (Ct L2) or D (Ct D), C. muridarum (MoPn [mouse pneumonitis agent]), or C. pneumoniae (Cp) as described in Materials and Methods. Cells were fixed after 24 h (Ct L2, Ct D, and MoPn) or 72 h (Cp) of infection and processed for indirect immunofluorescence assay as described in Materials and Methods to detect 3×FLAG constructs. To detect chlamydial inclusions and nuclei (blue), cells were stained with DAPI. The results shown are representative of three independent experiments. White stars denote chlamydial inclusions, and white arrows indicate areas of colocalization of 3×FLAG-VAMP4 with the chlamydial inclusion. Bars = 10 μm. 3×FLAG-VAMP4 localizes to inclusions of C. trachomatis serovar L2 and C. muridarum but not to inclusions of C. trachomatis serovar D and C. pneumoniae.
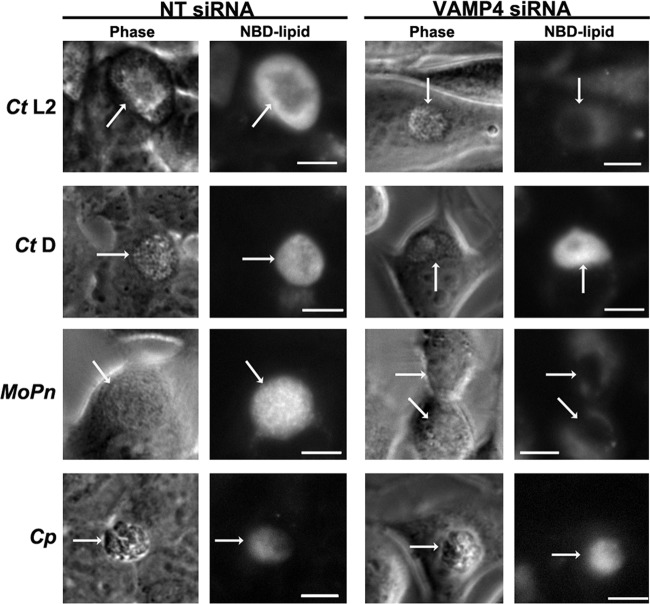
Given the role of VAMP4 in C. trachomatis serovar L2 sphingomyelin acquisition and the fact that sphingomyelin acquisition is conserved across chlamydial species (23, 48, 49), we examined the effect of VAMP4 knockdown on sphingomyelin trafficking to the different chlamydial species (Fig. 9). HeLa monolayers were treated with nontargeting and VAMP4 siRNA, as shown in Fig. 4, and infected with the indicated chlamydial species. Cells were labeled with C6-NBD-ceramide, back-exchanged, and imaged as described above. Knockdown was confirmed by Western blotting, and the knockdown efficiency was similar to that obtained in the experiment whose results are shown in Fig. 5B (data not shown). As expected, knockdown of VAMP4 inhibited sphingomyelin trafficking only to inclusions where VAMP4 was demonstrated to localize. Specifically, VAMP4 localized to inclusions containing C. trachomatis serovar L2 and C. muridarum, corresponding with VAMP4 siRNA knockdown inhibiting sphingomyelin trafficking to those inclusions. These data confirm a role of VAMP4 at the chlamydial inclusion and indicate that VAMP4 knockdown does not in and of itself alter trafficking within the cell that would lead to a general disruption in trafficking to the chlamydial inclusion.
Fig 9.
Live-cell imaging of NBD-lipid trafficking to the chlamydial inclusions of multiple species in VAMP4 knockdown cells. Cells were transfected with siRNA and infected with different chlamydial species, labeled with fluorescent C6-NBD-ceramide (NBD-lipid), back-exchanged, and imaged as described in Materials and Methods. The chlamydial species are denoted as follows: C. trachomatis serovar L2 (Ct L2) and D (Ct D), C. muridarum (MoPn), and C. pneumoniae (Cp). White arrows indicate chlamydial inclusions. Bars = 10 μm. siRNA knockdown of VAMP4 inhibits NBD-lipid trafficking only to inclusions in which VAMP4 localizes.
DISCUSSION
Previous studies demonstrated that the chlamydial inclusion actively intercepts basolaterally targeted Golgi apparatus-derived vesicles (24), which led to an examination of trans-Golgi-associated syntaxin proteins as potential candidates facilitating this process. It was determined that syntaxin 6 was recruited to the chlamydial inclusion in a manner that required chlamydial protein synthesis and was conserved across chlamydial species (17). Given the nature and conservation of syntaxin 6 recruitment to the chlamydial inclusion, we hypothesized that syntaxin 6 and syntaxin 6 binding partners play an important role in supporting chlamydial development. Previous studies demonstrated that VAMP4 is a syntaxin 6 binding partner and a component of syntaxin 6 SNARE complexes (30, 32, 50). In this study, we demonstrate that VAMP4 and syntaxin 6 colocalize and bind to one another at the chlamydial inclusion, corresponding with VAMP4 playing a significant role in chlamydial development and sphingomyelin acquisition.
Previous data have shown that syntaxin 6 localization to the chlamydial inclusion is dependent on a YGRL signal sequence (17). We hypothesized that the YGRL signal sequence would, by default, dictate syntaxin 6-mediated trafficking events and interactions at the chlamydial inclusion. We anticipated that knockdown of syntaxin 6 would abrogate VAMP4 localization to the chlamydial inclusion; however, the data refute this hypothesis, as knockdown of syntaxin 6 has little impact on VAMP4 localization but knockdown of VAMP4 virtually eliminates the retention of syntaxin 6 at the chlamydial inclusion (Fig. 2 and 3). Hence, there are two distinct mechanisms that are involved with syntaxin 6 localization to and retention at the chlamydial inclusion: (i) the presence of a eukaryotic signal sequence that specifically targets the chlamydial inclusion (17) and (ii) SNARE protein interactions that facilitate the retention of syntaxin 6 at the chlamydial inclusion (this study). Specifically, data in this study support the hypothesis that VAMP4 interacts with and is required for retention of syntaxin 6 at the chlamydial inclusion.
Given that syntaxin 6 recruitment to the chlamydial inclusion was shown to require chlamydial protein synthesis and is conserved across chlamydial species (17) and that syntaxin 6 and VAMP4 interact at the chlamydial inclusion (Fig. 3B and C), we hypothesized that similar requirements are necessary for VAMP4 localization to the chlamydial inclusion. The data presented confirm that VAMP4 localization to the chlamydial inclusion requires chlamydial protein synthesis (Fig. 7); however, of all of the species examined, VAMP4 only localized to C. trachomatis serovar L2 and C. muridarum inclusions (Fig. 8). These data are consistent with the results of Delevoye et al., who reported that GFP-VAMP4 does not localize to C. trachomatis serovar D inclusions (51). Interestingly, syntaxin 6 has been shown to bind VAMPs 3, 7, and 8 (52, 53), all of which were identified as localizing to serovar D-containing inclusions (51), suggesting that the VAMP, or R-SNARE, component of syntaxin 6 SNARE complexes may be interchangeable.
SNARE complexes are comprised of 3 Q-SNARE domains and 1 R-SNARE domain, and the classical neuronal synapse SNARE complex is comprised of 3 proteins—a syntaxin (single Q-SNARE), a SNAP protein (2 Q-SNARE domains), and a VAMP (R-SNARE) (54). In the Golgi apparatus, SNARE complexes are typically comprised of 4 proteins—3 different proteins containing a Qa-, Qb-, or Qc-SNARE domain, along with another protein containing an R-SNARE domain (29). Syntaxin 6 and VAMP4 are classified as a Qc-SNARE and an R-SNARE, respectively (55). Assuming these proteins are interacting within a SNARE complex at the chlamydial inclusion, the contributing Qa- and Qb-SNAREs remain to be identified. Syntaxin 6 and VAMP4 have been found in SNARE complexes with Vti1a and syntaxin 16 (56). Vti1a was classified as a Qb-SNARE and syntaxin 16 was classified as a Qa-SNARE (57), which would complete a SNARE Qabc-R complex. However, previous studies have shown that syntaxin 16 does not localize to the chlamydial inclusion (17), making this protein an unlikely candidate to be functioning in a SNARE complex with syntaxin 6 and VAMP4 at this microbial subcellular localization. Because Vti1a coimmunoprecipitates with syntaxin 6 in Chlamydia-infected cells (E. J. Kabeiseman and E. R. Moore, unpublished data), we are currently examining the localization of Vti1a with VAMP4 and syntaxin 6 at the chlamydial inclusion. Future studies will determine whether Vti1a is functioning as the Qb-SNARE at the chlamydial inclusion. Based on Duolink analysis, the data revealed that the VAMP4 proteins that localize to the inclusion, on average, outnumber the VAMP4-syntaxin 6 complexes (Fig. 3B and C), suggesting that VAMP4 interacts with additional binding partners and may function separately from syntaxin 6 at the chlamydial inclusion membrane. Future biochemical analyses will focus on identifying functional syntaxin 6-VAMP4 SNARE complexes which fuse with the chlamydial inclusion membrane.
Given that chlamydial protein synthesis is required for both syntaxin 6 and VAMP4 localization to the inclusion, we hypothesize that one (or two) of the proteins forming a SNARE complex at the chlamydial inclusion are chlamydial in origin. Previous studies demonstrated that syntaxin 16, a Qa-SNARE (27), does not localize to the chlamydial inclusion (17). Hence, we have yet to identify candidate proteins to fill the Qa-SNARE component of an active syntaxin 6-VAMP4 SNARE complex at the chlamydial inclusion. Nevertheless, the data support the hypothesis that a chlamydial protein may be filling the role of the Qa-SNARE. There is precedence for chlamydial proteins acting as SNAREs (46, 51). Chlamydiae encode a protein containing an SNARE domain, IncA (51). IncA, a member of the chlamydial inclusion membrane proteins known as Incs, localizes to the cytoplasmic face of the chlamydial inclusion (58) and is required for homotypic inclusion fusion (59). Alignments of IncA with other known SNARE proteins reveal that it is a Q-SNARE, specifically, a Qc-SNARE (51). Previous studies support the idea that IncA interacts with other SNARE proteins, including a weak interaction with VAMP4, at the chlamydial inclusion (51). However, we have not detected IncA in syntaxin 6 pulldowns (E. J. Kabeiseman and E. R. Moore, unpublished data), indicating that IncA and syntaxin 6 may be interacting in separate SNARE complexes. A recent study presented data which indicate that IncA acts as an inhibitory SNARE protein, effectively blocking membrane fusion with undesirable subcellular compartments, such as vesicles in the classical endosomal/lysosomal pathway (46). Combined, these data suggest that there are networks of SNARE complexes operating at the chlamydial inclusion membrane. This hypothesis is strengthened by recent data demonstrating that members of the conserved oligomeric Golgi (COG) complexes, which are involved in Golgi apparatus vesicle tethering by binding to specific Rab GTPases and SNARE, are recruited to the chlamydial inclusion and are also required for optimal chlamydial development (60).
Given that the chlamydial inclusion membrane is the interface between chlamydial organisms and the host cells that they rely upon for nutrients, such as lipids, it is remarkable that only within the last decade have eukaryotic proteins been demonstrated to localize to the chlamydial inclusion, the first being 14-3-3β (61). Additionally, concerted effort is taken to confirm the function of eukaryotic proteins at the chlamydial inclusion. One method used to screen for eukaryotic protein function at the chlamydial inclusion is to ascertain the protein's involvement in trafficking lipids to the chlamydial inclusion by using the fluorescent lipid C6-NBD-ceramide (21, 23, 39).
A critical process in chlamydial development is chlamydial acquisition of sphingomyelin (18, 19). This process can be studied by utilizing fluorescent C6-NBD-ceramide, a vital stain for the Golgi apparatus (43). Within the Golgi apparatus, C6-NBD-ceramide is metabolized into NBD-sphingomyelin and NBD-glucosylceramide, and these metabolites can be followed as they are trafficked from the Golgi apparatus to the plasma membrane, where they can be extracted or back-exchanged from the plasma membrane by an acceptor that can bind to the NBD moiety, such as a liposome or BSA (21, 44). In Chlamydia-infected cells, the inclusion intercepts fluorescent sphingomyelin but not fluorescent glucosylceramide, resulting in less sphingomyelin being back-exchanged to the plasma membrane (21, 23). Furthermore, sphingomyelin and cholesterol are taken from exocytic vesicles and the lipids are incorporated into chlamydial cell walls (20, 21); presumably, the contents of these vesicles are being emptied into the chlamydial inclusion and scavenged by the organisms. Hence, examining chlamydial acquisition of sphingomyelin by live-cell imaging and thin-layer chromatography after knockdown of key eukaryotic proteins is an effective strategy in determining the role of specific proteins recruited to the chlamydial inclusion (39).
In this study, we demonstrated that VAMP4 plays a pivotal role in chlamydial sphingomyelin acquisition (Fig. 4, 5, and 9), which correlates with a log decrease in chlamydial progeny from organisms propagated in VAMP4 knockdown cells (Fig. 6). Interestingly, knockdown of syntaxin 6 does not prevent sphingomyelin trafficking to the inclusion; if anything, it allows excess sphingomyelin to accumulate within the organisms. By comparison, knockdown of VAMP4 does inhibit chlamydial sphingomyelin trafficking to the inclusion. A caveat to these studies is that VAMP4 also inhibits syntaxin 6 localization to the chlamydial inclusion, and the impact of combined knockdown of syntaxin 6 and VAMP4 on chlamydial sphingomyelin acquisition was not examined. We attempted simultaneous knockdown of both proteins, which resulted in cytotoxic death of both HeLa and C2BBe1 cells (E. J. Kabeiseman and E. R. Moore, personal observation). Nevertheless, the data indicate that of the two proteins, localization of VAMP4 to the chlamydial inclusion corresponds with optimal chlamydial development and lipid acquisition. Importantly, as evident in live-cell imaging studies, VAMP4 knockdown results in a smaller chlamydial inclusion size later in infection, suggesting that VAMP4-mediated sphingolipid acquisition also supports chlamydial inclusion expansion. The role of VAMP4 in chlamydial inclusion expansion needs to be further defined by additional follow-up studies.
The syntaxin 6-VAMP4 interactions at the chlamydial inclusion demonstrated by positive Duolink reactions suggest that the proteins are cooperating in a SNARE complex. Given that syntaxin 6 knockdown results in an accumulation of sphingomyelin in the chlamydial inclusion, we hypothesize that syntaxin 6 cycles away from the inclusion and is returned by the YGRL signal sequence as a mechanism to maintain balance between the chlamydial inclusion and the host cell. SNARE complexes between syntaxin 6 and VAMP4 at the chlamydial inclusion may be a checkpoint allowing a limited number of exocytic vesicles to recycle to the host cell surface instead of fusing with the chlamydial inclusion membrane. The mechanisms that allow the chlamydial inclusion to parasitize host cell nutrients while maintaining an otherwise healthy cell are poorly understood.
Because sphingolipid acquisition is a critical step in the development of obligate intracellular chlamydial organisms (18, 19), there are multiple compensatory mechanisms, as evident in the role of Src family kinase Fyn (62) and CERT (63, 64), an endoplasmic reticulum-associated ceramide transfer protein, in chlamydial sphingomyelin acquisition. Data in this study identify the first SNARE protein involved in sphingomyelin trafficking to the chlamydial inclusion; however, there are other proteins involved in vesicle fusion that have been demonstrated to play a role in chlamydial sphingomyelin acquisition, such as small GTPase Rab proteins, which are involved in vesicle formation and trafficking (65). Specifically, Rabs 6, 11, and 14 have been identified as important mediators of chlamydial sphingomyelin acquisition (66, 67). Interestingly, the recruitment of Rab proteins is not conserved across chlamydial species (68–70); however, a unique facet of VAMP4's recruitment to the chlamydial inclusion is that it is not conserved across chlamydial species or even C. trachomatis serovars, implying that VAMP4 may be recruited only to LGV-containing inclusions, potentially contributing to the more-invasive nature of LGV strains compared to the invasiveness of other C. trachomatis serovars. Further studies need to be performed to test this hypothesis.
In conclusion, we have identified a syntaxin 6-binding partner, VAMP4, and characterized syntaxin 6-VAMP4 interactions at the chlamydial inclusion. Importantly, for the first time, a SNARE protein has been implicated in a likely fusion event with the chlamydial inclusion, exhibited by the requirement of VAMP4 for chlamydial sphingomyelin acquisition. Further study is required to define the proteins involved in functional SNARE complexes at the chlamydial inclusion, including the identification of chlamydial proteins involved in these complexes. Additionally, by examining discrete fusion events at the chlamydial inclusion, the mechanism by which the organisms are directly obtaining, utilizing, and incorporating eukaryotic lipids into their cell walls will be elucidated. It is likely that these eukaryotic lipids aid the organisms in their ability to limit host response to infection, as well as play a role in chlamydial membrane fluidity. We hypothesize that SNARE proteins are recruited to the chlamydial inclusion to deliver nutrients, such as lipids, to the chlamydial organisms by way of fusion with the inclusion membrane. Furthermore, data in this study support the hypothesis that multiple independent SNARE complexes act at the inclusion membrane, indicating that the coordination of these complexes optimizes chlamydial nutrient acquisition.
ACKNOWLEDGMENTS
We acknowledge the technical assistance of Christine Rinehart (University of South Dakota) and Tina Clark, Cheryl Dooley, David Mead, and Janet Sager (Rocky Mountain Laboratories).
This work was supported by an NIAID K22 award (1 K22 AI089856-01A1) and USD/SSOM/BBS start-up funds both awarded to E.R.M. and an award from the Intramural Research Program at NIAID/NIH to T.H.
Footnotes
Published ahead of print 24 June 2013
REFERENCES
- 1.Schachter J. 1999. Infection and disease epidemiology, p 139–169 In Stephens RS. (ed), Chlamydia: intracellular biology, pathogenesis, and immunity. American Society for Microbiology, Washington, DC [Google Scholar]
- 2.Datta SD, Sternberg M, Johnson RE, Berman S, Papp JR, McQuillan G, Weinstock H. 2007. Gonorrhea and chlamydia in the United States among persons 14 to 39 years of age, 1999 to 2002. Ann. Intern. Med. 147:89–97 [DOI] [PubMed] [Google Scholar]
- 3.Darville T, Hiltke TJ. 2010. Pathogenesis of genital tract disease due to Chlamydia trachomatis. J. Infect. Dis. 201:S114–S125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hatch TP. 1975. Utilization of L-cell nucleoside triphosphates by Chlamydia psittaci for ribonucleic acid synthesis. J. Bacteriol. 122:393–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McClarty G. 1994. Chlamydiae and biochemistry of intracellular parasitism. Trends Microbiol. 2:157–164 [DOI] [PubMed] [Google Scholar]
- 6.Wylie JL, Hatch GM, McClarty G. 1997. Host cell phospholipids are trafficked to and then modified by Chlamydia trachomatis. J. Bacteriol. 179:7233–7242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fields KA, Mead DJ, Dooley CA, Hackstadt T. 2003. Chlamydia trachomatis type III secretion: evidence for a functional apparatus during early-cycle development. Mol. Microbiol. 48:671–683 [DOI] [PubMed] [Google Scholar]
- 8.Clausen JD, Christiansen G, Holst HU, Birkelund S. 1997. Chlamydia trachomatis utilizes the host cell microtubule network during early events of infection. Mol. Microbiol. 25:441–449 [DOI] [PubMed] [Google Scholar]
- 9.Grieshaber SS, Grieshaber NA, Hackstadt T. 2003. Chlamydia trachomatis uses host cell dynein to traffic to the microtubule-organizing center in a p50 dynamitin-independent process. J. Cell Sci. 116:3793–3802 [DOI] [PubMed] [Google Scholar]
- 10.Hackstadt T. 1999. Cell biology, p 101–138 In Stephens R. (ed), Chlamydia: intracellular biology, pathogenesis, and immunity. American Society for Microbiology, Washington, DC [Google Scholar]
- 11.Heinzen RA, Scidmore MA, Rockey DD, Hackstadt T. 1996. Differential interaction with endocytic and exocytic pathways distinguish parasitophorous vacuoles of Coxiella burnetii and Chlamydia trachomatis. Infect. Immun. 64:796–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scidmore MA, Fischer ER, Hackstadt T. 2003. Restricted fusion of Chlamydia trachomatis vesicles with endocytic compartments during the initial stages of infection. Infect. Immun. 71:973–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taraska T, Ward DM, Ajioka RS, Wyrick PB, Davis-Kaplan SR, Davis CH, Kaplan J. 1996. The late chlamydial inclusion membrane is not derived from the endocytic pathway and is relatively deficient in host proteins. Infect. Immun. 64:3713–3727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Ooij C, Apodaca G, Engel J. 1997. Characterization of the Chlamydia trachomatis vacuole and its interaction with the host endocytic pathway in HeLa cells. Infect. Immun. 65:758–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wyrick PB. 2000. Intracellular survival by Chlamydia. Cell. Microbiol. 2:275–282 [DOI] [PubMed] [Google Scholar]
- 16.Mital J, Miller NJ, Fischer ER, Hackstadt T. 2010. Specific chlamydial inclusion membrane proteins associate with active Src family kinases in microdomains that interact with the host microtubule network. Cell. Microbiol. 12:1235–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moore ER, Mead DJ, Dooley CA, Sager J, Hackstadt T. 2011. The trans-Golgi SNARE syntaxin 6 is recruited to the chlamydial inclusion membrane. Microbiology 157:830–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Ooij C, Kalman L, vanIjzendoorn S, Nishijima M, Hanada K, Mostov K, Engel JN. 2000. Host cell-derived sphingolipids are required for the intracellular growth of Chlamydia trachomatis. Cell. Microbiol. 2:627–637 [DOI] [PubMed] [Google Scholar]
- 19.Robertson DK, Gu L, Rowe RK, Beatty WL. 2009. Inclusion biogenesis and reactivation of persistant Chlamydia trachomatis requires host cell sphingolipid biosynthesis. PLoS Pathog. 5:e1000664. 10.1371/journal.ppat.1000664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carabeo RA, Mead DJ, Hackstadt T. 2003. Golgi-dependent transport of cholesterol to the Chlamydia trachomatis inclusion. Proc. Natl. Acad. Sci. U. S. A. 100:6771–6776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hackstadt T, Scidmore MA, Rockey DD. 1995. Lipid metabolism in Chlamydia trachomatis-infected cells: directed trafficking of Golgi-derived sphingolipids to the chlamydial inclusion. Proc. Natl. Acad. Sci. U. S. A. 92:4877–4881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scidmore MA, Fischer ER, Hackstadt T. 1996. Sphingolipids and glycoproteins are differentially trafficked to the Chlamydia trachomatis inclusion. J. Cell Biol. 134:363–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hackstadt T, Rockey DD, Heinzen RA, Scidmore MA. 1996. Chlamydia trachomatis interrupts an exocytic pathway to acquire endogenously synthesized sphingomyelin in transit from the Golgi apparatus to the plasma membrane. EMBO J. 15:964–977 [PMC free article] [PubMed] [Google Scholar]
- 24.Moore ER, Fischer ER, Mead DJ, Hackstadt T. 2008. The chlamydial inclusion preferentially intercepts basolaterally directed sphingomyelin-containing exocytic vacuoles. Traffic 9:2130–2140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fasshauer D, Sutton RB, Brunger AT, Jahn R. 1998. Conserved structural features of the synaptic fusion complex: SNARE proteins reclassified as Q- and R-SNAREs. Proc. Natl. Acad. Sci. U. S. A. 95:15781–15786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bock JB, Matern HT, Peden AA, Scheller RH. 2001. A genomic perspective on membrane compartment organization. Nature 409:839–841 [DOI] [PubMed] [Google Scholar]
- 27.Uemura T, Ueda T, Ohniwa RL, Nakano A, Takeyasu K, Sato MH. 2004. Systematic analysis of SNARE molecules in Arabidopsis: dissection of the post-Golgi network in plant cells. Cell Struct. Funct. 29:49–65 [DOI] [PubMed] [Google Scholar]
- 28.Paul TR, Knight ST, Raulston JE, Wyrick PB. 1997. Delivery of azithromycin to Chlamydia trachomatis-infected polarized human endometrial epithelial cells by polymorphonuclear leucocytes. J. Antimicrob. Chemother. 39:623–630 [DOI] [PubMed] [Google Scholar]
- 29.Zwilling D, Cypionka A, Pohl WH, Fasshauer D, Walla PJ, Wahl MC, Jahn R. 2007. Early endosomal SNAREs form a structurally conserved SNARE complex and fuse liposomes with multiple topologies. EMBO J. 26:9–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watson RT, Pessin JE. 2000. Functional cooperation of two independent targeting domains in syntaxin 6 is required for its efficient localization in the trans-golgi network of 3T3L1 adipocytes. J. Biol. Chem. 275:1261–1268 [DOI] [PubMed] [Google Scholar]
- 31.Wendler F, Tooze S. 2001. Syntaxin 6: the promiscuous behavior of a SNARE protein. Traffic. 2:606–611 [DOI] [PubMed] [Google Scholar]
- 32.Antonin W, Holroyd C, Fasshauer D, Pabst S, von Mollard GF, Jahn R. 2000. A SNARE complex mediating fusion of late endosomes defines conserved properties of SNARE structure and function. EMBO J. 19:6453–6464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cocucci E, Racchetti G, Rupnik M, Meldolesi J. 2008. The regulated exocytosis of enlargeosomes is mediated by a SNARE machinery that includes VAMP4. J. Cell Sci. 121:2983–2991 [DOI] [PubMed] [Google Scholar]
- 34.Tran THT, Zeng Q, Hong W. 2007. VAMP4 cycles from the cell surface to the trans-Golgi network via sorting and recycling endosomes. J. Cell Sci. 120:1028–1041 [DOI] [PubMed] [Google Scholar]
- 35.Caldwell HD, Kromhout J, Schachter J. 1981. Purification and partial characterization of the major outer membrane protein of Chlamydia trachomatis. Infect. Immun. 31:1161–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scidmore MA. 2005. Cultivation and laboratory maintenance of Chlamydia trachomatis. Curr. Protoc. Microbiol. Chapter 11:Unit 11A.1. 10.1002/9780471729259.mc11a01s00 [DOI] [PubMed] [Google Scholar]
- 37.Furness G, Graham DM, Reeve P. 1960. The titration of trachoma and inclusion blennorrhoea viruses in cell culture. J. Gen. Microbiol. 23:613–619 [DOI] [PubMed] [Google Scholar]
- 38.Lippincott-Schwartz J, Yuan L, Bonifacino J, Klausner R. 1989. Rapid redistribution of Golgi proteins in the ER in cells treated with brefeldin A: evidence for membrane cycling from Golgi to ER. Cell 56:801–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moore ER. 2012. Sphingolipid trafficking and purification in Chlamydia trachomatis-infected cells. Curr. Protoc. Microbiol. Chapter 11:Unit 11A.2. 10.1002/9780471729259.mc11a02s27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bligh EG, Dyer WJ. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37:911–917 [DOI] [PubMed] [Google Scholar]
- 41.Soderberg O, Gullberg M, Jarvius M, Ridderstrale K, Leuchowius KJ, Jarvius J, Wester K, Hydbring P, Bahram F, Larsson LG, Landegren U. 2006. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods 3:995–1000 [DOI] [PubMed] [Google Scholar]
- 42.Lipsky NG, Pagano RE. 1983. Spingolipid metabolism in cultured fibroblasts: microscopic and biochemical studies employing a fluorescent ceramide analogue. Proc. Natl. Acad. Sci. U. S. A. 80:2608–2612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lipsky NG, Pagano RE. 1985. A vital stain for the Golgi apparatus. Science 228:745–747 [DOI] [PubMed] [Google Scholar]
- 44.Lipsky NG, Pagano RE. 1985. Intracellular translocation of fluorescent sphingolipids in cultured fibroblasts: endogenously synthesized sphingomyelin and glucocerebroside analogues pass through the Golgi apparatus en route to the plasma membrane. J. Cell Biol. 100:27–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shaw EI, Dooley CA, Fischer ER, Scidmore MA, Fields KA, Hackstadt T. 2000. Three temporal classes of gene expression during the Chlamydia trachomatis developmental cycle. Mol. Microbiol. 37:913–925 [DOI] [PubMed] [Google Scholar]
- 46.Paumet F, Wesolowski J, Garcia-Diaz A, Delevoye C, Aulner N, Shuman HA, Subtil A, Rothman JE. 2009. Intracellular bacteria encode inhibitory SNARE-like proteins. PLoS One 4:e7375. 10.1371/journal.pone.0007375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xiong Q, Rikihisa Y. 2012. Subversion of NPC1 pathway of cholesterol transport by Anaplasma phagocytophilum. Cell. Microbiol. 14:560–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rockey DD, Fischer ER, Hackstadt T. 1996. Temporal analysis of the developing Chlamydia psittaci inclusion by use of fluorescence and electron microscopy. Infect. Immun. 64:4269–4278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wolf K, Hackstadt T. 2001. Sphingomyelin trafficking in Chlamydia pneumoniae-infected cells. Cell. Microbiol. 3:145–152 [DOI] [PubMed] [Google Scholar]
- 50.Steegmaier M, Klumperman J, Foletti DL, Yoo JS, Scheller RH. 1999. Vesicle-associated membrane protein 4 is implicated in trans-Golgi network vesicle trafficking. Mol. Biol. Cell 10:1957–1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Delevoye C, Nilges M, Dehoux P, Paumet F, Perrinet S, Dautry-Varsat A, Subtil A. 2008. SNARE protein mimicry by an intracellular bacterium. PLoS Pathog. 4:e1000022. 10.1371/journal.ppat.1000022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gerrard SR, Levi BP, Stevens TH. 2000. Pep12p is a multifunctional yeast syntaxin that controls entry of biosynthetic, endocytic and retrograde traffic into the prevacuolar compartment. Traffic 1:259–269 [DOI] [PubMed] [Google Scholar]
- 53.Hanson PI, Otto H, Barton N, Jahn R. 1995. The N-ethylmaleimide-sensitive fusion protein and alpha-SNAP induce a conformational change in syntaxin. J. Biol. Chem. 270:16955–16961 [DOI] [PubMed] [Google Scholar]
- 54.Sollner T, Bennett MK, Whiteheart SW, Scheller RH, Rothman JE. 1993. A protein assembly-disassembly pathway in vitro that may correspond to sequential steps of synaptic vesicle docking, activation, and fusion. Cell 75:409–418 [DOI] [PubMed] [Google Scholar]
- 55.Malsam J, Sollner TH. 2011. Organization of SNAREs within the Golgi stack, p 123–139 In Warren G, Rothman J. (ed), The Golgi. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kreykenbohm V, Wenzel D, Antonin W, Atlachkine V, von Mollard GF. 2002. The SNAREs vti1a and vti1b have distinct localization and SNARE complex partners. Eur. J. Cell Biol. 81:273–280 [DOI] [PubMed] [Google Scholar]
- 57.Kloepper TH, Kienle CN, Fasshauer D. 2007. An elaborate classification of SNARE proteins sheds light on the conservation of the eukaryotic endomembrane system. Mol. Biol. Cell 18:3463–3471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bannantine JP, Stamm WE, Suchland RJ, Rockey DD. 1998. Chlamydia trachomatis IncA is localized to the inclusion membrane and is recognized by antisera from infected humans and primates. Infect. Immun. 66:6017–6021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hackstadt T, Scidmore-Carlson MA, Shaw EI, Fischer ER. 1999. The Chlamydia trachomatis IncA protein is required for homotypic vesicle fusion. Cell. Microbiol. 1:119–130 [DOI] [PubMed] [Google Scholar]
- 60.Pokrovskaya ID, Szwedo JW, Goodwin A, Lupashina TV, Nagarajan UM, Lupashin VV. 2012. Chlamydia trachomatis hijacks intra-Golgi COG complex-dependent vesicle trafficking pathway. Cell. Microbiol. 14:656–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Scidmore MA, Hackstadt T. 2001. Mammalian 14-3-3beta associates with the Chlamydia trachomatis inclusion membrane via its interaction with IncG. Mol. Microbiol. 39:1638–1650 [DOI] [PubMed] [Google Scholar]
- 62.Mital J, Hackstadt T. 2011. Role for the SRC family kinase Fyn in sphingolipid acquisition by chlamydiae. Infect. Immun. 79:4559–4568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Derre I, Swiss R, Agaisse H. 2011. The lipid transfer protein CERT interacts with the Chlamydia inclusion protein IncD and participates to ER-Chlamydia inclusion membrane contact sites. PLoS Pathogens 7:e1002092. 10.1371/journal.ppat.1002092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Elwell CA, Jiang S, Kim JH, Lee A, Wittmann T, Hanada K, Melancon P, Engel JN. 2011. Chlamydia trachomatis co-opts GBF-1 and CERT to acquire host sphingomyelin for distinct roles during intracellular development. PLoS Pathog. 7:e1002198. 10.1371/journal.ppat.1002198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stenmark H. 2009. Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 10:513–525 [DOI] [PubMed] [Google Scholar]
- 66.Capmany A, Damiani MT. 2010. Chlamydia trachomatis intercepts Golgi-derived sphingolipids through a rab 14-mediated transport required for bacterial development and replication. PLoS One 5:e14084. 10.1371/journal.pone.0014084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lipinski AR, Heymann J, Meissner C, Karlas A, Brinkmann V, Meyer TF, Heuer D. 2009. Rab6 and Rab11 regulate Chlamydia trachomatis development and Golgin-84-dependent Golgi fragmentation. PLoS Pathog. 5:e1000615. 10.1371/journal.ppat.1000615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Moorhead AR, Rzomp KA, Scidmore MA. 2007. The Rab6 effector bicaudal D1 associates with Chlamydia trachomatis inclusions in a biovar-specific manner. Infect. Immun. 75:781–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rzomp KA, Moorhead AR, Scidmore MA. 2006. The GTPase Rab4 interacts with Chlamydia trachomatis inclusion membrane protein CT229. Infect. Immun. 74:5362–5373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rzomp KA, Scholtes LD, Briggs BJ, Whittaker GR, Scidmore MA. 2003. Rab GTPases are recruited to chlamydial inclusions in both a species-dependent and species-independent manner. Infect. Immun. 71:5855–5870 [DOI] [PMC free article] [PubMed] [Google Scholar]