Abstract
Tumor cells with donor genotype have been identified in human skin cancer after allogeneic transplantation; however, the donor contribution to the malignant epithelium has not been established. Kidney transplant recipients have an increased risk of invasive skin squamous cell carcinoma (SCC), which is associated with accumulation of the tumor suppressor p53 and TP53 mutations. In 21 skin SCCs from kidney transplant recipients, we systematically assessed p53 expression and donor/recipient origin in laser-microdissected p53+ tumor cells. In one patient, molecular analyses demonstrated that skin tumor cells had the donor genotype and harbored a TP53 mutation in codon 175. In a kidney graft biopsy performed 7 years before the skin SCC diagnosis, we found p53+ cells in the renal tubules. We identified the same TP53 mutation in these p53+ epithelial cells from the kidney transplant. These findings provide evidence for a donor epithelial cell contribution to the malignant skin epithelium in the recipient in the setting of allogeneic kidney transplantation. This finding has theoretical implications for cancer initiation and progression and clinical implications in the context of prolonged immunosuppression and longer survival of kidney transplant patients.
Introduction
The concept of adult stem cell plasticity (1, 2) has opened new fields for scientific and clinical developments. Allogeneic transplantations offer a rare clinical opportunity to study homing and in situ development of stem cells in humans. In addition, the main long-term complication of allogeneic transplantations (3) is the occurrence of cancers (4, 5). Patients with kidney transplants are at risk for skin epithelial tumors (4), which preferentially develop on sun-exposed areas (6). Accumulation of tumor-suppressing p53 is significantly associated with these squamous cell carcinomas (SCCs) (7), and TP53 mutations are found in tumor cells (8). In a series of kidney transplant recipients, tumor cells of donor genotype have been identified in 1 skin basal cell carcinoma (9), but the type of donor cell that homed to the skin remains to be characterized. In addition, there was no evidence of a donor epithelial cell contributing to the malignant epithelium in the recipient.
Systematic study of p53 expression and TP53 mutations in 21 skin SCCs from kidney transplant recipients enabled us to identify a case (referred to herein as Patient 1) with p53+ tumor cells of donor origin and a mutation on codon 175. In a kidney graft biopsy performed on Patient 1 7 years before the skin SCC diagnosis, we found p53+ cells in the renal tubules and laser microdissected them for further molecular study.
Results and Discussion
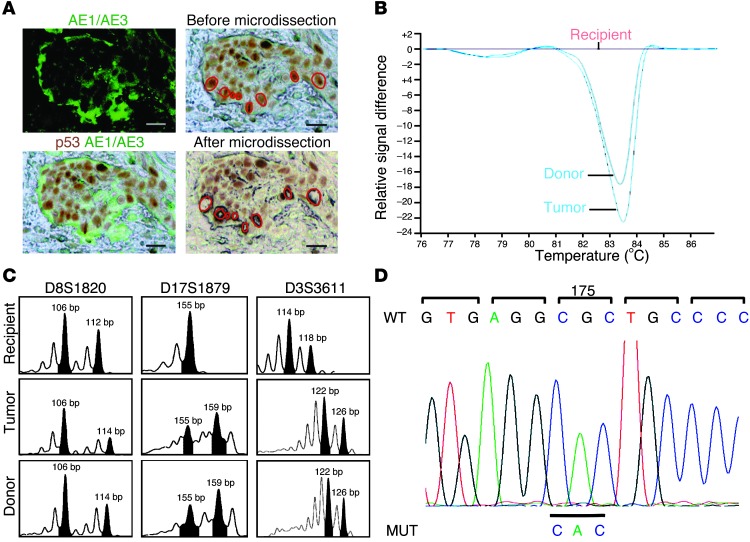
21 kidney transplant patients diagnosed with skin SCC between 2000 and 2007 in the same university hospital had a minimum 5-year follow-up. For this study, SCC histological diagnosis was independently reviewed by 2 pathologists. As expected in organ transplant recipients, SCCs occurred after a long immunosuppression period (mean, 10.55 years; range, 3–21 years) and on sun-exposed areas (3, 6). Systematic p53 analyses showed that all SCCs strongly expressed p53 in the tumor cells. In nonlesional sun-exposed areas, skin basal cells weakly expressed p53. In SCC, p53 expression also predominated in the basal layer, particularly at sites of dermal invasion (Supplemental Figure 1; supplemental material available online with this article; doi: 10.1172/JCI66721DS1). We therefore performed laser microdissection of p53+ tumor cells from these areas that coexpressed the cytokeratin AE1/AE3 (Figure 1A), in order to select representative material for genetic analyses (10).
Figure 1. Recipient skin SCC.
(A) Laser microdissection of p53+AE1/AE3+ basal cells from invasive areas. Scale bars: 15 μm. (B) mtDNA-HRM of laser-microdissected p53+ tumor cells and donor and recipient normal cells. Laser-microdissected p53+ tumor cells and donor normal cells showed similar profiles. (C) DNA profiles from laser-microdissected p53+ tumor cells and donor and recipient normal cells. Microsatellite analysis at the D8S1820 locus showed the donor origin of the p53+ tumor cells (106 and 114 bp), similar to donor DNA, but different from recipient DNA (106 and 112 bp). At the D17S1879 locus, microdissected p53+ tumor cell and donor DNA were heterozygous (155 and 159 bp), but recipient DNA was homozygous (155 bp). (D) Sequencing of TP53 exon 5 PCR product identified a base substitution G>A on codon 175, corresponding to a R>H amino acid change in DNA, from laser-microdissected p53+ tumor cells.
Tumor cell origin was studied on DNA extracted from laser-microdissected p53+ tumor cells, using mitochondrial DNA–high-resolution melting (mtDNA-HRM) and polymorphic microsatellite markers. 2 female patients with previous male pregnancies were excluded from this study. The 2 molecular methods used gave interpretable results in 14 of the remaining 19 patients.
In Patient 1, mtDNA-HRM showed that laser-microdissected p53+ tumor cells had curves similar to the donor curves and different from the recipient curves (Figure 1B). Since widespread heterogeneity (heteroplasmy) has been found in normal and cancer cell mtDNA in humans (11), we checked our HRM results using Sanger sequencing: no heteroplasmy was detected. We next studied 7 polymorphic microsatellite markers for their informative value, showing different donor and recipient profiles. Of these, 3 (D8S1820, D17S1879, and D3S3611) again clearly established the donor-derived origin of the skin SCC (Figure 1C). In all, 12 of the 14 patients with interpretable molecular results had a SCC of recipient origin, and 2 had a SCC with presence of donor cells.
A donor-derived malignant proliferation after allogeneic transplantation was first identified in 1971, in a leukemia of donor origin after bone marrow transplantation (12). More than 50 cases have since been reported and are considered to result from oncogenic transformation of apparently normal donor hematopoietic cells in the transplant recipient (12). More surprising was the identification of a donor-derived genotype in epithelial cells in 4 cases of oral SCC after bone marrow transplantation (13), because this implied that donor-derived bone marrow cells, either hematopoietic or mesenchymal, yielded epithelial tumors. Donor-derived cells were also found in a skin basal cell carcinoma in a kidney transplant recipient (9), but the donor cell contribution to the malignant epithelium was not established.
The skin carcinoma in Patient 1 was, as in a large series of kidney transplant recipients, a SCC (3, 6) that occurred in UV-exposed areas (4, 6) and expressed p53 (7). The predominance of SCC, a fully differentiated type of skin carcinoma, over basal cell carcinoma in kidney transplant recipients contrasts with the predominance of basal cell carcinoma in the general population (3, 4). The specific distribution of SCC in UV-exposed areas could be linked to UV-induced keratinocyte damage, an event able to trigger stem cell mobilization in the skin (14). In addition, the role of UV-induced TP53 mutations in skin SCC has long been established (15). More than 10 TP53 mutations in skin SCC in kidney transplant recipients have been characterized (8), including the UV-specific tandem CC>TT, a mutation more linked to the UVA effect in the epidermal basal layer (16).
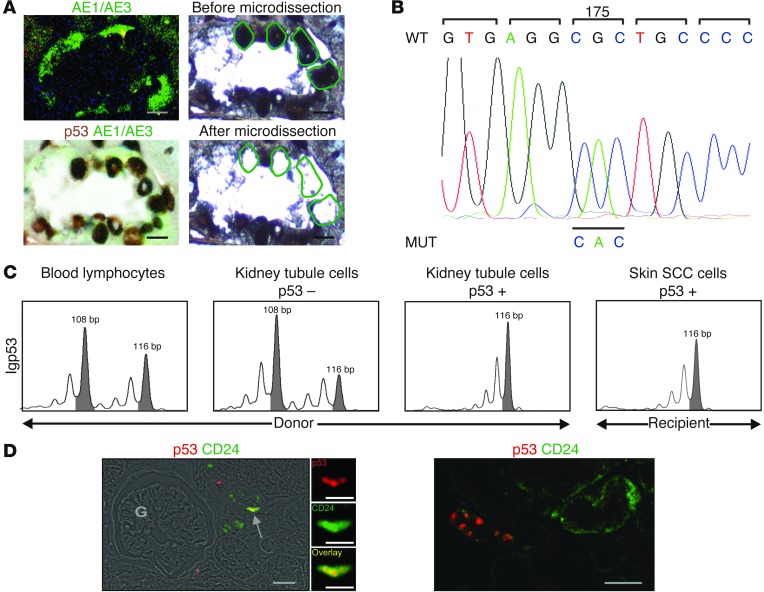
In the 2 patients with donor-derived p53+cells in their skin SCC, PCR-HRM screening was performed to compare DNA from microdissected p53+ tumor cells and a control DNA, including a wild-type TP53 gene. In Patient 1, PCR-HRM screening showed a shift fragment for exons 5–8 of TP53. Sequencing of this shift fragment identified a G>A base substitution on codon 175, corresponding to a R>H amino acid (Figure 1D). This mutation did not belong to the group of well-characterized UV-induced TP53 mutations. We therefore decided to study the previous available biopsies of Patient 1. In a biopsy performed 7 years before this patient’s SCC diagnosis, we found p53+ cells in around 10% of kidney tubules. Using laser microdissection, we selected p53+AE1/AE3+ cells from sections of the kidney biopsy (Figure 2A). After DNA extraction, sequencing enabled us to identify an identical mutation of TP53 (codon 175) in epithelial tubular cells from the kidney transplant (Figure 2B). The p53+ cells in the kidney tubules had no morphological sign of malignancy. Possible primitive renal tumors or metastatic cells transferred in the kidney graft (17) were excluded via pathological analyses, including HPV typing (Supplemental Figure 2 and Supplemental Methods) and careful MRI. Over the 7-year follow-up, systematic MRI was also found to be normal. In addition, sequencing showed that the TP53 mutation in p53+ cells from both kidney tubules and skin SCC was not heterozygous. We therefore used a different molecular method to check this result. A study of the polymorphic microsatellite marker IGP53, located in intron 1 of TP53, was performed to compare the profiles of the donor and the microdissected p53+ cells from skin SCC and from kidney graft tubules. This demonstrated a loss of heterozygosity in the skin SCC and kidney graft tubule microdissected cells (Figure 2C). In the case of Patient 1, these TP53 changes were not linked to a cancer predisposition (Li-Fraumeni syndrome) (18), since there was no family history of cancer under 45 years, and this patient had no other cancer than his skin cancer.
Figure 2. Donor kidney.
(A) Kidney graft biopsy performed 7 years before skin SCC diagnosis. Laser microdissection was used to select p53+AE1/AE3+ cells in renal tubules. Scale bars: 5 μm. (B) TP53 sequencing identified the same base substitution in laser-microdissected p53+ epithelial cells from the kidney graft tubules as in skin SCC tumor cells. (C) Since sequencing showed an homozygous mutation in both skin SCC and kidney graft tubules, we checked this result. Polymorphic microsatellite marker IGP53, located in intron 1, showed loss of heterozygosity for this locus in recipient skin SCC and p53+ kidney graft tubule cells, but not in donor blood lymphocytes or p53– kidney graft tubule cells. (D) Kidney graft biopsy with staining for p53 as well as the renal stem/progenitor cell marker CD24. Double-stained cells were few and were located in kidney tubules (arrow), not in the glomerular area (G). Scale bars: 25 μm; 10 μm (insets, enlarged ×3).
We thus provided evidence of a donor renal epithelial cell contribution to the malignant epithelium in the recipient skin SCC. In adult human kidney, different types of progenitor cells have been characterized in tubules. Cells with a CD133+CD24+CD106– phenotype are committed toward tubular lineage (19). In Patient 1, p53+CD24+ cells were found in tubules in the kidney biopsy (Figure 2D and Supplemental Methods). In addition, TP53 expression increases in kidney allografts when cold ischemia is prolonged beyond 24 hours, a common time lapse when kidneys from deceased donors are transplanted (20), as in Patient 1. Different TP53 mutations have been reported in renal carcinomas, including mutations in exons 5–8, where we found the codon 175 mutation.
Our finding of an identical TP53 mutation in the p53+ cells in the tubular epithelium from the kidney graft and in tumor cells from the recipient skin SCC demonstrated a contribution of donor cells to the recipient malignant epithelium. However, the question remains why, in the 7-year follow-up, Patient 1 developed a carcinoma in the skin and not in the grafted kidney. This could be explained by additional oncogenic events in the recipient skin. Extended genomic analysis via the molecular inversion probe-based genotyping system enabled us to detect a KRAS mutation in the skin SCC (Supplemental Figure 3). However, we used a specific KRAS probe to perform PCR analysis and did not detect this mutation in the kidney biopsy (Supplemental Methods). RAS mutations have been recently identified in skin SCC in patients treated with BRAF inhibitors (21). This was not the case for Patient 1. Apart from the KRAS mutation, UV radiation could also instigate additional oncogenic events in the recipient skin. In this patient, a deleterious synergistic effect between UV and azathioprine could have occurred, since azathioprine causes 6-thioguanine accumulation in patient DNA and, in cell cultures, 6-thioguanine and UVA are synergistically mutagenic (22).
Our results provide the first evidence for donor contribution to the malignant epithelium of skin SCC in a kidney transplant recipient. This finding has theoretical implications for the model of cancer initiation and/or progression as well as clinical implications in the context of prolonged immunosuppression and longer survival of kidney transplant patients.
Methods
Patients and samples.
From 2000 to 2007, skin SCC were diagnosed in 21 kidney transplant recipients who had available archival samples and donor and recipient DNA. Patients had received kidneys from deceased donors for extramembranous glomerulonephritis, malformative uropathy, or Henoch-Schönlein purpura. After kidney transplantation, immunosuppression included corticosteroids, tacrolimus, mycophenolate-mofetil, cyclosporine, and azathiopine. All biopsy samples, taken for diagnostic purposes, had been formalin fixed and paraffin embedded. SCC histological diagnosis was reviewed by 2 pathologists. From samples remaining after diagnosis had been established, sister sections were subsequently taken from each block.
p53 immunostaining and laser microdissection of skin p53+AE1/AE3+ tumor cells.
p53 immunostaining was performed on 5-μm-thick skin SCC sections using indirect immunoperoxidase staining, with anti–human p53 mouse antibody (clone DO7, Dako). Skin SCC areas with strong p53 expression in basal layers and invasive areas were selected, and 7-μm-thick paraffin sections from the corresponding tissue blocks were spread on membrane-coated slides. Double immunostaining was then performed using anti-p53 antibody with immunoperoxidase staining and anti-cytokeratin antibody (clone AE1/AE3, an epithelial marker, Roche Diagnostics) with FITC staining.
Using a PALM-Microbeam/Zeiss system, laser microdissection was performed on p53+AE1/AE3+ cells in invasive areas of skin SCC with a pulsed UV-A nitrogen laser (337 nm), used to cut and catapult microdissected cells directly into the buffer-containing cap of a microfuge tube. At least 1,000 p53+ tumor cells were laser microdissected, and DNA was extracted using a Qiagen kit. DNA quality was checked by spectrometric assay (Nanodrop 1000, Thermo Scientific).The cell population selected by laser microdissection was checked using quantitative RT-PCR (qRT-PCR) on an ABI PRISM 7700 system and predeveloped TaqMan assay reagents specific to KRT14 (Hs00265033_m1) and PTPRC (Hs00365634_g1). Laser-microdissected cells expressed KRT14 mRNA, an epithelial marker, and not PTPRC mRNA, a pan-leucocyte marker.
mtDNA-HRM analyses of microdissected p53+ tumor cells.
mtDNA profiles from SCC laser-microdissected p53+ tumor cells were compared with those from donor and recipient cells. Primers were designed to flank the hypervariable regions I of the noncoding displacement loop (D-loop) of mtDNA from NCBI reference sequence NC_012920.1 (23), and synthesized (Eurogentec). Primers flanking the hypervariable regions I were used to amplify an initial 243-bp PCR product using the forward primer I 16105F (5′-TGCCAGCCACCATGAATA-3′) and reverse primer I 16348R (5′-GACTGTAATGTGCTATGTACGGTAAA-3′). See Supplemental Methods for PCR cycling, mix, and HRM steps. All reactions were completed in triplicate.
Polymorphic microsatellite marker analyses of microdissected p53+ tumor cells.
7 highly polymorphic microsatellites were evaluated: D3S3611, D3S1597, D5S2095, D8S1820, D9S162, D17S1879, and IGP53. Primer sequences were obtained from UCSC Genome Bioinformatics ( http://genome.ucsc.edu/). See Supplemental Methods for PCR. Fluorescent allelic profiles from SCC laser-microdissected p53+ tumor cells were compared with those from donor and recipient cells.
TP53 screening and sequencing in p53+ tumor cells from 1 skin SCC with cells of donor origin.
Sequencing of the shift fragment for exons 5–8 of TP53, determined by PCR-HRM screening, was performed using the Sanger method (24). Primers were designed from NCBI reference sequence X54156. Amplicons 80–150 bp in length covered the coding sequence and exon-intron boundaries. All forward primers were tailed with M13-Universal nucleotidic sequence for sequencing standardization (Supplemental Table 1). See Supplemental Methods for PCR, HRM steps, and sequencing.
Genotyping of skin SCC.
DNA from laser-microdissected p53+ tumor cells in skin SCC was studied using the molecular inversion probe-based genotyping system OncoScan Express (Affymetrix), which determines genotype of 330,000 SNPs. Analyses for mutation detection were performed using Nexus 7 software (BioDiscovery). See Supplemental Methods for details.
Laser microdissection of p53+ renal tubule cells and TP53 sequencing.
In Patient 1, with donor-derived tumor cells and TP53 mutation in the skin SCC, we studied kidney biopsies performed on the graft before the skin SCC diagnosis. Immunohistochemistry using anti-human p53 antibody enabled us to identify areas with p53+ renal tubule cells. On the corresponding tissue blocks, double immunostaining was performed using p53 antibody with immunoperoxidase staining and anti-cytokeratin (clone AE1/AE3, an epithelial marker, Roche Diagnostics) with FITC staining. p53+AE1/AE3+ cells in renal tubules were laser microdissected and DNA extracted, and quality was checked. The cell population selected by laser microdissection was also checked, and qRT-PCR showed that it expressed KRT14 mRNA, not PTPRC mRNA. PCR and TP53 sequencing were then carried out as for laser-microdissected tumor cells from skin SCC in the same patient.
Study approval.
The study was approved by the Institutional Review Board of Hôpital Saint Louis, and informed consent was obtained in accordance with the Declaration of Helsinki.
Supplementary Material
Acknowledgments
This work was supported by Région Ile de France, Cancéropôle Ile de France, INCa, ANR, Tumorothèque Hopital Saint Louis, and Société Française Dermatologie. A. Swaine reviewed English usage and structure.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2013;123(9):3797–3801. doi:10.1172/JCI66721.
See the related Commentary beginning on page 3708.
References
- 1.Krause DS, et al. Multi-organ, multi-lineage engraftment by a single bone marrow-derived stem-cell. Cell. 2001;105(3):369–377. doi: 10.1016/S0092-8674(01)00328-2. [DOI] [PubMed] [Google Scholar]
- 2.Korbling M, et al. Hepatocytes and epithelial cells of donor origin in recipients of peripheral-blood stem-cells. N Engl J Med. 2002;346(10):738–746. doi: 10.1056/NEJMoa3461002. [DOI] [PubMed] [Google Scholar]
- 3.Euvrard S, Kanitakis J, Claudy A. Skin cancers after organ transplantation. N Engl J Med. 2003;348(17):1681–1691. doi: 10.1056/NEJMra022137. [DOI] [PubMed] [Google Scholar]
- 4.Moloney FJ, Comber H, O’Lorcain P, O’Kelly P, Conlon PJ, Murphy GM. A population-based study of skin cancer incidence and prevalence in renal transplant recipients. Br J Dermatol. 2006;154(3):498–504. doi: 10.1111/j.1365-2133.2005.07021.x. [DOI] [PubMed] [Google Scholar]
- 5.Ingvar A, et al. Solid organ transplantation and risk of post-transplant cutaneous squamous cell carcinoma. Nephrol Dial Transplant. 2010;25(8):2764–2771. doi: 10.1093/ndt/gfp425. [DOI] [PubMed] [Google Scholar]
- 6.Terhorst D, Drecoll U, Stockfleth E, Ulrich C. Organ transplant recipients and skin cancer: assessment of risk factors with focus on sun exposure. Br J Dermatol. 2009;161(suppl 3):85–89. doi: 10.1111/j.1365-2133.2009.09454.x. [DOI] [PubMed] [Google Scholar]
- 7.Stark LA, Arends MJ, McLaren KM, Shahidullah H, Hunter JA, Bird CC. Accumulation of p53 is associated with tumour progression in cutaneous lesions of renal allograft recipients. Br J Cancer. 1994;70(4):662–667. doi: 10.1038/bjc.1994.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Queille S, et al. Skin cancer risk factors in renal transplant patients show high levels of UV-specific tandem CC to TT mutations of the p53 gene. Carcinogenesis. 2007;28(3):724–731. doi: 10.1093/carcin/bgl191. [DOI] [PubMed] [Google Scholar]
- 9.Aractingi S, et al. Skin carcinoma arising from donor cells in a kidney transplant recipient. Cancer Res. 2005;65(5):1755–1760. doi: 10.1158/0008-5472.CAN-04-2783. [DOI] [PubMed] [Google Scholar]
- 10.Backvall H, Asplund A, Gustafsson A, Sivertsson A, Lundeberg J, Ponten F. Microdissection and genetic heterogeneity in squamous and basal cell carcinoma. Mutat Res. 2005;571(1–2):65–79. doi: 10.1016/j.mrfmmm.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 11.He Y, et al. Heteroplasmic mitochondrial DNA mutations in normal and tumour cells. Nature. 2010;464(7288):610–614. doi: 10.1038/nature08802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flynn CM, Kaufman DS. Donor cell leukemia: insight into cancer stem cells and the stem cell niche. Blood. 2007;109(7):2688–2692. doi: 10.1182/blood-2006-07-021980. [DOI] [PubMed] [Google Scholar]
- 13.Janin A, et al. Donor-derived oral squamous cell carcinoma after allogeneic bone marrow transplantation. Blood. 2009;113(8):1834–1840. doi: 10.1182/blood-2008-07-171702. [DOI] [PubMed] [Google Scholar]
- 14.Beachy PA, Karhadkar SS, Berman DM. Tissue repair and stem cell renewal in carcinogenesis. Nature. 2004;432(7015):324–331. doi: 10.1038/nature03100. [DOI] [PubMed] [Google Scholar]
- 15.Brash DE, et al. UV-induced p53 mutations in squamous cell carcinoma. Proc Natl Acad Sci U S A. 1991;88(22):10124–10128. doi: 10.1073/pnas.88.22.10124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Agar NS, Halliday GM, Barnetson RS, Ananthaswamy HN, Wheeler M, Jones AM. The basal layer in human squamous tumors harbors more UVA than UVB fingerprint mutations. Proc Natl Acad Sci U S A. 2004;101(14):4954–4959. doi: 10.1073/pnas.0401141101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.MacKie RM, Reid R, Junor B. Fatal melanoma transferred in a donated kidney 16 years after melanoma surgery. N Engl J Med. 2003;348(6):567–568. doi: 10.1056/NEJM200302063480620. [DOI] [PubMed] [Google Scholar]
- 18.Malkin D. Li-fraumeni syndrome. Genes Cancer. 2011;2(4):475–484. doi: 10.1177/1947601911413466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Angelotti ML, et al. Characterization of renal progenitors committed toward tubular lineage and their regenerative potential in renal tubular injury. Stem Cells. 2012;30(8):1714–1725. doi: 10.1002/stem.1130. [DOI] [PubMed] [Google Scholar]
- 20.lznerowicz A, et al. Duration of brain death and cold ischemia time increases expression of genes associated with apoptosis in transplanted kidneys from deceased donors. Transplant Proc. 2011;43(8):2887–2890. doi: 10.1016/j.transproceed.2011.08.013. [DOI] [PubMed] [Google Scholar]
- 21.Su F, et al. RAS mutations in cutaneous squamous-cell carcinomas in patients treated with BRAF inhibitors. N Engl J Med. 2012;366(3):207–215. doi: 10.1056/NEJMoa1105358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O’Donovan P, et al. Azathioprine and UVA light generate mutagenic oxidative DNA damage. Science. 2005;309(5742):1871–1874. doi: 10.1126/science.1114233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gidlof O, Burvall S, Edvinsson L, Montelius M, Allen M, Molin M. Complete discrimination of six individuals based on high-resolution melting of hypervariable regions I and II of the mitochondrial genome. Biotechniques. 2009;47(2):671–672. doi: 10.2144/000113197. [DOI] [PubMed] [Google Scholar]
- 24.Bastien R, et al. High-throughput amplicon scanning of the TP53 gene in breast cancer using high-resolution fluorescent melting curve analyses. Hum Mutat. 2008;29(5):757–764. doi: 10.1002/humu.20726. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.