Abstract
Mice deficient in Schnurri-3 (SHN3; also known as HIVEP3) display increased bone formation, but harnessing this observation for therapeutic benefit requires an improved understanding of how SHN3 functions in osteoblasts. Here we identified SHN3 as a dampener of ERK activity that functions in part downstream of WNT signaling in osteoblasts. A D-domain motif within SHN3 mediated the interaction with and inhibition of ERK activity and osteoblast differentiation, and knockin of a mutation in Shn3 that abolishes this interaction resulted in aberrant ERK activation and consequent osteoblast hyperactivity in vivo. Additionally, in vivo genetic interaction studies demonstrated that crossing to Lrp5–/– mice partially rescued the osteosclerotic phenotype of Shn3–/– mice; mechanistically, this corresponded to the ability of SHN3 to inhibit ERK-mediated suppression of GSK3β. Inducible knockdown of Shn3 in adult mice resulted in a high–bone mass phenotype, providing evidence that transient blockade of these pathways in adults holds promise as a therapy for osteoporosis.
Introduction
Adult bone mass reflects the balance between production of bone by osteoblasts and resorption of bone by osteoclasts, and disturbance of this balance results in bone pathology such as osteoporosis. As osteoblasts have a limited ability to directly perceive the thickness of the bone they overlay, they rely on the ability to integrate extracellular cues to appropriately adjust the rate of bone formation. One such extracellular cue, the WNT/β-catenin pathway, is well established as a positive regulator of osteoblast differentiation, appearing to divert early progenitors away from a chondrocyte fate into becoming osteoblasts (1, 2). Deletion or inhibition of individual WNT signaling components results in dysregulation of osteoblast activity and differentiation and, by extension, altered anabolic bone formation in mice (3). Under basal conditions, cytosolic β-catenin undergoes constitutive ubiquitination and proteasome-dependent degradation via the destruction complex that consists of adenomatous polyposis coli (APS), Axin, CK1α, and glycogen synthase kinase 3-β (GSK3β) (4). A subset of ligands of the WNT family binds to the 7-pass transmembrane receptor Frizzled and the single-pass low-density lipoprotein receptor–related protein 5 (LRP5) or LRP6. This inhibits the constitutive phosphorylation of β-catenin by GSK3β, which in turn prevents β-catenin ubiquitination and proteasome-dependent degradation. Subsequently, β-catenin translocates into the nucleus to activate its transcriptional program (5). Notably, humans or mice deficient for a secreted inhibitor of WNT signaling, sclerostin (SOST), have substantially increased rates of bone formation, demonstrating that regulation of WNT signaling is central to the control of bone mass (4). Thus, understanding novel pathways involved in the regulation of WNT/β-catenin signaling is an area of major interest in skeletal biology.
MAPK pathways are essential for skeletal development and bone mass maintenance, mediating the response to a wide range of extracellular ligands relevant to osteoblast activity, such as BMPs, WNTs, PTH, TNF, insulin, IGF, and FGFs (6–8). In particular, ERK MAPK phosphorylates a range of substrates critical to osteoblast differentiation, including the transcription factor RUNX2 and the kinase RSK2, which in turn phosphorylates ATF4, a transcriptional regulator of late-stage osteoblast synthetic functions (7, 9). Accordingly, a germline deletion of Erk1 and a conditional deletion of Erk2 in limb mesenchyme (Erk1–/–Erk2Prx1 mice) results in substantially reduced bone mineralization (10). While these findings underscore the importance of ERK in osteoblasts, ultimately, the contexts in which ERK is activated and how ERK activity is regulated in osteoblasts remain unclear.
The Schnurri family of large zinc finger proteins consists of 3 mammalian homologs, Schnurri-1 (SHN1), SHN2, and SHN3 (also known as HIVEP3). Recent studies using genetic approaches have shown that Schnurri proteins are critical regulators of postnatal bone mass. Shn2–/– mice display a mildly reduced bone turnover due to reduced activity of both osteoblasts and osteoclasts (11, 12). In contrast, Shn3–/– mice exhibit a profound osteosclerotic bone phenotype due to augmented osteoblast activity (13–15). This phenotype was exacerbated in mice lacking both Shn2 and Shn3, which suggests that SHN2 and SHN3 cooperate to regulate bone mass (16). Despite these genetic studies, the molecular mechanism through which Schnurri proteins control osteoblast activity remains to be fully elucidated.
Here we address the mechanism by which SHN3 regulates bone mass. Our findings showed that SHN3 controlled osteoblast function via regulating WNT/LRP5-induced ERK activation. SHN3 dampened ERK kinase activity, thereby preventing excessive activation of downstream substrates in response to WNT stimulation. These pathways operated in vivo, as evidenced by recapitulation of the Shn3–/– high–bone mass phenotype in mice bearing a Shn3 allele with a mutation of the ERK interaction domain, and by observation of a genetic interaction between null alleles of Shn3 and the WNT coreceptor Lrp5. Finally, inducible knockdown of Shn3 in adult mice increased bone mass, which suggests that compounds blocking SHN3 expression or activity would be attractive therapeutic agents for the treatment of osteoporosis.
Results
Identification of a functional motif in SHN3.
We previously demonstrated that mice lacking SHN3 display an osteosclerotic phenotype with profoundly increased lamellar bone mass due to augmented osteoblast activity. In vitro analysis revealed that SHN3 controls protein levels of RUNX2, the master regulator of osteoblast differentiation (17), by promoting its proteasome-dependent degradation via WWP1 E3 ubiqutin ligase–mediated ubiquitination (13). To investigate the relevance of this mechanism in vivo, Wwp1–/– mice were examined and found to have no obvious alteration in bone mass (J.-H. Shim, unpublished observations). Similarly, the osteosclerotic bone phenotype in Shn3+/– and Shn3–/– mice could not be reversed by haploinsufficency of Runx2, and the clavicular hypoplasia observed in Runx2+/– mice was not reversed by SHN3 deficiency (Supplemental Figure 1; supplemental material available online with this article; doi: 10.1172/JCI69443DS1). This unexpected finding may be explained by redundancy between WWP1 and another HECT domain E3 ligase or by the existence of additional substrates of a WWP1/SHN3 ubiquitin ligase complex beyond RUNX2. Additionally, it is possible that compensatory alterations in osteoclast activity may mask changes in bone formation in Runx2+/–Shn3–/– mice. Nevertheless, these results prompted us to perform a systematic structure-function study of the mechanism by which SHN3 regulates bone mass.
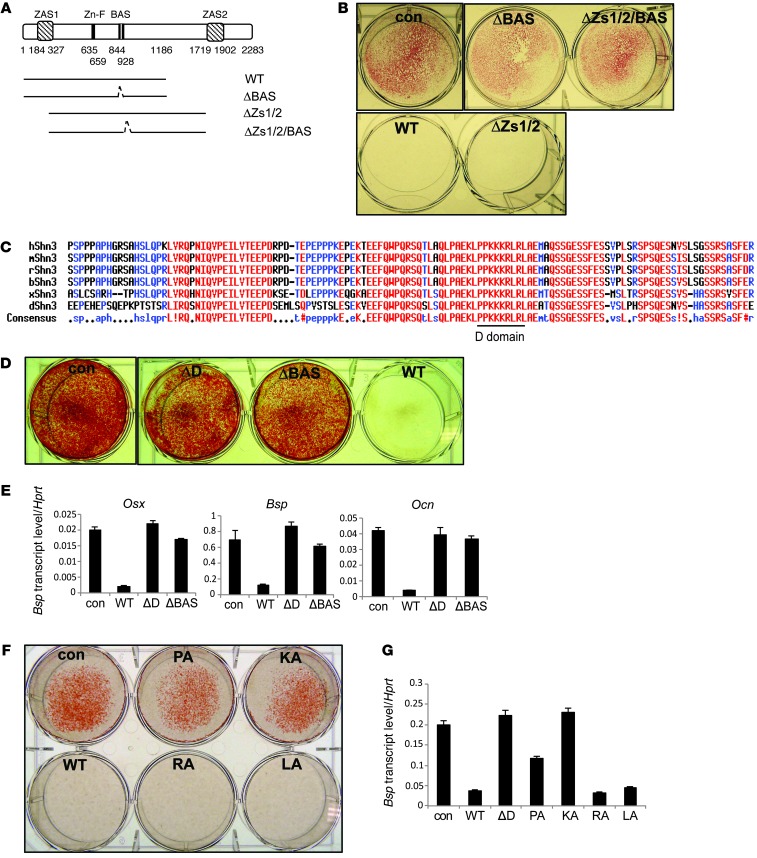
Mammalian Schnurri proteins are large (>260 KD) and contain few known structural motifs. Among these are 2 highly conserved zinc finger and acidic/serine-rich region (ZAS) domains and a single basic aa stretch and acidic/serine-rich region (BAS) domain (Figure 1A and ref. 18). To identify which of these contribute to SHN3 function, mutant SHN3 expression constructs with various deletions (Figure 1A) were assessed for their ability to repress osteoblast differentiation. Due to the large size of SHN3 and the limitations of the lentiviral expression systems used, a fragment of SHN3 consisting of the first 1,186 aas of SHN3 (referred to herein as SHN3-WT) was studied, and its functional competence was further confirmed by luciferase assay with various reporter genes whose regulation by SHN3 has been previously validated (Supplemental Figure 2). Lentivirus-mediated expression of SHN3-WT blocked the differentiation of human mesenchymal stem cells (hMSCs) into osteoblasts, as measured by the reduced formation of mineralized matrix and reduced expression of osteoblast marker genes (Figure 1B and Supplemental Figure 3, B and C). SHN3-WT was also able to inhibit osteoblast differentiation in primary murine calvarial osteoblast precursors (COBs) isolated from Shn3–/– neonates (Supplemental Figure 3, D and E), which indicates that this SHN3 construct is functionally intact. Consistent with the phenotype of Shn3–/– COBs, expression of SHN3-WT had only a modest effect on ALP activity (Supplemental Figure 3A). Deletion of the central BAS domain, but not the other domains assayed, rendered SHN3 incapable of repressing osteoblast differentiation (Figure 1B and Supplemental Figure 3, B–E). The BAS domain was required for neither interaction of SHN3 with WWP1 and RUNX2 nor SHN3-mediated RUNX2 ubiquitination via the ubiquitin E3 ligase WWP1 (Supplemental Figure 4). These data demonstrated that the BAS domain of SHN3 is crucial for its inhibition of osteoblast functions, and that the mechanism by which the BAS domain acts is distinct from those previously described.
Figure 1. Screening for functional motifs in SHN3.
All aa numbering is relative to the reference sequence NP_001121186.1. (A) Truncated SHN3 mutants (1–2,283 aa). WT, 1–1,186 aa; ΔBAS, Δ844–928 aa in 1–1,186 aa; ΔZs1/2 (ZAS1 and ZAS2 deletion), 327–1,719 aa; ΔZs1/2/BAS (ZAS1, ZAS2, and BAS deletion), Δ844–928 in 327–1,719 aa. (B, D, and F) hMSCs were infected with control lentivirus or lentiviruses encoding the indicated SHN3 constructs and cultured in differentiation medium for 21 days. Mineralization activity was analyzed by alizarin red staining. ΔD, D-domain deletion. (C) The aa sequence of the critical region of the BAS domain (p.810–933 aa), containing the D-domain (p.902–910 aa). (E and G) RNA levels of Osx, Bsp, and Ocn, analyzed by RT-PCR, in hMSCs infected by lentiviruses expressing the indicated SHN3 constructs or a control. Values in G are normalized to Hprt. Results are presented as mean + SD.
To identify the specific region within the BAS domain essential for SHN3 function, a series of shorter truncation mutants of SHN3-WT were generated (Supplemental Figure 5A). A 1–900 aa fragment of SHN3, but not a shorter 1–850 aa fragment, was able to block osteoblast differentiation, identifying a critical 50-aa stretch within the BAS domain. To understand the function of these 50 aas, ELM protein motif analysis (19, 20) was conducted and was notable for the presence of a D-domain (PPKKKRLRL), a potential MAPK docking site (Figure 1, A and C, and refs. 21–23) Deletion of this D-domain abolished the ability of SHN3 to repress osteoblast differentiation (Figure 1, D and E). The function of the D-domain was further assessed using alanine mutagenesis (with all aa numbering relative to the reference sequence NP_001121186.1): 2 prolines were substituted with alanines (p.[P884A;P885A]; referred to herein as SHN3-PA), as were 3 lysines (p.[K886A;K887A;K888A]; SHN3-KA), 2 arginines (p.[R889A;R891A]; SHN3-RA), and 2 leucines (p.[L890A;L892A]; SHN3-LA). Mineralization and osteoblast gene expression were significantly decreased by SHN3-WT, SHN3-RA, and SHN3-LA, but not by SHN3-KA or SHN3-PA (Figure 1, F and G, and Supplemental Figure 5, B–D). Thus, the D-domain of SHN3 was identified as being essential for the in vitro function of SHN3 in osteoblasts.
The D-domain is critical for SHN3 function in bone in vivo.
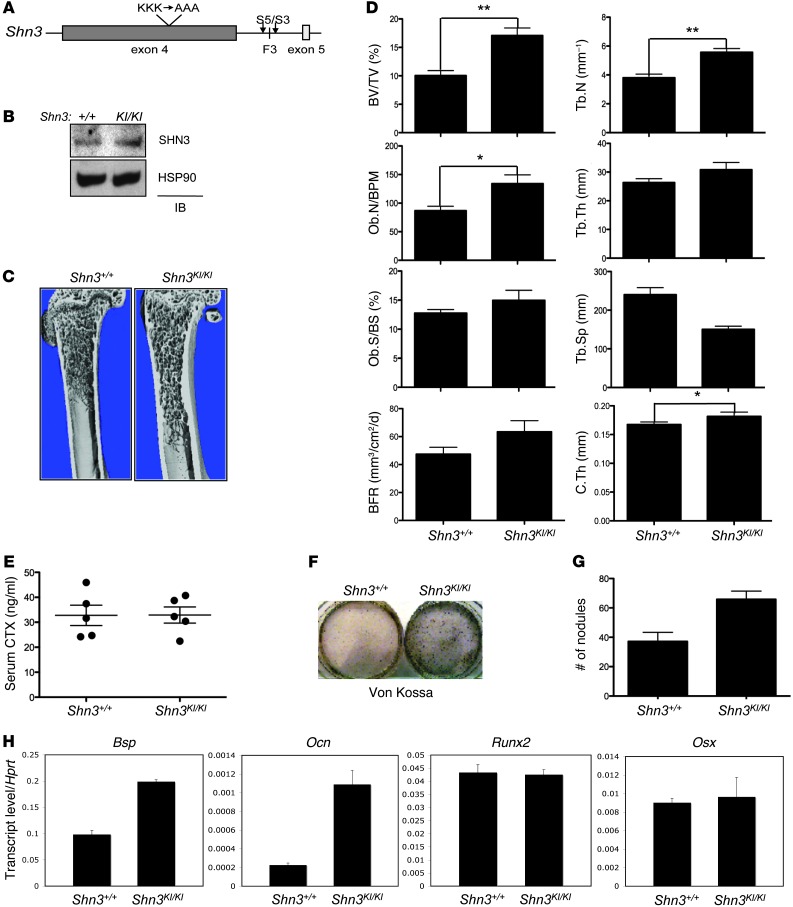
As proline 884 and 885 are likely to serve primarily to position lysines 886, 887, and 888 in space, SHN3-KA (the mutant targeting lysines 886, 887, and 888) was selected for further study. To determine whether these 3 lysine residues are critical for the function of SHN3 in vivo, we generated mice bearing a knockin allele of SHN3-KA in the endogenous Shn3 locus (referred to herein as Shn3KI/KI mice; Figure 2A). Shn3KI/KI mice were born at expected Mendelian ratios and did not display any gross developmental abnormalities. Analysis of WT and Shn3KI/KI mice revealed comparable Shn3 transcript and protein levels (Figure 2B and Supplemental Figure 6A). Remarkably, μCT and histologic analysis of 8-week-old male and female Shn3KI/KI mice revealed an increase in femoral bone mass compared with WT littermate controls (Figure 2, C and D, and Supplemental Figure 6, B and C). Shn3KI/KI mice also exhibited a progressive increase in bone mass (Figure 2E), recapitulating the key feature of the Shn3–/– phenotype (21). Additionally, no apparent change in osteoclast number was observed (Supplemental Figure 6B and J.-H. Shim, unpublished observations). Previous studies established that SHN3 functions intrinsically in osteoblasts to regulate bone mass (13). To directly examine whether Shn3KI/KI osteoblasts also exhibit augmented activity, bone marrow stromal cells (BMSCs) were isolated from Shn3KI/KI mice and littermate controls and cultured under osteoblast differentiation conditions. Consistent with the phenotype of Shn3–/– osteoblasts, Shn3KI/KI osteoblasts exhibited increased mineralization and expression of Bsp and Ocn (Figure 2, F–H, and ref. 13), which suggests that the 3 lysine residues in the D-domain are critical for the ability of SHN3 to regulate osteoblast-mediated bone formation both in vitro and in vivo. Additionally, RUNX2 protein levels were not altered in Shn3K/KI osteoblasts (Supplemental Figure 6D), which indicates that the 3-lysine motif does not function to regulate the posttranslational stability of RUNX2.
Figure 2. In vivo function of the 3-lysine motif of SHN3 in bone formation.
(A) The 3 lysine-to-alanine alleles knocked in to the endogenous Shn3 locus. Arrowheads denote position of primers (S5 and S3) used for PCR genotyping. (B) Primary Shn3+/+ and Shn3KI/KI BMSCs were lysed and immunoblotted with anti-SHN3 antibody. (C) μCT of 8-week-old Shn3+/+ and Shn3KI/KI mouse proximal femurs. (D) Histomorphometric and μCT analysis of 8-week-old Shn3+/+ and Shn3KI/KI mouse tibias. BV/TV, bone volume fraction; Ob.N/BPM, osteoblast number per bone perimeter; Ob.S/BS, osteoblast surface relative to bone surface; BFR, bone formation rate; Tb.N, trabecular number; Tb.Th, trabecular thickness; Tb.Sp, trabecular space; C.Th, cortical thickness. (E) Serum CTX levels in 8-week-old Shn3+/+ and Shn3KI/KI mice were determined by ELISA. (F–H) Shn3+/+ and Shn3KI/KI BMSCs were cultured in differentiation medium, and mineralization was analyzed by Von Kossa staining (F) and quantization of mineralized nodules (G). Alternatively, total RNA was extracted for RT-PCR analysis (H), and values normalized to Hprt. Results are presented as mean + SD. *P < 0.05, **P < 0.005, Student’s t test.
The D-domain of SHN3 regulates ERK activity.
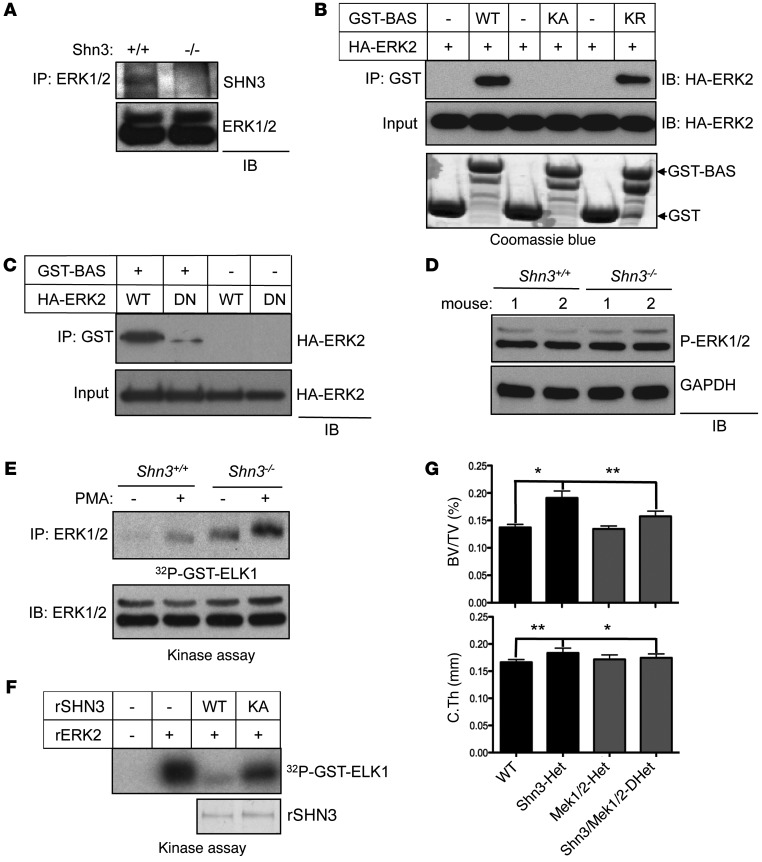
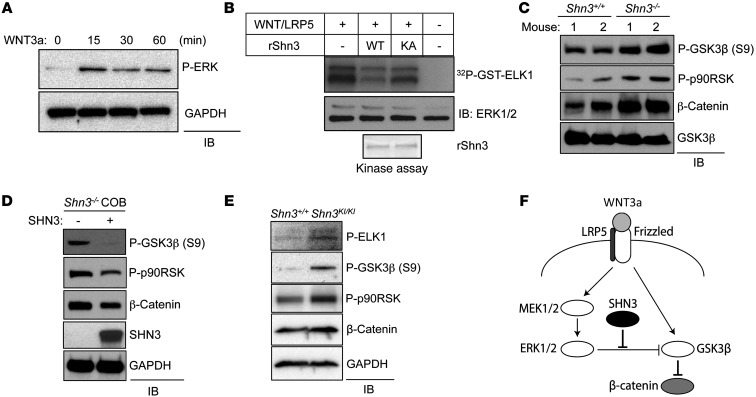
Given that the D-domain is known to serve as a MAPK docking site (22), we tested whether the D-domain of SHN3 serves as a point of interaction with MAPKs. Intriguingly, SHN3 bound selectively to ERK MAPK, but not other MAPKs, including JNK or p38 MAPK (Supplemental Figure 7A). Furthermore, endogenous SHN3 interacted with ERK in osteoblasts (Figure 3A), and this interaction occurred through the D-domain of SHN3 (Supplemental Figure 7B). To determine whether these residues are important for the interaction of SHN3 with ERK, the interaction between a recombinant GST-BAS fragment of SHN3 containing the D-domain and a purified HA-ERK2 protein was assessed in a cell-free system. WT-BAS was able to interact with HA-ERK2, but a BAS mutant in which the 3 lysines of the D-domain were mutated to alanine (referred to herein as the KA-BAS mutant) was not (Figure 3B). Additionally, the effect of a lysine-to-arginine substitution in the BAS domain of SHN3 (p.[K886R/K887R/K888R] in the BAS domain; KR-BAS) was assessed; this biochemically conservative substitution prevented ubiquitination of the altered residues, but generally did not perturb protein-protein interactions (Figure 3B and Supplemental Figure 7, C and D). Unlike KA-BAS, KR-BAS was able to bind to ERK2 and inhibit osteoblast differentiation, consistent with the model that these 3 lysines function by regulating the interaction of SHN3 with ERK2. The common docking domain (CD-domain) of MAPKs mediates the interactions with the D-domain of MAPK substrates (22, 24, 25). To examine whether the D-domain of SHN3 interacts with the CD-domain of ERK2, in vitro interaction analysis using an ERK2 containing a CD-domain mutation (p.[D319N]) was performed (Figure 3C and refs. 22, 26). GST-BAS interacted strongly with WT-ERK2, but not with D319N-ERK2. Likewise, interaction of GST-BAS with WT-ERK2 decreased in a dose-dependent manner by addition of an ERK peptide inhibitor (ERK PI; Supplemental Figure 7E) that was previously demonstrated to interfere with binding of the ERK CD-domain to D-domain–containing proteins (27). Taken together, these results demonstrate that the D-domain within the BAS domain of SHN3 binds specifically to the CD-domain of ERK2.
Figure 3. SHN3 inhibits ERK MAPK activity in osteoblasts via the D-domain 3 lysine motif.
(A) Immortalized Shn3+/+ and Shn3–/– COBs were lysed, immunoprecipitation was performed with anti-ERK1/2 antibody and protein A–agarose, and the resulting complex was immunoblotted with an anti-SHN3 antibody. (B and C) HA-ERK2 (WT or DN [D319N]) was incubated with recombinant GST or with GST-tagged BAS, KA-BAS, or KR-BAS. GST-containing proteins were immunoprecipitated with glutathione-agarose, and the resulting complex was immunoblotted with anti-HA antibody. Input indicates loading controls of HA-ERK2, GST, or GST-BAS protein. (D) Primary COBs were isolated from Shn3+/+ and Shn3–/– mice, lysed, and immunoblotted with anti–phospho-ERK1/2 antibody. GAPDH was used as a cytosolic protein control. (E) Immortalized Shn3+/+ and Shn3–/– COBs were stimulated with PMA for 30 minutes, and then ERK1/2 was immunoprecipitated. The immunoprecipitates were mixed with recombinant ELK1, and ERK kinase activity was analyzed by in vitro kinase assay. (F) Recombinant ERK2 (rERK2) was incubated with or without the indicated recombinant His-SHN3 (rSHN3) along with GST-ELK1, and ERK kinase activity was analyzed by in vitro kinase assay. Coomassie blue staining indicates loading control of recombinant His-SHN3. (G) μCT analysis of 5-week-old WT (Shn3fl/+), Shn3-Het, Mek1/2-Het, and Shn3/Mek1/2-DHet mouse femurs. Bone volume fraction and cortical thickness are shown. Results are presented as mean + SD. *P < 0.05, **P < 0.005, Student’s t test.
To explore how the interaction with SHN3 affects ERK activity in osteoblasts, phosphorylation levels and kinase activity of ERK were examined in WT and Shn3–/– COBs (Figure 3, D and E). While phosphorylation levels of ERK itself were comparable between Shn3–/– and WT COBs, Shn3–/– COBs exhibited elevated phosphorylation of the ERK1/2-substrate ELK1, which was further increased upon stimulation with PMA. Likewise, in an in vitro kinase assay using purified recombinant proteins, His-SHN3-WT, but not His-SHN3-KA, ablated ERK2-induced phosphorylation of GST-ELK1 (Figure 3F), while JNK1-induced phosphorylation of GST-cJUN and p38α-induced phosphorylation of GST-ATF2 were little affected by His-SHN3-WT (Supplemental Figure 7F). Thus, SHN3 appears to directly dampen the ability of ERK to phosphorylate its substrates, while leaving the activation of ERK by its upstream MAP2Ks intact.
To extend the evidence that SHN3 regulates the ERK MAPK pathway in osteoblasts from in vitro to in vivo systems, the genetic interaction among Shn3 and Mek1 and Mek2, the MAP2Ks upstream of ERK, was examined by comparing the bone mass of littermate WT, Shn3fl/+Prx1-cre (referred to herein as Shn3-Het), Mek1fl/+;Mek2+/–Prx1-cre (Mek1/2-Het), and triple-heterozygous Shn3fl/+;Mek1fl/+;Mek2+/–Prx1-cre (Shn3/Mek1/2-DHet) mice (Figure 3G). As expected, Shn3-Het mice displayed increased bone mass in the trabecular and cortical compartments of the appendicular skeleton (14), and Mek1/2-Het mice displayed near-normal bone mass. However, Shn3/Mek1/2-DHet mice displayed a reversal of the high–bone mass phenotype imparted by Shn3 haploinsufficency. Thus, reduction in the activity of ERK imparted by haploinsufficiency for Mek1 and Mek2 can partially normalize the effects of Shn3 haploinsufficency. Likewise, inhibition of ERK activity by the MEK inhibitor U0216 significantly reduced the mineralization activity of Shn3–/– COBs (Supplemental Figure 7G), validating the model that SHN3 acts via suppressing ERK activity in vitro and in vivo.
Shn3 regulates the WNT signaling pathway in osteoblasts.
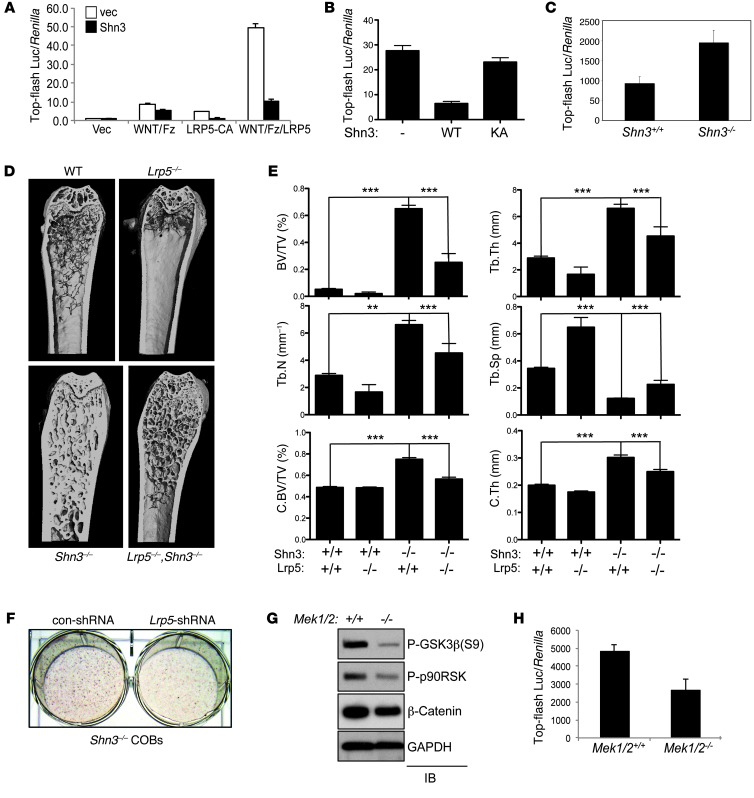
In order to determine which pathways function upstream of SHN3 in osteoblasts, C3H10T1/2 cells infected by lentivirus expressing either vector or SHN3 were stimulated with various ligands relevant to osteoblast function, including FGF18, BMP2, TGF-β, and a canonical WNT activator (xWNT8/Fz5 and/or LRP5) (Figure 4A, Supplemental Figure 8A, and ref. 28). Overexpression of SHN3-WT ablated β-catenin transcriptional activity in response to WNT pathway stimulation, as measured by top-flash luc in osteoblasts (Figure 4, A and B). This activity required the 3-lysine motif, as expression of SHN3-KA had little to no effect on top-flash luc activity. In contrast, gene induction by other stimuli was relatively unaffected by SHN3 expression (Supplemental Figure 8A). Likewise, β-catenin transcriptional activity was enhanced in Shn3–/– versus WT COBs (Figure 4C). β-catenin transcript levels were not altered by the absence of SHN3 (Supplemental Figure 8B, top), which suggests that SHN3 acts to repress the posttranslational activity of β-catenin. Thus, SHN3 is likely to function as a dampener of β-catenin activity downstream of WNT signaling in osteoblasts.
Figure 4. SHN3 inhibits the WNT-mediated β-catenin pathway via ERK regulation.
(A and B) C3H10T1/2 cells were infected by lentivirus expressing vector or SHN3-WT (A) or SHN3-KA (B) and transfected with top-flash luc and Renilla along with a xWNT8/Fz5 fusion protein, a constitutively active mutant of LRP5 (LRP5-CA), or xWNT8/Fz5 fusion protein plus LRP5. Relative luciferase activity normalized to Renilla is shown. (C) Shn3+/+ and Shn3–/– COBs were transfected with top-flash luc along with Renilla. Luciferase activity was analyzed 6 days after culture in differentiation medium. Values are normalized to Renilla. (D and E) μCT analysis of 8-week-old Shn3+/+, Lrp5–/–, Shn3–/–, and Lrp5–/–;Shn3–/– mouse femurs. (D) 3-dimensional reconstructions of proximal femur. (E) Bone volume fraction, trabecular number, trabecular thickness, cortical bone volume fraction (C.BV/TV), trabecular space, and cortical thickness. (F) Primary Shn3–/– COBs were infected by control or Lrp5 shRNA–expressing lentivirus and cultured in differentiation medium, and mineralization activity was analyzed by alizarin red staining. (G) Primary Mek1fl/fl;Mek2–/– COBs were infected by lentivirus expressing vector (Mek1/2+/+) or Cre recombinase (Mek1/2–/–), cultured in differentiation medium, lysed, and immunoblotted with the indicated antibodies. (H) Primary Mek1fl/fl;Mek2–/– COBs infected as in G were transfected with top-flash luc and Renilla. Luciferase activity was analyzed 6 days after culture in differentiation medium. Values are normalized to Renilla. Results are presented as mean + SD. **P < 0.005, ***P < 0.0005, Student’s t test.
To demonstrate that SHN3 regulates WNT signaling in vivo, the ability of a deletion of LRP5, a coreceptor of the WNT family of ligands, to reverse the Shn3–/– bone phenotype was assessed by comparing the bone mass of WT, Shn3–/–, Lrp5–/–, and Shn3–/–;Lrp5–/– mice (Figure 4, D and E). As expected, Lrp5–/– mice displayed low bone mass in both trabecular and cortical bones (29). Notably, Lrp5 deletion partially reversed the osteosclerotic bone phenotype exhibited by Shn3–/– mice. Mirroring these in vivo findings, shRNA-mediated knockdown of Lrp5 partially rescued the increased mineralization activity of Shn3–/– COBs in vitro (Figure 4F and Supplemental Figure 8, C and D). Thus, both in vivo and in vitro approaches suggested that SHN3 is a key regulator of WNT signaling in osteoblasts.
SHN3 controls GSK3β activity and β-catenin levels via ERK.
β-catenin is a positive regulator of osteoblast differentiation, and limb-specific deletion of β-catenin results in a complete loss of bony structure in mouse embryos (1, 2, 30). In the absence of WNT stimulation, β-catenin is constitutively phosphorylated by GSK3β, which results in β-catenin ubiquitination and proteasome-dependent degradation (4). This GSK3β-mediated suppression of β-catenin is relieved by ERK-induced phosphorylation of GSK3β T43. ERK may additionally contribute to GSK3β suppression by phosphorylating p90RSK, which in turn can promote GSK3β suppression via phosphorylation of S9 (31). To examine whether ERK activation is important for regulation of GSK3β activity in osteoblasts, phosphorylation levels of GSK3β (S9) and p90RSK were analyzed in COBs lacking MEK1 and MEK2 (referred to herein as Mek1/2–/– COBs). Phosphorylation levels of GSK3β (S9) and p90RSK were markedly decreased in Mek1/2–/– versus WT COBs (Figure 4G). This was accompanied by a decrease in β-catenin levels and transcriptional activity in Mek1/2–/– COBs (Figure 4, G and H). β-catenin transcript levels were not altered by deletion of Mek1 and Mek2 (Supplemental Figure 8B, bottom), demonstrating that ERK activation acts to promote the posttranslational activity of β-catenin in osteoblasts.
Given that SHN3 is able to suppress ERK activity and WNT-mediated β-catenin transcriptional activity, we examined the possibility that SHN3 suppresses ERK activity downstream of WNT signaling in osteoblasts. Stimulation of primary COBs or C3H10T1/2 cells (MSC-like cell line) with WNT3a induced ERK phosphorylation (Figure 5A and Supplemental Figure 8E). Additionally, ERK1/2 immunocomplexes extracted from cells overexpressing a WNT8/Fz5 fusion protein and LRP5 were able to phosphorylate GST-ELK1, demonstrating that WNTs induce ERK activity. ERK-induced phosphorylation of GST-ELK1 was suppressed by His-SHN3-WT, and this suppression was reduced by His-SHN3-KA (Figure 5B), which indicates that the 3-lysine motif of the D-domain in SHN3 is responsible for suppressing ERK activity in the context of WNT signaling.
Figure 5. SHN3 regulates GSK3β activity and β-catenin level in osteoblasts.
(A) Primary WT COBs were stimulated with murine WNT3a (200 ng/ml) for the indicated time points after 12 hour serum starvation. Cells were lysed and immunoblotted with antibodies specific to phospho-ERK and GAPDH. (B) HEK293 cells were transfected with a xWNT8/Fz5 fusion protein along with LRP5, lysed, and immunoprecipitated by anti-ERK1/2 antibody and protein A–agarose. The immunoprecipitates were incubated with GST-ELK1 and either His-SHN3-WT or His-SHN3-KA, and ERK kinase activity was analyzed by in vitro kinase assay. Inputs indicate loading control of recombinant His-SHN3. (C and E) Primary Shn3+/+ and Shn3–/– (C) or Shn3KI/KI (E) COBs were cultured in differentiation medium, lysed, and immunoblotted with the indicated antibodies. (D) Primary Shn3–/– COBs were infected with lentivirus expressing vector or SHN3 and cultured in differentiation medium containing puromycin for 6 days. Cells were lysed and immunoblotted with the indicated antibodies. (F) SHN3 regulation of ERK activity in the context of WNT/β-catenin signaling.
Supporting these observations, basal phosphorylation levels of the ERK substrates GSK3β and p90RSK and overall levels of β-catenin were increased in mature Shn3–/– COBs (Figure 5C) and were relatively normalized by reconstitution of mature Shn3–/– COBs with exogenous expression of SHN3 (Figure 5D). Likewise, mature Shn3KI/KI COBs showed an increase in basal phosphorylation levels of the ERK substrates ELK1, GSK3β, and p90RSK and an increase in β-catenin levels (Figure 5E). As expected, SHN3-KA failed to repress GSK3β S9 phosphorylation (Supplemental Figure 8F). Intriguingly, this suppressive activity of SHN3 did not alter phosphorylation levels of other known ERK substrates, such as BIM and MNK1 (Supplemental Figure 8F), which suggests that SHN3-mediated suppression of ERK activity displays substrate specificity. In mature Shn3–/– COBs, LRP5 deficiency reduced the basal phosphorylation levels of MEK1/2 and ERK1/2 (Supplemental Figure 8G). Similarly, phosphorylation of ERK substrates and β-catenin protein levels were reduced. This suggests that WNT/LRP5 contributes to ERK activation in osteoblasts and that the increase in ERK activity seen in the absence of SHN3 is downstream of LRP5.
We conclude that engagement of Frizzled and LRP5/6 by WNTs activates the ERK MAPK pathway in osteoblasts, which phosphorylates and suppresses GSK3β activity, thereby promoting β-catenin activity (5). SHN3 represses ERK-induced phosphorylation of GSK3β via the 3 lysines within the D-domain of the BAS domain. Therefore, SHN3 functions as a dampener of WNT signaling and, by extension, osteoblast function.
Inducible knockdown of SHN3 enhances bone mass in adult mice.
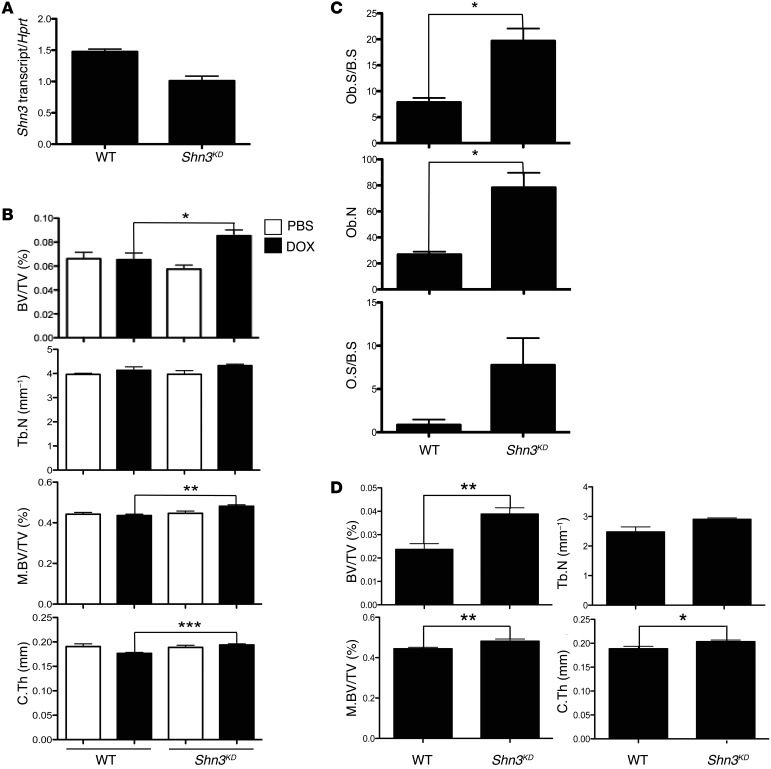
Haploinsufficiency of Shn3 results in a substantial increase of bone mass in adult mice (13). Similarly, Shn3 knockdown enhances osteoblast activity in vitro (13). To examine whether in vivo knockdown of SHN3 is able to increase bone mass in adult mice, we generated transgenic mice bearing a germline doxycycline-inducible (DOX-inducible) Shn3 shRNA construct (referred to herein as Shn3KD mice). 8-week-old Shn3KD and littermate control mice were fed chow containing PBS or DOX for 6 weeks, and bone RNAs were extracted from femurs to measure Shn3 knockdown efficiency. After 6 weeks of induction, Shn3KD mice displayed a 30% reduction in Shn3 mRNA levels in bone relative to DOX-treated WT mice (Figure 6A). Remarkably, this modest reduction of Shn3 transcripts was sufficient to enhance bone mass up to 25% in trabecular and cortical bones (Figure 6B). Likewise, histomorphometric analysis revealed that Shn3 knockdown increased osteoblast number and activity on the bone surface (Figure 6C). Therefore, even modest levels of reduction of SHN3 activity in adult mice resulted in enhanced bone formation. To examine the effect of Shn3 knockdown on aging-associated bone loss, the same experiment was performed, but mice were instead examined at 6 months of age, which revealed a greater than 50% increase in bone mass in Shn3KD mice (Figure 6D). These results indicate that compounds designed to block SHN3 expression or activity are attractive therapeutic agents for the treatment of osteoporosis.
Figure 6. Inducible knockdown of SHN3 increases bone mass in adult mice.
(A) 8-week-old WT and Shn3KD mice were fed DOX-containing chow for 6 weeks, and total RNA was extracted from long bones for RT-PCR analysis. Values are normalized to Hprt. (B) 8-week-old WT and Shn3KD mice were fed PBS- or DOX-containing chow for 6 weeks. Femurs from 14-week-old mice were analyzed by μCT. Shown are bone volume fraction, trabecular number, midshaft bone volume fraction (M.BV/TV), and cortical thickness. (C) 8-week-old WT and Shn3KD mice were fed DOX-containing chow for 6 weeks, and femurs from 14-week-old mice were analyzed by histomorphometric analysis. (D) 18-week-old WT and Shn3KD mice were fed DOX-containing chow for 6 weeks, and femurs from 24-week-old mice were analyzed by μCT. Results are presented as mean + SD. *P < 0.05, **P < 0.005, ***P < 0.0005, Student’s t test.
Discussion
Osteoblasts overlying a bone surface have no direct means to sense the thickness of the bone beneath them. Thus, they rely on the integration of extrinsic signals to modulate their activity in order to maintain consistent architecture throughout a given bone. In this respect, SHN3 appears to function as a rheostat that acts to interpret and transmit signals governing bone production. Here we demonstrated that SHN3 accomplishes this, at least in part, by dampening ERK activity in the context of WNT stimulation in osteoblasts (Figure 5F). SHN3 acts distal to the activation of ERK itself to prevent the phosphorylation of ERK substrates such as p90RSK and GSK3β. Notably, this dampening of ERK activity only occurs with respect to a subset of ERK substrates, which argues that SHN3 does not simply sequester ERK in an inactive complex, but rather acts to selectively “filter” ERK activity with respect to specific substrates. The physiologic importance of this mechanism was validated by the high bone mass of mice bearing a knockin of a Shn3 allele unable to inhibit ERK (Figure 2, C and D). Additionally, enforced reduction of ERK activity by the Mek1/2-Het genotype was able to reverse the high–bone mass phenotype imparted by Shn3 haploinsufficiency (Figure 3G). These findings provide strong evidence that SHN3 acts in vivo by suppressing ERK activity.
Genetic interaction and biochemical studies similarly suggest that SHN3 modulation of ERK occurs in the context of LRP5-dependent WNT signaling. Given that previous evidence demonstrates that SHN3 function is intrinsic to osteoblasts, these data are most consistent with a model of LRP5 acting directly in osteoblasts to modulate bone mass (29). Notably, crossing to other alleles, such as a null allele of Runx2 (Supplemental Figure 1) or null alleles of the regulator of bone formation Atf4 (Supplemental Figure 9), did not evoke the same degree of reversal of the Shn3–/– phenotype as did crossing to Lrp5-null alleles. However, these results must be interpreted with caution, as confounding results due to compensatory alterations in osteoclast activity cannot be excluded without performing histomorphometry.
It is notable that the osteosclerotic phenotype of Shn3–/– mice was not completely reversed by crossing to a Lrp5–/– background. This might be explained by functional redundancy of LRP6, the additional coreceptor of WNT ligands in skeletal system (32). Thus, double deletion of Lrp5 and Lrp6 is likely to be required for a complete reverse of the Shn3–/– phenotype. Alternatively, SHN3 may regulate other pathways in addition to WNT signaling, possibly including the growth factor–mediated signaling pathways — such as FGF2, FGF18, EGF, IGF1, BMP2, and TGF-β — that are able to activate ERK in osteoblasts. Likewise, the observation that the Shn3KI/KI phenotype is less severe than that of Shn3–/– mice suggests that SHN3 also works via other mechanisms in addition to those described here (Supplemental Figure 10). Similarly, the difference between the effects of the Shn3KI allele and those of either the germline-null allele or Prx1-mediated deletion of the Shn3fl allele indicate that there are additional domains outside of the 3-lysine motif that contribute to SHN3 function, for example those domains that interact with WWP1 or RUNX2 (Supplemental Figure 4A). The absence of a detectable cortical phenotype likely reflects the milder overall phenotype imparted by the Shn3KI allele compared with the Prx1-mediated deletion. The ability of SHN3 to regulate osteoblast/osteoclast crosstalk also appears to map outside of the BAS domain, based on the absence of basal alterations in osteoclasts in Shn3KI/KI mice. However, detailed testing of osteoclast function was not performed, and a stress model of osteoclast-induced bone loss may reveal defects not apparent at baseline.
SOST is an osteocyte-secreted antagonist of WNTs that suppresses bone formation by osteoblasts. Thus, SOST functions as a means for osteocytes to orchestrate the maintenance of a consistent bone thickness through the modulation of osteoblast activity (4). Our observation that SHN3 also regulated the interpretation of WNT signals suggests parallels between SHN3 and SOST function, in that both may impinge on the same pathway at different levels. This suggests that the profound osteosclerotic phenotype shared by SHN3- and SOST-deficient mice has a basis in their shared regulation of WNT signaling. However, at this time, there are no data supporting a specific mechanistic link between SHN3 and SOST. Interestingly, β-catenin appears to have diverse functions at various stages of osteoblast differentiation. At the level of very early osteoblasts or osteoblast progenitors, β-catenin serves to promote osteoblast differentiation at the expense of chondrocytic differentiation (1, 2). In mature osteoblasts, β-catenin acts to suppress the production of signals that promote osteoclast differentiation (33). Similarly, stabilization of β-catenin can have disparate effects on fracture healing, depending on the timing relative to injury (34). Given the complexity of the differentiation stage–specific functions of β-catenin in osteoblasts, it will be of special interest to further define the stage of osteoblast differentiation at which SHN3 functions.
In providing nonenzymatic regulation of the ERK pathway, SHN3 can be assigned to the category of regulatory MAPK scaffold proteins, including JNK-interacting proteins (JIPs). In this regard, the large adaptor protein SHN3 (2,283 aa) bound to many key regulators of osteoblast differentiation in a selective manner. Interestingly, these interactions did not require the D-domain in the BAS domain (Supplemental Figure 11). This class of scaffold proteins has been postulated to represent a mechanism by which nearly ubiquitous MAPK pathways can mediate tissue-specific functions, and the observation that the phenotype of Shn3–/– mice showed a high degree of specificity for the skeletal system suggests that SHN3 imprints osteoblast-specific functionality on the ERK pathway (35).
Finally, transient, inducible knockdown of Shn3 in adult mice resulted in a significant increase in bone mass. Building on previous studies of SHN3 in bone, the present observations offer further evidence that the suppressive effect of SHN3 on bone formation reflects a continual function of SHN3 in adult osteoblasts and is not a secondary consequence of an unappreciated developmental abnormality. The evidence that osteoblasts in adults were primarily populated from a short-lived population of progenitors implies that SHN3 acts either in this population or in its progeny, in light of observations that inducible deletion of Shn3 in adults produces high bone mass (36). This may indicate that ERK mediates a pathway for osteoblast differentiation parallel to the established RUNX2 pathway, although further investigation is needed to determine the range of substrates targeted by ERK and the stage of differentiation at which SHN3 acts. Moreover, these studies provide proof of principle that the SHN3 signaling axis is an attractive therapeutic target for the treatment of osteoporosis. Small molecules that reduce Shn3 expression or inhibit its activity should increase bone mass in the adult by increasing anabolic bone formation.
Methods
Cells, plasmids, and antibodies.
hMSCs were cultured and differentiated into osteoblasts as described by the manufacturer (Lonza). Primary COBs, C3H10T1/2 cells (mouse mesenchymal fibroblast–like cell line), immortalized COBs, and BMSCs were cultured in α-MEM medium (Cellgro) containing 10% FBS, 2 mM l-glutamine, 1% penicillin/streptomycin, 1% HEPES, and 1% nonessential aas and differentiated with ascorbic acid and β-glycerophosphate. Primary COBs were differentiated under osteoblast differentiation (OBD) conditions for 14 days and immortalized by transfecting with SV40 large T Ag using Amaxa nucleofector (Lonza). HEK293 cells and HEK293FT cells were purchased from ATCC and Invitrogen, respectively. Plasmids encoding HA-ERK2 (WT, D319N) and HA-GSK3β were gifts from J. Blenis (Harvard Medical School, Boston, Massachusetts, USA) and J. Woodgett (Samuel Lunenfeld Research Institute, Toronto, Ontario, Canada), respectively. Lentiviral plasmid expressing mouse Lrp5 shRNA hairpins were obtained from the Broad Institute. Anti–β-catenin, anti–phospho-BIM (S69), anti–phospho-MNK1 (T197/202), anti–phospho-GSK3β (S9), and anti–phospho-ERK1/2 were obtained from Cell Signaling. Anti–phospho-ERK, anti-HSP90, anti-HA (agarose conjugated), anti–c-Myc (HRP conjugated), and anti-HA (HRP conjugated) were obtained from Santa Cruz. Anti–phospho-p90RSK (S380, Calbiochem), anti-Flag (M2, Sigma-Aldrich), anti-ELK1 (Abcam), and anti-GAPDH (Affinity Bioreagents) were also used. An anti-SHN3 polyclonal antibody was generated by immunizing rabbits with recombinant GST-SHN3 corresponding to the C-terminal sequence of mouse SHN3 (2,011–2,083 aa).
Shn3 plasmid constructs.
Full-length murine Shn3 cDNA, or truncated or deletion mutants, were PCR amplified and cloned into pEF-Nuc mammalian expression vector (Invitrogen) or pHASE/PGK-PURO lentiviral vector. Constructs were as follows (all aa numbering relative to the reference sequence NP_001121186.1): N-terminal fragments (1–1,186 aa, 1–1,084 aa, 1–900 aa, 1–850 aa, and 1–817 aa); BAS deletion (Δ844–928 of 1–1,186 aa); ZAS1 and ZAS2 deletion (327–1,719 aa); ZAS1, ZAS2, and BAS deletion (Δ844–928 of 327–1,719 aa). BAS domain (844–928 aa) and His-SHN3 (50–936 aa) were PCR amplified and cloned into pGEX5x-1 (Clontech) and pFastBac vectors (Invitrogen), respectively.
Generation and breeding of Shn3-knockin mice.
To generate Shn3-knockin mice, DNA changes corresponding to SHN3-KA in exon 4 of the Shn3 reference sequence NP_001121186.1 were made via homologous recombination between a targeting vector and the Shn3 genomic locus (Figure 2A). These mutations were then validated by PCR-based DNA sequencing analysis. All mice analyzed (Shn3+/+, Shn3–/–, and Shn3KI/KI) were maintained on the C57BL/6 background.
Generation of inducible Shn3-knockdown mice.
The TetR/O system uses a reverse tet trans-repressor protein that responds to DOX. In the absence of DOX, TetR protein recognizes the sequence of a tet-inducible promoter (TetO) and blocks gene transcription. Upon addition of DOX, DOX-bound TetR protein is unable to bind to the TetO sequence, resulting in gene transcription. Using this inducible system, the shRNA of mouse Shn3 (5′-CCGGCCTGCTCTCAAGTAGTTTGTACTCGAGTACAAACTACTTGAGAGCAGGTTTTTG-3′) was cloned into the transgenic vector, placing the transgene downstream of the TetO sequence. For reproducible expression of mouse Shn3 shRNA in mice, the transgenic vector was placed into the Rosa26 locus and transmitted through the germline of chimeric mice. Treatment of these transgenic mice with DOX prevented binding of the TetR protein to the TetO sequence, resulting in transactivation of mouse Shn3 shRNA.
Skeletal preparations and histology.
For skeletal preparation, mice were skinned, eviscerated, and fixed in 95% ethanol. Then skeletons were stained by Alizarin Red S/Alcian Blue and sequentially cleared in 1% potassium hydroxide. For histological analyses, paraffin sections of bones were produced from 8-week-old mice. Limb tissues were dissected and fixed in 4% PFA in PBS. They were then decalcified by daily changes of 15% tetrasodium EDTA until soft and pliable. Tissues were dehydrated by passage through an ethanol series, cleared twice in xylene, embedded in paraffin, and sectioned. For morphological analyses, tissue sections were stained with hematoxylin and eosin.
μCT analysis.
For μCT analysis, a Scanco Medical μCT 35 system with an isotropic voxel size of 7 μm was used to image the distal femur. Scans were conducted in 70% ethanol and used an X-ray tube potential of 55 kVp, an X-ray intensity of 0.145 mA, and an integration time of 600 ms. For analysis of femoral bone mass, a region of trabecular bone 2.1 mm wide was contoured, starting 280 μm from the proximal end of the distal femoral growth plate. Femoral trabecular bone was thresholded at 211 per mille. Femoral cortical bone was thresholded at 350 per mille. A Gaussian noise filter optimized for murine bone was applied to reduce noise in the thresholded 2-dimensional image. 3-dimensional reconstructions were created by stacking the thresholded, 2-dimensional images from the contoured region.
Cell culture and osteoblast differentiation analysis.
Primary COBs were isolated from 5-day-old Shn3+/+ and Shn3–/– neonates by triple collagenase/dispase II. Alternatively, BMSCs were isolated from 2-month-old Shn3+/+ and Shn3KI/KI mice. Cells were cultured in differentiation medium containing ascorbic acid and β-glycerophosphate, and osteoblast differentiation analyses were performed at days 6, 14, and 21. For Von Kossa staining of extracellular matrix mineralization, cells were fixed with 10% neutral buffered formalin and stained with 2.5% silver nitrate (Sigma-Aldrich). To quantify mineralization activity, cells were fixed with cold 70% ethanol and stained with alzarin red (20 mg/ml), and alizarin red levels were measured by colorimetric analysis (Millipore).
Luciferase reporter assay.
C3H10T1/2 or HEK293 cells grown on 12-well plates were transiently transfected using Effectene (QIAGEN) with the RUNX2-responsive (6xOSE2-luc) or NF-κB–responsive (PBII-luc) reporter gene and the Renilla luciferase vector (Promega), together with plasmids expressing various Shn3 mutants in the absence or presence of RUNX2. HEK293 cells were stimulated with TNF (10 ng/ml) for 24 hours 1 day after transfection with PBII-luc, Renilla reporter genes, and constructs expressing various Shn3 mutants. Jurkat T cells were transiently transfected using Amaxa nucleofector (Lonza) with AP1-luc and the Renilla reporter genes, and cells were stimulated with PMA (20 ng/ml) and ionomycin (1 μM) for 24 hours 1 day after transfection with AP1-responsive reporter gene (2xAP1-luc) and the Renilla reporter genes along with various Shn3 mutants. Alternatively, C3H10T1/2 cells grown on 10-cm petri dishes were infected by lentivirus expressing either vector or SHN3, and after puromycin selection, cells were transfected using Effectene (QIAGEN) with the β-catenin–responsive reporter gene (Top-flash luc) and the Renilla luciferase vector (Promega) in the absence or presence of a WNT/Frizzed fusion protein (xWNT8/Fz5), a constitutively active mutant of LRP5 (LRP5-CA), or xWNT8/Fz5 plus LRP5. Total DNA concentration in each experiment was maintained by adding the appropriate control vector to the DNA mixture. 48 hours after transfection, cells were lysed, and luciferase activity was measured using the dual luciferase assay kit (Promega). Alternatively, primary COBs were cells were transfected with Top-flash luc and the Renilla reporter genes and cultured in differentiation medium for 6 days.
Immunoprecipitation and immunoblot analysis.
After transfection of the plasmids using Effectene (QIAGEN), HEK293 cells were lysed in TNT lysis buffer (10 mM Tris, 50 mM NaCl, 5 mM EDTA, 2 mM NaF, 30 mM sodium pyrophosphate, 100 mM Na3VO4, 0.5 mM PMSF, 1 μg/ml leupeptin, 5 μg/ml aprotinin, 1% Triton X-100). Cell lysates were incubated with Flag-, HA-, or Myc-conjugated agarose beads, fractionated by SDS-PAGE, and transferred to Immobilon-P membranes (Millipore). Protein levels were normalized by immunoblotting with anti-GAPDH or -HSP90 antibody. Membranes were blocked in TTBS buffer (100 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.1% Tween-20) with 4% skim milk and then incubated with the indicated antibodies. Finally, membranes were washed, incubated with HRP-conjugated secondary antibodies, and developed with ECL (Thermo Scientific). For endogenous interaction of SHN3 with ERK1/2, immortalized Shn3+/+ and Shn3–/– COBs were lysed and incubated with IgG control or anti-ERK1/2 antibody along with protein A–agarose beads for immunoprecipitation. Alternatively, immortalized WT COBs were stimulated with FGF2 (10 ng/ml) for 30 minutes, and nuclear proteins were fractionated and immunoprecipitated by either rabbit IgG or anti–phospho-ERK1/2 antibody and protein A–agarose.
Kinase assay.
Immortalized Shn3+/+ and Shn3–/– COBs grown on 10-cm Petri dishes were stimulated with PMA (20 ng/ml) for 30 minutes and then lysed in TNT lysis buffer (50 mm Tris, 150 mm NaCl, and 1% Triton X-100, pH 7.4). Cell lysates were incubated with anti-ERK1/2 antibody and protein A–agarose overnight at 4°C, and the immunoprecipitates were incubated for 30 minutes at 30°C in kinase buffer (20 mM HEPES, pH 7.5, 20 mM MgCl2, 1 mM EDTA, 2 mM NaF, 2 mM-glycerophosphate, 1 mM DTT, 10 μM ATP) containing appropriate GST-ELK1 (Cell Signaling) and 10 μCi [γ32P]ATP (PerkinElmer). The substrates were then precipitated using glutathione-agarose (Novagen) and resolved by SDS-PAGE, and phosphorylated proteins were visualized by autoradiography. Alternatively, HEK293 cells were transfected with a construct expressing a xWNT8/Fz5 fusion protein and LRP5 and lysed and immunoprecipitated with anti-ERK1/2 antibody and protein A–agarose. The immunoprecipitates were incubated with GST-ELK1 in the absence or presence of recombinant His-SHN3.
For in vitro kinase assay, 200 ng recombinant ERK2, HA-ERK2 protein eluted from HEK293 cells by anti-HA (agarose conjugated), or the indicated recombinant proteins were incubated for 30 minutes at 30°C in kinase buffer (20 mM HEPES, pH 7.5, 20 mM MgCl2, 1 mM EDTA, 2 mM NaF, 2 mM-glycerophosphate, 1 mM DTT, 10 μM ATP) containing either GST, GST-ELK1, GST-BAS (844–928 aa), or His-SHN3 (50–930 aa) and 10 μCi [γ32P]ATP (Perkin Elmer).
Statistics.
Statistical analysis was performed using 2-tailed, unpaired Student’s t test. A P value less than 0.05 was considered significant.
Study approval.
All animals were maintained in accordance with the NIH Guide for the Care and Use of Laboratory Animals and were handled according to protocols approved by the Weill Cornell Medical College subcommittee on animal care (IACUC).
Supplementary Material
Acknowledgments
We thank Henry Kronenberg and Antonios Aliprantis for helpful discussions and Michelle Schweitzer, Pamela Okerholm, Rebecca Drapp, Chantel Lester, Heather De Rivera, Christina Raekyung Lee, Yeon-Suk Yang, Alfred Zullo, and Kirsten Sigrist for technical support. We also thank the many individuals who provided valuable reagents. This work was supported by NIH grants HD055601 (to L.H. Glimcher) and K99AR055668 (to D.C. Jones). J.-H. Shim was supported by a fellowship from the Arthritis Foundation and by an NRSA postdoctoral fellowship (DFCI/NCI T32 CA070083).
Footnotes
Conflict of interest: Laurie H. Glimcher is on the Board of Directors and holds equity in Bristol Myers Squibb Pharmaceutical Company.
Citation for this article: J Clin Invest. 2013;123(9):4010–4022. doi:10.1172/JCI69443.
References
- 1.Hill TP, Spater D, Taketo MM, Birchmeier W, Hartmann C. Canonical Wnt/β-catenin signaling prevents osteoblasts from differentiating into chondrocytes. Dev Cell. 2005;8(5):727–738. doi: 10.1016/j.devcel.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Day TF, Guo X, Garrett-Beal L, Yang Y. Wnt/β-catenin signaling in mesenchymal progenitors controls osteoblast and chondrocyte differentiation during vertebrate skeletogenesis. Dev Cell. 2005;8(5):739–750. doi: 10.1016/j.devcel.2005.03.016. [DOI] [PubMed] [Google Scholar]
- 3.Kolpakova E, Olsen BR. Wnt/β-catenin — a canonical tale of cell-fate choice in the vertebrate skeleton. Dev Cell. 2005;8(5):626–627. doi: 10.1016/j.devcel.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 4.Clevers H, Nusse R. Wnt/β-catenin signaling and disease. Cell. 2012;149(6):1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 5. MacDonald BT, He X. Frizzled and LRP5/6 receptors for Wnt/β-catenin signaling. Cold Spring Harb Perspect Biol. 2012. 4 (12).pii: a007880 10.1101/cshperspect.a007880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greenblatt MB, et al. The p38 MAPK pathway is essential for skeletogenesis and bone homeostasis in mice. J Clin Invest. 2010;120(7):2457–2473. doi: 10.1172/JCI42285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zou W, et al. MLK3 regulates bone development downstream of the faciogenital dysplasia protein FGD1 in mice. J Clin Invest. 2011;121(11):4383–4392. doi: 10.1172/JCI59041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ge C, Xiao G, Jiang D, Franceschi RT. Critical role of the extracellular signal-regulated kinase-MAPK pathway in osteoblast differentiation and skeletal development. J Cell Biol. 2007;176(5):709–718. doi: 10.1083/jcb.200610046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang X, et al. ATF4 is a substrate of RSK2 and an essential regulator of osteoblast biology; implication for Coffin-Lowry Syndrome. Cell. 2004;117(3):387–398. doi: 10.1016/S0092-8674(04)00344-7. [DOI] [PubMed] [Google Scholar]
- 10.Matsushita T, Chan YY, Kawanami A, Balmes G, Landreth GE, Murakami S. Extracellular signal-regulated kinase 1 (ERK1) and ERK2 play essential roles in osteoblast differentiation and in supporting osteoclastogenesis. Mol Cell Biol. 2009;29(21):5843–5857. doi: 10.1128/MCB.01549-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nagao M, et al. Schnurri-2 deficiency counteracts against bone loss induced by ovariectomy. J Cell Physiol. 2011;226(3):573–578. doi: 10.1002/jcp.22521. [DOI] [PubMed] [Google Scholar]
- 12.Saita Y, et al. Lack of Schnurri-2 expression associates with reduced bone remodeling and osteopenia. J Biol Chem. 2007;282(17):12907–12915. doi: 10.1074/jbc.M611203200. [DOI] [PubMed] [Google Scholar]
- 13.Jones DC, Wein MN, Oukka M, Hofstaetter JG, Glimcher MJ, Glimcher LH. Regulation of adult bone mass by the zinc finger adapter protein Schnurri-3. Science. 2006;312(5777):1223–1227. doi: 10.1126/science.1126313. [DOI] [PubMed] [Google Scholar]
- 14.Wein MN, et al. Control of bone resorption in mice by Schnurri-3. Proc Natl Acad Sci U S A. 2012;109(21):8173–8178. doi: 10.1073/pnas.1205848109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones DC, Wein MN, Glimcher LH. Schnurri-3: a key regulator of postnatal skeletal remodeling. Adv Exp Med Biol. 2007;602:1–13. doi: 10.1007/978-0-387-72009-8_1. [DOI] [PubMed] [Google Scholar]
- 16.Jones DC, et al. Uncoupling of growth plate maturation and bone formation in mice lacking both Schnurri-2 and Schnurri-3. Proc Natl Acad Sci U S A. 2010;107(18):8254–8258. doi: 10.1073/pnas.1003727107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lou Y, et al. A Runx2 threshold for the cleidocranial dysplasia phenotype. Hum Mol Genet. 2009;18(3):556–568. doi: 10.1093/hmg/ddn383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu LC. ZAS: C2H2 zinc finger proteins involved in growth and development. Gene Expr. 2002;10(4):137–152. doi: 10.3727/000000002783992479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dinkel H, et al. ELM — the database of eukaryotic linear motifs. Nucleic Acids Res. 2012;40(Database issue):D242–D251. doi: 10.1093/nar/gkr1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weatheritt RJ, Jehl P, Dinkel H, Gibson TJ. iELM — a web server to explore short linear motif-mediated interactions. Nucleic Acids Res. 2012;40(Web Server issue):W364–W369. doi: 10.1093/nar/gks444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharrocks AD, Yang SH, Galanis A. Docking domains and substrate-specificity determination for MAP kinases. Trends Biochem Sci. 2000;25(9):448–453. doi: 10.1016/S0968-0004(00)01627-3. [DOI] [PubMed] [Google Scholar]
- 22.Tanoue T, Adachi M, Moriguchi T, Nishida E. A conserved docking motif in MAP kinases common to substrates, activators and regulators. Nat Cell Biol. 2000;2(2):110–116. doi: 10.1038/35000065. [DOI] [PubMed] [Google Scholar]
- 23.Barsyte-Lovejoy D, Galanis A, Sharrocks AD. Specificity determinants in MAPK signaling to transcription factors. J Biol Chem. 2002;277(12):9896–9903. doi: 10.1074/jbc.M108145200. [DOI] [PubMed] [Google Scholar]
- 24.Liu S, Sun JP, Zhou B, Zhang ZY. Structural basis of docking interactions between ERK2 and MAP kinase phosphatase 3. Proc Natl Acad Sci U S A. 2006;103(14):5326–5331. doi: 10.1073/pnas.0510506103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Akella R, Moon TM, Goldsmith EJ. Unique MAP Kinase binding sites. Biochim Biophys Acta. 2008;1784(1):48–55. doi: 10.1016/j.bbapap.2007.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bott CM, Thorneycroft SG, Marshall CJ. The sevenmaker gain-of-function mutation in p42 MAP kinase leads to enhanced signalling and reduced sensitivity to dual specificity phosphatase action. FEBS Lett. 1994;352(2):201–205. doi: 10.1016/0014-5793(94)00958-9. [DOI] [PubMed] [Google Scholar]
- 27.Kelemen BR, Hsiao K, Goueli SA. Selective in vivo inhibition of mitogen-activated protein kinase activation using cell-permeable peptides. J Biol Chem. 2002;277(10):8741–8748. doi: 10.1074/jbc.M108459200. [DOI] [PubMed] [Google Scholar]
- 28.Kratchmarova I, Blagoev B, Haack-Sorensen M, Kassem M, Mann M. Mechanism of divergent growth factor effects in mesenchymal stem cell differentiation. Science. 2005;308(5727):1472–1477. doi: 10.1126/science.1107627. [DOI] [PubMed] [Google Scholar]
- 29.Cui Y, et al. Lrp5 functions in bone to regulate bone mass. Nat Med. 2011;17(6):684–691. doi: 10.1038/nm.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hill TP, Taketo MM, Birchmeier W, Hartmann C. Multiple roles of mesenchymal beta-catenin during murine limb patterning. Development. 2006;133(7):1219–1229. doi: 10.1242/dev.02298. [DOI] [PubMed] [Google Scholar]
- 31.Ding Q, et al. Erk associates with and primes GSK-3β for its inactivation resulting in upregulation of β-catenin. Mol Cell. 2005;19(2):159–170. doi: 10.1016/j.molcel.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 32.Joeng KS, Schumacher CA, Zylstra-Diegel CR, Long F, Williams BO. Lrp5 and Lrp6 redundantly control skeletal development in the mouse embryo. Dev Biol. 2011;359(2):222–229. doi: 10.1016/j.ydbio.2011.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Glass DA, et al. Canonical Wnt signaling in differentiated osteoblasts controls osteoclast differentiation. Dev Cell. 2005;8(5):751–764. doi: 10.1016/j.devcel.2005.02.017. [DOI] [PubMed] [Google Scholar]
- 34.Chen Y, et al. β-Catenin signaling plays a disparate role in different phases of fracture repair: implications for therapy to improve bone healing. PLoS Med. 2007;4(7):e249. doi: 10.1371/journal.pmed.0040249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Whitmarsh AJ, Cavanagh J, Tournier C, Yasuda J, Davis RJ. A mammalian scaffold complex that selectively mediates MAP kinase activation. Science. 1998;281(5383):1671–1674. doi: 10.1126/science.281.5383.1671. [DOI] [PubMed] [Google Scholar]
- 36.Park D, et al. Endogenous bone marrow MSCs are dynamic, fate-restricted participants in bone maintenance and regeneration. Cell Stem Cell. 2012;10(3):259–272. doi: 10.1016/j.stem.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.