Abstract
Multidrug resistance in Gram-negative bacteria has become so threatening to human health that new antibacterial platforms are desperately needed to combat these deadly infections. The concept of siderophore conjugation, which facilitates compound uptake across the outer membrane by hijacking bacterial iron acquisition systems, has received significant attention in recent years. While standard in vitro MIC and resistance frequency methods demonstrate that these compounds are potent, broad-spectrum antibacterial agents whose activity should not be threatened by unacceptably high spontaneous resistance rates, recapitulation of these results in animal models can prove unreliable, partially because of the differences in iron availability in these different methods. Here, we describe the characterization of MB-1, a novel siderophore-conjugated monobactam that demonstrates excellent in vitro activity against Pseudomonas aeruginosa when tested using standard assay conditions. Unfortunately, the in vitro findings did not correlate with the in vivo results we obtained, as multiple strains were not effectively treated by MB-1 despite having low MICs. To address this, we also describe the development of new in vitro assays that were predictive of efficacy in mouse models, and we provide evidence that competition with native siderophores could contribute to the recalcitrance of some P. aeruginosa isolates in vivo.
INTRODUCTION
As the incidence of infections with multidrug-resistant (MDR) Gram-negative pathogens continues to grow, the list of therapeutic options available for effective treatment is dwindling. This, coupled with the fact that very few pharmaceutical companies remain invested in this disease area, paints a very dismal picture about what lies ahead for this significant, unmet medical need. While there are candidates currently in development that will hopefully address some populations of these resistant pathogens (1–4), each of them still lacks activity against certain organisms and/or resistance phenotypes. This deficiency in spectrum, coupled with historical evidence that new and spreading resistance mechanisms will inevitably become a widespread reality, compels those committed to antibacterial drug discovery to continue to pursue new antibiotic scaffolds. Standing in their way is the remarkable number of efficient antibiotic resistant determinants that the target pathogen population produces, each of which presents a different set of chemical and biological challenges that hinder the design of an effective antibiotic. Reduction of cell permeability through porin downregulation/mutation is a common and effective way that Gram-negative bacteria prevent certain antibiotics from gaining access to the cell to impart their antimicrobial activity (5–7). In response to this, scientists have designed new molecules that avoid this resistance mechanism by exploiting bacterial dependence on iron acquisition for survival (8, 9).
Gram-negative bacteria secrete iron-chelating molecules called siderophores, which are principally responsible for acquiring iron and bringing it into the cell through specific outer membrane receptors (10). The expression of siderophore biosynthesis and transport genes, which is directly regulated by the level of iron in the surrounding environment, requires significant upregulation in an infection, where concentrations of free iron are typically 10−9 M or lower (11). This process requires tight regulation, however, as the toxic threat of free oxygen radical generation via the Fenton reaction (12, 13) warrants a constant monitoring of intracellular iron levels and, when appropriate, the downregulation of siderophore-mediated iron uptake mechanisms. Recognizing the utility of this system as a potential means for circumventing decreases in outer membrane permeability, researchers have designed novel antimicrobial compounds that physically link siderophores to pharmacologically active molecules (9, 14), thereby promoting compound import into cells when iron acquisition functions are being expressed. To provide a broader spectrum of activity against Gram-negative organisms, as well as to facilitate the complex chemical synthesis required to assemble these conjugates, the siderophore moieties incorporated are often relatively simple small molecules which mimic catechol and related iron-binding groups found in a number of native siderophore molecules produced endogenously by these pathogens.
Pseudomonas aeruginosa exemplifies highly problematic MDR Gram-negative pathogens that contribute to the significantly high mortality rates seen in immunocompromised patients. While its frequency and persistence within the lungs of cystic fibrosis (CF) patients have been the focus of a substantial amount of research effort, its virulence has also been linked to other non-CF disease states or conditions such as burns (15), wound infections (16), and nosocomial pneumonia (17). The pathogenic versatility of P. aeruginosa can be attributed to the myriad of virulence factors that it is capable of producing, including multiple toxins (18, 19), degradative enzymes (20), a functional type III secretion system (21), and several different polysaccharides (22–24), each of which have been demonstrated to protect this pathogen in various stages of its life cycle. Also included in that list of important virulence factors is the extensive iron acquisition system encoded within the genome of P. aeruginosa strains. The overlapping, redundant roles of the many siderophores and siderophore receptors it can produce provide a comprehensive system for importing iron via both its own endogenous siderophores, pyoverdine and pyochelin, and stealing other organisms' siderophores through the production of the cognate outer membrane receptors. Examination of the genome of the P. aeruginosa prototype strain, PAO1, reveals that it is predicted to encode >25 different siderophore receptors, the majority of which are not specific for the native siderophores which it produces. Interestingly, many of these nonnative siderophore receptors are highly conserved with the primary siderophores of members of the Enterobacteriaceae, including other problematic Gram-negative pathogens such as Escherichia coli and Klebsiella pneumoniae. In many cases, it is through these receptors that siderophore-conjugated antibiotics gain access to the periplasmic space, thus making them attractive moieties for ensuring a broad spectrum of activity against Gram-negative organisms (8, 25).
We have previously described the in-depth in vitro characterization of MC-1, a novel siderophore-conjugated monocarbam (8) (Fig. 1), which included a thorough evaluation of the impact of clinically relevant resistance mechanisms on the activity of this novel compound. In addition, we determined the frequency of resistance (FOR) for MC-1 with P. aeruginosa PAO1 using standard methods and subsequently identified the primary route of entry of this compound into P. aeruginosa. In this report, we present both an in vitro and in vivo evaluation of MB-1, a siderophore-conjugated monobactam that has broad-spectrum activity against Gram-negative organisms and is unaffected by prevalent resistance mechanisms such as porin loss, efflux pumps, and β-lactamase-mediated hydrolysis. Through the course of our assessment of in vivo efficacy, however, we discovered an unexpected adaptation-type resistance to MB-1, one which went undetected through our use of standard, traditional antibiotic discovery methodologies. During the course of our investigation into the mechanism underlying this adaptation phenotype, we developed new methods that not only helped to define this phenomenon mechanistically but also generated data that were highly correlative with in vivo outcome across a panel of MDR P. aeruginosa clinical isolates.
Fig 1.
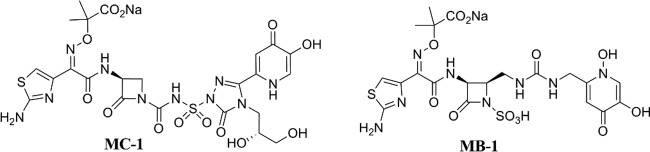
Chemical structures of siderophore conjugates MC-1 and MB-1.
MATERIALS AND METHODS
Bacterial strains, medium conditions, and reagents used.
The P. aeruginosa clinical isolates used in this study were selected from either the Pfizer or Hartford Hospital internal culture collection. All bacterial strains were routinely grown on Luria-Bertani (LB) broth and agar. The purified siderophores pyoverdine and enterobactin (catalog numbers P8124 and E3910, respectively) were purchased from Sigma. All antibiotics were purchased from USP. MIC assays were conducted according to CLSI guidelines (26, 27) in cation-adjusted Mueller-Hinton broth (MHB; Difco) unless otherwise indicated. Chelexed, dialyzed MHB (CDMHB), the iron-depleted version of MHB, was made as described previously (8), with the exception that 40 g of Chelex resin (Bio-Rad) was used instead of 20 g to ensure maximal iron removal. CDMHB was used for certain MIC experiments and for resistance frequency determinations. In the case of the latter, 1.5% UltraPure agarose (Life Technologies) was used as the solidifying agent instead of agar. Conditioned medium was prepared by growing overnight cultures of P. aeruginosa strains in CDMHB, removing the cells by centrifugation, and filter sterilizing the supernatant using a 0.22-μm filter.
Hollow-fiber assay conditions.
Bioreactors were primed for 72 h according to the manufacturer's instructions (Fibercell, Inc., Frederick, MD) using sterile phosphate-buffered saline (PBS). P. aeruginosa UC12120 was streaked onto a blood agar plate and grown overnight at 37°C. The resulting colonies were used to prepare a 0.5 McFarland standard, 100 μl of which was added to 100 ml of prewarmed MHB to achieve a starting inoculum of 2 × 105 CFU/ml. Ten microliters of this suspension was loaded into the extra capillary space of each hollow fiber cartridge, which was then incubated at 37°C for 2 h prior to initiating the flow of fresh MHB. MB-1 was suspended in 100% dimethyl sulfoxide (DMSO), and 500-mg doses (each infused over a 1-h period) were delivered at 0, 8, and 16 h after the initiation of MHB flow. Five-hundred-microliter samples were taken from the extra capillary space using a 1-ml syringe at the 0-, 2-, 4-, 6-, 8-, 10-, 12-, 24-, and 26-h time points. One hundred microliters of each sample was serially diluted in sterile PBS and plated on blood agar plates to determine CFU/ml, while an additional 100 μl was plated on MB-1-containing MHB plates to assess resistance development, and the remaining sample volume was used to determine MB-1 concentrations. Cartridges were run in duplicate in order to determine the reproducibility of MB-1 activity.
MB-1 in vivo efficacy time course.
All animal studies were conducted in compliance with approved animal use protocols by the Pfizer and/or Hartford Hospital institutional animal care and use committee. To evaluate the efficacy of MB-1 kinetically, female CF-1 mice were first rendered neutropenic by oral administration of 150 and 100 mg/kg (of body weight) of cyclophosphamide (Alfa Aesar, Ward Hill, MA) at 4 and 1 day prior to infection, respectively. Approximately 1.8 × 105 CFU (in 100 μl of PBS) of overnight-grown P. aeruginosa UC12120 was injected intramuscularly into the left femoral bicep of each mouse. Untreated mice (n = 3) were sacrificed by CO2 asphyxiation, thighs were aseptically removed, and the resulting starting bacterial burden was determined by tissue homogenization and subsequent dilution in PBS followed by plating on 5% sheep blood agar. Two hours postinfection, mice were dosed with 50 mg/kg of MB-1, and this dosing regimen continued every 3 h for 24 h. After administration of the first MB-1 dose, groups of 3 mice were sacrificed at 1, 2, 3, 6, 9, 12, 15, 18, 21, and 24 h to determine CFU values. To monitor resistance development over the course of the study, 100 μl of undiluted thigh homogenate was directly plated onto MHB plates containing 0.5, 1, and 2 μg/ml of MB-1, and plates were incubated overnight at 37°C before observation of colony formation.
Pharmacokinetic (PK) analysis and determination of in vivo dosing regimen.
Pathogen-free, female ICR mice weighing approximately 25 g were obtained from Harlan Sprague-Dawley, Inc. (Indianapolis, IN), and utilized throughout these experiments. Two hours after bacterial inoculation, a group of 48 mice were given either single or the first of multiple subcutaneous doses. At 8 time points ranging from 0.25 to 8 h after dosing, groups of 6 mice were euthanized by CO2 exposure, followed by cervical dislocation and blood collection via intracardiac puncture. Blood samples were collected in vials containing potassium EDTA, and plasma was separated after centrifugation. Plasma samples were stored at −80°C until concentrations of MB-1 were determined using a validated methodology. Briefly, 50-μl aliquots of samples and standards were subjected to protein precipitation with 200 μl of acetonitrile containing 100 ng/ml of aztreonam, which served as an internal standard. Samples were vortexed prior to transfer of 120 μl to a 96-well plate, after which they were dried under nitrogen and reconstituted with 240 μl of 5% acetonitrile containing 0.1% formic acid. Samples were analyzed using an API5500 mass spectrometer (Applied Biosystems, Foster City, CA) equipped with Turbo V sources and TurboIonSpray interface. PK parameters for single doses of MB-1 were calculated using first-order input and elimination, by nonlinear least-squares techniques (WinNonlin, version 5.0.1; Pharsight, Mountain View, CA). Compartment model selection and weighting schemes were based on visual inspection of the fit and use of the correlation between the observed and calculated concentrations. After PK characterization of single doses, these data were used to determine regimens in mice that simulated the free drug exposure profile seen in humans for MB-1. For these simulations, MB-1 protein binding values of 9% and 0% in mice and humans, respectively, were utilized. Confirmatory PK studies were undertaken with infected mice prior to the use of the regimen in the efficacy analyses. For these studies, infected neutropenic mice were dosed with the previously determined regimen and groups of 6 mice were euthanized at 8 time points throughout the dosing. To ensure equivalent exposures in neutropenic and immunocompetent animals, 3 groups of 6 immunocompetent animals were dosed with the same regimen as neutropenic animals, described above, and sampled at 3 of the 8 time points.
Thigh infection models with immunosuppressed and immunocompetent mice.
Female ICR mice were maintained and used in accordance with National Research Council recommendations and provided food and water ad libitum. Mice were rendered neutropenic with 100- and 150-mg/kg intraperitoneal injections of cyclophosphamide (Cytoxan; Bristol-Myers Squibb, Princeton, NJ) given 1 and 4 days prior to inoculation, respectively. Two hours prior to the initiation of antimicrobial therapy, each thigh was inoculated intramuscularly with a 0.1-ml solution containing approximately 107 CFU/ml of the test isolate. Immunocompetent mice underwent the same procedures as outlined above for neutropenic animals, except that cyclophosphamide was not given and an inoculum of 108 CFU/ml was used to produce thigh infection.
In vivo efficacy of MB-1 against P. aeruginosa using humanized PK regimen.
The efficacy of human simulated MB-1 against P. aeruginosa isolates was assessed in both the neutropenic- and immunocompetent-mouse thigh infection models. For these studies, groups of 3 mice were administered human simulated regimens of MB-1 beginning 2 h after inoculation (see Fig. S2 in the supplemental material). This required doses of 10, 13.25, 23.25, 23.25, 25, 20, 13.25, 13.25, and 6.75 mg/kg given at 0, 0.75, 1.5, 2.5, 3.25, 4, 4.75, 5.75, and 6.75 h after initiation of therapy, respectively. All doses were administered as 0.2-ml subcutaneous injections and consisted of three 8-h dosing intervals (i.e., 24 h total). To serve as control animals, an additional group of mice were administered normal saline at the same volume, route, and frequency as in the treatment regimen. Thighs from all animals were harvested 24 h after the initiation of therapy; mice that failed to survive for 24 h were harvested at the time of expiration. The harvesting procedure for all study mice began with euthanization by CO2 exposure, followed by cervical dislocation. After sacrifice, thighs were removed and individually homogenized in normal saline. Serial dilutions of the thigh homogenates were plated on Trypticase soy agar with 5% sheep blood using spiral plating techniques for CFU determination. In addition to the above-mentioned treatment and control groups, another group of 3 infected, untreated mice were harvested at the initiation of dosing and served as 0-h controls. Efficacy, defined as the change in bacterial density, was calculated as the change in log10CFU obtained for treated mice after 24 h from that of that starting densities observed in 0-h control animals.
To assess the role of pyoverdine in in vivo efficacy, MB-1 was evaluated against 4 parent P. aeruginosa strains (PAO1, JJ4-36, JJ11-54, and JJ8-16) and their isogenic pyoverdine knockouts (PAO1 ΔpvdA, JJ4-36 ΔpvdA, JJ11-54 ΔpvdA, and JJ8-16 ΔpvdA) in the neutropenic-mouse thigh infection model using the methodology described above. Comparative activities between parent and knockout strains were evaluated using a Student t test, with a P value of ≤0.05 identified as statistically significant (SigmaStat, version 2.03; SPSS Inc., Chicago, IL).
Construction of pyoverdine biosynthetic site-directed mutants in P. aeruginosa.
The E. coli strain SM10/pEX100T-pvdA::Gm, which includes a gentamicin resistance cassette in place of a significant internal portion of the pvdA coding region, was provided by Mike Vasil (University of Colorado at Denver). This construct was introduced into clinical isolates of P. aeruginosa via triparental mating using the helper plasmid pRK2013 (28), and the resulting exconjugates were screened for successful double-crossover recombination as described previously (8). The ability of clinical isolates, along with their corresponding pyoverdine-deficient mutants, to produce pyoverdine was tested by growing strains overnight in CDMHB, removing the cells by centrifugation, and using spectrophotometry to measure the absorbance at 405 nm.
Frequency of resistance assessments.
Resistance frequencies were determined using two different methods. First, standard FOR determinations were made using P. aeruginosa UC12120 as described previously (8), with 1 × 108 cells plated on each MB-1-containing plate. To assess the resistance emergence potential of P. aeruginosa strains using in vivo-relevant iron conditions, ∼5 × 106 mid-log-phase cells were plated onto CDMHB plates containing MB-1 at concentrations ranging from 0.25 to 32 μg/ml. Plates were incubated at 37°C for 40 h prior to determination of the highest MB-1 concentration which supported bacterial growth. For conditioned-medium supplementation FOR assays, 1% conditioned medium (described above) was added to the molten CDMHB agarose and allowed to solidify in the plate prior to spreading of bacteria on the plate surface.
Supplemental material.
The potential for β-lactamase-mediated MB-1 hydrolysis was assessed by standard MIC testing (see Table S1 in the supplemental material) using an isogenic panel of individual β-lactamases representing each of the 4 Ambler classes of these enzymes as described previously (8). The humanized PK dosing regimen utilized in neutropenic- and immunocompetent-mouse thigh models was determined through single-dose and multidose PK studies as described above, and the predicted versus observed MB-1 concentrations are shown in Fig. S2 in the supplemental material. The MICs for MB-1 and a number of clinically relevant antibiotics were measured against the MDR P. aeruginosa clinical isolates used in this study using standard methods (26, 27) (see Table S3 in the supplemental material).
RESULTS
MB-1 has activity against a broad spectrum of Gram-negative pathogens.
We began our in vitro characterization of MB-1 (Fig. 1) by performing MIC50 and MIC90 assays against a variety of Gram-negative pathogens. Table 1 shows that MB-1 demonstrates activity against several clinically relevant genera that is equivalent or superior to those of existing antibiotics representing the cephalosporin, monobactam, carbapenem, quinolone, and aminoglycoside classes. Of particular interest was the 4-fold improvement in P. aeruginosa MIC90 for MB-1 compared to meropenem. As mentioned previously, the activity of a siderophore-conjugated compound, unlike clinically utilized carbapenem antibiotics, does not require functional outer membrane porins on the cell surface of Gram-negative organisms. It was also encouraging to note the improved activity of MB-1 against E. coli, which included several CTX-M-producing isolates, as well as its superior coverage of Stenotrophomonas maltophilia relative to all comparator agents.
Table 1.
MIC50s and MIC90s for MB-1 and other drugs against clinically relevant Gram-negative pathogens
| Drug | MIC50/MIC90 (μg/ml)a |
|||||||
|---|---|---|---|---|---|---|---|---|
| P. aeruginosa (101) | K. pneumoniae (100) | E. coli (80) | Enterobacter spp. (27) | Citrobacter spp. (26) | Serratia marcescens (30) | Proteus spp. (26) | S. maltophilia (26) | |
| MB-1 | 0.25/1 | 0.06/4 | 0.12/1 | 1/8 | 0.25/4 | 0.25/1 | ≤0.03/0.12 | 0.06/0.25 |
| Cefepime | 2/16 | ≤0.06/32 | ≤0.06/>64 | 0.12/2 | ≤0.06/2 | 0.25/8 | ≤0.06/0.12 | 32/64 |
| Aztreonam | 4/>16 | ≤0.12/>16 | 0.25/>16 | 0.5/>16 | ≤0.12/>16 | 0.25/>16 | ≤0.12/≤0.12 | >16/>16 |
| Meropenem | 0.5/4 | ≤0.12/≤0.12 | ≤0.12/≤0.12 | ≤0.12/0.25 | ≤0.12/≤0.12 | ≤0.12/8 | ≤0.12/≤0.12 | >8/>8 |
| Ciprofloxacin | ≤0.5/4 | ≤0.5/4 | ≤0.5/>4 | ≤0.5/≤0.5 | ≤0.5/1 | ≤0.5/>4 | ≤0.5/4 | 4/>4 |
| Amikacin | 2/8 | 1/2 | 2/8 | 1/2 | 2/2 | 2/4 | 4/8 | >32/>32 |
Numbers of strains tested are in parentheses.
Resistance frequency assays identify a first-step resistance mechanism and lack of β-lactamase-mediated hydrolysis.
The MB-1 FOR was determined using standard methodology and plating on MHB agarose plates as described previously (8). In this study, P. aeruginosa UC12120 showed a frequency of 8.1 × 10−7 at 4× the MIC, with no recoverable colonies observed at 8× the MIC. Resistant mutants that were confirmed to be stable were shown to harbor mutations within the piuA coding region (data not shown), which was consistent with MC-1-resistant mutants identified and characterized previously (8). No changes to porin expression, efflux pump activity, or β-lactamase production were detected in any of these isolates, again consistent with data generated with MC-1. The discovery that the major first-step resistance mechanism appeared to be involved in siderophore receptor-mediated uptake across the outer membrane prompted us to evaluate MB-1 MICs against the P. aeruginosa PAO1 isogenic receptor mutant panel that we used previously to determine MC-1 entry mechanisms. The results of these MICs suggested that the same two outer membrane siderophore receptors (PiuA and PirA) are responsible for MB-1 uptake into P. aeruginosa and that high-level MB-1 resistance (64 μg/ml) can be achieved only by mutation of both of these receptors (M. F. Brown, unpublished observations).
Next, we probed for any β-lactamase-mediated hydrolysis liabilities that MB-1 may have by testing an extensive isogenic panel of individual β-lactamases, each of which was expressed in a naive E. coli background. Just as we observed with MC-1 previously (8), none of the β-lactamases tested demonstrated any significant cleavage activity, as evidenced by the lack of MIC shift seen when various enzymes were expressed in E. coli (see Table S1 in the supplemental material). Further supporting this conclusion was the lack of any decrease in MIC when 4 μg/ml of the β-lactamase inhibitor tazobactam was included with MB-1. Of particular significance was the inclusion of several metallo-β-lactamases (including NDM-1) and KPC variants, none of which demonstrated any hydrolytic activity against MB-1. This lack of activity was confirmed biochemically by measuring kinetic properties of MB-1 hydrolysis by these and other purified β-lactamases using isothermal titration calorimetry (data not shown).
MB-1 hollow-fiber and in vivo efficacy studies fail to demonstrate the anticipated bactericidal effect predicted by standard in vitro antibacterial models.
Given the encouraging data generated in MIC90, standard resistance frequency, mechanism of entry, and β-lactamase stability experiments, we next chose to evaluate the pharmacokinetic/pharmacodynamic (PK/PD) profile of MB-1 in an in vitro dynamic model using hollow-fiber technology. This flow cell-based system allows for drug administration that mimics actual human dosing regimens, thus enabling both the monitoring of compound effects on bacterial growth and, more importantly, the development of resistant subpopulations within the system over time at various drug concentrations. Based on its susceptible MIC (0.25 μg/ml) and limited propensity for spontaneous resistance development, we elected to use P. aeruginosa UC12120 and an MB-1 dosing regimen designed to mimic an efficacious human dosing schedule. Taking into account predicted human PK parameters such as protein binding, half-life, and clearance, we designed a strategy (500 mg dosed every 8 h) that would provide unbound MB-1 concentrations above the MIC for >90% of the duration of the experiment (24 h). Based on this PK/PD modeling, we anticipated that a >2-log reduction in bacterial burden would be observed at the conclusion of the study. As shown in Fig. 2A, both hollow-fiber cartridges showed a significant degree of bacterial killing after administration of the first MB-1 dose, although regrowth was detected prior to infusion of the second dose. Interestingly, while the bacterial population in one of the replicates was reduced after receiving the second dose, the other replicate did not respond with a similar reduction. In both cases, however, delivery of the third dose did not result in any detectable bacterial killing, with the 24-h burden being equivalent to the starting inoculum (Fig. 2A). Given our expectation of a substantial reduction in bacterial burden using this dosing regimen, we were surprised that MB-1 only achieved a static endpoint with this fully susceptible strain, particularly since no stably resistant colonies were recovered on MB-1-containing MHB plates at any time point.
Fig 2.
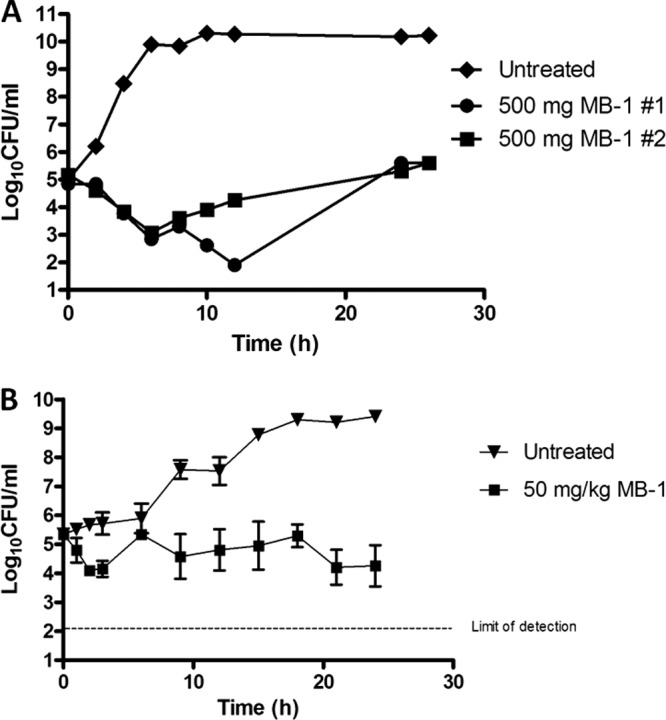
In vitro and in vivo assessment of MB-1 activity against P. aeruginosa UC12120. (A) Hollow-fiber results after 26 h of MB-1 dosing (500 mg infused over 1 h every 8 h) demonstrate a lack of bacterial responsiveness after 2 doses. Cartridges were run in duplicate. (B) Neutropenic-mouse thigh model of infection indicates that dosing MB-1 every 3 h (50 mg/kg) results in an ∼1-log reduction in bacterial burden over 24 h. Mice (n = 3) were sacrificed at the indicated time points, and thighs were harvested for bacterial burden determination.
Since MB-1 is a siderophore-containing compound, it is possible that the lack of sustained killing observed in the hollow-fiber model was due to the iron-replete nature of MHB, and we speculated that the low free-iron concentrations in mammalian systems would promote MB-1 activity. Therefore, we performed a multidose efficacy study in a standard neutropenic-mouse thigh model of infection and determined the bacterial burden over the course of the experiment. As with our hollow-fiber experiments, we used P. aeruginosa strain UC12120, but in order to account for the moderately high clearance of MB-1 in mice, we designed a dosing regimen to provide drug concentrations exceeding the MIC for >90% of the study duration (50 mg/kg dosed every 3 h). Again expecting a reduction in the bacterial burden of at least 2 logs, we were surprised that MB-1 reduced the UC12120 population only by ∼1 log at 24 h (Fig. 2B). However, unlike in the hollow-fiber study, MB-1 did demonstrate some killing effects after repeated dosing (compare bacterial burdens at the 18- and 21-h time points), which gave us confidence that this compound could still be an efficacious antibacterial agent. Further bolstering this confidence was the lack of any recoverable MB-1-resistant colonies when mouse thigh samples were plated onto drug-containing MHB plates.
An in vivo evaluation of MB-1 against multiple P. aeruginosa clinical isolates reveals a lack of correlation between MIC and in vivo efficacy.
In an attempt to refine our PK/PD models and achieve the desired degree of bacterial killing against a panel of clinical isolates, we performed both single-dose and multidose murine PK studies and analyzed the resulting free drug levels in plasma over time (see Fig. S2 in the supplemental material). From these analyses, we determined that a regimen simulating a human 1-g dose administered as a 4-h infusion every 8 h would provide unbound-drug concentrations exceeding the MIC of UC12120 as well as 8 other MDR P. aeruginosa strains (see Table S3 in the supplemental material) for >90% of the time. Again using the neutropenic-mouse thigh model, we measured MB-1 efficacy against this panel of isolates, and Fig. 3A shows the 24-h endpoint bacterial burden for each strain. It should first be noted that the degree of MB-1 activity against P. aeruginosa UC12120 did achieve the predicted >2-log kill, suggesting that the efficacy deficiencies seen against this strain in both the hollow-fiber and preliminary time course mouse thigh models were attributable to insufficient exposure derived from the from the lower simulated human dose (500 mg dosed every 8 h). And while we were encouraged by this specific result, we were disappointed to see how variable the efficacy of MB-1 was against the other 8 strains that were tested in this model (Fig. 3A). Of particular concern was the lack of correlation between the in vitro MIC for each strain and the corresponding level of MB-1 efficacy against it in vivo. While the bacterial burdens from both strains with MICs of 0.25 μg/ml (UC12120 and PA-3581) were effectively reduced by MB-1 after 24 h of treatment, we observed mixed efficacy results with the 3 P. aeruginosa strains that had MICs of 0.5 μg/ml (PAO1, JJ8-16, and JJ5-35). A similar inconsistency existed with the 3 strains with MICs of 1 μg/ml, as the burden from strain AZ8-18 was reduced ∼1.5 log, while strains AZ32-13 and JJ4-36 showed ∼1.5- to 2-log increases in growth (Fig. 3A). Lastly, strain JJ11-54, which possessed the highest MB-1 MIC (2 μg/ml) of all the clinical isolates tested, demonstrated ∼3-log growth over the course of this experiment. While the in vitro and in vivo behaviors of this highly recalcitrant strain did seem to correlate, as did the results seen with the 2 most susceptible strains, our inability to consistently treat different P. aeruginosa strains that had equivalent MICs was an area of significant concern. These results were especially perplexing after we confirmed that the dosing regimen provided unbound MB-1 concentrations above the MIC for >90% of the time for all strains tested in the study (see Fig. S2 in the supplemental material), particularly since other β-lactam antibiotics, such as meropenem, have been described to have optimal efficacy when free concentrations above the MIC are achieved with exposures for >40% of the time (29).
Fig 3.
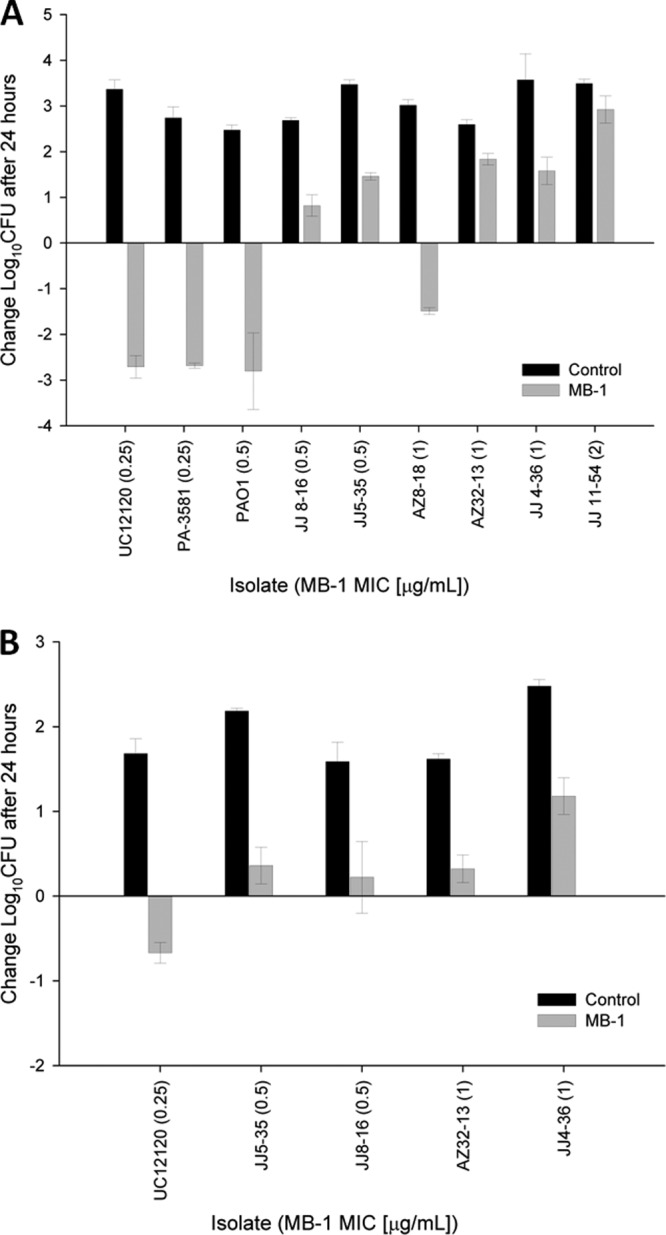
Thigh models for immunosuppressed (A) and immunocompetent (B) mice show variable MB-1 efficacies across a collection of P. aeruginosa clinical isolates. MB-1 was dosed at concentrations and frequencies necessary to mimic a human efficacious regimen (1 g every 8 h). Mice (n = 6) were sacrificed either at time zero or at 24 h, and thighs were harvested for bacterial burden determination.
Next, we chose to evaluate a subset of these clinical P. aeruginosa strains in an immunocompetent-mouse model of infection, to determine if an intact immune system would effectively contribute to bacterial burden reduction during MB-1 treatment. Using the same dosing regimen as was used in the neutropenic-mouse model described above, we again observed inconsistent results in terms of the degree of MB-1-mediated killing over the course of the 24-h study (Fig. 3B). It should be noted that the starting inocula in this immunocompetent model were 10-fold higher than they were in the immunosuppressed model, which perhaps explains why the magnitude of bacterial killing or growth was not as significant as in the previous study. It should also be noted, however, that the 24-h bacterial burdens from the untreated mice in the immunocompetent-mouse model were roughly equivalent to those at 24 h in the immunosuppressed-mouse model (data not shown), which, given the differences in starting inoculum densities, indicates that a functional immune system does respond to challenges with P. aeruginosa infection. Irrespective of those contributions, however, we did not see any substantial improvements in MB-1 in vivo efficacy when an intact immune system was present.
Alteration of the FOR protocol to more closely mimic the in vivo environment provides some insight into the mechanism of P. aeruginosa adaptation seen in mouse models.
Our unexpected in vivo results required us to identify the mechanism(s) by which certain strains of P. aeruginosa can resist the activity of MB-1. Seeing as we did not recover any stably resistant colonies from either the hollow-fiber or murine thigh model of infection, we were left to consider transient, adaptation-type mechanisms rather than constitutive phenotypes. Recognizing that MB-1 includes a siderophore moiety, and that its uptake through the PiuA and PirA outer membrane receptors may be affected by the amount of iron in the surrounding environment, we elected to reexamine MB-1's in vitro activity using a medium that more appropriately reflected the extremely low free-iron levels present in vivo. First, we conducted standard MIC assays with each of the 9 strains profiled in the immunosuppressed-mouse model of infection, but we replaced the CLSI standard MHB with CDMHB, an iron-depleted version of MHB. Using the same starting density of cells as is specified in the CLSI guidelines (26, 27), we found all 9 strains to have MICs of <0.06 μg/ml, which is likely the result of increased siderophore receptor expression and subsequent compound uptake. Thus, this assay did not identify any differences in MICs that correlated with the in vivo failure observed with these clinical isolates.
Next, we revisited the FOR methodology we used to initially characterize spontaneous resistance development against MB-1. We replaced the standard MHB agarose with our iron-deficient CDMHB agarose medium and plated ∼5 × 106 cells on a series of plates containing a range of MB-1 concentrations. Much to our surprise, we observed significantly different patterns of P. aeruginosa regrowth than we had seen in our standard FOR experiments. In particular, we first noticed that colonies that emerged on MB-1-containing plates were surrounded by a fluorescent yellow-green haze as depicted in Fig. 4A. But more importantly, we identified strains that had regrowth on plates containing MB-1 concentrations as high as 32 μg/ml (Table 2), and the colonies that emerged were always clustered together on the agar surface (Fig. 4A). This pattern of growth made accurate colony counting impossible, thereby preventing us from calculating reliable resistance frequencies. We were able to estimate, however, that the FOR values were in the 10−6 range, based on the low cell density plated and the emergence of even 5 to 10 colonies per plate. While these FOR values were abnormally high and not in agreement with the value obtained using standard FOR methods, it was the drug concentrations at which these strains demonstrated regrowth that correlated perfectly with their in vivo outcome (Table 2). The dosing regimen used in the neutropenic-mouse thigh model resulted in drug concentrations that exceeded 4 μg/ml for >90% of the study duration (see Fig. S2 in the supplemental material), and as discussed previously, this would predict a bactericidal outcome for all strains based on their MICs. The 5 strains that did not respond to MB-1 treatment in vivo, however, all showed CDMHB FOR regrowth at concentrations above 4 μg/ml, suggesting that significantly higher drug concentrations would be required to effectively treat infections with these isolates. Interestingly, after surveying multiple colonies that had emerged at the highest MB-1 concentration for each respective strain, we were unable to detect any stable resistance to MB-1.
Fig 4.
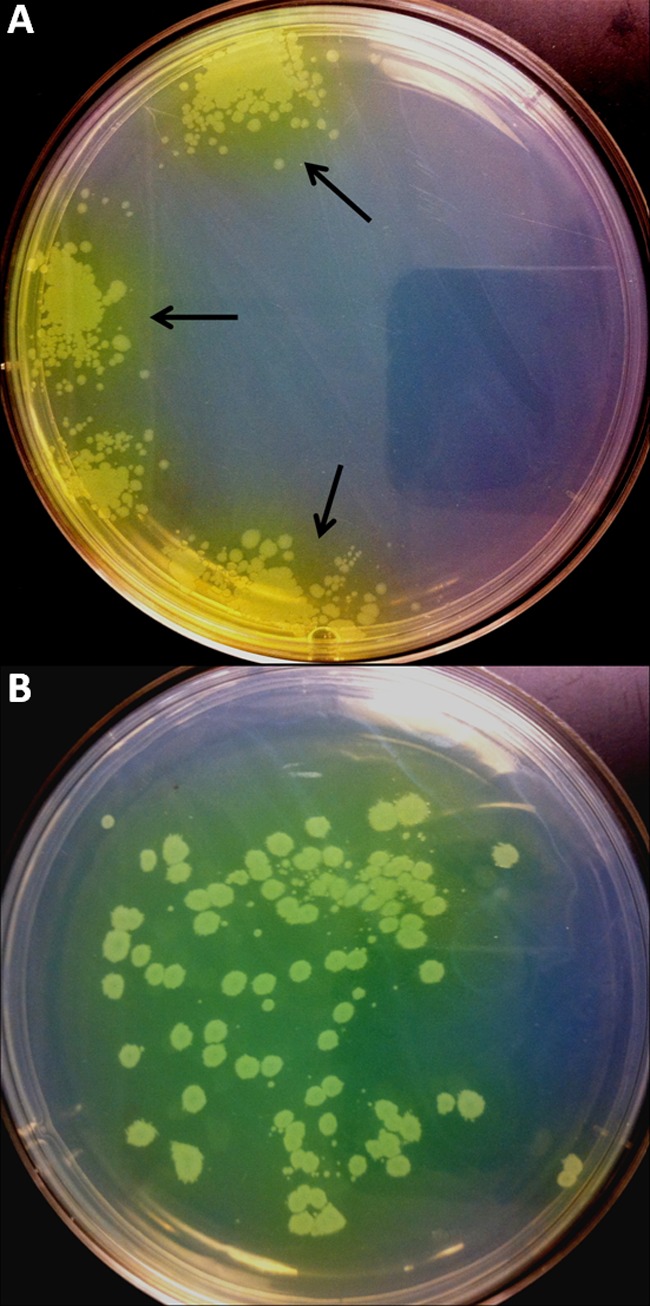
Detection of pyoverdine diffusion surrounding MB-1-resistant colonies in CDMHB FOR experiments. Plates, either unsupplemented (A) or supplemented with 1% pyoverdine-containing conditioned medium (B), were grown for 40 h at 37°C prior to being photographed with a digital camera. Arrows indicate the yellow-green haze observed surrounding all colonies that grew in the presence of MB-1.
Table 2.
MB-1 FOR results in CDMHB are highly correlative with compound efficacy in vivo as assessed by change in bacterial burden at 24 h relative to starting inoculum
| Strain | In vitro MB-1 MIC (μg/ml)a | Highest [MB-1] (μg/ml) with growth on CDMHB agarose platesb | In vivo outcome in immunosuppressed-mouse model |
|---|---|---|---|
| PAO1 | 0.5 | 0.25 | Reduction |
| UC12120 | 0.25 | 0.25 | Reduction |
| PA-3581 | 0.25 | 0.25 | Reduction |
| AZ8-18 | 1 | 2 | Reduction |
| JJ8-16 | 0.5 | 8 | Growth |
| AZ32-13 | 1 | 8 | Growth |
| JJ5-35 | 0.5 | 32 | Growth |
| JJ4-36 | 1 | 32 | Growth |
| JJ11-54 | 2 | 32 | Growth |
MIC determinations were performed using standard, CLSI-compliant MHB.
Determination of accurate resistance frequencies was not possible given the clumped distribution of colony growth on CDMHB plates.
Endogenously produced P. aeruginosa siderophores influence MB-1 activity both in vitro and in vivo.
Our newly developed in vivo-relevant FOR assay provided us with some preliminary clues into the mechanism behind the adaptation seen in our in vivo models. We were particularly interested in exploring the yellow-green haze that we found surrounding the colonies which emerged on our MB-1-containing plates, and we suspected that this substance was pyoverdine, the pigmented native siderophore produced by most P. aeruginosa strains. Given pyoverdine's extremely high affinity for iron (30), we formulated the hypothesis that under low-iron conditions, P. aeruginosa increases production and reliance on pyoverdine for iron uptake, thereby reducing use of other outer membrane siderophore receptors, including those shown to be involved in MB-1 uptake (PiuA and PirA). To test this, we first constructed a pyoverdine-deficient mutant of P. aeruginosa PAO1 and prepared conditioned-medium samples from both the parental strain and its isogenic ΔpvdA mutant. As this conditioned medium is the exhausted, cell-free supernatant harvested from overnight cultures grown in the low-iron CDMHB medium, it was not surprising to see pyoverdine accumulation in the wild-type PAO1 sample, whereas the ΔpvdA sample lacked this endogenous siderophore (data not shown). We conducted CDMHB MIC experiments with wild-type PAO1 in which we added back various concentrations of each conditioned medium sample and monitored the resulting MIC of MB-1 as well as the non-siderophore-conjugated antibiotic cefepime. The results, shown in Table 3, demonstrated that the PAO1 MB-1 MIC shifted 32-fold through the addition of 0.25% of the wild-type PAO1 conditioned medium and >256-fold when 2.5% was added. These sharp increases in MIC were not seen when the ΔpvdA conditioned medium was added in equivalent concentrations, indicating that the presence of pyoverdine in the wild-type conditioned medium was sufficient to significantly increase the PAO1 MIC to levels that were not achieved in our mouse models of infection. In further support of this finding, we used purified pyoverdine and enterobactin, the primary native siderophore produced by members of the Enterobacteriaceae which P. aeruginosa can utilize (31), in similar CDMHB MIC assays, and we observed 16- and 4-fold increases in MIC, respectively, when as little as a 200 nM concentration of each siderophore was added.
Table 3.
Supplementation with pyoverdine-proficient conditioned medium or purified Gram-negative siderophores leads to elevated MB-1 MICs in CDMHB
| Source of conditioned medium | Amt added (%) | MB-1 MIC (μg/ml) | Cefepime MIC (μg/ml) | Siderophore | Amt added | MB-1 MIC (μg/ml) | Cefepime MIC (μg/ml) |
|---|---|---|---|---|---|---|---|
| None | NAa | 0.06 | 4 | None | NA | 0.06 | 4 |
| Wild-type PAO1 | 0.025 | 0.125 | 2 | Pyoverdine | 20 pM | 0.06 | 4 |
| 0.25 | 2 | 4 | Pyoverdine | 2 nM | 0.125 | 2 | |
| 2.50 | >16 | 4 | Pyoverdine | 200 nM | 1 | 4 | |
| PAO1 ΔpvdA | 0.025 | 0.06 | 2 | Enterobactin | 20 pM | 0.06 | 4 |
| 0.25 | 0.06 | 2 | Enterobactin | 2 nM | 0.06 | 4 | |
| 2.50 | 0.06 | 2 | Enterobactin | 200 nM | 0.25 | 2 |
NA, not applicable.
In light of our discovery that native siderophores can affect the activity of MB-1 in MIC assays, we next wanted to explore the impact of pyoverdine in our CDMHB FOR assay. After constructing pyoverdine-deficient derivatives of 3 of the other 8 strains utilized for in vivo experiments (JJ8-16, JJ4-36, and JJ11-54) to accompany PAO1 ΔpvdA, we conducted resistance frequency experiments in our iron-depleted medium as described earlier. Plating nearly equivalent numbers of cells from both the ΔpvdA strains and their respective parental strains, it was clearly noted that each of the 4 mutants was severely impacted in the ability to grow on MB-1 concentrations as high as for their corresponding wild-type strains (Table 4). Additionally, we noted that in the few instances where ΔpvdA colonies did emerge, they were not surrounded by the yellow-green haze we observed with each of the 4 parental strains, providing confidence that this diffusible material was indeed pyoverdine. Furthermore, in an effort to recapitulate our conditioned-medium supplementation MIC results, we also attempted add-back-type experiments in the CDMHB FOR assay, in which we supplemented the molten agar with conditioned medium from either the wild type or ΔpvdA mutants from multiple P. aeruginosa strains. As it was the most sensitive strain used in these studies, we elected to use the PAO1 ΔpvdA strain as the test organism spread on the surfaces of these CDMHB plates. As shown in Table 5, the incorporation of 1% conditioned medium from each of the 3 pyoverdine-producing strains tested led to a significant increase in MB-1 resistance by PAO1 ΔpvdA, a phenotype that was completely absent when ΔpvdA conditioned medium from any of the 3 strains was incorporated into the agar. We also noted that when pyoverdine-containing conditioned medium was used for supplementation, the even distribution of this native siderophore throughout the agar plate resulted in a nonclustered pattern of P. aeruginosa colony growth (Fig. 4B), which supports our hypothesis that pyoverdine cross-feeding contributes to the clustered nature of colony growth when CDMHB plates are not supplemented with conditioned medium (Fig. 4A).
Table 4.
Pyoverdine-deficient mutant variants of MB-1-resistant P. aeruginosa isolates are unable to grow on the same MB-1 concentrations as their respective parent strains in the CDMHB FOR assay
| Strain | Highest [MB-1] (μg/ml) with growth | [MB-1] fold decrease relative to parental strain |
|---|---|---|
| PAO1 ΔpvdA | <0.25 | >2 |
| JJ8-16 ΔpvdA | 0.25 | 8 |
| JJ4-36 ΔpvdA | <0.25 | >32 |
| JJ11-54 ΔpvdA | 0.25 | 16 |
Table 5.
Supplementation with 1% conditioned medium from pyoverdine-proficient strains has significant effects on the CDMHB FOR of P. aeruginosa PAO1 ΔpvdA
| Source of conditioned medium | Highest [MB-1] (μg/ml) with growth |
|---|---|
| NSa | <0.5 |
| PAO1 wild type | 2 |
| PAO1 ΔpvdA | <0.5 |
| JJ4-36 wild type | 16 |
| JJ4-36 ΔpvdA | <0.5 |
| JJ11-54 wild type | 4 |
| JJ11-54 ΔpvdA | <0.5 |
NS, no supplementation.
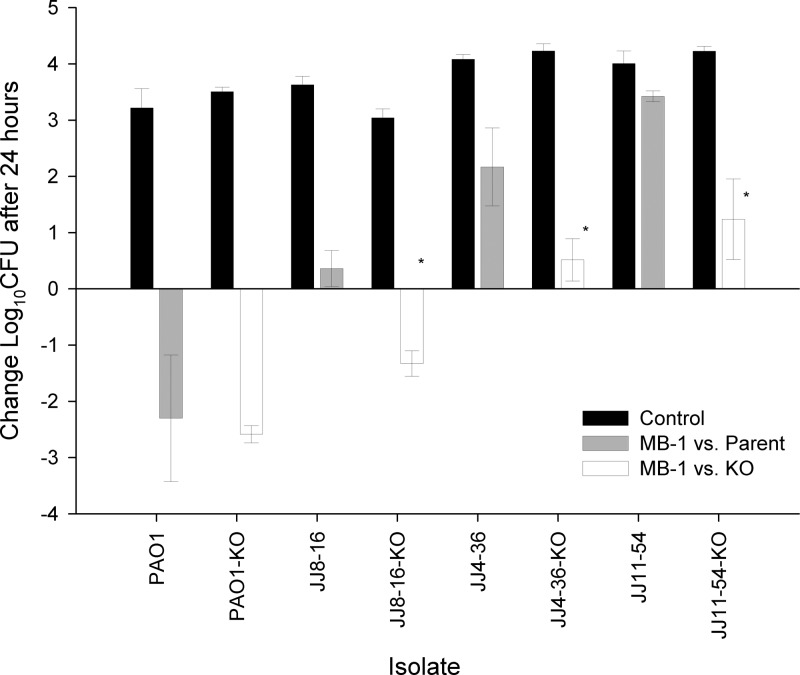
With in vitro data supporting our hypothesis that native siderophore production can impact MB-1 activity, we sought to complete this analysis with an in vivo evaluation of MB-1 efficacy comparing the wild-type P. aeruginosa clinical isolates to their respective ΔpvdA mutants. Using the neutropenic-mouse thigh model and dosing regimen we described earlier, we observed a 1.5- to 2-log improvement in MB-1 efficacy against each of the 3 ΔpvdA mutants whose pyoverdine-proficient parents showed an overall net growth after 24 h of dosing (Fig. 5). We also noted that the ΔpvdA variant from strain PAO1 was not impacted more significantly than its parent strain, although wild-type PAO1 was fully susceptible to MB-1-mediated killing to begin with. From all of these results, we were able to assign a role for pyoverdine in both our in vitro and in vivo assays, thereby confirming our suspicions that this endogenously produced siderophore was capable of attenuating the activity of this novel antibacterial agent against a number of representative clinical isolates of P. aeruginosa.
Fig 5.
Comparison of MB-1 in vivo efficacies against both wild-type and isogenic pyoverdine-deficient knockout mutants (KO) of 4 clinical isolates in the neutropenic-mouse thigh model. Mice (n = 6) were sacrificed either at time zero or at 24 h, and thighs were harvested for bacterial burden determination. Asterisks indicate statistically significant (P < 0.001) differences in MB-1 efficacy when comparing ΔpvdA strains with their respective parental strains.
DISCUSSION
Siderophore conjugation represents one of the approaches with which antibacterial drug discoverers have attempted to circumvent multiple Gram-negative resistance mechanisms that commonly thwart the activity of new drug candidates. While this avenue of research has demonstrated potential (9), it also requires a thorough appreciation for the nature of the in vivo environment in which these compounds would be utilized. This, in turn, requires the development and use of the appropriate in vitro model systems to effectively recapitulate the host environment in order to properly assess the strengths and potential liabilities of siderophore conjugates. From the results generated in this study, it is clear that standard CLSI-compliant MIC and FOR methodologies, known to be highly predictive of in vivo efficacy for numerous antibacterial drug classes, do not effectively predict the potential of the siderophore conjugate MB-1 (and potentially other such conjugates). We have shown not only that standard MHB provides a very different picture of MB-1 activity than CDMHB but also that the standard number of bacterial cells used in MIC experiments (5 × 104) falls below the threshold for this adaptation-type phenomenon we see in CDMHB FOR assays and, correspondingly, in in vivo models. In addition, these discrepancies have confounded our ability to effectively predict the PK/PD relationship of MB-1, as adaptation-type resistance along with the corresponding MIC shifts of adapted cells have proven difficult to incorporate into our PK/PD models. And while our in vivo results were disappointing and unexpected, they also underscore the importance of testing a variety of clinical isolates in animal models to definitively prove that a preclinical antibacterial candidate is likely to demonstrate the spectrum of in vivo activity required to be an effective drug. This is particularly evident when one considers that 8 of the 9 clinical isolates utilized in the neutropenic-mouse thigh model had standard MICs that fell within the MIC90 of 1 μg/ml. And although we still do not fully understand the differences between the P. aeruginosa strains that MB-1 was effective against versus those that resisted its activity, we likely would not have observed the adaptation phenotype had we chosen to profile only 1 or 2 strains in our in vivo models.
In the context of the mechanism underlying P. aeruginosa adaptation to MB-1, we are particularly intrigued by the pattern of bacterial growth seen in the CDMHB FOR assays (Fig. 4). The fact that the cells which resist MB-1 activity tend to grow in groups surrounded by the fluorescent yellow-green haze that is undeniably pyoverdine, together with our inability to recover any stably resistant colonies from those clusters, suggests a mechanism similar to satellite colony formation with β-lactamase-producing colonies. In our case, we envision a small subpopulation of pyoverdine-hyperproducing cells that provide sufficient quantities of this native siderophore to support the growth of neighboring cells. This cross-feeding phenomenon, particularly in light of pyoverdine's extremely high affinity for iron, therefore provides both the hyperproducers and the neighboring bystander cells with a highly efficient means for acquiring this micronutrient, which likely deemphasizes their dependence on other, less efficient outer membrane siderophore receptors, including those used for MB-1 uptake. For reference, it should be noted that a recent survey of sputum samples collected from the lungs of cystic fibrosis patients revealed pyoverdine concentrations that ranged from 0.3 to 51 μM (32). It is also important to note that throughout this entire study, we did not test the effects or contributions of other endogenous P. aeruginosa siderophores, such as pyochelin, on MB-1 activity and adaptation. While we opted to profile the role of pyoverdine, which has a much higher affinity for iron than pyochelin (30), MB-1's iron affinity is currently unknown and could prove to be lower than that of pyochelin as well. If that is indeed the case, it could explain why the ΔpvdA variants of multiple P. aeruginosa strains were not fully susceptible to MB-1-mediated killing in vivo (Fig. 5).
Based on our results, it appears that some adjustments to the overall approach and strategy are needed in order to maximize the chance for success of siderophore-conjugated antibacterial compounds. In terms of a path forward, we propose strategies below that may restore the functionality of these compounds in light of the adaptation-type resistance phenomenon we have described. One approach requires that the iron-binding moiety used be able to effectively compete with the native siderophores produced by the target organisms for the small amount of free iron available in the host environment. Indeed, previous studies have demonstrated that the pyoverdine outer membrane receptor FpvA discriminates between the iron-loaded versus apo-forms of this native siderophore, leading to a substantially higher transport rate of the former over the latter (33). While it remains to be determined if simple siderophore mimics, such as pyridones, hydroxymates, and catecholates, require iron binding for effective uptake, there is no doubt that these mimics would rapidly lose the ability to compete for free iron when faced with rising levels of native siderophores such as pyoverdine. Fortunately, the concept of native siderophore conjugation for antibacterial drug delivery is currently being explored (34), and early proof-of-concept studies suggest that the increased molecular weight associated with utilization of the entire siderophore does not interfere with active transport across the outer membrane of Gram-negative organisms. In addition, if expression of secondary and tertiary siderophore receptors (such as piuA and pirA) is downregulated as a consequence of pyoverdine-mediated iron acquisition, siderophore mimics that instead target FpvA and FpvB, the outer membrane receptors that pyoverdine uses to bring iron into the cell (35, 36), could circumvent this issue if they prove to be competitive with pyoverdine for receptor binding.
Another approach to circumventing endogenous siderophore competition could be the combination of siderophore conjugates with other compounds active against Gram-negative bacteria. If the MB-1 adaptation described in this report is truly dependent upon a subpopulation of cells hyperproducing endogenous siderophores, thereby providing protection to the entire surrounding population, then a compound whose efficacy is not affected by this hyperproduction could effectively eliminate this subpopulation and prevent the beneficial bystander effect we propose in this study. It is possible that marketed antibiotics that impact protein synthesis could be effective combination partners, as the inhibition of pyoverdine synthesis and transport could circumvent the issues we faced with MB-1. Alternatively, and in the context of preventing transport of native siderophores, one combination approach could include the utilization of efflux pump inhibitors. Recent research efforts have identified a specific efflux pump in P. aeruginosa that is responsible for export of newly synthesized pyoverdine as well as for recycling of apo-pyoverdine once it has completed an iron delivery cycle inside the cell (37, 38). This efflux system, comprised of the PvdRT-OpmQ proteins, is a tripartite pump that shares features with other ATP-binding cassette transporters produced by Gram-negative bacteria. And while a significant amount of research investment has focused on the identification of inhibitors of the resistance-nodulation-division (RND)-type efflux pumps (39, 40), it would be worthwhile to study their ability to inhibit PvdRT-OpmQ activity in parallel, as the latter could potentially synergize with siderophore-conjugated compounds by avoiding competition by native siderophore systems.
In summary, we have described our in vitro and in vivo characterizations of MB-1, a novel siderophore-conjugated monobactam that initially appeared to have broad spectrum in vitro activity against a number of MDR Gram-negative pathogens. The extent of this activity was explained, at least in part, by the fact that clinically relevant antibiotic resistance mechanisms commonly employed by these resistant pathogenic species do not impact the activity of MB-1. Despite these encouraging in vitro results, however, we have demonstrated that the manner in which this compound was tested is not necessarily predictive of its in vivo efficacy against multiple strains of P. aeruginosa. Through the development of new in vitro assays that we have demonstrated to correlate with in vivo outcome, we have exposed a potentially serious adaptation-based resistance mechanism to this compound. As the concept of siderophore conjugation to facilitate compound penetration into Gram-negative cells is becoming a more commonly explored platform, we hope that this initial work will encourage others to seek to further understand the complex interplay of native siderophores, their receptors, and their influence on the efficacy of siderophore conjugates such as MB-1.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Lisa Aschenbrenner and M. Megan Lemmon for assisting with the design, construction, and MIC testing of the β-lactamase library, and we thank Hank Christensen, Jennifer Hull, Lucinda Lamb, Deborah Santini, Pamela Tessier, and Lindsay Tuttle for their assistance with in vivo experimentation. We also thank Hongying Gao for pharmacokinetic sample analysis and Hillary Workman for critically evaluating the manuscript.
A.P.T., C.J.M., S.M.F., R.L.I., M.F.B., and J.P.O. are currently or were previously employed by Pfizer and may own stock in the company. D.P.N. has been the recipient of speaker agreements and research grants from Pfizer.
Footnotes
Published ahead of print 17 June 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.00629-13.
REFERENCES
- 1.Ehmann DE, Jahic H, Ross PL, Gu RF, Hu J, Kern G, Walkup GK, Fisher SL. 2012. Avibactam is a covalent, reversible, non-beta-lactam beta-lactamase inhibitor. Proc. Natl. Acad. Sci. U. S. A. 109:11663–11668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sader HS, Rhomberg PR, Farrell DJ, Jones RN. 2011. Antimicrobial activity of CXA-101, a novel cephalosporin tested in combination with tazobactam against Enterobacteriaceae, Pseudomonas aeruginosa, and Bacteroides fragilis strains having various resistance phenotypes. Antimicrob. Agents Chemother. 55:2390–2394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aggen JB, Armstrong ES, Goldblum AA, Dozzo P, Linsell MS, Gliedt MJ, Hildebrandt DJ, Feeney LA, Kubo A, Matias RD, Lopez S, Gomez M, Wlasichuk KB, Diokno R, Miller GH, Moser HE. 2010. Synthesis and spectrum of the neoglycoside ACHN-490. Antimicrob. Agents Chemother. 54:4636–4642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grossman TH, Starosta AL, Fyfe C, O'Brien W, Rothstein DM, Mikolajka A, Wilson DN, Sutcliffe JA. 2012. Target- and resistance-based mechanistic studies with TP-434, a novel fluorocycline antibiotic. Antimicrob. Agents Chemother. 56:2559–2564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Farra A, Islam S, Strålfors A, Sörberg M, Wretlind B. 2008. Role of outer membrane protein OprD and penicillin-binding proteins in resistance of Pseudomonas aeruginosa to imipenem and meropenem. Int. J. Antimicrob. Agents 31:427–433 [DOI] [PubMed] [Google Scholar]
- 6.Luo L, Jiang X, Wu Q, Wei L, Li J, Ying C. 2011. Efflux pump overexpression in conjunction with alternation of outer membrane protein may induce Acinetobacter baumannii resistant to imipenem.Chemotherapy 57:77–84 [DOI] [PubMed] [Google Scholar]
- 7.Tsai Y-K, Fung C-P, Lin J-C, Chen J-H, Chang F-Y, Chen T-L, Siu LK. 2011. Klebsiella pneumoniae outer membrane porins OmpK35 and OmpK36 play roles in both antimicrobial resistance and virulence. Antimicrob. Agents Chemother. 55:1485–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McPherson CJ, Aschenbrenner LM, Lacey BM, Fahnoe KC, Lemmon MM, Finegan SM, Tadakamalla B, O'Donnell JP, Mueller JP, Tomaras AP. 2012. Clinically relevant Gram-negative resistance mechanisms have no effect on the efficacy of MC-1, a novel siderophore-conjugated monocarbam. Antimicrob. Agents Chemother. 56:6334–6342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Möllmann U, Heinisch L, Bauernfeind A, Köhler T, Ankel-Fuchs D. 2009. Siderophores as drug delivery agents: application of the “Trojan Horse” strategy. Biometals 22:615–624 [DOI] [PubMed] [Google Scholar]
- 10.Wooldridge KG, Williams PH. 1993. Iron uptake mechanisms of pathogenic bacteria. FEMS Microbiol. Rev. 12:325–348 [DOI] [PubMed] [Google Scholar]
- 11.Ratledge C, Dover LG. 2000. Iron metabolism in pathogenic bacteria. Annu. Rev. Microbiol. 54:881–941 [DOI] [PubMed] [Google Scholar]
- 12.Zhao G, Ceci P, Ilari A, Giangiacomo L, Laue TM, Chiancone E, Chasteen ND. 2002. Iron and hydrogen peroxide detoxification properties of DNA-binding protein from starved cells. A ferritin-like DNA-binding protein of Escherichia coli. J. Biol. Chem. 277:27689–27696 [DOI] [PubMed] [Google Scholar]
- 13.Andrews SC, Robinson AK, Rodriguez-Quinones F. 2003. Bacterial iron homeostasis. FEMS Microbiol. Rev. 27:215–237 [DOI] [PubMed] [Google Scholar]
- 14.Miller MJ, Walz AJ, Zhu H, Wu C, Moraski G, Möllmann U, Tristani EM, Crumbliss AL, Ferdig MT, Checkley L, Edwards RL, Boshoff HI. 2011. Design, synthesis, and study of a mycobactin-artemisinin conjugate that has selective and potent activity against tuberculosis and malaria. J. Am. Chem. Soc. 133:2076–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krishnan P, Frew Q, Green A, Martin R, Dziewulski P. 2013. Cause of death and correlation with autopsy findings in burns patients. Burns 39:583–588 [DOI] [PubMed] [Google Scholar]
- 16.Bessa LJ, Fazii P, Di Giulio M, Cellini L. 24 February 2013. Bacterial isolates from infected wounds and their antibiotic susceptibility pattern: some remarks about wound infection. Int. Wound J. [Epub ahead of print.] 10.1111/iwj.12049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.El Solh AA, Alhajhusain A. 2009. Update on the treatment of Pseudomonas aeruginosa pneumonia. J. Antimicrob. Chemother. 64:229–238 [DOI] [PubMed] [Google Scholar]
- 18.Vasil ML, Berka RM, Gray GL, Pavlovskis OR. 1985. Biochemical and genetic studies of iron-regulated (exotoxin A) and phosphate-regulated (hemolysin phospholipase C) virulence factors of Pseudomonas aeruginosa. Antibiot. Chemother. 36:23–39 [DOI] [PubMed] [Google Scholar]
- 19.de Lima CD, Calegari-Silva TC, Pereira RM, Santos SA, Lopes UG, Plotkowski MC, Saliba AM. 2012. ExoU activates NF-kappaB and increases IL-8/KC secretion during Pseudomonas aeruginosa infection. PLoS One 7:e41772. 10.1371/journal.pone.0041772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stonehouse MJ, Cota-Gomez A, Parker SK, Martin WE, Hankin JA, Murphy RC, Chen W, Lim KB, Hackett M, Vasil AI, Vasil ML. 2002. A novel class of microbial phosphocholine-specific phospholipases C. Mol. Microbiol. 46:661–676 [DOI] [PubMed] [Google Scholar]
- 21.Engel J, Balachandran P. 2009. Role of Pseudomonas aeruginosa type III effectors in disease. Curr. Opin. Microbiol. 12:61–66 [DOI] [PubMed] [Google Scholar]
- 22.Mishra M, Byrd MS, Sergeant S, Azad AK, Parsek MR, McPhail L, Schlesinger LS, Wozniak DJ. 2012. Pseudomonas aeruginosa Psl polysaccharide reduces neutrophil phagocytosis and the oxidative response by limiting complement-mediated opsonization. Cell. Microbiol. 14:95–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mann EE, Wozniak DJ. 2012. Pseudomonas biofilm matrix composition and niche biology. FEMS Microbiol. Rev. 36:893–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Franklin MJ, Nivens DE, Weadge JT, Howell PL. 2011. Biosynthesis of the Pseudomonas aeruginosa extracellular polysaccharides, alginate, Pel, and Psl. Front. Microbiol. 2:167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fung-Tomc J, Bush K, Minassian B, Kolek B, Flamm R, Gradelski E, Bonner D. 1997. Antibacterial activity of BMS-180680, a new catechol-containing monobactam. Antimicrob. Agents Chemother. 41:1010–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.CLSI 2009. Performance standards for antimicrobial susceptibility testing: 19th informational supplement. Clinical and Laboratory Standards Institute document M100-S19 CLSI, Wayne, PA [Google Scholar]
- 27.CLSI 2009. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically: approved standard. Clinical and Laboratory Standards Institute document M7-A10 CLSI, Wayne, PA [Google Scholar]
- 28.Figurski DH, Helinski DR. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. U. S. A. 76:1648–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mattoes HM, Kuti JL, Drusano GL, Nicolau DP. 2004. Optimizing antimicrobial pharmacodynamics: dosage strategies for meropenem. Clin. Ther. 26:1187–1198 [DOI] [PubMed] [Google Scholar]
- 30.Visca P, Imperi F, Lamont IL. 2007. Pyoverdine siderophores: from biogenesis to biosignificance. Trends Microbiol. 15:22–30 [DOI] [PubMed] [Google Scholar]
- 31.Poole K, Young L, Neshat S. 1990. Enterobactin-mediated iron transport in Pseudomonas aeruginosa. J. Bacteriol. 172:6991–6996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martin LW, Reid DW, Sharples KJ, Lamont IL. 2011. Pseudomonas siderophores in the sputum of patients with cystic fibrosis. Biometals 24:1059–1067 [DOI] [PubMed] [Google Scholar]
- 33.Schalk IJ, Hennard C, Dugave C, Poole K, Abdallah MA, Pattus F. 2001. Iron-free pyoverdin binds to its outer membrane receptor FpvA in Pseudomonas aeruginosa: a new mechanism for membrane iron transport. Mol. Microbiol. 39:351–360 [DOI] [PubMed] [Google Scholar]
- 34.Zheng T, Bullock JL, Nolan EM. 2012. Siderophore-mediated cargo delivery to the cytoplasm of Escherichia coli and Pseudomonas aeruginosa: syntheses of monofunctionalized enterobactin scaffolds and evaluation of enterobactin-cargo conjugate uptake. J. Am. Chem. Soc. 134:18388–18400 [DOI] [PubMed] [Google Scholar]
- 35.Meyer JM, Stintzi A, Poole K. 1999. The ferripyoverdine receptor FpvA of Pseudomonas aeruginosa PAO1 recognizes the ferripyoverdines of P. aeruginosa PAO1 and P. fluorescens ATCC 13525. FEMS Microbiol. Lett. 170:145–150 [DOI] [PubMed] [Google Scholar]
- 36.Ghysels B, Dieu BT, Beatson SA, Pirnay JP, Ochsner UA, Vasil ML, Cornelis P. 2004. FpvB, an alternative type I ferripyoverdine receptor of Pseudomonas aeruginosa. Microbiology 150:1671–1680 [DOI] [PubMed] [Google Scholar]
- 37.Hannauer M, Yeterian E, Martin LW, Lamont IL, Schalk IJ. 2010. An efflux pump is involved in secretion of newly synthesized siderophore by Pseudomonas aeruginosa. FEBS Lett. 584:4751–4755 [DOI] [PubMed] [Google Scholar]
- 38.Imperi F, Tiburzi F, Visca P. 2009. Molecular basis of pyoverdine siderophore recycling in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 106:20440–20445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Askoura M, Mottawea W, Abujamel T, Taher I. 2011. Efflux pump inhibitors (EPIs) as new antimicrobial agents against Pseudomonas aeruginosa. Libyan J. Med. 10.3402/ljm.v6i0.5870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lomovskaya O, Warren MS, Lee A, Galazzo J, Fronko R, Lee M, Blais J, Cho D, Chamberland S, Renau T, Leger R, Hecker S, Watkins W, Hoshino K, Ishida H, Lee VJ. 2001. Identification and characterization of inhibitors of multidrug resistance efflux pumps in Pseudomonas aeruginosa: novel agents for combination therapy. Antimicrob. Agents Chemother. 45:105–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.