Abstract
The dynamics of occurrence and the genetic basis of ciprofloxacin resistance were studied in a long-term evolution experiment (940 generations) in wild-type, reference strain (PAO1) and hypermutable (PAOΔmutS and PAOMY-Mgm) P. aeruginosa populations continuously exposed to sub-MICs (1/4) of ciprofloxacin. A rapid occurrence of ciprofloxacin-resistant mutants (MIC of ≥12 μg/ml, representing 100 times the MIC of the original population) were observed in all ciprofloxacin-exposed lineages of PAOΔmutS and PAOMY-Mgm populations after 100 and 170 generations, respectively, and in one of the PAO1 lineages after 240 generations. The genetic basis of resistance was mutations in gyrA (C248T and G259T) and gyrB (C1397A). Cross-resistance to beta-lactam antibiotics was observed in the bacterial populations that evolved during exposure to sublethal concentrations of ciprofloxacin. Our study shows that mutants with high-level ciprofloxacin resistance are selected in P. aeruginosa bacterial populations exposed to sub-MICs of ciprofloxacin. This can have implications for the long-term persistence of resistant bacteria and spread of antibiotic resistance by exposure of commensal bacterial flora to low antibiotic concentrations.
INTRODUCTION
In the airways of patients with cystic fibrosis (CF), antibiotic treatment is an important selective pressure that influences the adaptation and evolution of Pseudomonas aeruginosa. As a result of this adaptation, the bacteria persist for long periods of time, undergoing up to 200,000 generations in the CF lung (1). Patterns of parallel evolution at the levels of phenotype, gene expression, and genotype have been documented in CF, indicating repeatable patterns of long-term adaptation.
One of the common evolutionary traits is the development of antibiotic resistance, which is based on genetic variation and the selection of genotypes that this generates. P. aeruginosa strains with a hypermutator phenotype (conferring increased rates of mutation) are frequently encountered in bacterial populations from the CF lung (2, 3), and acquisition of this phenotype will increase the chances of acquiring antibiotic resistance by mutational events.
Exposure of P. aeruginosa to ciprofloxacin, which is a DNA-damaging antibiotic, stimulates bacteria to produce reactive oxygen species (ROS) that can directly damage DNA and lead to accumulation of mutations (4). The role of ciprofloxacin at inhibitory concentrations in development of resistance has been well established in bacterial populations with normal or increased mutation frequencies (5) and in evolution experiments (6, 7). In clinical settings, however, compartments where ciprofloxacin does not reach bacterial MIC are created, such as inside the sputum in the conductive zones of the airways (8). The anaerobic conditions inside the thick CF mucus might also decrease the efficacy of quinolones (9), requiring higher local concentrations. In addition, ciprofloxacin is excreted in sweat and present on the skin at low concentrations for long periods of time (10), with rapid development of resistance in skin bacteria. Epidemiological studies have shown an association between the use of quinolones and the occurrence of methicillin-resistant Staphylococcus aureus (11).
The role of long-term exposure of P. aeruginosa to sub-MICs of ciprofloxacin for the development of resistance has not been investigated before but is clinically relevant and can reveal important bacterial adaptive pathways. It has been shown in Escherichia coli that sublethal levels of bactericidal antibiotics induce mutagenesis that can lead to multidrug resistance (12). Recently, it has been shown in E. coli that very low antibiotic concentration can select de novo mutants, which would take over in a susceptible population (13). Long-term evolution experiments with microorganisms have been used to study the dynamics of evolutionary adaptation (14).
Therefore, we decided to investigate the evolution of P. aeruginosa during many (more than 900) bacterial generations exposed to sublethal concentrations of ciprofloxacin and to compare it to the evolution without selective pressure, focusing on the development of resistance to ciprofloxacin. To investigate the role of the mutation rates in the dynamics of bacterial evolution under the described conditions, two mutator strains due to mutations in DNA mismatch repair and oxidative damage repair were employed, along with the reference strain PAO1. Both types of mutators have been described in CF patients (2, 15, 16).
MATERIALS AND METHODS
Bacterial strains.
Three P. aeruginosa strains were used for this study: the wild-type PAO1 (mutation rate, 6.75 × 10−9) and the two mutator strains PAO1ΔmutS and PAOMY-Mgm (17). PAO1ΔmutS is deficient in the DNA mismatch repair system (mutation rate, 5 × 10−6), and the PAOMY-Mgm mutant is unable to repair oxidative DNA damage (mutation rate, 1.9 × 10−7) (17).
Experimental setup of the long-term evolution study.
The three strains were tested in triplicates (lineages A, B, and C) with or without 0.05 μg/ml ciprofloxacin, representing 1/4 the MIC of ciprofloxacin (0.19 μg/ml), which increases the doubling time from 24.5 (±1.94) min to 32 (±1.25) min. For each strain, a single colony was inoculated in Luria-Bertani (LB) medium and grown overnight at 37°C. One hundred microliters of each starter culture was inoculated into six different flasks each with 100 ml fresh preheated LB medium, allowing the growth of approximately 10 generations/day (106 CFU/ml starting inoculum to 109 CFU/ml ending inoculum). Ciprofloxacin (0.05 μg/ml) was added to three of the six cultures. The cultures were propagated daily for 94 days, allowing for triplicates to grow with and without addition of 0.05 μg/ml ciprofloxacin. The ciprofloxacin concentration remained the same, and a steady CFU count was maintained throughout the study. Daily streaking of the overnight cultures on 5% blood agar plates (5% horse blood; SSI Diagnostica, Hillerød, Denmark) was performed to check for contamination, and the incubated cultures were stored for two nights at 4°C to restart the experiment in case of contamination. Matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) spectrometry was applied to confirm the identity of the strains as Pseudomonas aeruginosa at days 73, 80, 87, and 94. Three colonies representative of each strain were selected from the CFU plate at the 10−7 dilution and subjected to MALDI-TOF analysis. To confirm that the three P. aeruginosa lineages (A, B, and C) belonged to PAO1, PAO1ΔmutS, or PAOMY-Mgm, respectively, confirmatory PCR was performed.
Bacterial population analysis.
To characterize the evolution of the bacterial populations grown for 940 generations in the presence or absence of subinhibitory concentrations of ciprofloxacin (1/4 the MIC), bacterial populations of PAO1, PAO1ΔmutS, and PAOMY-Mgm at different generations (days 3, 10, 17, 24, and 94) were tested for susceptibility to ciprofloxacin. For each of the three lineages of the three strains, a large inoculum (108 CFU/ml; a 10−1 dilution of an overnight culture) was plated on blood agar plates, and an Etest strip of ciprofloxacin was applied. The concentration of ciprofloxacin on the Etest that inhibited the bacterial population was defined as the “population MIC.” To distinguish between different sizes of the resistant subpopulations that grew in the inhibition zone (Fig. 1), four descriptive entities were defined: (i) “double zone” if the resistant colonies exhibited confluent growth, (ii) +++ if the number of colonies was higher than 100, (iii) ++ if the number of colonies was between 10 and 100, and (iv) + if fewer than 10 colonies were present. Figure 1 shows an example of the bacterial population analysis. Colonies grown in the inhibition zone were harvested for further analysis.
Fig 1.
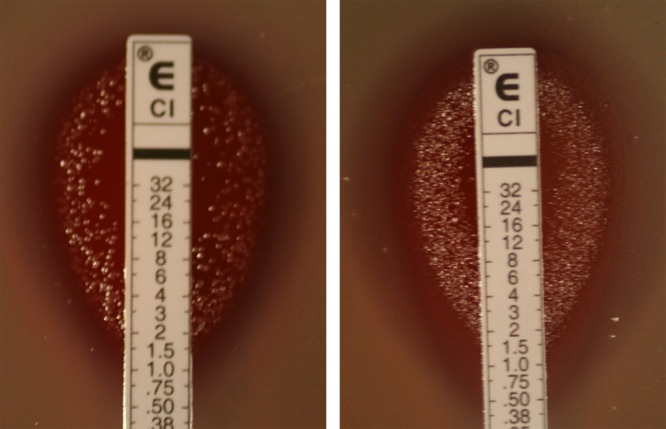
Characterization of the susceptibility to ciprofloxacin of the bacterial population and assignment of population MIC values. To distinguish between different sizes of the resistant subpopulations that grew in the inhibition zone, four descriptive entities were defined: (i) “double zone” if the resistant colonies exhibit a confluent growth, (ii) +++ if the number of colonies was higher than 100, (iii) ++ if the number of colonies was between 10 and 100, and (iv) + if fewer than 10 colonies were present. The descriptions of the two bacterial populations in this figure are thus as follows: in the left panel, the population MIC is 1.5 μg/ml, there is no double zone (dz−), and the size of the resistant subpopulation is +++; in the right panel, the population MIC is 1.5 μg/ml, a double zone is present until 12 μg/ml (dz 12), and resistant colonies in the inhibition zone are also present (+).
Antibiotic susceptibility.
Ciprofloxacin-resistant subpopulations (+, ++, or +++) from days 3, 10, 17, 24, and 94 of the three lineages of each strain evolved in the presence or absence of ciprofloxacin were harvested by scraping from the inhibition zone and retested for ciprofloxacin susceptibility after 2 passages in antibiotic-free medium to confirm the mutagenic nature of their phenotype. To investigate the homogeneity of the harvested resistant subpopulations, antibiotic susceptibility and sequence variability of quinolone resistance determinants were determined for 6 to 9 independent single resistant colonies for each of the three strains.
Sequencing of the quinolone resistance determinants (nfxB, gyrA, gyrB, and parC) on all these resistant subpopulations (a total of 127 colonies) was performed. The MIC of ciprofloxacin for the selected ciprofloxacin-resistant subpopulations was determined by the dilution method in microtiter plates as previously described (18). To determine the occurrence of cross-resistance, the MICs of ciprofloxacin, meropenem, ceftazidime, and tobramycin against each of the bacterial populations evolved in the presence and absence of ciprofloxacin were investigated by Etest according to the instructions of the manufacturer (inoculum, 105 CFU/ml).
Formation of reactive oxygen species under treatment with 0.05 μg/ml ciprofloxacin.
As the bactericidal effect of ciprofloxacin has been shown to be hydroxyl radical dependent (4), we wanted to test if the sub-MIC level of ciprofloxacin used in our study led to formation of oxygen radicals. This was tested as previously described (4). In short, exponentially grown P. aeruginosa was exposed to 0.05 μg/ml ciprofloxacin and 5 μM hydroxyl radical indicator 3′-(p-hydroxyphenyl)fluorescein (HPF) for 3 h. To confirm the specificity of the assay, thiourea (150 mM) (Sigma) was used as a scavenger of hydroxyl radicals. After incubation, hydroxyl radical formation was measured in a FACScanto flow cytometer (BD Biosciences, San Jose, CA) with a 488-nm argon laser and a 530/30-nm band pass emission filter for recording of HPF fluorescence as the area of the pulse in FL1. To maximize resolution, samples were analyzed at low flow rate corresponding to 10 μl/min. A least 10,000 events were recorded for each sample. The following photomultiplier tube (PMT) voltage settings were used for collection of linear amplified signals: 200 (forward scatter [FSC]), 420 (side scatter [SSC]), and 550 (FL1). Cytometer Setup and Tracking Beads (BD) were used for instrument calibration, The HPF fluorescence intensity is expressed as the mean channel in FL1-A.
Measurements of mutation frequency in P. aeruginosa mutant isolates.
The mutation frequencies were investigated on LB plates containing rifampin (300 mg/liter) and streptomycin (500 mg/liter) as previously described (3). An isolate was considered hypermutable when the mutation frequency was 20-fold higher than the mutation frequency of the reference strain PAO1 (≥3 × 10−7).
Sequencing of quinolone resistance-determining regions (QRDRs).
Sequencing of ciprofloxacin resistance-determining genes nfxB, gyrA, gyrB, parC, and parE was performed on purified total DNA (Qiagen) of the selected resistant isolates and of the starting bacterial strains (PAO1_start, mutS_start, and mutMY_start). The genes were amplified by PCR using previously described primers (17). The obtained sequences were compared to the sequence of the reference strain PAO1 by applying DNASIS Max version 2.0 (Hitachi Software Engineering).
Fitness cost analysis.
To investigate if the type of mutations in gyrA and gyrB has a fitness cost for the growth of the mutants, growth curves (optical density at 600 nm [OD600]) were constructed in LB and LB with 0.05 μg/ml ciprofloxacin, and the doubling time was calculated on the exponential phase of the curve.
Nutritional status determination.
The nutritional status of the evolved populations was determined by spreading a 10−6 dilution of an overnight culture on “blue” plates. After incubation at 37°C overnight, 50 colonies/lineage were replica plated to both glucose minimal medium and LB. Possible auxotrophs were scored as cells forming colonies on the LB plates but not on the minimal medium plates after 48 h of incubation at 37°C.
RESULTS
Selection of preexisting resistant subpopulations occurs during exposure to sub-MICs of ciprofloxacin.
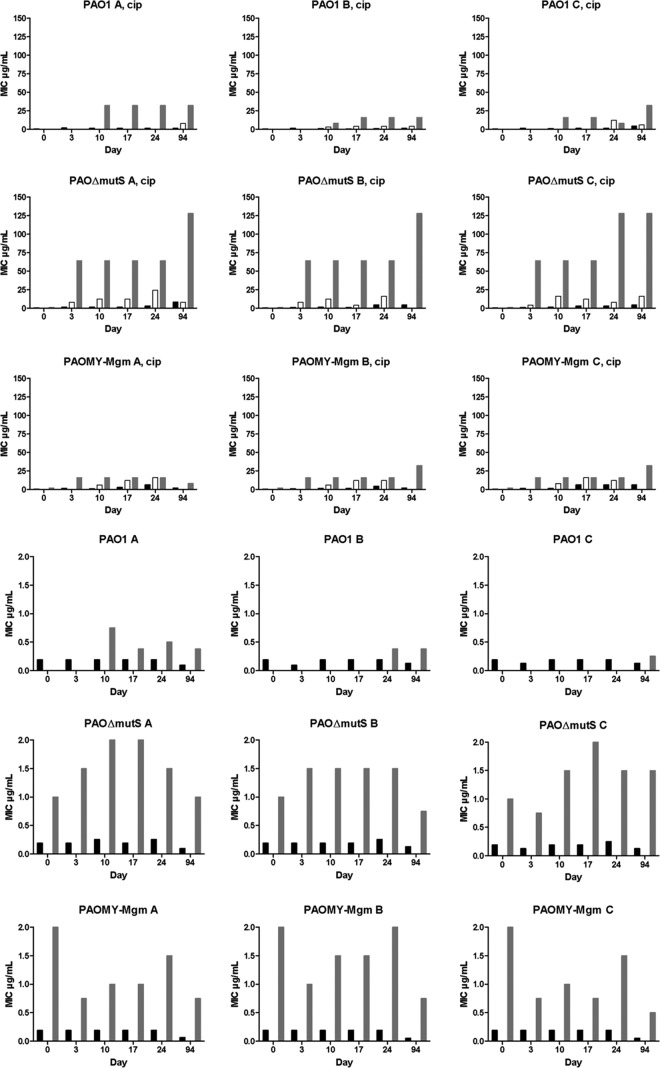
Analysis of the bacterial populations of PAO1, PAOΔmutS, and PAOMY-Mgm showed a gradual increase in the size of resistant subpopulations during 940 generations in the lineages treated with a sublethal concentration of ciprofloxacin (1/4 the MIC), while no increase in resistance was observed for the populations that evolved in the absence of selective pressure (Fig. 2). It is important to underline that at this concentration the bacteria are not killed but grow slightly slower (the doubling time increases from 24.5 to 32 min).
Fig 2.
Development of resistance to ciprofloxacin in ciprofloxacin-treated (top three rows) and untreated (bottom three rows) PAO1, PAOΔmutS, and PAOMY-Mgm during the evolution experiment. Resistant mutants occurred faster in the mutator populations than in PAO1, with an especially drastic increase in MICs for the ciprofloxacin-treated PAOΔmutS strains. Black bars, MICs of population; white bars, MICs of double zone; gray bars, MICs of resistant colonies. Notice the difference in the numerical size of the ordinates in the top three and bottom three rows.
The resistant mutants occurred and were enriched faster in the mutator populations than in PAO1, as seen in Fig. 2 and Table S1 in the supplemental material. The fastest occurrence of resistance and the highest MIC were observed in PAOΔmutS, followed by PAOMY-Mgm and PAO1. This shows an association between the mutation rate of the strain and the size and speed of the occurrence of the resistant subpopulation (Fig. 2).
Resistant subpopulations creating double zones on Etests inhibited by 12 μg/ml ciprofloxacin (100 times the MIC) were observed in all ciprofloxacin-exposed lineages of PAOΔmutS and PAOMY-Mgm after 100 and 170 generations, respectively, and in one of the PAO1 lineages after 240 generations (see “MIC and susceptibility of population” in Table S1 in the supplemental material). No resistant subpopulations were observed in the starting population of PAO1, showing that the ciprofloxacin-resistant mutants that occurred in the three PAO1 lineages during ciprofloxacin exposure were present in a very small proportion in the starting wild-type population or occurred de novo. In contrast, resistant colonies were observed in the starting populations of the two mutator strains (PAOΔmutS and PAOMY-Mgm), showing the preexistence of antibiotic-resistant mutants in these bacterial populations (see Tables S1 and S2 in the supplemental material).
Type of mutations responsible for high-level ciprofloxacin resistance.
In all three lineages of PAOΔmutS, the gyrA C248T mutants leading to T83I were selected by 1/4 the MIC of ciprofloxacin during the evolution experiment. The MICs of the resistant colonies to ciprofloxacin increased from 3 and 4 μg/ml in the starting population to 64 μg/ml at day 3 and 128 μg/ml at day 94 (see Table S1 in the supplemental material).
This gyrA mutation is in accordance with the type of mutations (transitions and frameshifts) encountered in mutators with an impaired mismatch repair (MMR) system (19). The same mutation was found in resistant PAOΔmutS isolates of control populations after 24 days of passage in LB medium (see Table S1b in the supplemental material) and in single resistant colonies of the initial population, confirming the selection of the preexisting mutation in the original population (see Table S2 in the supplemental material).
The ciprofloxacin-resistant mutants which emerged already at day 3 in the PAOMY-Mgm population exposed to ciprofloxacin were caused by a G-to-T transversion at position 259 in gyrA in two of the lineages (A and B) and by a C-to-A transversion at position 1397 in gyrB in the third lineage (C) (Table 1). The same gyrA mutation was found in ciprofloxacin-resistant colonies from the control populations at day 94, suggesting that this mutation was preexisting in the original PAOMY-Mgm population.
Table 1.
Overview of mutations found in quinolone resistance-determining regions (nfxB, gyrA, gyrB, and parC) in ciprofloxacin-resistant subpopulations selected at days 3, 10, 24, and 94 of the evolution experiment in the three lineages of PAO1, PAOΔmutS, and PAOMY-Mgm under exposure to 1/4 the MIC of ciprofloxacina
| Lineage (MIC [μg/ml] on days 3, 10, 17, 24, 94) | Gene | Mutation on day: |
||||
|---|---|---|---|---|---|---|
| 3 | 10 | 17 | 24 | 94 | ||
| PAO1-A-cip (ND, 32, 32, 32, 32) | nfxB | — | Del C pos 6 | — | — | — |
| gyrA | — | C248T | C248T | C248T | C248T | |
| gyrB | — | — | — | — | — | |
| parC | — | — | — | — | — | |
| PAO1-B-cip (ND, 8, 16, 16, 16) | nfxB | — | — | Del C pos 6 | G436A | — |
| gyrA | — | G259T | G259T | G259T | G259T | |
| gyrB | — | — | — | — | — | |
| parC | — | — | — | — | — | |
| PAO1-C-cip (ND, 16, 16, 8, 32) | nfxB | — | 336 ins G | G461A | G461A | — |
| gyrA | — | — | — | — | G259T | |
| gyrB | — | C1397A | C1397A | C1397A | C1397A | |
| parC | — | — | — | — | — | |
| PAOΔmutS-A-cip (64, 64, 64, 64, 128) | nfxB | C434T | — | — | G461A | — |
| gyrA | C248T | C248T | C248T | C248T | C248T | |
| gyrB | — | — | — | — | — | |
| parC | — | — | — | — | C260T | |
| PAOΔmutS-B-cip (64, 64–128, 64, 64, 128) | nfxB | — | G538A | G538A | — | — |
| gyrA | C248T | C248T | C248T | C248T | C248T | |
| gyrB | — | — | — | — | — | |
| parC | — | — | — | — | — | |
| PAOΔmutS-C-cip (64, 64, 64, 64, 128) | nfxB | — | G461A | — | — | T499C |
| gyrA | C248T | C248T | C248T | C248T | C248T | |
| gyrB | — | — | — | — | — | |
| parC | — | — | — | — | C260T | |
| PAOMY-Mgm-A-cip (16, 16, 16, 16, 8) | nfxB | C473A | — | G538T | G539T | G140T |
| gyrA | — | — | — | ND | — | |
| gyrB | C1397A | C1397A | C1397A | ND | C1397A | |
| parC | — | — | — | — | — | |
| PAOMY-Mgm-B-cip (16, 16, 16, 16, 32) | nfxB | G538T | G538T | G538T | — | — |
| gyrA | G259T | G259T | G259T | G259T | G259T | |
| gyrB | — | — | — | — | — | |
| parC | — | — | — | — | — | |
| PAOMY-Mgm-C-cip (16, 16, 16, 16, 32) | nfxB | — | — | — | — | C495A |
| gyrA | G259T | G259T | G259T | G259T | G259T | |
| gyrB | — | — | — | — | — | |
| parC | — | — | — | — | — | |
ND, not determined; —, no change; del, deletion; pos, position; ins, insertion.
Transversions are the specific type of mutations to be expected in mutants of the DNA oxidative repair system (GO) due to mispairing with the uncorrected oxidative lesion of guanine, 8-oxo-2′-deoxyguanosine (8oxodG). The ciprofloxacin MICs of the preexisting resistant mutants in the starting population were 4 μg/ml and 6 μg/ml (see Table S2 in the supplemental material) and increased to 16 μg/ml at day 3 and 32 μg/ml at day 94 during the evolution experiment (Table 1; see Table S1 in the supplemental material).
The fitness cost analysis of the three types of mutations identified in our study to cause ciprofloxacin resistance, i.e., C248T (T83I) and G259T (D87Y) in gyrA and C1397A (S465Y) in gyrB, showed that these mutations did not reduce the growth rate (no fitness cost) compared to that of the starting strains (Table 2). The occurrence of these mutations restored the slightly lower growth rates of the starting strains observed in the presence of the 1/4 the MIC of ciprofloxacin.
Table 2.
Doubling times of PAO1, PAOΔmutS, PAOMY-Mgm, and the ciprofloxacin-resistant mutants in the presence and absence of 0.05 μg/ml ciprofloxacin
| Isolate | Doubling time, min (mean ± SD) |
|
|---|---|---|
| Without ciprofloxacin | With 0.05 μg/ml ciprofloxacin | |
| Starting strains | ||
| PAO1 | 24.5 ± 1.9 | 32 ± 1.2 |
| PAOΔmutS | 26.2 ± 0.6 | 31.4 ± 0.5 |
| PAOMY-Mgm | 25.8 ± 0.6 | 32.3 ± 0 |
| Ciprofloxacin-resistant mutants | ||
| Day 10 | ||
| PAO1-A-cip (GyrA T83I) | 27.15 ± 0.5 | 26 ± 0.2 |
| PAO1-B-cip (GyrA D87Y) | 26.9 ± 0.07 | 28 ± 1.9 |
| PAO1-C-cip (GyrB S466Y) | 27.2 ± 0.2 | 27.8 ± 1.2 |
| Day 3 | ||
| PAOΔmutS-A-cip (GyrA T83I) | 28.9 ± 0 | 27.8 ± 2.5 |
| PAOΔmutS-B-cip (GyrA T83I) | 29.7 ± 1.1 | 28.2 ± 0.4 |
| PAOΔmutS-C-cip (GyrA T83I) | 29.9 ± 1.4 | 28.6 ± 0.4 |
| PAOMY-Mgm-A-cip (GyrB S466Y) | 29.7 ± 1.4 | 25.7 ± 0.3 |
| PAOMY-Mgm-B-cip (GyrA D87Y) | 29.5 ± 0 | 27.8 ± 0.7 |
| PAOMY-Mgm-C-cip (GyrA D87Y) | 28.3 ± 0.2 | 27.8 ± 0.8 |
gyrA parC double mutants (gyrA, C248T; parC, C260T) were identified only in the PAOΔmutS A and C lineages. The mutation in parC leading to replacement of Ser 87 with Leu occurred at a late stage during the evolutionary experiment (940 generations), confirming that parC mutations are second-step mutations. A gyrA gyrB double mutant was also identified at day 94 in PAO1 lineage C (Table 1), which presented only a gyrB mutation until day 87 (data not shown), showing the late occurrence of double mutations during the evolution study.
Fitness in secondary environments.
To test whether adaptation in the rich LB medium resulted in the loss of metabolic fitness in secondary environments, we determined the frequency of possible auxotrophs (lack of growth on glucose minimal media) in all the populations that evolved in the presence or absence of ciprofloxacin at days 24 and 94 of the long-term experiment. We found an increased proportion of colonies with lack of growth on minimal medium in mutator, compared to nonmutator, lineages (see Table S4 in the supplemental material).
Reactive oxygen species are produced during treatment with sub-MICs of ciprofloxacin.
Measurement of reactive oxygen species under treatment with 0.05 μg/ml ciprofloxacin showed a 10-fold increase in the level of hydroxyl radicals (FL1-A mean, 2,564) compared to that in untreated bacterial cells (FL1-A mean, 233). No fluorescence was measured when thiourea, which is a hydroxyl radical scavenger, was added (data not shown).
No changes in the mutation frequencies of the bacterial populations of the three strains after evolution in the presence of sublethal concentrations of ciprofloxacin were observed.
Cross-resistance to beta-lactam antibiotics after exposure to sub-MICs of ciprofloxacin.
Selection of subpopulations with decreased susceptibility to the beta-lactam antibiotics ceftazidime and meropenem was observed in the bacterial populations that evolved during exposure to sublethal concentrations of ciprofloxacin (see Table S3 in the supplemental material). This was in contrast to the evolution in the absence of ciprofloxacin, where no increase in resistance was observed, suggesting that the changes in the susceptibility to beta-lactams are related to the ciprofloxacin treatment. For example, for PAO1, ceftazidime- and meropenem-resistant subpopulations (++) growing up to 4 μg/ml and 0.5 μg/ml, respectively, were observed after 240 generations of growth in the presence of 1/4 the MIC of ciprofloxacin, but such subpopulations were not observed in bacterial cultures that evolved in the absence of the quinolone.
The efficacy of tobramycin seemed not to be affected by the decrease in the sensitivity to ciprofloxacin and beta-lactams.
DISCUSSION
The preexistence of antibiotic-resistant mutants in PAOΔmutS hypermutable bacterial populations and the rapid selection of these under monotherapy treatment with ciprofloxacin at high antibiotic concentrations (0.5 and 1 μg/ml) have been reported previously (20). The resistance mechanisms responsible for the resistance phenotype involved mutations in the quinolone resistance-determining regions (QRDR) gyrA and gyrB and upregulation of the efflux pumps MexAB-OprM and MexCD-OprJ (21).
Here, we show that exposure at a sub-MIC of ciprofloxacin (0.05 μg/ml, representing 1/4 the MIC) also rapidly selects for preexisting mutants with high-level resistance from P. aeruginosa populations. The faster occurrence of ciprofloxacin-resistant mutants in the mutator populations compared to PAO1 is not surprising, as the rate of adaptation depends on both the mutation rate and the population size (22) and only a bottleneck of 108 cells was tested in our study. The high mutation rates that initially confer a benefit because of a faster generation of adaptive mutations might present drawbacks during evolution, probably because the bacteria can accumulate deleterious mutations at a higher rate. The occurrence of trade-offs with disadvantages in secondary environments during the evolution of Salmonella in an animal model has previously been reported (22).
In accordance with these results, colonies unable to grow on glucose minimal medium (indicating possible auxotrophs) were found in the mutator populations during the 940 generations in rich LB medium. This indicates that in our long-term evolution study, accumulation of mutations caused a growth disadvantage of the evolved populations in secondary environments. This might explain the lack of transmissibility of mutator strains among CF patients (16) and the reduced potential for colonization of new environments (23, 24). Although the rich LB medium used in our study has a composition different from that of the CF sputum, it is worth mentioning that auxotrophy is a common P. aeruginosa phenotype selected during growth in the amino acid-rich environment in the CF sputum (25).
Generally, it is assumed that enrichment of resistant mutants is a consequence of using antibiotic concentrations placed within the mutant selection windows, i.e., the range between the concentration which blocks growth of the majority of susceptible pathogens (MIC) and the concentration that blocks growth of first-step resistant mutants (mutation prevention concentration) (13). However, our results imply that the biologically relevant sub-MIC selective window is much wider and has to include antibiotic concentrations below the MIC of the susceptible population. These concentrations have been shown to be several hundredfold lower than the MIC in E. coli (13).
At selection above the MIC, the main driving force of the selection is antibiotic resistance, while the fitness cost of the mutation is less critical. Even mutations with a very high cost will be selected, since competitors in the form of susceptible bacteria will be eliminated. At sub-MIC levels, however, the situation is different, since the susceptible bacteria will not die; they will only grow slower. Because of this, resistance mutations conferring a high fitness cost will not be enriched; only mutations where the fitness cost is lower than the growth reduction caused by the antibiotic in the susceptible bacteria will be competitive. This suggests that a new spectrum of low-cost or no-cost resistance mutations may be enriched under such conditions (13).
However, at concentrations that only slightly reduce and do not inhibit the bacterial growth, we found the same spectrum of mutations in gyrA and gyrB as observed under treatment with inhibitory concentrations (21, 26). Combined with the findings of no growth disadvantage of these mutants, these data suggest that the bacterial fitness is not affected by these mutations or that accumulation of compensatory mutations at other loci occurs fast. This is in accordance with previous studies showing that only 3 out of 8 P. aeruginosa gyrA mutants were affected in their growth rate (26). Identification of possible loci for compensatory mutations has recently been investigated by Wong et al. (6) by whole-genome sequencing. In that study, the authors found that a possible locus of compensatory mutations could be nusA, encoding an elongation factor, a putative kinase, and ate1, which encodes an arginyl-tRNA protein.
Enrichment of mutants with a high resistance level has been observed for Salmonella and E. coli exposed to low-antibiotic concentrations as well (13), probably due to their low (or lack of) fitness cost. Interestingly, the same type of mutations has been identified both in vitro and in vivo as well in a large number of ciprofloxacin-resistant clinical P. aeruginosa isolates, including strains from patients with cystic fibrosis (21, 27, 28).
Sequencing of the efflux pump regulator gene nfxB in ciprofloxacin-resistant isolates selected during the evolution study identified various mutations, especially in the mutator population (Table 1; see Table S1 in the supplemental material). However, no nfxB mutants were fixed during the long-term evolution experiment, probably due to the small advantage conferred by these mutations compared to the mutations in gyrA and gyrB. Mutations in nfxB, though, have been frequently found in clinical CF isolates (27), suggesting that these mutants are selected under in vivo conditions. The method used to isolate ciprofloxacin-resistant mutants, namely, isolation of resistant colonies in the inhibition zone of ciprofloxacin Etests, which enriches for ciprofloxacin-resistant mutants with high MIC levels, could have created a selection bias for gyrA and gyrB mutations. A population analysis of the whole bacterial population would have identified bacterial subpopulations with low MIC levels due to upregulation of efflux pumps as previously described (5, 17).
The MICs of resistant mutants selected from the PAOΔmutS mutator population (MIC = 128 μg/ml) was higher than that of the resistant mutants selected in PAO1 (MIC = 32 μg/ml). This is probably due to accumulation of mutations in different genes, such as seen with the gyrA parC double mutant that was selected late (after 940 generations) during the evolution experiment in the A and C lineages of PAOΔmutS. The late occurrence of the parC mutation confirms previous reports showing that the parC mutations are second-step mutations occurring in strains already having alterations in gyrA (29). It is noteworthy that strains with two alterations (Thr83Ile in GyrA plus Ser87Leu in ParC) were most frequently identified in clinical isolates (48 isolates) (29).
These findings suggest that mutators have an adaptive advantage also at sub-MIC levels of antibiotics and that the mutant selection window is wider than for wild-type strains, not only toward high (30) but also toward low antibiotic concentrations. A clear correlation between the type of mutations identified and the MIC was, however, difficult to assess, and this is in accordance with previous reports where the same mutations were identified in isolates with a wide range of MICs, probably due to the simultaneous presence of mutations in unidentified loci (21).
Worrisomely, cross-resistance to beta-lactams was observed in bacterial populations treated with ciprofloxacin at sub-MIC levels. This concentration of ciprofloxacin causes formation of reactive oxygen species as shown by measurement of HPF fluorescence. These reactive oxygen species are potentially mutagenic and can cause increased mutability, as previously suggested for strains under ciprofloxacin treatment (12). However, we did not observe an increase in the mutation frequency in the studied bacterial populations, which would especially be expected in the PAOMY-Mgm mutant. This mutant is unable to repair the oxidative DNA lesions that might occur during ciprofloxacin-related ROS exposure.
In conclusion, exposure of P. aeruginosa to sub-MICs of ciprofloxacin rapidly selects for mutants with high-level ciprofloxacin resistance and for subpopulations with an increased MIC to unrelated antibiotics. This raises the possibility of long-term persistence of resistant bacteria and spread of antibiotic resistance by exposure of commensal bacterial flora to low antibiotic concentrations.
Supplementary Material
Footnotes
Published ahead of print 17 June 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.00493-13.
REFERENCES
- 1.Yang L, Jelsbak L, Marvig RL, Damkiaer S, Workman CT, Rau MH, Hansen SK, Folkesson A, Johansen HK, Ciofu O, Høiby N, Sommer MO, Molin S. 2011. Evolutionary dynamics of bacteria in a human host environment. Proc. Natl. Acad. Sci. U. S. A. 108:7481–7486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oliver A, Canton R, Campo P, Baquero F, Blazquez J. 2000. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 288:1251–1254 [DOI] [PubMed] [Google Scholar]
- 3.Ciofu O, Riis B, Pressler T, Poulsen HE, Høiby N. 2005. Occurrence of hypermutable Pseudomonas aeruginosa in cystic fibrosis patients is associated with the oxidative stress caused by chronic lung inflammation. Antimicrob. Agents Chemother. 49:2276–2282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. 2007. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130:797–810 [DOI] [PubMed] [Google Scholar]
- 5.Morero NR, Monti MR, Argarana CE. 2011. Effect of ciprofloxacin concentration on the frequency and nature of resistant mutants selected from Pseudomonas aeruginosa mutS and mutT hypermutators. Antimicrob. Agents Chemother. 55:3668–3676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wong A, Rodrigue N, Kassen R. 2012. Genomics of adaptation during experimental evolution of the opportunistic pathogen Pseudomonas aeruginosa. PLoS Genet. 8:e1002928. 10.1371/journal.pgen.1002928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wong A, Kassen R. 2011. Parallel evolution and local differentiation in quinolone resistance in Pseudomonas aeruginosa. Microbiology 157:937–944 [DOI] [PubMed] [Google Scholar]
- 8.Moriarty TF, McElnay JC, Elborn JS, Tunney MM. 2007. Sputum antibiotic concentrations: implications for treatment of cystic fibrosis lung infection. Pediatr. Pulmonol. 42:1008–1017 [DOI] [PubMed] [Google Scholar]
- 9.Kohanski MA, Dwyer DJ, Collins JJ. 2010. How antibiotics kill bacteria: from targets to networks. Nat. Rev. Microbiol. 8:423–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Høiby N, Jarlov JO, Kemp M, Tvede M, Bangsborg JM, Kjerulf A, Pers C, Hansen H. 1997. Excretion of ciprofloxacin in sweat and multiresistant Staphylococcus epidermidis. Lancet 349:167–169 [DOI] [PubMed] [Google Scholar]
- 11.Weber SG, Gold HS, Hooper DC, Karchmer AW, Carmeli Y. 2003. Fluoroquinolones and the risk for methicillin-resistant Staphylococcus aureus in hospitalized patients. Emerg. Infect. Dis. 9:1415–1422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kohanski MA, DePristo MA, Collins JJ. 2010. Sublethal antibiotic treatment leads to multidrug resistance via radical-induced mutagenesis. Mol. Cell 37:311–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gullberg E, Cao S, Berg OG, Ilback C, Sandegren L, Hughes D, Andersson DI. 2011. Selection of resistant bacteria at very low antibiotic concentrations. PLoS Pathog. 7:e1002158. 10.1371/journal.ppat.1002158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elena SF, Lenski RE. 2003. Evolution experiments with microorganisms: the dynamics and genetic bases of adaptation. Nat. Rev. Genet. 4:457–469 [DOI] [PubMed] [Google Scholar]
- 15.Mandsberg LF, Ciofu O, Kirkby N, Christiansen LE, Poulsen HE, Høiby N. 2009. Antibiotic resistance in Pseudomonas aeruginosa strains with increased mutation frequency due to inactivation of the DNA oxidative repair system. Antimicrob. Agents Chemother. 53:2483–2491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ciofu O, Mandsberg LF, Bjarnsholt T, Wassermann T, Høiby N. 2010. Genetic adaptation of Pseudomonas aeruginosa during chronic lung infection of patients with cystic fibrosis: strong and weak mutators with heterogeneous genetic backgrounds emerge in mucA and/or lasR mutants. Microbiology 156:1108–1119 [DOI] [PubMed] [Google Scholar]
- 17.Mandsberg LF, Macia MD, Bergmann KR, Christiansen LE, Alhede M, Kirkby N, Høiby N, Oliver A, Ciofu O. 2011. Development of antibiotic resistance and up-regulation of the antimutator gene pfpI in mutator Pseudomonas aeruginosa due to inactivation of two DNA oxidative repair genes (mutY, mutM). FEMS Microbiol. Lett. 324:28–37 [DOI] [PubMed] [Google Scholar]
- 18.Hengzhuang W, Wu H, Ciofu O, Song Z, Høiby N. 2011. Pharmacokinetics/pharmacodynamics of colistin and imipenem on mucoid and nonmucoid Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 55:4469–4474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chopra I, O'Neill AJ, Miller K. 2003. The role of mutators in the emergence of antibiotic-resistant bacteria. Drug Resist. Updat. 6:137–145 [DOI] [PubMed] [Google Scholar]
- 20.Oliver A, Levin BR, Juan C, Baquero F, Blazquez J. 2004. Hypermutation and the preexistence of antibiotic-resistant Pseudomonas aeruginosa mutants: implications for susceptibility testing and treatment of chronic infections. Antimicrob. Agents Chemother. 48:4226–4233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Macia MD, Borrell N, Segura M, Gomez C, Perez JL, Oliver A. 2006. Efficacy and potential for resistance selection of antipseudomonal treatments in a mouse model of lung infection by hypermutable Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 50:975–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nilsson AI, Kugelberg E, Berg OG, Andersson DI. 2004. Experimental adaptation of Salmonella typhimurium to mice. Genetics 168:1119–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mena A, Macia MD, Borrell N, Moya B, de Francisco T, Perez JL, Oliver A. 2007. Inactivation of the mismatch repair system in Pseudomonas aeruginosa attenuates virulence but favors persistence of oropharyngeal colonization in cystic fibrosis mice. J. Bacteriol. 189:3665–3668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Montanari S, Oliver A, Salerno P, Mena A, Bertoni G, Tummler B, Cariani L, Conese M, Döring G, Bragonzi A. 2007. Biological cost of hypermutation in Pseudomonas aeruginosa strains from patients with cystic fibrosis. Microbiology 153:1445–1454 [DOI] [PubMed] [Google Scholar]
- 25.Barth AL, Pitt TL. 1996. The high amino-acid content of sputum from cystic fibrosis patients promotes growth of auxotrophic Pseudomonas aeruginosa. J. Med. Microbiol. 45:110–119 [DOI] [PubMed] [Google Scholar]
- 26.Kugelberg E, Lofmark S, Wretlind B, Andersson DI. 2005. Reduction of the fitness burden of quinolone resistance in Pseudomonas aeruginosa. J. Antimicrob. Chemother. 55:22–30 [DOI] [PubMed] [Google Scholar]
- 27.Jalal S, Ciofu O, Høiby N, Gotoh N, Wretlind B. 2000. Molecular mechanisms of fluoroquinolone resistance in Pseudomonas aeruginosa isolates from cystic fibrosis patients. Antimicrob. Agents Chemother. 44:710–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee JK, Lee YS, Park YK, Kim BS. 2005. Alterations in the GyrA and GyrB subunits of topoisomerase II and the ParC and ParE subunits of topoisomerase IV in ciprofloxacin-resistant clinical isolates of Pseudomonas aeruginosa. Int. J. Antimicrob. Agents 25:290–295 [DOI] [PubMed] [Google Scholar]
- 29.Akasaka T, Tanaka M, Yamaguchi A, Sato K. 2001. Type II topoisomerase mutations in fluoroquinolone-resistant clinical strains of Pseudomonas aeruginosa isolated in 1998 and 1999: role of target enzyme in mechanism of fluoroquinolone resistance. Antimicrob. Agents Chemother. 45:2263–2268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Macia MD, Perez JL, Molin S, Oliver A. 2011. Dynamics of mutator and antibiotic-resistant populations in a pharmacokinetic/pharmacodynamic model of Pseudomonas aeruginosa biofilm treatment. Antimicrob. Agents Chemother. 55:5230–5237 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.