Abstract
Trimethoprim-sulfamethoxazole (co-trimoxazole) is the primary drug used for oral eradication therapy of Burkholderia pseudomallei infections (melioidosis). Here, we demonstrate that trimethoprim resistance is widespread in clinical and environmental isolates from northeast Thailand and northern Australia. This resistance was shown to be due to BpeEF-OprC efflux pump expression. No dihydrofolate reductase target mutations were involved, although frequent insertion of ISBma2 was noted within the putative folA transcriptional terminator. All isolates tested remained susceptible to trimethoprim-sulfamethoxazole, suggesting that resistance to trimethoprim alone in these strains probably does not affect the efficacy of co-trimoxazole therapy.
INTRODUCTION
Burkholderia pseudomallei is a saprophytic Gram-negative bacterium found mostly in soil and water in many subtropical and tropical regions of the world, including northern Australia and southeast Asia (1, 2). B. pseudomallei is the etiologic agent of the multifacted disease melioidosis (2–6). Treatment of melioidosis is complicated by the intrinsic resistance of B. pseudomallei to many classes of antimicrobials (7, 8). The current recommended therapy includes an initial intensive phase followed by a lengthy eradication phase to prevent relapse (6, 9, 10). Most patients require at least 10 to 14 days of parenteral ceftazidime or a carbapenem followed by 12 to 20 weeks of oral trimethoprim-sulfamethoxazole with or without doxycycline. Trimethoprim and sulfamethoxazole inhibit the folic acid biosynthetic pathway by targeting dihydrofolate reductase (FolA) and dihydropteroate synthase (FolP), respectively (11). The synergistic trimethoprim-sulfamethoxazole combination, co-trimoxazole, has a potent antimicrobial effect. B. pseudomallei co-trimoxazole resistance was previously documented in regions where the disease is endemic (12–16), and rates range from 2.5% in Australia (13) to 13 to 16% in Thailand (12, 14). Previous studies have identified and characterized trimethoprim resistance mechanisms including resistant dihydrofolate reductases in other organisms, such as Escherichia coli (11, 17), but in B. pseudomallei, trimethoprim resistance has been studied only indirectly in surrogate (18) or closely related (19) bacteria, showing that efflux could play an important role in resistance. The objective of this study was to identify and characterize the mechanism responsible for trimethoprim resistance in clinical and environmental B. pseudomallei isolates from northeast Thailand and northern Australia.
MATERIALS AND METHODS
Bacterial strains.
B. pseudomallei strain 1026b was used as a prototype strain for all experiments in this study (20, 21). Additionally, a collection of 30 clinical and 30 environmental isolates from Thailand (isolated in 2001 and 1990 to 2001, respectively) and 4 clinical isolates and 1 environmental isolate from Australia (isolated between 1994 and 1997) were examined. All procedures involving B. pseudomallei were performed in select-agent-approved biosafety level 3 (BSL3) facilities in the Rocky Mountain Regional Biosafety Laboratory at Colorado State University, using approved select-agent-compliant procedures and protocols.
Antimicrobial susceptibility testing.
Trimethoprim, sulfamethoxazole, and co-trimoxazole susceptibilities were assessed by determining MICs using the Etest method according to the manufacturer's instructions (AB bioMérieux, Marcy l'Etoile, France). Briefly, strains were grown to mid-log phase (optical density at 600 nm [OD600] = 0.6 to 0.8) and diluted to a 0.5 McFarland standard in 0.85% sterile saline. The resulting bacterial cell suspension was then used to swab Mueller-Hinton II agar plates (Becton, Dickinson and Company, Sparks, MD) to which the Etest strips were applied. MIC results were determined following 16 to 20 h of incubation at 37°C. Results were read at 80% inhibition, again according to the manufacturer's guidelines. Since there are no established breakpoints for non-Enterobacteriaceae, the following Enterobacteriaceae MIC cutoffs were used to define susceptibility and resistance for trimethoprim alone (≤8 μg/ml for susceptible and >8 μg/ml for resistant) according to CLSI guidelines (see Table 2A in reference 22). The non-Enterobacteriaceae sulfonamide cutoffs were used for sulfamethoxazole alone (≤256 μg/ml for susceptible and >256 μg/ml from resistant) according to CLSI guidelines (see Table 2B-5 in reference 22), while CLSI standard MIC cutoffs for B. pseudomallei were used for co-trimoxazole (trimethoprim-sulfamethoxazole, ≤2/38 μg/ml for susceptible and >2/38 μg/ml for resistant) (see Table 2K in reference 22).
Amplification and sequencing of folA.
The folA coding sequence was PCR amplified in four independent PCRs from genomic DNA isolated with PureGene Core kit A (Qiagen, Valencia, CA), using primers P1966 (5′-CTTCCGGCCTCTTTTCTTTC) (Integrated DNA Technologies, Coralville, IA) and P1967 (5′-GTGCTGATCGAGCAGATGAC) and Platinum Taq DNA Polymerase High Fidelity (Life Technologies Corporation, Grand Island, NY). The PCR products were pooled for each strain and purified from agarose gels by using the GenElute gel extraction kit (Sigma-Aldrich, St. Louis, MO). The PCR products were sequenced using P1966 and P1967 at the Colorado State University Proteomics and Metabolomics facility. Alignments of folA sequences from experimental samples and comparison with the 1026b folA sequence were performed by using ClustalW2 (23).
Multiplex ISBma2 PCR.
Primers P2578 (5′-CCAACGATTTCACGTACGC), P2569 (5′-CCGTACAGCACGACCAATC), and P2579 (5′-GACGTTGACCTGGACCTCAC) were designed and used in a multiplex PCR to determine the orientation of ISBma2 in the clinical and environmental strains. P2578, P2569, and P2579 were all added at final concentrations of 0.6 pmol/μl, and standard Taq polymerase (New England BioLabs, Waltham, MA) was used. PCR conditions were an initial denaturation step at 95°C for 2 min and 30 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 1.5 min, followed by a final extension step at 72°C for 10 min.
Markerless deletion of bpeEF-oprC.
A 4,314-bp region of the bpeEF-oprC operon was deleted in several of the clinical and environmental isolates by allelic exchange using the pEXKm5 vector system and sucrose counterselection, as described previously (24).
Complementation of bpeEF-oprC deletions.
Genetic complementation was accomplished by utilizing the mini-Tn7 system, which allows for stable and site-specific single-copy insertions into the B. pseudomallei genome at three possible glmS-associated sites (25). The inducible E. coli lac operon Ptac promoter was used to express the bpeEF-oprC operon, which originated from strain 1026b (18). BpeEF-OprC expression in the complemented strains was induced by addition of isopropyl-β-d-thiogalactopyranoside (IPTG; Gold Biotechnology, St. Louis, MO) at a final concentration of 1 mM.
RT-qPCR.
Expression levels of bpeF and bpeT mRNAs were analyzed in bacteria grown to mid-log phase (OD600 = 0.6 to 0.8) in Lennox Luria broth (Mo Bio Laboratories, Carlsbad, CA), at which point bpeEF-oprC expression either remained uninduced or was induced for 1 h by the addition of trimethoprim to a final concentration of 32 μg/ml. RNA extraction, cDNA synthesis, and reverse transcription-quantitative PCR (RT-qPCR) were done as previously described (26, 27), except that the RNeasy Protect Bacteria minikit (Qiagen, Valencia, CA) was used for RNA extraction. 23S rRNA was used as the housekeeping control. The primer sets used were Bp23S_F and Bp23S_R for 23S rRNA, bpeF-F1_RT and bpeF-R1_RT for bpeF (26), and bpeT_RT_for (P1814) (5′-GAGCTTTCAGGTCAACAACC) and bpeT_RT_rev (P1815) (5′-GTGAGTGGAATTCGCAGAG) for bpeT.
RESULTS AND DISCUSSION
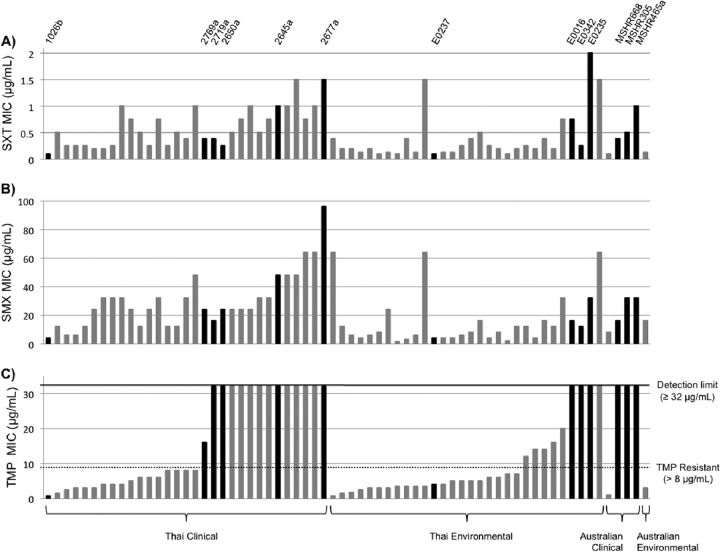
To evaluate the innate frequency of trimethoprim, sulfamethoxazole, and co-trimoxazole resistance in B. pseudomallei clinical and environmental isolates, MICs were determined for a collection of 60 clinical and environmental isolates from northeast Thailand and 5 clinical isolates and 1 environmental isolate from northern Australia. Using the susceptibility criteria described in Materials and Methods, MIC testing of strains from Thailand showed that 47% of clinical strains (14/30 strains) and 30% of environmental strains (9/30) were trimethoprim resistant, with MICs ranging from 12 μg/ml to the detection limit of ≥32 μg/ml (Fig. 1). None of these isolates were resistant to sulfamethoxazole or co-trimoxazole. The frequency of trimethoprim resistance in Australian isolates was 60% (3/5 isolates), and again, these 5 isolates were all susceptible to sulfamethoxazole and co-trimoxazole. These data show that although trimethoprim resistance was highly prevalent in our collection of B. pseudomallei isolates, none of the 65 strains tested showed clinically significant co-trimoxazole resistance. We would therefore expect treatment with the standard regimen to still be effective for infections caused by the strains investigated in this study.
Fig 1.
Distribution of trimethoprim, sulfamethoxazole, and trimethoprim-sulfamethoxazole susceptibilities in B. pseudomallei clinical and environmental isolates. MICs of the trimethoprim-sulfamethoxazole drug combination (SXT) (A), sulfamethoxazole (SMX) (B), and trimethoprim (TMP) (C) were determined for a collection of 35 clinical and 31 environmental isolates from Thailand and Australia. All 66 isolates tested were susceptible to trimethoprim-sulfamethoxazole (A) and sulfamethoxazole (B). However, isolates in panel C with values above the dotted line are classified as trimethoprim resistant, while others below the dotted line are trimethoprim susceptible. Several isolates from this collection were selected for further studies (black bars), and the names of these 13 strains are indicated at the top of panel A.
Mutations affecting the trimethoprim target dihydrofolate reductase (FolA) can cause resistance to this antibiotic. We selected nine Thai isolates (eight of which are trimethoprim resistant) to compare to the trimethoprim-susceptible 1026b reference strain (20, 21). Two mutations, V77A and A144T, were found in five of the eight trimethoprim-resistant isolates compared to reference strain 1026b. However, these conservative mutations were not linked to trimethoprim resistance in these isolates.
While attempting to PCR amplify the folA region in the nine isolates, we noticed that four strains (2665a, 2769a, E0016, and E0342) yielded the same 694-bp DNA fragment as reference strain 1026b (Fig. 2A). The remaining four isolates (2650a, 2677a, 2719a, and E0235), however, showed a substantially larger (≈2.3-kb) fragment when using the same primers. DNA sequencing of this fragment with P1967 revealed the presence of an ISBma2 insertion sequence (28). Further analyses were conducted by using representative strains E0016 (694-bp fragment) and 2650a (≈2.3-kb fragment). Employing genomic DNAs as templates and PCR amplification with the subset of primers shown in Fig. 2A for E0016 and Fig. 2B for 2650a followed by sequencing confirmed for E0016 the identical sequence and gene order found in 1026b. In strain 2650a, the 1,596-bp ISBma2 sequence was inserted into the 130-bp folA-pmbA intergenic region in a palindromic sequence located between nucleotides 47 and 75 downstream of folA that most likely serves as this gene's transcriptional terminator. The insertion site and orientation of ISBma2 were identical to those found in the genome of strain K96243 (29). In both 1026b and K96243, the folA gene and neighboring genes are located on chromosome 1 but in different locations and in reverse order (Fig. 2). The two PCR amplification patterns observed for the isolates analyzed in this study indicate two distinct populations of strains, one similar to 1026b and another similar to K96243. The frequency of the ISBma2 insertion at this site was determined for the 65 clinical and environmental isolates using PCR amplification of the folA region with P1966 and P1967. This analysis showed that 38% of clinical and 45% of environmental isolates contain the ISBma2 insertion at this locus. Additionally, DNA sequencing of this region in strain MSHR305 and comparison to the whole-genome shotgun sequence (30) revealed that the ISBma2 element was in the opposite orientation, relative to K96243, and thus can insert at this site in either orientation. For the strains that contained ISBma2, a multiplex PCR was used to determine the orientation of the insertion element. There was a near-equal distribution of ISBma2 orientation, with 52% having the same orientation as K96243, resulting in a 440-bp product, and 48% having the opposite orientation, resulting in a 647-bp product. There was a slight bias in the orientation when environmental isolates were compared to clinical isolates, where 64% of environmental and 38% of clinical isolates were in the same orientation as in strain K96243. However, a significantly larger sample size would be needed to determine the true orientation bias. The location of ISBma2 most likely does not affect folA expression but has implications regarding the design of PCR primers for folA amplification for diagnostic purposes.
Fig 2.
Genomic location and organization of the folA region of B. pseudomallei in the two sequenced prototypes 1026b (A) and K96243 (B). Chromosome 1 sequence coordinates and gene nomenclatures are taken from the GenBank entries for strains 1026b (accession number NC_017831.1) and K96243 (accession number NC_006350.1). Gene annotations are as follows: _I0835 (short for BP1026B_I0835) and BPSL2475, σ54-dependent transcriptional regulators; folA, dihydrofolate reductase; pmbA, protein belonging to the peptidase U62 family; tnpA, transposase. The same sequences and gene organizations were confirmed for Thai isolates E0016 (1026b-like) and 2650a (K96243-like). Horizontal black lines indicate PCR fragments obtained with the indicated primers. Primers pairs used were P1966 and P1967, which amplify the folA gene with and without ISBma2, and P2578 and P2579, which amplify a fragment containing the folA-ISBma2 junction sequences. In isolates where ISBma2 is in the opposite orientation, primer set P2578 and P2579 is replaced by P2578 and P2569, and this combination yields a 647-bp fragment containing the folA-ISBma2 junction sequences (not shown). All primer sequences are given in the text. The folA upstream region and 5′ coding sequences can be amplified by using primer pair P2182 (5′-CTGTATCGGCTGATGGTGTC) and P2183 (5′-AGGCCTTCCTCGTACAGTTG). In panel A, the sequence with underlined inverted repeats indicates the putative folA transcriptional terminator. In panel B, sequences that compose the 5′ and 3′ ISBma2 inverted repeats (IR) (underlined) and the 23-bp duplicated genomic DNA segments (in boldface type) are indicated. The sequences of ISBam2 elements and their organization as well as those of the duplicated target DNA segments are consistent with previously reported data on ISBam2 (28).
The BpeEF-OprC efflux pump is encoded by the bpeEF-oprC genes, which are the distal genes of the llpE-bpeEF-oprC operon located on B. pseudomallei chromosome 2 (26, 29). Expression of this operon is governed by the LysR-type regulator BpeT, which is encoded by a gene located immediately upstream of the llpE-bpeEF-oprC operon. BpeEF-OprC has been shown to efflux trimethoprim when expressed in Pseudomonas aeruginosa (18), in Burkholderia thailandensis isolates that are resistant to chloramphenicol (19), and in B. pseudomallei isolates that express BpeEF-OprC as a result of a BpeT truncation (21). To investigate a potential contribution of this efflux pump to trimethoprim resistance in clinical and environmental B. pseudomallei isolates, a 4,314-bp segment of the llpE-bpeEF-oprC operon was deleted from 11 trimethoprim-resistant isolates and trimethoprim-susceptible reference strains 1026b and E0237. Deletion of bpeEF-oprC from the trimethoprim-resistant strains resulted in susceptible mutant derivatives with at least 10-fold decreases in MIC. A lower trimethoprim MIC was observed for E0237, but the susceptibility of 1026b remained unchanged(Table 1). No significant trimethoprim susceptibility changes were observed in uninduced complemented strains, but induction of BpeEF-OprC expression resulted in significant MIC increases such that all of the isolates, except for 1026b, became trimethoprim resistant (Table 1). Similar results were observed with strains containing deleted and complemented bpeT, respectively (data not shown). These results suggest that expression of the BpeEF-OprC pump is required for trimethoprim resistance in these isolates.
Table 1.
Trimethoprim, sulfamethoxazole, and trimethoprim-sulfamethoxazole susceptibilities of selected clinical and environmental isolates with and without the BpeEF-OprC efflux pumpa
| Strain | MIC (μg/ml) |
|||||
|---|---|---|---|---|---|---|
| SXT (bpeE+F+-oprC+) | SMX (bpeE+F+-oprC+) | TMP |
||||
| bpeE+F+-oprC+ | Δ(bpeEF-oprC) | Δ(bpeEF-oprC), complemented,b uninduced | Δ(bpeEF-oprC), complemented,b +1 mM IPTG | |||
| 1026b | 0.094/1.79 | 4 | 0.75 | 0.5 | 0.5 | 6 |
| Thailand Clinical | ||||||
| 2650a | 1/19 | 8 | ≥32 | 1.5 | 1.5 | ≥32 |
| 2665a | 1/19 | 16 | ≥32 | 1.5 | 1.5 | 16 |
| 2677a | 1.5/28.5 | 24 | ≥32 | 2 | 2 | ≥32 |
| 2719a | 1/19 | 12 | ≥32 | 1 | 1 | 16 |
| 2769a | 0.75/14.25 | 12 | 16 | 1.5 | 2 | ≥32 |
| Thailand environmental | ||||||
| E0016 | 0.75/14.25 | 12 | ≥32 | 1 | 1.5 | 16 |
| E0235 | 1.5/28.5 | 24 | ≥32 | 1.5 | 3 | ≥32 |
| E0237 | 0.5/9.5 | 8 | 4 | 1 | 2 | 16 |
| E0342 | 0.75/14.25 | 24 | ≥32 | 1 | 1.5 | 16 |
| Australia clinical | ||||||
| MSHR305 | 0.5/9.5 | 32 | ≥32 | 2 | 2 | 16 |
| MSHR668 | 0.38/7.22 | 16 | ≥32 | 2 | 2 | 32 |
| MSHR465a | 1/19 | 32 | ≥32 | 3 | 2 | ≥32 |
Abbreviations: TMP, trimethoprim; SMX, sulfamethoxazole; SXT, trimethoprim-sulfamethoxazole.
The respective strains containing the Δ(bpeEF-oprC) mutation have a mini-Tn7T-Ptac-bpeE+F+-oprC+ element integrated at the chromosomal glmS2 locus, except for the MSHR668 derivative, which has the same element integrated at glmS3. The bpeE+F+-oprC+ operon contained on this mini-Tn7 element is derived from prototype strain 1026b (18).
To assess whether mutations in structural pump components were contributing to trimethoprim resistance, we sequenced the bpeT-llpE-bpeE and oprC regions in nine Thai isolates and the bpeF gene in three of those isolates using a primer-walking strategy. These analyses showed that the DNA sequence of the entire region was highly conserved, with few synonymous and conservative mutations. For example, compared to the 1026b sequence, the following mutations were found in ≤22% of the sequenced trimethoprim-resistant isolates: A401T in BpeE and V78A, A207Q, and T508A in OprC. There were only a few conservative mutations in the bpeT gene and no mutations in the bpeT-llpE intergenic region containing the putative promoter regions for bpeT and the llpE-bpeEF-oprC operon. Given the conservative nature of the observed mutations and the results of the complementation study which showed that the 1026b operon sequence was sufficient for the trimethoprim-resistant phenotype, it is unlikely that changes in structural BpeEF-OprC components are root causes for resistance in any of the tested isolates.
To gauge expression levels of the BpeT regulator and BpeEF-OprC, RT-qPCR was used to determine the relative bpeT and bpeF mRNA expression levels. Interestingly, in uninduced cells, bpeF mRNA levels were very similar between the nine Thai isolates tested and 1026b. However, when induced with 32 μg/ml trimethoprim, isolates with trimethoprim MICs of ≥32 μg/ml had increased bpeF mRNA levels, with fold increases ranging from 3.2 to 6.6. In contrast, two isolates, E0237 and 2769a, with respective MICs of 4 μg/ml and 16 μg/ml, and 1026b exhibited no increase in bpeF expression levels when induced with 32 μg/ml of trimethoprim. As this high concentration of trimethoprim may adversely affect strains with MICs lower than 32 μg/ml, the experiment was repeated by performing induction with trimethoprim at one-half the respective MICs for 1026b, 2769a, and E0237 for 1 h. However, even when using one-half the MICs of trimethoprim for induction, no increases in bpeF expression levels were observed. These data suggest that high-level trimethoprim resistance results from overexpression of the BpeEF-OprC efflux pump. Repetition of these experiments for bpeT showed no changes in regulatory gene expression both with and without trimethoprim induction. Since DNA sequencing revealed only conservative mutations in bpeT and RT-qPCR analysis indicated no changes in expression levels of this regulator, we conclude that BpeEF-OprC expression in the trimethoprim-resistant clinical and environmental isolates tested is governed by a to-date-unidentified regulatory mechanism(s).
This is the first study aimed at elucidation of the molecular mechanisms governing trimethoprim resistance in clinical and environmental B. pseudomallei isolates. The results show that resistance to trimethoprim alone is frequent in B. pseudomallei strains. However, resistance to sulfamethoxazole and co-trimoxazole was not detected in any of these clinical and environmental isolates tested. In all of the Australian and Thai isolates assessed, trimethoprim resistance was attributed to expression of BpeEF-OprC but not changes in the dihydrofolate reductase target, indicating that efflux is the predominant trimethoprim resistance mechanism in B. pseudomallei. Increased transcription of the bpeEF-oprC structural genes in the presence of trimethoprim led to high-level resistance, with MICs above the detection limit of 32 μg/ml. The regulatory mechanism(s) governing BpeEF-OprC expression in these isolates remains unknown. In the course of these studies, we discovered that the genomic location and organization of genes in the immediate folA region follow a distinct pattern that, in reference to three sequenced prototypes, allows grouping of strains into K96243-like, MSHR305-like, and 1026b-like strains.
ACKNOWLEDGMENTS
Funding for this project was provided through the Rocky Mountain Regional Center of Excellence for Biodefense and Emerging Infectious Diseases Research (funded by NIH NIAID grant AI065357). Vanaporn Wuthiekanun was supported by the Wellcome Trust, and Sharon J. Peacock was supported by the NIHR Cambridge Biomedical Research Centre.
We acknowledge the support of Takehiko Mima in the initial phases of this study. We thank Bart Currie, Menzies School of Heath Research, Darwin, Australia, and Apichai Tuanyok and Paul Keim from Northern Arizona University for providing the Australian B. pseudomallei isolates used in this research and for their insightful comments.
Footnotes
Published ahead of print 1 July 2013
REFERENCES
- 1.Currie BJ, Dance DAB, Cheng AC. 2008. The global distribution of Burkholderia pseudomallei and melioidosis: an update. Trans. R. Soc. Trop. Med. Hyg. 102(Suppl 1):S1–S4. 10.1016/S0035-9203(08)70002-6 [DOI] [PubMed] [Google Scholar]
- 2.Wiersinga WJ, Currie BJ, Peacock SJ. 2012. Melioidosis. N. Engl. J. Med. 367:1035–1044 [DOI] [PubMed] [Google Scholar]
- 3.Wiersinga WJ, van der Poll T, White NJ, Day NP, Peacock SJ. 2006. Melioidosis: insights into the pathogenicity of Burkholderia pseudomallei. Nat. Rev. Microbiol. 4:272–282 [DOI] [PubMed] [Google Scholar]
- 4.Cheng AC, Currie BJ. 2005. Melioidosis: epidemiology, pathophysiology, and management. Clin. Microbiol. Rev. 18:383–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Currie BJ, Ward L, Cheng AC. 2010. The epidemiology and clinical spectrum of melioidosis: 540 cases from the 20 year Darwin prospective study. PLoS Negl. Trop. Dis. 4:e900. 10.1371/journal.pntd.0000900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Limmathurotsakul D, Peacock SJ. 2011. Melioidosis: a clinical overview. Br. Med. Bull. 99:125–139 [DOI] [PubMed] [Google Scholar]
- 7.Wuthiekanun V, Peacock SJ. 2006. Management of melioidosis. Expert Rev. Anti Infect. Ther. 4:445–455 [DOI] [PubMed] [Google Scholar]
- 8.Schweizer HP. 2012. Mechanisms of antibiotic resistance in Burkholderia pseudomallei: implications for treatment of melioidosis. Future Microbiol. 7:1389–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peacock SJ, Schweizer HP, Dance DAB, Smith TL, Gee JE, Wuthiekanun V, DeShazer D, Steinmetz I, Tan P, Currie BJ. 2008. Accidental laboratory exposure to Burkholderia pseudomallei and Burkholderia mallei. Emerg. Infect. Dis. 14:e2. 10.3201/eid1407.071501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lipsitz R, Garges S, Aurigemma R, Baccam P, Blaney DD, Cheng AC, Currie BJ, Dance D, Gee JE, Larsen J, Limmathurotsakul D, Morrow MG, Norton R, O'Mara E, Peacock SJ, Pesik N, Rogers LP, Schweizer HP, Steinmetz I, Tan G, Tan P, Wiersinga WJ, Wuthiekanun V, Smith TL. 2012. Workshop on treatment of and postexposure prophylaxis for Burkholderia pseudomallei and B. mallei infection, 2010. Emerg. Infect. Dis. 18:e2. 10.3201/eid1812.120638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walsh C. 2003. Antibiotics: actions, origins, resistance. ASM Press, Washington, DC [Google Scholar]
- 12.Lumbiganon P, Tattawasatra U, Chetchotisakd P, Wongratanacheewin S, Thinkamrop B. 2000. Comparison between the antimicrobial susceptibility of Burkholderia pseudomallei to trimethoprim-sulfamethoxazole by standard disk diffusion method and by minimal inhibitory concentration determination. J. Med. Assoc. Thai. 83:856–860 [PubMed] [Google Scholar]
- 13.Piliouras P, Ulett GC, Ashurst-Smith C, Hirst RG, Norton RE. 2002. A comparison of antibiotic susceptibility testing methods for cotrimoxazole with Burkholderia pseudomallei. Int. J. Antimicrob. Agents 19:427–429 [DOI] [PubMed] [Google Scholar]
- 14.Wuthiekanun V, Cheng AC, Chierakul W, Amornchai P, Limmathurotsakul D, Chaowagul W, Simpson AJ, Short JM, Wongsuvan G, Maharjan B, White NJ, Peacock SJ. 2005. Trimethoprim/sulfamethoxazole resistance in clinical isolates of Burkholderia pseudomallei. J. Antimicrob. Chemother. 55:1029–1031 [DOI] [PubMed] [Google Scholar]
- 15.Raja NS. 2008. Cases of melioidosis in a university teaching hospital in Malaysia. J. Microbiol. Immunol. Infect. 41:174–179 [PubMed] [Google Scholar]
- 16.Paveenkittiporn W, Apisarnthanarak A, Dejsirilert S, Trakulsomboon S, Thongmali O, Sawanpanyalert P, Aswapokee N. 2009. Five-year surveillance for Burkholderia pseudomallei in Thailand from 2000 to 2004: prevalence and antimicrobial susceptibility. J. Med. Assoc. Thai. 9(Suppl 4):S46–S52 [PubMed] [Google Scholar]
- 17.Sköld O. 2001. Resistance to trimethoprim and sulfonamides. Vet. Res. 32:261–273 [DOI] [PubMed] [Google Scholar]
- 18.Kumar A, Chua K-L, Schweizer HP. 2006. Method for regulated expression of single-copy efflux pump genes in a surrogate Pseudomonas aeruginosa strain: identification of the BpeEF-OprC chloramphenicol and trimethoprim efflux pump of Burkholderia pseudomallei 1026b. Antimicrob. Agents Chemother. 50:3460–3463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Biot FV, Valade E, Garnotel E, Chevalier J, Villard C, Thibault FM, Vidal DR, Pages JM. 2011. Involvement of the efflux pumps in chloramphenicol selected strains of Burkholderia thailandensis: proteomic and mechanistic evidence. PLoS One 6:e16892. 10.1371/journal.pone.0016892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DeShazer D, Brett P, Carlyon R, Woods D. 1997. Mutagenesis of Burkholderia pseudomallei with Tn5-OT182: isolation of motility mutants and molecular characterization of the flagellin structural gene. J. Bacteriol. 179:2116–2125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hayden HS, Lim R, Brittnacher MJ, Sims EH, Ramage ER, Fong C, Wu Z, Crist E, Chang J, Zhou Y, Radey M, Rohmer L, Haugen E, Gillett W, Wuthiekanun V, Peacock SJ, Kaul R, Miller SI, Manoil C, Jacobs MA. 2012. Evolution of Burkholderia pseudomallei in recurrent melioidosis. PLoS One 7:e36507. 10.1371/journal.pone.0036507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clinical and Laboratory Standards Institute 2010. Performance standards for antimicrobial susceptibility testing; twentieth informational supplement, CSLI document M100-S20 Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 23.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 24.Lopez CM, Rholl DA, Trunck LA, Schweizer HP. 2009. Versatile dual-technology system for markerless allele replacement in Burkholderia pseudomallei. Appl. Environ. Microbiol. 75:6496–6503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi K-H, Mima T, Casart Y, Rholl D, Kumar A, Beacham IR, Schweizer HP. 2008. Genetic tools for select-agent-compliant manipulation of Burkholderia pseudomallei. Appl. Environ. Microbiol. 74:1064–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar A, Mayo M, Trunck LA, Cheng AC, Currie BJ, Schweizer HP. 2008. Expression of resistance-nodulation-cell division efflux pumps in commonly used Burkholderia pseudomallei strains and clinical isolates from Northern Australia. Trans. R. Soc. Trop. Med. Hyg. 102(Suppl 1):S145–S151. 10.1016/S0035-9203(08)70032-4 [DOI] [PubMed] [Google Scholar]
- 27.Mima T, Schweizer HP. 2010. The BpeAB-OprB efflux pump of Burkholderia pseudomallei 1026b does not play a role in quorum sensing, virulence factor production, or extrusion of aminoglycosides but is a broad-spectrum drug efflux system. Antimicrob. Agents Chemother. 54:3113–3120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song H, Hwang J, Yi H, Ulrich RL, Yu Y, Nierman WC, Kim HS. 2010. The early stage of bacterial genome-reductive evolution in the host. PLoS Pathog. 6:e1000922. 10.1371/journal.ppat.1000922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holden MTG, Titball RW, Peacock SJ, Cerdeno-Tarraga AM, Atkins TP, Crossman LC, Pitt TL, Churcher C, Mungall KL, Bentley SD, Sebaihia M, Thomson NR, Bason N, Beacham IR, Brooks K, Brown KA, Brown NF, Challis GL, Cherevach I, Chillingworth T, Cronin A, Crossett B, Davis P, DeShazer D, Feltwell T, Fraser A, Hance Z, Hauser H, Holroyd S, Jagels K, Keith KE, Maddison M, Moule S, Price C, Quail MA, Rabbinowitsch E, Rutherford K, Sanders M, Simmonds M, Songsivilai S, Stevens K, Tumapa S, Vesaratchavest M, Whitehead S, Yeats C, Barrell BG, Oyston PCF, Parkhill J. 2004. Genomic plasticity of the causative agent of melioidosis, Burkholderia pseudomallei. Proc. Natl. Acad. Sci. U. S. A. 101:14240–14245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tuanyok A, Leadem BR, Auerbach RK, Beckstrom-Sternberg SM, Beckstrom-Sternberg JS, Mayo M, Wuthiekanun V, Brettin TS, Nierman WC, Peacock SJ, Currie BJ, Wagner DM, Keim P. 2008. Genomic islands from five strains of Burkholderia pseudomallei. BMC Genomics 9:566. 10.1186/1471-2164-9-566 [DOI] [PMC free article] [PubMed] [Google Scholar]