Abstract
The CmeABC efflux pump in Campylobacter jejuni confers resistance to structurally divergent antimicrobials, and inhibition of CmeABC represents a promising strategy to control antibiotic-resistant Campylobacter. Antisense peptide nucleic acids (PNAs) targeting the three components of CmeABC were evaluated for inhibition of CmeABC expression. The result revealed a synergistic effect of the PNAs targeting CmeA and CmeB on sensitizing C. jejuni to antibiotics. This finding further demonstrates the feasibility of using PNAs to potentiate antibiotics against antibiotic-resistant Campylobacter.
TEXT
Campylobacter jejuni is a major food-borne pathogen accounting for 400 to 500 million cases of diarrhea each year worldwide (1). This organism is increasingly resistant to clinically important antibiotics, particularly fluoroquinolone (FQ) antimicrobials (2). The CmeABC efflux pump in Campylobacter confers resistance to a broad range of antimicrobials and toxic compounds (3). In addition, this efflux pump functions synergistically with other mechanisms (e.g., spontaneous mutations in antibiotic targets) in mediating high-level resistance to antibiotics (3). CmeABC is also important for bile resistance and is essential for Campylobacter colonization in the intestinal tract of animal hosts (4).
Due to the significant role of CmeABC in antibiotic resistance, inhibition of CmeABC represents a promising strategy to control antibiotic-resistant Campylobacter. Efflux pump inhibitors, such as phenyl-arginine-β-naphthylamide (PAβN), have been evaluated to inhibit antibiotic efflux in Campylobacter (5). The general findings are that PAβN has a significant impact on erythromycin (Ery) but has limited or no effect on the MICs of FQ antimicrobials in Campylobacter (5–7). Peptide nucleic acids (PNAs) are synthetic homologs of nucleic acids in which the phosphate backbone of polynucleotides is replaced by a flexible pseudopeptide polymer (8). PNAs function as antisense agents by binding specifically to complementary sequences in DNA and RNA and inhibiting gene expression and/or translation (9). Recently, we showed that a CmeA-specific PNA reduced the expression of CmeA and increase the susceptibility of C. jejuni strains resistant to both ciprofloxacin (Cipro) and erythromycin (Ery) (10). However, it remains unknown if PNAs against other components of CmeABC are also effective in inhibiting the function of the efflux pump and if combinatorial use of PNAs against different components of the efflux system enhances the inhibitory effect.
In this study, we designed multiple PNAs against all three components of the CmeABC efflux pump based on the genome sequence of C. jejuni NCTC 11168 and evaluated their activities individually and in combination using a wild-type strain (NCTC 11168), a Cipro-resistant mutant (62301R33), and an Ery-resistant mutant (JL272). The CmeA-specific PNA sequence is TCATGGTTTTGC, the CmeB-specific PNA sequence is ATTATTGTGCTC, and the CmeC-specific PNA sequences are CATGAACCTTAC, CCTTACCTCTTT, and TATTCATGAACC. A negative-control PNA (ACACACACACAC) was also synthesized. All PNAs were conjugated to the oligopeptide KFFKFFKFFK to improve PNA entry into bacterial cells (11).
The PNAs were added to Campylobacter cultures in Mueller-Hinton (MH) broth at different concentrations (0, 1, 2, and 4 μM). To detect if the PNAs inhibited CmeABC expression, SDS-PAGE and Western blotting were performed with antibodies against CmeA, CmeB, and CmeC as described previously (10). Addition of the CmeA PNA to culture media reduced the expression of CmeA, as well as that of CmeB and CmeC (Fig. 1A). The CmeB PNA reduced the expression of CmeB, but it did not affect the expression of CmeC and CmeA (Fig. 1A). Unlike the CmeA and CmeB PNAs, none of the three CmeC PNAs examined in this study altered the expression of CmeC as determined by Western blotting (partly shown in Fig. 1A). Combination of the CmeA and CmeB PNAs reduced the expression of all three proteins of the efflux pump. Compared with the individual PNAs, the combination of CmeA and CmeB PNAs produced stronger inhibition of CmeABC expression (Fig. 1A). Densitometric analysis of the blotting results also confirmed the synergistic effect of the PNA combination on the expression of CmeABC (Fig. 1B). The negative-control PNA did not affect the expression of CmeABC (data not shown). None of the examined PNAs affected the expression of the major outer membrane protein (MOMP), which was used as an internal control (Fig. 1A).
Fig 1.
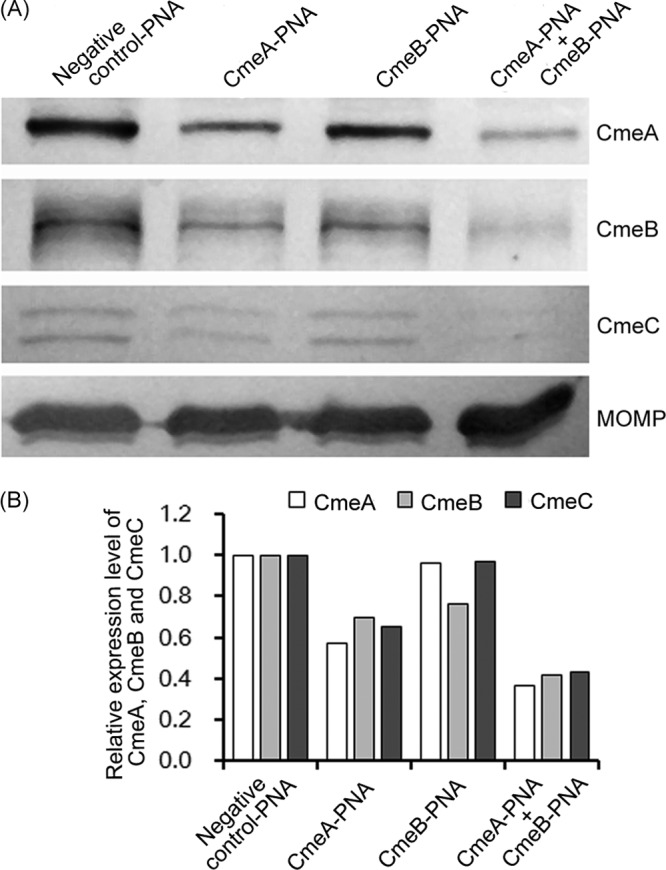
Effect of PNAs on the production of CmeA, CmeB, and CmeC in C. jejuni. (A) Western immunoblotting showing the inhibition by various PNAs (labeled on top of the panel). C. jejuni NCTC 11168 was treated with the PNAs before the analysis, and the bacterial cells were blotted with specific antibodies against CmeA, CmeB, and CmeC. The CmeC protein is shown as a doublet due to glycosylation. The same amount of total proteins was loaded in each lane, and MOMP was used as an internal control. (B) Densitometric analysis of the immunoblotting results. The CmeA, CmeB, and CmeC levels in the samples treated with the specific PNAs were normalized against the sample treated with the negative-control PNA. Each bar represents the average of two independent immunoblots.
It should be pointed out that CmeC is an N-glycosylated protein and shows as two bands, resulting from different glycosylated forms (3). The finding that the tested CmeC PNAs had no effect on the translation of CmeC was surprising, and the exact reason for this observation is unknown. One possibility is that the CmeC mRNA has unique secondary structures that prevent the binding of PNAs. Alternatively, the ribosome binding site (RBS) of CmeC is embedded in the coding sequence of CmeB, and the translation from CmeB might alleviate the inhibition of CmeC by PNAs.
To assess whether the PNAs against CmeA, CmeB, and CmeC affected the susceptibility of C. jejuni to antimicrobials, we measured the MICs of Cipro and Ery in the presence of the PNAs either individually or in combination using a microtiter broth dilution method described previously (10). The key results are shown in Table 1. At 1 and 2 μM, none of the PNAs altered the susceptibility of NCTC 11168 to Cipro and Ery. At 4 μM, the CmeA-specific PNA and the CmeB-specific PNA increased the susceptibility of NCTC 11168 to Cipro and Ery by 2-fold, but the CmeC PNAs did not show any effects, consistent with the results from Western blotting. Notably, the combination of the two PNAs against CmeA and CmeB at 1 μM each resulted in 16- and 4-fold decreases in the MICs of Cipro and Ery, respectively, in NCTC 11168 (Table 1). At higher concentrations (2 and 4 μM), this combination further sensitized NCTC 11168 to the antibiotics (Table 1). We also evaluated the PNAs in a Cipro-resistant strain (62301R33) and an Ery-resistant strain (JL272). Isolate 62301R33 has the C257T mutation in gyrA, which contributes to the high-level resistance to Cipro (12), while JL272 has the A2074G mutation in 23S rRNA, mediating high resistance to erythromycin (12, 13). In both cases, the target mutations work synergistically with CmeABC in conferring high-level resistance to the antibiotics. In 62301R33, the PNAs against CmeA and CmeB reduced the Cipro MIC 8- and 2-fold, respectively, while the combination of the two resulted in 16-fold reduction in the MIC. In JL272, the two PNAs each produced a 2-fold reduction in Ery MIC and combination of the two reduced the MIC by 4-fold.
Table 1.
Effect of CmeABC PNAs on the susceptibility of C. jejuni to ciprofloxacin and erythromycin
| C. jejuni strain and treatment | PNA concn (μM) | MIC (mg/liter)a |
|
|---|---|---|---|
| Ciprofloxacin | Erythromycin | ||
| NCTC 11168 | |||
| No PNA | 0.125 | 0.25 | |
| Negative-control PNA | 4 | 0.125 | 0.25 |
| CmeA PNA | 4 | 0.0625 (2) | 0.125 (2) |
| CmeB PNA | 4 | 0.0625 (2) | 0.125 (2) |
| CmeC PNAs | 4 | 0.125 | 0.25 |
| CmeA + CmeB PNAs | 1 | 0.0075 (16) | 0.0625 (4) |
| 2 | <0.00375 (>32) | 0.0156 (16) | |
| 4 | <0.00375 (>32) | <0.0075 (>32) | |
| 62301R33 | |||
| No PNA | 16 | NT | |
| CmeA PNA | 2 | 2 (8) | NT |
| CmeB PNA | 2 | 8 (2) | NT |
| CmeA + CmeB PNAs | 2 | 1 (16) | NT |
| JL272 | |||
| No PNA | NT | 512 | |
| CmeA PNA | 2 | NT | 256 (2) |
| CmeB PNA | 2 | NT | 256 (2) |
| CmeA + CmeB PNAs | 2 | NT | 128 (4) |
The numbers in parentheses indicate the fold changes in the MIC compared with the MIC for cells with no PNAs. NT, not tested.
These results indicate that CmeABC PNAs potentiate the activities of antibiotics in both wild-type and antibiotic-resistant strains of C. jejuni. The results also suggest that the inhibition of CmeABC expression and function depends on the target gene, with CmeA and CmeB PNAs showing the highest efficiency. Importantly, this study demonstrates a synergistic effect of the PNAs against CmeA and CmeB in sensitizing C. jejuni to antibiotic when used in combination. These findings further illustrate the feasibility of using PNAs to potentiate antibiotics against antibiotic-resistant Campylobacter. Considering that PNAs are resistant to proteases and nucleases and are stable in acidic pH (12, 14), anti-CmeABC PNAs may be potentially used as an adjunctive therapy for antibiotic-resistant Campylobacter in vivo.
ACKNOWLEDGMENTS
This study was supported by National Institutes of Health grant R21AI098742. Y.M. was supported by a scholarship from the China Scholarship Council.
The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Published ahead of print 1 July 2013
REFERENCES
- 1.Ruiz-Palacios GM. 2007. The health burden of Campylobacter infection and the impact of antimicrobial resistance: playing chicken. Clin. Infect. Dis. 44:701–703 [DOI] [PubMed] [Google Scholar]
- 2.Luangtongkum T, Jeon B, Han J, Plummer P, Logue CM, Zhang Q. 2009. Antibiotic resistance in Campylobacter: emergence, transmission and persistence. Future Microbiol. 4:189–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin J, Michel LO, Zhang Q. 2002. CmeABC functions as a multidrug efflux system in Campylobacter jejuni. Antimicrob. Agents Chemother. 46:2124–2131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin J, Sahin O, Michel LO, Zhang Q. 2003. Critical role of multidrug efflux pump CmeABC in bile resistance and in vivo colonization of Campylobacter jejuni. Infect. Immun. 71:4250–4259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Quinn T, Bolla JM, Pages JM, Fanning S. 2007. Antibiotic-resistant Campylobacter: could efflux pump inhibitors control infection? J. Antimicrob. Chemother. 59:1230–1236 [DOI] [PubMed] [Google Scholar]
- 6.Hannula M, Hanninen ML. 2008. Effect of putative efflux pump inhibitors and inducers on the antimicrobial susceptibility of Campylobacter jejuni and Campylobacter coli. J. Med. Microbiol. 57:851–855 [DOI] [PubMed] [Google Scholar]
- 7.Mamelli L, Amoros JP, Pages JM, Bolla JM. 2003. A phenylalanine-arginine beta-naphthylamide sensitive multidrug efflux pump involved in intrinsic and acquired resistance of Campylobacter to macrolides. Int. J. Antimicrob. Agents 22:237–241 [DOI] [PubMed] [Google Scholar]
- 8.Corradini R, Sforza S, Tedeschi T, Totsingan F, Manicardi A, Marchelli R. 2011. Peptide nucleic acids with a structurally biased backbone. Updated review and emerging challenges. Curr. Top. Med. Chem. 11:1535–1554 [DOI] [PubMed] [Google Scholar]
- 9.Paulasova P, Pellestor F. 2004. The peptide nucleic acids (PNAs): a new generation of probes for genetic and cytogenetic analyses. Ann. Genet. 47:349–358 [DOI] [PubMed] [Google Scholar]
- 10.Jeon B, Zhang Q. 2009. Sensitization of Campylobacter jejuni to fluoroquinolone and macrolide antibiotics by antisense inhibition of the CmeABC multidrug efflux transporter. J. Antimicrob. Chemother. 63:946–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kilk K, Langel U. 2005. Cellular delivery of peptide nucleic acid by cell-penetrating peptides. Methods Mol. Biol. 298:131–141 [DOI] [PubMed] [Google Scholar]
- 12.Luo N, Pereira S, Sahin O, Lin J, Huang S, Michel L, Zhang Q. 2005. Enhanced in vivo fitness of fluoroquinolone-resistant Campylobacter jejuni in the absence of antibiotic selection pressure. Proc. Natl. Acad. Sci. U. S. A. 102:541–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luangtongkum T, Shen Z, Seng VW, Sahin O, Jeon B, Liu P, Zhang Q. 2012. Impaired fitness and transmission of macrolide-resistant Campylobacter jejuni in its natural host. Antimicrob. Agents Chemother. 56:1300–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hatamoto M, Ohashi A, Imachi H. 2010. Peptide nucleic acids (PNAs) antisense effect to bacterial growth and their application potentiality in biotechnology. Appl. Microbiol. Biotechnol. 86:397–402 [DOI] [PubMed] [Google Scholar]


