Abstract
The same plasmid carrying blaCTX-M-14b was identified from an Escherichia coli isolate and an Enterobacter cloacae isolate collected from cattle in the United Kingdom by complete plasmid sequencing. This 35,341-bp plasmid, pSAM7, had an IncX4 backbone that is 99% identical to that of pJIE143 from a human isolate in Australia. PCR screening identified pSAM7-like plasmids in three other E. coli isolates of different multilocus sequence types isolated from cattle on different farms in the United Kingdom.
TEXT
The dissemination of extended-spectrum-β-lactamase (ESBL) genes via plasmids is a worldwide problem, with genes of the blaCTX-M group being the most widespread plasmid-mediated genes encoding ESBL (1, 2). blaCTX-M genes have been found in various members of Enterobacteriaceae isolated from both humans and animals. The blaCTX-M-14 gene is frequently found in human isolates in Asia (3, 4) and some parts of Europe (5), but in the United Kingdom it is more prevalent in animal isolates (6). blaCTX-M-14 has been associated with pCT-like IncK plasmids in the United Kingdom and around the world, with apparent transmission of this plasmid between human and animal isolates (6, 7). Here, small (35-kb) plasmids carrying blaCTX-M14b from Escherichia coli and Enterobacter cloacae isolates collected from cattle in the United Kingdom were sequenced and annotated.
E. coli isolate SAM7 was obtained from cattle feces in 2008 and Enterobacter cloacae ECR528 from waste milk (milk from cows treated with antibiotics that was not suitable for human consumption) from a different farm in 2012, during routine screening for ESBL genes. The bacterial species were identified using matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) analysis (8). ESBL plasmids from both bacterial species were conjugated into Salmonella enterica serovar Typhimurium 26R, using a surface mating method with a 10:1 recipient-to-donor ratio, and transconjugants were selected on Rambach agar containing 100 μg/ml rifampin and 1 μg/ml cefotaxime. Transconjugants from both isolates were found to carry a 35-kb plasmid by S1 nuclease digestion and pulsed-field gel electrophoresis (PFGE) (9 and data not shown). They could not be typed by the original PCR-based replicon typing (PBRT) scheme available at the time (10), so both plasmids were sequenced for further characterization. Total plasmid DNA was extracted from each isolate using a Qiagen Hi Speed plasmid Midi kit following the manufacturer's protocol and electroporated into Electromax DH10B E. coli cells (Invitrogen), and transformants were selected with 4 μg/ml cefotaxime. Plasmid DNA was isolated from transformants using a Qiagen large construct kit. A DNA library prepared from 500 ng of DNA following Roche protocol 2.3 was sequenced on one-eighth of a plate using a Roche 454 GS-FLX system. Sequences were assembled using Newbler version 2.3 (Roche), and >240-fold coverage was achieved. Single contigs were closed by PCR (Table 1) and ABI sequencing.
Table 1.
Primers used in this study to close pSAM7, genetic markers, and environment
| Primer | Primer sequence (5′–3′) | Amplicon size (bp) | Position in pSAM7 (bp) |
|---|---|---|---|
| Rpir FW | CAGTGTGGATTTTGAGCAT | 774 | 822–1595 |
| Rpir RV | GCCCCTATTGTATAAAGATTCA | ||
| Rpilx5 FW | CTTAGTTCATTTGTGAATGCC | 1,060 | 20925–21984 |
| Rpilx5 RV | GAAAGTGTTGATGCTGTGAT | ||
| RhicA FW | CCAGTTTTCCATACAGGACA | 350 | 28212–28561 |
| RhicA RV | GTTGCATATCTATAGGGGATG | ||
| Hyex FW | CAAAGGGAGGGTGTGAAT | 841 | 10553–11393 |
| Hyex RV | GGAATGGCGATACAAACA | ||
| Smet FW | CGATGGCCTTAAGACCTT | 471 | 31316–31786 |
| Smet RV | CGGACACGGTATTTGTTG | ||
| ISEC | GAAAAGCGTGGTAATGCT | 739 | 29776–30514 |
| CTXISEC | GCACCTGCGTATTATCTGC | ||
| CTXSMETHa | GTCGTGGACTGTAGGTGATA | 767 | 31020–31786 |
| ISECHICa | GCAAATTGGATATTGTAGCA | 1,098 | 28212–29309 |
| pSAM7 FW | GCACGCATTAAAAGCCTTAT | 307 | 508–814 |
| pSAM7 RW | GGCAGATTAACAACAGATTCAA | ||
| pSAM7-2 FW | GAGTGGGGATCAAGTTTACG | 327 | 1364–1690 |
| pSAM7-2 RV | CTTCCGTATGTTTCATGATTTC |
CTXSMETH was used with Smet RV and ISECHIC was used with RhicA FW.
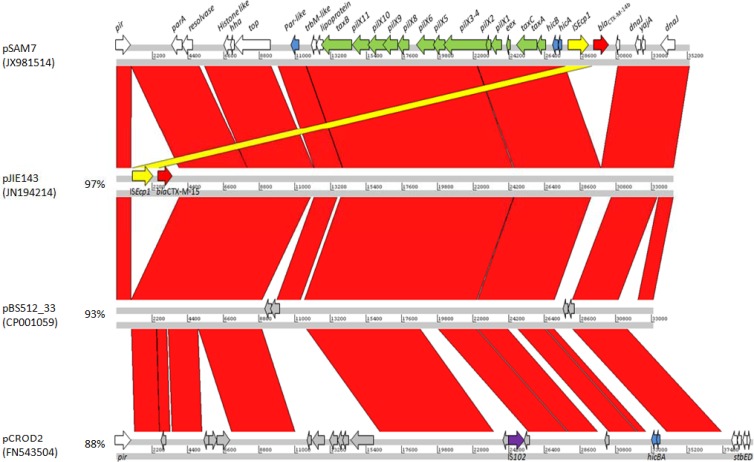
Plasmids from E. coli and Enterobacter cloacae were found to be 100% identical, and the 35,341-bp plasmid was designated pSAM7. BLAST searches of GenBank indicated that the backbone of pSAM7 was closely related to the newly defined IncX4 group of plasmids (11), with the highest identity (99%) to pJIE143 (GenBank accession no. JN194214) harboring blaCTX-M-15 from an E. coli isolated from a human in Australia in 2006 (12). pSAM7 was also related to pJEG012 (KC354802) (93%) from Klebsiella pneumoniae, also isolated in Australia and carrying the antimicrobial resistance genes aacA4, aadA1, and blaOXA-9 (13), and other IncX4 plasmids, pBS512_33 (CP001059) (93%) from Shigella boydii, pSHS696_34 (JX258654) (91%) from Salmonella enterica serovar Heidelberg, and pCROD2 (FN543504) (88%) from Citrobacter rodentium (14), none of which harbor any antimicrobial resistance genes. Annotation using RAST (15) and Artemis and comparisons with pJIE143 and pBS512_33 identified 51 open reading frames (ORFs), of which 20 were hypothetical. Alignments carried out using the WebACT comparison tool (16) revealed high levels of identity in large parts of the plasmid backbones (Fig. 1). As described for pJIE143, the initiation of replication in pSAM7 is likely to be mediated by the pir-encoded π replication initiation protein and several iterons (12). All iterons in pSAM7 were identical to those in pJIE143, apart from γ3, which differed by two nucleotides (bold) (AAACATGATAACTTCCTCGGTT in pSAM7 and AAACATGAGAGCTTCCTCGGTT in pJIE143).
Fig 1.
Artemis Comparison Tool (ACT) comparison of pSAM7 with related plasmids pJIE143, pBS512_33, and pCROD2 (which has an unrelated replication protein). Green arrows, genes relating to conjugation; blue arrows, stability genes; yellow arrows, ISEcp1; red arrows, blaCTX-M genes. Other genes are shown in white, and conserved hypothetical genes are shown in gray. Red areas show nucleotide identity > 93%, the highlighted yellow area indicates ISEcp1, and the percentages shown indicate the levels of identity of the complete plasmids to the pSAM7 backbone.
Conjugation of IncX plasmids involves the products of the 11 traX (pilX) genes, three tax genes (taxA, taxB, and taxC), and oriTα (17). Like other IncX4 plasmids, pSAM7 contains the hicA-hicB addiction system (18) and a partitioning gene, parB, adjacent to a resolvase. pSAM7 has two short insertions absent from pJIE143, the first between positions (bp) 6061 and 6377 in a conserved hypothetical gene in pJIE143 (positions 8971 to 9222) (12). This insertion is flanked by nearly perfect direct repeat sequences GGACAGAATCACCTGTATGTC (positions 6040 to 6060) and GGACAAAATGACCTGTATGTC (6377 to 6397). A similar insertion that includes these repeats is present in pCROD2 (91% identical) (14). The other insertion is between positions 10489 and 10925 of pSAM7, part of a hypothetical coding sequence (10268 to 10843). This region is also present in the IncX4 plasmids pSH696_34 and pBS512_33 and the IncX1 plasmid pE001 (19).
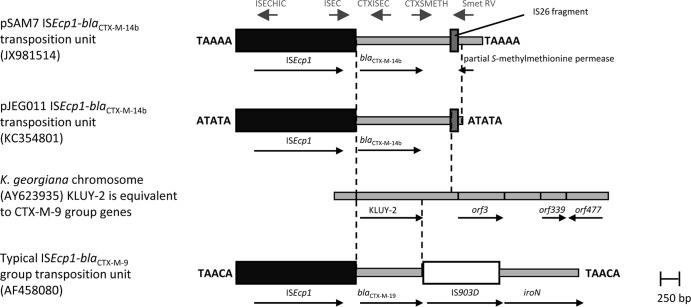
ISEcp1 transposition units carrying blaCTX-M-9 group genes usually contain a 920-bp region corresponding to the Kluyvera georgiana chromosome with IS903 2 bp after the stop codon of blaCTX-M (Fig. 2) (20, 21). In pSAM7, a 1,256-bp region matching K. georgiana is followed by a 108-bp fragment of the IS26 insertion sequence. Downstream of this is a region consisting of 177 bp corresponding to an internal part of a gene encoding an S-methylmethionine permease found in several E. coli chromosomes. This 3,246-bp transposition unit is flanked by 5-bp direct repeats (TAAAA) characteristic of ISEcp1 transposition. This combination of different components suggests that the transposition unit in pSAM7 may have been compiled from segments acquired in different ISEcp1 transposition events (22). A similar transposition unit with the IS26 fragment but only 73 bp of the S-methylmethionine permease gene is also present, flanked by direct repeats of ATATA, in the IncL/M plasmid pJEG011 (KC354801) from the same K. pneumoniae isolate as pJEG012 (13). The transposition unit in pSAM7 is inserted 97 bp upstream of the hicA gene, while in pJIE143 the ISEcp1-blaCTX-M-15-orf477Δ transposition unit is inserted 22 bp downstream of the pir gene, flanked by direct repeats of GGATA (12), indicating different insertions of blaCTX-M genes in the same backbone. No other known antimicrobial resistance or virulence genes were identified in pSAM7.
Fig 2.
Comparison of the ISEcp1 transposition units in pSAM7 and pJEG011 with a typical ISEcp1-blaCTX-M-9 group transposition unit and with the chromosome of Kluyvera georgiana. Solid black boxes indicate transposons, black arrows indicate extents and directions of genes, and 5-bp direct repeats flanking insertions are indicated. Dashed lines indicated the boundaries of regions common to two or more sequences. Gray arrows indicate primers used for investigating the genetic environment of the blaCTX-M-14b gene in pSAM7.
Sequence comparisons were used to design primers to detect pSAM7-like plasmids (Table 1). The genetic markers selected were the pir (Rpir), hicA (RhicA), and pilX5 (Rpilx5) regions, a hypothetical gene (Hyex), and the transposition unit (Smet). Sequence differences between pir genes would be expected to prevent binding of the Rpir primers to pCROD2 and pSH696_34, the RhicA region is absent from pBS512_32 and pJEG012, and Hyex is present only in pSAM7 and pSH696_34. The original isolates, transconjugants, and transformants carrying pSAM7 produced all five amplicons, while JIE143 carrying pJIE143 (12) produced Rpir, Rpilx5, and RhicA amplicons only, as expected.
The same primers were used to screen 42 E. coli isolates, collected from the United Kingdom, The Netherlands, or Germany between 2004 and 2009, which had been identified as having small plasmids carrying ESBL genes (unpublished data). Isolates carried blaCTX-M-1 (human n = 9, cattle n = 7, poultry n = 5, pig n = 4), blaCTX-M-14 (human n = 2, cattle n = 5), blaCTX-M-15 (cattle n = 2), blaTEM-52 (poultry n = 7), or blaSHV-12 (human n = 1) as determined by PCR and sequencing. Six of the 42 isolates had at least three of the markers pir (Rpir), hicA (RhicA), and pilX5 (Rpilx5) (Table 2). Transconjugants carrying ESBL genes from three isolates, collected from cattle at different farms in the United Kingdom, had an ∼35-kb plasmid by S1/PFGE (data not shown) and all of the pSAM7 markers and carried blaCTX-M-14 in the same genetic environment as in pSAM7 (Table 2). Comparison of these isolates by PFGE and multilocus sequence typing (MLST) (following the protocol available at http://mlst.ucc.ie/mlst/dbs/Ecoli) (23) showed that they were not related to each other (ST117, ST559, and ST2177) or to SAM7 (ST10). Thus, pSAM7-like plasmids are moving between E. coli sequence types.
Table 2.
Screening of ESBL isolates and transconjugants for pSAM7-like plasmidsa
| Isolate | Speciesb | Host | Origin | Yr | CTX-M | ST | pSAM7 markersc |
blaCTX-M-14 TUd | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Rpir | Rpil | RhicA | Hyex | Smet | ||||||||
| SAM7 | E. coli | Cattle | United Kingdom | 2008 | 14 | 10 | Y | Y | Y | Y | Y | Y |
| ECR528 | E. cloacae | Cattle | United Kingdom | 2012 | 14 | Y | Y | Y | Y | Y | Y | |
| ESBL487 | E. coli | Cattle | United Kingdom | 2008 | 14 | 117 | Y | Y | Y | Y | Y | Y |
| ESBL562 | E. coli | Cattle | United Kingdom | 2007 | 14 | 559 | Y | Y | Y | Y | Y | Y |
| ESBL592 | E. coli | Cattle | United Kingdom | 2008 | 14 | 2177 | Y | Y | Y | Y | Y | Y |
| ESBL219 | E. coli | Human | Germany | Unknown | 1 | ND | Y | Y | Y | N | N | ND |
| ESBL220 | E. coli | Human | Germany | Unknown | 1 | ND | Y | Y | Y | N | N | ND |
| ESBL484 | E. coli | Cattle | United Kingdom | 2007 | 1 | ND | Y | Y | Y | N | N | ND |
ST, serotype; ND, not determined.
Confirmed by MALDI-TOF.
pSAM7 transformed into DH10 was used as a positive control and Salmonella Typhimurium strain 26R was used as a negative control in PCR. Y, amplicon obtained; Y, amplicon also obtained from transconjugant; N, no amplicon.
blaCTX-M-14 TU, ISEcp1-blaCTX-M-14 transposition unit found in pSAM7.
The original PBRT scheme (10) included primers to detect IncX2 plasmids, typified by R6K, but not IncX1 plasmids, typified by R485 (24). Primers are now available to detect IncX1 and IncX2 plasmids and two additional groups, IncX3 and IncX4 (11). The IncX4 primers in the taxC gene are designed to detect all IncX4 plasmids (11), while the markers used here differentiated pSAM7 from other IncX4 plasmids. The similarity between pSAM7 and pJIE143 suggests that these plasmids share a ancestor which has been in the bacterial population for some time, as is evident from their identification from isolates with no epidemiological links on separate continents. To date, pSAM7-like plasmids have been found only in isolates from cattle in the United Kingdom, which may act as a reservoir, but the similarity to pJIE143 suggests that pSAM7 may be able to facilitate spread of blaCTX-M-14b from animal to human isolates. These plasmids are capable of conjugation, as is evident from the occurrence of pSAM7 in two different bacterial genera and its ability to transfer to S. enterica serovar Typhimurium from both original isolates and transformants. pSAM7 and pJIE143 and other IncX4 backbones may represent emerging vectors for blaCTX-M genes, conferring resistance to extended-spectrum cephalosporins in Enterobacteriaceae. This work shows that IncX4 plasmids play a role in disseminating blaCTX-M-14b between different species of bacteria. Several other IncX plasmids bearing important β-lactamase genes, including IncX1 plasmids carrying blaTEM-52 in E. coli and Salmonella found in meat imported into Denmark from The Netherlands and Germany, have been reported recently (19). blaKPC carbapenemase genes have also been identified on the IncX3 plasmid pKpS90 (blaKPC-2) (25) and a novel IncX5 plasmid (blaKPC-5) (26), both isolated from K. pneumoniae.
Nucleotide sequence accession number.
The sequence of pSAM7 from E. coli SAM7 has been submitted to GenBank under accession number JX981514.
ACKNOWLEDGMENTS
We thank Richard Ellis for sequencing of the plasmids and Onyi Diribe, Matthew Hayward, Hannah Preedy, Roberto La Ragione, Luke Randall, Robert Horton, Chris Teale, Jonathan Iredell, and Alison Kelly for technical help and useful discussions. We also acknowledge Beatriz Guerra, Reiner Helmuth, and Irene Rodriguez, who work on the ESBL-Safefoodera Consortium and kindly supplied isolates.
This work was funded by the Department for Environment, Food and Rural Affairs (project grant OD2028) and by Kingston University BPSRG funding.
Footnotes
Published ahead of print 8 July 2013
REFERENCES
- 1.Bonnet R. 2004. Growing group of extended-spectrum β-lactamases: the CTX-M enzymes. Antimicrob. Agents Chemother. 48:1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cantón R, Coque TM. 2006. The CTX-M β-lactamase pandemic. Curr. Opin. Microbiol. 9:466–475 [DOI] [PubMed] [Google Scholar]
- 3.Chanawong A, M'Zali FH, Heritage J, Xiong JH, Hawkey PM. 2002. Three cefotaximases, CTX-M-9, CTX-M-13 and CTX-M-14, among Enterobacteriaceae in the People's Republic of China. Antimicrob. Agents Chemother. 46:630–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim J, Bae IK, Jeong SH, Chang CL, Lee CH, Lee K. 2011. Characterization of IncF plasmids carrying the blaCTX-M-14 gene in clinical isolates of Escherichia coli from Korea. J. Antimicrob. Chemother. 66:1263–1268 [DOI] [PubMed] [Google Scholar]
- 5.Weill FX, Lailler R, Praud K, Kerouanton A, Fabre L, Brisabois A, Grimont PAD, Cloeckaert A. 2004. Emergence of extended-spectrum-β-lactamase (CTX-M-9)-producing multiresistant strains of Salmonella enterica serotype Virchow in poultry and humans in France. J. Clin. Microbiol. 42:5767–5773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stokes MO, Cottell JL, Piddock LVJ, Wu G, Wootton M, Mevius DJ, Randall LP, Teale CJ, Fielder MD, Coldham NG. 2012. Detection and characterization of pCT-like plasmid vectors for blaCTX-M-14 in Escherichia coli isolates from humans, turkeys and cattle in England and Wales. J. Antimicrob. Chemother. 67:1639–1644 [DOI] [PubMed] [Google Scholar]
- 7.Cottell JL, Webber MA, Coldham NG, Taylor DL, Cerdeno-Tarraga AM, Hauser H, Thomson NR, Woodward MJ, Piddock LJV. 2011. Complete sequence and molecular epidemiology of IncK epidemic plasmid encoding blaCTX-M-14. Emerg. Infect. Dis. 17:645–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eigner U, Holfelder M, Oberdorfer K, Betz-Wild U, Bertsch D, Fahr A. 2009. Performance of a matrix-assisted laser desorption ionization-time-of-flight mass spectrometry system for the identification of bacterial isolates in the clinical routine laboratory. Clin. Lab. 55:289–296 [PubMed] [Google Scholar]
- 9.Barton BM, Harding GP, Zuccarelli AJ. 1995. A general method for detecting and sizing large plasmids. Anal. Biochem. 226:235–240 [DOI] [PubMed] [Google Scholar]
- 10.Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. 2005. Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63:219–228 [DOI] [PubMed] [Google Scholar]
- 11.Johnson TJ, Bielak EM, Fortini D, Hansen LH, Hasman H, Debroy C, Nolan LK, Carattoli A. 2012. Expansion of the IncX plasmid family for improved identification and typing of novel plasmids in drug-resistant Enterobacteriaceae. Plasmid 68:43–50 [DOI] [PubMed] [Google Scholar]
- 12.Partridge SR, Ellem JA, Tetu SG, Zong Z, Paulsen IT, Iredell JR. 2011. Complete sequence of pJIE143, a pir-type plasmid carrying ISEcp1-blaCTX-M-15 from an Escherichia coli ST131 isolate. Antimicrob. Agents Chemother. 55:5933–5935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Espedido BA, Steen JA, Ziochos H, Grimmond SM, Cooper MA, Gosbell IB, van Hal SJ, Jensen SO. 2013. Whole genome sequence analysis of the first Australian OXA-48-producing outbreak-associated Klebsiella pneumoniae isolates: the resistome and in vivo evolution. PLoS One 8:e59920. 10.1371/journal.pone.0059920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petty NK, Bulgin R, Crepin VF, Cerdeno-Tarraga AM, Schroeder GN, Quail MA, Lennard N, Corton C, Barron A, Clark L, Toribio AL, Parkhill J, Dougan G, Frankel G, Thomson NR. 2010. The Citrobacter rodentium genome sequence reveals convergent evolution with human pathogenic Escherichia coli. J. Bacteriol. 192:525–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. 10.1186/1471-2164-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abbott JC, Aanensen DM, Rutherford K, Butcher S, Spratt BG. 2005. WebACT—an online companion for the Artemis Comparison Tool. Bioinformatics 21:3665–3666 [DOI] [PubMed] [Google Scholar]
- 17.Núñez B, Avila P, de la Cruz F. 1997. Genes involved in conjugative DNA processing of plasmid R6K. Mol. Microbiol. 24:1157–1168 [DOI] [PubMed] [Google Scholar]
- 18.Jørgensen MG, Pandey DP, Jaskolska M, Gerdes K. 2009. HicA of Escherichia coli defines a novel family of translation-independent mRNA interferases in bacteria and archaea. J. Bacteriol. 191:1191–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bielak E, Bergenholtz RD, Jorgensen MS, Sorensen SJ, Hansen LH, Hasman H. 2011. Investigation of diversity of plasmids carrying the blaTEM-52 gene. J. Antimicrob. Chemother. 66:2465–2474 [DOI] [PubMed] [Google Scholar]
- 20.Olson AB, Silverman M, Boyd DA, McGeer A, Willey BM, Pong-Porter V, Daneman N, Mulvey MR. 2005. Identification of a progenitor of the CTX-M-9 group of extended-spectrum β-lactamases from Kluyvera georgiana isolated in Guyana. Antimicrob. Agents Chemother. 49:2112–2115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poirel L, Decousser JW, Nordmann P. 2003. Insertion sequence ISEcp1B is involved in expression and mobilization of a blaCTX-M β-lactamase gene. Antimicrob. Agents Chemother. 47:2938–2945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Partridge SR. 2011. Analysis of antibiotic resistance regions in Gram-negative bacteria. FEMS Microbiol. Rev. 35:820–855 [DOI] [PubMed] [Google Scholar]
- 23.Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, Karch H, Reeves PR, Maiden MC, Ochman H, Achtman M. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 60:1136–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones CS, Osborne DJ, Stanley J. 1993. Molecular comparison of the IncX plasmids allows division into IncX1 and IncX2 subgroups. J. Gen. Microbiol. 139:735–741 [DOI] [PubMed] [Google Scholar]
- 25.Kassis-Chikhani N, Frangeul L, Drieux L, Sengelin C, Jarlier V, Brisse S, Arlet G, Decre D. 2013. Complete nucleotide sequence of the first KPC-2- and SHV-12-encoding IncX plasmid, pKpS90, from Klebsiella pneumoniae. Antimicrob. Agents Chemother. 57:618–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen L, Chavda KD, Fraimow HS, Mediavilla JR, Melano RG, Jacobs MR, Bonomo RA, Kreiswirth BN. 2013. Complete nucleotide sequences of blaKPC-4- and blaKPC-5-harboring IncN and IncX plasmids from Klebsiella pneumoniae strains isolated in New Jersey. Antimicrob. Agents Chemother. 57:269–276 [DOI] [PMC free article] [PubMed] [Google Scholar]