Abstract
There exist numerous genes disseminated by mobile elements that can confer cross-resistance to lincosamides and streptogramin A compounds in staphylococci. This study investigated the nature and means of dissemination of genes responsible for LSA resistance among 24 French clinical isolates screened for reduced susceptibility to lincomycin. The vga(A)v gene was found to be the most prevalent determinant of LSA resistance, while Tn5406 appeared to be its exclusive gene support.
TEXT
Combined resistance to lincosamides and streptogramin A (SA) compounds is referred to as the LSA phenotype in clinical microbiology laboratories. The resistance phenotype relies on the presence of the ARE subfamily of class 2 ATP-binding cassette (ABC) ATPases, a class of ABC proteins made up of two homologous ABC ATPase domains separated by a flexible linker without any identifiable transmembrane domains. Despite the efforts of several laboratories, the resistance mechanism remains elusive. Two hypotheses have been proposed: protection of the ribosome by preventing access to the antibiotic binding site or antibiotic efflux (1). Direct evidence supporting the first hypothesis is still lacking, while many experimental data have recently sustained the second one. Reports on membrane localization of ARE proteins Vga(A) and VmlR, formerly ExpZ (2), in bacteria showing either acquired or intrinsic resistance to LSA antibiotics (3, 4), along with transport assays using the radiolabeled lincomycin compound (5), have given credit to the hijacking hypothesis, wherein the ABC protein alters the specificity of at least one transporter (1). The flexible linker between each ATPase domain is presumed to be the drug-binding region of the ARE proteins (5, 6).
Contrary to Cfr- or Erm-based methylations of 23S rRNA that confer resistance to numerous translation inhibitors (7, 8), the ABC-mediated resistance mechanism is limited to only a few antimicrobials, macrolides and streptogramin B (SB) compounds or lincosamides and SA compounds, depending on what kind of ARE protein is present in the bacterial strain (see http://faculty.washington.edu/marilynr/ for any detailed information). Due to their drug specificity, ARE proteins can be divided into two separate clusters: those in the first cluster confer various degrees of resistance to lincosamides, SA compounds, and pleuromutilins, while those in the second cluster confer various degrees of resistance to macrolides and SB compounds. Within each cluster, the linker regions of the ARE proteins may be strongly dissimilar, pointing to their possible involvement, outside the drug resistance spectrum, in certain interactions with membrane partners (4, 9). Mapping of the diversity within ARE proteins is thus helpful for deciphering the resistance mechanism but also for epidemiological surveys.
The goal of the present study was to analyze the gene diversity occurring among various staphylococcal species with respect to the LSA phenotype. Here we also report the manner in which the most prevalent gene is disseminated and resistance levels are conferred by a series of Vga(A) isoforms detected in a 3-month sampling procedure.
(Parts of this work were presented at the 48th Interscience Conference on Antimicrobial Agents and Chemotherapy [ICAAC], Washington, DC, 24 to 28 October 2008.)
Screening for the LSA phenotype was initially carried out at five French military hospitals among clinical isolates identified to be erythromycin-susceptible (Erys) staphylococci. This was done by disk diffusion susceptibility tests which incorporated lincomycin as the best indicator of lincosamide resistance (10). The susceptible/intermediate/resistant categorization for the 14 other tested antibiotics was obtained using a SIRscan automated system and EUCAST interpretive criteria (11). For lincomycin, breakpoint values were provided by the Comité de l'Antibiogramme de la Société Francaise de Microbiologie (CA-SFM). Strains with diameters of less than 17 mm were defined as resistant, while those with diameters equal to or greater than 17 mm but less than 21 mm were considered intermediate. Following these French guidelines, 36 lincomycin-resistant (Linr) or lincomycin-intermediate (Lini) strains were collected and included in the study for further characterization.
Bacterial DNA of the 36 screened strains was extracted using a MagNA Pure LC DNA isolation kit III on a MagNA Pure LC instrument (Roche Diagnostics) and eluted in 50 μl of elution buffer. Various DNA targets were amplified using different PCR primer sets already described or specifically designed for this study (Table 1). Accurate species identification was provided by rRNA gene sequencing as described previously (12) by running the SeqMatch algorithm (http://rdp.cme.msu.edu) for taxonomic assignment. Detection of genes coding for lincomycin resistance was positive for all but one strain. The only negative sample corresponded to a Staphylococcus sciuri strain. A total of 10 putative duplicates, which either were from the same patient or shared the same antibiotype at a given hospital, were excluded.
Table 1.
Oligodeoxyribonucleotides used in PCR experiments for diagnosis and cloning purposesa
| Primer | Nucleotide sequence (5′ to 3′) | DNA target | Programb | Amplicon size |
|---|---|---|---|---|
| V1 | AAGTGGTGGTGAAGTAACACG | vga(A) | P1 diagnosis | 1 kbp |
| V2 | TCAAGAAAGTTTGTTGGTTCATC | |||
| T1 | ATATCCGACATCGCCAACTC | Tn5406 | P1 diagnosis | 740 bp |
| T2 | TTCGTGTTACTTCACCACCAC | |||
| X | ATTTCATTATCGCCATCTGTC | EES57021 | P2 cloning | 2.4 kbp |
| Y | TCTTCCTTCTTCAATTTCCC | |||
| lnuACDf | GGTTWGATGGWGGYTGGGG | lnu(A) | P3 diagnosis | 230 bp |
| lnuAr | AATTGCCACCTTCTGGGTTTGC | |||
| lnuBFf | AGARGGTGACSARTWCTCTGA | lnu(B) | P3 diagnosis | 400 bp |
| lnuBFr | AKGMRCGAGCATAYTCTCC |
The primer pairs described here were not found in the literature. Other primer pairs were vgaB1 and vgaB2 for detection of vga(B) (24), vgaC-1 and vgaC-2 for detection of vga(C) (21), vgaE_fw and vgaE_rv for detection of vga(E) (22), lsaB-1 and lsaB-2 for detection of lsa(B) (21), cfr_fw and cfr_rv for detection of cfr (23), vatA1 and vatA2 for detection of vat(A) (24), and 16S-27F and 16S-907R for detection of rrs (12).
Thermocycling programs were as follows: P1, 1 cycle of 5 min at 94°C and 2 min at 54°C; 30 cycles of 60 s at 72°C, 60 s at 94°C, and 60 s at 54°C; and 1 cycle of 10 min at 72°C; P2, 1 cycle of 5 min at 94°C and 2 min at 54°C; 30 cycles of 2 min at 72°C, 1 min at 94°C, and 1 min at 54°C; and 1 cycle of 10 min at 72°C); P3, 1 cycle of 5 min at 94°C and 2 min at 50°C; 30 cycles of 40 s at 72°C, 50 s at 94°C, and 50 s at 50°C; and 1 cycle of 10 min at 72°C.
The 26 remaining strains were tested for growth on brain heart infusion plates supplemented with 6 μg/ml pristinamycin IIA and replica plated with 2 μg/ml lincomycin for storage. Only two of them did not grow on test plates, demonstrating that a lack of susceptibility to lincomycin in Erys staphylococci, at least in France, is often linked to co- or cross-resistance to SA compounds. These two strains harbored an lnu(A) gene and thus exhibited the L phenotype, but not the LSA phenotype. The MICs of clindamycin (CLI) and pristinamycin (PT) for the 24 strains displaying the LSA phenotype were determined using Etest strips (AB Biodisk) (Table 2). According to identical EUCAST and CA-SFM breakpoint tables, only 1 strain was found to be resistant to PT (MIC > 2 μg/ml), while 12 others were defined to be resistant to CLI (with 3 having MICs of >0.5 μg/ml) or intermediate (with 9 having MICs of 0.5 μg/ml). Once again and unlike the findings for Enterococcus faecalis, it was verified that the LSA phenotype detected among staphylococci did not impair the efficacy of the streptogramin mixtures typified by PT.
Table 2.
Relevant characteristics of the 24 LSA strains
| Strain (hospitala) | Speciesb | Antibiotypec | MLS genotyped | CLI |
PT |
||
|---|---|---|---|---|---|---|---|
| MIC (μg/ml) | Susceptibilitye | MIC (μg/ml) | Susceptibility | ||||
| S3 (A) | S. aureus | Oxar Kanr Tobr Pefr Lini | vga(A) allele 1 | 0.25 | S | 0.75 | S |
| S4 (A) | S. aureus | Oxar Kanr Tobr Pefr Lini | vga(A)v | 0.25 | S | 0.75 | S |
| S5 (A) | S. aureus | Oxar Kanr Tobr Genr Pefr Sulr Rifr Linr | vga(A)v | 1 | R | 0.5 | S |
| S6 (E) | S. aureus | Oxar Kanr Tobr Pefr Lini | vga(A)v | 0.25 | S | 0.5 | S |
| S7 (D) | S. epidermidis | Penr Linr | vga(A) allele 2 | 0.75 | R | 0.19 | S |
| S8 (B) | S. aureus | Oxar Pefr Lini | vga(A)v | 0.25 | S | 0.38 | S |
| S9 (B) | S. aureus | Oxar Tetr Sxtr Lini | vga(A)v | 0.5 | I | 0.5 | S |
| S10 (B) | S. aureus | Oxar Kanr Tobr Pefr Lini | vga(A)v | 0.25 | S | 0.5 | S |
| S11 (B) | S. aureus | Penr Linr | lnu(A) + vga(A) | 0.19 | S | 0.38 | S |
| S12 (C) | S. aureus | Oxar Kanr Tobr Pefr Lini | vga(A)v | 0.25 | S | 0.5 | S |
| S13 (B) | S. aureus | Penr Kanr Tobr Tetr Sxtr Linr | lnu(A) + vga(A) | 0.19 | S | 0.38 | S |
| S14 (A) | S. sciuri | Lini | Unknown | 0.38 | S | 0.75 | S |
| S15 (E) | S. epidermidis | Oxar Kanr Tobr Genr Pefr Sxtr Lini | vga(A)v | 0.5 | I | 0.75 | S |
| S17 (C) | S. aureus | Oxar Pefr Sulr Lini | vga(A) | 0.19 | S | 0.5 | S |
| S19 (C) | S. epidermidis | Oxar Kanr Tobr Genr Pefr Sxtr Lini | vga(A)v | 0.5 | I | 0.5 | S |
| S22 (E) | S. aureus | Oxar Kanr Tobr Genr Pefr Sxtr Lini | vga(A)v | 0.5 | I | 0.5 | S |
| S23 (E) | Staphylococcus hominis | Oxar Kanr Tobr Genr Pefr Sxtr Teir Lini | vga(A) | 0.25 | S | 0.5 | S |
| S25 (D) | S. aureus | Oxar Kanr Tobr Pefr Lini | vga(A)v | 0.25 | S | 0.75 | S |
| S26 (D) | S. epidermidis | Oxar Kanr Tobr Genr Pefr Sxtr Lini | vga(A)v | 0.5 | I | 1 | S |
| S27 (B) | S. aureus | Oxar Kanr Tobr Pefr Lini | vga(A)v + vat(A) | 0.25 | S | 12 | R |
| S29 (B) | S. aureus | Oxar Kanr Tobr Pefr Linr | vga(A)v | 0.5 | I | 1 | S |
| S31 (C) | S. haemolyticus | Fosr Linr | vga(A)LC | 3 | R | 0.38 | S |
| S33 (A) | S. epidermidis | Oxar Kanr Tobr Genr Pefr Sxtr Lini | vga(A)v | 0.5 | I | 0.75 | S |
| S36 (E) | S. epidermidis | Oxar Kanr Tobr Genr Pefr Sxtr Fusi Lini | vga(A)v | 0.5 | I | 0.5 | S |
Hospitals at which strains were collected: A, HIA Val-de-Grace, Paris, France; B, HIA Robert Picqué, Bordeaux, France; C, HIA Clermont-Tonerre, Brest, France; D, HIA Percy, Clamart, France; E, HIA Legouest, Metz, France.
Species identification was done by 16S rRNA gene sequencing.
Antibiotype was determined according to EUCAST and CA-SFM table breakpoints. Fos, fosfomycin; Fus, fusidic acid; Gen, gentamicin; Kan, kanamycin; Lin, lincomycin; Oxa, oxacillin; Pef, pefloxacin; Pen, penicillin; Rif, rifampin; Sul, sulfonamide; Sxt, co-trimoxazole; Tei, teicoplanin; Tet, tetracyclin; Tob, tobramycin.
The macrolide-lincosamide-streptogramin (MLS) genotype was obtained by PCR amplification and direct sequencing.
S, susceptible; R, resistant; I, intermediate.
Except for the S. sciuri strain, all 23 other strains were PCR positive for vga(A) detection using primers V1 and V2. To analyze allele diversity within vga(A) in the linker region known to be important for antibiotic substrate specificity (6), 900 bp of each amplicon was sequenced with both or either of the two PCR primers. Comparison with protein sequences in the GenBank database yielded the following results: 4 linkers were exactly the same as Vga(A), 16 linkers were exactly the same as the Vga(A) variant [Vga(A)v], 1 linker was exactly the same as Vga(A)LC, 1 linker (allele 1) had two of the four mutations (L212S and A220T) reported for Vga(A)LC plus another newly described one (G215S), and 1 linker (allele 2) was exactly the same as EES57021, for which no functional information is available in the literature.
Using primers X and Y (Table 1), which point away from the EES57021 linker sequence, an inverse PCR cloning procedure was conducted to obtain the whole sequence of this vga(A) variant [vga(A)v] gene, along with additional information about its flanking regions. After digestion of bacterial DNA with HpaII, the corresponding restricted fragments were ligated and subjected to PCR amplification. The UV agarose-detectable amplicon was cleaned up by use of a Qiagen QIAquick PCR purification kit, cloned into a pCR4-Blunt TOPO vector (Invitrogen), and subsequently transformed into chemically competent Escherichia coli TOP10 cells. Completion of the sequence of the DNA insert was obtained using M13 universal and reverse primers, as well as primers X and Y. The 3.05-kbp sequence identified the EES57021 vga(A) variant in a gene linkage to be identical to that recently described for mobilizable plasmid pUR3036 (13).
To gain information on the functionality of EES57021 gene, plasmid DNA of strain S7 was isolated according to a modified Qiagen alkaline lysis procedure (14). The 2.7-kbp PvuII-EcoRI restriction fragment encompassing the presumed resistance gene was then cloned into SmaI-EcoRI-digested shuttle vector pRB474 (15). The recombinant plasmid was electroporated into Staphylococcus aureus RN4220. In comparison with S. aureus RN4220 carrying the empty plasmid, S. aureus RN4220 transformants carrying the EES57021 vga(A) variant cloned into pRB474 exhibited 4-fold- and 32-fold increased MIC values of clindamycin (1 μg/ml) and pristinamycin IIA (32 μg/ml), respectively. It would be useful to evaluate whether this variant confers resistance to pleuromutilin antibiotics.
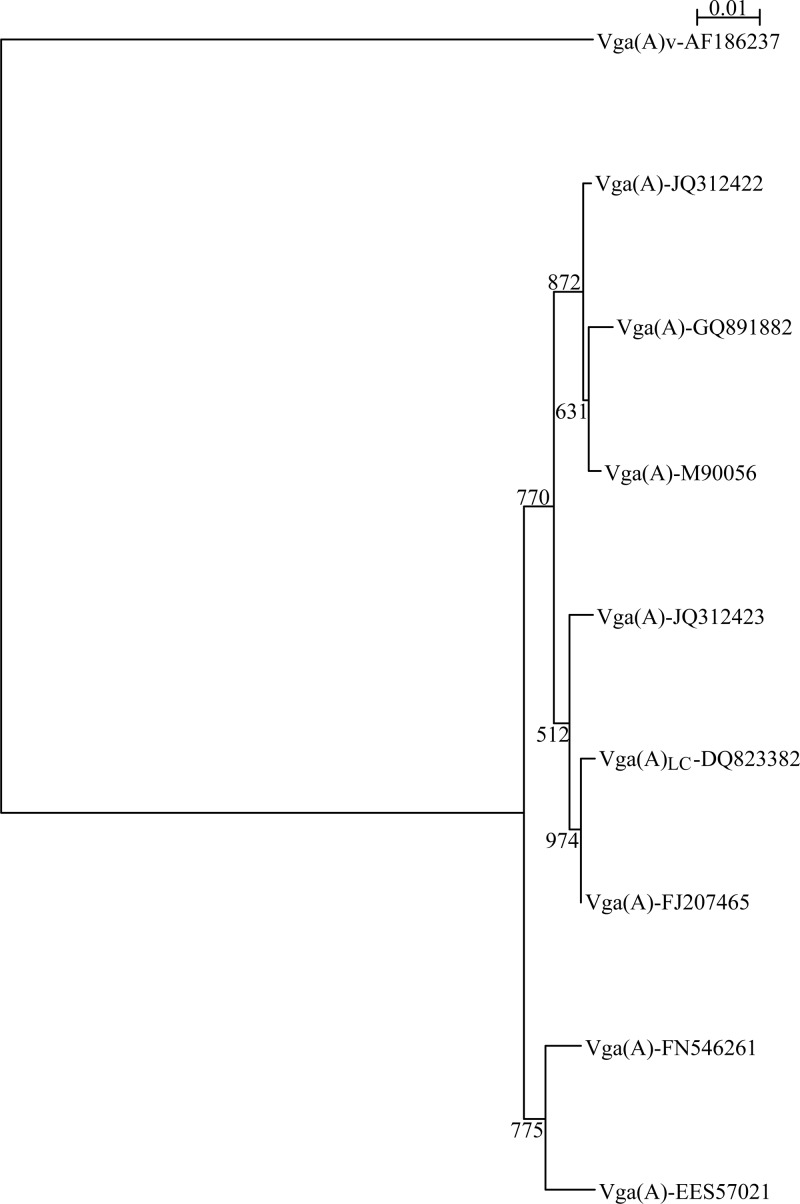
Following functional analysis of the EES57021 gene, nine Vga(A) variant proteins were characterized for their resistance properties (6, 13, 16–18). Their phylogenetic relationships distinguished Vga(A)v from all others (Fig. 1). The vga(A)v gene was shown to be disseminated by a chromosomal transposon rather than by plasmids (19, 20). In light of data collected in the United States and Europe, the allelic variability of vga(A) seems to be greater when a plasmid constitutes its genetic support. Most of the Vga(A) protein polymorphism was located within 20 amino acids of the linker region. This is clearly a hot spot for modulating the substrate specificity of the Vga(A) ABC proteins. There would appear to be an increased level of lincosamide resistance when leucine, glycine, and alanine residues of the Vga(A) allele linker sequence are mutated to serine or threonine residues. This was demonstrated by the study of Novotna and Janata in 2006 with Vga(A)LC (6) and by our present results with EES57021. Of note, the S7 strain of Staphylococcus epidermidis that bears an EES57021-carrying plasmid was one of the three clindamycin-resistant strains: the two others were the S5 strain of S. aureus, which carries a vga(A)v gene, and the S31 strain of Staphylococcus haemolyticus, which carries a vga(A)LC gene. The infrequent and confirmed plasmid location of the vga(A)v gene within strain S5 might explain why the levels of clindamycin resistance are higher than those usually observed for the other isolates (Table 2).
Fig 1.
Phylogenetic relatedness among Vga(A) isoforms. Multiple-sequence alignment was carried out with the ClustalX program. The phylogenetic tree was constructed using bootstrap neighbor joining and visualized by NJ plot tree drawing software.
A Tn5406 transposon structure was detected by PCR using primers T1 and T2 for all vga(A)v-carrying strains (Tables 1 and 2). To the best of our knowledge, this is the first report of the presence of Tn5406 within S. epidermidis isolates. Even though several PCR assays using various control strains and validated primers have been performed (21–24), no known resistance gene responsible for the LSA phenotype was evidenced in the unique S. sciuri strain. Since the time of this study, only one other possible resistance gene has been described among staphylococci. It corresponds to lsa(E) (25). This gene was found to be in close association with lnu(B) on plasmids that disseminate LSA resistance. If present in S. sciuri, the novel gene would be disseminated alone or in a remote gene linkage. Indeed, no trace of DNA homologous to lnu sequences (Table 1) was evidenced, apart from lnu(A) (Table 2).
In conclusion, this study does not show a wide diversity of resistance genes underlying the LSA phenotype among French staphylococcal isolates. The vga(A)v gene was found to be the most prevalent, and Tn5406 was first detected in S. epidermidis. Due to its chromosomal location, this resistance gene may not evolve as rapidly as the other vga(A) variant genes.
Nucleotide sequence accession number.
The 3.05-kbp sequence was deposited in GenBank under accession number KC539823.
ACKNOWLEDGMENTS
This work was supported by a French military health care PRC grant.
We are grateful to all providers of strains: C. Soler from HIA Percy, B. Soullie from HIA Robert Picqué, P. Le Guen from HIA Clermont-Tonnerre, and J.-M. Puyhardy from HIA Legouest.
Footnotes
Published ahead of print 24 June 2013
REFERENCES
- 1.Kerr ID, Reynolds ED, Cove JH. 2005. ABC proteins and antibiotic drug resistance: is it all about transport? Biochem. Soc. Trans. 33:1000–1002 [DOI] [PubMed] [Google Scholar]
- 2.Bunai K, Ariga M, Inoue T, Nozaki M, Ogane S, Kakeshita H, Nemoto T, Nakanishi H, Yamane K. 2004. Profiling and comprehensive expression analysis of ABC transporter solute-binding proteins of Bacillus subtilis membrane based on a proteomic approach. Electrophoresis 25:141–155 [DOI] [PubMed] [Google Scholar]
- 3.Ohki R, Tateno K, Takizawa T, Aiso T, Murata M. 2005. Transcriptional termination control of a novel ABC transporter gene involved in antibiotic resistance in Bacillus subtilis. J. Bacteriol. 187:5946–5954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chesneau O, Ligeret H, Hosan-Aghaie N, Morvan A, Dassa E. 2005. Molecular analysis of resistance to streptogramin A compounds conferred by the Vga proteins of staphylococci. Antimicrob. Agents Chemother. 49:973–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jacquet E, Girard JM, Ramaen O, Pamlard O, Lévaique H, Betton JM, Dassa E, Chesneau O. 2008. ATP hydrolysis and pristinamycin IIA inhibition of the Staphylococcus aureus Vga(A), a dual ABC protein involved in streptogramin A resistance. J. Biol. Chem. 283:25332–25339 [DOI] [PubMed] [Google Scholar]
- 6.Novotna G, Janata J. 2006. A new evolutionary variant of the streptogramin A resistance protein, Vga(A)LC from Staphylococcus haemolyticus with shifted substrate specificity towards lincosamides. Antimicrob. Agents Chemother. 50:4070–4076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwendener S, Perreten V. 2012. New MLSB resistance gene erm(43) in Staphylococcus lentus. Antimicrob. Agents Chemother. 56:4746–4752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shen J, Wang Y, Schwarz S. 29 March 2013. Presence and dissemination of the multi-resistance gene cfr in Gram-positive and Gram-negative bacteria. J. Antimicrob. Chemother. [Epub ahead of print.] 10.1093/jac/dkt092 [DOI] [PubMed] [Google Scholar]
- 9.Nunez-Samudio V, Chesneau O. 2013. Functional interplay between the ATP binding cassette Msr(D) protein and the membrane facilitator superfamily Mef(E) transporter for macrolide resistance in Escherichia coli. Res. Microbiol. 164:226–235 [DOI] [PubMed] [Google Scholar]
- 10.Leclercq R, Canton R, Brown DFJ, Giske CG, Heisig P, MacGowan AP, Mouton JW, Nordmann P, Rodloff AC, Rossolini GM, Soussy CJ, Steinbakk M, Winstanley TG, Kahlmeter G. 2013. EUCAST expert rules in antimicrobial susceptibility testing. Clin. Microbiol. Infect. 19:141–160 [DOI] [PubMed] [Google Scholar]
- 11.Leclercq R, Courvalin P. 1991. Intrinsic and unusual resistance to macrolide, lincosamide, and streptogramin A antibiotics in bacteria. Antimicrob. Agents Chemother. 35:1273–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Becker K, Harmsen D, Mellmann A, Meier C, Schumann P, Peters G, von Eiff C. 2004. Development and evaluation of a quality-controlled ribosomal sequence database for 16S ribosomal DNA-based identification of Staphylococcus species. J. Clin. Microbiol. 42:4988–4995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lozano C, Aspiroz C, Rezusta A, Gomez-Sanz E, Simon C, Gomez P, Ortega C, Revillo MJ, Zarazaga M, Torres C. 2012. Identification of novel vga(A)-carrying plasmids and a Tn5406-like transposon in meticillin-resistant Staphylococcus aureus and Staphylococcus epidermidis of human and animal origin. Int. J. Antimicrob. Agents 40:306–312 [DOI] [PubMed] [Google Scholar]
- 14.Schwarz S, Spies U, Cardoso M. 1991. Cloning and sequence analysis of a plasmid-encoded chloramphenicol acetyltransferase gene from Staphylococcus intermedius. J. Gen. Microbiol. 137:977–981 [DOI] [PubMed] [Google Scholar]
- 15.Bruckner R. 1992. A series of shuttle vectors for Bacillus subtilis and Escherichia coli. Gene 122:187–192 [DOI] [PubMed] [Google Scholar]
- 16.Gentry DR, McCloskey L, Gwynn MN, Rittenhouse SF, Scangarella N, Shawar R, Holmes DJ. 2008. Genetic characterization of Vga ABC proteins conferring reduced susceptibility to pleuromutilins in Staphylococcus aureus. Antimicrob. Agents Chemother. 52:4507–4509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mendes RE, Smith TC, Deshpande L, Diekema DJ, Sader HS, Jones RN. 2011. Plasmid-borne vga(A)-encoding gene in methicillin-resistant Staphylococcus aureus ST398 recovered from swine and a swine farmer in the United States. Diagn. Microbiol. Infect. Dis. 71:177–180 [DOI] [PubMed] [Google Scholar]
- 18.Qin X, Poon B, Kwong J, Niles D, Schmidt BZ, Rajagopal L, Gantt S. 2011. Two paediatric cases of skin and soft-tissue infections due to clindamycin-resistant Staphylococcus aureus carrying a plasmid-encoded vga(A) allelic variant for a putative efflux pump. Int. J. Antimicrob. Agents 38:81–83 [DOI] [PubMed] [Google Scholar]
- 19.Haroche J, Allignet J, El Solh N. 2002. Tn5406, a new staphylococcal transposon conferring resistance to streptogramin A and related compounds including dalfopristin. Antimicrob. Agents Chemother. 46:2337–2343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haroche J, Morvan A, Davi M, Allignet J, Bimet F, El Solh N. 2003. Clonal diversity among streptogramin A-resistant Staphylococcus aureus isolates collected in French hospitals. J. Clin. Microbiol. 41:586–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gomez-Sanz E, Torres C, Lozano C, Fernandez-Perez R, Aspiroz C, Ruiz-Larrea F, Zarazaga M. 2010. Detection, molecular characterization, and clonal diversity of Staphylococcus aureus CC398 and CC97 in Spanish slaughter pigs of different age groups. Foodborne Pathog. Dis. 7:1269–1277 [DOI] [PubMed] [Google Scholar]
- 22.Hauschild T, Fessler AT, Kadlec K, Billerbeck C, Schwarz S. 2012. Detection of the novel vga(E) gene in methicillin-resistant Staphylococcus aureus CC398 isolates from cattle and poultry. J. Antimicrob. Chemother. 67:503–504 [DOI] [PubMed] [Google Scholar]
- 23.Kehrenberg C, Schwarz S. 2006. Distribution of florfenicol resistance genes fexA and cfr among chloramphenicol-resistant Staphylococcus isolates. Antimicrob. Agents Chemother. 50:1156–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Werner G, Cuny C, Schmitz FJ, Witte W. 2001. Methicillin-resistant, quinupristin-dalfopristin-resistant Staphylococcus aureus with reduced sensitivity to glycopeptides. J. Clin. Microbiol. 39:3586–3590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wendlandt S, Lozano C, Kadlec K, Gomez-Sanz E, Zarazaga M, Torres M, Schwarz S. 2013. The enterococcal ABC transporter gene lsa(E) confers combined resistance to lincosamides, pleuromutilins and streptogramin A antibiotics in methicillin-susceptible and methicillin-resistant Staphylococcus aureus. J. Antimicrob. Chemother. 68:473–475 [DOI] [PubMed] [Google Scholar]