Abstract
The spread of pandemic methicillin-resistant Staphylococcus aureus (MRSA) clones such as USA300 and EMRSA-15 is a global health concern. As a part of a surveillance study of three long-term care facilities in the Greater Chicago area, phenotypic and molecular characterization of nasal MRSA isolates was performed. We report a cluster of pandemic EMRSA-15, an MRSA clone rarely reported from the United States, detected during this study.
TEXT
The global spread of methicillin-resistant Staphylococcus aureus (MRSA) is one of the most serious public health challenges worldwide. MRSA-related hospitalizations have increased by an estimated 118% between 1999 and 2005 (1) and continue to rise at many U.S. academic hospitals (2). Long-term care facility (LTCF) residents are at high risk for MRSA carriage (3). Rates of MRSA infection may increase faster in nursing home residents than in hospital inpatients (4), possibly due to unique associated risk factors, such as greater social interaction and multiple interactions with health care workers, including repeated hospital admissions (5). Recent evidence suggests that infection control strategies in the LTCF, such as identification of MRSA carriage and prevention of disease, could have a long-term positive impact on regional MRSA control (6). Infection control policies are often guided by, and benefit from, knowledge of the local MRSA epidemiology, especially identification of the reservoirs of epidemic clones in the population or of major changes in the existing MRSA clones (7). We report the detection of epidemic MRSA-15 (EMRSA-15), a clone that is widespread in the United Kingdom and 15 countries worldwide, in LTCF residents (8), which has hitherto been reported to account for only a very small number of surveillance isolates across the United States (9).
The isolates included in this study were part of an active MRSA surveillance program to reduce colonization at three Chicago area LTCFs, between March 2011 and November 2012. Identification of nasal MRSA carriage was done using Cepheid Xpert MRSA (Cepheid, Sunnyvale, CA) according to the manufacturer's instructions. A positive result was confirmed by growth on selective agar, BBL CHROMagar II (BD Diagnostics, Sparks, MD), and a positive Staphaurex agglutination test (Remel, Lenexa, KS). Testing of antimicrobial susceptibility to ciprofloxacin, trimethoprim-sulfamethoxazole, gentamicin, minocycline, and clindamycin was performed by disk diffusion on Mueller-Hinton agar (BD Diagnostics) and interpreted according to the Clinical and Laboratory Standards Institute (CLSI) guidelines (10). Isolates were typed by pulsed-field gel electrophoresis (PFGE) using SmaI as previously described (11, 12). The patterns were identified on BioNumerics version 6.6 (Applied Maths Inc., Austin, TX) using a dendrogram generated by the unweighted-pair group method with arithmetic mean based on Dice coefficients, where optimization and band position tolerance were set at 0.8 and 1.5%, respectively. A similarity coefficient of 80% was selected to define the patterns (9, 12). Assignment of pulsotype was correlated by comparison to published literature (13, 14). All MRSA isolates were tested for genes encoding high-level mupirocin resistance (mupA) and the Panton-Valentine leukocidin (PVL) toxin as previously described (15, 16). The isolates included in this study also underwent spa typing (17). This study was approved by the Institutional Review Board of NorthShore University HealthSystem.
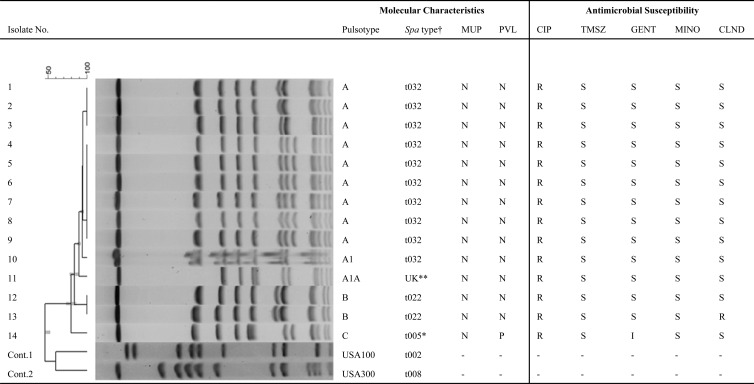
Among the 803 MRSA isolates identified, 22 (2.7%) isolates from 14 patients were recognized as belonging to the EMRSA-15 pulsotype (18). These isolates could be divided into three major PFGE patterns, with one pattern being predominant (Fig. 1). All except one isolate (spa type unknown) belonged to spa types included in the ST22 group, a characteristic of EMRSA-15 isolates (Ridom SpaServer database; http://www3.ridom.de/spa-server/). All isolates were uniformly resistant to ciprofloxacin but susceptible to trimethoprim-sulfamethoxazole and minocycline. One isolate was resistant to clindamycin, and one had intermediate susceptibility to gentamicin. One isolate tested positive for the PVL gene, and mupA was not detected in any of the isolates (Fig. 1). The susceptibility observed matches the EMRSA-15 susceptibility pattern: susceptible to trimethoprim-sulfamethoxazole and gentamicin but resistant to ciprofloxacin (7). Evidence in the literature suggests that ciprofloxacin resistance confers a survival advantage to EMRSA-15 and could be a factor enabling it to supplant other existing clones (19, 20). Whole-genome sequencing studies show that ciprofloxacin resistance is a recently acquired genetic trait in EMRSA-15, which has led to the separation of clade ST22A2 (19). The emergence of this clade has been traced to shortly after the introduction of fluoroquinolones into clinical medicine in the United Kingdom (19). It appears therefore that the EMRSA-15 isolates observed in this study are comparable to the ones described in Europe (7, 19, 20).
Fig 1.
Molecular characterization and antibiogram of isolates showing EMRSA-15 pulsotype. The PFGE dendrogram compares fingerprint patterns of the related isolates from 14 patients. Columns marked “MUP” and “PVL” indicate the results for genetic tests performed to detect the mupA and PVL genes, respectively. CIP, ciprofloxacin; TMSZ, trimethoprim-sulfamethoxazole; GENT, gentamicin; MINO, minocycline; CLIND, clindamycin; N, negative; P, positive; S, susceptible; R, resistant; I, intermediate; †, all isolates belong to spa types included in the cluster ST22, a recognized cluster for EMRSA-15; **, this isolate has a unique pulsotype, although it is closely related to pulsotype A; the spa sequence was unique and not listed in the Ridom SpaServer database; *, this isolate had a distinct PFGE profile and belongs to spa type t005; it is also the only isolate positive for the PVL gene; of note, spa type t005 has been reported to be PVL positive (29).
The epidemiology of MRSA is unique in being able to spread in pandemic waves with successful global dissemination of certain MRSA clones, such as USA300 between Australia (21) and Europe (22), which is of significant concern to health care professionals around the world. The mechanisms leading to the relative success of these pandemic clones in being able to effectively displace other existing ones remain poorly understood. Epidemic MRSA or EMRSA-15 is one such clone; emerging in the United Kingdom in the 1990s, it has not only successfully spread in hospitals within the United Kingdom but also repeatedly replaced other established MRSA strains locally in several different countries in Europe, Australia, and Asia (19). In recent years, EMRSA-15 and EMRSA-16 strains combined have accounted for 93 to 95% of MRSA bacteremias in the United Kingdom, with EMRSA-15 alone accounting for greater than 60% of infections (23, 24). Recent surveillance studies in the United States, however, found EMRSA-15 to account for only 0.5% of MRSA isolated from the blood (1 of 194 isolates) and 0.3% of MRSA isolated from the nares (1 of 299) in the United States (25). Earlier U.S. national surveillance reported an EMRSA-15 prevalence of 0.2% among 1,984 invasive isolates (9). We identified 22 EMRSA-15 isolates from 14 patients in three nursing homes that share patients with some 40 acute care facilities in the region. Further, 12 of these 14 patients were residents of one nursing home, and their nasal surveillance isolates accounted for 5.8% of all MRSA isolates identified at that facility. In fact, to the best of our knowledge this represents the largest identified cluster of EMRSA-15 isolates observed in the United States, which is of concern since EMRSA-15 has been shown to replace previously predominant MRSA strains in Germany, Spain, Portugal, and Singapore (26–28). Further studies are required to monitor the rates of colonization by EMRSA-15 in these LTCFs and other hospitals in the area to determine whether this is an isolated event or an indication of the introduction of a new pandemic clone.
ACKNOWLEDGMENTS
We thank Colleen Siemen at the Microbiology Laboratory, NorthShore University HealthSystem, for help with spa sequencing.
This work was funded by the Agency for Healthcare Research and Quality (Grant 1R18HS019968 award).
Footnotes
Published ahead of print 24 June 2013
REFERENCES
- 1.Klein E, Smith DL, Laxminarayan R. 2007. Hospitalizations and deaths caused by methicillin-resistant Staphylococcus aureus, United States, 1999–2005. Emerg. Infect. Dis. 13:1840–1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.David MZ, Medvedev S, Hohmann SF, Ewigman B, Daum RS. 2012. Increasing burden of methicillin-resistant Staphylococcus aureus hospitalizations at US academic medical centers, 2003-2008. Infect. Control Hosp. Epidemiol. 33:782–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mody L, Kauffman CA, Donabedian S, Zervos M, Bradley SF. 2008. Epidemiology of Staphylococcus aureus colonization in nursing home residents. Clin. Infect. Dis. 46:1368–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Delorme T, Rose S, Senita J, Callahan C, Nasr P. 2009. Epidemiology and susceptibilities of methicillin-resistant Staphylococcus aureus in Northeastern Ohio. Am. J. Clin. Pathol. 132:668–677 [DOI] [PubMed] [Google Scholar]
- 5.Murphy CR, Quan V, Kim D, Peterson E, Whealon M, Tan G, Evans K, Meyers H, Cheung M, Lee BY, Mukamel DB, Huang SS. 2012. Nursing home characteristics associated with methicillin-resistant Staphylococcus aureus (MRSA) burden and transmission. BMC Infect. Dis. 12:269. 10.1186/1471-2334-12-269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee BY, Singh A, Bartsch SM, Wong KF, Kim DS, Avery TR, Brown ST, Murphy CR, Yilmaz SL, Huang SS. 2013. The potential regional impact of contact precaution use in nursing homes to control methicillin-resistant Staphylococcus aureus. Infect. Control Hosp. Epidemiol. 34:151–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amorim ML, Faria NA, Oliveira DC, Vasconcelos C, Cabeda JC, Mendes AC, Calado E, Castro AP, Ramos MH, Amorim JM, de Lencastre H. 2007. Changes in the clonal nature and antibiotic resistance profiles of methicillin-resistant Staphylococcus aureus isolates associated with spread of the EMRSA-15 clone in a tertiary care Portuguese hospital. J. Clin. Microbiol. 45:2881–2888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grundmann H, Aanensen DM, van den Wijngaard CC, Spratt BG, Harmsen D, Friedrich AW. 2010. Geographic distribution of Staphylococcus aureus causing invasive infections in Europe: a molecular-epidemiological analysis. PLoS Med. 7:e1000215. 10.1371/journal.pmed.1000215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Limbago B, Fosheim GE, Schoonover V, Crane CE, Nadle J, Petit S, Heltzel D, Ray SM, Harrison LH, Lynfield R, Dumyati G, Townes JM, Schaffner W, Mu Y, Fridkin SK. 2009. Characterization of methicillin-resistant Staphylococcus aureus isolates collected in 2005 and 2006 from patients with invasive disease: a population-based analysis. J. Clin. Microbiol. 47:1344–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clinical and Laboratory Standards Institute 2010. Performance standards for antimicrobial susceptibility testing. Approved standard M100-S17, vol 30 Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 11.Jaggi P, Paule SM, Peterson LR, Tan TQ. 2007. Characteristics of Staphylococcus aureus infections, Chicago Pediatric Hospital. Emerg. Infect. Dis. 13:311–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McDougal LK, Steward CD, Killgore GE, Chaitram JM, McAllister SK, Tenover FC. 2003. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J. Clin. Microbiol. 41:5113–5120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goering RV, Shawar RM, Scangarella NE, O'Hara FP, Amrine-Madsen H, West JM, Dalessandro M, Becker JA, Walsh SL, Miller LA, van Horn SF, Thomas ES, Twynholm ME. 2008. Molecular epidemiology of methicillin-resistant and methicillin-susceptible Staphylococcus aureus isolates from global clinical trials. J. Clin. Microbiol. 46:2842–2847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Golding GR, Campbell JL, Spreitzer DJ, Veyhl J, Surynicz K, Simor A, Mulvey MR. 2008. A preliminary guideline for the assignment of methicillin-resistant Staphylococcus aureus to a Canadian pulsed-field gel electrophoresis epidemic type using spa typing. Can. J. Infect. Dis. Med. Microbiol. 19:273–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hacek DM, Robb WJ, Paule SM, Kudrna JC, Stamos VP, Peterson LR. 2008. Staphylococcus aureus nasal decolonization in joint replacement surgery reduces infection. Clin. Orthop. Relat. Res. 466:1349–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paule SM, Pasquariello AC, Hacek DM, Fisher AG, Thomson RB, Jr, Kaul KL, Peterson LR. 2004. Direct detection of Staphylococcus aureus from adult and neonate nasal swab specimens using real-time polymerase chain reaction. J. Mol. Diagn. 6:191–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mathema B, Mediavilla J, Kreiswirth BN. 2008. Sequence analysis of the variable number tandem repeat in Staphylococcus aureus protein A gene: spa typing. Methods Mol. Biol. 431:285–305 [DOI] [PubMed] [Google Scholar]
- 18.Wolter DJ, Chatterjee A, Varman M, Goering RV. 2008. Isolation and characterization of an epidemic methicillin-resistant Staphylococcus aureus 15 variant in the central United States. J. Clin. Microbiol. 46:3548–3549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holden MT, Hsu LY, Kurt K, Weinert LA, Mather AE, Harris SR, Strommenger B, Layer F, Witte W, de Lencastre H, Skov R, Westh H, Zemlickova H, Coombs G, Kearns AM, Hill RL, Edgeworth J, Gould I, Gant V, Cooke J, Edwards GF, McAdam PR, Templeton KE, McCann A, Zhou Z, Castillo-Ramirez S, Feil EJ, Hudson LO, Enright MC, Balloux F, Aanensen DM, Spratt BG, Fitzgerald JR, Parkhill J, Achtman M, Bentley SD, Nubel U. 2013. A genomic portrait of the emergence, evolution and global spread of a methicillin resistant Staphylococcus aureus pandemic. Genome Res. 23:653–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horvath A, Dobay O, Kardos S, Ghidan A, Toth A, Paszti J, Ungvari E, Horvath P, Nagy K, Zissman S, Fuzi M. 2012. Varying fitness cost associated with resistance to fluoroquinolones governs clonal dynamic of methicillin-resistant Staphylococcus aureus. Eur. J. Clin. Microbiol. Infect. Dis. 31:2029–2036 [DOI] [PubMed] [Google Scholar]
- 21.Gottlieb T, Su WY, Merlino J, Cheong EY. 2008. Recognition of USA300 isolates of community-acquired methicillin-resistant Staphylococcus aureus in Australia. Med. J. Aust. 189:179–180 [DOI] [PubMed] [Google Scholar]
- 22.Ruppitsch W, Stoger A, Schmid D, Fretz R, Indra A, Allerberger F, Witte W. 2007. Occurrence of the USA300 community-acquired Staphylococcus aureus clone in Austria. Euro Surveill. 12:E071025 071021 http://www.eurosurveillance.org/viewarticle.aspx?articleid=3294 [DOI] [PubMed] [Google Scholar]
- 23.Ellington MJ, Hope R, Livermore DM, Kearns AM, Henderson K, Cookson BD, Pearson A, Johnson AP. 2010. Decline of EMRSA-16 amongst methicillin-resistant Staphylococcus aureus causing bacteraemias in the UK between 2001 and 2007. J. Antimicrob. Chemother. 65:446–448 [DOI] [PubMed] [Google Scholar]
- 24.Johnson AP, Aucken HM, Cavendish S, Ganner M, Wale MC, Warner M, Livermore DM, Cookson BD. 2001. Dominance of EMRSA-15 and -16 among MRSA causing nosocomial bacteraemia in the UK: analysis of isolates from the European Antimicrobial Resistance Surveillance System (EARSS). J. Antimicrob. Chemother. 48:143–144 [DOI] [PubMed] [Google Scholar]
- 25.Tenover FC, Tickler IA, Goering RV, Kreiswirth BN, Mediavilla JR, Persing DH. 2012. Characterization of nasal and blood culture isolates of methicillin-resistant Staphylococcus aureus from patients in United States Hospitals. Antimicrob. Agents Chemother. 56:1324–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aires-de-Sousa M, Correia B, de Lencastre H. 2008. Changing patterns in frequency of recovery of five methicillin-resistant Staphylococcus aureus clones in Portuguese hospitals: surveillance over a 16-year period. J. Clin. Microbiol. 46:2912–2917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Albrecht N, Jatzwauk L, Slickers P, Ehricht R, Monecke S. 2011. Clonal replacement of epidemic methicillin-resistant Staphylococcus aureus strains in a German university hospital over a period of eleven years. PLoS One 6:e28189. 10.1371/journal.pone.0028189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hsu LY, Loomba-Chlebicka N, Koh YL, Tan TY, Krishnan P, Lin RT, Tee NW, Fisher DA, Koh TH. 2007. Evolving EMRSA-15 epidemic in Singapore hospitals. J. Med. Microbiol. 56:376–379 [DOI] [PubMed] [Google Scholar]
- 29.Boakes E, Kearns AM, Ganner M, Perry C, Warner M, Hill RL, Ellington MJ. 2011. Molecular diversity within clonal complex 22 methicillin-resistant Staphylococcus aureus encoding Panton-Valentine leukocidin in England and Wales. Clin. Microbiol. Infect. 17:140–145 [DOI] [PubMed] [Google Scholar]