Abstract
Y143C,R substitutions in HIV-1 integrase define one of three primary raltegravir (RAL) resistance pathways. Here we describe clinical isolates with alternative substitutions at position 143 (Y143A, Y143G, Y143H, and Y143S [Y143A,G,H,S]) that emerge less frequently, and we compare the genotypic and phenotypic profiles of these viruses to Y143C,R viruses to reconcile the preferential selection of Y143C,R variants during RAL treatment. Integrase amino acid sequences and RAL susceptibility were characterized in 117 patient isolates submitted for drug resistance testing and contained Y143 amino acid changes. The influence of specific Y143 substitutions on RAL susceptibility and their preferential association with particular secondary substitutions were further defined by evaluating the composition of patient virus populations along with a large panel of site-directed mutants. Our observations demonstrate that the RAL resistance profiles of Y143A,G,H,S viruses and their association with specific secondary substitutions are similar to the well-established Y143C profile but distinct from the Y143R profile. Y143R viruses differ from Y143A,C,G,H,S viruses in that Y143R confers a greater reduction in RAL susceptibility as a single substitution, consistent with a lower resistance barrier. Among Y143A,C,G,H,S viruses, the higher prevalence of Y143C viruses is the result of a lower genetic barrier than that of the Y143A,G,S viruses and a lower resistance barrier than that of the Y143H viruses. In addition, Y143A,C,G,H,S viruses require multiple secondary substitutions to develop large reductions in RAL susceptibility. Patient-derived viruses containing Y143 substitutions exhibit cross-resistance to elvitegravir.
INTRODUCTION
HIV-1 integrase inhibitors act by blocking the integration of viral double-stranded DNA into the host cell chromosomal DNA. Different classes of integrase inhibitors have been described and act by interfering with the 3′ processing of viral DNA long terminal repeats, preventing the strand transfer of viral DNA into the host genome, or targeting integrase allosterically (1–6). Two strand transfer inhibitors have progressed to clinical treatment settings: raltegravir (RAL), approved by the FDA in 2007 for the treatment of HIV-1 infection in antiretroviral (ARV) treatment-experienced patients and in 2009 for the treatment of ARV-naive patient populations (7–11); and elvitegravir (EVG) coformulated with cobicistat, tenofovir disoproxil fumarate, and emtricitabine, recently approved by the FDA in 2012 for the treatment of ARV treatment-naive patients (12, 13). A third strand transfer inhibitor, dolutegravir, is currently in phase III clinical evaluation (14).
Three major mutation pathways that confer RAL resistance have been reported in patients failing RAL-containing treatment regimens. Each of these pathways is defined by one or more substitutions at specific amino acid positions within the integrase coding region: Y143C or R (here denoted Y143C,R), Q148H,K,R, and N155H (8, 15–19). Cross-resistance between RAL and EVG has been described for the viruses containing Q148H,K,R and N155H substitutions (20–22). However, it has been reported that viruses containing amino acid changes at position 143 retain susceptibility to EVG (23).
RAL-resistant viruses that contain amino acid substitutions at position 143 emerge less frequently than viruses with substitutions at position 148 or 155 (8, 16, 24). However, longitudinal evaluations have demonstrated that RAL-resistant viruses with substitutions at position 148 or 155 may be replaced by variants with Y143 substitutions upon continued RAL treatment (25–27). In a majority of cases, Y143 substitutions comprise C and R, both of which can confer significant reductions in RAL susceptibility (17, 18, 25, 27). RAL-resistant viruses with Y143H substitutions have also been reported, albeit much less frequently (27, 28).
Several studies have demonstrated significant associations between specific primary and secondary integrase inhibitor resistance substitutions. For example, E92Q and G140A, S are associated with N155H and Q148H,K,R, respectively (16, 29), whereas T97A is associated with Y143C,R (27, 30). In addition, L74I,M, E92Q, E138K, V151I, G163R, I203M, and S230R have been associated with RAL treatment failure (31). G140S and T97A substitutions rescue the catalytic defect conferred by Q148H and Y143C,R substitutions, respectively (29, 30).
During the inspection of viruses submitted for routine RAL resistance testing, we have observed several substitutions at position 143 other than Y143C or Y143R. To better understand the influence of distinct resistance and genetic barriers of alternative Y143 substitutions (Y143X) and RAL resistance-associated secondary substitutions on RAL susceptibility, we analyzed 117 Y143X patient virus populations and clones derived from a subset of these virus populations as well as a series of site-directed mutants (SDMs) containing resistance-associated substitutions. We also used patient isolates and SDMs to ascertain the impact of Y143X resistance profiles on EVG susceptibility.
MATERIALS AND METHODS
Patient samples.
One hundred seventeen HIV-1-positive patient samples that were submitted to the Monogram Biosciences Clinical Reference Laboratory for routine integrase inhibitor resistance testing and that contained amino acid substitutions at position 143 of the HIV-1 integrase coding region were selected for this study. Molecular clones (n = 20 to 48 per sample) were isolated from each of a subset of 23 patient virus populations that contained amino acid mixtures at integrase amino acid position 143.
Site-directed mutagenesis.
Single Y143A,C,G,H,R,S substitutions were introduced into the integrase coding region of an HIV-1 resistance test vector (RTV), as previously described (16, 32). Secondary substitutions L74I, M, T97A, G163R, and S230R were introduced into Y143A,C,G,H,R,S single-site mutant vectors to create double-, triple-, or quadruple-site mutants.
Integrase inhibitor susceptibility assays.
Susceptibilities of patient-derived integrase samples and SDMs were assessed using the PhenoSense Integrase assay (16). The C-terminal connection and RNase H domains of the reverse transcriptase coding regions along with the integrase coding regions (collectively referred to as RH/IN) of patient plasma viruses were amplified and transferred into RTVs containing a luciferase reporter gene. HEK293 cells were transfected with RTVs containing patient-derived RH/IN sequences together with an amphotropic murine leukemia virus envelope expression vector to generate pseudovirus stocks that were used to infect fresh HEK293 cells in the presence and absence of integrase inhibitors RAL (Merck Research Labs) and EVG (Gilead Sciences). Susceptibility to RAL or EVG was calculated by plotting the percent inhibition of virus replication (luciferase activity) at increasing drug concentrations. Reductions in susceptibility were represented as the fold changes (FC) in 50% effective concentrations (EC50) of the test virus relative to the EC50 of the reference virus. Validation studies have demonstrated that the variation in FC measurements among multiple replicates is less than 2-fold (validation report on file). A Wilcoxon test was performed to compare the number of secondary substitutions in Y143X viruses. Correlation of FC values between RAL and EVG was evaluated with nonparametric tests (Spearman's rank correlation coefficient) using GraphPad Prism 5 (GraphPad, San Diego, CA).
Integrase-mediated RC.
Integrase-mediated replication capacity (RC) was determined and expressed as a percentage of viral infectivity (luciferase activity) in the absence of drug relative to the wild-type reference strain. The selective advantage of mutant (resistant) and wild-type (susceptible) variants was assessed by comparing the ratio of luciferase activity of the mutant and wild-type variants in the absence of RAL and in the presence of increasing concentrations of RAL.
Integrase DNA sequencing.
The complete nucleotide sequences of the integrase coding regions of patient virus populations, molecular clones derived from patient viruses, and SDMs were determined using conventional Sanger dye-dideoxy chain terminator chemistry (ABI, Foster City, CA). Differences in nucleotide and deduced amino acid sequences were recorded relative to the NL4-3 reference sequence. Validation studies have demonstrated that the detection of minor variants ranges from 10 to 20% (validation report on file).
RESULTS
Identification of clinical isolates containing Y143A, G, H, or S substitutions.
We evaluated 117 HIV-1 clinical isolates containing substitutions at integrase amino acid position 143. Among these samples, 77 displayed a single homogeneous substitution at position 143, 17 displayed mixed amino acid substitutions at position 143, and 23 displayed a second primary substitution at either position 148 or position 155 in addition to position 143 (Table 1). The majority of the 77 samples with single substitutions at position 143 harbored either Y143R (n = 44) or Y143C (n = 23) substitutions; however, 10 samples contained other substitutions: Y143A (n = 1), Y143G (n = 3), Y143H (n = 2), and Y143S (n = 4). Among the 17 samples with mixed amino acids at position 143 (wild-type and/or other RAL resistance-associated substitutions), nearly one-half displayed mixtures of Y143Y/C/H/R (n = 8), while the remainder displayed mixtures including Y143C/R (n = 3), Y143Y/C (n = 3), Y143C/G (n = 2), and Y143H/R (n = 1) (Table 1). Among the 23 samples displaying combinations of primary substitutions, the majority of samples were comprised of 143 and 155 (n = 18) variants while the remaining samples were comprised of 143 and 148 (n = 5) variants. Based on population sequencing, 13/23 samples were suggestive of mixed subpopulations of distinct 143 and 148 variants or distinct 143 and 155 variants, whereas the remaining 10 samples were more suggestive of direct linkages among 143 and 148 or 143 and 155 substitutions, which was confirmed by the analysis of molecular clones derived from a subset of these virus populations (Table 1).
Table 1.
Analysis of 117 pseudoviruses containing patient-derived HIV-1 integrase sequences harboring Y143X substitutions
| Sample group and no. of samplesa | Substitution(s) at primary positions as determined by: |
|
|---|---|---|
| Population sequenceb | Clonal analysisc | |
| Y143 (n = 77) | ||
| 44 | Y143R | NP |
| 23 | Y143C | NP |
| 4 | Y143S | NP |
| 3 | Y143G | NP |
| 2 | Y143H | NP |
| 1 | Y143A | NP |
| Y143 mixtures (n = 17) | ||
| 6 | Y143Y/C/H/R | Y143Y,R |
| 1 | Y143Y/C/H/R | Y143Y,C,R |
| 1 | Y143Y/C/H/R | Y143Y,C,G,R |
| 3 | Y143C/R | NP |
| 3 | Y143Y/C | NP |
| 2 | Y143C/G | Y143C,G |
| 1 | Y143H/R | Y143H,R |
| Y143 and N155 (n = 18) | ||
| 4 | Y143Y/C/H/R, N155N/H | Y143R, N155H |
| 4 | Y143Y/C, N155N/H | NP |
| 2 | Y143Y/C, N155H | NP |
| 2 | Y143R, N155H | NP |
| 1 | Y143H/R, N155H | Y143H,R+N155H |
| 1 | Y143Y/C/S, N155N/H | Y143C,S, N155H |
| 1 | Y143Y/C/D/G, N155N/H | Y143C,G, N155H |
| 1 | Y143C, N155H | NP |
| 1 | Y143H, N155H | NP |
| 1 | Y143R/C/G, N155G | Y143C,G,R+N155G |
| Y143 and Q148 (n = 5) | ||
| 2 | Y143Y/C/H/R, Q148Q/H | Y143R, Q148H |
| 1 | Y143Y/H, Q148H | Y143H+Q148H, Q148H |
| 1 | Y143Y/C, Q148H | NP |
| 1 | Y143Y/C/D/G, Q148Q/R | Y143C,G, Q148R |
One hundred seventeen patient-derived HIV-1 integrase sequences with Y143X substitutions were grouped based upon the absence or presence of mixed substitutions at position 143 or the presence of other RAL primary substitutions (Y143 and N155; or Y143 and Q148). Numbers in boldface in this column indicate that Y143H and Y143D substitutions predicted based on population sequences were not identified in clonal analyses.
IN population sequences containing predicted substitutions at positions 143, 148, and 155 are indicated. Substitutions in boldface in this column are present at positions 143 and 155 or 148 without mixtures at one or both positions, which suggests that these viruses contain two linked primary substitutions.
IN clonal sequences isolated from 23 samples (20 to 48 clones/each sample) predicted to contain alternative substitution Y143D, H, G, or S were analyzed. NP, not performed.
Based on derived amino acid sequences, 17/36 virus populations containing mixtures at position 143 were predicted to contain Y143H (Y143Y/C/H/R, Y143Y/H, and Y143H/R). This notably contrasted with the representation of Y143H among the subset of viruses lacking mixtures at position 143 (2.6%, 2/77) (Table 1). To attempt to verify Y143H and other alternative substitutions, we performed clonal analysis on 23 virus populations containing alternative amino acid mixtures at position 143 based on the population sequences (Table 1). The clonal analysis of 14 virus populations with predicted Y143Y/C/H/R mixtures demonstrated that 12 virus populations were comprised solely of Y143 and Y143R variants, 1 population was comprised of Y143, Y143C, and Y143R variants, and 1 population was comprised of Y143, Y143C, Y143G, and Y143R variants. Notably, Y143H clones were identified only in the three virus populations with predicted amino acid mixtures Y143H/R (n = 2) and Y143Y/H (n = 1). These data indicate that Y143H substitutions occur far less frequently in patient viruses than predicted based on the amino acid sequences that are deduced from the nucleotide sequences of virus populations. Similarly, Y143C variants were not identified in 12/14 virus populations with predicted Y143Y/C/H/R sequences, nor were Y143D variants in two Y143Y/C/D/G sequences. Based on the genetic code, both Y143H (CAC, CAT) and Y143C (TGC, TGT) substitutions are predicted when Y143 (TAC, TAT) and Y143R (CGC, CGT) variants are both present within a virus population, whereas Y143D (GAC, GAT) substitutions are predicted when Y143 (TAC, TAT) and Y143G (GGC, GGT) variants are both present in a virus population. The presence of Y143G variants in four virus populations and Y143S variants in one virus population was confirmed by clonal analysis (Table 1). Notably, one virus population contained a previously unreported N155G substitution in addition to Y143 substitutions. The ability of N155G to reduce RAL susceptibility was confirmed by SDM (data not shown).
Patient-derived viruses containing Y143X substitutions display broad reductions in susceptibility to RAL.
To define the effects of different Y143X substitutions on RAL susceptibility, we analyzed 77 viruses containing a single substitution at position 143 (without mixtures). Reductions in RAL susceptibility of these viruses spanned a broad range, and large reductions (FC > 150) were observed, regardless of the specific substitution at position 143 (Fig. 1). As a group, larger reductions in RAL susceptibility were observed for viruses containing Y143R substitutions (median FC, >150; range, 13 to >150) than for viruses containing Y143C substitutions (median FC, 100; range, 3.9 to >150) or Y143A,G,H,S substitutions (median FC, 115; range, 42 to >150).
Fig 1.
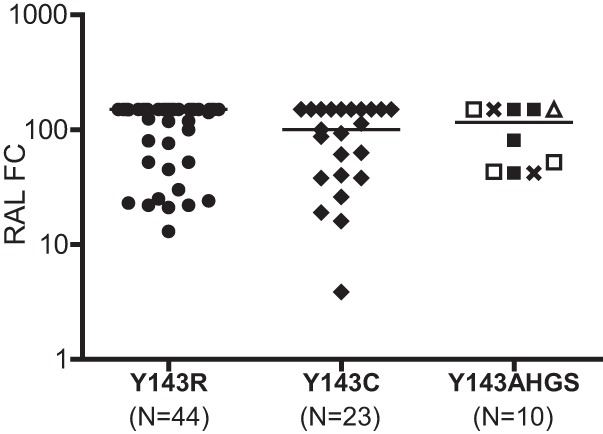
RAL susceptibility of 77 pseudoviruses containing patient-derived HIV-1 integrase sequences with Y143X substitutions. Susceptibility was measured using the Phenosense Integrase assay and is expressed as the fold change (FC) in the EC50 of patient-derived viruses relative to the wild-type reference virus. Solid circles and diamonds depict the RAL susceptibility of viruses containing Y143R and Y143C substitutions, respectively. Open triangle, open squares, crosses, and filled squares depict viruses containing Y143A (n = 1), Y143G (n = 3), Y143H (n = 2), and Y143S (n = 4), respectively. The median FC for each virus group is indicated by a solid line. The highest RAL concentration evaluated in these assays prevented exact quantitation of EC50 FC measurements that exceeded the reference virus EC50 150-fold. Viruses with mixtures at position 143 were excluded.
Secondary substitutions associated with specific Y143X substitutions.
The broad variation in RAL susceptibility of viruses containing specific Y143X substitutions indicates that other amino acid changes in the integrase region also contribute to reductions in RAL susceptibility. A number of the secondary substitutions (L74I or M, E92Q, T97A, E138K, G140S or A, V151I, G163R, I203M, and S230R) have been associated with RAL resistance based on phase III clinical trial data (31). Among the 77 viruses listed in Table 1 that lacked predicted amino acid mixtures at position 143, 72 viruses contained at least one of following substitutions: L74I or M, E92Q, T97A, E138K, G163R, I203M, and S230R. As a group, Y143R viruses harbored fewer secondary substitutions (53 substitutions among 44 viruses) than did Y143C viruses (53 substitutions among 23 viruses) or Y143A,G,H,S viruses (21 substitutions among 10 viruses). The average number of secondary substitutions per virus was 1.2 for Y143R, 2.3 for Y143C, and 2.1 for Y143A,G,H,S viruses (Table 2). T97A was the most prevalent of all secondary substitutions irrespective of the Y143R, Y143C, and Y143A,G,H,S substitutions (73%, 74%, and 80%, respectively). In contrast, S230R was preferentially identified in Y143C viruses (78%) and some Y143A,G,H,S viruses (20%) but was not observed in any of the 44 Y143R viruses. L74I,M and G163R substitutions were less frequently observed than T97A, and L74I,M substitutions were more often observed in Y143A,C,G,H,S viruses than Y143R viruses. E92Q, E138K, and I203M were identified only in a small percentage (≤10%) of virus populations (Table 2).
Table 2.
Frequency of the secondary substitutions among 77 viruses containing Y143R, Y143C, and Y143A,G,H,Sa
| Y143 substitution (no. of samples) | No. (%) of samples with secondary substitution: |
Avg no. of secondary substitutions per sample | ||||||
|---|---|---|---|---|---|---|---|---|
| L74M(I) | E92Q | T97A | E138K | G163R | I203M | S230R | ||
| Y143R (n = 44) | 7 (16) | 0 (0) | 32 (73) | 3 (7) | 8 (18) | 3 (7) | 0 (0) | 1.2 |
| Y143C (n = 23) | 10 (43) | 2 (9) | 17 (74) | 0 (0) | 5 (22) | 1 (4) | 18 (78) | 2.3 |
| Y143A/H/G/S (n = 10) | 6 (60) | 1 (10) | 8 (80) | 0 (0) | 3 (33) | 1 (10) | 2 (20) | 2.1 |
| Total no. of substitutions (n = 77) | 23 (30) | 3 (4) | 57 (74) | 3 (4) | 16 (21) | 5 (6) | 20 (26) | 1.6 |
Bold font designates a high prevalence of secondary substitutions found in specific Y143X patient virus groups. Y143C and Y143A,H,G,S viruses contain more secondary substitutions than do Y143R viruses (P < 0.001; Wilcoxon's test).
Effects of Y143X and associated secondary substitutions on RAL susceptibility.
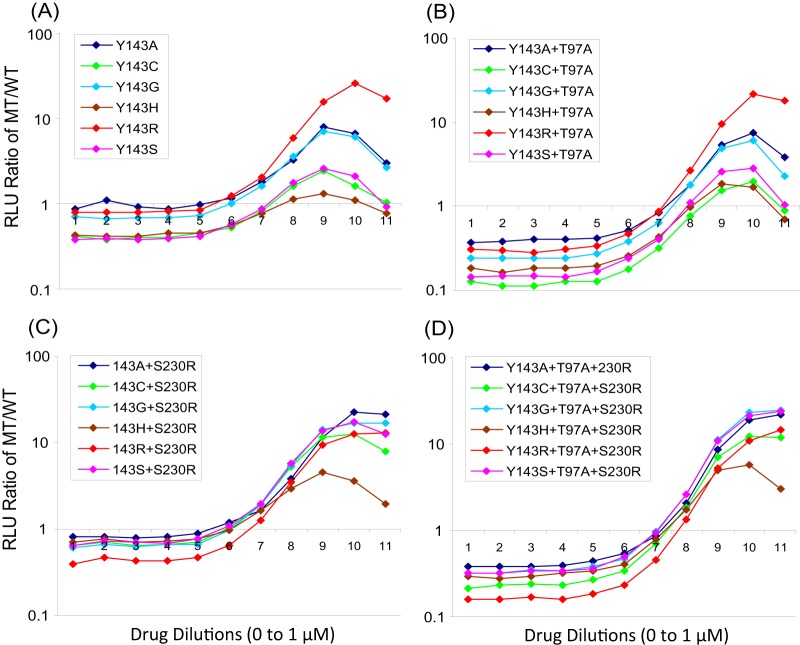
To further evaluate the impact of Y143X and associated secondary substitutions on RAL susceptibility, we created a large panel of SDMs that included six distinct Y143X substitutions (Y143A,C,G,H,R,S) alone and in combination with T97A, L74I or M, G163R, and S230R (Table 3). SDMs containing Y143X substitutions alone exhibited modest reductions (FC < 10) in RAL susceptibility (FC for Y143A, 8.0; for Y143C, 4.4; for Y143G, 6.2; for Y143H, 2.6; and for Y143S, 4.8), with the exception of Y143R, which was associated with a more notable reduction (FC = 19.6). The addition of T97A, the most common secondary substitution, further reduced RAL susceptibility relative to the corresponding Y143X single-site mutant (Table 3). The double-site mutant Y143R+T97A was associated with a larger reduction in RAL susceptibility (FC = 70) than the other Y143X+T97A double-site mutants (FC = 6.8 to 27.2). The addition of L74I, L74M, G163R, or S230R substitutions also conferred larger reductions in RAL susceptibility than did single Y143X mutants. The most notable reduction in RAL susceptibility by addition of a secondary substitution to Y143X was conferred by S230R (FC = 3.9 to 39), except in the context of Y143R (FC = 24.2), in which it was similar to that of Y143R alone (FC = 19.6). These data are consistent with the observation that S230R was not identified in any of the 44 Y143R patient viruses (Table 2). Since most viruses contained more than one secondary substitution, we next examined each Y143X in the context of multiple secondary substitutions. We observed larger reductions in RAL susceptibility when L74I or M, G163R, and S230R were added to Y143X+T97A. The combination containing Y143X, T97A, and S230R exhibited the largest reductions in RAL susceptibility among triple-site mutants, and addition of L74M or L74I to triple-site mutants further decreased RAL susceptibility (Table 3). It is worth noting that L74I or M, T97A, G163R, and S230R alone resulted in little to no reduction in RAL susceptibility (FC = 1.2 to 1.6; data not shown). Based on the data in Table 3, it is not surprising that Y143R-resistant viruses, which exhibit larger reductions in RAL susceptibility requiring fewer secondary substitutions, are more prevalent than Y143A,C,G,H,S-resistant viruses, which exhibit more-modest reductions in RAL susceptibility requiring more secondary substitutions. However, it is not immediately obvious why Y143C viruses are more prevalent than Y143A,G,S viruses since the latter all exhibit similar or larger reductions in RAL susceptibility. To explore this apparent discrepancy, we analyzed the replication capacity of Y143X mutants alone and in combination with the two most common secondary substitutions, T97A and S230R (Fig. 2). In the presence of RAL, the Y143R mutant exhibited the largest replication advantage, followed by Y143A,G mutants. In the absence of RAL, the addition of T97A to Y143X substitutions equally decreased the replication capacity of all Y143X mutants. In the presence of RAL, the Y143R+T97A double-site mutant replicated more efficiently than the other double-site mutants, followed by the Y143A+T97A and Y143G+T97A double-site mutants; replication was also improved when S230R was combined with Y143X. Notably, Y143R+S230R replicated least well among the double-site mutants in the absence of drug, which again is consistent with the observed absence of S230R in patient Y143R viruses studied here. Similarly the Y143R+S230R+T97A triple-site mutant replicated less efficiently than the other Y143X+S230R+T97A triple-site mutants in the absence of RAL, and with an efficiency similar to or lesser than that of all triple-site mutants in the presence of RAL, excluding Y143H+S230R+T97A. It is important to note that the Y143C mutants did not exhibit higher replication capacity than the Y143A,G,S mutants in single, double, or triple combinations, either in the absence or in the presence of RAL.
Table 3.
RAL susceptibility of viruses containing site-directed Y143X substitutions alone and in combination with secondary substitutionsa
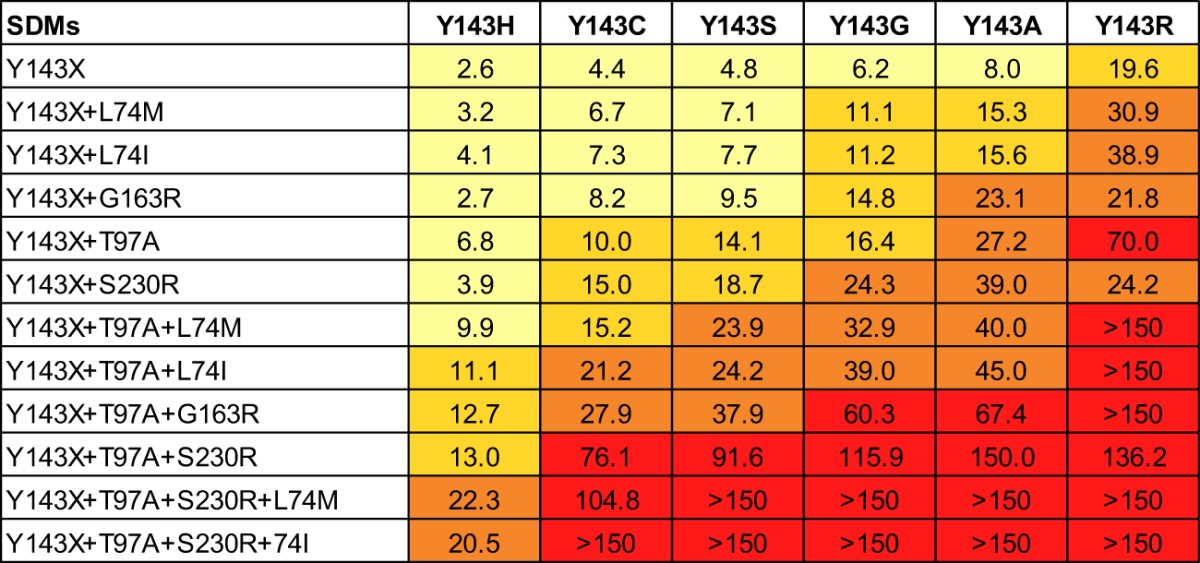
RAL susceptibility of pseudoviruses engineered by site-directed mutagenesis containing Y143X substitutions alone or in combination with additional RAL resistance-associated secondary substitutions. Susceptibility was measured using the PhenoSense Integrase assay and expressed as FC in EC50 relative to the wild-type reference virus. RAL FC results are expressed as means of duplicates of each mutant. FC variation between duplicates of each mutant was <1.5. FC values are color coded as follows: light yellow, >2 to <10; yellow, 10 to <20; brown, 20 to <50; red, ≥50.
Fig 2.
Replication profiles of site-directed mutants carrying a specific Y143 substitution alone and in combination with secondary substitutions. The ratios of mutant (MT) to wild-type (WT) replication (relative light units [RLU]) were assessed as single determinations in the absence of RAL (designated 1 on the x axis) and in the presence of 10 serial 3-fold dilutions of RAL (0.000051 μM to 1 μM, designated 2 to 11 on the x axis). (A) Y143X single-site mutants (A, C, G, H, R, and S); (B) Y143X plus T97A double-site mutants; (C) Y143X plus S230R double-site mutants; (D) Y143X plus T97A and S230R triple-site mutants.
Nucleotide changes in Y143 codons that result in amino acid substitutions Y143A, C, G, H, R, and S.
To attempt to further explain the high prevalence of Y143C substitutions, we next analyzed nucleotide differences in the codons for all Y143 substitutions. The nucleotide changes required to transition from the wild-type amino acid Y to either A, C, G, H, R, or S at position 143 were inspected and are summarized in Fig. 3. Y143C,H,S can emerge from a single nucleotide change (transitions for Y143C,H; transversion for Y143S), whereas Y143A,G,R require two nucleotide changes. Both Y143C and Y143H can serve as intermediates for Y143R, and all first and second nucleotide changes leading to Y143R represent nucleotide transitions. Y143C is also intermediate for Y143G; however, in this case, the first change is a transition and the second change is a transversion. Similarly, Y143S is intermediate for Y143A, although in this case, both the first and second nucleotide changes are transversions.
Fig 3.
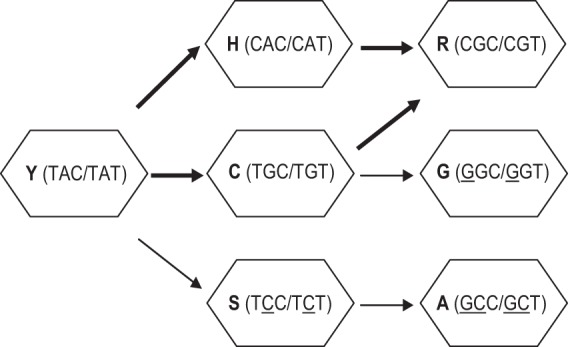
Nucleotide changes in Y143 codons that result in observed amino acid substitutions. Single nucleotide changes within either of two possible Y143 codons (TAC or TAT) are sufficient to produce Y143H, Y143C, or Y143S substitutions. Two nucleotide changes are required to produce Y143R, Y143G, or Y143A substitutions. Transitions (purine to purine or pyrimidine to pyrimidine changes) and transversions (purine to pyrimidine or pyrimidine to purine) are indicated with thick or thin arrows, respectively. Nucleotides involved in transversions are underlined.
Y143X containing viruses exhibit cross-resistance to EVG.
In previous in vitro and in vivo studies, Y143C and Y143R virus populations emerged in response to RAL exposure but not EVG exposure (20, 33, 34). We therefore investigated the susceptibility of patient viruses and a panel of Y143X SDMs with and without various RAL resistance-associated secondary substitutions (Fig. 4). Among 77 viruses containing a single substitution at position 143 (no mixtures based on population sequences), EVG FC values varied from 1.96 to >150 (median FC = 20). The observed reductions in EVG susceptibility were notably smaller than the corresponding reductions in RAL susceptibility, which ranged from 3.86 to >150 (median FC > 150). Reduced EVG susceptibility was observed for almost all patient-derived viruses, regardless of the specific amino acid substitution at position 143 (Fig. 4A). The median EVG FCs for Y143R viruses (n = 44), Y143C viruses (n = 23), and Y143A,G,H,S viruses (n = 10) were 20, 14, and 27, respectively. A pairwise comparison of the RAL and EVG FC values for each virus clearly illustrates the ability of Y143X viruses that emerge under RAL selective pressure to confer a degree of cross-resistance to EVG, and measurements of EVG and RAL susceptibility for this patient virus population were highly correlated (R2 = 0.833, P < 0.001) (Fig. 4A). These observations from patient-derived viruses are supported by the reductions in EVG susceptibility that were observed in the evaluation of a panel of isogenic viruses containing specific Y143X substitutions with and without secondary substitutions L74M, L74I, T97A, G163R, and S230R (Fig. 4B and 5). The impact on EVG susceptibility of each Y143X substitution alone and in the presence of specific secondary substitutions correlated strongly with the impact each profile had on RAL susceptibility (R2 = 0.949, P < 0.001), although the magnitudes of reductions in EVG susceptibility were lower than in RAL susceptibility. Some single Y143X substitutions conferred subtle but reproducible decreases in EVG susceptibility (Y143A FC = 2.5, Y143C FC = 1.5, Y143G FC = 2.1, Y143H FC = 1.4, Y143S FC = 1.7, Y143R FC = 2.9). The incremental addition of RAL resistance-associated secondary substitutions further reduced EVG susceptibility to 10- to 20-fold or higher (Fig. 5).
Fig 4.
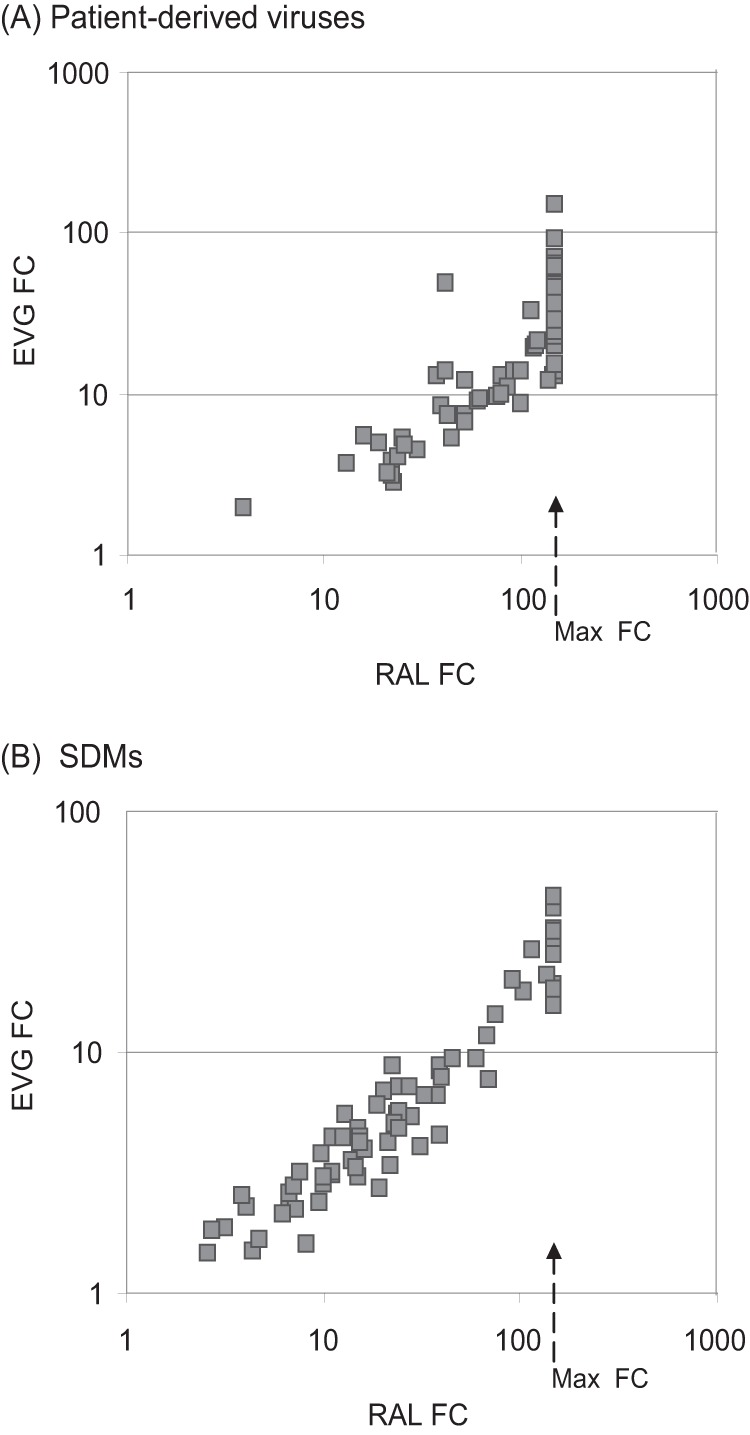
Comparison of RAL and EVG susceptibility of patient-derived viruses and site-directed-mutants (SDMs) containing Y143X substitutions using the PhenoSense Integrase assay and expressed as FC in EC50 relative to the wild-type reference virus. (A) Correlation of RAL and EVG susceptibility among 77 patient viruses (see Fig. 1). (B) Correlation of RAL and EVG susceptibility of SDMs containing Y143X substitutions alone or in combination with additional RAL resistance-associated secondary substitutions (see Table 3 and Fig. 5). Arrows indicate the highest RAL FC in EC50 that was measured in the assay.
Fig 5.
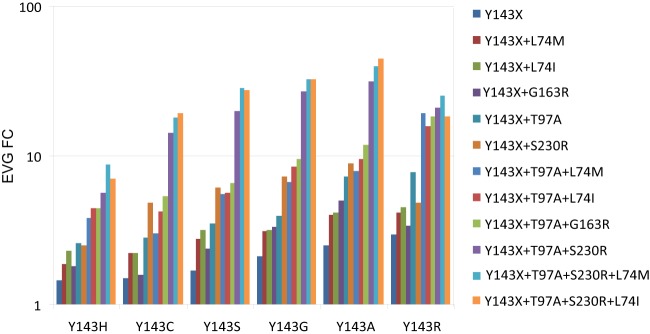
EVG susceptibility of pseudoviruses engineered by site-directed mutagenesis (SDMs) containing Y143X substitutions alone or in combination with additional RAL resistance-associated secondary substitutions. Susceptibility was measured using the PhenoSense Integrase assay and expressed as FC in EC50 relative to the wild-type reference virus. EVG FC results are expressed as the means of duplicates of each mutant. The FC variation between duplicates of each mutant was <1.5.
DISCUSSION
Y143C,R substitutions represent one of three major resistance pathways for RAL. In this study, we have surveyed and categorized viruses containing amino acid substitutions at position 143 of HIV-1 integrase and associated secondary substitutions using patient samples submitted for routine RAL resistance testing. In addition to the more prevalent and well-documented Y143C and R viruses, this survey identified viruses containing Y143A, G, H, and S substitutions, which also confer reduced susceptibility to RAL. The analysis of patient-derived viruses and a series of SDMs demonstrated that the RAL resistance profiles conferred by these alternative substitutions are similar to those of Y143C viruses, which exhibit more modest reductions in RAL susceptibility than do Y143R viruses, which display notably larger reductions in RAL susceptibility.
Analysis of integrase sequences of 77 patient samples bearing a homogeneous substitution at amino acid position 143 identified Y143R substitutions in 44 samples (57%), Y143C substitutions in 23 samples (30%), and Y143A, G, H, or S substitutions in 10 samples (13% in total: A, 1 sample [1%]; H, 2 samples [3%]; G, 4 samples [5%]; S, 3 samples [4%]). Based on the analyses of molecular clones derived from virus populations with predicted mixed amino acid substitutions at position 143, we demonstrate that mixtures of Y143 (TAC or TAT) and Y143R (CGC or CGT) variants result in the overestimation of Y143H (CAC or CAT) and Y143C (TGC or TGT) variants. Similarly, the sequences derived from mixed populations of Y143 plus Y143G (GGC or GGT) variants predicted Y143D (GAC or GAT) variants that were not detected upon clonal inspection. The extremely low replication capacity of a Y143D SDM (0.7%, data not shown) is consistent with the observed absence of this substitution in the patient viruses studied here.
Based on the evaluation of viruses containing SDMs at amino acid position 143 of HIV-1 integrase, Y143R conferred larger reductions in RAL susceptibility (FC = 19.6) than did Y143A,C,G,H,S (FC = 2.6 to 8.0), consistent with a lower resistance barrier for Y143R substitutions. Correspondingly larger reductions in RAL susceptibility were observed in patient-derived viruses containing different substitutions at position 143, suggesting that additional integrase substitutions contribute to larger reductions in RAL susceptibility. As a group, we observed more secondary substitutions in Y143A,C,G,H,S viruses than in Y143R viruses, which implied that Y143A,C,G,H,S viruses require more secondary substitutions to develop these larger reductions in RAL susceptibility than do Y143R viruses. The most commonly observed secondary substitution was T97A, which was identified at high frequency in all Y143X viruses irrespective of the particular amino acid substitution (Y143R, 73%; Y143C, 74%; Y143A,G,H,S, 80%). In contrast, S230R was observed preferentially in Y143C viruses (78%) relative to Y143A,G,H,S viruses and was never identified in Y143R viruses. These observed associations between Y143X substitutions and secondary substitutions are consistent with RAL susceptibility profiles and the replicative abilities of the SDM panel. For example, while the addition of S230R to Y143A,C,G,H,S conferred further reductions in RAL susceptibility, along with an improved replication advantage relative to the wild-type parental virus in the presence of RAL, the same was not observed for Y143R. Although the introduction of the T97A to each of the Y143X SDM viruses did not improve their replication advantage relative to the wild type, this substitution did confer further reductions in RAL susceptibility compared to the corresponding single-site mutant. Notably, the observed RAL susceptibilities and replication advantages of the Y143C+T97A+S230R and Y143R+T97A SDMs were consistent with the high representation of these profiles in the viruses studied here. Although comparable reductions in RAL susceptibility and improvements in replication advantage generally were also observed for Y143A,G,S variants, the underrepresentation of these profiles in the viruses studied here prompted the investigation of additional factors.
The emergence of all of the Y143C, H, and S substitutions requires only a single nucleotide change to either of two wild-type tyrosine codons; however, both Y143C and Y143H substitutions are the result of transition events (i.e., lower genetic barrier), while Y143S substitutions require transversion events (i.e., higher genetic barrier). Taken together with the fact that the Y143H substitution confers more-modest reductions in RAL susceptibility than do Y143C and Y143S, it seems reasonable that Y143C would emerge more often than either Y143H or Y143S. Likewise, although the emergence of Y143A, G, or R requires two nucleotide changes, Y143R emerges as a result of two nucleotide transitions (i.e., lower genetic barrier), while Y143G is produced from a transition followed by a transversion and Y143A arises from two transversion events (i.e., higher genetic barrier). Taken together with the facts that Y143R confers larger reductions in RAL susceptibility than does Y143A or Y143G and, furthermore, that both Y143C and Y143H serve as intermediates on the path to Y143R, it again seems reasonable that Y143R would emerge more often than either Y143A or Y143G because this substitution can more easily overcome both resistance and genetic barriers. The impact of genetic barriers on resistance development observed in our study is consistent with the reported observation of other laboratories (see the recent review by Gotte et al. [35]).
Although the high prevalence of Y143R can be reasonably explained based on a more favorable selective advantage (greatest reduction in RAL susceptibility) combined with multiple pathways with lower genetic barriers than other observed Y143X substitutions, these factors alone appear insufficient to explain the high prevalence of Y143C. In this case, it appears that a strong and preferential association of Y143C with S230R, which when linked confers significant reductions in RAL susceptibility, coupled with the fact that Y143C also serves as an intermediate for Y143R, provides additional explanation for the apparent overrepresentation of Y143C variants. Thus, the emergence of Y143C variants may give rise to viruses that exhibit large reductions in RAL susceptibility either by acquiring additional secondary substitutions, most often T97A and S230R, or by serving as intermediates on the path to Y143R and Y143R+T97A variants.
In contrast to reports citing that HIV-1 isolates harboring Y143X substitutions retain susceptibility to EVG, the detailed evaluation of pseudoviruses containing patient virus-derived integrase sequences along with SDMs reported here unambiguously and incontrovertibly demonstrates the ability of Y143X viruses to confer reduced susceptibility (i.e., cross-resistance) to EVG, albeit to a lesser degree than RAL. This is consistent with the intasome structure analyses that showed that Y143 substitutions affect RAL susceptibility to a much greater extent than EVG (36). Our study also reveals that RAL-selected secondary substitutions play a critical role in EVG cross-resistance among Y143X viruses.
In summary, the high prevalence of Y143R viruses can be explained by the following observations: (i) this substitution alone confers larger reductions in RAL susceptibility than do other single substitutions (Y143A, C, G, H, and S), and (ii) it requires fewer secondary substitutions (typically T97A alone) to establish a high level of resistance to RAL. Although Y143A, G, and S all confer similar or larger reductions in RAL susceptibility than Y143C, the high representation of Y143C viruses is likely a consequence of several factors that include (i) the need for only a single nucleotide transition to convert either of two Y143 codons to Y143C codons, (ii) a strong and preferential association with S230R, which confers larger reductions in RAL susceptibility than any other secondary substitution alone, and (c) the fact that Y143C codons serve as intermediates in pathways that convert wild-type Y143 codons to the strongly selected Y143R codons. In conclusion, the disproportionate representation of various amino acid substitutions at position 143 (Y143R > C > A, G, H, and S) of HIV-1 integrase is a reflection of the combined effects of differential selective advantages for HIV-1 replication in the face of RAL drug pressure and genetic barriers. The Y143X RAL resistance pathway distinctly illustrates the influence and importance of both resistance and genetic barriers on the emergence of drug-resistant HIV-1 variants under antiretroviral drug pressure.
ACKNOWLEDGMENTS
This study was supported in part by National Institutes of Health (NIH) U.S. National Institute of Allergy and Infectious Disease (NIAID) Small Business Innovation Research (SBIR) grant R44 AI057074.
We thank the Monogram Biosciences Clinical Reference Laboratory for performing the PhenoSense Integrase and GeneSeq Integrase assays, Cynthia Sedik for administrative assistance, and Mojgan Haddad for input related to statistical analysis.
Footnotes
Published ahead of print 3 June 2013
REFERENCES
- 1.Bonnenfant S, Thomas CM, Vita C, Subra F, Deprez E, Zouhiri F, Desmaele D, D'Angelo J, Mouscadet JF, Leh H. 2004. Styrylquinolines, integrase inhibitors acting prior to integration: a new mechanism of action for anti-integrase agents. J. Virol. 78:5728–5736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pannecouque C, Pluymers W, Van Maele B, Tetz V, Cherepanov P, De Clercq E, Witvrouw M, Debyser Z. 2002. New class of HIV integrase inhibitors that block viral replication in cell culture. Curr. Biol. 12:1169–1177 [DOI] [PubMed] [Google Scholar]
- 3.Bushman FD, Craigie R. 1991. Activities of human immunodeficiency virus (HIV) integration protein in vitro: specific cleavage and integration of HIV DNA. Proc. Natl. Acad. Sci. U. S. A. 88:1339–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bushman FD, Fujiwara T, Craigie R. 1990. Retroviral DNA integration directed by HIV integration protein in vitro. Science 249:1555–1558 [DOI] [PubMed] [Google Scholar]
- 5.Hazuda DJ, Felock P, Witmer M, Wolfe A, Stillmock K, Grobler JA, Espeseth A, Gabryelski L, Schleif W, Blau C, Miller MD. 2000. Inhibitors of strand transfer that prevent integration and inhibit HIV-1 replication in cells. Science 287:646–650 [DOI] [PubMed] [Google Scholar]
- 6.Christ F, Debyser Z. 2013. The LEDGF/p75 integrase interaction, a novel target for anti-HIV therapy. Virology 435:102–109 [DOI] [PubMed] [Google Scholar]
- 7.Grinsztejn B, Nguyen BY, Katlama C, Gatell JM, Lazzarin A, Vittecoq D, Gonzalez CJ, Chen J, Harvey CM, Isaacs RD. 2007. Safety and efficacy of the HIV-1 integrase inhibitor raltegravir (MK-0518) in treatment-experienced patients with multidrug-resistant virus: a phase II randomised controlled trial. Lancet 369:1261–1269 [DOI] [PubMed] [Google Scholar]
- 8.Cooper DA, Steigbigel RT, Gatell JM, Rockstroh JK, Katlama C, Yeni P, Lazzarin A, Clotet B, Kumar PN, Eron JE, Schechter M, Markowitz M, Loutfy MR, Lennox JL, Zhao J, Chen J, Ryan DM, Rhodes RR, Killar JA, Gilde LR, Strohmaier KM, Meibohm AR, Miller MD, Hazuda DJ, Nessly ML, DiNubile MJ, Isaacs RD, Teppler H, Nguyen BY. 2008. Subgroup and resistance analyses of raltegravir for resistant HIV-1 infection. N. Engl. J. Med. 359:355–365 [DOI] [PubMed] [Google Scholar]
- 9.Steigbigel RT, Cooper DA, Kumar PN, Eron JE, Schechter M, Markowitz M, Loutfy MR, Lennox JL, Gatell JM, Rockstroh JK, Katlama C, Yeni P, Lazzarin A, Clotet B, Zhao J, Chen J, Ryan DM, Rhodes RR, Killar JA, Gilde LR, Strohmaier KM, Meibohm AR, Miller MD, Hazuda DJ, Nessly ML, DiNubile MJ, Isaacs RD, Nguyen BY, Teppler H. 2008. Raltegravir with optimized background therapy for resistant HIV-1 infection. N. Engl. J. Med. 359:339–354 [DOI] [PubMed] [Google Scholar]
- 10.Hicks C, Gulick RM. 2009. Raltegravir: the first HIV type 1 integrase inhibitor. Clin. Infect. Dis. 48:931–939 [DOI] [PubMed] [Google Scholar]
- 11.Lennox JL, Dejesus E, Berger DS, Lazzarin A, Pollard RB, Ramalho Madruga JV, Zhao J, Wan H, Gilbert CL, Teppler H, Rodgers AJ, Barnard RJ, Miller MD, Dinubile MJ, Nguyen BY, Leavitt R, Sklar P. 2010. Raltegravir versus Efavirenz regimens in treatment-naive HIV-1-infected patients: 96-week efficacy, durability, subgroup, safety, and metabolic analyses. J. Acquir. Immune Defic. Syndr. 55:39–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeJesus E, Rockstroh JK, Henry K, Molina JM, Gathe J, Ramanathan S, Wei X, Yale K, Szwarcberg J, White K, Cheng AK, Kearney BP. 2012. Co-formulated elvitegravir, cobicistat, emtricitabine, and tenofovir disoproxil fumarate versus ritonavir-boosted atazanavir plus co-formulated emtricitabine and tenofovir disoproxil fumarate for initial treatment of HIV-1 infection: a randomised, double-blind, phase 3, non-inferiority trial. Lancet 379:2429–2438 [DOI] [PubMed] [Google Scholar]
- 13.Sax PE, DeJesus E, Mills A, Zolopa A, Cohen C, Wohl D, Gallant JE, Liu HC, Zhong L, Yale K, White K, Kearney BP, Szwarcberg J, Quirk E, Cheng AK. 2012. Co-formulated elvitegravir, cobicistat, emtricitabine, and tenofovir versus co-formulated efavirenz, emtricitabine, and tenofovir for initial treatment of HIV-1 infection: a randomised, double-blind, phase 3 trial, analysis of results after 48 weeks. Lancet 379:2439–2448 [DOI] [PubMed] [Google Scholar]
- 14.Walmsley S, Antela A, Clumeck N, Duiculescu D, Eberhard A, Gutierrez F, Hocqueloux L, Maggiolo F, Sandkovsky U, Granier C, Wynne B, Pappa K. 2012. Dolutegravir (DTG; S/GSK1349572)+abacavir/lamivudine once daily statistically superior to tenofovir/emtricitabine/efavirenz: 48-week results-SINGLE (ING114467). 52th Annu. Intersci. Conf. Antimicrob. Agents Chemother. (ICAAC) and the Infectious Diseases Society of America (IDSA) 46th Annu. Meet., San Francisco, CA [Google Scholar]
- 15.Malet I, Delelis O, Valantin MA, Montes B, Soulie C, Wirden M, Tchertanov L, Peytavin G, Reynes J, Mouscadet JF, Katlama C, Calvez V, Marcelin AG. 2008. Mutations associated with failure of raltegravir treatment affect integrase sensitivity to the inhibitor in vitro. Antimicrob. Agents Chemother. 52:1351–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fransen S, Gupta S, Danovich R, Hazuda D, Miller M, Witmer M, Petropoulos CJ, Huang W. 2009. Loss of raltegravir susceptibility by human immunodeficiency virus type 1 is conferred via multiple nonoverlapping genetic pathways. J. Virol. 83:11440–11446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reigadas S, Anies G, Masquelier B, Calmels C, Stuyver LJ, Parissi V, Fleury H, Andreola ML. 2010. The HIV-1 integrase mutations Y143C/R are an alternative pathway for resistance to Raltegravir and impact the enzyme functions. PLoS One. 5:e10311. 10.1371/journal.pone.0010311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Delelis O, Thierry S, Subra F, Simon F, Malet I, Alloui C, Sayon S, Calvez V, Deprez E, Marcelin AG, Tchertanov L, Mouscadet JF. 2010. Impact of Y143 HIV-1 integrase mutations on resistance to Raltegravir in vitro and in vivo. Antimicrob. Agents Chemother. 54:491–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Canducci F, Marinozzi MC, Sampaolo M, Boeri E, Spagnuolo V, Gianotti N, Castagna A, Paolucci S, Baldanti F, Lazzarin A, Clementi M. 2010. Genotypic/phenotypic patterns of HIV-1 integrase resistance to raltegravir. J. Antimicrob. Chemother. 65:425–433 [DOI] [PubMed] [Google Scholar]
- 20.Goethals O, Clayton R, Van Ginderen M, Vereycken I, Wagemans E, Geluykens P, Dockx K, Strijbos R, Smits V, Vos A, Meersseman G, Jochmans D, Vermeire K, Schols D, Hallenberger S, Hertogs K. 2008. Resistance mutations in human immunodeficiency virus type 1 integrase selected with elvitegravir confer reduced susceptibility to a wide range of integrase inhibitors. J. Virol. 82:10366–10374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garrido C, Villacian J, Zahonero N, Pattery T, Garcia F, Gutierrez F, Caballero E, Van Houtte M, Soriano V, de Mendoza C. 2012. Broad phenotypic cross-resistance to elvitegravir in HIV-infected patients failing on raltegravir-containing regimens. Antimicrob. Agents Chemother. 56:2873–2878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abram ME, Hluhanich RM, Goodman DD, Andreatta KN, Margot NA, Ye L, Nlidziela-Majka A, Barnes TL, Novikov N, Chen X, Svarovskaia ES, McColl DJ, White KL, Miller MD. 2013. Impact of primary elvitegravir resistance-associated mutations in HIV-1 integrase on drug susceptibility and viral replication fitness. Antimicrob. Agents Chemother. 57:2654–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Metifiot M, Vandegraaff N, Maddali K, Naumova A, Zhang X, Rhodes D, Marchand C, Pommier Y. 2011. Elvitegravir overcomes resistance to raltegravir induced by integrase mutation Y143. AIDS 25:1175–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malet I, Delelis O, Soulie C, Wirden M, Tchertanov L, Mottaz P, Peytavin G, Katlama C, Mouscadet JF, Calvez V, Marcelin AG. 2009. Quasispecies variant dynamics during emergence of resistance to raltegravir in HIV-1-infected patients. J. Antimicrob. Chemother. 63:795–804 [DOI] [PubMed] [Google Scholar]
- 25.da Silva D, Van Wesenbeeck L, Breilh D, Reigadas S, Anies G, Van Baelen K, Morlat P, Neau D, Dupon M, Wittkop L, Fleury H, Masquelier B. 2010. HIV-1 resistance patterns to integrase inhibitors in antiretroviral-experienced patients with virological failure on raltegravir-containing regimens. J. Antimicrob. Chemother. 65:1262–1269 [DOI] [PubMed] [Google Scholar]
- 26.Mukherjee R, Jensen ST, Male F, Bittinger K, Hodinka RL, Miller MD, Bushman FD. 2011. Switching between raltegravir resistance pathways analyzed by deep sequencing. AIDS 25:1951–1959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fransen S, Gupta S, Frantzell A, Petropoulos CJ, Huang W. 2012. Substitutions at amino acid positions 143, 148, and 155 of HIV-1 integrase define distinct genetic barriers to raltegravir resistance in vivo. J. Virol. 86:7249–7255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blanco JL, Varghese V, Rhee SY, Gatell JM, Shafer RW. 2011. HIV-1 integrase inhibitor resistance and its clinical implications. J. Infect. Dis. 203:1204–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Delelis O, Malet I, Na L, Tchertanov L, Calvez V, Marcelin AG, Subra F, Deprez E, Mouscadet JF. 2009. The G140S mutation in HIV integrases from raltegravir-resistant patients rescues catalytic defect due to the resistance Q148H mutation. Nucleic Acids Res. 37:1193–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reigadas S, Masquelier B, Calmels C, Laguerre M, Lazaro E, Vandenhende M, Neau D, Fleury H, Andreola ML. 2011. Structure-analysis of the HIV-1 integrase Y143C/R raltegravir resistance mutation in association with the secondary mutation T97A. Antimicrob. Agents Chemother. 55:3187–3194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cooper D, Gatell JM, Rockstroh J, Katlama C, Yeni P, Lazzarin A, Chen J, Isaacs R, Teppler H, Nguyen B, Group Benchmark Study Team 2007. Results of BENCHMRK-1, a phase III study evaluating the efficacy and safety of raltegravir (MK-0518), a novel HIV- integrase inhibitor, in patients with triple-class resistant virus, abstr. 105a LB. 14th Conf. Retrovir. Opportun. Infect., Los Angeles, CA [Google Scholar]
- 32.Sarkar G, Sommer SS. 1990. The “megaprimer” method of site-directed mutagenesis. Biotechniques 8:404–407 [PubMed] [Google Scholar]
- 33.Winters MA, Lloyd RM, Jr, Shafer RW, Kozal MJ, Miller MD, Holodniy M. 2012. Development of elvitegravir resistance and linkage of integrase inhibitor mutations with protease and reverse transcriptase resistance mutations. PLoS One. 7:e40514. 10.1371/journal.pone.0040514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.White KL, Kulkarni R, Szwarcberg J, Quirk E, Cheng AK, Miller MD. 2012. Integrated analysis of emergent drug resistance from the HIV-1 phase 3 QUAD studies through week 48. Antivir. Ther. 17:A12 [Google Scholar]
- 35.Gotte M. 2012. The distinct contributions of fitness and genetic barrier to the development of antiviral drug resistance. Curr. Opin. Virol. 2:644–650 [DOI] [PubMed] [Google Scholar]
- 36.Hare S, Gupta SS, Valkov E, Engelman A, Cherepanov P. 2010. Retroviral intasome assembly and inhibition of DNA strand transfer. Nature 464:232–236 [DOI] [PMC free article] [PubMed] [Google Scholar]