Abstract
Polymorphisms in the Plasmodium falciparum multidrug resistance 1 (pfmdr1) gene impact sensitivity to multiple antimalarials. In Africa, polymorphisms at N86Y and D1246Y are common and have various impacts on sensitivity to different drugs. To gain insight into the fitness consequences of these polymorphisms, we cultured parasites isolated from children with malaria in Tororo, Uganda, where the multiplicity of infection is high, and used pyrosequencing to follow polymorphism prevalences in culture over time. Of 71 cultures, parasites in 69 were successfully analyzed at N86Y and parasites in 68 were successfully analyzed at D1246Y over 3 to 36 days of culture. For position 86, the sequences of 39/69 (56.5%) parasites remained stable (>90% prevalence over 2 to 17 time points), with 82.1% of these being stable for the 86Y mutation. For position 1246, the sequences of 31/68 (45.6%) parasites remained stable, with 64.5% of these being stable for the wild-type D1246 sequence (P = 0.0002 for comparison of stable mutant genotypes for the two alleles). Defining allele selection as a ≥15% change in prevalence between the first and last samples assessed, for position 86, 11 samples showed selection, with selection toward 86Y occurring in 72.7% of alleles; for position 1246, 14 samples showed selection, with selection toward D1246 occurring in 64.3% of alleles (P = 0.11 for comparison of selection of mutations at the two alleles). Among the 7 samples with selection at both alleles, 5 showed selection for both 86Y and D1246. Overall, consistent trends in the direction of selection were seen, although differences were not statistically significant. Our results suggest fitness advantages for parasites with the pfmdr1 86Y mutation and wild-type D1246, highlighting the complex interplay between drug resistance and fitness in malaria parasites. (This study has been registered at ClinicalTrials.gov under registration no. NCT00948896 and NCT00993031.)
INTRODUCTION
Resistance of Plasmodium falciparum to widely used antimalarials is one of the greatest challenges to the control of malaria. Increasing resistance has been thwarted somewhat by the advent of highly effective artemisinin-based combination therapies (ACTs) for the treatment of falciparum malaria, but potential resistance to both artemisinins and partner drugs is of great concern (1, 2). Mechanisms of drug resistance in malaria parasites are incompletely understood. For 4-aminoquinolines, polymorphisms in the P. falciparum crt (pfcrt) gene, in particular, the 76T mutation, are the principal mediators of resistance to chloroquine and amodiaquine (3), but for most other antimalarials, resistance mechanisms are more complex (4).
Polymorphisms in the P. falciparum multidrug resistance 1 (pfmdr1) gene impact sensitivity to a diverse set of antimalarial drugs (5, 6). This gene encodes the P-glycoprotein homolog protein, which is homologous to transporter proteins in other organisms. In humans, P-glycoprotein polymorphisms are associated with resistance to cancer drugs (7). In P. falciparum, pfmdr1 localizes to the food vacuole membrane (8). The N86Y and D1246Y mutations in pfmdr1 (N86 and D1246 are considered the wild-type sequences on the basis of the sequence of the 3D7 reference strain), which are common in Africa, have been linked to decreased sensitivity to chloroquine and amodiaquine but increased sensitivity to lumefantrine, halofantrine, mefloquine, and artemisinins (9–12). Other polymorphisms primarily seen outside Africa (1034C, 1042D, and increased gene copy number) are associated with altered sensitivity to halofantrine, mefloquine, and artemisinins (12–15). An additional polymorphism, Y184F, is common but of uncertain significance (6). Additional insight into the roles of pfmdr1 polymorphisms in drug sensitivity can be gained by evaluation of the selection of specific alleles in infections that emerge soon after prior therapy with long-acting drugs. Of particular interest are studies with artesunate-amodiaquine and artemether-lumefantrine, the two most widely used ACTs in Africa. In studies in Tanzania (16), Burkina Faso (17), and Uganda (18), therapy with amodiaquine-containing combination regimens selected for the 86Y and (in East Africa) 1246Y alleles in subsequent infections. In contrast, therapy with artemether-lumefantrine selected for the N86 and D1246 polymorphisms in subsequent infections within 60 days of prior therapy (16, 17, 19–22). Importantly, the impacts of pfmdr1 polymorphisms on drug sensitivity are modest, correlations between particular polymorphisms and treatment efficacy have not been seen, and artesunate-amodiaquine and artemether-lumefantrine remain highly efficacious for the treatment of uncomplicated falciparum malaria in Africa (23, 24). However, as seen for chloroquine and amodiaquine, pfmdr1 polymorphisms may contribute, with additional polymorphisms, to higher-level resistance to the component drugs of ACTs, which are now increasingly used.
An important consideration is the impact of P. falciparum polymorphisms selected by particular drugs on parasite fitness. For pfcrt, the impact of resistance-mediating mutations on fitness is clear. In Malawi, chloroquine was discontinued as the treatment of choice for malaria in 1993, and parasites harboring the wild-type K76 sequence reappeared (25), with a return to excellent treatment efficacy for chloroquine (26). For pfmdr1, the impacts of common polymorphisms on parasite drug sensitivity are less pronounced, and impacts on fitness have been little studied. In one study, parasites with replacement of wild-type pfmdr1 with a gene with 4 mutations (184F, 1034C, 1042D, 1246Y) demonstrated a loss of fitness in coculture experiments, whereby wild-type parasites outgrew those with mutant pfmdr1 (27). In parasites selected in vitro for resistance to mefloquine, amplification of the pfmdr1 gene was seen without any single nucleotide polymorphisms, and in coculture experiments, wild-type parasites outgrew those selected with mefloquine (28). These experiments provide the most direct demonstration of the fitness consequences of pfmdr1 polymorphisms, but they did not consider the pfmdr1 86Y mutation, which is common and appears to play a key role in drug sensitivity in Africa. In a study in the Gambia, the prevalence of pfmdr1 N86 wild-type parasites was greater at the beginning of the transmission season than later in that season, consistent with a selective advantage for the wild-type parasites when selective drug pressure is limited (29). In contrast, in Uganda, ex vivo culture of fresh isolates with mixed genotypes at the pfmdr1 position 86 locus demonstrated selection of the mutant 86Y allele (30). In summary, limited available data demonstrate apparent fitness advantages for parasites with certain pfmdr1 sequences, but the specific relevance of the most common polymorphisms in Africa is uncertain.
To better characterize the impacts of the most important pfmdr1 polymorphisms in Africa, we cultured parasites from Ugandan children and followed the prevalence of pfmdr1 N86Y and D1246Y genotypes over time. We posited that many infections in this high-transmission area would be polyclonal and that the tendencies of certain genotypes to overgrow others in culture would highlight differences in fitness. We found that selection was uncommon but that when it occurred, two pfmdr1 polymorphisms, one considered a mutation and associated with decreased sensitivity to chloroquine and amodiaquine (86Y) and one considered wild type and associated with decreased sensitivity to mefloquine, lumefantrine, and other drugs (D1246), were selected. These results suggest complex determinants of fitness and drug resistance.
MATERIALS AND METHODS
Parasite sampling and culture.
All samples were from children enrolled in randomized trials of malaria chemoprevention and presenting with uncomplicated falciparum malaria in Tororo, Uganda. Details of both trials are described elsewhere, and protocols are available at clinicaltrials.gov (NCT00948896 and NCT00993031). Both trials were approved by the Makerere University School of Medicine Research and Ethics Committee, the Uganda National Council for Science and Technology, and the University of California, San Francisco, Committee for Human Research. A total of 58 samples were from a trial comparing the incidence of malaria in children randomized to receive, beginning at 6 months of age, daily trimethoprim-sulfamethoxazole, monthly sulfadoxine-pyrimethamine, monthly dihydroartemisinin-piperaquine, or no therapy for 18 months (V. Bigira et al., submitted for publication). An additional 13 samples were from a trial comparing the incidence of malaria in HIV-infected children aged 2 months to 5 years at enrollment and randomized to receive antiretroviral therapy with either a nonnucleoside reverse transcriptase inhibitor (NNRTI; nevirapine or efavirenz)-based or protease inhibitor (lopinavir-ritonavir)-based regimen (31). All episodes in children providing samples for this study occurred between July 2010 and June 2011. Subjects who presented to the study clinic with fever were evaluated by examination of Giemsa-stained smears.
At the time of malaria diagnosis and before initiation of therapy, blood from cultures with ≥1% parasitemia of P. falciparum and without other plasmodial species detected by thin smear was collected in heparinized tubes, spotted onto filter paper (Whatman 3MM), and also used to inoculate cultures. Cultures were initiated by centrifuging samples at 3,000 × g for 5 min and removing the plasma supernatant and buffy coat. The pellet was washed 3 times with RPMI 1640 medium. Parasites were then diluted with group O uninfected erythrocytes and cultured at 1% parasitemia in 10 ml RPMI 1640 medium supplemented with 25 mM HEPES, 0.2% NaHCO3, 0.1 mM hypoxanthine, 100 μg/ml gentamicin, and 0.5% Albumax II serum substitute to produce a packed cell volume of ∼2%. Cultures were maintained with replacement of medium with fresh medium every 2 days and dilution of erythrocytes with fresh cells as needed to maintain parasitemias at ∼1 to 2%. Culture aliquots were spotted onto filter paper at regular intervals, generally every 1 to 3 days.
DNA isolation and amplification.
Parasite genomic DNA was isolated from filter paper blood spots placed into 100 μl of water by saponin-Chelex extraction (32). The pfmdr1 and pfcrt genes were then amplified using primers designed to span the N86Y, D1246Y, and K76T alleles (Table 1); for each reaction, one of the PCR primers (ADS2388FPB for pfmdr1 N86Y, ADS2389FPB for pfmdr1 D1246Y, and ADS2555FPB for pfcrt K76T) was biotinylated. PCR was performed using 1 μl of extracted DNA in a 15-μl final reaction volume containing 3 mM MgCl2,200 μM each deoxynucleoside triphosphate, 6 pmol each of forward and reverse primers, and 0.75 U HotStar Taq polymerase (Qiagen). The samples were amplified in a Bio-Rad C1000 thermal cycler with initial activation at 95°C for 15 min and 45 cycles of denaturation at 95°C for 30 s, annealing at 60°C for 30 s, and extension at 72°C for 30 s, followed by a final incubation at 72°C for 10 min. Amplicons were assessed by electrophoresis before sequencing.
Table 1.
Primers used for PCR and pyrosequencing
| Name | Description | Sequence |
|---|---|---|
| ADS2388FPB | pfmdr1 position 86 forward PCR primer | TTACCTTTTTTTATATCTGTGTTTG |
| ADS2388RP | pfmdr1 position 86 reverse PCR primer | CGCTGTACTAAACCTATAGATACTAATGATAA |
| ADS2388RS | pfmdr1 position 86 pyrosequencing primer | ATTAATATCATCACCTAAAT |
| ADS2389FPB | pfmdr1 position 1246 forward PCR primer | TTTTCAAACCAATCTGGATCTGC |
| ADS2389RP | pfmdr1 position 1246 reverse PCR primer | CGCATGGGTTCTTGACTAACTATTGAA |
| ADS2389RS | pfmdr1 position 1246 pyrosequencing primer | TTGAAAATAAGTTTCTAAGA |
| ADS2555FPB | pfcrt position 76 forward PCR primer | ATGTGCTCATGTGTTTAAACTTA |
| ADS2555RP | pfcrt position 76 reverse PCR primer | GAGTTTCGGATGTTACAAAAC |
| ADS2555RS | pfcrt position 76 pyrosequencing primer | AGTTCTTTTAGCAAAAATT |
Pyrosequencing.
Reactions were carried out at EpigenDX. Biotinylated PCR products (10 μl of a 15-μl reaction mixture) were bound to streptavidin beads and converted to single-stranded DNA sequencing templates using a PyroMark Q96 vacuum workstation (Qiagen) (33). The single-stranded DNA was sequenced with a specific primer (Table 1) using a pyrosequencing PSQ96 HS system following the manufacturer's instructions (Qiagen). Every pyrosequencing run included negative controls and positive controls with known sequence; as pyrosequencing data are provided within a sequence context, the likelihood of false reads is low. The sequences of each sample were analyzed using PSQ software, and alleles were quantified as the percentage of each base at sites of interest.
Determination of copy number.
Estimates of pfmdr1 gene copy number were carried out by quantitative PCR, as previously reported (34, 35), using a 7500 real-time PCR system (Applied Biosystems). Results were normalized to those for control strains 3D7 (1 copy) and Dd2 (4 copies).
Statistical analysis.
Fisher's exact test was used to compare outcomes. A two-tailed P value of ≤0.05 was considered statistically significant. Selection coefficients for all samples that were not stable throughout culture were calculated assuming a generation time of 48 h, using a model of haploid inheritance (36).
RESULTS
Collection of samples during culture.
At the time of diagnosis of malaria (day 0), blood was collected on filter paper, samples were placed in culture, and the cultures were maintained for 3 to 36 days. A total of 69 fresh isolates of P. falciparum were successfully followed in culture. At frequent intervals (generally 1 to 3 days), culture specimens were spotted onto filter paper. Results for at least 4 separate time points were available for all but 5 cultures (results for 2 time points were available for one and results for 3 time points were available for 4). The mean number of time points for which results were available was >6 for both alleles (Table 2). One culture was followed for 3 days, but all others were followed for ≥6 days, with 80% being followed for >10 days.
Table 2.
Description of study samples
| Allele | No. of isolates assayed | No. of time points analyzed per isolate |
No. of days of culture |
||
|---|---|---|---|---|---|
| Mean | Range | Mean | Range | ||
| N86Y | 69 | 6.7 | 2–17 | 14.3 | 3–36 |
| D1246D | 68 | 6.1 | 2–17 | 14.0 | 3–36 |
Selection of parasite genotypes in culture.
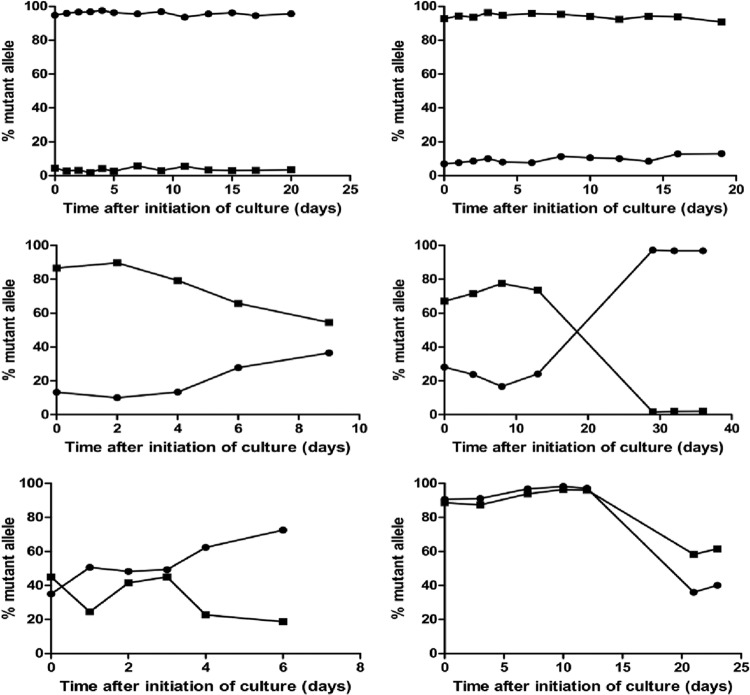
For each time point of isolate collection from cultures of samples derived from individuals in the two clinical trials described above, parasite DNA was isolated, pfmdr1 regions of interest were amplified, and the sequences at the N86Y and D1246Y alleles were quantified by pyrosequencing. A total of 460 successful pyrosequencing reactions were performed for the pfmdr1 86 allele and 431 successful pyrosequencing reactions were performed for the pfmdr1 1246 allele. Varied patterns of genotype prevalence were seen over time (Fig. 1). Genotypes were stable at position 86 for 56% of samples and at position 1246 for 46% of samples over 3 to 36 days of culture, with over 90% of sequences being either wild type or mutant for every time point (Table 3). Many of these samples were likely homogeneous at the alleles of interest at the time of collection. Results at the two loci differed: for position 86, most samples with stable sequences were mutant (82% 86Y), while for position 1246, most samples with stable sequences were wild type (65% D1246) (P = 0.0002 for comparison of stable mutant genotypes for the two alleles).
Fig 1.
Representative examples of selection in culture over time. Each graph shows the prevalence of mutant alleles at pfmdr1 positions 86 (86Y; circles) and 1246 (1246Y; squares) in a single culture over time.
Table 3.
Selection of pfmdr1 alleles in culturea
| Codon | No. of isolates | No. (%) of isolates with >90% of sequences of the following genotypes at all time points: |
Mixed without selection | No. (%) of isolates with selectionc |
||
|---|---|---|---|---|---|---|
| Mutant | Wild type | Selection of mutant | Selection of wild type | |||
| 86 | 69 | 32b (82.1) | 7 (17.9) | 19 | 8 (72.7) | 3 (27.3) |
| 1246 | 68 | 11b (35.5) | 20 (64.5) | 23 | 5 (35.7) | 9 (64.3) |
Wild-type alleles are N86 and D1246; mutant alleles are 86Y and 1246Y. Percentages were calculated for each category.
P = 0.0002 for comparisons of outcomes between alleles using two-tailed Fisher's exact test.
P = 0.11 for comparisons of outcomes between alleles using two-tailed Fisher's exact test.
The remainder of the samples showed mixed patterns, allowing us to assess the likelihood of selection of either wild-type or mutant sequence at the 86 and 1246 positions (Table 3). We defined selection as a change in prevalence of an allele by ≥15% between the first and last available sequencing result. With this definition, 28% of sequences at the 86 position and 34% of sequences at the 1246 position showed no change in prevalence of wild-type and mutant alleles over the course of culture. At the 86 position, 11 samples showed selection, and 8 of these had selection for the mutant 86Y sequence. At the 1246 position, 14 samples showed selection, and 9 of these had selection for the wild-type D1246 sequence (P = 0.11 for comparison of selection of the mutant sequence at the two alleles). Selection was seen at both the 86 and 1246 positions in 7 samples: 5 selected for 86Y and D1246 and 1 each selected for N86/D1246 and N86/1246Y. With more stringent definitions for selection, the numbers of isolates with selection decreased, but trends remained the same. For example, with selection defined as a change in prevalence of 20% or greater at two or more time points, at position 86 selection was toward 86Y in 3 of 4 samples and at position 1246 selection was toward D1246 in 7 of 9 samples. Lastly, for cultures either stable at or selected for a particular allele, for N86 the sequence was mutant 86Y in 80% and for D1246 it was wild type D1246 in 64% (P = 0.0001 for the mutant sequence at the two alleles). Comparison of selection coefficients was limited by the small sample size, but for samples that showed any changes in culture, selection coefficients varied greatly and were not significantly different from zero (median trend toward selection of the mutant sequence for pfmdr1 position 86, 0.03 [interquartile range {IQR}, −0.09 to 0.19]; median trend toward selection of the mutant sequence for pfmdr1 position 1246, 0.05 [IQR, −0.10 to 0.20]).
It was also of interest to determine if changes in pfmdr1 copy number were selected in culture. The pfmdr1 copy number was successfully estimated for 57 day 0 samples. For all but 1 sample, the copy number was estimated to be 1, and for 1 sample it was estimated to be 2 (mean, 1.95, on the basis of 8 duplicate or triplicate readings yielding 20 total data points). That sample, which had stable 86Y and D1246 sequences, had a decrease in copy number from 2 to 1 over 19 days of culture.
We also assessed the pfcrt K76T polymorphism, which was successfully assessed for 57 day 0 samples; 48 (84%) had the mutant sequence, 1 (2%) had mixed sequences, and 8 (14%) had the wild-type sequence (considering 10 to 90% of either allele to be mixed). Of 27 samples evaluated at the last culture time point, 19% had mixed or wild-type sequences, whereas 16% had mixed or wild-type sequences on day 0, indicating a slight and nonsignificant trend toward selection of wild-type parasites. We also considered the potential impact of the pfcrt genotype on selection of pfmdr1 genotypes, as linkage disequilibrium has been noted between pfcrt 76T and pfmdr1 86Y (37). For the 55 samples successfully evaluated at pfmdr1 position 86 and pfcrt position 76 at day 0, 2 had wild-type or mixed (<90% mutant) sequences at both alleles, 6 had the mutant sequence at position 86 and wild-type or mixed sequences at position 76, 25 had the mutant sequence at position 76 and wild-type or mixed sequences at position 86, and 22 had mutant sequences at both alleles (P = 0.25 for the association between the two alleles). Of the 44 evaluable samples that had the pfcrt 76T genotype at initiation of culture, 6 selected for the pfmdr1 86Y genotype, 2 selected for the N86 genotype, 10 had mixed genotypes during culture, and the remainder were stable. Of the 8 samples that had the pfcrt K76 genotype or a mixed genotype at initiation of culture, none selected for a different pfmdr1 genotype.
Impacts of recent antimalarial therapy on selection of genotypes in culture.
In both of our longitudinal trials, uncomplicated malaria was treated with artemether-lumefantrine, and many children were treated prior to the episode for which samples were collected. It was considered unlikely that prior therapy would impact selection in culture, but we considered this possibility. Samples from children with prior treatment with artemether-lumefantrine within 60 days were more likely than those from children without recent treatment to have a stable mutant genotype at position 86 and a stable wild-type genotype at position 1246, but prior treatment history was not associated with differences in selection in culture (Table 4). Considering regular chemoprevention regimens, most of the samples from the chemoprevention trial were from children not receiving therapy at the time of sample collection, either because they had not yet been randomized or because they were randomized to the no-chemoprevention control arm; for the trial comparing antiretroviral regimens, samples were equally distributed between the two arms. The sample sizes for specific comparisons were small, but in all cases, there were no obvious associations between treatment arm and selection of particular genotypes in culture (see Tables S1 and S2 in the supplemental material).
Table 4.
Impact of prior therapy on selection of pfmdr1 alleles in culturea
| Codon | Treatment history | No. of isolates | No. (%) of samples with >90% of sequences of the following genotypes at all time points: |
Mixed without selection | No. (%) of isolates with selection |
||
|---|---|---|---|---|---|---|---|
| Mutant | Wild type | Selection of mutant | Selection of wild type | ||||
| 86 | AL ≤60 days | 35 | 21b (95.5) | 1b (4.5) | 10 | 3c (100) | 0c |
| No tx | 32 | 12b (63.2) | 7b (36.8) | 9 | 2c (50.0) | 2c (50.0) | |
| 1246 | AL ≤60 days | 32 | 4d (22.2) | 14d (77.8) | 9 | 1e (20.0) | 4e (80.0) |
| No tx | 33 | 6d (50.0) | 6d (50.0) | 13 | 3e (37.5) | 5e (62.5) | |
Percentages were calculated for each category. For each allele, results for samples from subjects treated with artemether-lumefantrine within 60 days (AL ≤ 60 daus) were compared to those from subjects without prior treatment (No tx).
P = 0.016 for comparisons of outcomes between alleles using two-tailed Fisher's exact test.
P = 0.43 for comparisons of outcomes between alleles using two-tailed Fisher's exact test.
P = 0.14 for comparisons of outcomes between alleles using two-tailed Fisher's exact test.
P = 1.0 for comparisons of outcomes between alleles using two-tailed Fisher's exact test.
DISCUSSION
We used pyrosequencing to evaluate the likelihood for malaria parasites cultured from Ugandan children to undergo selection toward wild-type or mutant sequences at two alleles in the pfmdr1 gene that are commonly polymorphic in Africa. Many samples did not demonstrate selection over 3 to 36 days of culture, either because they consistently demonstrated only a single allele or because, despite containing both alleles, they did not change the proportions of the alleles over time. For samples with a stable high prevalence of a single allele, the pattern was most commonly pfmdr1 86Y and D1246. When selection was evident, it was most commonly toward pfmdr1 86Y and D1246. Thus, in a setting with a high prevalence of mixed infections and a high baseline prevalence of the pfcrt 76T allele, our results suggest selective advantages independent of drug pressure for parasites with the mutant allele at pfmdr1 position 86 and the wild-type allele at position 1246. Selection of pfmdr1 sequences by key ACT partner drugs is quite well characterized. Prior use of amodiaquine selects in subsequent infections for the mutant sequence at both the 86 and the 1246 positions, while prior use of lumefantrine selects in the opposite direction at both alleles (16, 17, 19–22). The impacts of the position 86 and 1246 polymorphisms on in vitro drug sensitivity are modest but generally as predicted by selection experiments, with N86 and D1246 leading to improved sensitivity to chloroquine and amodiaquine but decreased sensitivity to mefloquine, halofantrine, lumefantrine, and artemisinins (9–12). An increased copy number of pfmdr1 has a more pronounced effect on drug sensitivity, but this polymorphism is not commonly detected in Africa (38–40).
Our results are consistent with most of the limited available literature on pfmdr1 polymorphisms and parasite fitness. In studies of coculture of parasites that had the wild-type sequence or 4 polymorphisms (184F, 1034C, 1042D, 1246Y) in pfmdr1, wild-type parasites outgrew the quadruple mutants (27). In a prior study from our group in Uganda, ex vivo culture of fresh isolates with mixed genotypes at the pfmdr1 86 locus demonstrated selection of the mutant 86Y allele (30); in that study, selection was not seen at the 1246 allele. These limited results suggest selective advantages for the 86Y mutant and D1246 wild-type sequences. In contrast, in the Gambia, a greater prevalence of pfmdr1 N86 wild-type parasites was seen at the beginning of the transmission season than later in that season, suggesting a selective advantage for parasites with the wild-type N86 sequence, as fitness might be a more dominant selective pressure during the dry season, when drug use is low (29). A selection in the Gambia opposite that seen in Uganda might be explained by the very different means by which selection was assessed or by the very low prevalence of the 1246Y polymorphism in West Africa, which might affect selective pressure at the 86 allele. In all of these cases, the impacts of pfmdr1 single nucleotide polymorphisms on parasite fitness have appeared to be modest and probably less than the effects of alterations in pfmdr1 copy number (28). We found only one sample with an increased copy number of pfmdr1 at day 0, and in that case, the parasites had only a single copy after 19 days of culture, consistent with the fitness disadvantage of increased copy number demonstrated previously (28). Most parasites had the pfcrt 76T mutation that mediates chloroquine resistance and has been widespread in Uganda for many years, and minimal selection toward the wild-type sequence was seen in culture. The high prevalence of the pfcrt 76T genotype may have facilitated our observed selection of pfmdr1 86Y, as linkage disequilibrium has been observed between these two mutant genotypes (36). However, analysis of day 0 genotypes did not identify an association between pfcrt position 76 and pfmdr1 position 86 polymorphisms. Considering selection, all parasites that underwent selection of the pfmdr1 86Y genotype also harbored the pfcrt 76T mutation, but selection of the wild-type N86 sequence was also seen in parasites with 76T, and a small sample size and low prevalence of the pfcrt K76 genotype limited our ability to ascertain the importance of the pfcrt genotype in driving selection at pfmdr1 position 86.
Our study had important limitations. First, logistical constraints necessitated maintenance of cultures for varied lengths of time and collection of DNA at varied sampling intervals. Comparison of selection coefficients was limited by the small and varied sample sizes for each culture interval. Thus, we could not quantifiably assess the contributions of genotypes to fitness; rather, we could assess only the consequences of varied times in culture on parasite genotypes. Second, our study was designed to assess changes in cultures of clinical samples and was thereby limited by the makeup of samples from study subjects. Most samples did not show any change in the prevalence of polymorphisms of interest during culture, limiting the sample size for key comparisons. Third, because of limited success in adaptation of low-parasitemia cultures, only those with parasitemias of ≥1% were studied. This design may have introduced bias, as results for parasites not successfully cultured are unknown. Fourth, we did not assess the full sequences of pfmdr1 or sequences of other portions of the genomes of parasites under study. In multiple prior studies, we did not see variation at other known polymorphisms in pfmdr1, except for the 184 allele, in which the polymorphism has uncertain relevance, and so survey of the full sequence of pfmdr1 for each sample was not deemed necessary. We did analyze for changes in pfmdr1 copy number and the pfcrt K76T allele, but the low prevalence of parasites with an increased copy number or the wild-type pfcrt genotype limited this analysis. Fifth, related to the points identified above, our available sample size did not allow us to demonstrate statistically significant selection of pfmdr1 polymorphisms of interest. This result is not surprising, as numerous genetic variations, in addition to those assessed in pfmdr1, likely impact parasite fitness. Nonetheless, our consistent results suggest that the pfmdr1 86Y and D1246 alleles exert modest selective fitness advantages for P. falciparum.
Our results offer insight into the complex interplay between drug resistance and fitness in malaria parasites. Two single nucleotide polymorphisms associated with the same impacts on resistance to different antimalarials appear to have opposite effects on fitness, with pfmdr1 86Y and D1246 appearing to have fitness advantages. This situation may explain the lack of strong selection for pfmdr1 genotypes seen in Africa. In most of Africa, widespread use of chloroquine selected strongly for the mutant pfcrt 76T genotype, despite a clear fitness disadvantage. In contrast, pfmdr1 genotypes have generally remained complex, with a high prevalence of wild-type and mutant sequences. Currently in Africa, with widespread use of two ACTs, artesunate-amodiaquine and artemether-lumefantrine, parasites are subject to complex selective pressures at pfmdr1 alleles, with artemisinins and lumefantrine selecting for wild-type N86 and D1246 sequences, amodiaquine selecting for mutant 86Y and 1246Y sequences, and fitness constraints apparently selecting for the 86Y and D1246 sequences. Selective effects on other portions of the P. falciparum genome are undoubtedly also occurring, and continued observation of the impacts of chemotherapy on the drug sensitivity and fitness of malaria parasites is warranted. In addition, policy makers may consider adoption of treatment regimens to limit the selection of parasites with decreased drug sensitivity, by alternating first-line regimens with opposite selective pressures and/or by utilizing ACTs whose components have opposite selective pressures (41).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the National Institutes of Health (AI075045, AI089674, TW007375, HD059454) and the Doris Duke Charitable Foundation. Control strains of P. falciparum were from the Malaria Research and Reference Reagent Resource Center.
We thank the participants in the clinical trials from which samples were collected, their parents and guardians, and our clinical study team. We also thank Andrew Walakira, Christine Nakazibwe, Melisa Kortan, and Kaitlin Hall for assistance with culture and collection of clinical samples; Sam Nsobya, Moses Kiggundu, Fred Baliraine, Roland Cooper, and Melissa Conrad for helpful advice; and Grant Dorsey for assistance with correlation of results with clinical parameters.
Footnotes
Published ahead of print 24 June 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.00161-13.
REFERENCES
- 1.Dondorp AM, Nosten F, Yi P, Das D, Phyo AP, Tarning J, Lwin KM, Ariey F, Hanpithakpong W, Lee SJ, Ringwald P, Silamut K, Imwong M, Chotivanich K, Lim P, Herdman T, An SS, Yeung S, Singhasivanon P, Day NP, Lindegardh N, Socheat D, White NJ. 2009. Artemisinin resistance in Plasmodium falciparum malaria. N. Engl. J. Med. 361:455–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Petersen I, Eastman R, Lanzer M. 2011. Drug-resistant malaria: molecular mechanisms and implications for public health. FEBS Lett. 585:1551–1562 [DOI] [PubMed] [Google Scholar]
- 3.Fidock DA, Nomura T, Talley AK, Cooper RA, Dzekunov SM, Ferdig MT, Ursos LM, Sidhu AB, Naude B, Deitsch KW, Su XZ, Wootton JC, Roepe PD, Wellems TE. 2000. Mutations in the P. falciparum digestive vacuole transmembrane protein PfCRT and evidence for their role in chloroquine resistance. Mol. Cell 6:861–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ekland EH, Fidock DA. 2007. Advances in understanding the genetic basis of antimalarial drug resistance. Curr. Opin. Microbiol. 10:363–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sanchez CP, Dave A, Stein WD, Lanzer M. 2010. Transporters as mediators of drug resistance in Plasmodium falciparum. Int. J. Parasitol. 40:1109–1118 [DOI] [PubMed] [Google Scholar]
- 6.Valderramos SG, Fidock DA. 2006. Transporters involved in resistance to antimalarial drugs. Trends Pharmacol. Sci. 27:594–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharom FJ. 2011. The P-glycoprotein multidrug transporter. Essays Biochem. 50:161–178 [DOI] [PubMed] [Google Scholar]
- 8.Cowman AF, Karcz S, Galatis D, Culvenor JG. 1991. A P-glycoprotein homologue of Plasmodium falciparum is localized on the digestive vacuole. J. Cell Biol. 113:1033–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duraisingh MT, Jones P, Sambou I, von Seidlein L, Pinder M, Warhurst DC. 2000. The tyrosine-86 allele of the pfmdr1 gene of Plasmodium falciparum is associated with increased sensitivity to the anti-malarials mefloquine and artemisinin. Mol. Biochem. Parasitol. 108:13–23 [DOI] [PubMed] [Google Scholar]
- 10.Duraisingh MT, Roper C, Walliker D, Warhurst DC. 2000. Increased sensitivity to the antimalarials mefloquine and artemisinin is conferred by mutations in the pfmdr1 gene of Plasmodium falciparum. Mol. Microbiol. 36:955–961 [DOI] [PubMed] [Google Scholar]
- 11.Mwai L, Kiara SM, Abdirahman A, Pole L, Rippert A, Diriye A, Bull P, Marsh K, Borrmann S, Nzila A. 2009. In vitro activities of piperaquine, lumefantrine, and dihydroartemisinin in Kenyan Plasmodium falciparum isolates and polymorphisms in pfcrt and pfmdr1. Antimicrob. Agents Chemother. 53:5069–5073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reed MB, Saliba KJ, Caruana SR, Kirk K, Cowman AF. 2000. Pgh1 modulates sensitivity and resistance to multiple antimalarials in Plasmodium falciparum. Nature 403:906–909 [DOI] [PubMed] [Google Scholar]
- 13.Pickard AL, Wongsrichanalai C, Purfield A, Kamwendo D, Emery K, Zalewski C, Kawamoto F, Miller RS, Meshnick SR. 2003. Resistance to antimalarials in Southeast Asia and genetic polymorphisms in pfmdr1. Antimicrob. Agents Chemother. 47:2418–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sidhu AB, Uhlemann AC, Valderramos SG, Valderramos JC, Krishna S, Fidock DA. 2006. Decreasing pfmdr1 copy number in Plasmodium falciparum malaria heightens susceptibility to mefloquine, lumefantrine, halofantrine, quinine, and artemisinin. J. Infect. Dis. 194:528–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sidhu AB, Valderramos SG, Fidock DA. 2005. pfmdr1 mutations contribute to quinine resistance and enhance mefloquine and artemisinin sensitivity in Plasmodium falciparum. Mol. Microbiol. 57:913–926 [DOI] [PubMed] [Google Scholar]
- 16.Humphreys GS, Merinopoulos I, Ahmed J, Whitty CJ, Mutabingwa TK, Sutherland CJ, Hallett RL. 2007. Amodiaquine and artemether-lumefantrine select distinct alleles of the Plasmodium falciparum mdr1 gene in Tanzanian children treated for uncomplicated malaria. Antimicrob. Agents Chemother. 51:991–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zongo I, Dorsey G, Rouamba N, Tinto H, Dokomajilar C, Guiguemde RT, Rosenthal PJ, Ouedraogo JB. 2007. Artemether-lumefantrine versus amodiaquine plus sulfadoxine-pyrimethamine for uncomplicated falciparum malaria in Burkina Faso: a randomised non-inferiority trial. Lancet 369:491–498 [DOI] [PubMed] [Google Scholar]
- 18.Nsobya SL, Dokomajilar C, Joloba M, Dorsey G, Rosenthal PJ. 2007. Resistance-mediating Plasmodium falciparum pfcrt and pfmdr1 alleles after treatment with artesunate-amodiaquine in Uganda. Antimicrob. Agents Chemother. 51:3023–3025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baliraine FN, Rosenthal PJ. 2011. Prolonged selection of pfmdr1 polymorphisms after treatment of falciparum malaria with artemether-lumefantrine in Uganda. J. Infect. Dis. 204:1120–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Happi CT, Gbotosho GO, Folarin OA, Sowunmi A, Hudson T, O'Neil M, Milhous W, Wirth DF, Oduola AM. 2009. Selection of Plasmodium falciparum multidrug resistance gene 1 alleles in asexual stages and gametocytes by artemether-lumefantrine in Nigerian children with uncomplicated falciparum malaria. Antimicrob. Agents Chemother. 53:888–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sisowath C, Stromberg J, Martensson A, Msellem M, Obondo C, Bjorkman A, Gil JP. 2005. In vivo selection of Plasmodium falciparum pfmdr1 86N coding alleles by artemether-lumefantrine (Coartem). J. Infect. Dis. 191:1014–1017 [DOI] [PubMed] [Google Scholar]
- 22.Some AF, Sere YY, Dokomajilar C, Zongo I, Rouamba N, Greenhouse B, Ouedraogo JB, Rosenthal PJ. 2010. Selection of known Plasmodium falciparum resistance-mediating polymorphisms by artemether-lumefantrine and amodiaquine-sulfadoxine-pyrimethamine but not dihydroartemisinin-piperaquine in Burkina Faso. Antimicrob. Agents Chemother. 54:1949–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dorsey G, Staedke S, Clark TD, Njama-Meya D, Nzarubara B, Maiteki-Sebuguzi C, Dokomajilar C, Kamya MR, Rosenthal PJ. 2007. Combination therapy for uncomplicated falciparum malaria in Ugandan children: a randomized trial. JAMA 297:2210–2219 [DOI] [PubMed] [Google Scholar]
- 24.Four Artemisinin-Based Combinations (4ABC) Study Group 2011. A head-to-head comparison of four artemisinin-based combinations for treating uncomplicated malaria in African children: a randomized trial. PLoS Med. 8:e1001119. 10.1371/journal.pmed.1001119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kublin JG, Cortese JF, Njunju EM, Mukadam RA, Wirima JJ, Kazembe PN, Djimde AA, Kouriba B, Taylor TE, Plowe CV. 2003. Reemergence of chloroquine-sensitive Plasmodium falciparum malaria after cessation of chloroquine use in Malawi. J. Infect. Dis. 187:1870–1875 [DOI] [PubMed] [Google Scholar]
- 26.Laufer MK, Thesing PC, Eddington ND, Masonga R, Dzinjalamala FK, Takala SL, Taylor TE, Plowe CV. 2006. Return of chloroquine antimalarial efficacy in Malawi. N. Engl. J. Med. 355:1959–1966 [DOI] [PubMed] [Google Scholar]
- 27.Hayward R, Saliba KJ, Kirk K. 2005. pfmdr1 mutations associated with chloroquine resistance incur a fitness cost in Plasmodium falciparum. Mol. Microbiol. 55:1285–1295 [DOI] [PubMed] [Google Scholar]
- 28.Preechapornkul P, Imwong M, Chotivanich K, Pongtavornpinyo W, Dondorp AM, Day NP, White NJ, Pukrittayakamee S. 2009. Plasmodium falciparum pfmdr1 amplification, mefloquine resistance, and parasite fitness. Antimicrob. Agents Chemother. 53:1509–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ord R, Alexander N, Dunyo S, Hallett R, Jawara M, Targett G, Drakeley CJ, Sutherland CJ. 2007. Seasonal carriage of pfcrt and pfmdr1 alleles in Gambian Plasmodium falciparum imply reduced fitness of chloroquine-resistant parasites. J. Infect. Dis. 196:1613–1619 [DOI] [PubMed] [Google Scholar]
- 30.Nsobya SL, Kiggundu M, Joloba M, Dorsey G, Rosenthal PJ. 2008. Complexity of Plasmodium falciparum clinical samples from Uganda during short-term culture. J. Infect. Dis. 198:1554–1557 [DOI] [PubMed] [Google Scholar]
- 31.Achan J, Kakuru A, Ikilezi G, Ruel T, Clark TD, Nsanzabana C, Charlebois E, Aweeka F, Dorsey G, Rosenthal PJ, Havlir D, Kamya MR. 2012. Antiretroviral agents and prevention of malaria in HIV-infected Ugandan children. N. Engl. J. Med. 367:2110–2118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Plowe CV, Djimde A, Bouare M, Doumbo O, Wellems TE. 1995. Pyrimethamine and proguanil resistance-conferring mutations in Plasmodium falciparum dihydrofolate reductase: polymerase chain reaction methods for surveillance in Africa. Am. J. Trop. Med. Hyg. 52:565–568 [DOI] [PubMed] [Google Scholar]
- 33.Zhou Z, Poe AC, Limor J, Grady KK, Goldman I, McCollum AM, Escalante AA, Barnwell JW, Udhayakumar V. 2006. Pyrosequencing, a high-throughput method for detecting single nucleotide polymorphisms in the dihydrofolate reductase and dihydropteroate synthetase genes of Plasmodium falciparum. J. Clin. Microbiol. 44:3900–3910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dokomajilar C, Nsobya SL, Greenhouse B, Rosenthal PJ, Dorsey G. 2006. Selection of Plasmodium falciparum pfmdr1 alleles following therapy with artemether-lumefantrine in an area of Uganda where malaria is highly endemic. Antimicrob. Agents Chemother. 50:1893–1895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Price RN, Uhlemann AC, Brockman A, McGready R, Ashley E, Phaipun L, Patel R, Laing K, Looareesuwan S, White NJ, Nosten F, Krishna S. 2004. Mefloquine resistance in Plasmodium falciparum and increased pfmdr1 gene copy number. Lancet 364:438–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wootton JC, Feng X, Ferdig MT, Cooper RA, Mu J, Baruch DI, Magill AJ, Su XZ. 2002. Genetic diversity and chloroquine selective sweeps in Plasmodium falciparum. Nature 418:320–323 [DOI] [PubMed] [Google Scholar]
- 37.Adagu IS, Warhurst DC. 2001. Plasmodium falciparum: linkage disequilibrium between loci in chromosomes 7 and 5 and chloroquine selective pressure in northern Nigeria. Parasitology 123:219–224 [DOI] [PubMed] [Google Scholar]
- 38.Gadalla NB, Adam I, Elzaki SE, Bashir S, Mukhtar I, Oguike M, Gadalla A, Mansour F, Warhurst D, El-Sayed BB, Sutherland CJ. 2011. Increased pfmdr1 copy number and sequence polymorphisms in Plasmodium falciparum isolates from Sudanese malaria patients treated with artemether-lumefantrine. Antimicrob. Agents Chemother. 55:5408–5411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Holmgren G, Bjorkman A, Gil JP. 2006. Amodiaquine resistance is not related to rare findings of pfmdr1 gene amplifications in Kenya. Trop. Med. Int. Health 11:1808–1812 [DOI] [PubMed] [Google Scholar]
- 40.Uhlemann AC, Ramharter M, Lell B, Kremsner PG, Krishna S. 2005. Amplification of Plasmodium falciparum multidrug resistance gene 1 in isolates from Gabon. J. Infect. Dis. 192:1830–1835 [DOI] [PubMed] [Google Scholar]
- 41.Sutherland CJ, Babiker H, Mackinnon MJ, Ranford-Cartwright L, El Sayed BB. 2011. Rational deployment of antimalarial drugs in Africa: should first-line combination drugs be reserved for paediatric malaria cases? Parasitology 138:1459–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.