Summary
The unfolded protein response (UPR) maintains endoplasmic reticulum (ER) proteostasis through the activation of transcription factors such as XBP1s and ATF6. The functional consequences of these transcription factors for ER proteostasis remain poorly defined. Here, we describe methodology that enables orthogonal, small-molecule-mediated activation of the UPR-associated transcription factors XBP1s and/or ATF6 in the same cell independent of stress. We employ transcriptomics and quantitative proteomics to evaluate ER proteostasis network remodeling owing to the XBP1s and/or ATF6 transcriptional programs. Furthermore, we demonstrate that the three ER proteostasis environments accessible by activating XBP1s and/or ATF6 differentially influence the folding, trafficking, and degradation of destabilized ER client proteins without globally affecting the endogenous proteome. Our data reveal how the ER proteostasis network is remodeled by the XBP1s and/or ATF6 transcriptional programs at the molecular level and demonstrate the potential for selective restoration of aberrant ER proteostasis of pathologic, destabilized proteins through arm-selective UPR activation.
Introduction
One-third of the human proteome is directed to the endoplasmic reticulum (ER) for partitioning between folding and trafficking versus ER-associated degradation (ERAD), a decision primarily dictated by the exact composition of the ER protein homeostasis (or proteostasis) network (Balch et al., 2008; Braakman and Bulleid, 2011; Hartl et al., 2011; McClellan et al., 2005). This partitioning protects the integrity of downstream proteomes by ensuring that only folded, functional proteins are trafficked from the ER (Brodsky and Skach, 2011; Smith et al., 2011b; Wiseman et al., 2007).
The folding, trafficking, and degradation capacity of the ER is dynamically adjusted to meet demand by the unfolded protein response (UPR)—a stress-responsive signaling pathway comprising three integrated signaling cascades emanating from the ER transmembrane proteins IRE1, ATF6, and PERK (Schröder and Kaufman, 2005; Walter and Ron, 2011). UPR signaling is activated by the accumulation of misfolded or aggregated proteins within the ER lumen. UPR activation causes transient, PERK-mediated translational attenuation and activation of the basic leucine zipper transcription factors ATF4, XBP1s, and the cleaved N-terminal fragment of ATF6 downstream of the ER stress sensors PERK, IRE1, and full-length ATF6, respectively. These transcription factors increase expression of distinct but overlapping sets of genes comprising both ER-specific and general cellular proteostasis pathways (Adachi et al., 2008; Lee et al., 2003; Okada et al., 2002; Yamamoto et al., 2004, 2007). The three mechanistically distinct arms of the metazoan UPR presumably evolved to provide cells with flexibility to adapt to tissue-specific environmental and metabolic demands, creating a mechanism to restore ER proteostasis in response to a wide array of cellular insults (Gass et al., 2008; Harding et al., 2001; Kaser et al., 2008; Wu et al., 2007).
Pharmacologic activation of the UPR offers the potential to adapt ER proteostasis and rescue misfolded, aberrantly degraded, or aggregation-prone ER client proteins without significantly affecting the healthy, wild-type proteome (Balch et al., 2008; Walter and Ron, 2011). For example, activation of a UPR signaling pathway that increases ER protein folding capacity could decrease the aberrant ERAD and increase the ER folding and export of destabilized, mutant proteins, thereby ameliorating loss-of-function diseases such as cystic fibrosis or lysosomal storage diseases (Chiang et al., 2012; Mu et al., 2008; Wang et al., 2006). Alternatively, increasing ERAD activity could attenuate the secretion of destabilized, aggregation-prone proteins that undergo concentration-dependent extracellular aggregation into amorphous aggregates and amyloid fibrils (Braakman and Bulleid, 2011; Brodsky and Skach, 2011; Luheshi and Dobson, 2009; Sitia and Braakman, 2003), providing a potential strategy to ameliorate amyloid disease pathology.
Concomitant pharmacologic activation of the PERK, IRE1, and ATF6 UPR arms can be achieved by the application of toxic small molecules such as tunicamycin (Tm; inhibits protein N-glycosylation) or thapsigargin (Tg; disrupts ER calcium homeostasis) that induce ER protein misfolding and aggregation, ultimately causing apoptosis (Schröder and Kaufman, 2005; Walter and Ron, 2011). These global UPR activators have proven useful for delineating the molecular underpinnings of UPR signaling pathways. Unfortunately, the pleiotropic effects and acute toxicity of global UPR activation complicate studies focused on understanding how UPR activation (either global or arm selective) remodels the ER proteostasis network in the absence of an acute ER stress or how the partitioning between folding and trafficking versus degradation of ER client proteins can be influenced by arm-selective UPR activation. Thus, despite the considerable effort focused on understanding the signaling mechanisms of IRE1, ATF6, and PERK activation, the functional implications of activating these pathways on ER proteostasis pathway composition and function remain poorly defined.
Herein, we introduce small molecule-regulated, genetically encoded transcription factors that enable orthogonal activation of UPR transcriptional programs in the same cell. Using our methodology, we characterize the three distinct ER proteostasis environments accessible by activating XBP1s and/or ATF6 to physiologically relevant levels in the absence of stress. We also evaluate the functional consequences of activating XBP1s and/or ATF6 on the folding and trafficking versus degradation of destabilized ER client proteins, including transthyretin (TTR). Ultimately, we demonstrate that arm-selective UPR activation selectively reduces secretion of a destabilized, aggregation-prone TTR variant without affecting the analogous wild-type protein and without globally altering the endogenous intracellular or secreted proteomes. Our results demonstrate, in molecular detail, how the XBP1s and/or ATF6 transcriptional programs integrate to adapt ER proteostasis pathways and highlight the capacity of functionally distinct ER proteostasis environments accessed by arm-selective UPR activation to restore the aberrant ER proteostasis of destabilized protein variants.
Results
To characterize the ER proteostasis environments accessible by the selective or combined activity of the UPR-associated transcription factors XBP1s and ATF6, we required methodology for the small molecule-mediated, orthogonal regulation of two transcription factors in the same cell. Tetracycline (tet)-repressor technology can be applied to allow doxycycline (dox)-dependent control of XBP1s levels in the physiologic range (Lee et al., 2003). However, we have found that tet-repressor regulation of ATF6 activity within the physiologically relevant range is difficult. Even after careful optimization and single-colony stable cell selection of HEK293T-REx cells expressing constitutively active ATF6(1–373) (henceforth termed ATF6) under the tet repressor, we observed nonphysiologic levels of ATF6 target gene expression and significant off-target effects including strong upregulation of established XBP1s target genes, following ATF6 induction at all permissive dox doses (Figures S1A and S1B). We required, therefore, an alternative strategy to regulate the ATF6 transcription factor that would be dosable and orthogonal to tet-repressor technology.
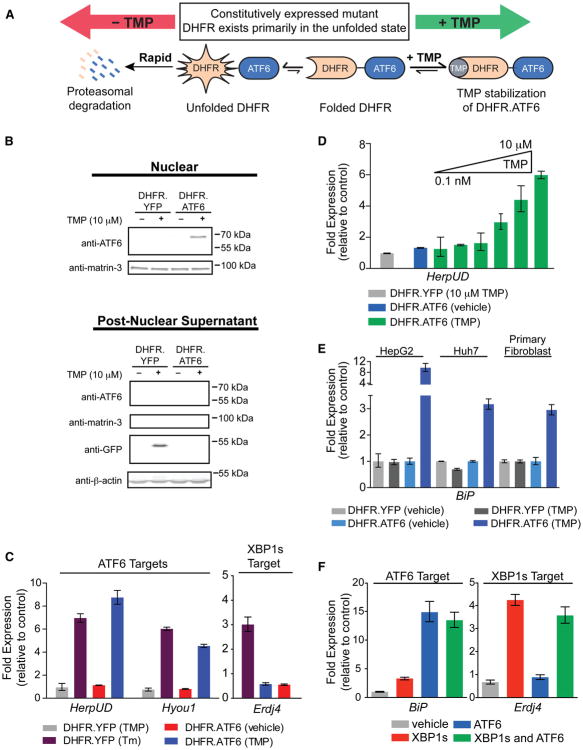
We envisioned that destabilized domain (DD) technology (Figure 1A) (Banaszynski et al., 2006; Iwamoto et al., 2010) could be adapted to prepare a dose-dependent, ligand-regulated ATF6 transcription factor whose activity would be inducible to levels more consistent with those observed in human physiology. We fused a destabilized variant of E. coli dihydrofolate reductase (DHFR) to the N terminus of ATF6 via a short Gly-Ser linker. The poorly folded DHFR domain directs the entire constitutively expressed DHFR.ATF6 fusion protein to rapid proteasomal degradation. Administration of the DHFR-specific pharmacologic chaperone, trimethoprim (TMP), stabilizes the folded DHFR conformation, increasing the initially poorly populated folded DHFR population, attenuating proteasomal degradation, and inducing the ATF6 transcriptional program (Figure 1A).
Figure 1. Orthogonal, Ligand-Dependent Control of XBP1s and ATF6 Transcriptional Activity.
(A) Model illustrating the TMP-mediated, posttranslational regulation of DHFR.ATF6.
(B) Immunoblot of nuclear (top) and postnuclear (bottom) fractions from HEK293T-REx cells expressing DHFR.YFP or DHFR.ATF6 treated 12 hr with TMP (10 μM). The immunoblot of matrin-3 shows the efficiency of the nuclear extraction.
(C) qPCR analysis of Hyou1, HerpUD, and Erdj4 in HEK293T-REx cells expressing DHFR.YFP or DHFR.ATF6 following a 12 hr treatment with TMP (10 μM) or a 6 hr treatment with Tm (10 μg/ml). qPCR data are reported relative to vehicle-treated cells expressing DHFR.YFP. qPCR data are reported as the mean ± 95% confidence interval.
(D) TMP dose dependence of HerpUD upregulation in HEK293T-REx cells expressing DHFR.ATF6 (12 hr treatments with TMP). qPCR data are reported as the mean ± 95% confidence interval.
(E) qPCR analysis of the ATF6 target gene BiP in HepG2, Huh7, or primary fibroblast cells transiently transduced with DHFR.YFP- or DHFR.ATF6-expressing adenoviruses and treated for 12 hr with 100 μM TMP or vehicle. qPCR data are reported relative to the corresponding vehicle-treated cells. qPCR data are reported as the mean ± 95% confidence interval.
(F) qPCR analysis of BiP and Erdj4 in HEK293DAX cells following a 12 hr activation of XBP1s (dox; 1 μg/ml), DHFR.ATF6 (TMP; 10 μM), or both. qPCR data are reported relative to vehicle-treated HEK293DYG cells. qPCR data are reported as the mean ± 95% confidence interval. See also Figure S1 and Table S4.
The addition of TMP stabilizes DHFR.ATF6 in nuclear fractions isolated from HEK293T-REx cells expressing DHFR.ATF6 (Figure 1B). DHFR.ATF6 is not detected in the absence of TMP. Furthermore, TMP induces expression of the ATF6 target genes HerpUD and Hyou1 (Adachi et al., 2008) in cells expressing DHFR.ATF6 to levels consistent with those observed following global UPR-dependent activation induced by Tm (Figure 1C). We observe no increased expression of these genes inuntreated cells expressing DHFR.ATF6 or TMP-treated cells expressing DHFR.YFP. The TMP-dependent activation of DHFR.ATF6 is rapid, causing significant upregulation of HerpUD in <2 hr (Figure S1C). Importantly, TMP treatment does not induce expression of the XBP1s-selective target gene Erdj4 (Lee et al., 2003) (Figure 1C). Increasing concentrations of TMP reveal a linear dose-dependent upregulation of ATF6 target genes, demonstrating a significant dynamic range for activation of DHFR.ATF6 by TMP (Figure 1D). Because DHFR.ATF6 is a single gene product, it similarly enables the straightforward, ligand-dependent activation of the ATF6 transcriptional program at physiologic levels in a wide variety of other cellular model systems (Figure 1E).
In order to activate both XBP1s and ATF6 in the same cell, we incorporated DHFR.ATF6 and tet-inducible XBP1s into a HEK293T-REx cell line stably expressing the tet repressor. Selection of a single colony resulted in the HEK293DAX cell line in which XBP1s is induced by dox, and DHFR.ATF6 is activated by TMP (TMP-dependent DHFR.ATF6 activation in HEK293DAX cells will henceforth be referred to as ATF6 activation for simplicity). We confirmed ligand-dependent regulation of XBP1s and ATF6 by immunoblotting (Figure S1D). qPCR analysis of HEK293DAX cells demonstrates the orthogonal, ligand-dependent activation of the XBP1s and/or ATF6 transcriptional programs (Figures 1F and S1E) (Lee et al., 2003). An analogous HEK293DYG control cell line expressing tet-inducible EGFP and DHFR.YFP was also prepared as a control (Figure S1D).
The addition of activating ligands to HEK293DAX cells neither alters the incorporation of [35S]-labeled methionine into the cellular proteome (Figures S1F and S1G) nor increases eIF2α phosphorylation (Figure S1H), demonstrating that selective XBP1s and/or ATF6 activation within the physiologically relevant regime does not cause PERK-mediated translational attenuation through stress-induced global UPR activation. Independent activation of XBP1s or ATF6 also does not significantly reduce cellular viability (unlike global UPR activators such as Tm or Tg; Figure S1I). Thus, HEK293DAX cells enable orthogonal control of the transcriptional programs regulated by XBP1s and ATF6 in the same cell independent of stress.
Transcriptional and Proteomic Profiling of XBP1s and/or ATF6 Activation in the Same Cell
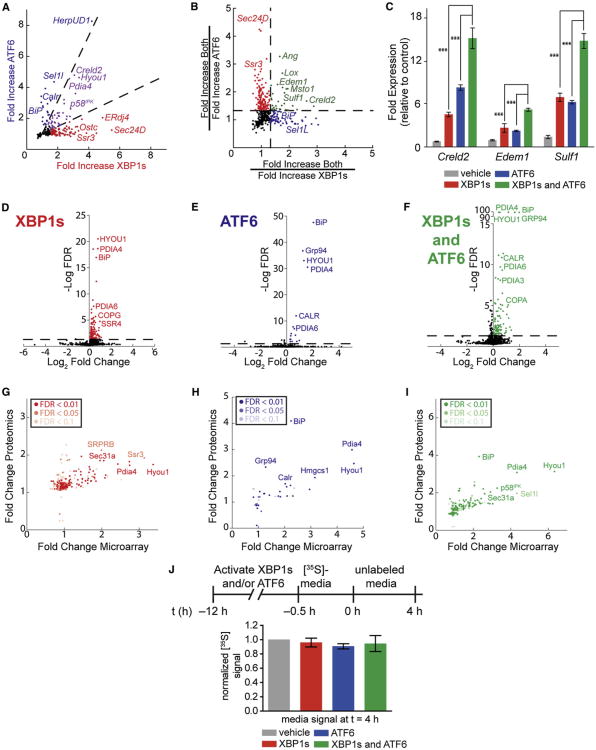
We assessed changes in the transcriptome of HEK293DAX cells following activation of XBP1s and/or ATF6 using Affymetrix whole-genome arrays. Defining a false discovery rate (FDR) <0.05, XBP1s activation increases the expression of 180 genes, ATF6 activation increases the expression of 41 genes, and activation of both XBP1s and ATF6 increases the expression of 351 genes (a smaller number of genes are modestly downregulated; see Table S1). Importantly, the addition of TMP and dox to HEK293DYG cells did not induce these genes (Table S1). Gene sets transcriptionally induced by XBP1s and/or ATF6 are highly enriched for proteins localized to the ER and involved in proteostasis (see Table S2) (Wu et al., 2008; Zhou et al., 2005).
The activation of XBP1s or ATF6 results in the upregulation of overlapping but divergent gene sets (Figure 2A), reflecting two distinct ER proteostasis environments accessible by activating these transcription factors independently. The transcriptional targets induced by XBP1s or ATF6 largely overlap with those previously identified by Adachi et al. (2008), Lee et al. (2003), Okada et al. (2002), and Yamamoto et al. (2004, 2007). Interestingly, activating both XBP1s and ATF6 affords a third, previously inaccessible, ER proteostasis environment that is not simply the sum of the transcriptional consequences of activating XBP1s or ATF6 independently. This third ER proteostasis environment includes genes upregulated to similar levels by activating either XBP1s or ATF6 in comparison to the combination (Figure 2B, red and blue, respectively). In addition, 31 genes display cooperative upregulation owing to combined XBP1s and ATF6 activation (Figure 2B, green). We have validated the cooperative induction of several of these genes by qPCR (Figure 2C). This cooperative induction likely reflects the binding of both XBP1s and ATF6 to promoter regions or the preferential binding of XBP1s/ATF6 heterodimers to select promoters (Yamamoto et al., 2007) and represents a unique transcriptional profile only accessible by our ability to activate both XBP1s and ATF6 in the same cell independent of stress.
Figure 2. Transcriptional and Proteomic Profiling of Stress-Independent XBP1s and/or ATF6 Activation in HEK293DAX Cells.
Data derived from Affymetrix whole-genome arrays and SILAC-MuDPIT whole-cell proteomic analyses of HEK293DAX cells following a 12 hr activation of XBP1s (dox; 1 μg/ml), DHFR.ATF6 (TMP; 10 μM), or both. Only genes with a FDR <0.05 are described (n = 3), unless otherwise indicated.
(A) Plot depicting mRNA fold increase owing to XBP1s activation versus ATF6 activation. Dashed lines indicate a 1.9-fold filter to assign genes as selectively induced by XBP1s (red), ATF6 (blue), or lacking selectivity (purple). Only genes upregulated ≥1.5-fold are colored.
(B) Plot depicting a ratio-of-ratios comparison to identify genes cooperatively induced by XBP1s and ATF6. Genes colored green are cooperatively induced ≥1.33-fold by the combined activation of XBP1s and ATF6 relative to activating either transcription factor individually.
(C) qPCR analysis validating select genes cooperatively upregulated by the combination of XBP1s and ATF6 in HEK293DAX cells. qPCR data are reported relative to vehicle-treated HEK293DAX cells. Error bars indicate SE from biological replicates (n = 3). ***p < 0.005.
(D–F) Plots of log2 fold change versus log FDR for all proteins identified in our SILAC-MuDPIT analysis following activation of (D) XBP1s, (E) ATF6, or (F) both. FDRs in (D) and (E) were calculated from ANOVA p values. FDRs in (F) were calculated using p values from t test distributions.
(G–I) Plots of fold change in microarray experiments versus fold change in proteomics experiments for HEK293DAX cells following activation of (G) XBP1s, (H) ATF6, or (I) both. In each panel, the significance of the SILAC-MuDPIT quantification is indicated by shading.
(J) Quantification of autoradiograms of media from HEK293DAX cells collected after a 4 hr chase in nonradioactive media. The metabolic-labeling protocol employed is shown. Error bars represent SE from biological replicates (n = 3).
See also Tables S1 and S3.
To characterize proteome remodeling upon XBP1s and/or ATF6 activation, we applied quantitative proteomics using Stable Isotope Labeling by Amino Acids in Cell Culture (SILAC)-Multi-Dimensional Protein Identification Technology (MuDPIT) (Ong et al., 2002; Washburn et al., 2001). The conditions used were identical to those employed in our transcriptome analyses (see Extended Experimental Procedures for details). Select data are reported in Table 1, and the complete proteomic data set is provided in Table S3.
Table 1. Characterization of XBP1s- and/or ATF6-Remodeled ER Proteostasis Environments.
| Fold Change in Arraysa | Fold Change in Proteomicsb,c | |||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| Protein | XBP1s | ATF6 | Both | XBP1s | ATF6 | Both |
| ER Import | ||||||
| Sec61A1 | 2.14 | NS | 1.95 | ND | ND | ND |
| SRPR | 1.98 | NS | 1.84 | NS | NS | NS |
| SRPRB | 1.99 | NS | 1.86 | 2.1 | NS | 1.7 |
| SRP19 | 2.43 | NS | 2.11 | NS | NS | NS |
| Hsp40/70/90 | ||||||
| GRP78 (BiP) | NS | 2.24 | 2.30 | 1.6 (1.6) | 4.1 (4.5) | 3.9 (4.5) |
| GRP94 | NS | NS | 1.26 | 1.3 (2.0) | 2.3 (4.7) | 2.2 (4.1) |
| P58/IPK | 2.34 | 2.36 | 3.26 | NS | 1.5 | 2.2 |
| ERDJ3 | 1.51 | 1.94 | 1.91 | ND (1.3) | ND (1.2) | ND (1.5) |
| ERDJ4 | 4.89 | NS | 5.07 | ND | ND | ND |
| ERDJ5 | 1.69 | NS | 1.92 | NS | NS | 1.8 |
| HYOU1 | 3.37 | 4.64 | 6.32 | 1.7 (1.7) | 2.5 (2.0) | 3.1 (2.6) |
| N-Glycosylation and Lectin-Assisted Folding | ||||||
| STT3A | 1.80 | NS | 1.75 | NS | NS | NS |
| DDOST | NS | NS | NS | 1.3 | NS | NS |
| RPN1 | 1.64 | NS | 1.76 | 1.4 | NS | 1.2 |
| CANX | 1.21 | NS | 1.29 | 1.2 | NS | NS |
| CRT | 1.38 | 2.22 | 2.09 | 1.2 | 1.6 | 1.6 |
| LMAN1 (ERGIC53) | 1.66 | NS | 1.52 | 1.7 | NS | 1.5 |
| Disulfide Redox | ||||||
| PDIA3 (ERP57) | 1.62 | 1.76 | 1.80 | 1.2 | 1.2 | 1.3 |
| PDIA4 | 2.74 | 4.57 | 4.32 | 1.7 | 3.0 | 3.1 |
| PDIA6 | 1.62 | NS | 1.98 | 1.3 | 1.4 | 1.4 |
| ERO1L | NS | 1.28 | 1.38 | NS | NS | 1.2 |
| ERO1LB | 2.78 | 2.13 | 3.44 | ND | ND | ND |
| Quality Control and Degradation | ||||||
| EDEM1 | NS | NS | 4.18 | ND (1.4) | ND (0.9) | ND (2.5) |
| EDEM2 | 2.08 | NS | 2.13 | ND | ND | ND |
| EDEM3 | 1.76 | NS | 1.82 | ND | ND | ND |
| DERLIN2 | NS | NS | 2.76 | ND | ND | ND |
| SYVN1 (HRD1) | 2.07 | NS | 2.67 | ND | ND | ND |
| VCP (p97) | NS | NS | 1.77 | 1.3 | 1.3 | 1.3 |
| HERPUD1 | 4.19 | 8.27 | 8.62 | ND | ND | ND |
| XTP3B | 2.27 | NS | 2.32 | ND (1.6) | ND (1.0) | ND (1.9) |
| OS9 | NS | 1.60 | 2.15 | ND (0.9) | ND (1.6) | ND (1.6) |
| SEL1L | 1.76 | 4.36 | 4.32 | NS | NS | 2.0 |
| Anterograde Trafficking/Golgi | ||||||
| SEC23B | 1.77 | NS | 1.71 | 1.6 | NS | 1.5 |
| SEC24D | 5.48 | NS | 5.60 | NS (3.7) | NS (0.9) | NS (3.1) |
| SEC31A | 2.05 | NS | 2.60 | NS | NS | 2.0 |
| GOLGB1 | 1.86 | NS | 1.81 | 1.6 | NS | 1.5 |
| Miscellaneous | ||||||
| DDIT3 (CHOP) | NS | NS | 2.67 | ND | ND | ND |
| WIPI1 | 3.14 | 3.61 | 5.71 | ND | ND | ND |
| MSTO1 | NS | NS | 4.63 | ND | ND | ND |
| SULF1 | NS | 5.17 | ND | ND | ND | |
| CRELD2 | 3.07 | 4.80 | 8.04 | ND | ND | ND |
NS, not significant; ND, not detected.
FDR <0.05.
FDR <0.1.
Quantity in parentheses is from immunoblotting (see also Figure S2).
SILAC-MuDPIT analysis demonstrates that activation of XBP1s and/or ATF6 does not significantly influence the vast majority of the proteins that comprise the cellular proteome (Figures 2D–2F). Rather, most proteins significantly upregulated by activation of XBP1s and/or ATF6 are transcriptional targets of XBP1s, ATF6, or both (Figures 2G–2I; Table 1). We used quantitative immunoblotting to confirm the increased protein levels for selected proteins shown to be increased by SILAC-MuDPIT quantification and for specific ER proteostasis network pathways underrepresented in our SILAC-MuDPIT analyses (Table 1; Figures S2A and S2B). Comparison of the fold change observed in mRNA levels with that in protein levels demonstrates a qualitative correlation between mRNA and protein levels, with the fold change in transcript levels being generally higher than at the protein level (Figures 2G–2I). Thus, combined microarray and quantitative proteomic characterization of ER proteostasis pathways in HEK293DAX cells more accurately reflects the impact of activating XBP1s and/or ATF6 on the molecular composition of the ER proteostasis network than does transcriptomics alone.
Remodeling the ER proteostasis network through activation of these transcription factors could have an impact on the secretion of endogenous, wild-type proteins to the extracellular space. Interestingly, we found that there was no significant difference in the accumulation of [35S]-labeled proteins in the extracellular space following activation of XBP1s and/or ATF6 (Figures 2J and S2C). These findings strongly suggest that activating XBP1s and/or ATF6 does not posttranscriptionally alter either the composition of the intracellular proteome or the folding and trafficking of endogenous, wild-type proteins through the secretory pathway, although we cannot conclusively rule out alterations in the secretion of low-abundance proteins that are not detected in our SILAC-MuDPIT or [35S]-labeling experiments.
Integration of Transcriptomics and Proteomics Reveals Three Distinct ER Proteostasis Environments Accessible upon Activation of XBP1s and/or ATF6
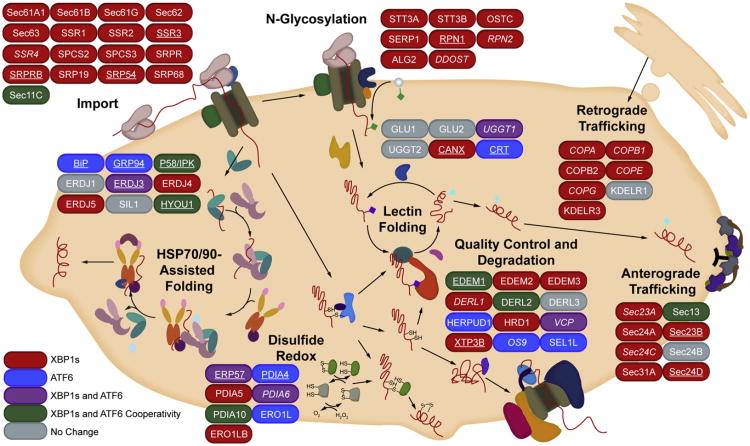
Our transcriptional and proteomic profiling of HEK293DAX cells reveals how the composition of the ER proteostasis network is differentially remodeled by activation of the XBP1s and/or ATF6 transcriptional programs (Figure 3). Consistent with the IRE1-XBP1s signaling cascade being the only UPR pathway conserved from yeast to humans, XBP1s activation has a broader impact on the composition of ER proteostasis pathways than does ATF6. XBP1s activation upregulates entire ER proteostasis pathways, including those involved in ER protein import, N-linked glycosylation, and anterograde/retrograde vesicular trafficking (Figure 3, red). The induction of these pathways is similarly observed by enrichment analysis (Table S2). In contrast, although ATF6 is responsible for upregulating only a select subset of ER proteostasis network proteins, these ATF6-selective targets represent critical hub proteins in the ER proteostasis network, including BiP, Sel1L, and calreticulin (Figure 3, blue).
Figure 3. Predictive Pathway Analysis for Stress-Independent XBP1s- and/or ATF6-Mediated Remodeling of the ER Proteostasis Network.
Cartoon depicting the impact of activating XBP1s, ATF6, or both XBP1s and ATF6 on the composition of ER proteostasis pathways obtained by integrating transcriptional, proteomic, and biochemical results. XBP1s (red) and ATF6 (blue)-selective genes are genes where activating either XBP1s (but not ATF6) or ATF6 (but not XBP1s) independently results in >75% of the induction observed when both XBP1s and ATF6 are activated (“max induction”). Genes induced >75% of the max induction by activating XBP1s in isolation and induced >75% of the max induction by activating ATF6 in isolation (i.e., lacking selectivity) are colored purple. Genes cooperatively induced >1.33-fold upon activation of both XBP1s and ATF6 relative to the activation of either transcription factor alone are colored green. Plain type indicates results from array data. Italicized type indicates results from proteomics data. Underlined type indicates results confirmed at both the transcript and the protein levels. Thresholds for transcriptional analyses were set at a FDR of <0.05. Thresholds for proteomic analyses were set at a FDR of 0.1.
Some proteins are upregulated to similar levels by activating XBP1s in isolation, ATF6 in isolation, or both XBP1s and ATF6 (Figure 3, purple). Alternatively, a number of proteins primarily involved in ER quality control and degradation are cooperatively upregulated when both XBP1s and ATF6 are activated (Figure 3, green). These results are consistent with the biological pathways predicted to be transcriptionally enhanced by XBP1s:ATF6 heterodimers (Yamamoto et al., 2007) and clearly demonstrate that the impact of the combined activation of XBP1s and ATF6 on the composition of the ER proteostasis network is greater than the sum of activating XBP1s or ATF6 individually.
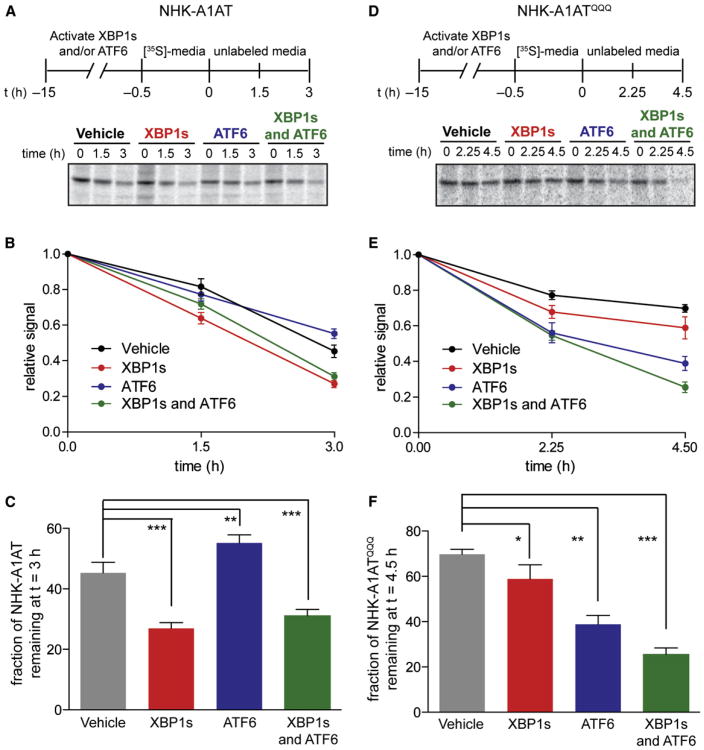
Selective Activation of the XBP1s and/or ATF6 Transcriptional Programs Differentially Influences the Degradation of Null Hong Kong α1-Antitrypsin Variants
To evaluate the functional consequences of activating XBP1s and/or ATF6 on the degradation of terminally misfolded ER client proteins, we employed the nonsecreted ERAD substrate null Hong Kong α1-antitrypsin (NHK-A1AT) (Christianson et al., 2008) and [35S]-metabolic labeling (Figures 4A–4C). XBP1s activation increases the rate of NHK-A1AT degradation. Activation of ATF6 has the opposite effect—resulting in a small but statistically significant delay in NHK-A1AT degradation (also observed by cycloheximide chase experiments; see Figure S3A). The effect of activating both XBP1s and ATF6 on NHK-A1AT degradation is similar to that observed for activation of XBP1s alone. Notably, we observed no significant impact on NHK-A1AT degradation in HEK293DYG cells using the same experimental conditions (Figure S3B). Thus, XBP1s and ATF6 activation have distinct impacts on NHK-A1AT degradation, consistent with their differing transcriptional adaptations of the ER proteo-stasis network (Figure 3).
Figure 4. XBP1s and/or ATF6 Activation Differentially Influences the Degradation of NHK-A1AT and NHK-A1ATQQQ.
(A) Representative autoradiogram of [35S]-labeled NHK-A1AT immunopurified from transfected HEK293DAX cells following a 15 hr preactivation of XBP1s (dox; 1 μg/ml), DHFR.ATF6 (TMP; 10 μM), or both. The metabolic-labeling protocol employed is shown.
(B) Quantification of autoradiograms in (A) monitoring the degradation of [35S]-labeled NHK-A1AT. The fraction of NHK-A1AT remaining was calculated by normalizing the recovered [35S] signal to the total amount of labeling observed at 0 hr. Error bars represent SE from biological replicates (n = 18).
(C) Bar graph depicting the normalized fraction of NHK-A1AT remaining at 3 hr calculated as in (B).
(D) Representative autoradiogram of [35S]-labeled NHK-A1ATQQQ immunopurified from transfected HEK293DAX cells following a 15 hr preactivation of XBP1s (dox; 1 μg/ml), DHFR.ATF6 (TMP; 10 μM), or both. The metabolic-labeling protocol employed is shown.
(E) Quantification of autoradiograms in (D) monitoring the degradation of [35S]-labeled NHK-A1ATQQQ. The fraction of NHK-A1ATQQQ remaining was calculated as in (B). Error bars represent SE from biological replicates (n = 6).
(F) Bar graph depicting the normalized fraction of NHK-A1ATQQQ remaining at 4.5 hr calculated as in (B).
*p < 0.05, **p < 0.01, and ***p < 0.001. See also Figure S3.
Stress-independent transcriptional remodeling of the ER proteostasis environment to enhance the clearance of terminally misfolded proteins is likely to be client dependent. Because proteostasis network interactions with NHK-A1AT are primarily mediated by glycan-binding lectins (Christianson et al., 2008), removing the three N-linked glycans in the NHK-A1AT structure by mutating glycosylated asparagines to glutamines changes the ER proteostasis pathways traversed by NHK-A1AT and, thus, could alter the consequences of arm-specific UPR activation for NHK-A1AT degradation. We found that XBP1s activation moderately enhances degradation of the nonglycosylated NHK-A1ATQQQ variant, whereas ATF6 activation dramatically increases the degradation rate of NHK-A1ATQQQ (Figures 4D–4F). Activating both XBP1s and ATF6 further increases the rate of NHK-A1ATQQQ degradation (Figures 4D–4F). As in the case of NHK-A1AT, we observed no significant impact on NHK-A1ATQQQ degradation in HEK293DYG cells using the same experimental conditions (Figure S3C). Clearly, the NHK-A1AT and NHK-A1ATQQQ degradation pathways are uniquely affected by arm-selective UPR activation, reflecting discrete, substrate-specific functional differences between the ER proteostasis environments illustrated in Figure 3.
Activation of the ATF6 Transcriptional Program Reduces Secretion of Destabilized Amyloidogenic TTR Variants without Affecting Wild-Type TTR
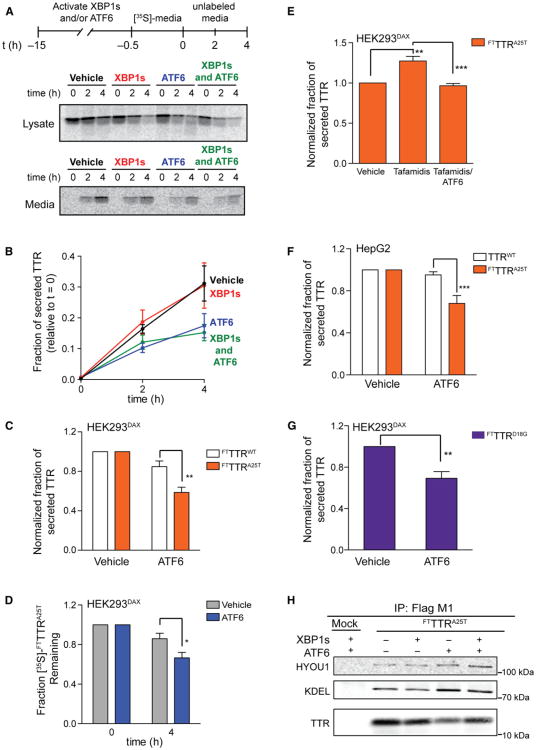
We speculated that arm-selective UPR activation could provide a means to selectively reduce the secretion of destabilized proteins because quality control and degradation are differentially regulated by the three accessible proteostasis environments (Figure 3). To test this hypothesis, we evaluated the secretion of the destabilized, aggregation-prone A25T TTR (FTTTRA25T) variant (Sekijima et al., 2005) in HEK293DAX cells using [35S]-metabolic labeling.
FTTTRA25T secretion is decreased 40% following activation of the ATF6 transcriptional program but is unaffected by XBP1s activation (Figures 5A and 5B). Activating both XBP1s and ATF6 decreases FTTTRA25T secretion to a level identical to that observed for ATF6 activation alone. The ATF6-dependent decrease in FTTTRA25T secretion is also observed by quantitative immunoblotting (Figure S4A). FTTTRA25T secretion is not sensitive to treatment with dox, TMP, or both in HEK293DYG cells (Figure S4B). The decrease in FTTTRA25T secretion observed upon ATF6 activation is similar to that observed upon Tg-dependent global UPR activation (Figure S4C). Arm-selective ATF6 activation, however, avoids the negative consequences of a concomitant decrease in cell viability caused by global UPR activation (Figure S1I). The effects of ATF6 are selective for destabilized TTR variants because FTTTRA25T secretion is selectively reduced relative to FTTTRWT following ATF6 activation, as measured by either pulse-chase (Figure 5C) or quantitative immunoblotting (Figure S4A). A 50% decrease in TTRA25T concentration dramatically reduces its rate and extent of aggregation at pH 6.0 (Figure S4D) (Sekijima et al., 2003). Thus, ATF6-dependent reductions in mutant TTR secretion would likely attenuate extracellular, concentration-dependent mutant TTR aggregation.
Figure 5. ATF6 Activation Selectively Attenuates the Secretion of Amyloidogenic TTR.
(A) Autoradiogram of [35S]-labeled FTTTRA25T immunopurified from media and lysates collected from transfected HEK293DAX cells following a 15 hr preactivation of XBP1s (dox; 1 μg/ml), DHFR.ATF6 (TMP; 10 μM), or both. The metabolic-labeling protocol employed is shown.
(B) Quantification of autoradiograms as shown in (A). Fraction secreted was calculated as previously described by Sekijima et al. (2005). Error bars represent SE from biological replicates (n = 4).
(C) Graph depicting the normalized fraction secreted of [35S]-labeled FTTTRWT (white bars) or FTTTRA25T (orange bars) at 4 hr following a 15 hr preactivation of DHFR.ATF6 (TMP; 10 μM) in HEK293DAX cells. Error bars represent SE from biological replicates (n = 8 for FTTTRA25T, and n = 9 for FTTTRWT).
(D) Graph depicting the total [35S]-labeled FTTTRA25T remaining in HEK293DAX cells (combined media and lysate protein levels as in A). The fraction remaining was calculated as reported previously by Sekijima et al. (2005). Error bars represent SE from biological replicates (n = 8).
(E) Graph depicting the normalized fraction secreted of [35S]-labeled FTTTRA25T (orange bars) at 4 hr following preactivation of DHFR.ATF6 (TMP; 10 μM; 15 hr) inthe presence or absence of tafamidis (10 μM; 15 hr) in HEK293DAX cells. Error bars represent SE from biological replicates (n = 4).
(F) Bar graph depicting the normalized fraction secreted of FTTTRA25T and endogenous TTRWT at 4 hr following a 13 hr pretreatment with TMP (100 μM) in HepG2 cells stably expressing DHFR.ATF6. Error bars represent SE from biological replicates (n = 4).
(G) Bar graph depicting the normalized fraction secreted of [35S]-labeled FTTTRD18G at 4 hr following a 15 hr pretreatment with TMP (10 μM) from HEK293DAX cellstransfected with both FTTTRD18G and TTRWT. Error bars represent SE from biological replicates (n = 4).
(H) Immunoblot of α-FLAG M1FTTTRA25T immunoisolations from DSP-crosslinked lysates prepared from HEK293DAX cells expressing FTTTRA25T following 15 hr activation of XBP1s (dox; 1 μg/ml), DHFR.ATF6 (TMP; 10 μM), or both. HEK293DAX cells expressing GFP are shown as a negative control (Mock). The KDEL immunoblot shows BiP.
*p < 0.05, **p < 0.01, ***p < 0.005. See also Figure S4.
ATF6 activation also leads to a significant decrease in total [35S]-labeledFTTTRA25T relative to vehicle-treated controls (Figure 5D). This decrease in total FTTTRA25T cannot be attributed to increased intracellular TTR aggregation because only ∼10% of intracellular TTR is found in detergent-insoluble pellets, and this fraction is not significantly affected by ATF6 activation (Figure S4E). Intracellular levels of the nonsecreted, highly destabilized TTR variant FTTTRD18G are similarly decreased following ATF6 activation relative to controls (Figures S4F and S4G), suggesting that the observed decrease in mutant TTR can be attributed to increased degradation. Consistent with this prediction, the ATF6-dependent decrease in FTTTRD18G protein levels is attenuated by the proteasome inhibitor MG-132 (Figure S4G).
Previously, small molecule TTR ligands including the endogenous ligand thyroxine were shown to increase secretion of destabilized TTR mutants, such as TTRA25T (Sekijima et al., 2005). The thyroxine-dependent increase in mutant TTR secretion is predicted to contribute to the deposition of highly destabilized TTR mutants in the CNS (Sekijima et al., 2005). We explored whether ATF6 activation could attenuate the ligand-dependent increase in mutant TTR secretion using the highly selective small molecule TTR stabilizer tafamidis (Bulawa et al., 2012). As expected, both pulse-chase experiments and quantitative immunoblotting show that the addition of tafamidis increases FTTTRA25T secretion (Figures 5E, S4H, and S4I). ATF6 activation attenuates this tafamidis-mediated increase in FTTTRA25T secretion (Figures 5E, S4H, and S4I). Notably, tafamidis also attenuates the ATF6-dependent increase in mutant TTR degradation (cf. Figures 5D and S4I), suggesting that tafamidis stabilizes FTTTRA25T in the ER lumen. These results indicate that ATF6 activation prevents the increase in mutant TTR secretion afforded by ligand-dependent stabilization of the TTR tetramer.
To further explore the potential for ATF6 activation to decrease mutant TTR secretion in a physiologically relevant model system, we leveraged the high transportability (see Figure 1E) of our methodology to activate ATF6 to establish a liver-derived HepG2 cell line stably expressing DHFR.ATF6 (the liver produces 95% of the TTR in human serum). TMP-dependent activation of DHFR.ATF6 in HepG2 cells was confirmed by qPCR (Figure S4J). We overexpressed FTTTRA25T in these HepG2 cells, which already secrete high levels of endogenous TTRWT. ATF6 activation reduced FTTTRA25T secretion by ∼35% in HepG2 cells but did not affect secretion of endogenous TTRWT (Figures 5F and S4K). Thus, ATF6 activation selectively reduces the secretion of destabilized TTR mutants from physiologically relevant human liver-derived cells.
We further explored the secretion of TTR heterotetramers comprising TTRD18G subunits and TTRWT subunits from HEK293DAX cells. Although TTRD18G is too destabilized to be significantly secreted as a homotetramer (Figure S4F) (Hammarström et al., 2003a), the expression of both TTRD18G and TTRWT enables heterotetramers composed of TTRD18G and TTRWT subunits to be secreted (Hammarström et al., 2003b; Sekijima et al., 2005). Activating ATF6 in HEK293DAX cells expressing both FTTTRD18G and TTRWT decreases the secretion of the amyloidogenic FTTTrD18G subunit that is permitted by heterotetramer formation (Figures 5G and S4L).
We next explored the impact of ATF6 activation on the interactions between FTTTRA25T and the central ER chaperone BiP, which was previously shown to selectively bind destabilized TTR variants and affect their ER proteostasis (Sörgjerd et al., 2006; Susuki et al., 2009). We found that activating ATF6 increases the amount of BiP and the BiP cochaperone HYOU1 that coimmunoprecipitates with FTTTRA25T (Figure 5H). Notably, the extent of HYOU1 association withFTTTRA25T following XBP1s activation did not increase, despite XBP1s increasing HYOU1 protein levels (Table 1). These results suggest that the progression of FTTTRA25T through the ER proteostasis network is altered by ATF6-dependent remodeling of the ER environment.
Discussion
Herein, we establish methodology that allows for the orthogonal, small molecule-mediated regulation of the UPR-associated transcription factors XBP1s and/or ATF6 in the same cell independent of stress. We employ our methodology to reveal the molecular composition of the three distinct ER proteostasis environments accessible by activating the XBP1s and/or ATF6 transcriptional programs. Furthermore, we show that selectively activating XBP1s and/or ATF6 differentially influences the ER partitioning of destabilized protein variants between folding and trafficking versus degradation. Our results provide molecular insights into how the XBP1s and/or ATF6 transcriptional programs remodel the ER proteostasis environment and demonstrate the potential to influence the ER proteostasis of destabilized protein variants via physiologic levels of arm-selective UPR activation.
Our quantitative transcriptional and proteomic profiling of HEK293DAX cells provides an experimentally validated, conceptual framework to identify specific ER proteins and/or pathways that can be adapted to alter the fate of disease-associated ER client proteins (Figure 3). Critical pathways directly responsible for the partitioning of ER client proteins between folding and trafficking versus degradation are differentially impacted by XBP1s and/or ATF6 activation. For example, the levels of BiP and BiP cochaperones, which are known to modulate folding versus degradation decisions of client proteins in the ER lumen, are differentially influenced by XBP1s and/or ATF6 activation (Figure 3) (Kampinga and Craig, 2010). Considering the importance of BiP cochaperones in defining BiP function, these findings suggest that the fates of BiP clients are distinctly influenced by XBP1s and ATF6 activation. Consistent with this prediction, we show that BiP and HYOU1 have increased association with TTRA25T only when ATF6 is activated, even though HYOU1 is also upregulated by XBP1s (Table 1).
Analogously, XBP1s- or ATF6-dependent remodeling of ER client protein folding pathways can be deconvoluted from our bioinformatic characterization of HEK293DAX cells. For example, XBP1s-selective transcriptional upregulation of the ERAD-associated proteins ERMan1, ERdj5, and EDEM-3 may explain the enhanced degradation of NHK-A1AT upon XBP1s activation because overexpression of these three proteins enhances NHK-A1AT ERAD (Hosokawa et al., 2003, 2006; Ushioda et al., 2008). Alternatively, ATF6 selectively enhances the expression of the ERAD-associated protein Sel1L, which when overexpressed, accelerates degradation of the nonglycosylated protein NHK-A1ATQQQ (Iida et al., 2011). Thus, our transcriptional and proteomic profilesofcellsremodeledbyXBP1s and/or ATF6 activation enable hypothesis generation to dissect the contributions of ER proteostasis proteins and/or pathways involved in altering the folding, trafficking, or degradation of ER client proteins.
We used HEK293DAX cells to explore the potential for ER proteostasis environments accessed through arm-selective UPR activation to reduce the secretion of a destabilized, amyloidogenic TTR variant. We found that ATF6 activation selectively reduces secretion of the destabilized, aggregation-prone TTRA25T, but not the secretion of TTRWT or the global endogenous secreted proteome. Previously, we and others have demonstrated that the efficient secretion of destabilized TTR variants through the hepatic secretory pathway is a contributing factor to the extracellular aggregation and distal deposition of TTR as amyloid in the pathology of numerous TTR amyloid diseases (Hammarström et al., 2002, 2003a; Holmgren et al., 1993; Sekijima et al., 2003, 2005; Suhr et al., 2000; Susuki et al., 2009; Tan et al., 1995). Thus, our discovery that ATF6-dependent remodeling of the ER proteostasis environment selectively reduces secretion of destabilized TTRA25T reveals a potential mechanism to attenuate the secretion and subsequent pathologic extracellular aggregation of the >100 destabilized TTR variants involved in TTR amyloid diseases (Sekijima et al., 2008). Furthermore, the establishment and characterization of the DHFR.ATF6 construct (which we demonstrate can be rapidly incorporated into any cellular model) and the HEK293DAX cell line provide invaluable resources to evaluate the functional impact of arm-selective UPR activation to physiologic levels on the aberrant ER proteostasis of destabilized mutant proteins involved in the pathology of many other protein misfolding-related diseases. Consistent with the potential to correct pathologic imbalances in destabilized protein ER proteostasis, recent studies that employ global UPR activation using toxic small molecules or the unregulated overexpression of XBP1s or ATF6 have suggested that remodeling ER proteostasis pathways through arm-selective UPR activation could correct the aberrant ER proteostasis of pathologic destabilized protein mutants involved in protein misfolding diseases (Chiang et al., 2012; Mu et al., 2008; Smith et al., 2011a).
Finally, we note that despite clear functional roles for XBP1s and ATF6 in adapting the composition of ER proteostasis pathways highlighted herein, organisms have distinct dependencies on these transcription factors. XBP1s is critical for biological processes including plasma cell differentiation and development (XBP1s knockout mice are not viable; Reimold etal., 2000). Alternatively, mice lacking ATF6α, the primary ATF6 homolog involved in UPR-dependent remodeling of the ER proteostasis environment, develop normally, although deletion of both mammalian ATF6 homologs, ATF6α and ATF6β, is embryonic lethal (Adachi et al., 2008; Wu et al., 2007; Yamamoto et al., 2007). Thus, whereas XBP1s is required for organismal development, our results suggest that functional roles for ATF6 in remodeling the ER proteostasis environment are adaptive—adjusting ER proteostasis capacity to match demand under conditions of cellular or organismal stress. Therefore, modulation of ATF6 may provide a unique opportunity to sensitively “tune” the ER proteostasis environment without globally influencing the folding, trafficking, or degradation of the secreted proteome.
In summary, we show that the application of DD methodology to control ATF6 transcriptional activity provides an experimental strategy to characterize the impact of stress-independent activation of XBP1s and/or ATF6 on ER proteostasis pathway composition and ER function. Adapting the underlying biology of the proteostasis network through the activation of specific UPR transcriptional programs reveals emergent functions of the proteostasis network, including a window to alter the ER proteostasis of destabilized mutant proteins without significantly affecting the proteostasis of the vast majority of the endogenous, wild-type proteome. Our transcriptional, proteomic, and functional characterization of the ER proteostasis environments accessible by activating XBP1s and/or ATF6 in a single cell validates targeting specific pathways within the proteostasis network as a potential therapeutic approach for adapting the aberrant ER proteostasis associated with numerous protein misfolding diseases, strongly motivating the development of arm-selective small molecule activators of the UPR.
Experimental Procedures
Plasmids
DHFR.YFP.pBMN was a generous gift from Professor T.J. Wandless at Stanford University. ATF6 was amplified from human cDNA, substituted for YFP in DHFR.YFP.pBMN between the SphI and NotI sites, and then transferred to pENTR1A vectors using the BamH1 and Not1 sites. XBP1s was amplified from human cDNA and cloned between the BamHI and Not1 sites in pENTR1A. EGFP and ATF6 were similarly cloned into empty pENTR1A vectors using the BamHI and Not1 sites. Genes of interest in pENTR1A were shuttled into pcDNA-DEST40, pT-REx-DEST30, pLenti4/TO/V5-DEST, or pAd/CMV/V5-DEST vectors, as appropriate, using LR clonase II (Invitrogen) recombination. A1AT-NHK.pcDNA 3.1 was a generous gift from Professor R.R. Kopito at Stanford University.FTTTRWTwas cloned from FTTTRWT.pET3C (Sekijima et al., 2005) into pcDNAI using the BamHI and BstB1 sites. FTTTRA25T.pcDNAI and A1AT-NHKQQQ.pcDNA3.1 were prepared by site-directed mutagenesis. All constructs were sequenced to confirm their identity.
Immunoblotting
Postnuclear supernatants and nuclear lysates (prepared as described in the Extended Experimental Procedures) were separated by SDS-PAGE and analyzed by immunoblotting using the Li-COR Biosciences Odyssey System. Details of immunoprecipitation experiments and antibodies used are provided in the Extended Experimental Procedures.
Quantitative RT-PCR
The relative mRNA expression levels of target genes were measured using quantitative RT-PCR (see the Extended Experimental Procedures for details and Table S4 for a list of the primers used).
Cell Culture and Transfections
HEK293T-REx cells (Invitrogen) were cultured in complete DMEM (CellGro) supplemented with 10% fetal bovine serum (FBS) (CellGro). Transient transfections of DHFR.ATF6, ATF6, DHFR.YFP, EGFP,A1AT, and TTR constructs were performed by calcium phosphate transfection. Lentiviruses encoding dox-inducibleXBP1sand EGFP were transduced into HEK293T-REx cells expressing DHFR.ATF6 or DHFR.YFP, respectively, using 1-5 μl of virus in media containing 5 μg/ml polybrene. Adenoviruses encoding DHFR.ATF6 or DHFR.YFP were transduced into HepG2, Huh7, and primary human fibroblasts at identical multiplicities of infections, experimentally determined to transduce ∼90% of cells. Detailed protocols for lentivirus and adenovirus production are provided in the Extended Experimental Procedures. Stable cell lines were selected by culturing in blasticidin (10 μg/ml), geneticin sulfate (G-418; 500 μg/ml), orzeocin(50 μg/ml), as appropriate, prior to single-colony selection and characterization.
Whole-Genome Array Analyses
HEK293DAX or HEK293DYG cells were treated for 12 hr with vehicle, 1 μg/ml dox, 10 μM TMP, or both in biological triplicate. Cells were harvested, and RNA was extracted using the RNeasy Mini Kit (QIAGEN). Genomic DNA was removed by on-column digestion using the RNase-free DNase Set (QIAGEN). Detailed protocols for the whole-genome array analyses are provided in the Extended Experimental Procedures. Data from HEK293DYG cells showed no significant overlap in the ligand-treated transcriptomes obtained from HEK293DAX and HEK293DYG cells (see Table S1).
SILAC-Assisted Liquid Chromatography-Mass Spectrometry Analyses
HEK293DAX cells were grown for >6 passages in either light or heavy SILAC media (heavy media were supplemented with lysine-2HCl-[U-13C6, 97%–99%] and arginine-2HCl [U-13C6, 99%, and U-15N4, 99%]; Cambridge Isotopes). Samples were prepared from “heavy” HEK293DAX cells treated for 12 hr with dox (1 μg/ml), TMP (10 μM), or both and “light” cells treated with vehicle. Detailed protocols for cell lysis and mass spectrometric analyses are provided in the Extended Experimental Procedures.
Pulse-Chase Experiments
HEK293DAX or HEK293DYG cells plated on poly-D-lysine-coated plates were metabolically labeled in pulse medium ([35S]-Translabel [MP Biomedical], DMEM –Cys/–Met [CellGro] with supplemented glutamine, penicillin/streptomycin, and dialyzed FBS) for 30 min. Cells were then washed with complete media and incubated in prewarmed DMEM for the indicated chase times. Media or lysates were harvested at the indicated times. Lysates were prepared in either 20 mM HEPES (pH 7.5), 100 mM NaCl, 1% Triton X-100, and fresh protease inhibitor cocktail (Roche) for NHK-A1AT experiments or standard RIPA buffer for TTR experiments. Proteins were immunopurified using either anti-HA antibody conjugated to protein-G Sepharose or M1 anti-Flag beads (Sigma-Aldrich), respectively. The resin was eluted by boiling beads in Laemili buffer, and the samples were separated by SDS-PAGE. The gels were then dried, exposed to phosphorimager plates (GE Healthcare), and imaged with a Typhoon imager. Band intensities were quantified by densitometry in ImageQuant.
Supplementary Material
Acknowledgments
The authors are grateful to the Center for Physiological Proteomics at TSRI and to Professor Thomas Wandless, Dr. Laura Banaszynski, and Dr. Patrick Braun for helpful discussion. We thank NIH (DK075295, DK046335, and AG036634), Arlene and Arnold Goldstein, the Ellison Medical Foundation, the Skaggs Institute for Chemical Biology, the Lita Annenberg Hazen Foundation, and the Scripps Research Institute for financial support. M.D.S. was supported by an American Cancer Society postdoctoral fellowship. J.C.G. was supported by the NHLBI (F32-HL099245). J.J.M., P.G.T., and J.R.Y. were supported by the National Center for Research Resources (P41RR011823), National Institute of General Medical Sciences (P41GM103533), and National Institute on Aging (AG027463).
Footnotes
Accession Numbers: The Gene Expression Omnibus accession number for the data reported herein is GSE44951.
Supplemental Information: Supplemental Information includes Extended Experimental Procedures, four figures, and four tables and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2013.03.024
Licensing Information: This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
References
- Adachi Y, Yamamoto K, Okada T, Yoshida H, Harada A, Mori K. ATF6 is a transcription factor specializing in the regulation of quality control proteins in the endoplasmic reticulum. Cell Struct Funct. 2008;33:75–89. doi: 10.1247/csf.07044. [DOI] [PubMed] [Google Scholar]
- Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- Banaszynski LA, Chen LC, Maynard-Smith LA, Ooi AGL, Wandless TJ. A rapid, reversible, and tunable method to regulate protein function in living cells using synthetic small molecules. Cell. 2006;126:995–1004. doi: 10.1016/j.cell.2006.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braakman I, Bulleid NJ. Protein folding and modification in the mammalian endoplasmic reticulum. Annu Rev Biochem. 2011;80:71–99. doi: 10.1146/annurev-biochem-062209-093836. [DOI] [PubMed] [Google Scholar]
- Brodsky JL, Skach WR. Protein folding and quality control in the endoplasmic reticulum: recent lessons from yeast and mammalian cell systems. Curr Opin Cell Biol. 2011;23:464–475. doi: 10.1016/j.ceb.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulawa CE, Connelly S, Devit M, Wang L, Weigel C, Fleming JA, Packman J, Powers ET, Wiseman RL, Foss TR, et al. Tafamidis, a potent and selective transthyretin kinetic stabilizer that inhibits the amyloid cascade. Proc Natl Acad Sci USA. 2012;109:9629–9634. doi: 10.1073/pnas.1121005109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang WC, Hiramatsu N, Messah C, Kroeger H, Lin JH. Selective activation of ATF6 and PERK endoplasmic reticulum stress signaling pathways prevent mutant rhodopsin accumulation. Invest Ophthalmol Vis Sci. 2012;53:7159–7166. doi: 10.1167/iovs.12-10222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson JC, Shaler TA, Tyler RE, Kopito RR. OS-9 and GRP94 deliver mutant alpha1-antitrypsin to the Hrd1-SEL1L ubiquitin ligase complex for ERAD. Nat Cell Biol. 2008;10:272–282. doi: 10.1038/ncb1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gass JN, Jiang HY, Wek RC, Brewer JW. The unfolded protein response of B-lymphocytes: PERK-independent development of antibody-secreting cells. Mol Immunol. 2008;45:1035–1043. doi: 10.1016/j.molimm.2007.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammarström P, Jiang X, Hurshman AR, Powers ET, Kelly JW. Sequence-dependent denaturation energetics: a major determinant in amyloid disease diversity. Proc Natl Acad Sci USA. 2002;99(Suppl 4):16427–16432. doi: 10.1073/pnas.202495199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammarström P, Sekijima Y, White JT, Wiseman RL, Lim A, Costello CE, Altland K, Garzuly F, Budka H, Kelly JW. D18G trans-thyretin is monomeric, aggregation prone, and not detectable in plasma and cerebrospinal fluid: a prescription for central nervous system amyloidosis? Biochemistry. 2003a;42:6656–6663. doi: 10.1021/bi027319b. [DOI] [PubMed] [Google Scholar]
- Hammarström P, Wiseman RL, Powers ET, Kelly JW. Prevention of transthyretin amyloid disease by changing protein misfolding energetics. Science. 2003b;299:713–716. doi: 10.1126/science.1079589. [DOI] [PubMed] [Google Scholar]
- Harding HP, Zeng H, Zhang Y, Jungries R, Chung P, Plesken H, Sabatini DD, Ron D. Diabetes mellitus and exocrine pancreatic dysfunction in perk-/- mice reveals a role for translational control in secretory cell survival. Mol Cell. 2001;7:1153–1163. doi: 10.1016/s1097-2765(01)00264-7. [DOI] [PubMed] [Google Scholar]
- Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- Holmgren G, Ericzon BG, Groth CG, Steen L, Suhr O, Andersen O, Wallin BG, Seymour A, Richardson S, Hawkins PN, et al. Clinical improvement and amyloid regression after liver transplantation in hereditary transthyretin amyloidosis. Lancet. 1993;341:1113–1116. doi: 10.1016/0140-6736(93)93127-m. [DOI] [PubMed] [Google Scholar]
- Hosokawa N, Tremblay LO, You Z, Herscovics A, Wada I, Nagata K. Enhancement of endoplasmic reticulum (ER) degradation of mis-folded Null Hong Kong alpha1-antitrypsin by human ER mannosidase I. J Biol Chem. 2003;278:26287–26294. doi: 10.1074/jbc.M303395200. [DOI] [PubMed] [Google Scholar]
- Hosokawa N, Wada I, Natsuka Y, Nagata K. EDEM accelerates ERAD by preventing aberrant dimer formation of misfolded alpha1-anti-trypsin. Genes Cells. 2006;11:465–476. doi: 10.1111/j.1365-2443.2006.00957.x. [DOI] [PubMed] [Google Scholar]
- Iida Y, Fujimori T, Okawa K, Nagata K, Wada I, Hosokawa N. SEL1L protein critically determines the stability of the HRD1-SEL1L endoplasmic reticulum-associated degradation (ERAD) complex to optimize the degradation kinetics of ERAD substrates. J Biol Chem. 2011;286:16929–16939. doi: 10.1074/jbc.M110.215871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto M, Björklund T, Lundberg C, Kirik D, Wandless TJ. A general chemical method to regulate protein stability in the mammalian central nervous system. Chem Biol. 2010;17:981–988. doi: 10.1016/j.chembiol.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampinga HH, Craig EA. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Biol. 2010;11:579–592. doi: 10.1038/nrm2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaser A, Lee AH, Franke A, Glickman JN, Zeissig S, Tilg H, Nieuwenhuis EES, Higgins DE, Schreiber S, Glimcher LH, Blumberg RS. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell. 2008;134:743–756. doi: 10.1016/j.cell.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AH, Iwakoshi NN, Glimcher LH. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol. 2003;23:7448–7459. doi: 10.1128/MCB.23.21.7448-7459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luheshi LM, Dobson CM. Bridging the gap: from protein mis-folding to protein misfolding diseases. FEBS Lett. 2009;583:2581–2586. doi: 10.1016/j.febslet.2009.06.030. [DOI] [PubMed] [Google Scholar]
- McClellan AJ, Tam S, Kaganovich D, Frydman J. Protein quality control: chaperones culling corrupt conformations. Nat Cell Biol. 2005;7:736–741. doi: 10.1038/ncb0805-736. [DOI] [PubMed] [Google Scholar]
- Mu TW, Ong DS, Wang YJ, Balch WE, Yates JR, 3rd, Segatori L, Kelly JW. Chemical and biological approaches synergize to ameliorate protein-folding diseases. Cell. 2008;134:769–781. doi: 10.1016/j.cell.2008.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada T, Yoshida H, Akazawa R, Negishi M, Mori K. Distinct roles of activating transcription factor 6 (ATF6) and double-stranded RNA-activated protein kinase-like endoplasmic reticulum kinase (PERK) in transcription during the mammalian unfolded protein response. Biochem J. 2002;366:585–594. doi: 10.1042/BJ20020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- Reimold AM, Etkin A, Clauss I, Perkins A, Friend DS, Zhang J, Horton HF, Scott A, Orkin SH, Byrne MC, et al. An essential role in liver development for transcription factor XBP-1. Genes Dev. 2000;14:152–157. [PMC free article] [PubMed] [Google Scholar]
- Schröder M, Kaufman RJ. The mammalian unfolded protein response. Annu Rev Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- Sekijima Y, Hammarström P, Matsumura M, Shimizu Y, Iwata M, Tokuda T, Ikeda S, Kelly JW. Energetic characteristics of the new transthyretin variant A25T may explain its atypical central nervous system pathology. Lab Invest. 2003;83:409–417. doi: 10.1097/01.lab.0000059937.11023.1f. [DOI] [PubMed] [Google Scholar]
- Sekijima Y, Wiseman RL, Matteson J, Hammarström P, Miller SR, Sawkar AR, Balch WE, Kelly JW. The biological and chemical basis for tissue-selective amyloid disease. Cell. 2005;121:73–85. doi: 10.1016/j.cell.2005.01.018. [DOI] [PubMed] [Google Scholar]
- Sekijima Y, Kelly JW, Ikeda S. Pathogenesis of and therapeutic strategies to ameliorate the transthyretin amyloidoses. Curr Pharm Des. 2008;14:3219–3230. doi: 10.2174/138161208786404155. [DOI] [PubMed] [Google Scholar]
- Sitia R, Braakman I. Quality control in the endoplasmic reticulum protein factory. Nature. 2003;426:891–894. doi: 10.1038/nature02262. [DOI] [PubMed] [Google Scholar]
- Smith SE, Granell S, Salcedo-Sicilia L, Baldini G, Egea G, Teckman JH, Baldini G. Activating transcription factor 6 limits intracellular accumulation of mutant α(1)-antitrypsin Z and mitochondrial damage in hepa-toma cells. J Biol Chem. 2011a;286:41563–41577. doi: 10.1074/jbc.M111.280073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MH, Ploegh HL, Weissman JS. Road to ruin: targeting proteins for degradation in the endoplasmic reticulum. Science. 2011b;334:1086–1090. doi: 10.1126/science.1209235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sörgjerd K, Ghafouri B, Jonsson BH, Kelly JW, Blond SY, Hammarström P. Retention of misfolded mutant transthyretin by the chaperone BiP/GRP78 mitigates amyloidogenesis. J Mol Biol. 2006;356:469–482. doi: 10.1016/j.jmb.2005.11.051. [DOI] [PubMed] [Google Scholar]
- Suhr OB, Herlenius G, Friman S, Ericzon BG. Liver transplantation for hereditary transthyretin amyloidosis. Liver Transpl. 2000;6:263–276. doi: 10.1053/lv.2000.6145. [DOI] [PubMed] [Google Scholar]
- Susuki S, Sato T, Miyata M, Momohara M, Suico MA, Shuto T, Ando Y, Kai H. The endoplasmic reticulum-associated degradation of transthyretin variants is negatively regulated by BiP in mammalian cells. J Biol Chem. 2009;284:8312–8321. doi: 10.1074/jbc.M809354200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan SY, Pepys MB, Hawkins PN. Treatment of amyloidosis. Am J Kidney Dis. 1995;26:267–285. doi: 10.1016/0272-6386(95)90647-9. [DOI] [PubMed] [Google Scholar]
- Ushioda R, Hoseki J, Araki K, Jansen G, Thomas DY, Nagata K. ERdj5 is required as a disulfide reductase for degradation of misfolded proteins in the ER. Science. 2008;321:569–572. doi: 10.1126/science.1159293. [DOI] [PubMed] [Google Scholar]
- Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- Wang XD, Venable J, LaPointe P, Hutt DM, Koulov AV, Coppinger J, Gurkan C, Kellner W, Matteson J, Plutner H, et al. Hsp90 cochaperone Aha1 downregulation rescues misfolding of CFTR in cystic fibrosis. Cell. 2006;127:803–815. doi: 10.1016/j.cell.2006.09.043. [DOI] [PubMed] [Google Scholar]
- Washburn MP, Wolters D, Yates JRI., 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- Wiseman RL, Powers ET, Buxbaum JN, Kelly JW, Balch WE. An adaptable standard for protein export from the endoplasmic reticulum. Cell. 2007;131:809–821. doi: 10.1016/j.cell.2007.10.025. [DOI] [PubMed] [Google Scholar]
- Wu J, Rutkowski DT, Dubois M, Swathirajan J, Saunders T, Wang J, Song B, Yau GDY, Kaufman RJ. ATF6alpha optimizes long-term endoplasmic reticulum function to protect cells from chronic stress. Dev Cell. 2007;13:351–364. doi: 10.1016/j.devcel.2007.07.005. [DOI] [PubMed] [Google Scholar]
- Wu C, Delano DL, Mitro N, Su SV, Janes J, McClurg P, Batalov S, Welch GL, Zhang J, Orth AP, et al. Gene set enrichment in eQTL data identifies novel annotations and pathway regulators. PLoS Genet. 2008;4:e1000070. doi: 10.1371/journal.pgen.1000070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K, Yoshida H, Kokame K, Kaufman RJ, Mori K. Differential contributions of ATF6 and XBP1 to the activation of endoplasmic reticulum stress-responsive cis-acting elements ERSE, UPRE and ERSE-II. J Biochem. 2004;136:343–350. doi: 10.1093/jb/mvh122. [DOI] [PubMed] [Google Scholar]
- Yamamoto K, Sato T, Matsui T, Sato M, Okada T, Yoshida H, Harada A, Mori K. Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6alpha and XBP1. Dev Cell. 2007;13:365–376. doi: 10.1016/j.devcel.2007.07.018. [DOI] [PubMed] [Google Scholar]
- Zhou YY, Young JA, Santrosyan A, Chen KS, Yan SF, Winzeler EA. In silico gene function prediction using ontology-based pattern identification. Bioinformatics. 2005;21:1237–1245. doi: 10.1093/bioinformatics/bti111. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.