Abstract
In the present study, early stage apoptosis is explored with high temporal resolution. In addition to monitoring early apoptosis induction in single cells by ultrasensitive confocal fluorescence microscopy (UCFM), the mitochondrial proteins release kinetics was explored. The current study shows development and optimization of a novel, rapid apoptosis assay to explore the earliest changes in cells by the intrinsic apoptosis pathway. We show that early apoptotic changes in the mitochondria begin nearly simultaneously with the addition of an apoptosis-inducing drug, such as staurosporine. With a temporal resolution of five minutes, this non-invasive analytical technique can elucidate the earliest apoptotic events in living cells. Moreover, our results show that the mitochondrial inter-membrane proteins are not involved in the extrinsic pathway of Ramos cells mediated by an anti-CD95 antibody. Additional techniques such as light microscopy and flow cytometry were employed to confirm the results obtained by ultrasensitive confocal fluorescence microscopy. The results of this study help to understand the earliest mechanisms of apoptosis induction in cells, enabling new methods of drug testing and dose-response analyses.
Introduction
The detection of apoptotic events plays an important role in diagnosing and treating several diseases such as cancer, heart diseases, muscular dystrophy, aging, etc.1–4 Most chemotherapeutic agents mediate their effects by induction of apoptosis in cancer cells.5 The early detection and characterization of individual cells is an important clinical finding to initiate early treatments for cancer. A highly sensitive and high throughput single cell detection technique would be capable of diagnosing the earliest stages of diseases in a cell population and help track patient response to treatments. Therefore, there is a clear need to develop technologies supporting single cell analysis with high temporal resolution. Since mitochondria-driven apoptosis has been the main target for many drugs,6 this mechanism has been explored by various analytical techniques.
Early stages of apoptosis can be detected by monitoring the mitochondrial dysfunction caused by the depolarization of the mitochondrial membrane.7 Mitochondrial Outer Membrane Permeabilization (MOMP) is the major cause of the release of mitochondrial proteins that are situated in the inter-membrane space.8–9 The spatial and temporal dynamics of the mitochondrial membrane permeability have been reported to fluctuate due to the differential release dynamics of these proteins7. Therefore, there has been a continuing need for apoptosis assays that can show a steady response of the mitochondria-driven process at early stages.
Depolarization of the mitochondrial membrane post apoptosis induction have been measured using several fluorophores in the past, such as 3,3′-dihexyloxacarbocyanine iodide (DiOC6),10 JC-1,11 MitoTracker Red (CMX-Ros),12 and many more.13 Protein release from mitochondria has been reported to happen through opening of the mitochondrial permeability transition pore (MPTP) located in between the inner and outer membranes.14 JC-1 assays are commonly employed for measuring mitochondrial membrane potentials.11 However, J-aggregate formation in the JC-1 assay need a high ΔΨ and are not suitable for fixed samples.19 Our technique measures changes in the mitochondrial membrane at times before it is reasonably depolarized for J-aggregate formation.
Most of the standard apoptotic assays20–25 lack sufficient temporal resolution to study apoptosis in real time in living cells. In order to elucidate earliest stages of apoptosis completely, rapid assays with high temporal resolution are required to minimize the time between induction and detection26–27 Release of cytochrome c in single cells can be completed within few minutes,28 thus it is necessary to be able to obtain measurements with high temporal resolution. Our method measures rapid changes in the mitochondria by potent apoptosis inducing drugs with a temporal resolution of five minutes.
Although the change in membrane potentials can be measured by flow cytometry,15 and plate readers,31,32 these methods is incapable of collecting single cell response repeatedly within a short time frame. Moreover, a wash step is required to remove the excess dye that can contribute to overlapping of small signals that can otherwise be easily detected in the absence of high background.33 Ultrasensitive Confocal Fluorescence Microscopy (UCFM) is an ideal analytical technique to study internal dynamics of living cells at sub-nanomolar concentrations with sub-microsecond temporal resolution34. Table 1 illustrates a comparison of some most commonly employed instruments and their specifications. In addition to detection of concealed events or events overlapped by bulk signal, UCFM also has the capability to be integrated to microfluidic devices.35
Table 1.
Comparison of instrumental capabilities commonly used for cellular apoptosis studies.
| Instrument Specification | Flow Cytometry | Plate-Based Assays | UCFM |
|---|---|---|---|
| Temporal Resolution | Moderate | High | High |
| Wash Step Integration | No | No | Yes |
| Repetitive Single Cell Analysis | No | No | Yes |
| Integration with Microfluidics | Yes | No | Yes |
| Subpopulation Analysis | Yes | Yes | Yes |
| Extended Fluorescence Parameter Measurement* | No | No | Yes |
| Cost Effectiveness Per Assay | Low | Low | Low |
Extended fluorescence parameters include molecular brightness, molecular diffusion, triplet state fractions, free vs. bound states, etc.
In the present study, the accumulation of MitoTracker Deep Red apoptosis probe in the mitochondria of Ramos cells was monitored after apoptosis induction. The temporal resolution and release kinetics of the red probe inside the cellular mitochondria have been explored and the dye release was observed to occur within 5 minutes after staurosporine induction. Induction kinetics varied with the dose of staurosporine. The high temporal resolution, simplicity, capability of integration with microfluidic devices and rapid analysis make our technique amenable to facilitate earliest detection of apoptotic events in living cells.
Experimental
Materials and Reagents
The mitochondrion-selective probe MitoTracker Deep Red FM (MTDR) and sterile Phosphate-Buffered Saline (PBS, pH = 7.4) were purchased from Invitrogen. Bovine Serum Albumin (BSA) was purchased from Sigma- Aldrich. The apoptosis inducing agents, staurosporine and anti-human CD95 (APO-1/Fas) were purchased from Calbiochem and eBioscience, respectively. Annexin V-FITC apoptosis detection kit that includes Annexin V-FITC conjugate probe and Propidium Iodide (PI) was purchased from BioVision and the Annexin-binding buffer (1X) was purchased from SouthernBiotech.
Cells and Cell Culture
The human cell line Ramos (B lymphocyte) was obtained from American Type Culture Collection (ATCC, Rockville, MD). The cells were sub-cultured at 37° C and 5% CO2 in RPMI 1640 growth medium (Hyclone) with 10% Fetal Bovine Serum (Hyclone) and 20 mL/L antibiotic (penicillin-streptomycin by Sigma-Aldrich) solution twice a week.
Induction of Apoptosis by Staurosporine
Prior to induction of apoptosis using staurosporine, the cells were centrifuged (4500 rpm for 3 minutes) and re-suspended in two petri dishes (control + sample) in growth medium at a density of ~3 × 106 cells/mL measured by a haemocytometer. MitoTracker deep red probe was dissolved in dimethylsulfoxide (DMSO) to prepare a 1 mM stock solution and was stored at −4° C. The cells in both petri dishes were stained with 0.1 μM MitoTracker deep red probe by incubating the cells in the dark at 37° C for 40 minutes. Apoptosis in the sample petri dish was induced with 4 μM staurosporine. 200 μL of the cell suspension was drawn from each of the petri dish (sample and control) at each time interval, centrifuged, re-suspended in PBS, and was assayed immediately by microscopy and flow cytometry from 0th minute to 4 hours. Both control and sample measurements were recorded at 5 or 15-minute intervals, depending on the experiment.
Induction of Apoptosis by Anti CD95
The same procedure as that of staurosporine induction described above was carried out for apoptosis induction by the extrinsic pathway using 0.8 and 1.6 μg/mL of anti-CD95 antibody instead of staurosporine. Control and sample measurements were obtained every hour for a total of 4-hours.
The cell culturing procedures, cell staining protocols and safety considerations followed during the experiments confirms to existing lab protocols.36
Microscopy Imaging
Fluorescence imaging was carried out using an inverted microscope (IX-71, Olympus). A metal halide lamp was used for fluorescence excitation using filters appropriate for MitoTracker Deep Red excitation and emission. The images were acquired using a 0.3 NA 10X objective with a 12-bit CCD camera (Orca-285, Hamamatsu). The images were then processed using ImageJ (v. 1.41, National Institutes of Heath) for intensity measurements.
Ultrasensitive Confocal Fluorescence Microscopy (UCFM) Measurements
A custom-built single molecule detection system was employed to acquire ultrasensitive fluorescence intensity measurements. The instrumental setup has been described in details by Dong et al.37 A red diode laser (635 nm, Edmund Optics) was incident on the back port of a microscope base (IX51, Olympus) through a mirror periscope.38 A handheld power meter (Laser Check, Edmund Optics) was used to measure the laser power before the cells were exposed to the laser beam. The sample consisted of a 30 μL droplet of Ramos cells (105–106 cells / mL) in PBS placed on a 150 μm glass coverslip. An oil-immersion objective (100X, 1.3 NA, Olympus) was used to focus the laser beam inside individual cells. One cell was aligned within the confocal volume at a given time, and in this manner five cells were serially illuminated. One spot in each cell was measured for 10 seconds and the time lag between subsequent cell measurements was around 5 seconds. In our measurements, the placement of the laser beam focal point in the cell did not statistically influence fluorescence signals. Fluorescence from MitoTracker Deep Red (Ex: 644 nm, Em: 665 nm) was collected by the objective, passed through the dichroic mirror and then via a 676 nm (Omega Optics) interference filter. Finally, the fluorescence intensity was detected by a single-photon avalanche photodiode (SPCM-AQR 14 Single Photon Counting Module, Perkin-Elmer) via an aspheric lens (Newport).39 The integration time of the avalanche photodiode was 1 ms.
Immediately after the addition of the sample/control drop on the coverslip, the cells were exposed to 50–90 μW of laser beam and fluorescent counts were acquired with a continuous exposure of cells to the laser beam for 10 s. No significant photo bleaching was observed during data collection. Data was monitored and acquired simultaneously using a Labview program (version 8, National Instruments). A photon counting histogram was obtained for every measurement and then imported to Origin for analysis. The median fluorescence intensity values obtained for five cells were averaged and plotted as a function of time.
Flow Cytometry
Suspensions of stained cells, with and without apoptosis inducers, were analyzed using FACSCalibur flow cytometer (Becton–Dickinson). The apoptosis induction by both mitochondrial pathway and extrinsic pathway was verified. Two molecular probes were employed for verification of apoptotic cell population induced by anti-CD95. Staining with MitoTracker deep red probe: Cell samples (500 μL from each petri dish) treated with 1.6 μg/mL of anti-CD95 and control cells were centrifuged, re-suspended in PBS and analyzed by forward scatter (FSC), side scatter (SSC) and MitoTracker deep red fluorescence (FL 4, ~661/16 nm long pass filter) using CellQuest software every hour for four hours duration. Staining with Annexin V-FITC/PI: Cell samples treated with 1.6 μg/mL as mentioned above were centrifuged and re-suspended in 200 μL of 1X Annexin binding buffer. The cells in this buffer were treated with 2 μL Annexin V-FITC probe and 2 μL PI (50 μg/mL). The sample was incubated at room temperature in dark for 5 minutes before re-suspending the cells in PBS for analysis by flow cytometer. Cells were characterized by forward and side scatter (scatter plot) as well as the fluorescence of Annexin V-FITC and PI. The scatter plots were subjected to quadrant analysis to discriminate between apoptotic cells, live cells and dead cells.
Results and Discussion
Staurosporine Induction of Ramos Cells
In order to observe the change in fluorescence intensity of apoptotic cells, white light and red fluorescence images were acquired every 15 minutes over 2 hours for controls and samples using light microscopy. The decrease in intensity for the sample cells as compared to the control cells (Supp Info Fig. S1) indicates that apoptosis was induced in the cells with acceptable spatial resolution. However, the temporal resolution of 15 minutes and the sensitivity of detection on an inverted microscope with a halide lamp were insufficient to observe fast changes in cell fluorescence (Supp. Info. Fig. S2). Mitra et. al.40 have demonstrated use of fluorescent dyes for obtaining high resolution time lapse images of mitochondria but it lacks the high sensitivity offered by a confocal system. On the other hand, for high spatial resolution, cells require labeling with comparatively higher concentration of probe (<100 nM). At these concentrations, there is a risk of over-staining that may increase the probability of labeling other cell organelles. UCFM is a very sensitive technique due to extremely low concentration of fluorescent molecules needed for measurement using a single photon detecting system. Therefore, UCFM was employed for rapid intensity measurements in the mitochondria with sensitivity good enough to show considerable change in magnitude over a short time period.
Temporal Resolution of MTDR by UCFM Measurements
The intensity decline observed by microscopy imaging was confirmed by flow cytometry. Scatter plots and histograms were analyzed to monitor the change in MTDR intensity over time. MTDR fluorescence of non-apoptotic (control) cells did not change over time and showed higher intensity as compared to the induced cell population (Supp. Info. Fig S3). This observation agrees with the MTDR fluorescence data obtained by F. Martinez-Pastor et al.41 on apoptotic spermatozoa. The steady decrease in fluorescence intensity of apoptotic cells over an hour gave rise to the need to monitor the intensity change at the shortest time interval possible. To achieve this goal, we employed UCFM methods to detect small changes in MTDR intensity.
To indirectly monitor the rapid release of proteins from the mitochondria post induction, fluorescence measurements were acquired by focusing the red laser beam inside individual cells at five-minute intervals. Data from five random cells were collected as photon counting histograms and median intensity (determined by Gaussian fit) were averaged. Five cells were chosen for each measurement because it was the fastest way possible to collect data from five static cells during each measurement post cell workup, which includes washing and centrifuging the aliquots before each measurement. For this analysis, the wash step was found to be critical to increasing sensitivity.
Another barrier in fluorescent measurements is autofluorescence from green fluorescent molecules present in cells such as NADH and FADH2 that lowers the S/N ratio; in turn affecting spatial resolution.29 Most of the commercial dyes that are excited at 488 nm overlap with the autofluorescence spectral region (>500 nm) that interferes with the dye fluorescence considerably. The use of deep red probe eliminates the possibility of cellular autofluorescence interference and increases the signal to noise ratio.
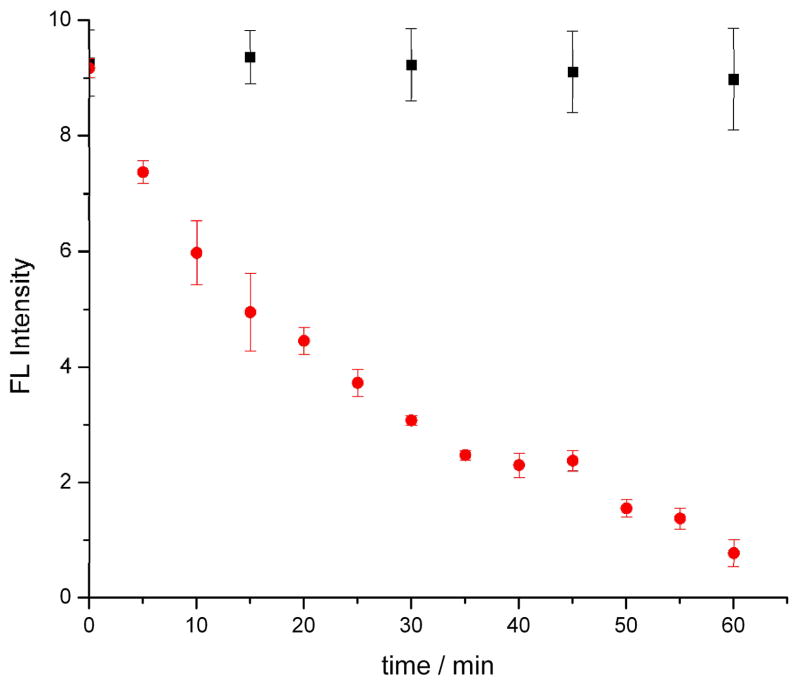
Figure 1 demonstrates the steady, exponential decrease in photon-counts with time with a 1/e time (τ) of ~28 minutes. Control cells show stable fluorescence intensity over the measurement period (Supp. Info. Fig S4). The intensity difference between the apoptotic and control cells are statistically significant at each time interval, based on t-tests at 95% confidence interval. Based on these results, we propose that protein release from the mitochondria is a near-instantaneous process that initiates within few minutes post induction by the intrinsic pathway.
Fig. 1.
Mean MitoTracker deep red fluorescence intensity of five Ramos cells at each time interval by ultrasensitive confocal fluorescence microscopy (error bars represent the standard deviation of the mean). Control cells are represented as black squares and drug induced samples as red circles. All cells were stained with 0.1 μM MitoTracker deep red and samples were induced with 4 μM staurosporine. The decay in cell fluorescence was measured with a 1/e time = 28 minutes. The five-minute temporal resolution enables early apoptosis events to be analyzed.
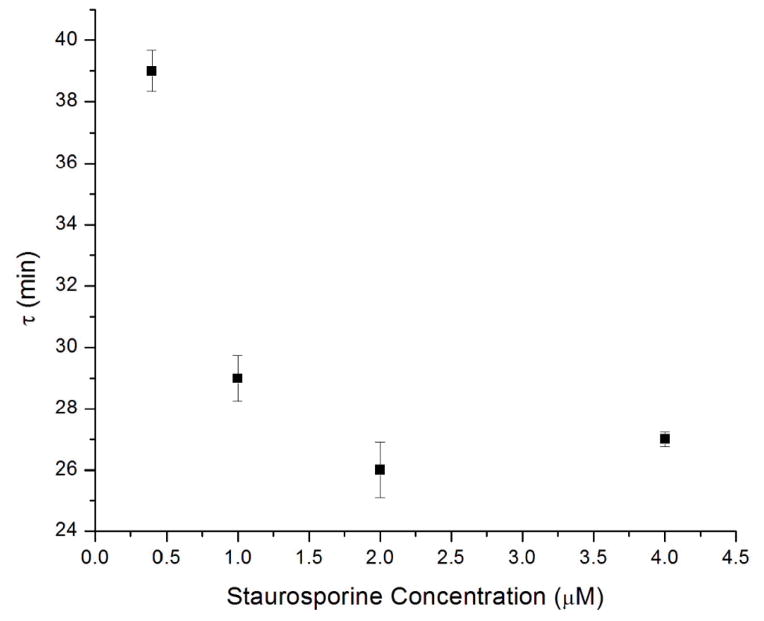
We also carried out a staurosporine dose-dependent study to verify that the rapid decrease in MTDR fluorescence was due to apoptosis induction (Figure 2). Cells induced with 0.4 μM staurosporine (10x staurosporine dilution) showed a slower rate of fluorescence intensity decay with a 1/e time of ~38 minutes as compared to the 4 μM dose (Fig. 2). However, a 100x dilution of the drug doses (0.04 μM) did not show significant changes between control and sample cells over the experiment duration (Fig 2b). Our methods are therefore capable of determining differences in apoptosis kinetics based on the target compound dose. The 1/e (t) decay time for fluorescence intensity (Figure 3) follows a dose-response curve, with decreasing τ values with increasing staurosporine concentration. Staurosporine is a potent apoptosis inducer, but this approach can be used with less potent compounds as well, elucidating the earliest stages of programmed cell death. Cell response can therefore be identified earlier than extant assays, and the same cell can be tracked over time to observe later cell response as well. This methodology can therefore be used to probe the temporal dynamics of an anti-cancer drug, and can compliment methods such as flow cytometry that are used to assay viability and toxicity at later time points after drug administration.
Figure 2.
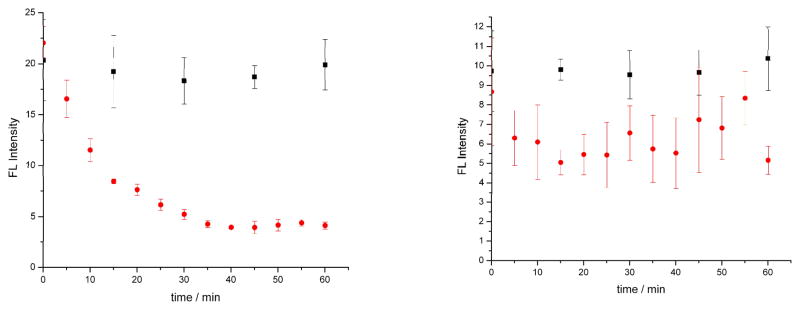
Mean MitoTracker deep red (MTDR) fluorescence intensity by as a function of time for five Ramos cells at each time interval by ultrasensitive confocal microscopy. Control cells are represented as black squares and drug induced samples as red circles. All cells were stained with 0.1 μM MTDR and samples were induced with (a) 0.4 μM staurosporine or (b) 0.04 μM staurosporine. The 1/e time for the 0.4 μM sample was 38 minutes, and was not measured for the 0.04 μM sample.
Figure 3.
The change in decay time (t) with staurosporine concentration. The decay of fluorescence (τ) measured by UCFM occurs more rapidly with increasing staurosporine concentration.
Receptor-Mediated Apoptosis Measurements
To ensure that the intrinsic apoptosis pathway was being triggered with staurosporine, we used our methods with a receptor-mediated apoptosis inducer. There is a limit to the number of Fas receptors present on a cell membrane; hence the time of induction through the extrinsic mechanism can be longer as compared to the mitochondrial pathway. Since receptor-mediated apoptosis does not directly involve mitochondria, we did not anticipate a rapid change in MTDR fluorescence after the induction event. However, when the receptor-mediated pathway also triggers mitochondrial apoptosis, there is a lag between apoptosis induction and a decrease in MTDR fluorescence. Almost no significant intensity decline was observed between the apoptotic and control cells (Supp. Info. Fig S5). To confirm the results obtained by UCFM, cells were stained with MTDR and induced with anti-CD95 for monitoring using flow cytometry. Figure S6 (Supp. Info.) shows no decline in the MTDR fluorescence for apoptotic and non-apoptotic Ramos cells. These results prove that the mitochondrial inter-membrane proteins are not involved in the extrinsic apoptotic pathway of Ramos cells using anti-CD95 antibodies.
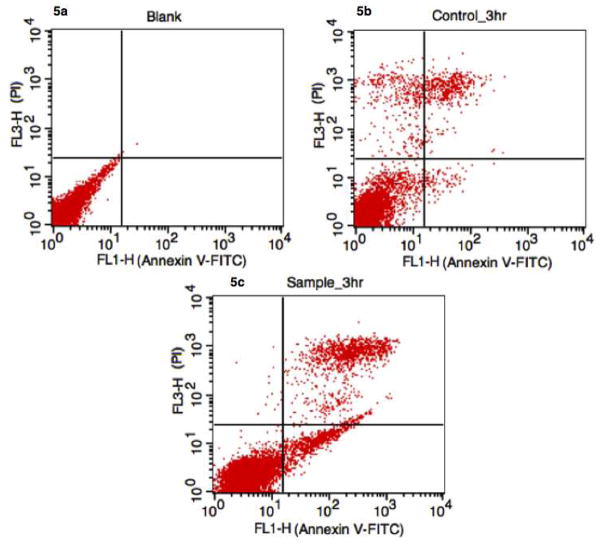
To ensure that apoptosis is still being induced by anti-CD95, an Annexin V-FITC / PI assay was carried out using flow cytometry to study downstream apoptosis markers. Induction of apoptosis was observed from the third hour, which showed ~8% apoptotic cells (staining Annexin+/PI−), ~78% viable cells (Annexin−/PI−) and ~14% necrotic cells (Annexin+/PI+) (Fig. 4). Similarly, the control measured at 3h had ~90% viable cells, ~1% apoptotic cells and ~9% necrotic cells (Fig. 4b). The observations confirm induction of apoptosisin Ramos cells by anti-CD95 via the extrinsic pathway, and that the intrinsic pathway is not triggered during receptor mediated apoptosis in this cell line.
Figure 4.
Flow cytometry analysis of anti-CD95 induced apoptosis using Annexin V-FITC/PI. Apoptotic cells are located in the lower right quantrant. (a) Control (probe without anti-CD95) and (b) blank (no probe or antibody) showed normal levels of nectrotic and apoptotic cells. However, when receptor mediated apoptosis was initiated, apoptotic cells were observed at 3 hours (c). Given the rapid temporal resolution of our mitochondrial-based assay, it is possible to exclude mitochondrial protein release as a co-mechanism during receptor-mediated apoptosis in Ramos cells.
The dynamics of early apoptotic changes by the intrinsic pathway start from intact mitochondrial membrane potential. Depolarization of the membrane potential gives rise to formation of Mitochondrial Permeability Transition Pore (MPTP), followed by release of mitochondrial inter-membrane proteins, and lastly activating caspases leading to cell death. The results of this study agrees with the results found by F. Martinez-Pastor et. al.40 for early apoptotic changes observed in sperm cells using MTDR fluorescence, assuming the apoptosis in sperm cells partly resemble most of the other cell types. However, we propose that these early apoptotic changes in the mitochondria begin rapidly with the addition of inducing drugs like staurosporine. Commonly used Membrane-potential-dependent dyes such, as Rhodamine 123 and TMRM/TMRE are useful as long as the mitochondrion maintains its negative membrane potential. Tait et. al.23 have demonstrated successful use of TMRE to measure changes in ΔΨ in living cells due to its low toxicity. However, TMRE also has a tendency to redistribute itself across the plasma membrane.24 Once the process of cell death becomes irreversible after apoptosis induction, the plasma membrane lysis begins within 30 minutes25 and therefore fluorescent measurements using plasma membrane potential sensitive dyes like Rhodamine 123, TMRM/TMRE can give spurious results after 30 mins. Therefore, it is advantageous to use MTDR in multiple labeling experiments for steady monitoring of intensities for longer time periods. The high temporal resolution and simplicity of our methods allows new insights into the mechanisms of early stage apoptosis.
Conclusion
In this study, we employed UCFM for measurements with high temporal resolution, which facilitated rapid time based analyses of the earliest stage of apoptosis. We validated our UCFM assay and instrumental response by comparing the results with other commonly used techniques such as flow cytometry and fluorescence microscopy. The five-minute temporal resolution enabled us to observe near-instantaneous release of mitochondrial contents during apoptosis induction. The kinetics of mitochondrial release followed a dose-dependent response, even though the process started rapidly. In the case of receptor-mediated apoptosis, our method can observe if mitochondrial release occurs, although a lag between induction and detection is expected in these cases. When Ramos cells were used, the receptor mediated pathway did not trigger mitochondrial-driven apoptosis.
We have developed a unique high-resolution assay to study the earliest stages of apoptosis in the intrinsic pathway using a novel analytical tool. The diffusion of mitochondrial proteins in apoptotic cells occurs rapidly after induction and therefore a high temporal resolution technique is important to monitor these early biological changes in cells non-invasively. Our future efforts will be focused on increasing cell throughput by automation of cell analysis.
Supplementary Material
Acknowledgments
R.D. Ray acknowledges support from the Undergraduate Research Fellowship from Texas Tech University. This work was supported by grants from the Robert A. Welch Foundation (Grant D-1667) and the National Institutes of Health (Grants RR025782 and GM103550).
References
- 1.Yoshinori O, Zhonglian L, Masa-Ai S. J Hisochem Cytochem. 2003;38:275–340. [Google Scholar]
- 2.Gill C, Mestril R, Samali A. FASEB J. 2002;16:135–146. doi: 10.1096/fj.01-0629com. [DOI] [PubMed] [Google Scholar]
- 3.Dupont-Versteegden EE. J Appl Physiol. 1994;77:1736–1741. doi: 10.1152/jappl.1994.77.4.1736. [DOI] [PubMed] [Google Scholar]
- 4.Fadeel B, Orrenius S. J Intern Med. 2005;258:479–517. doi: 10.1111/j.1365-2796.2005.01570.x. [DOI] [PubMed] [Google Scholar]
- 5.Debatin KM, Poncet D, Kroemer G. Oncogene. 2002;21:8786–803. doi: 10.1038/sj.onc.1206039. [DOI] [PubMed] [Google Scholar]
- 6.Martinez MM, Reif RD, Pappas D. Anal Methods. 2010;2:996–1004. [Google Scholar]
- 7.Bhola PD, Mattheyses AL, Simon SM. Biophysical Journal. 2009;97:2222–2231. doi: 10.1016/j.bpj.2009.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wlodkowic D, Telford W, Skommer J, Darzynkiewicz Z. Methods Cell Biol. 2011;103:55–98. doi: 10.1016/B978-0-12-385493-3.00004-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Munoz-Pinedo C, Guio-Carrio A, Goldstein JC, Fitzgerald P, Newmeyer DD, Green DR. PNAS. 2006;103:11573–11578. doi: 10.1073/pnas.0603007103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Petit PX, O-Connor JE, Grunwald D, Brown SC. Eur J Biochem. 1990;194:389–397. doi: 10.1111/j.1432-1033.1990.tb15632.x. [DOI] [PubMed] [Google Scholar]
- 11.Ceccarelli CA, Massini A. Exp Cell Res. 1996;222:84–94. doi: 10.1006/excr.1996.0011. [DOI] [PubMed] [Google Scholar]
- 12.Poot M, Zhang YZ, Krämer JA, Wells KS, Jones LJ, Hanzel DK, Lugade AG, Singer VL, Haugland RP. J Histochem Cytochem. 1996;44:1363–1372. doi: 10.1177/44.12.8985128. [DOI] [PubMed] [Google Scholar]
- 13.Gilmore K, Wilson M. Cytometry. 1999;36:355–358. doi: 10.1002/(sici)1097-0320(19990801)36:4<355::aid-cyto11>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 14.Machado NG, Alves MG, Carvalho RA, Oliveira PJ. Cardiovasc Toxicol. 2009;9:211–227. doi: 10.1007/s12012-009-9055-1. [DOI] [PubMed] [Google Scholar]
- 15.Xue X, Song Y, Zhang Q, Wu Y, Guo-zhang Z, Wang PC, Zhao Y, Jia YX, Zhang X, Liang XJ. Mol Pharmaceutics. 2012;9:634–644. doi: 10.1021/mp200571k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bouchier-Hayes L, Munoz-Pinedo C, Connell C, Green DR. Methods. 2008;44:222–228. doi: 10.1016/j.ymeth.2007.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roy S, Hajnoczky G. Methods. 2008;46:213–223. doi: 10.1016/j.ymeth.2008.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perry SW, Norman JP, Barbieri J, Brown EB, Gelbard AH. Biotechniques. 2011;50:98–115. doi: 10.2144/000113610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cossarizza A, Salvioli S. Purdue Cytometry CD-ROM series. :4. Retrieved from www.cyto.purdue.edu/archive/flowcyt/research/cytotech/apopto/data/cossar1/cossariz.htm.
- 20.Goldstein JC, Waterhouse NJ, Juin P, Evan GI, Green DR. Nat Cell Biol. 2000;2:156–162. doi: 10.1038/35004029. [DOI] [PubMed] [Google Scholar]
- 21.Waterhouse NJ, Goldstein JC, Ahsen O, Schuler M, Newmeyer DD, Green DR. J Cell Biol. 2001;153:319–328. doi: 10.1083/jcb.153.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gottlieb E, Armour SM, Harris MH, Thompson CB. Cell Death Differ. 2003;10:709. doi: 10.1038/sj.cdd.4401231. [DOI] [PubMed] [Google Scholar]
- 23.Tait SWG, Bouchier-Hayes L, Oberst A, Connel C, Green DR. Apoptosis. 2009;559:33–48. doi: 10.1007/978-1-60327-017-5_3. [DOI] [PubMed] [Google Scholar]
- 24.Wong A, Cortopassi GA. Biochem Biophy Res Commun. 2002;298:750–754. doi: 10.1016/s0006-291x(02)02546-9. [DOI] [PubMed] [Google Scholar]
- 25.Nicholls DG, Ward NW. Mitochondria and excitotoxicity TINS. 2000;23:166–174. doi: 10.1016/s0166-2236(99)01534-9. [DOI] [PubMed] [Google Scholar]
- 26.Martinez MM, Reif RD, Pappas D. Anal Bioanal Chem. 2010;396:1177–1185. doi: 10.1007/s00216-009-3298-3. [DOI] [PubMed] [Google Scholar]
- 27.Li P, Nijhawan D, Budihardjo I, Srinivasula SM, Ahmad M, Alnemri ES, Wang X. Cell. 1997;91:479–489. doi: 10.1016/s0092-8674(00)80434-1. [DOI] [PubMed] [Google Scholar]
- 28.Goldstein JC, Waterhouse NJ, Juin P, Evan GI, Green DR. Cell Biol. 2000;2:156–162. doi: 10.1038/35004029. [DOI] [PubMed] [Google Scholar]
- 29.Chazotte B. Cold Spring Harb Protoc. 2011:990–992. doi: 10.1101/pdb.prot5648. [DOI] [PubMed] [Google Scholar]
- 30.Reif RD, Martinez MM, Pappas D. Anal Bioanal Chem. 2010;397:3387–3396. doi: 10.1007/s00216-010-3567-1. [DOI] [PubMed] [Google Scholar]
- 31.FLIPR application note, Molecular Devices. Retrieved from http://www.moleculardevices.com/Documents/general-documents/mkt-appnotes/flipr-appnotes/FLIPR_App_Note_mito_membrane_rev_B.pdf.
- 32.FLIPR High Throughput Cellular Screening, Molecular Devices. Retrieved from http://www.moleculardevices.com/Products/Instruments/FLIPR-Systems.html.
- 33.Florian AE, Lepensky CK, Kwon O, Haynes MK, Sklar LA, Zweifach A. J Biomol Screen. 2012 doi: 10.1177/1087057112466697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim SA, Heinze KG, Schwille P. Nature Methods. 2007;4:963–973. doi: 10.1038/nmeth1104. [DOI] [PubMed] [Google Scholar]
- 35.Pappas D, Burrows SM, Reif RD. Trends in Analytical Chemistry. 2007;26:884–894. [Google Scholar]
- 36.Pappas D. Practical Cell Analysis. 1. Wiley; 2010. [Google Scholar]
- 37.Dong M, Martinez MM, Meyer MF, Pappas D. Analyst. 2012;137:2997–3003. doi: 10.1039/c2an16173g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tian Y, Pappas D. Appl Spectrosc. 2010;64:324–327. doi: 10.1366/000370210790918391. [DOI] [PubMed] [Google Scholar]
- 39.Reif RD, Martinez MM, Wang K, Pappas D. Anal Bioanal Chem. 2009;395:787–795. doi: 10.1007/s00216-009-3024-1. [DOI] [PubMed] [Google Scholar]
- 40.Mitra K, Lippincott-Schwartz J. Curr Protoc Cell Biol. 2010;Chapter 4:1–21. doi: 10.1002/0471143030.cb0425s46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martínez-Pastor F, Fernández-Santos MR, Olmo E, Domínguez-Rebolledo AE, Esteso MC, Montoro V, Garde JJ. Reprod Fertil Dev. 2008;20:547–556. doi: 10.1071/rd08002. [DOI] [PubMed] [Google Scholar]
- 42.Wachsmuth M, Waldeck W, Langowski J. J Mol Biol. 2000;298:677–689. doi: 10.1006/jmbi.2000.3692. [DOI] [PubMed] [Google Scholar]
- 43.Ruan L, Xu Z, Lan T, Wang J, Liu H, Li C, Dong C, Ren J. Anal Chem. 2012;84:7350–7358. doi: 10.1021/ac301654g. [DOI] [PubMed] [Google Scholar]
- 44.Perevoshchikova IV, Zorov DB, Antonenko YN. Biochimica et Biophysica Acta-Biomembranes. 2008;1778(10):2182–2190. doi: 10.1016/j.bbamem.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 45.Tanaka S. Soft Matter. 2012;8:8936–8943. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.