Abstract
This work describes efficient manipulation of bacteriophage virus particles using a nanostructured dielectrophoresis (DEP) device. The non-uniform electric field for DEP is created by utilizing a nanoelectrode array (NEA) made of vertically aligned carbon nanofibers (VACNFs) versus a macroscopic indium tin oxide electrode in a “points-and-lid” configuration integrated in a microfluidic channel. The capture of the virus particles has been systematically investigated versus the flow velocity, sinusoidal AC frequency, peak-to-peak voltage, and virus concentration. The DEP capture at all conditions is reversible and the captured virus particles are released immediately when the voltage is turned off. At the low virus concentration (8.9×104 pfu·ml−1), the DEP capture efficiency up to 60% can be obtained. The virus particles are individually captured at isolated nanoelectrode tips and accumulate linearly with time. Due to the comparable size, it is more effective to capture virus particles than larger bacterial cells with such NEA based DEP devices. This technique can be potentially utilized as a fast sample preparation module in a microfluidic chip to capture, separate, and concentrate viruses and other biological particles in small volumes of dilute solutions in a portable detection system for field applications.
Keywords: Bacteriophage, Virus detection, Dielectrophoresis, Nanoelectrode Array, Vertically Aligned Carbon Nanofibers
1 Introduction
The development of microdevices for the on-chip capture, sorting, and concentration of microbes is critical for developing rapid pathogen detection technologies based on microfluidics [1, 2] and other miniaturized biosensor chips [3, 4]. Dielectrophoresis (DEP) is one of the most effective techniques for interacting with any polarizable particle using easily generated electric fields [5, 6]. Extensive studies employing various DEP devices have demonstrated the ability to precisely manipulate single cells with sizes varying from over 10 microns (mammalian cells) down to about 1 micron (bacteria) [7-9]. However, applying DEP to capture virus particles remains difficult due to their much smaller size (tens to hundreds of nanometers). Studies have demonstrated that it is possible to trap and accumulate an ensemble of virus particles from a static solution using microDEP [10-12], but capture and manipulation of individual virus particles, particularly those from a high-flow-rate solution (as needed for the rapid concentration of dilute virus samples) have not been developed. A few studies have demonstrated that single virus particles in a flow can be trapped between the gap of two nanoelectrodes [13] or using optical tweezers coupled with a microDEP concentrator [14]. However, these techniques are expensive and can only be used in low-throughput studies. Here we demonstrate a simple DEP device based on a nanoelectrode array (NEA) made of vertically aligned carbon nanofibers (VACNFs), which can capture single virus particles or large ensembles of particles at the carbon nanofiber tip from a high-velocity fluidic flow. Bacteriophages T4r and T1 were used as the testing agents to demonstrate the capability of this nanostructured DEP device.
The principles of DEP manipulation and separation of microparticles were previously described by Pohl [6]. When a polarizable particle (charged or neutral) is placed in a non-uniform electric field, a net electrical force will be generated on the particle, causing it to move along the field. In most studies, an alternating current (AC) voltage is applied to a pair of electrodes to generate the electric field. The time averaged DEP force (FDEP) exerted on a spherical particle is given by [6]:
| (1) |
where r is the radius of the particle, εm is the absolute permittivity of the suspending medium, ∇E2 is the gradient of the square of the applied electric field strength, and Re[K(ω)] is the real component of the complex Clausius-Mossotti (CM) factor given by [6]:
| (2) |
with ε* representing the complex permittivity and the indices p and m referring to the particle and medium respectively. σ is the conductivity, ω is the angular frequency (ω = 2πf) of the applied electric field, and j = √-1. In this study, the proper medium is chosen to give Re[K(ω)] > 0 so that the particles experience a DEP force which moves the particles toward higher electric field strength, a phenomenon called positive DEP (pDEP) [6].
By Equation 1, the DEP force is proportional to the volume or cube of the radius (r3) of the particle. On the other hand, the hydrodynamic force to carry the particles with flow (i.e. drag force FDrag) is directly proportional to the radius of the particle with
| (3) |
where η is the dynamic viscosity, k is a factor accounts for the wall effects, and ʋ is the linear flow rate (flow velocity) [8]. In addition, Brownian motion of a particle increases as the particle size is reduced. These reasons together make DEP capture of nanoparticles (such as viruses) more difficult than that of microparticles (such as mammalian cells and bacteria), particularly in a high-velocity fluidic flow [11]. One approach to compensate for the smaller size is to utilize nanostructured DEP devices that enhance the magnitude of ∇E2 when properly designed. Here we demonstrate the use of a NEA as ‘point’ electrodes vs. a macroscopic indium tin oxide (ITO) electrode used as the ‘lid’ electrode in a “points-and-lid” configuration [7] to create a highly non-uniform electric field for DEP capture of virus particles. From our previous work, ∇E2 within a distance of 3 μm above the embedded VACNF nanoelectrode (NE) tip can be as high as ~1.2×1018 V2m−3 [15], about 200 times higher than the microscale “points-and-lid” devices. The size of the CNFs (~50-200 nm in dia.) is comparable to that of common virus particles.
2 Materials and Methods
2.1 DEP Device Fabrication
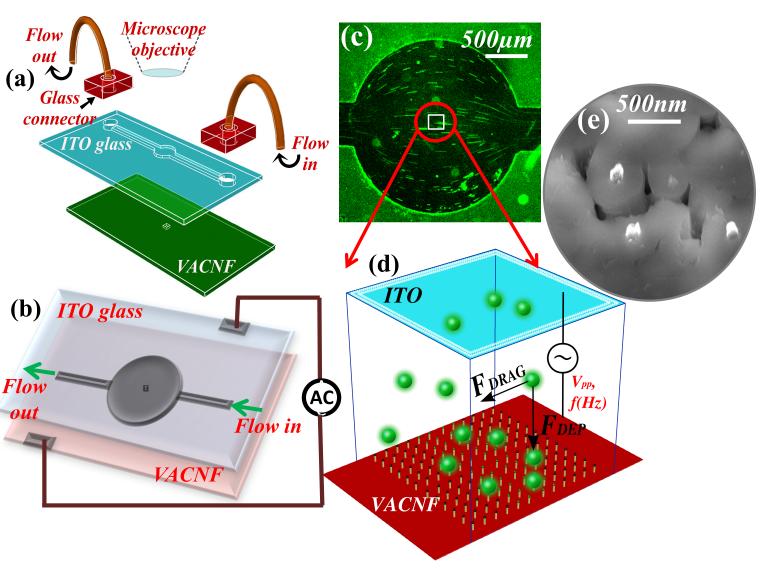
The device fabrication was similar to that described previously by Syed et. al [16]. Briefly, the device consisted of two chips coated with different thicknesses of SU-8 photoresist; namely a lid electrode made of ITO-coated glass (~1 mm in thickness) and a points array electrode made of an embedded VACNF NEA on a Si(100) chip (0.5 mm in thickness) (Fig. 1a). The embedded VACNF were fabricated as described previously [15-18]. Briefly, a 4” silicon (100) wafer (Si-Tech, Topsfield, MA) was diced into small chips of 1 cm × 2 cm. These chips were then coated with 100 nm of chromium as a conductive layer and a 22.5 nm thick nickel catalyst layer by ion sputtering (Gatan, PA). The nickel film was broken into randomly distributed nanoparticles during a thermal treatment at ~500° C in a plasma-enhanced chemical vapor deposition (PECVD) system (Aixtron, CA). The VACNFs were then grown at 775° C in a PECVD with a mixture of C2H2 and NH3 precursors under a DC bias. This process reliably produced VACNFs with an average diameter of ~100 nm and an average length of ~5 μm. Each of the CNFs was directly anchored on the Si chip and aligned vertically forming a brush-like structure with average spacing of ~300-400 nm. These brush-like randomly arranged VACNFs were then encapsulated with SiO2 by thermal chemical vapor deposition (CVD) using tetraethyorthosilicate (TEOS) as a precursor. The embedded randomly arranged VACNF array was then polished with 1 μm and 0.3 μm alumina slurries (Buehler, Lake Bluff, IL) to planarize the top surface and then subjected to reactive ion etching (RIE) (Nano-Master, NRE3000) with CHF3 etchant to etch the SiO2 until desired number of CNF tips were exposed (as shown in Fig. 5e). The average spacing between the exposed CNF tips was ~1-2 μm in this study. A fluidic channel (500 μm in width and 2 cm in length) and a circular chamber (~2 mm in diameter) were made by UV-lithography in the ~18 μm thick spin-coated SU-8 2010 photoresist (Microchem, Newton, MA) on the 1 cm × 2 cm diced ITO glass chip (Delta Technologies, MN). Two holes of ~ 0.75 mm diameter were drilled through the glass piece at the two ends of the microchannel for fluidic inlet and outlet. The planarized NEAs made of VACNFs embedded in SiO2 were fabricated by the method described before [17, 18]. The top surface of the NEA chip was spin-coated with a layer of ~2.0 μm thick SU-8 2002 photoresist (Microchem, Newton, MA) with square area of 200 μm × 200 μm at the position corresponding to the center of the circular microchamber in the glass chip. The two chips were then aligned under 4X objective lens of a stereo microscope, pressed against each other with a mechanical force, and placed in a preheated vacuum oven (Curtin Matheson Scientific, Inc.) at 175°C for ~20 min to form a permanent bond. Finally, thirty gauge wires were attached using conductive silver epoxy (MG Chemicals, Ontario) on the chips for electrical connections and microbore PTFE (polytetrafluoroethylene) tubes were used to connect the syringe pump (NE-1000, New Era Pump Systems, Inc.) with the DEP device for fluidic control (Fig. 1b).
Figure 1.
Schematic of the DEP device. (a) The components of the device, including a lid electrode (indium tin oxide coated glass) with a SU layer containing a microfluidic channel, a Si chip consisting of a nanoelectrode array in a 200 μm × 200 μm area, glass fluidic connectors, and microbore tubes. (b) The electrical connections made to the device. (c) A low-magnification optical microscope image showing the flow profile of the fluorescent labeled bacteriophage solution through the circular microchamber. The stretched lines represent the length of particle movement under the hydrodynamic flow. (d) Schematic diagram of the virus particles in the active 200 μm × 200 μm NEA area, which are subjected to the hydrodynamic drag force (F Drag) in the flow direction and the dielectrophoretic force (FDEP) mostly perpendicular to the flow solution. The NEA could be fabricated with any stable metal materials and ideally in a regular array. (e) A scanning electron microscopy image of the exposed tips of carbon nanofibers which are embedded in the SiO2 matrix. The electrode tips were randomly distributed in the present study.
Figure 5.
DEP capture at different concentrations. (a) The kinetic DEP capture curves when AC voltage is turned on and off with Bacteriophage T4r concentration at the normal concentration (5×109 pfu·ml−1) and two diluted concentrations (5.5×108 and 2.5×107 pfu·ml−1). (b) The kinetic curve of DEP capture with a very low concentration (8.9×104 pfu·ml−1) of Bacteriophage T1 flow above the 200 μm × 200 μm active NEA area at 0.87 mm·sec−1. The captured virus particles are well separated from each other and thus can be precisely counted. (c) and (d) are representative snapshots from the video with the AC voltage off and on, respectively. The flowing virus particles in solution are seen as streaks while the captured ones are focused bright spots. The AC voltage is fixed at 10 kHz and 10 Vpp in this set of experiments.
2.2 Bacteriophage T4r Culture and Labeling
The bacteriophage culture was carried out in an assembly consisting of two conical flasks (50ml, Pyrex®) connected with rubber stoppers and glass tubes (O.D. = 3.12 mm). The first flask was used for culturing and the second one for trapping any overflow of the media and allowing passage of outgoing air. The flasks and tubes were wet-sterilized at 121°C for 20 min., and rubber stoppers were dry-heat sterilized at 160°C for 4 hr. After cooling, 30 ml of the sterilized nutrient peptone broth was added into the flask, incubated at 37°C for 12 hr. The broth remained clear to confirm the sterility. The stock solutions (~ 3 ml) of Bacteriophage T4r and host bacterium E. coli B on a Tryptone agar slant was obtained from Carolina Biological Supply Company (Burlington, NC). Bacteriophage T1 were obtained as pellets from ATCC (Manassas, VA) with the same E. coli B as host bacterium. About 15 ml of nutrient peptone broth medium (Fisher, Pittsburg, PA) was added to the bacterial agar slant and incubated at 37° C for 12 hr. The concentration of the bacteria was measured with a counting chamber (Hausser Scientific Partnership, Horsham, PA) and found to be ~2.25×109 cfu·ml−1. Next, 0.1 ml of cultured E. coli was used to inoculate 30 ml of sterile medium in the first flask and incubated for ~2 hr. at 37°C for bacteria to reach mid logarithmic phase. 1 ml of high titer solution of Bacteriophage (phage) at a concentration of 1×109 pfu·ml−1 was added into the host bacteria solution. The virus-host culture was periodically examined by checking the turbidity of the solution. Lysis of the bacteria in the medium began after an hour of incubation, as indicated by frothing in the solution. Turbidity of the solution decreased as the lysis progressed and eventually turned clear after 3 hr. The solution was filtered with a 0.2 μm filter from Fisher (Pittsburg, PA) to remove live bacteria or bacterial debris. The phage culture was stored in the refrigerator at 4°C with chloroform to kill and to ensure that no live bacteria were in the solution, as suggested by the vendor.
The cultured bacteriophages were characterized with both microbiological and microscopic methods. Double layer agar (DLA) method was used to determine the titer of the phages (see Supporting Information Fig. S1). All culture broths and agar were sterilized at 121°C for 20 min. Transmission electron microscope (TEM) images were taken with FEI CM 100 (Philips, Eindhoven, Holland) with an AMT digital capturing system to illustrate the size and shape of Bacteriophage T4r and Bacteriophage T1 (see Supporting Information Fig. S2). In TEM sample preparation, 5 μl of phage suspension and 5 μl of 2% uranyl acetate onto a 400-mesh copper grid coated with a carbon film. The droplet was partially wicked off after 45 secs and the grid was air-dried before measurements.
Washing and labeling were carried out by centrifugation using Amicon® Ultra 0.5 centrifugal filter devices (Millipore, Billerica, MA). Phage suspensions were also centrifuged and re-dispensed into lower volumes to make up the lost phages during removal of the medium and the following washing procedures. In general, 400 μl of phage solution was added in the filter device consisting of a detachable inner filter tube and a larger outer tube and the set was placed in the centrifuge (Minispin® plus, Eppendorf, Hauppauge, NY) at 14,000 rpm for 5 min. The device was then separated and the solution in the outer tube was discarded. The inner filter tube was flipped upside down and placed in a new outer tube. The set was centrifuged at 14,000 rpm for 10 min to spin down the concentrated phage solutions into the outer tube. 400 μl of Tris EDTA (TE) buffer was then added to dilute the phage solution. The washing steps were repeated three times to remove the remains of the nutrient peptone media.
The phages were labeled with SYBR® Green I nucleic acid gel stain (Lonza, Rockland, ME). A commercial stock solution of 10,000X concentration in dimethylsulfoxide (DMSO) was thawed in dark at room temperature. A 500X working solution was prepared by diluting with TE buffer. For the optimal staining, 1 ml of the phage solution and 500 μl of the SYGR® green-I working solution were mixed at 80° C in dark conditions for 10 min. After which, the samples were allowed to cool down for 5 min. The excess dye was removed and washed three times with TE buffer using the above described centrifugal filter device. The labeled phages were dispensed in double deionized water (Barnstead Easypure II, Thermo scientific Asheville, Pittsburg, PA) containing 280 mM D-mannitol (Fisher, Pittsburg, PA). Addition of mannitol was found necessary to enhance the efficiency of pDEP capture of virus particles [10, 19]. The final concentration of the phages for the normal DEP experiments was ~5×109 pfu·ml−1 except in some concentration-dependent experiments. All the solutions were filtered with 0.2μm filtration membrane and sterilized at 121°C for 20 min.
2.3 DEP Experiments
The setup for DEP-facilitated virus capture is schematically illustrated in Fig. 1, similar to the previous study of DEP capture of bacteria by Syed et al. [16]. An upright fluorescence optical microscope equipped with an Axio Cam MRm digital camera (Axioskop 2 FS plus; Carl Zeiss) was focused at the NEA surface through the ITO-glass window while the virus solution was pushed through the channel between them by a syringe pump (NE-1000, New Era Pump Systems Inc.). The field of view covers the whole 2 mm dia. microchamber using a 10X objective lens (see Fig. 1c) or just the 200 μm × 200 μm active NEA area using a 50X objective lens. The filter set allowed an excitation wavelength of 475-495 nm and an emission wavelength of 515-565 nm. The fluorescence videos were recorded with an exposure time of 0.2-0.5 secs and 0-1.0 secs waiting time per frame using multi-dimensional acquisition mode in the Axio-vision 4.7.1 release software (Carl Zeiss MicroImaging, Inc) for up to 95 secs. A sinusoidal AC voltage was turned on and off during the period while the fluorescence image of Bacteriophage was recorded. The frequency (f) of the AC voltage was varied from 100 Hz to 100 kHz and peak-to-peak amplitude (Vpp) ranged from 4V to 10V. The flow rate was changed from 0.33 to 3.06 mm·sec−1, and the concentration of bacteriophages was systematically varied. The linear flow velocity (ʋ) at the NEA surface was calculated from the track length of virus particles in the video frames. The fluorescence videos were analyzed using the interactive measurement module in Axio vision 4.7.1 software (Carl Zeiss) to quantify the intensity or number of virus particles in the active region before and after capture.
3 Results and Discussions
The flow of the labeled Bacteriophage T4r was first examined at low magnification (with a 10X objective lens) as shown in Fig. 1c. The streaks represent the movement of individual bacteriophage particles carried by the hydrodynamic flow of the media during the exposure time. The figure gives a good indication of the distribution of the particles as they enter from the narrow straight channel (500 μm in width) into the larger circular microchamber (2 mm in diameter) and back into the narrow channel at the other side. Only a fraction of the bacteriophage particles pass above the active NEA area. It should be noted that the VACNF NEA in this study consists of randomly distributed CNF tips with an average spacing of >1 μm (as shown in Fig. 1e), though such arrays can be fabricated into regular patterns using more expensive e-beam lithography processes [20]. Fluorescence videos of the labeled virus over the 200 μm × 200 μm active NEA area were recorded at higher magnification (with a 50X objective lens) (see Supplementary Information for representative videos). At high capture density, it was difficult to distinguish individual bacteriophages. Thus the capture efficiency was first quantified using the increase in the integrated fluorescence intensity (ΔF) over the 200 μm × 200 μm active NEA area during the kinetic DEP process after the AC voltage was applied.
Key factors affecting DEP capture include flow velocity, frequency, voltage and concentration. As shown in Fig. 2, the integrated fluorescence intensity rose to a saturation level (ΔFmax) in less than 10 secs as a 10 Vpp AC voltage was applied on the DEP device while a 5×109 pfu·ml−1 Bacteriophage T4r solution was flowing through the channel at a velocity ʋ varying from 0.085 to 3.06 mm·sec−1. Interestingly, a plot of the captured amount vs. the flow velocity showed a maximum at 0.73 mm·sec−1 (see Fig. 2b). At ≤ 0.73 mm·sec−1, the maximum captured bacteriophage particles (as represented by ΔFmax) approximately increased with ʋ in a linear fashion, indicating that the capture process was limited by the mass transport. However, at ≥ 0.73 mm·sec−1, the captured amount decreased vs. ʋ, clearly due to a different mechanism.
Figure 2.
The effects of flow velocity on DEP capture at 10 kHz and 10 V 9 pp with 5×10 pfu·ml−1 Bacteriophage T4r. (a) The kinetic curves of the integrated fluorescence intensity over the 200 μm × 200 μm NEA area as the AC voltage is turned on (at ~10 sec) and off (at ~ 80 sec). (b) The quantity of DEP capture, represented by the maximum increase of the integrated fluorescence intensity (ΔF max), versus the flow velocity, which is peaked at 0.73 mm·sec−1. (c)-(e) are the representative snapshots from the videos just before the AC voltage was turned off at flow velocity of 0.33, 0.73 and 2.72 mm·sec−1, respectively.
When a virus particle is flowing in the microfluidic channel, the lateral hydrodynamic drag force FDrag acting on the particle linearly increases with ʋ as shown in eq 3. This drag force competes with the DEP force (FDEP) to determine whether a virus particle will be captured at the NE point or continue flowing downstream. It is not surprising that fewer virus particles are retained at the active area of the NEA at the highest flow rates. It is noteworthy that the relative direction of FDEP vs. FDrag changes with the particle’s position at the NEA, defined by the nonuniform distribution of ∇E2. As we described in the finite element modeling (FEM) simulation in a previous work [15], the DEP force is dominated by the vertical component (caused by ∇Ez2) when the particle is more than 3 μm above the NEA. At such distances the particles are nearly orthogonal to the lateral FDrag and this facilitates the downward deflection of the particles even at high ʋ. Once they come down at the NE points, a larger lateral DEP force (caused by ∇Ex2) will dominate, which cancels the lateral FDrag and leads to the capture of the virus particle at the NEA surface. As ʋ is increased beyond a certain threshold value, fewer particles are brought sufficiently close to the NE points so that they can be captured.
Interestingly, the captured virus particles showed very different patterns as a function of flow rate in the captured frames of the fluorescence video. At < 0.73 mm·sec−1, isolated bright spots were seen, likely corresponding to individual virus particles (Fig. 2c). At ≥ 0.73 mm·sec−1, however, the captured frames showed fractal-like lightening patterns (Figs. 2d&2e). These patterns are called Lichtenberg figures and are commonly generated under conditions where a high electric field is produced between a point and planar electrode separated by a dielectric containing charged or polarizable materials as is the case here. The generation of such patterns requires a relatively high concentration of polarizable particles and so is seen only when the particle flux is sufficiently high. Even though similar “pearl-chain-like” patterns were observed by Suehiro et al. in DEP trapping of E. coli cells between interdigitated microelectrodes[21], our previous DEP studies showed that only isolated E. coli cells were captured at the NE sites [15, 16]. The larger size and higher internal conductivity of bacteria may have screened the high electric field at the NE tip and reduced the electrical interaction with additional cells.
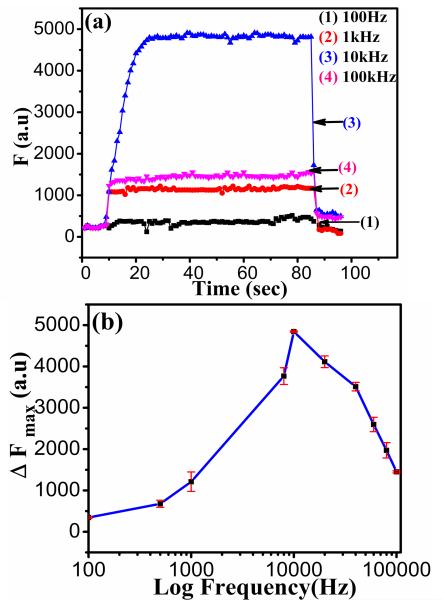
From Eq. (1), the DEP force depends on the frequency of the AC bias through the complex CM factor. This force is associated with each bioparticle’s structure and molecular composition and may be used to capture particular bioparticles at specific frequencies and to separate them from other types of bioparticles. Fig. 3 shows the frequency dependence of DEP capture of Bacteriophage T4r from 100 Hz to 1 MHz, measured with fixed AC voltage (at 10 Vpp) and flow velocity (at 0.73 mm·sec−1). Similar kinetic curves were obtained at all frequencies after application of Vpp. The capture action was observed over a wide frequency range from ~500 Hz to 100 kHz with the maximum capture efficiency at about 10 kHz. This optimum frequency is much lower than 100 kHz to 1 MHz observed in previous studies with E. coli bacteria. Considering that mannitol had to be added to adjust the permittivity and conductivity of the media (i.e. water) [10, 19], it is clear that the small virus particles (Bacteriophage T4r, 80-200 nm in size) have very different CM factor from much larger bacterial cells (E. coli, ~1-2 μm in size).
Figure 3.
The frequency dependence of DEP capture of 5×109 pfu·ml −1 Bacteriophage T4r at a flow velocity of 0.73 mm·sec−1 with the AC bias fixed at 10 Vpp. (a) The kinetic DEP capture curves of the integrated fluorescence intensity (F) versus time when the AC voltage at different frequencies was turned on (at ~10 sec) and off (at ~85 sec). (b) The maximum DEP (ΔFmax) versus the applied AC frequency from 100 Hz to 1 MHz. The optimum capture was obtained with ~10 kHz AC voltage.
The magnitude of ∇E2 is another critical factor that depends on both the DEP device design and the amplitude of the applied AC voltage (represented by Vpp). As shown in Fig. 4, the amount of captured Bacteriophage T4r particles linearly increased with the applied Vpp at all three flow velocities. However, the DEP capture became unreliable at Vpp < 6 V and no capture was observed for the 75 secs after Vpp was dropped to 4 V. The snapshots in Figs. 4b and 4c clearly show the difference between these two scenarios, representing a rapid transition from stretched lines of the free hydrodynamic flow at 4 Vpp to captured Lichtenberg pattern at 6 Vpp. This may be attributed to the fact that the DEP force is smaller than the combination of drag force and Brownian motions at low electric field strengths. To ensure efficient capture, all other DEP experiments in this study were carried out with 10 Vpp.
Figure 4.
(a) DEP capture of Bacteriophage T4r versus the applied AC voltage. The DEP capture is represented by the maximum increase in the integrated fluorescence intensity ΔF max over the active NEA area during the ~75 secs of DEP period. The frequency is fixed at 10 kHz, the flow velocity is varied at 0.33, 0.73, and 1.35 mm·sec−1, and the concentration of Bacteriophage T4r is 5×109 pfu·ml−1. (b) and (c) are snapshots of the video taken at 4 Vpp and 6 Vpp, respectively with the flow velocity at 0.73 mm·sec−1.
The ultimate goal of this study is to develop a rapid sample preparation method for further identification of specific virus particles that can be integrated with other methods on a microfluidic device. Ultimately, the capture and analysis of single virus particles is desired. To assess this potential, a study of the concentration dependence of DEP capture was carried out first by diluting the normal 5×109 pfu·ml−1 Bacteriophage T4r solution to 5.5×108 pfu·ml−1 and then to 2.5×107 pfu·ml−1, respectively. As shown in Fig. 5a, the magnitude of ΔF during DEP capture monotonically decreases as the virus solution was diluted. The kinetics of DEP capture also became slower with dilution and ΔF was not able to reach a saturated level after about 75 secs of DEP period.
More interestingly, the DEP kinetics dramatically changed when a very dilute solution of Bacteriophage T1 (8.7×104 pfu·ml−1) was passed through the DEP device. At such low concentrations, the DEP capture was found to be fully limited by mass transport. The capture of individual virus particles onto the NEA was observed as random single events and the number of captured virus particles linearly increased with time over the ~30 secs timeframe (as shown in Fig. 5b). Similar results were obtained at various flow velocities from 0.59 to 0.94 mm·sec−1. The representative snapshot in Fig. 5c shows the streaks over the active NEA area when the AC voltage was off, indicating the distance that the phage particles moved at 0.87 mm·sec−1 flow velocity in the 0.2 s exposure time. The longest streak was used to calculate the accurate flow velocity of the particles within the focus depth (~0.6 μm [16]) from the NEA surface. In contrast, the snapshot in Fig. 5d shows focused isolated spots corresponding to the captured virus particles at exposed tips of CNF after the voltage was turned on for ~30 secs. The captured virus particles were clearly separated from each other and can be precisely counted. The calculation (as listed in Table 2 in Supplementary Information) indicates that up to 60% of virus particles flowing through the active NEA area were captured at 0.87 mm·sec−1 flow velocity. It is noted that the DEP device design in this study was focused on understanding of fundamental phenomena rather than achieving optimum capture efficiency. The capture efficiency can be significantly enhanced by fabricating elongated active NEA area across the full width of a straight microfluidic channel so that all virus particles are forced to pass through the strong electric field. Nevertheless, the reported results clearly demonstrate that, with proper design, the NEA based DEP device can capture virus particles at concentrations potentially approaching 1-10 cfu·ml−1. By coupling with highly sensitive detection methods (such as surface enhanced Raman spectroscopy), it is very promising to develop an ultrasensitive portable microfluidic system for rapid viral pathogen detection.
Comparing the above virus capture results to the previous reports on the DEP capture of E. coli cells with similar NEA devices [15, 16], four novel observations were made: (1) the optimum AC frequency was found to be ~10 kHz for virus as compared to 100 kHz to 1 MHz for E. coli. (2) The DEP capture of virus particles was observed even at ~8.9×104 pfu·ml−1, more than 4 orders of magnitude lower than the concentration (~1×109 cfu·ml−1) used in previous E. coli studies. (3) The capture efficiency was quantitatively assessed to be 60% from the reliable data at low virus concentration (~8.9×104 pfu·ml−1). (4) The formation of Lichtenberg figures by the captured virus particles after being captured at high flow velocities (≥ 0.73 mm·sec−1) with high virus concentration. The general capture efficiency of virus particles was found to be much higher than that for E. coli cells. This was quite surprising since the ratio of DEP force to drag force was expected to drop significantly as E. coli cells were replaced with ~10 times smaller virus particles. However, the observed higher DEP capture efficiency for smaller virus particles can be well explained with the highly focused electric field at the CNF NEA tip whose size is closer to that of virus particles than bacteria. As shown by the FEM simulation in our previous study [15], the magnitude of E2 is highly focused at the nanoelectrode tip with an elongated distribution in vertical direction like a spear. The magnitude of E2 dropped quickly from ~1.2×1014 V2m−2 at the center of a disk-shaped nanoelectrode to ~1.8×107 V2 m−2 at ~600 nm above [15]. E2 dropped by the same magnitude in lateral direction only ~50 nm from the outside the edge of the nanoelectrode [15]. Such highly focused electric fields provide the extremely large magnitude of ∇E2 which generates the desired DEP force to capture bioparticles. However, it is only available within hundreds of nanometers from the nanoelectrode tip. This length scale is comparable to the size of the virus particles but much smaller than that of E. coli cells. As a result, the large ∇E2 likely interacts with entire virus particles but only affects small portions of E. coli cells. The DEP force relative to the particle volume thus was higher in virus than in bacteria. Further study with FEM simulation by incorporating the localized E2 at the nanoelectrode is necessary to quantitatively assess the size dependence. Mathematical models may be applied in the future to determine the contour of ∇Ez2 and ∇Ex2 near the capture region to better understand how the vertical and lateral components of the field affect the capture of virus particles. These simulations will also be used to define the transition from “trapping zone” for Lichtenberg patterns and “capture zone” for the isolated spots pattern.[22]
4 Concluding remarks
In summary, it can be concluded that the high electric field gradient at the CNF NEA tip can generate strong DEP force to capture virus particles. It is more effective to capture virus particles than larger bacterial cells due to the comparable sizes of virus and CNF tip. Single virus particles can be observed as they are captured at isolated spots at the 200 × 200 μm2 active NEA surface as a low concentration (8.9×104 pfu·ml−1) was flowing by. At this condition, up to 60% of the total virus particles can be captured. At high concentrations (~ 1×109 pfu·ml−1), the DEP capture shows two possible mechanisms depending on the flow velocity. Isolated virus particles were observed at low flow velocities (< 0.73 mm·sec−1) while virus particles form Lichtenberg figures in the flow direction at high flow velocities (≥ 0.73 mm·sec−1). The DEP method with NEA can be used as a rapid microfluidic sample preparation module for further specific analyses.
Supplementary Material
Acknowledgement
We thank the financial support by the Department of Homeland Security and Kansas Bioscience Authority through the Center of Excellence for Emerging Zoonotic Animal Disease (CEEZAD) at Kansas State University. Some instrument was acquired with a grant R15CA159250 from the National Cancer Institute.
Abbreviations
- CNF
carbon nanofiber
- DEP
dielectrophoresis
- AC
alternating current
- CM
Clausius-Mossotti factor
- NE
nanoelectrode
- NEA
nanoelectrode array
- VACNF
vertically aligned carbon nanofiber
- Vpp
peak to peak voltage amplitude
- DLA
double-layer agar
- pfu·ml−1
plaque forming units per milliliter
References
- [1].Cabrera CR, Yager P. Electrophoresis. 2001;22:355–362. doi: 10.1002/1522-2683(200101)22:2<355::AID-ELPS355>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- [2].Hughes MP. Electrophoresis. 2002;23:2569–2582. doi: 10.1002/1522-2683(200208)23:16<2569::AID-ELPS2569>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- [3].Fung DYC. Comprehensive Reviews in Food Science and Food Safety. 2002;1:3–22. doi: 10.1111/j.1541-4337.2002.tb00003.x. [DOI] [PubMed] [Google Scholar]
- [4].Lazcka O, Del Campo FJ, Munoz FX. Biosens. Bioelectron. 2007;22:1205–1217. doi: 10.1016/j.bios.2006.06.036. [DOI] [PubMed] [Google Scholar]
- [5].Pohl HA. J. Electrochem. Soc. 1960;107:386–390. [Google Scholar]
- [6].Pohl HA. Dielectrophoresis: The Behavior of Neutral. Matter in Non-uniform Electric Fields. Cambridge University Press; New York: 1978. [Google Scholar]
- [7].Voldman J. Annu. Rev. Biomed. Eng. 2006;8:425–454. doi: 10.1146/annurev.bioeng.8.061505.095739. [DOI] [PubMed] [Google Scholar]
- [8].Li H, Zheng Y, Akin D, Bashir R. J. Microelectromech. Syst. 2005;14:103–112. [Google Scholar]
- [9].Markx GH, Dyda PA, Pethig R. Journal of Biotechnology. 1996;51:175–180. doi: 10.1016/0168-1656(96)01617-3. [DOI] [PubMed] [Google Scholar]
- [10].Hughes MP, Morgan H, Rixon FJ. Eur Biophys J. 2001;30:268–272. doi: 10.1007/s002490100144. [DOI] [PubMed] [Google Scholar]
- [11].Morgan H, Green NG. Journal of Electrostatics. 1997;42:279–293. [Google Scholar]
- [12].Ermolina I, Milner J, Morgan H. Electrophoresis. 2006;27:3939–3948. doi: 10.1002/elps.200500928. [DOI] [PubMed] [Google Scholar]
- [13].Akin D, Li H, Bashir R. Nano Letters. 2003;4:257–259. [Google Scholar]
- [14].Maruyama H, Kotani K, Masuda T, Honda A, et al. Microfluidics and Nanofluidics. 2011;10:1109–1117. [Google Scholar]
- [15].Arumugam PU, Chen H, Cassell AM, Li J. J. Phys. Chem. A. 2007;111:12772–12777. doi: 10.1021/jp076346e. [DOI] [PubMed] [Google Scholar]
- [16].Syed LU, Liu J, Price AK, Li Y.-f., et al. Electrophoresis. 2011;32:2358–2365. doi: 10.1002/elps.201100020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Li J, Ng HT, Cassell A, Fan W, et al. Nano Letters. 2003;3:597–602. [Google Scholar]
- [18].Li J, Koehne JE, Cassell AM, Chen H, et al. Electroanalysis. 2005;17:15–27. [Google Scholar]
- [19].Hughes MP, Morgan H, Rixon FJ, Burt JPH, Pethig R. Biochimica et Biophysica Acta (BBA) - General Subjects. 1998;1425:119–126. doi: 10.1016/s0304-4165(98)00058-0. [DOI] [PubMed] [Google Scholar]
- [20].Arumugam PU, Chen H, Siddiqui S, Weinrich JAP, et al. Biosens. Bioelectron. 2009;24:2818–2824. doi: 10.1016/j.bios.2009.02.009. [DOI] [PubMed] [Google Scholar]
- [21].Suehiro J, Yatsunami R, Hamada R, Hara M. J. Phys. D-Appl. Phys. 1999;32:2814–2820. [Google Scholar]
- [22].Baylon-Cardiel JL, Lapizco-Encinas BH, Reyes-Betanzo C, Chavez-Santoscoy AV, Martinez-Chapa SO. Lab on a Chip. 2009;9:2896–2901. doi: 10.1039/b906976c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.