Abstract
The immune system has crucial roles in the pathogenesis of multiple sclerosis. While the adaptive immune cell subsets, T and B cells, have been the main focus of immunological research in multiple sclerosis, it is now important to realize that the innate immune system also has a key involvement in regulating autoimmune responses in the central nervous system. Natural killer cells are innate lymphocytes that play vital roles in a diverse range of infections. There is evidence that they influence a number of autoimmune conditions. Recent studies in multiple sclerosis and its murine model, experimental autoimmune encephalomyelitis, are starting to provide some understanding of the role of natural killer cells in regulating inflammation in the central nervous system. Natural killer cells express a diverse range of polymorphic cell surface receptors, which interact with polymorphic ligands; this interaction controls the function and the activation status of the natural killer cell. In this review, we discuss evidence for the role of natural killer cells in multiple sclerosis and experimental autoimmune encephalomyelitis. We consider how a change in the balance of signals received by the natural killer cell influences its involvement in the ensuing immune response, in relation to multiple sclerosis.
Keywords: natural killer cells, multiple sclerosis, killer cell immunoglobulin-like receptors, natural killer cell receptors, human leukocyte antigen
Introduction
Multiple sclerosis is an inflammatory and degenerative disease of the CNS with variable disease course. While in the majority of patients, multiple sclerosis manifests as a relapsing–remitting disease consisting of attacks followed by periods of clinical stability, a small fraction (<10%) show a gradual disease progression from onset, with an inability to recover neurological function. Furthermore, most of the relapsing–remitting patients proceed to develop a progressive form after a period of 15–20 years, as shown in large epidemiological studies (Confavreux and Vukusic, 2006; Compston and Coles, 2008). The basis for this disease heterogeneity remains unknown. One explanation might be that there is a changing balance between multiple effector and regulatory immune cells that contribute to the chronic inflammation of the CNS (Sospedra and Martin, 2005; Fugger et al., 2009; Goverman, 2009). Immunological research in multiple sclerosis has mainly focused on T and B cells (Weber and Hemmer, 2010), which belong to the adaptive immune system. Several studies have explored the recognition by T cell receptors of myelin-derived epitopes presented by predisposing major histocompatibility complex (MHC) molecules (Madsen et al., 1999; Gregersen et al., 2006; Friese et al., 2008). The role of T cells in the pathogenesis of multiple sclerosis has been strengthened by findings of recent genome-wide association studies that have identified some 60 non-MHC risk loci for multiple sclerosis, which also point towards T cells as critical drivers of disease pathology (Hafler et al., 2007; De Jager et al., 2009; Sawcer et al., 2011). There is also now an appreciation of the role of B cells in multiple sclerosis, which influence T cell activation, secrete immune-modulatory cytokines and act as a source of antibody secreting plasma cells (Townsend et al., 2010). The importance of T and B cells in multiple sclerosis has been suggested by the beneficial effects of immunomodulatory therapies, with drugs such as alemtuzumab, natalizumab and rituximab, which target these adaptive immune cell subsets (Polman et al., 2006; Hauser et al., 2008; Bar-Or et al., 2010; Radue et al., 2010; Coles et al., 2011). However, it is now known that a complex network of immune mechanisms consisting of both adaptive and innate immune cells is likely to be involved in the pathogenesis of multiple sclerosis. Until recently, little was known about involvement of the innate immune system in disease pathogenesis (Batoulis et al., 2010) but it is now clear that innate cell types such as dendritic cells, macrophages and microglia amongst others, can influence CNS inflammation. It has been shown that dendritic cells play a critical role in immune invasion of the CNS by presenting antigen to activate autoreactive T cells and in epitope spreading, hence implicating these cells in CNS inflammation and disease development (Greter et al., 2005; McMahon et al., 2005; Bailey et al., 2007). Activation of microglia and macrophages also plays an essential role in pathogenesis of CNS inflammatory disease (Heppner et al., 2005; Adams et al., 2007). A detailed discussion of the function of these cells in multiple sclerosis and its animal model, experimental autoimmune encephalomyelitis (EAE) has been published elsewhere and is beyond the scope of this review (Wu and Laufer, 2007; Comabella et al., 2010; Chastain et al., 2011; Gao and Tsirka, 2011). Another innate cell type, which is receiving increasing attention, is the natural killer cell. These lymphocytes, which are part of the innate immune system, have vital roles in immune-regulation. Natural killer T cells are distinct from natural killer cells, even though they express some of the natural killer cell surface antigens. They represent innate-like T cells that mostly express the semi-invariant T cell receptor and recognize glycolipid antigens in context of the CD1d molecule (van Kaer, 2007). In vivo activation of natural killer T cells has been shown to limit CNS tissue damage in EAE by induction of Th2 responses, changing the cytokine secretion profile of autoreactive T cells (Jahng et al., 2001; Singh et al., 2001) or by inhibiting differentiation of naïve CD4+ T cells towards the Th17 cell lineage (Mars et al., 2009). Nevertheless, there are conflicting results, perhaps due to differences in dose, timing and ligand used for natural killer T cell activation (Jahng et al., 2001; Miyamoto et al., 2001; Pal et al., 2001). A reduction in the proportion of circulating natural killer T cells has also been observed in patients with multiple sclerosis, with increased interleukin (IL)4 production from CD4+ natural killer T cells isolated and expanded from patients in remission in comparison to relapse or healthy individuals (van der Vliet et al., 2001; Araki et al., 2003). Further discussion about natural killer T cell function and defects in multiple sclerosis, and varied approaches used to study natural killer T cell defects in human disease can be found in other recent reviews (Sakuishi et al., 2010; Berzins et al., 2011; Novak and Lehuen, 2011). In this review, we focus on the highly complex and interesting role of natural killer cells and their receptors in multiple sclerosis and EAE. The ability of natural killer cells (and their receptors) to respond to MHC class I molecules makes them ideal candidates for playing a crucial role in CNS disease (Lanier, 2005; Batoulis et al., 2010). MHC class I molecules may be highly expressed in various cell types in the CNS such as oligodendrocytes, astrocytes, microglia and macrophages, as observed in active demyelinating multiple sclerosis lesions (Hoftberger et al., 2004), or even in CNS neurons where the expression of MHC class I can be altered by cytokines such as interferon (IFN)γ (Neumann et al., 1995, 1997; Corriveau et al., 1998). As discussed later in this review, MHC class I molecules have also been genetically implicated in multiple sclerosis. To develop effective treatment of multiple sclerosis, there is a need to target multiple disease pathways and to have a better understanding of both the individual components and the interplay between the innate and adaptive immune systems (Batoulis et al., 2010). Recent studies have begun to address the potential importance of natural killer cells in modifying autoimmune responses. However, it is critical to take into account the wide range and complexity of the different cell surface receptors employed by natural killer cells and the effector mechanisms by which they interact with other cell types, both of which we review here in the context of multiple sclerosis and EAE.
Natural killer cells
Natural killer cells are large granular lymphocytes, which have front-line defensive actions against a variety of infections and tumours (Smyth et al., 2001; Cooper et al., 2009a). In contrast to B and T lymphocytes, natural killer cells can mediate host defences without any prior sensitization by antigen (Lanier et al., 1986b; Anegon et al., 1988). Unlike B and T lymphocyte receptors, natural killer cell receptors do not undergo somatic rearrangement, but instead vary at the germline in terms of allelic sequence, copy number and expression levels (Vivier et al., 2008; Orr and Lanier, 2010). Natural killer cells are generally identified as CD3−, to distinguish them from T cells. In humans, mature natural killer cells are subdivided into functionally distinct cell subsets based upon their expression levels of CD56 (high or low expression, i.e. CD56hi/CD56bright or CD56low/CD56dim) and presence or absence of CD16, as summarized in Table 1. As CD56 is not expressed in mice, it is difficult to draw a direct comparison of human and mouse natural killer cell subsets (Lanier et al., 1986a; Frey et al., 1998; Cooper et al., 2001; Jacobs et al., 2001; Vosshenrich et al., 2006; Huntington et al., 2007; Poli et al., 2009; Marquardt et al., 2010). A marker generally used for natural killer cell identification in both human and mouse is NKp46, although the expression of NKp46 is not fully specific as it can be observed in other cell types such as some rare populations of γδ+ T cells, a mucosal population of innate lymphoid cells and in human astrocytes (Stewart et al., 2007; Satoh-Takayama et al., 2008; Cella et al., 2009; Luci et al., 2009; Reynders et al., 2011; Durrenberger et al., 2012).
Table 1.
Human natural killer cell subsets
| CD16+CD56low natural killer cells | CD16−CD56hi natural killer cells | |
|---|---|---|
| Relative abundance | Constitute ∼90% of natural killer cells in blood | Constitute ∼10% of natural killer cells in blood; predominant in lymphoid organs |
| KIR expression | High | Low |
| Cytotoxic activity | High | Low |
| Cytokine production upon stimulation | Low | High |
Natural killer cell surface receptors include both inhibitory and activating molecules, many of which are expressed in stochastic, variegated and overlapping patterns (Box 1). This allows for expression of different constellations of receptors on different natural killer cell clones, which are then capable of discriminating between cells expressing different ligands, in particular different MHC class I molecules. This creates a diverse repertoire of functionally distinct natural killer cells within an individual and between populations (Raulet et al., 2001; Orr and Lanier, 2010; Jamil and Khakoo, 2011). Engagement of these cell surface receptors with their respective ligands regulates natural killer cell activities, and the integration of signals received by these receptors dictates the activation status of the natural killer cell (Vivier et al., 2008; Orr and Lanier, 2010). Once activated, natural killer cells can themselves produce immunoregulatory cytokines and regulate the development of the ensuing immune response and tissue inflammation, as well as kill target cells (Moretta et al., 2006, 2008; Vivier et al., 2008; Lieberman, 2010; Sun and Lanier, 2011).
Box 1 Definition of terms.
Epigenetic mechanisms—Include heritable changes that regulate gene transcription or expression, and are not due to changes in DNA sequence itself (Shenker and Flanagan, 2012).
Dimorphism—Occurring in two different forms. For example, dimorphism at positions 77–80 in HLA-C alleles encodes for serine or asparagine amino acids at position 77, and asparagine or lysine amino acids at position 80, hence broadly classifying HLA-C alleles in two groups.
Overlapping receptor expression—There is overlap in the receptor expression of different natural killer cells, i.e. natural killer cells express different combinations of receptors with a different degree of overlap in receptor expression with other natural killer cells (Raulet et al., 2001; Joncker and Raulet, 2008).
Pseudogene—Non-functional copies of coding genes (Gregory, 2005).
Stochastic—Often refers to a series of random or probabilistic processes. The probability of natural killer cells co-expressing a given combination of receptors can be estimated by the product of frequencies of natural killer cells expressing each receptor (Raulet et al., 2001).
Synteny—Earlier used to define occurrence of two or more genes on the same chromosome. However, it is often used to refer to conserved blocks of homologous genes that might be located on different chromosome in another species (Ehrlich et al., 1997).
Variegated—Individual natural killer cells express a subset of the receptor genes or alternatively each receptor is expressed only on a subset of natural killer cells (Raulet et al., 2001; Joncker and Raulet, 2008).
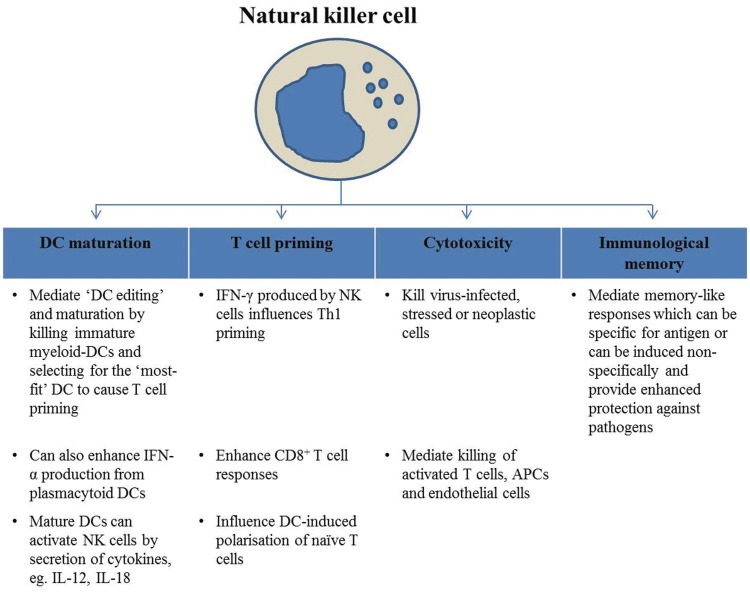
Alteration in natural killer cell number and function has been implicated in various human autoimmune diseases. For example, decreases in circulating natural killer cell number, reduced receptor expression or reduced natural killer activity has been observed in patients with type 1 diabetes and systemic lupus erythematosus, amongst others. Nevertheless, it is important to consider that there might be functional differences between natural killer cells studied in peripheral blood in comparison to the target site, and it is sometimes difficult to ascertain whether the reported natural killer cell alterations are a cause or consequence of disease (Erkeller-Yusel et al., 1993; Rodacki et al., 2007; Park et al., 2009). In inflammatory conditions, natural killer cells can be rapidly recruited from the blood to inflammatory sites in response to chemokine gradients and adhesion molecules, and mediate an immunoregulatory role (Fig. 1) (French and Yokoyama, 2004; O’Leary et al., 2006; Moretta et al., 2008; Perricone et al., 2008; Vivier et al., 2008; Cooper et al., 2009a, b; Sun et al., 2009, 2010; Lieberman, 2010; Sun and Lanier, 2011).
Figure 1.
Immunoregulatory functions of natural killer cells. APC = antigen presenting cell; DC = dendritic cell.
Killer cell immunoglobulin-like receptors (KIRs) represent one of the human natural killer cell receptor families that recognize MHC class I molecules as their ligands. Similarly, human natural killer cells also express the C-type lectin-like receptors, CD94/NKG2 heterodimers that recognize the non-classical MHC class I molecules [i.e. human leukocyte antigen (HLA)-E in humans and Qa1 in mouse]. They may also express leukocyte immunoglobulin like receptors (LILR), some of which also recognize MHC class I ligands. All these receptors consist of members with inhibitory or activating potential (Vilches and Parham, 2002; Lanier, 2005). Cells that physiologically express self-MHC class I molecules are resistant to natural killer cell-mediated killing, whereas loss of MHC class I expression (e.g. virally infected or neoplastic cells escaping conventional cytotoxic T cells) provokes natural killer cells. Thus, natural killer cells are activated by detecting ‘missing self’ (Karre et al., 1986; Ljunggren and Karre, 1990; Bix et al., 1991), so that the natural killer cell inhibitory receptors no longer restrain the activating receptors, which then initiate killing (Karlhofer et al., 1992; Raulet et al., 2001). The response of a natural killer cell is controlled by the expression of these multiple cell surface receptors. The activating receptors thus play an important role in promoting natural killer cell activation and cytotoxicity. There are many activating receptors, including the natural cytotoxicity receptors, represented by NKp46, NKp44 and NKp30, NKG2D, 2B4, CD2, LFA1 and co-receptors such as DNAM1 (Schleinitz et al., 2008; Pegram et al., 2011).
Interestingly, however, natural killer cells in hosts deficient in MHC class I expression are not spontaneously autoreactive in vivo (Bix et al., 1991; Hoglund et al., 1991; Liao et al., 1991; Yu et al., 1992; Zimmer et al., 1998; Vitale et al., 2002). This is because host MHC class I molecules are also required for the functional maturation of natural killer cells. Several models have been proposed to describe this as ‘natural killer cell licensing’ or ‘education’ (Brodin et al., 2009; Hoglund and Brodin, 2010; Orr and Lanier, 2010). Whereas licensing of natural killer cells was initially attributed to signalling via the inhibitory natural killer cell receptors, recent data suggest that interactions of activating receptors with their MHC ligands can decrease subsequent natural killer cell responsiveness (Oppenheim et al., 2005; Sun and Lanier, 2008; Tripathy et al., 2008; Fauriat et al., 2010). Nevertheless, licensing requirements can sometimes be bypassed, for instance, when natural killer cells are preactivated, stimulated by cytokines or exposed to inflammatory conditions (Kim et al., 2005; Yokoyama and Kim, 2006). Also, licensing can be reversible; mature natural killer cells can be reprogrammed to either gain or lose activity after transfer to or from MHC-deficient hosts (Elliott et al., 2010; Joncker et al., 2010).
The following sections provide an account of some of the prominent human natural killer cell receptors including KIR, CD94:NKG2, NKG2D, natural cytotoxicity receptors and LILR, focusing on evidence of their involvement in multiple sclerosis.
Killer cell immunoglobulin-like receptors
KIR genes encode polymorphic activating as well as inhibitory natural killer cell receptors that belong to the immunoglobulin superfamily of Type I transmembrane proteins and comprise 15 genes and two pseudogenes. They span 100–200 kb in the leukocyte receptor complex on human chromosome 19q13.4, where allelic and copy number variations encompass a variety of haplotypes (Trowsdale, 2001; Barrow and Trowsdale, 2008). KIR receptors are important regulators of natural killer cell function that must have co-evolved with polymorphic HLA class I ligands (chromosome 6p21.3) (Trowsdale, 2001; Marsh et al., 2003; Uhrberg, 2005; Norman et al., 2007; Single et al., 2007); it is therefore vital to first understand the properties of these receptors in general before discussing their involvement in multiple sclerosis.
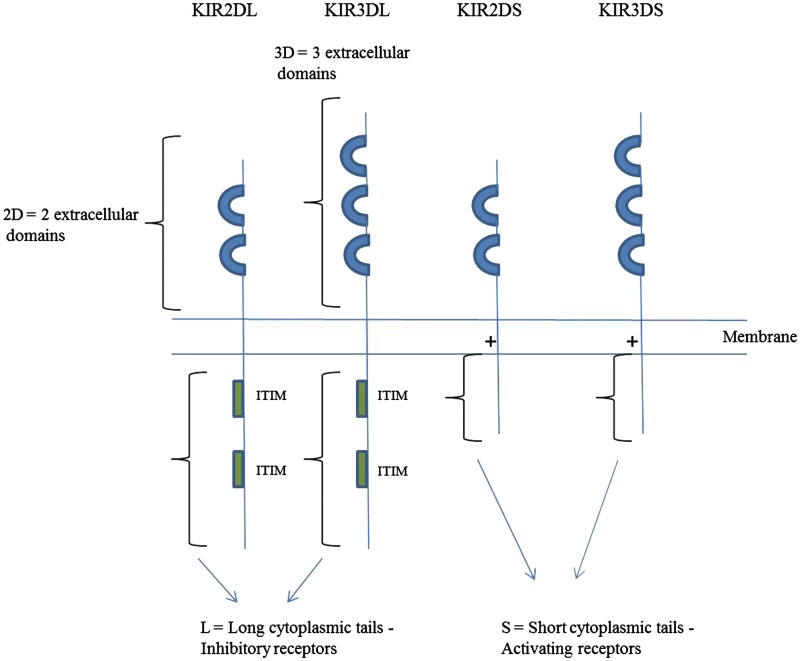
KIRs are classified according to their number of extracellular immunoglobulin-like domains, cytoplasmic tail length and sequence similarity (Fig. 2) (Vilches et al., 2000a; Vilches and Parham, 2002; Marsh et al., 2003; Purdy and Campbell, 2009). The inhibitory receptors have long cytoplasmic tails that contain one or more immunoreceptor tyrosine-based inhibitory motifs (ITIM), which become phosphorylated upon ligand binding. This leads to recruitment of such cytoplasmic phosphatases as Src homology 2 (SH2)-containing phosphatase-1 (SHP-1) or SHP-2. These enzymes then dephosphorylate protein substrates of tyrosine kinases that are linked to activating natural killer receptors, and suppress their signalling. In contrast, the activating receptors lack the intracellular signalling motifs and instead bind via positively charged arginine residues in their transmembrane regions—to complementary, negatively charged aspartate residues in adaptor proteins, such as DAP-12, that contain immunoreceptor tyrosine-based activating motifs (ITAMs). Both ITIM and ITAM are defined by different consensus sequences, which determine their specificity to bind different substrates. Therefore, once phosphorylated, the ITAM on the adaptor binds to an SH2 domain-containing kinase such as Syk or ZAP-70. This initiates a signalling cascade, culminating in actin-skeleton reorganization, degranulation and transcription of cytokine and chemokine genes (Lanier, 2005, 2008; Pegram et al., 2011).
Figure 2.
Killer cell immunoglobulin-like receptor structure and nomenclature. KIR genes consist of extracellular immunoglobulin-like domains, a stem region, a transmembrane region and a cytoplasmic tail. The inhibitory receptors contain one or two ITIM motifs in their long cytoplasmic tails, whereas the activating receptors have a charged residue in their transmembrane domains that allows them to bind and signal via adaptor proteins. KIR2DL4 is an exception and contains an ITIM in its cytoplasmic tail and a positively charged residue in its transmembrane domain.
In addition to the extensive allelic variation in the KIR genes, there is diversity in the number and type of KIR genes between individuals and populations. Some KIR alleles differ by level of expression, binding affinity to the HLA ligand, or access to the cell surface (Campbell and Purdy, 2011). Several KIR genes exhibit copy number variation; this is less prominent in flanking framework genes such as KIR3DL3 and KIR3DL2 (Jiang et al., 2012). Additional, more centrally placed framework genes include KIR2DL4 and KIR3DP1. Haplotypes with different complements of KIR loci are proposed to be products of unequal crossing-over, namely non-allelic homologous recombination, and may vary greatly (Trowsdale, 2001; Parham, 2003; O’Connor et al., 2007; Thananchai et al., 2007; Li et al., 2008; Traherne et al., 2010). They are broadly divided into Groups A and B, the prevalences of which differ in different populations. Group A haplotypes contain seven KIR genes and two pseudogenes, which are all inhibitory except for KIR2DS4. However, KIR2DS4 carries a deletion on the majority of A haplotypes (Hsu et al., 2002; Middleton et al., 2007). Group B haplotypes are more varied and encode more activating receptors, including KIR2DS1–3, KIR2DS5 and KIR3DS1, as well as inhibitory receptors such as KIR2DL2 and KIR2DL5 (Uhrberg et al., 1997, 2002; Parham, 2003). The KIR gene complex is unusually compact, its genes being only ∼2 kb apart. It has evolved rapidly, and differs considerably between humans, great apes and Old World monkeys (Khakoo et al., 2000; Volz et al., 2001; Guethlein et al., 2002; Parham, 2004; Hershberger et al., 2005; Sambrook et al., 2006).
KIRs are expressed mainly by natural killer cells, but also by subsets of γδ+ T cells, effector memory CD8+CD28− αβ+ T cells, chronically stimulated CD3+ T cells and some CD4+ memory T cells. Individual natural killer ‘clones’ express different numbers and combinations of KIRs, but always maintain the ability to sense and respond to dynamic changes in ligand expression such as those caused by viral infection or tumours. Once these variegated KIR expression patterns are formed, they are stabilized by epigenetic mechanisms over numerous cell divisions (Huard and Karlsson, 2000; Vilches and Parham, 2002; Young and Uhrberg, 2002; Parham, 2003; Vivier and Anfossi, 2004; Lanier, 2005).
HLA-C was once considered a minor MHC class I isotype because it is generally expressed at lower levels than HLA-A and HLA-B, but has since proved a key focus of recognition by natural killer cells (Barrow and Trowsdale, 2008). Polymorphic HLA-C alleles are broadly classified into two groups, based on the HLA-C1 (77Ser 80Asn), HLA-C2 (77Asn 80Lys) dimorphism at position 77–80, which defines their ability to bind KIR2D receptors. Table 2 summarizes the different HLA-C groups and their binding specificity for KIR2D receptors (Colonna et al., 1993; Moretta et al., 1993; Wagtmann et al., 1995; Winter et al., 1998; Boyington and Sun, 2002; Parham, 2005; Stewart et al., 2005; Moesta et al., 2008).
Table 2.
HLA-C ligand and KIR binding specificity
| HLA-C1 | HLA-C2 | |
|---|---|---|
| Alleles | HLA-Cw1, Cw3, Cw7, Cw8, Cw12, Cw13, Cw14, Cw1507, Cw1601/4 | HLA-Cw2, Cw4, Cw5, Cw6, Cw15, Cw1204/5, Cw0707/9, Cw1602, Cw17, Cw18 |
| Evolutionary age | Evolutionarily older | Arisen later in primate evolution |
| KIR recognition | KIR2DL2, KIR2DL3a | KIR2DL1a, KIR2DS1 |
| Strength of KIR-HLA binding and inhibition | KIR2DL1-HLA-C2 > KIR2DL2-HLA-C1 > KIR2DL3-HLA-C1 | |
a KIR2DL1 is specific in its binding to HLA-C2, but KIR2DL2 and KIR2DL3 can bind to HLA-C1, several HLA-C2 allotypes and two HLA-B allotypes that share key residues with HLA-C1.
By contrast, KIR3D receptors recognize some HLA-A and HLA-B alleles. The latter are split into HLA-Bw4 versus HLA-Bw6 ‘supratypes’, based on serological epitopes defined by positions 74–83 in their α1 domains, particularly their respective 80Ile or 80Thr versus 80Asn (near the C-termini of the bound peptides) (Muller et al., 1989). KIR3DL1 recognizes Bw4 alleles and Bw4+ HLA-A variants, and may show higher affinity and inhibition by those with 80Ile versus 80Thr (Cella et al., 1994; Wagtmann et al., 1995). Table 3 lists the different human KIRs and their respective ligands.
Table 3.
KIR and ligands
| Receptor | Type | Species | Ligand | References |
|---|---|---|---|---|
| KIR2DL1a,b | Inhibitory | Human | HLA-C2 (80Lys) | Colonna et al. (1993), Moretta et al. (1993), Wagtmann et al. (1995), Fan et al. (1997, 2001) |
| KIR2DL2a,b | Inhibitory | Human | HLA-C1 (80Asn), HLA-B*4601, *7301, Some HLA-C2 | Colonna et al. (1993), Moretta et al. (1993), Wagtmann et al. (1995), Snyder et al. (1999), Moesta et al. (2008) |
| KIR2DL3a | Inhibitory | Human | HLA-C1 (80Asn), HLA-B*4601, *7301, Some HLA-C2 | Colonna et al. (1993), Moretta et al. (1993), Wagtmann et al. (1995), Maenaka et al. (1999), Moesta et al. (2008) |
| KIR2DL4 | Inhibitory and activating | Human | HLA-G? | Rajagopalan and Long (1999) |
| KIR2DL5A,B | Inhibitory | Human | ? | Vilches et al. (2000b), Gomez-Lozano et al. (2002) |
| KIR2DS1 | Activating | Human | HLA-C2 (80Lys) | Moretta et al. (1995), Biassoni et al. (1997) |
| KIR2DS2a | Activating | Human | ? | Saulquin et al. (2003) |
| KIR2DS3,5 | Activating | Human | ? | |
| KIR2DS4a | Activating | Human | HLA-A11, HLA-C | Katz et al. (2001), Graef et al. (2009) |
| KIR3DL1b | Inhibitory | Human | HLA-Bw4, HLA-A23, A24, A32 | Cella et al. (1994), Gumperz et al. (1995), Stern et al. (2008), Vivian et al. (2011) |
| KIR3DL2 | Inhibitory | Human | HLA-A3, HLA-A11, CpGDNA/TLR9 | Hansasuta et al. (2004), Sivori et al. (2010) |
| KIR3DL3 | Inhibitory | Human | ? | |
| KIR3DS1 | Activating | Human | ? |
a Represents receptors whose crystal structures have been determined.
b Represents receptors whose crystal structures are determined in complex with ligand.
Because the KIRs and their HLA-class I ligands are so polymorphic, they are potential susceptibility factors for infections and autoimmune diseases, as well as obstetric complications such as pre-eclampsia. While HLA-KIR genotypes that favour natural killer cell or T cell activation might have evolved to enhance resistance to viruses or tumours, some combinations may concomitantly predispose to autoimmunity (Rajagopalan and Long, 2005). Interactive associations of HLA-KIR genotypes in autoimmune diseases, infectious models, reproductive failure, cancer and haemopoietic stem cell transplantation are reviewed elsewhere (Parham, 2005; Rajagopalan and Long, 2005; Williams et al., 2005; Kulkarni et al., 2008; Chazara et al., 2011; Jamil and Khakoo, 2011).
Killer cell immunoglobulin-like receptors and their human leukocyte antigen class I ligands in multiple sclerosis
The parallel recognition of HLA class I molecules by T cell receptor and KIRs has prompted a new look at susceptibility to multiple sclerosis. For example, HLA-A3 predisposes to multiple sclerosis (Fogdell-Hahn et al., 2000; Harbo et al., 2004). Since HLA-A3-restricted CD8+ T cells are implicated in its induction in a humanized mouse model (Friese et al., 2008), possible contributions of natural killer cell receptors like KIR3DL2, which recognize HLA-A3, demand further study. Recent studies in multiple sclerosis are beginning to increase our understanding of the importance of HLA-B and HLA-C alleles as ligands for KIRs. A comparison of 1201 multiple sclerosis cases and 3660 UK controls showed that HLA-Cw5 has a protective effect (relative risk ∼0.55), independent of HLA-DRB1*1501, *03 and *0103. However, grouping into HLA-C1 versus -C2 did not reveal further associations (Yeo et al., 2007). HLA-Cw5 can be recognized by a variety of receptors, i.e. KIR2DL1, KIR2DS1 and possibly also by KIR2DL2, and KIR2DL3 expressed on natural killer cells or a subpopulation of T cells, suggesting an increased potential for immune regulation via this HLA-KIR recognition pathway (Winter et al., 1998; Parham, 2005; Moesta et al., 2008). An Italian study reported that the protective effect of HLA-Cw5 was possibly synergistic with that of HLA-A2 (Bergamaschi et al., 2010). On the other hand, a Scandinavian study failed to confirm protection by HLA-Cw5, but instead showed a positive association with HLA-Cw8 in HLA-DRB1*15-negative subjects (Link et al., 2010).
There are similar reports of protective associations of HLA-Bw4 ligands for KIR3DL1. Thus HLA-B*44 (in the HLA-Bw4-80Thr group) is under-represented in multiple sclerosis, which might have confounded the implication of the linked Cw5 (Rioux et al., 2009; Healy et al., 2010). A Norwegian study also found protection by the HLA-Bw4 group, again independently of HLA-DRB1 alleles. Although differences in prevalence of inhibitory or activating KIR alleles did not reach significance, there were signs of interactions between KIR2DL1/S1 and HLA-C2 that warrant larger studies (Lorentzen et al., 2009). A small Italian study suggested a protective role or a decrease in the frequency of KIR2DS1 in patients with multiple sclerosis, an effect that was enhanced in the presence of its ligand group HLA-C2. On the contrary, frequency of another activating receptor KIR2DS4*001/002 allele was found to be higher in patients (Fusco et al., 2010). A recent study demonstrated a decrease in the frequency of the inhibitory gene KIR2DL3 in patients with multiple sclerosis (Jelcic et al., 2011). KIR2DL2 and KIR2DL3 segregate as alleles at the same locus (Uhrberg et al., 2002); therefore, the presence of two copies of KIR2DL2/S2 (KIR2DS2 is in high linkage disequilibrium with KIR2DL2) in the absence of KIR2DL3 was over-represented in the multiple sclerosis cohort (Jelcic et al., 2011).
A serious difficulty in many of these studies is the loss of statistical power after stratifying subjects according to combinations of KIR with HLA variants, excluding secondary effects of linked HLA-DRB1 alleles, and then correcting for multiple comparisons. Nevertheless, independent contributions of natural killer cells and their receptors to disease pathogenesis and progression clearly warrant investigation in larger studies. Furthermore, mechanistic studies that allow functional assessment of each of these genes in CNS disease are required.
CD94:NKG2, NKG2D and the natural cytotoxicity receptors
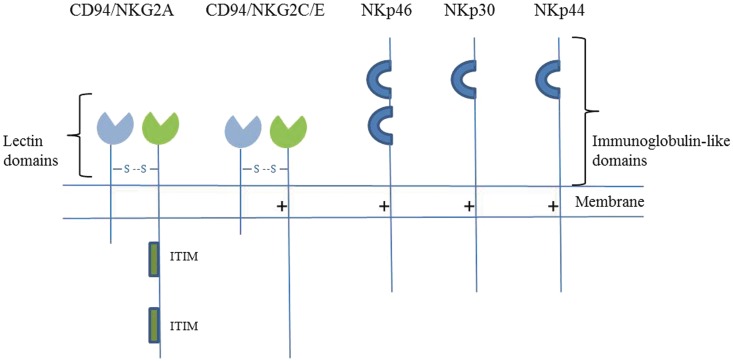
The CD94/NKG2 gene family is located in the natural killer gene complex on human chromosome 12 and the syntenic region on mouse chromosome 6. The CD94 gene is genetically linked to four NKG2 genes in humans (NKG2A, C, E and F) and three in mice (NKG2A, C and E). The common CD94 subunit forms heterodimers with NKG2A, E or C; the CD94:NKG2A heterodimer is an inhibitory receptor, whereas the CD94:NKG2C and CD94:NKG2E are activating (Fig. 3) (Borrego et al., 1998; Braud et al., 1998; Lanier, 2005; Pegram et al., 2011). The CD94/NKG2A receptors have two ITIM motifs in their cytoplasmic domains, whereas the CD94/NKG2C and CD94:NKG2E have a positively charged residue in their transmembrane regions and hence associate with DAP12 (Borrego et al., 2006). CD94:NKG2A/E/C molecules recognize non-classical MHC molecules, which carry grooves that normally bind peptides derived from the leader sequences of classical HLA class I molecules, hence allowing indirect monitoring of MHC class I expression on the target cell. CD94/NKG2A and CD94/NKG2E receptors bind HLA-E with higher affinity than CD94/NKG2C and minor sequence differences in the peptide bound to HLA-E can dramatically influence receptor binding affinity (Vales-Gomez et al., 1999; Kaiser et al., 2005; Hoare et al., 2008). The CD94/NKG2 receptors play a crucial role in modulating natural killer cell responses and have an indispensable role in protection against infectious diseases such as viral mousepox disease (Fang et al., 2011). These receptors have very limited polymorphism and are expressed on overlapping subsets of natural killer cells and T cells; unlike KIRs, expression of CD94/NKG2 receptors is not stable and can be modulated by cytokines or T cell receptor signals (Lanier, 2005; Pegram et al., 2011).
Figure 3.
CD94/NKG2 and natural cytotoxicity receptors. The inhibitory receptors contain ITIM motifs in their cytoplasmic tails, whereas the activating receptors have a positively charged residue in their transmembrane regions and associate with ITAM-bearing adaptor proteins.
The activating NKG2D molecule is conserved between humans and mice and is expressed as a disulphide-linked surface homodimer potentially on all mouse and human natural killer cells, human CD8+ αβ T cells, γδ T cells and a fraction of CD4+ T cells. Its expression on T cells is more selective in mice. Though its gene is also in the natural killer gene complex, it has so little homology with the rest the NKG2 family that ‘NKG2D’ is considered a misnomer (Raulet, 2003; Lanier, 2005). It associates with DAP-10 adaptor protein; its alternatively spliced short isoform, NKG2D-S, is found in mice (and not humans), and can associate with either DAP-10 or DAP-12. Stress-induced, MHC class I-related molecules act as ligands for NKG2D; they include MICA, MICB, ULBP1, ULBP2, ULBP3 and ULBP4 proteins in humans and RAE1, H60 and MULT proteins in mice. Most of these ligands are expressed at low levels by normal cells but are upregulated under inflammatory conditions, and upon cellular transformation or stress (Bauer et al., 1999; Diefenbach et al., 2000; Raulet, 2003; Lanier, 2005, 2008). NKG2D ligands are also induced in T cells upon activation and this interaction of NKG2D with its ligands plays an important role in the crosstalk between activated T cells and natural killer cells (Rabinovich et al., 2003; Cerboni et al., 2007). However, it has also been suggested that MICA that is induced in these activated T cells is mostly retained intracellularly, perhaps as a safeguard mechanism to protect them from NKG2D-dependent cytotoxicity (Molinero et al., 2006). In T cells, NKG2D can function as a co-stimulatory receptor and influence T cell receptor-dependent T cell activation (Groh et al., 2001); NKG2D conjugation along with the T cell receptor on CD8+ T cells can alter T cell function and decrease production of anti-inflammatory cytokines (Barber and Sentman, 2011). NKG2D is a powerful receptor and plays an important role in mediating immune surveillance against viral infections and tumours (Zafirova et al., 2011).
Natural cytotoxicity receptors, namely NKp30, NKp44 and NKp46, are another group of important activating receptors that belong to the Ig superfamily. Their reported ligands include viral haemagglutinins (NKp46 and NKp44), heparin sulphate proteoglycans (NKp46 and NKp30), HLA-B-associated transcript 3, i.e. BAT3 and B7-H6 (NKp30). They must have other unidentified endogenous ligands that stimulate natural killer cell responses, as antibody-mediated blocking of natural cytotoxicity receptors prevents lysis of various tumour cell types (Sivori et al., 1999; Moretta et al., 2001; Pegram et al., 2011). Some proteins block natural killer cell function and cytotoxicity by binding natural cytotoxicity receptor proteins. Human cytomegalovirus pp65 protein, for example binds to NKp30 (Arnon et al., 2005). Similarly, cancer-associated protein, proliferating cell nuclear antigen (PCNA) binds to NKp44 (Rosental et al., 2011). Natural cytotoxicity receptors associate with ITAM-bearing signal transduction molecules, which mediate activation, i.e. CD3ζ and FcεRIγ for NKp46 and NKp30, and DAP12 for NKp44 (Moretta et al., 2001; Pegram et al., 2011). NKp46 and NKp30 are expressed on activated and resting natural killer cells, while NKp44 is upregulated on IL2 stimulation. Additionally, NKp46 expression has recently been observed in astrocytes in human brain tissue and more so in white matter lesions from patients with multiple sclerosis, however, its function on astrocytes is unclear (Durrenberger et al., 2012). NKp46 is conserved between humans and mice; however, no functional mouse orthologue has been reported for NKp44 and NKp30 (Walzer et al., 2007). Natural cytotoxicity receptors appear to be key mediators of tumour cell killing by natural killer cells, and of natural killer recognition of immature dendritic cells (Barrow and Trowsdale, 2008; Pegram et al., 2011). An important role for NKp46 in protection against viral conditions and in the development of Type 1 diabetes has been demonstrated in vivo. Knock-in mice containing a non-functional NKp46 receptor demonstrated an enhanced susceptibility to lethal influenza virus infection (Gazit et al., 2006) and less development of type 1 diabetes (Gur et al., 2010). Some of these findings are contradictory and NKp46 is suggested to be involved in downregulation of natural killer cell responsiveness (Narni-Mancinelli et al., 2012). Nevertheless, NKp46 was shown to bind undetermined ligands on human and mouse pancreatic beta cells, and natural killer cells killed pancreatic beta cells in an NKp46-dependent manner. Moreover, induction of blocking NKp46-specific antibodies prevented diabetes development in non-obese diabetic mice (animal model of type 1 diabetes) (Gur et al., 2010, 2011). A summary of these receptors can be found in Table 4.
Table 4.
NKG2D, NKG2, natural cytotoxicity receptors and their ligands
| Receptor | Type | Species | Ligand | References |
|---|---|---|---|---|
| NKG2Da,b | Activating | Human and mouse | Mouse RAE-1, H60, MULT Human MICA, MICB, ULBP | Bauer et al. (1999), Li et al. (2001), Wolan et al. (2001) |
| NKp46a | Activating | Human and mouse | Viral haemagglutinins | Mandelboim et al. (2001), Foster et al. (2003) |
| NKp30a,b | Activating | Human | B7-H6, BAT3, HCMV pp65 | Arnon et al. (2005), Pogge von Strandmann et al. (2007), Brandt et al. (2009), Joyce et al. (2011), Li et al. (2011) |
| NKp44a | Activating | Human | Viral haemagglutinins, PCNA | Arnon et al. (2001), Cantoni et al. (2003), Rosental et al. (2011) |
| CD94/NKG2Aa,b | Inhibitory | Human and mouse | HLA-E, Qa-1 | Borrego et al. (1998), Braud et al. (1998), Lee et al. (1998), Vance et al. (1998), Sullivan et al. (2007), Petrie et al. (2008) |
| CD94/NKG2C | Activating | Human and mouse | HLA-E, Qa-1 | Borrego et al. (1998), Braud et al. (1998), Vance et al. (1999) |
| CD94/NKG2E | Activating | Human and mouse | HLA-E, Qa-1 | Borrego et al. (1998), Vance et al. (1999) |
a Represents receptors whose crystal structures have been determined.
b Represents receptors whose crystal structures are determined in complex with ligand.
CD94:NKG2 and NKG2D receptors in multiple sclerosis
The CD94/NKG2A receptors reportedly play an important role in regulating T cell activity in EAE. Inhibitory interactions between murine CD94:NKG2A on natural killer cells and Qa1 on activated T cells are important in protecting activated T cells from natural killer lysis, and thus in clonal expansion and memory generation by self-reactive T cells. Furthermore, EAE is ameliorated by antibody-mediated blockade of Qa1-NKG2A interactions, or by adoptive transfer of Qa1-deficient CD4+ T cells, because of potent natural killer killing of activated autoreactive T cells. Antibody-mediated blockage of Qa1-NKG2A interactions is associated with reduced cellular infiltrates and activated microglia, and an altered cytokine profile (i.e. decreased IL17 and IFNγ, and increased IL4 and IL10) of CD4+ T cells in the CNS (Lu et al., 2007; Leavenworth et al., 2010). In line with this, activated CD4+ T cells from Qa1 mutant knock-in mice (with a selective deficiency to bind the CD94/NKG2A receptor) are highly susceptible to natural killer cell lysis, reconfirming the role of this pathway in modulating adaptive/autoimmune responses (Lu et al., 2007). Furthermore, Qa1 engagement of CD94-NKG2A receptors on CD8+ T cells transmits an inhibitory signal that attenuates suppressive activity of CD8+ regulatory T cells; disruption of these Qa1-NKG2A interactions leads to robust CD8 regulatory activity and diminished development of EAE (Lu et al., 2008).
Interaction via NKG2D may be an alternative mechanism by which natural killer cells could suppress autoreactive T cells. Heat-shock protein 70 complexed with peptides isolated from EAE brains (Hsp70-pc) induced natural killer cell-dependent resistance to subsequent EAE induction in SJL/J mice, possibly due to upregulation of the NKG2D ligand, H60. NKG2D–H60 interactions seem to modulate dendritic cell function, leading to elimination of antigen-reactive T cells and induction of EAE tolerance. This was suggested by the reduced ability of dendritic cells preincubated with natural killer cells from Hsp70-pc mice to stimulate proliferation of proteolipid protein (PLP)-reactive cells in vitro, which also correlated with enhanced death of PLP-reactive cells. (Galazka et al., 2006, 2007). On the other hand, natural killer cells can also interact directly with brain-resident cell types; once activated, they can kill resting microglial cells in vitro via NKG2D- and NKp46-dependent pathways. Activated microglia are protected from lysis by these mechanisms by upregulating MHC class I (Lunemann et al., 2008). Additionally, inappropriate expression of NKG2D and its ligands can lead to activation of autoreactive effector cells. Tumour cells reduce NKG2D expression on natural killer and T cells, impairing their cytotoxic activity, by releasing soluble forms of MICA or MICB. Interestingly, in a recent study, serum levels of soluble MICB (but not MICA) were most elevated (above controls) in patients with multiple sclerosis during relapse (Groh et al., 2002; Fernandez-Morera et al., 2008).
Additional evidence suggests that NKG2D-expressing natural killer or T cells can contribute to tissue injury in multiple sclerosis by killing NKG2D-ligand bearing oligodendrocytes (Saikali et al., 2007) or astrocytes (Darlington et al., 2008). Similarly, dorsal root ganglion neurons are susceptible to natural killer cell-mediated lysis because they strongly express the NKG2D ligand, RAE1 (Backstrom et al., 2003). Nevertheless, this might also suggest differences in natural killer cell function in the CNS in comparison to the peripheral blood (Shi et al., 2011).
Leukocyte immunoglobulin-like receptors
Leukocyte immunoglobulin-like receptors (LILRs, LIR or CD85), also called immunoglobulin-like transcripts (ILTs), are encoded by genes in the leukocyte receptor complex on chromosome 19q13.4, closely linked to the KIR genes. The LILR family comprises six potentially activating LILRA, five inhibitory LILRB and two pseudogenes. LILRs can be expressed on various cells of the myeloid lineage, including dendritic cells; also on B cells, natural killer cells and T cells (Barrow and Trowsdale, 2008; Anderson and Allen, 2009). There are two main haplotypes, containing 13 LILR genes, one with a 6.7 kb deletion affecting the LILRA3 gene (Norman et al., 2003; Hirayasu et al., 2006). The ligands for some, but not all, LILRs are classical and non-classical HLA-class I molecules, as well as the human CMV HLA-class I homologue, UL18 (Brown et al., 2004; Barrow and Trowsdale, 2008; Anderson and Allen, 2009). A proposed ligand for LILRA4 is CD317, also known as tetherin (Cao et al., 2009). Like the other natural killer inhibitory receptors, the LILRBs transmit negative signals through their ITIM domains, whereas the activating LILRAs interact with ITAM-bearing adaptor proteins such as FcεRIγ to deliver positive signals. LILRB1 is variably expressed on subsets of blood natural killer cells and T cells and more uniformly on B cells and monocytes (Brown et al., 2004; Lanier, 2005; Barrow and Trowsdale, 2008; Anderson and Allen, 2009). Table 5 lists the different LILRs and their ligands.
Table 5.
LILR and ligands
| Receptor | Type | Species | Ligand | References |
|---|---|---|---|---|
| LILRA1 | Activating | Human | HLA-B27 | Allen et al. (2001) |
| LILRA2 | Activating | Human | ? | Chen et al. (2009) |
| LILRA3a | Activating | Human | Classical and non-classical HLA-Class I | Ryu et al. (2011) |
| LILRA4 | Activating | Human | Tetherin (CD317) | Cao et al. (2009) |
| LILRA5a | Activating | Human | ? | Shiroishi et al. (2006a) |
| LILRA6 | Activating | Human | ? | |
| LILRB1a,b | Inhibitory | Human | HLA-class I, HMCV UL18, HLA-F, HLA-G | Cosman et al. (1997), Colonna et al. (1998), Chapman et al. (1999, 2000), Vitale et al. (1999), Willcox et al. (2003) |
| LILRB2a,b | Inhibitory | Human | HLA-class I,HLA-F, HLA-G, | Colonna et al. (1998), Chapman et al. (1999), Willcox et al. (2002), Shiroishi et al. (2006b) |
| LILRB3 | Inhibitory | Human | ? | |
| LILRB4 | Inhibitory | Human | ? | Cheng et al. (2011) |
| LILRB5 | Inhibitory | Human | ? |
a Represents receptors whose crystal structures have been determined.
b Represents receptors whose crystal structures are determined in complex with ligand.
Given the wider expression of LILRs, their most important roles are probably in regulating leukocytes that lack KIR or other inhibitory receptors. Triggering of LILRs by interaction with ligands can modulate the activation status of dendritic cells, and thus their antigen-presenting functions, migration, cytokine secretion profile and capacity to induce or tolerize T cell responses (Chang et al., 2002; Young et al., 2008). LILRB1 mediates inhibition not only of natural killer cell killing and adhesion to target cells but also of T cell receptor signalling and T cell proliferation (Brown et al., 2004; Anderson and Allen, 2009). It binds with low affinity to HLA class I molecules and with >1000-fold higher affinity to human cytomegalovirus protein UL18, which acts as a decoy of LILRB1, suppressing its antiviral responses (Chapman et al., 1999). Alternative splicing of LILR messenger RNAs can generate soluble isoforms, suggesting further potential for regulating immune responses by blockade of inhibitory interactions with HLA-class I molecules (Jones et al., 2009). There is additional evidence that LILRB1 and LILRB2 can compete directly with CD8αα for binding to HLA class I, hence modulating T cell activation (Shiroishi et al., 2003). In keeping with these modulatory effects on immune responses, genetic polymorphisms and deletions in LILRs show association with disease (Brown et al., 2004; Anderson and Allen, 2009; Thomas et al., 2010).
Leukocyte immunoglobulin-like receptor association in multiple sclerosis
Moderately sized Spanish and German studies analysing 225 and 451 patients with relapsing–remitting multiple sclerosis, respectively, suggest disease predisposition by LILRA3 (ILT6) gene deletion (Koch et al., 2005; Ordonez et al., 2009). LILRA3 itself lacks transmembrane and cytoplasmic domains and is thus a potential soluble competitor. The disease-associated 6.7 kb gene deletion in the LILRA3 locus leads to a null LILRA3 allele, with seven of the eight LILRA3 exons being deleted (Torkar et al., 2000; Wilson et al., 2000).
Studies are now beginning to investigate expression differences of LILR and their ligands under inflammatory/autoimmune conditions. There appear to be higher numbers of circulating LILRB1+CD8+ T and LILRB1+ natural killer cells in patients with progressive multiple sclerosis than in patients with relapsing–remitting multiple sclerosis, perhaps suggesting accumulation of end-stage effector/memory T cells or experienced natural killer cells (Martinez-Rodriguez et al., 2010). HLA-G and one of its potential receptors, LILRB1, which are normally absent from the CNS, are reported to be abundantly co-expressed on macrophages and activated microglial cells in multiple sclerosis lesions, possibly suggesting counter-regulation of pathogenic T cells by HLA-G (Wiendl et al., 2005). Also, higher levels of soluble HLA-G have been observed in the CSF of patients with multiple sclerosis than in non-inflammatory controls (Wiendl et al., 2005; Fainardi et al., 2008). However, evidence for expression of HLA-G in tissues other than trophoblast has been questioned (Apps et al., 2008). On the other hand, another LILR receptor, LILRB4 is reduced on blood monocytes in active relapsing–remitting multiple sclerosis. Although its ligand is unidentified, when expressed on antigen presenting cell, LILRB4 can inhibit CD4+ T cell proliferation; hence modulating its expression by such therapeutics as IFNβ might be beneficial (Jensen et al., 2010). These studies are beginning to implicate LILR pathways in multiple sclerosis, although the exact contributions of different LILRs still remains unclear and LILR are expressed on cells other than natural killers.
Taken together, these studies support the view that natural killer cell receptors are involved in regulating autoreactive immune responses in the CNS. We will now discuss direct evidence for the involvement of natural killer cells in multiple sclerosis and recognize mechanisms by which they interact with other immune cells.
Involvement of natural killer cells in multiple sclerosis and experimental autoimmune encephalomyelitis
Accumulating evidence from murine models, ex vivo analysis of natural killer cells in patients with multiple sclerosis in both blood and brain sections and data from human clinical trials strongly implicate natural killer cells in modulating CNS inflammation.
Initiated by immunizing mice or rats with myelin antigens in complete Freund’s adjuvant, EAE shares clinical and neuropathological features with multiple sclerosis (Steinman, 1999; Friese et al., 2006). Several studies suggest that natural killer cells are involved in its regulation. Natural killer cell depletion prior to disease induction led to an increase in EAE severity and mortality. These animals exhibited pronounced cellular infiltrates, CNS inflammation and demyelination (Zhang et al., 1997; Matsumoto et al., 1998; Xu et al., 2005; Hao et al., 2010). There was also increased CD4+ T cell proliferation and production of Th1 cytokines such as IFNγ and TNFα (Zhang et al., 1997). These results imply a protective role for natural killer cells, consistent with the inhibitory effects of bone marrow-derived natural killer cells (from DA rats) on T cell proliferation and cytokine production (e.g. IL10 and IFNγ) (Smeltz et al., 1999). One suggested mechanism is direct killing of syngeneic myelin-specific encephalitogenic T cells, however, the molecular mechanism of this interaction is unclear (Zhang et al., 1997; Xu et al., 2005). Additionally, natural killer cells can themselves produce IFNγ and can promote and influence polarization of Th1 responses (Andoniou et al., 2008).
Recent work suggests that natural killer cells must localize to the CNS to regulate the development of autoimmune responses in EAE; the chemokine (fractalkine) receptor, CX3CR1 is critical for CNS natural killer cell recruitment, but not for that of T cells, natural killer T cells and monocytes/macrophages. Thus, CX3CR1−/− mice, which have fewer natural killer cells infiltrating the CNS, but normal numbers in the periphery, develop more severe EAE with persistent spastic paralysis and increased mortality. The disease phenotype is similar to that observed in natural killer cell depleted CX3CR1+/− mice; emphasizing the importance of locally infiltrating natural killer cells in controlling CNS autoimmunity (Huang et al., 2006). The concomitant increase in myelin-reactive CD4+ Th17 cell responses in the CNS (but not the lymph nodes)—in both settings—suggests that these are normally restrained by natural killer cells (Hao et al., 2010). Conversely, expansion of natural killer cells (by engaging IL2 receptor with IL2-IL2 monoclonal antibody complexes) reduced IL17 production by CD4+ T cells in the CNS and attenuated EAE. This protective effect apparently required natural killer cells in the CNS, as it was not seen in CX3CR1−/− mice. Since their microglia were an important source of Th17 polarizing cytokines in the absence of natural killer cells, perhaps interactions between natural killer cells, microglia and Th17 cells normally determine the magnitude of CNS inflammation in EAE (Hao et al., 2010, 2011).
Additional suggested mechanisms of natural killer cell-mediated control of CNS inflammation include expression of brain-derived neurotrophic factor and neurotrophin 3, which can contribute to neuronal survival and repair (Hammarberg et al., 2000). In line with this, immunomodulators that enhance natural killer cell activity, such as linomide and glatiramer acetate, ameliorated EAE (Karussis et al., 1993a, b; Arnon and Aharoni, 2004; Al-Falahi et al., 2009). Prior injection of glatiramer acetate enhanced killing of autologous immature or even mature dendritic cells by natural killer cells, whether activated or not with IL2 in vitro. So did exposure of human natural killer cells to glatiramer acetate in vitro. Therefore, one possible action of glatiramer acetate in EAE or multiple sclerosis is its enhancement of natural killer cell lysis of dendritic cells that might otherwise present autoantigens to pathogenic T cells (Al-Falahi et al., 2009; Sand et al., 2009).
Contrasting reports suggest that natural killer cells can be pathogenic and exacerbate EAE. Consistent with this, in C57BL/6 mice, myelin oligodendrocyte glycoprotein (MOG)-induced EAE was ameliorated after depletion of natural killer cells. There were parallel decreases in production of IFNγ and TNFα by CD4+ T cells in the draining lymph nodes (but not in the CNS), but, interestingly, IL17 production remained unaltered. There were concomitant changes in the maturational status of dendritic cells and in the T cell receptor Vβ usage of brain T cells (Winkler-Pickett et al., 2008). These discrepancies could reflect distinct natural killer cell localization patterns, or result from technical differences in antibody depletion regimes and doses, methods of immunization and EAE scoring, sub-strains of mice and/or subsets of natural killer cells. Further studies also suggest that the cytokine environment, and interactions of natural killer cells with other adaptive immune cell types, can facilitate the development of autoimmune responses. For example, release of IL18 by macrophages or dendritic cells can lead to increased IFNγ production by natural killer cells, promoting a Th1 response. Indeed, IL18−/− mice are resistant to MOG-induced EAE, an affect attributed to decreased cytotoxicity and IFNγ production by natural killer cells. Furthermore, this resistance was broken by injecting IL18, which also restored the defective Th1 responses if natural killer cells were present (Shi et al., 2000). Similarly, administration of IL21 before EAE induction enhanced inflammatory infiltration into the CNS and increased EAE severity by boosting IFNγ production and natural killer cell function (Vollmer et al., 2005). While these contradictions may eventually be resolved, natural killer cells are clearly important in inflammatory conditions in the CNS.
Human studies and clinical trials
While natural killer cells have been suggested to be present in demyelinating multiple sclerosis lesions (Traugott and Raine, 1984), this finding remains questionable as the antibodies used to identify natural killer cells (for example, Leu-7 or CD57) were not natural killer cell-specific but could identify both subsets of natural killer cells, T cells and possibly also oligodendrocytes (Lanier et al., 1983; McGarry et al., 1983). Similarly, other studies at the time suggested a reduced function or activity of natural killer cells in patients with multiple sclerosis, but these observations were mainly based on cytokine production or responsiveness and cytotoxicity assays done on patient peripheral blood mononuclear cells (Benczur et al., 1980; Uchida et al., 1982; Braakman et al., 1986) and in some cases were also limited in patient sample size (Oger et al., 1988). Natural killer cell numbers were reportedly decreased in peripheral blood of patients; while some studies used multiple cell surface markers (for example, CD3, CD56, CD16 and CD8) to define natural killer cell populations (Munschauer et al., 1995), others were more restricted (for example, CD56 only) in their selection of antibodies used for natural killer cell immunophenotyping (Vranes et al., 1989). Moreover, literature suggesting a relationship between natural killer cell deficiencies and disease status in multiple sclerosis is problematic due to widely different criteria and protocols that have been used to classify natural killer cell frequencies and activity, as well as differences in patient selection. Furthermore, defects in these lymphocyte populations could be an immune manifestation of the ongoing disease in patients. A recent phenotyping study that performed cytometric staining for multiple cell surface markers revealed lower frequencies of circulating CD8lowCD56+CD3−CD4− cells in untreated patients with relapsing–remitting multiple sclerosis or clinically isolated demyelination syndrome than in healthy controls (De Jager et al., 2008). Reduction in natural killer cell function in the periphery has also been correlated with the onset of clinical relapse in patients with multiple sclerosis (Kastrukoff et al., 1998, 2003). As in EAE, CX3CR1 expression on natural killer cells was apparently important in patients with relapsing–remitting or primary-progressive multiple sclerosis, in whom it was lower than in healthy controls. Indeed, high expression levels of CX3CR1 correlates with increased cytotoxicity of these cells. However, the proportions of circulating CX3CR1+ natural killer cells were shown to be high in patients with active disease/acute relapses than in those with stable disease (Infante-Duarte et al., 2005). It is not clear whether their CX3CR1low natural killer cells were defective in cytotoxicity or actively pathogenic. Alternatively, these CX3CR1high and CX3CR1low natural killer cells may represent distinct subsets or stages of natural killer cell maturation (Hamann et al., 2011).
Human natural killer cell subsets can be distinguished not only by their levels of CD56 and CD16 but also by their production of IL10 and IFNγ (NK1) or IL5 and IL13 (NK2). NK2 cells are reportedly increased in the blood during disease remissions. This is seen to correlate with increase in the proportion of natural killer cells expressing CD95 (Fas) on their surface. According to the frequency of CD95+ natural killer cells, patients are divided into CD95+ natural killer-high or CD95+ natural killer-low; natural killer cells from CD95+ natural killer-high patients were proposed to have a higher frequency of memory autoimmune T cells that are normally regulated by their natural killer cells (Takahashi et al., 2001, 2004). Similarly, cytokine-driven proliferation and IFNγ production (though not natural killer cell numbers) were selectively reduced by CD56hiCD16− natural killer cells in the blood of untreated patients with multiple sclerosis (Lunemann et al., 2011).
Various immuno-therapeutics are currently being tested in multiple sclerosis for their safety and efficacy in controlling brain inflammation and preventing further progression. One such is daclizumab, a humanized monoclonal antibody originally given to block the high affinity IL2 receptor α subunit (CD25), so as to inhibit T cell responses. In any event, inhibitions were only marginal, though there was a decline in circulating CD4+ and CD8+ T cells. In phase II clinical trials [initially in combination with interferon β (IFNβ) and then alone], daclizumab significantly inhibited the appearance of total and contrast-enhancing lesions, and improved clinical scores (Bielekova et al., 2004, 2009; Rose et al., 2004, 2007). During therapy with IFNβ or in combination with daclizumab, total circulating natural killer cell numbers declined slightly but there was a marked increase in the proportion of CD56hi natural killer cells (Perini et al., 2000; Saraste et al., 2007a; Vandenbark et al., 2009). The concomitant expansion of circulating CD56hi natural killer cells associated with decreased brain inflammation and with reduced survival of activated T cells. These changes in CD56hi natural killer cells and CD4+ and CD8+ T cells became more pronounced after the patients progressed to taking daclizumab alone (Bielekova et al., 2004, 2006, 2009). While therapies such as glatiramer acetate or IFNβ do not primarily target natural killer cells but have effects on multiple immune cell types, it appears that expansion of these regulatory CD56hi population of natural killer cells is the most important biological effect of daclizumab treatment (Kala et al., 2011; Kieseier, 2011; Martin, 2012). Interestingly, it was recently shown that treatment of patients with relapsing–remitting multiple sclerosis with daclizumab led to a significant expansion of CD56hi natural killer cells not only in the blood, but also in the CSF of treated patients, suggesting that natural killer cells can suppress immune responses directly in the CNS (Bielekova et al., 2011). It is also suggested that these therapies modulate receptor expression on natural killer cells, as demonstrated by the decrease in LILRB1+ natural killer cells and an increase in NKG2A+ natural killer cells following treatment with IFNβ (Martinez-Rodriguez et al., 2011).
The reduction in new contrast-enhancing lesions was confirmed in a recent phase II randomized, double-blind, placebo-controlled trial of additional daclizumab given to patients with active relapsing–remitting multiple sclerosis already receiving IFNβ, and correlated with the daclizumab dose. While the absolute numbers of T, B or total natural killer cells did not change significantly, there was again a 7- to 8-fold increase in the absolute CD56hi natural killer cell numbers (Wynn et al., 2010). In contrast with the above, frequencies of circulating CD4+CD25+Foxp3+ regulatory T cells declined modestly on daclizumab therapy (Oh et al., 2009), as their dependence on IL2 (Malek et al., 2002) might predict. The immunoregulatory potential of this CD56hi subset of natural killer cells is supported independently by their increase in the third trimester of pregnancies in patients with multiple sclerosis, which is when their relapse rates decline. There were concomitant decreases in the proportions of CD16+CD56low natural killer cells in patients’ blood, and in production of IFNγ by their peripheral blood mononuclear cells (Saraste et al., 2007b; Airas et al., 2008).
A recent study demonstrated that expansion of natural killer cells isolated from relapsing–remitting patients (using IL2 complexed with specific monoclonal antibody) enhanced cytokine production and cytotoxic activity from CD56hi and CD56low natural killer cells, respectively. Furthermore, transfer of CD56+ natural killer cells (pretreated with IL2-IL2 monoclonal antibody complexes) from patients with multiple sclerosis ameliorated EAE induced by the transfer of PLP-reactive human T cell lines in the human–mouse chimera model in RAG1−/− γc−/− mice (Hao et al., 2011).
Taken together, these studies indicate important immunoregulatory roles of human natural killer cells in multiple sclerosis. Some of the contradictions might be resolved by further studies on the differences in the natural killer cell subsets examined, and on their particular combinations of inhibitory and activating receptors.
Concluding remarks
While research in multiple sclerosis has focussed on the role of T and B lymphocytes in disease pathogenesis, there is now substantial evidence implicating natural killer cells in regulating tissue damage and autoimmune responses. Studies in both humans and in mouse models propose predisposing as well as protective effects of natural killer cells. It is, however, vital to take account of both the functional variability of their different subsets and the interplay between their receptors and their ligands. Before starting trials of natural killer-cell directed therapies in multiple sclerosis, further work is needed to clarify how natural killer cells can tip the balance between controlled and pathogenic autoimmune responses. The very polymorphic KIR genes that interact with HLA class I molecules have been the focus of recent work. While KIR/HLA interactions represent a rather interesting pathway involving natural killer cells in autoimmunity, these genes’ apparent associations in multiple sclerosis must be interpreted cautiously. The same HLA class I molecules that direct the licensing of a natural killer cell can also determine its responses and activation status. Additionally, the expression of natural killer cell receptors on other cell types (e.g. dendritic cells, monocytes, B cells and T cells) needs to be considered. In summary, there is no doubt that the many permutations of receptors on natural killer cells create numerous opportunities for their involvement in regulating autoimmune responses in multiple sclerosis. However, there is a major need for further mechanistic studies to clarify this complex network of cellular interactions between the innate and the adaptive arms of the immune system.
Funding
Medical Research Council (to G.K. and L.F., laboratory work); Multiple Sclerosis Society (to G.K. and L.F., laboratory work); Medical Research Council (to J.T., laboratory work); Wellcome Trust (to J.T., laboratory work) and National Institute for Health Research, Cambridge Biomedical Research Centre (to J.T., laboratory work, partial).
Acknowledgements
The authors would like to thank Nick Willcox for critical comments and careful reading of the manuscript, and Aiden Haghikia, James Traherne and Olympe Chazara for helpful discussions and comments.
Glossary
Abbreviations
- EAE
experimental autoimmune encephalomyelitis
- HLA
human leukocyte antigen
- IFN
interferon
- IL
interleukin
- ITAM
immunoreceptor tyrosine-based activating motif
- ITIM
immunoreceptor tyrosine-based inhibitory motif
- KIR
killer cell immunoglobulin-like receptor
- LILR
leukocyte immunoglobulin-like receptors
- MHC
major histocompatibility complex
References
- Adams RA, Bauer J, Flick MJ, Sikorski SL, Nuriel T, Lassmann H, et al. The fibrin-derived gamma377-395 peptide inhibits microglia activation and suppresses relapsing paralysis in central nervous system autoimmune disease. J Exp Med. 2007;204:571–82. doi: 10.1084/jem.20061931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Airas L, Saraste M, Rinta S, Elovaara I, Huang YH, Wiendl H. Immunoregulatory factors in multiple sclerosis patients during and after pregnancy: relevance of natural killer cells. Clin Exp Immunol. 2008;151:235–43. doi: 10.1111/j.1365-2249.2007.03555.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Falahi Y, Sand KL, Knudsen E, Damaj BB, Rolin J, Maghazachi AA. Splenic natural killer cell activity in two models of experimental neurodegenerative diseases. J Cell Mol Med. 2009;13:2693–703. doi: 10.1111/j.1582-4934.2008.00640.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen RL, Raine T, Haude A, Trowsdale J, Wilson MJ. Leukocyte receptor complex-encoded immunomodulatory receptors show differing specificity for alternative HLA-B27 structures. J Immunol. 2001;167:5543–7. doi: 10.4049/jimmunol.167.10.5543. [DOI] [PubMed] [Google Scholar]
- Anderson KJ, Allen RL. Regulation of T-cell immunity by leucocyte immunoglobulin-like receptors: innate immune receptors for self on antigen-presenting cells. Immunology. 2009;127:8–17. doi: 10.1111/j.1365-2567.2009.03097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andoniou CE, Coudert JD, Degli-Esposti MA. Killers and beyond: NK-cell-mediated control of immune responses. Eur J Immunol. 2008;38:2938–42. doi: 10.1002/eji.200838882. [DOI] [PubMed] [Google Scholar]
- Anegon I, Cuturi MC, Trinchieri G, Perussia B. Interaction of Fc receptor (CD16) ligands induces transcription of interleukin 2 receptor (CD25) and lymphokine genes and expression of their products in human natural killer cells. J Exp Med. 1988;167:452–72. doi: 10.1084/jem.167.2.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apps R, Gardner L, Moffett A. A critical look at HLA-G. Trends Immunol. 2008;29:313–21. doi: 10.1016/j.it.2008.02.012. [DOI] [PubMed] [Google Scholar]
- Araki M, Kondo T, Gumperz JE, Brenner MB, Miyake S, Yamamura T. Th2 bias of CD4+ NKT cells derived from multiple sclerosis in remission. Int Immunol. 2003;15:279–88. doi: 10.1093/intimm/dxg029. [DOI] [PubMed] [Google Scholar]
- Arnon R, Aharoni R. Mechanism of action of glatiramer acetate in multiple sclerosis and its potential for the development of new applications. Proc Natl Acad Sci USA. 2004;101(Suppl 2):14593–8. doi: 10.1073/pnas.0404887101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnon TI, Achdout H, Levi O, Markel G, Saleh N, Katz G, et al. Inhibition of the NKp30 activating receptor by pp65 of human cytomegalovirus. Nat Immunol. 2005;6:515–23. doi: 10.1038/ni1190. [DOI] [PubMed] [Google Scholar]
- Arnon TI, Lev M, Katz G, Chernobrov Y, Porgador A, Mandelboim O. Recognition of viral hemagglutinins by NKp44 but not by NKp30. Eur J Immunol. 2001;31:2680–9. doi: 10.1002/1521-4141(200109)31:9<2680::aid-immu2680>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- Backstrom E, Chambers BJ, Ho EL, Naidenko OV, Mariotti R, Fremont DH, et al. Natural killer cell-mediated lysis of dorsal root ganglia neurons via RAE1/NKG2D interactions. Eur J Immunol. 2003;33:92–100. doi: 10.1002/immu.200390012. [DOI] [PubMed] [Google Scholar]
- Bailey SL, Schreiner B, McMahon EJ, Miller SD. CNS myeloid DCs presenting endogenous myelin peptides ‘preferentially’ polarize CD4+ T(H)-17 cells in relapsing EAE. Nat Immunol. 2007;8:172–80. doi: 10.1038/ni1430. [DOI] [PubMed] [Google Scholar]
- Bar-Or A, Fawaz L, Fan B, Darlington PJ, Rieger A, Ghorayeb C, et al. Abnormal B-cell cytokine responses a trigger of T-cell-mediated disease in MS? Ann Neurol. 2010;67:452–61. doi: 10.1002/ana.21939. [DOI] [PubMed] [Google Scholar]
- Barber A, Sentman CL. NKG2D receptor regulates human effector T-cell cytokine production. Blood. 2011;117:6571–81. doi: 10.1182/blood-2011-01-329417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrow AD, Trowsdale J. The extended human leukocyte receptor complex: diverse ways of modulating immune responses. Immunol Rev. 2008;224:98–123. doi: 10.1111/j.1600-065X.2008.00653.x. [DOI] [PubMed] [Google Scholar]
- Batoulis H, Addicks K, Kuerten S. Emerging concepts in autoimmune encephalomyelitis beyond the CD4/T(H)1 paradigm. Ann Anat. 2010;192:179–93. doi: 10.1016/j.aanat.2010.06.006. [DOI] [PubMed] [Google Scholar]
- Bauer S, Groh V, Wu J, Steinle A, Phillips JH, Lanier LL, et al. Activation of NK cells and T cells by NKG2D, a receptor for stress-inducible MICA. Science. 1999;285:727–9. doi: 10.1126/science.285.5428.727. [DOI] [PubMed] [Google Scholar]
- Benczur M, Petranyl GG, Palffy G, Varga M, Talas M, Kotsy B, et al. Dysfunction of natural killer cells in multiple sclerosis: a possible pathogenetic factor. Clin Exp Immunol. 1980;39:657–62. [PMC free article] [PubMed] [Google Scholar]
- Bergamaschi L, Leone MA, Fasano ME, Guerini FR, Ferrante D, Bolognesi E, et al. HLA-class I markers and multiple sclerosis susceptibility in the Italian population. Genes Immun. 2010;11:173–80. doi: 10.1038/gene.2009.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berzins SP, Smyth MJ, Baxter AG. Presumed guilty: natural killer T cell defects and human disease. Nature reviews Immunology. 2011;11:131–42. doi: 10.1038/nri2904. [DOI] [PubMed] [Google Scholar]
- Biassoni R, Pessino A, Malaspina A, Cantoni C, Bottino C, Sivori S, et al. Role of amino acid position 70 in the binding affinity of p50.1 and p58.1 receptors for HLA-Cw4 molecules. Eur J Immunol. 1997;27:3095–9. doi: 10.1002/eji.1830271203. [DOI] [PubMed] [Google Scholar]
- Bielekova B, Catalfamo M, Reichert-Scrivner S, Packer A, Cerna M, Waldmann TA, et al. Regulatory CD56(bright) natural killer cells mediate immunomodulatory effects of IL-2Ralpha-targeted therapy (daclizumab) in multiple sclerosis. Proc Natl Acad Sci USA. 2006;103:5941–6. doi: 10.1073/pnas.0601335103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielekova B, Howard T, Packer AN, Richert N, Blevins G, Ohayon J, et al. Effect of anti-CD25 antibody daclizumab in the inhibition of inflammation and stabilization of disease progression in multiple sclerosis. Arch Neurol. 2009;66:483–9. doi: 10.1001/archneurol.2009.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielekova B, Richert N, Herman ML, Ohayon J, Waldmann TA, McFarland H, et al. Intrathecal effects of daclizumab treatment of multiple sclerosis. Neurology. 2011;77:1877–86. doi: 10.1212/WNL.0b013e318239f7ef. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielekova B, Richert N, Howard T, Blevins G, Markovic-Plese S, McCartin J, et al. Humanized anti-CD25 (daclizumab) inhibits disease activity in multiple sclerosis patients failing to respond to interferon beta. Proc Natl Acad Sci USA. 2004;101:8705–8. doi: 10.1073/pnas.0402653101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bix M, Liao NS, Zijlstra M, Loring J, Jaenisch R, Raulet D. Rejection of class I MHC-deficient haemopoietic cells by irradiated MHC-matched mice. Nature. 1991;349:329–31. doi: 10.1038/349329a0. [DOI] [PubMed] [Google Scholar]
- Borrego F, Masilamani M, Marusina AI, Tang X, Coligan JE. The CD94/NKG2 family of receptors: from molecules and cells to clinical relevance. Immunol Res. 2006;35:263–78. doi: 10.1385/IR:35:3:263. [DOI] [PubMed] [Google Scholar]
- Borrego F, Ulbrecht M, Weiss EH, Coligan JE, Brooks AG. Recognition of human histocompatibility leukocyte antigen (HLA)-E complexed with HLA class I signal sequence-derived peptides by CD94/NKG2 confers protection from natural killer cell-mediated lysis. J Exp Med. 1998;187:813–8. doi: 10.1084/jem.187.5.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyington JC, Sun PD. A structural perspective on MHC class I recognition by killer cell immunoglobulin-like receptors. Mol Immunol. 2002;38:1007–21. doi: 10.1016/s0161-5890(02)00030-5. [DOI] [PubMed] [Google Scholar]
- Braakman E, van Tunen A, Meager A, Lucas CJ. Natural cytotoxic activity in multiple sclerosis patients: defects in IL-2/interferon gamma-regulatory circuit. Clin Exp Immunol. 1986;66:285–94. [PMC free article] [PubMed] [Google Scholar]
- Brandt CS, Baratin M, Yi EC, Kennedy J, Gao Z, Fox B, et al. The B7 family member B7-H6 is a tumor cell ligand for the activating natural killer cell receptor NKp30 in humans. J Exp Med. 2009;206:1495–503. doi: 10.1084/jem.20090681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braud VM, Allan DS, O’Callaghan CA, Soderstrom K, D’Andrea A, Ogg GS, et al. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature. 1998;391:795–9. doi: 10.1038/35869. [DOI] [PubMed] [Google Scholar]
- Brodin P, Karre K, Hoglund P. NK cell education: not an on-off switch but a tunable rheostat. Trends Immunol. 2009;30:143–9. doi: 10.1016/j.it.2009.01.006. [DOI] [PubMed] [Google Scholar]
- Brown D, Trowsdale J, Allen R. The LILR family: modulators of innate and adaptive immune pathways in health and disease. Tissue Antigens. 2004;64:215–25. doi: 10.1111/j.0001-2815.2004.00290.x. [DOI] [PubMed] [Google Scholar]
- Campbell KS, Purdy AK. Structure/function of human killer cell immunoglobulin-like receptors: lessons from polymorphisms, evolution, crystal structures and mutations. Immunology. 2011;132:315–25. doi: 10.1111/j.1365-2567.2010.03398.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantoni C, Ponassi M, Biassoni R, Conte R, Spallarossa A, Moretta A, et al. The three-dimensional structure of the human NK cell receptor NKp44, a triggering partner in natural cytotoxicity. Structure. 2003;11:725–34. doi: 10.1016/s0969-2126(03)00095-9. [DOI] [PubMed] [Google Scholar]
- Cao W, Bover L, Cho M, Wen X, Hanabuchi S, Bao M, et al. Regulation of TLR7/9 responses in plasmacytoid dendritic cells by BST2 and ILT7 receptor interaction. J Exp Med. 2009;206:1603–14. doi: 10.1084/jem.20090547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cella M, Fuchs A, Vermi W, Facchetti F, Otero K, Lennerz JK, et al. A human natural killer cell subset provides an innate source of IL-22 for mucosal immunity. Nature. 2009;457:722–5. doi: 10.1038/nature07537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cella M, Longo A, Ferrara GB, Strominger JL, Colonna M. NK3-specific natural killer cells are selectively inhibited by Bw4-positive HLA alleles with isoleucine 80. J Exp Med. 1994;180:1235–42. doi: 10.1084/jem.180.4.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerboni C, Zingoni A, Cippitelli M, Piccoli M, Frati L, Santoni A. Antigen-activated human T lymphocytes express cell-surface NKG2D ligands via an ATM/ATR-dependent mechanism and become susceptible to autologous NK-cell lysis. Blood. 2007;110:606–15. doi: 10.1182/blood-2006-10-052720. [DOI] [PubMed] [Google Scholar]
- Chang CC, Ciubotariu R, Manavalan JS, Yuan J, Colovai AI, Piazza F, et al. Tolerization of dendritic cells by T(S) cells: the crucial role of inhibitory receptors ILT3 and ILT4. Nat Immunol. 2002;3:237–43. doi: 10.1038/ni760. [DOI] [PubMed] [Google Scholar]
- Chapman TL, Heikeman AP, Bjorkman PJ. The inhibitory receptor LIR-1 uses a common binding interaction to recognize class I MHC molecules and the viral homolog UL18. Immunity. 1999;11:603–13. doi: 10.1016/s1074-7613(00)80135-1. [DOI] [PubMed] [Google Scholar]
- Chapman TL, Heikema AP, West AP, Jr, Bjorkman PJ. Crystal structure and ligand binding properties of the D1D2 region of the inhibitory receptor LIR-1 (ILT2) Immunity. 2000;13:727–36. doi: 10.1016/s1074-7613(00)00071-6. [DOI] [PubMed] [Google Scholar]
- Chastain EM, Duncan DS, Rodgers JM, Miller SD. The role of antigen presenting cells in multiple sclerosis. Biochim Biophys Acta. 2011;1812:265–74. doi: 10.1016/j.bbadis.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chazara O, Xiong S, Moffett A. Maternal KIR and fetal HLA-C: a fine balance. J Leukoc Biol. 2011;90:703–16. doi: 10.1189/jlb.0511227. [DOI] [PubMed] [Google Scholar]
- Chen Y, Gao F, Chu F, Peng H, Zong L, Liu Y, et al. Crystal structure of myeloid cell activating receptor leukocyte Ig-like receptor A2 (LILRA2/ILT1/LIR-7) domain swapped dimer: molecular basis for its non-binding to MHC complexes. J Mol Biol. 2009;386:841–53. doi: 10.1016/j.jmb.2009.01.006. [DOI] [PubMed] [Google Scholar]
- Cheng H, Mohammed F, Nam G, Chen Y, Qi J, Garner LI, et al. Crystal structure of leukocyte Ig-like receptor LILRB4 (ILT3/LIR-5/CD85k): a myeloid inhibitory receptor involved in immune tolerance. J Biol Chem. 2011;286:18013–25. doi: 10.1074/jbc.M111.221028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coles AJ, Fox E, Vladic A, Gazda SK, Brinar V, Selmaj KW, et al. Alemtuzumab versus interferon beta-1a in early relapsing-remitting multiple sclerosis: post-hoc and subset analyses of clinical efficacy outcomes. Lancet Neurol. 2011;10:338–48. doi: 10.1016/S1474-4422(11)70020-5. [DOI] [PubMed] [Google Scholar]
- Colonna M, Borsellino G, Falco M, Ferrara GB, Strominger JL. HLA-C is the inhibitory ligand that determines dominant resistance to lysis by NK1- and NK2-specific natural killer cells. Proc Natl Acad Sci USA. 1993;90:12000–4. doi: 10.1073/pnas.90.24.12000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colonna M, Samaridis J, Cella M, Angman L, Allen RL, O’Callaghan CA, et al. Human myelomonocytic cells express an inhibitory receptor for classical and nonclassical MHC class I molecules. J Immunol. 1998;160:3096–100. [PubMed] [Google Scholar]
- Comabella M, Montalban X, Munz C, Lunemann JD. Targeting dendritic cells to treat multiple sclerosis. Nat Rev Neurol. 2010;6:499–507. doi: 10.1038/nrneurol.2010.112. [DOI] [PubMed] [Google Scholar]
- Compston A, Coles A. Multiple sclerosis. Lancet. 2008;372:1502–17. doi: 10.1016/S0140-6736(08)61620-7. [DOI] [PubMed] [Google Scholar]
- Confavreux C, Vukusic S. Natural history of multiple sclerosis: a unifying concept. Brain. 2006;129 (Pt 3):606–16. doi: 10.1093/brain/awl007. [DOI] [PubMed] [Google Scholar]
- Cooper MA, Colonna M, Yokoyama WM. Hidden talents of natural killers: NK cells in innate and adaptive immunity. EMBO Rep. 2009a;10:1103–10. doi: 10.1038/embor.2009.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper MA, Elliott JM, Keyel PA, Yang L, Carrero JA, Yokoyama WM. Cytokine-induced memory-like natural killer cells. Proc Natl Acad Sci USA. 2009b;106:1915–9. doi: 10.1073/pnas.0813192106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper MA, Fehniger TA, Turner SC, Chen KS, Ghaheri BA, Ghayur T, et al. Human natural killer cells: a unique innate immunoregulatory role for the CD56(bright) subset. Blood. 2001;97:3146–51. doi: 10.1182/blood.v97.10.3146. [DOI] [PubMed] [Google Scholar]
- Corriveau RA, Huh GS, Shatz CJ. Regulation of class I MHC gene expression in the developing and mature CNS by neural activity. Neuron. 1998;21:505–20. doi: 10.1016/s0896-6273(00)80562-0. [DOI] [PubMed] [Google Scholar]
- Cosman D, Fanger N, Borges L, Kubin M, Chin W, Peterson L, et al. A novel immunoglobulin superfamily receptor for cellular and viral MHC class I molecules. Immunity. 1997;7:273–82. doi: 10.1016/s1074-7613(00)80529-4. [DOI] [PubMed] [Google Scholar]
- Darlington PJ, Podjaski C, Horn KE, Costantino S, Blain M, Saikali P, et al. Innate immune-mediated neuronal injury consequent to loss of astrocytes. J Neuropathol Exp Neurol. 2008;67:590–9. doi: 10.1097/NEN.0b013e3181772cf6. [DOI] [PubMed] [Google Scholar]
- De Jager PL, Jia XM, Wang J, de Bakker PIW, Ottoboni L, Aggarwal NT, et al. Meta-analysis of genome scans and replication identify CD6, IRF8 and TNFRSF1A as new multiple sclerosis susceptibility loci. Nat Genet. 2009;41:776–U26. doi: 10.1038/ng.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jager PL, Rossin E, Pyne S, Tamayo P, Ottoboni L, Viglietta V, et al. Cytometric profiling in multiple sclerosis uncovers patient population structure and a reduction of CD8low cells. Brain. 2008;131 (Pt 7):1701–11. doi: 10.1093/brain/awn118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diefenbach A, Jamieson AM, Liu SD, Shastri N, Raulet DH. Ligands for the murine NKG2D receptor: expression by tumor cells and activation of NK cells and macrophages. Nat Immunol. 2000;1:119–26. doi: 10.1038/77793. [DOI] [PubMed] [Google Scholar]
- Durrenberger PF, Ettorre A, Kamel F, Webb LV, Sim M, Nicholas RS, et al. Innate Immunity in multiple sclerosis white matter lesions: expression of natural cytotoxicity triggering receptor 1 (NCR1) J Neuroinflammation. 2012;9:1. doi: 10.1186/1742-2094-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich J, Sankoff D, Nadeau JH. Synteny conservation and chromosome rearrangements during mammalian evolution. Genetics. 1997;147:289–96. doi: 10.1093/genetics/147.1.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott JM, Wahle JA, Yokoyama WM. MHC class I-deficient natural killer cells acquire a licensed phenotype after transfer into an MHC class I-sufficient environment. J Exp Med. 2010;207:2073–9. doi: 10.1084/jem.20100986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkeller-Yusel F, Hulstaart F, Hannet I, Isenberg D, Lydyard P. Lymphocyte subsets in a large cohort of patients with systemic lupus erythematosus. Lupus. 1993;2:227–31. doi: 10.1177/096120339300200404. [DOI] [PubMed] [Google Scholar]
- Fainardi E, Rizzo R, Melchiorri L, Stignani M, Castellazzi M, Tamborino C, et al. CSF levels of soluble HLA-G and Fas molecules are inversely associated to MRI evidence of disease activity in patients with relapsing-remitting multiple sclerosis. Mult Scler. 2008;14:446–54. doi: 10.1177/1352458507085137. [DOI] [PubMed] [Google Scholar]
- Fan QR, Long EO, Wiley DC. Crystal structure of the human natural killer cell inhibitory receptor KIR2DL1-HLA-Cw4 complex. Nat Immunol. 2001;2:452–60. doi: 10.1038/87766. [DOI] [PubMed] [Google Scholar]
- Fan QR, Mosyak L, Winter CC, Wagtmann N, Long EO, Wiley DC. Structure of the inhibitory receptor for human natural killer cells resembles haematopoietic receptors. Nature. 1997;389:96–100. doi: 10.1038/38028. [DOI] [PubMed] [Google Scholar]
- Fang M, Orr MT, Spee P, Egebjerg T, Lanier LL, Sigal LJ. CD94 is essential for NK cell-mediated resistance to a lethal viral disease. Immunity. 2011;34:579–89. doi: 10.1016/j.immuni.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauriat C, Ivarsson MA, Ljunggren HG, Malmberg KJ, Michaelsson J. Education of human natural killer cells by activating killer cell immunoglobulin-like receptors. Blood. 2010;115:1166–74. doi: 10.1182/blood-2009-09-245746. [DOI] [PubMed] [Google Scholar]
- Fernandez-Morera JL, Rodriguez-Rodero S, Lahoz C, Tunon A, Astudillo A, Garcia-Suarez O, et al. Soluble MHC class I chain-related protein B serum levels correlate with disease activity in relapsing-remitting multiple sclerosis. Hum Immunol. 2008;69:235–40. doi: 10.1016/j.humimm.2008.01.021. [DOI] [PubMed] [Google Scholar]
- Fogdell-Hahn A, Ligers A, Gronning M, Hillert J, Olerup O. Multiple sclerosis: a modifying influence of HLA class I genes in an HLA class II associated autoimmune disease. Tissue Antigens. 2000;55:140–8. doi: 10.1034/j.1399-0039.2000.550205.x. [DOI] [PubMed] [Google Scholar]
- Foster CE, Colonna M, Sun PD. Crystal structure of the human natural killer (NK) cell activating receptor NKp46 reveals structural relationship to other leukocyte receptor complex immunoreceptors. J Biol Chem. 2003;278:46081–6. doi: 10.1074/jbc.M308491200. [DOI] [PubMed] [Google Scholar]
- French AR, Yokoyama WM. Natural killer cells and autoimmunity. Arthritis Res Ther. 2004;6:8–14. doi: 10.1186/ar1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey M, Packianathan NB, Fehniger TA, Ross ME, Wang WC, Stewart CC, et al. Differential expression and function of L-selectin on CD56bright and CD56dim natural killer cell subsets. J Immunol. 1998;161:400–8. [PubMed] [Google Scholar]
- Friese MA, Jakobsen KB, Friis L, Etzensperger R, Craner MJ, McMahon RM, et al. Opposing effects of HLA class I molecules in tuning autoreactive CD8+ T cells in multiple sclerosis. Nat Med. 2008;14:1227–35. doi: 10.1038/nm.1881. [DOI] [PubMed] [Google Scholar]
- Friese MA, Montalban X, Willcox N, Bell JI, Martin R, Fugger L. The value of animal models for drug development in multiple sclerosis. Brain. 2006;129 (Pt 8):1940–52. doi: 10.1093/brain/awl083. [DOI] [PubMed] [Google Scholar]
- Fugger L, Friese MA, Bell JI. From genes to function: the next challenge to understanding multiple sclerosis. Nat Rev Immunol. 2009;9:408–17. doi: 10.1038/nri2554. [DOI] [PubMed] [Google Scholar]
- Fusco C, Guerini FR, Nocera G, Ventrella G, Caputo D, Valentino MA, et al. KIRs and their HLA ligands in remitting-relapsing multiple sclerosis. J Neuroimmunol. 2010;229:232–7. doi: 10.1016/j.jneuroim.2010.08.004. [DOI] [PubMed] [Google Scholar]
- Galazka G, Jurewicz A, Orlowski W, Stasiolek M, Brosnan CF, Raine CS, et al. EAE tolerance induction with Hsp70-peptide complexes depends on H60 and NKG2D activity. J Immunol. 2007;179:4503–12. doi: 10.4049/jimmunol.179.7.4503. [DOI] [PubMed] [Google Scholar]
- Galazka G, Stasiolek M, Walczak A, Jurewicz A, Zylicz A, Brosnan CF, et al. Brain-derived heat shock protein 70-peptide complexes induce NK cell-dependent tolerance to experimental autoimmune encephalomyelitis. J Immunol. 2006;176:1588–99. doi: 10.4049/jimmunol.176.3.1588. [DOI] [PubMed] [Google Scholar]
- Gao Z, Tsirka SE. Animal models of MS reveal multiple roles of microglia in disease pathogenesis. Neurol Res Int. 2011;2011:383087. doi: 10.1155/2011/383087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazit R, Gruda R, Elboim M, Arnon TI, Katz G, Achdout H, et al. Lethal influenza infection in the absence of the natural killer cell receptor gene Ncr1. Nat Immunol. 2006;7:517–23. doi: 10.1038/ni1322. [DOI] [PubMed] [Google Scholar]
- Gomez-Lozano N, Gardiner CM, Parham P, Vilches C. Some human KIR haplotypes contain two KIR2DL5 genes: KIR2DL5A and KIR2DL5B. Immunogenetics. 2002;54:314–9. doi: 10.1007/s00251-002-0476-2. [DOI] [PubMed] [Google Scholar]
- Goverman J. Autoimmune T cell responses in the central nervous system. Nat Rev Immunol. 2009;9:393–407. doi: 10.1038/nri2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graef T, Moesta AK, Norman PJ, Abi-Rached L, Vago L, Older Aguilar AM, et al. KIR2DS4 is a product of gene conversion with KIR3DL2 that introduced specificity for HLA-A*11 while diminishing avidity for HLA-C. J Exp Med. 2009;206:2557–72. doi: 10.1084/jem.20091010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregersen JW, Kranc KR, Ke X, Svendsen P, Madsen LS, Thomsen AR, et al. Functional epistasis on a common MHC haplotype associated with multiple sclerosis. Nature. 2006;443:574–7. doi: 10.1038/nature05133. [DOI] [PubMed] [Google Scholar]
- Gregory TR. Synergy between sequence and size in large-scale genomics. Nat Rev Genet. 2005;6:699–708. doi: 10.1038/nrg1674. [DOI] [PubMed] [Google Scholar]
- Greter M, Heppner FL, Lemos MP, Odermatt BM, Goebels N, Laufer T, et al. Dendritic cells permit immune invasion of the CNS in an animal model of multiple sclerosis. Nat Med. 2005;11:328–34. doi: 10.1038/nm1197. [DOI] [PubMed] [Google Scholar]
- Groh V, Rhinehart R, Randolph-Habecker J, Topp MS, Riddell SR, Spies T. Costimulation of CD8alphabeta T cells by NKG2D via engagement by MIC induced on virus-infected cells. Nat Immunol. 2001;2:255–60. doi: 10.1038/85321. [DOI] [PubMed] [Google Scholar]
- Groh V, Wu J, Yee C, Spies T. Tumour-derived soluble MIC ligands impair expression of NKG2D and T-cell activation. Nature. 2002;419:734–8. doi: 10.1038/nature01112. [DOI] [PubMed] [Google Scholar]
- Guethlein LA, Flodin LR, Adams EJ, Parham P. NK cell receptors of the orangutan (Pongo pygmaeus): a pivotal species for tracking the coevolution of killer cell Ig-like receptors with MHC-C. J Immunol. 2002;169:220–9. doi: 10.4049/jimmunol.169.1.220. [DOI] [PubMed] [Google Scholar]
- Gumperz JE, Litwin V, Phillips JH, Lanier LL, Parham P. The Bw4 public epitope of HLA-B molecules confers reactivity with natural killer cell clones that express NKB1, a putative HLA receptor. J Exp Med. 1995;181:1133–44. doi: 10.1084/jem.181.3.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gur C, Enk J, Kassem SA, Suissa Y, Magenheim J, Stolovich-Rain M, et al. Recognition and killing of human and murine pancreatic beta cells by the NK receptor NKp46. J Immunol. 2011;187:3096–103. doi: 10.4049/jimmunol.1101269. [DOI] [PubMed] [Google Scholar]
- Gur C, Porgador A, Elboim M, Gazit R, Mizrahi S, Stern-Ginossar N, et al. The activating receptor NKp46 is essential for the development of type 1 diabetes. Nat Immunol. 2010;11:121–8. doi: 10.1038/ni.1834. [DOI] [PubMed] [Google Scholar]
- Hafler DA, Compston A, Sawcer S, Lander ES, Daly MJ, De Jager PL, et al. Risk alleles for multiple sclerosis identified by a genomewide study. N Engl J Med. 2007;357:851–62. doi: 10.1056/NEJMoa073493. [DOI] [PubMed] [Google Scholar]
- Hamann I, Unterwalder N, Cardona AE, Meisel C, Zipp F, Ransohoff RM, et al. Analyses of phenotypic and functional characteristics of CX3CR1-expressing natural killer cells. Immunology. 2011;133:62–73. doi: 10.1111/j.1365-2567.2011.03409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammarberg H, Lidman O, Lundberg C, Eltayeb SY, Gielen AW, Muhallab S, et al. Neuroprotection by encephalomyelitis: rescue of mechanically injured neurons and neurotrophin production by CNS-infiltrating T and natural killer cells. J Neurosci. 2000;20:5283–91. doi: 10.1523/JNEUROSCI.20-14-05283.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansasuta P, Dong T, Thananchai H, Weekes M, Willberg C, Aldemir H, et al. Recognition of HLA-A3 and HLA-A11 by KIR3DL2 is peptide-specific. Eur J Immunol. 2004;34:1673–9. doi: 10.1002/eji.200425089. [DOI] [PubMed] [Google Scholar]
- Hao J, Campagnolo D, Liu R, Piao W, Shi S, Hu B, et al. Interleukin-2/interleukin-2 antibody therapy induces target organ natural killer cells that inhibit central nervous system inflammation. Ann Neurol. 2011;69:721–34. doi: 10.1002/ana.22339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao J, Liu R, Piao W, Zhou Q, Vollmer TL, Campagnolo DI, et al. Central nervous system (CNS)-resident natural killer cells suppress Th17 responses and CNS autoimmune pathology. J Exp Med. 2010;207:1907–21. doi: 10.1084/jem.20092749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbo HF, Lie BA, Sawcer S, Celius EG, Dai KZ, Oturai A, et al. Genes in the HLA class I region may contribute to the HLA class II-associated genetic susceptibility to multiple sclerosis. Tissue Antigens. 2004;63:237–47. doi: 10.1111/j.0001-2815.2004.00173.x. [DOI] [PubMed] [Google Scholar]
- Hauser SL, Waubant E, Arnold DL, Vollmer T, Antel J, Fox RJ, et al. B-cell depletion with rituximab in relapsing-remitting multiple sclerosis. N Engl J Med. 2008;358:676–88. doi: 10.1056/NEJMoa0706383. [DOI] [PubMed] [Google Scholar]
- Healy BC, Liguori M, Tran D, Chitnis T, Glanz B, Wolfish C, et al. HLA B*44: protective effects in MS susceptibility and MRI outcome measures. Neurology. 2010;75:634–40. doi: 10.1212/WNL.0b013e3181ed9c9c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heppner FL, Greter M, Marino D, Falsig J, Raivich G, Hovelmeyer N, et al. Experimental autoimmune encephalomyelitis repressed by microglial paralysis. Nat Med. 2005;11:146–52. doi: 10.1038/nm1177. [DOI] [PubMed] [Google Scholar]
- Hershberger KL, Kurian J, Korber BT, Letvin NL. Killer cell immunoglobulin-like receptors (KIR) of the African-origin sabaeus monkey: evidence for recombination events in the evolution of KIR. Eur J Immunol. 2005;35:922–35. doi: 10.1002/eji.200425408. [DOI] [PubMed] [Google Scholar]
- Hirayasu K, Ohashi J, Kashiwase K, Takanashi M, Satake M, Tokunaga K, et al. Long-term persistence of both functional and non-functional alleles at the leukocyte immunoglobulin-like receptor A3 (LILRA3) locus suggests balancing selection. Hum Genet. 2006;119:436–43. doi: 10.1007/s00439-006-0152-y. [DOI] [PubMed] [Google Scholar]
- Hoare HL, Sullivan LC, Clements CS, Ely LK, Beddoe T, Henderson KN, et al. Subtle changes in peptide conformation profoundly affect recognition of the non-classical MHC class I molecule HLA-E by the CD94-NKG2 natural killer cell receptors. J Mol Biol. 2008;377:1297–303. doi: 10.1016/j.jmb.2008.01.098. [DOI] [PubMed] [Google Scholar]
- Hoftberger R, Aboul-Enein F, Brueck W, Lucchinetti C, Rodriguez M, Schmidbauer M, et al. Expression of major histocompatibility complex class I molecules on the different cell types in multiple sclerosis lesions. Brain Pathol. 2004;14:43–50. doi: 10.1111/j.1750-3639.2004.tb00496.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoglund P, Brodin P. Current perspectives of natural killer cell education by MHC class I molecules. Nat Rev Immunol. 2010;10:724–34. doi: 10.1038/nri2835. [DOI] [PubMed] [Google Scholar]
- Hoglund P, Ohlen C, Carbone E, Franksson L, Ljunggren HG, Latour A, et al. Recognition of beta 2-microglobulin-negative (beta 2m-) T-cell blasts by natural killer cells from normal but not from beta 2m- mice: nonresponsiveness controlled by beta 2m- bone marrow in chimeric mice. Proc Natl Acad Sci USA. 1991;88:10332–6. doi: 10.1073/pnas.88.22.10332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu KC, Liu XR, Selvakumar A, Mickelson E, O’Reilly RJ, Dupont B. Killer Ig-like receptor haplotype analysis by gene content: evidence for genomic diversity with a minimum of six basic framework haplotypes, each with multiple subsets. J Immunol. 2002;169:5118–29. doi: 10.4049/jimmunol.169.9.5118. [DOI] [PubMed] [Google Scholar]
- Huang D, Shi FD, Jung S, Pien GC, Wang J, Salazar-Mather TP, et al. The neuronal chemokine CX3CL1/fractalkine selectively recruits NK cells that modify experimental autoimmune encephalomyelitis within the central nervous system. FASEB J. 2006;20:896–905. doi: 10.1096/fj.05-5465com. [DOI] [PubMed] [Google Scholar]
- Huard B, Karlsson L. KIR expression on self-reactive CD8+ T cells is controlled by T-cell receptor engagement. Nature. 2000;403:325–8. doi: 10.1038/35002105. [DOI] [PubMed] [Google Scholar]
- Huntington ND, Vosshenrich CA, Di Santo JP. Developmental pathways that generate natural-killer-cell diversity in mice and humans. Nat Rev Immunol. 2007;7:703–14. doi: 10.1038/nri2154. [DOI] [PubMed] [Google Scholar]
- Infante-Duarte C, Weber A, Kratzschmar J, Prozorovski T, Pikol S, Hamann I, et al. Frequency of blood CX3CR1-positive natural killer cells correlates with disease activity in multiple sclerosis patients. FASEB J. 2005;19:1902–4. doi: 10.1096/fj.05-3832fje. [DOI] [PubMed] [Google Scholar]
- Jacobs R, Hintzen G, Kemper A, Beul K, Kempf S, Behrens G, et al. CD56bright cells differ in their KIR repertoire and cytotoxic features from CD56dim NK cells. Eur J Immunol. 2001;31:3121–7. doi: 10.1002/1521-4141(2001010)31:10<3121::aid-immu3121>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- Jahng AW, Maricic I, Pedersen B, Burdin N, Naidenko O, Kronenberg M, et al. Activation of natural killer T cells potentiates or prevents experimental autoimmune encephalomyelitis. J Exp Med. 2001;194:1789–99. doi: 10.1084/jem.194.12.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamil KM, Khakoo SI. KIR/HLA interactions and pathogen immunity. J Biomed Biotechnol. 2011;2011:298348. doi: 10.1155/2011/298348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelcic I, Hsu KC, Kakalacheva K, Breiden P, Dupont B, Uhrberg M, et al. Killer immunoglobulin-like receptor locus polymorphisms in multiple sclerosis. Mult Scler. 2011 doi: 10.1177/1352458511431726. Advance Access published on December 20, 2011, doi:10.1177/1352458511431726. [DOI] [PubMed] [Google Scholar]
- Jensen MA, Yanowitch RN, Reder AT, White DM, Arnason BG. Immunoglobulin-like transcript 3, an inhibitor of T cell activation, is reduced on blood monocytes during multiple sclerosis relapses and is induced by interferon beta-1b. Mult Scler. 2010;16:30–8. doi: 10.1177/1352458509352794. [DOI] [PubMed] [Google Scholar]
- Jiang W, Johnson C, Jayaraman J, Trowsdale J, Traherne JA. Genome Res. 2012. Copy number variation leads to considerable diversity for B but not A haplotypes of the human KIR genes encoding NK cell receptors. in Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joncker NT, Raulet DH. Regulation of NK cell responsiveness to achieve self-tolerance and maximal responses to diseased target cells. Immunol Rev. 2008;224:85–97. doi: 10.1111/j.1600-065X.2008.00658.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joncker NT, Shifrin N, Delebecque F, Raulet DH. Mature natural killer cells reset their responsiveness when exposed to an altered MHC environment. J Exp Med. 2010;207:2065–72. doi: 10.1084/jem.20100570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DC, Roghanian A, Brown DP, Chang C, Allen RL, Trowsdale J, et al. Alternative mRNA splicing creates transcripts encoding soluble proteins from most LILR genes. Eur J Immunol. 2009;39:3195–206. doi: 10.1002/eji.200839080. [DOI] [PubMed] [Google Scholar]
- Joyce MG, Tran P, Zhuravleva MA, Jaw J, Colonna M, Sun PD. Crystal structure of human natural cytotoxicity receptor NKp30 and identification of its ligand binding site. Proc Natl Acad Sci USA. 2011;108:6223–8. doi: 10.1073/pnas.1100622108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser BK, Barahmand-Pour F, Paulsene W, Medley S, Geraghty DE, Strong RK. Interactions between NKG2x immunoreceptors and HLA-E ligands display overlapping affinities and thermodynamics. J Immunol. 2005;174:2878–84. doi: 10.4049/jimmunol.174.5.2878. [DOI] [PubMed] [Google Scholar]
- Kala M, Miravalle A, Vollmer T. Recent insights into the mechanism of action of glatiramer acetate. J Neuroimmunol. 2011;235:9–17. doi: 10.1016/j.jneuroim.2011.01.009. [DOI] [PubMed] [Google Scholar]
- Karlhofer FM, Ribaudo RK, Yokoyama WM. MHC class I alloantigen specificity of Ly-49+ IL-2-activated natural killer cells. Nature. 1992;358:66–70. doi: 10.1038/358066a0. [DOI] [PubMed] [Google Scholar]
- Karre K, Ljunggren HG, Piontek G, Kiessling R. Selective rejection of H-2-deficient lymphoma variants suggests alternative immune defence strategy. Nature. 1986;319:675–8. doi: 10.1038/319675a0. [DOI] [PubMed] [Google Scholar]
- Karussis DM, Lehmann D, Slavin S, Vourka-Karussis U, Mizrachi-Koll R, Ovadia H, et al. Inhibition of acute, experimental autoimmune encephalomyelitis by the synthetic immunomodulator linomide. Ann Neurol. 1993a;34:654–60. doi: 10.1002/ana.410340506. [DOI] [PubMed] [Google Scholar]
- Karussis DM, Lehmann D, Slavin S, Vourka-Karussis U, Mizrachi-Koll R, Ovadia H, et al. Treatment of chronic-relapsing experimental autoimmune encephalomyelitis with the synthetic immunomodulator linomide (quinoline-3-carboxamide) Proc Natl Acad Sci USA. 1993b;90:6400–4. doi: 10.1073/pnas.90.14.6400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastrukoff LF, Lau A, Wee R, Zecchini D, White R, Paty DW. Clinical relapses of multiple sclerosis are associated with ‘novel’ valleys in natural killer cell functional activity. J Neuroimmunol. 2003;145:103–14. doi: 10.1016/j.jneuroim.2003.10.001. [DOI] [PubMed] [Google Scholar]
- Kastrukoff LF, Morgan NG, Zecchini D, White R, Petkau AJ, Satoh J, et al. A role for natural killer cells in the immunopathogenesis of multiple sclerosis. J Neuroimmunol. 1998;86:123–33. doi: 10.1016/s0165-5728(98)00014-9. [DOI] [PubMed] [Google Scholar]
- Katz G, Markel G, Mizrahi S, Arnon TI, Mandelboim O. Recognition of HLA-Cw4 but not HLA-Cw6 by the NK cell receptor killer cell Ig-like receptor two-domain short tail number 4. J Immunol. 2001;166:7260–7. doi: 10.4049/jimmunol.166.12.7260. [DOI] [PubMed] [Google Scholar]
- Khakoo SI, Rajalingam R, Shum BP, Weidenbach K, Flodin L, Muir DG, et al. Rapid evolution of NK cell receptor systems demonstrated by comparison of chimpanzees and humans. Immunity. 2000;12:687–98. doi: 10.1016/s1074-7613(00)80219-8. [DOI] [PubMed] [Google Scholar]
- Kieseier BC. The mechanism of action of interferon-beta in relapsing multiple sclerosis. CNS Drugs. 2011;25:491–502. doi: 10.2165/11591110-000000000-00000. [DOI] [PubMed] [Google Scholar]
- Kim S, Poursine-Laurent J, Truscott SM, Lybarger L, Song YJ, Yang L, et al. Licensing of natural killer cells by host major histocompatibility complex class I molecules. Nature. 2005;436:709–13. doi: 10.1038/nature03847. [DOI] [PubMed] [Google Scholar]
- Koch S, Goedde R, Nigmatova V, Epplen JT, Muller N, de Seze J, et al. Association of multiple sclerosis with ILT6 deficiency. Genes Immun. 2005;6:445–7. doi: 10.1038/sj.gene.6364187. [DOI] [PubMed] [Google Scholar]
- Kulkarni S, Martin MP, Carrington M. The Yin and Yang of HLA and KIR in human disease. Semin Immunol. 2008;20:343–52. doi: 10.1016/j.smim.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanier LL. NK cell recognition. Annu Rev Immunol. 2005;23:225–74. doi: 10.1146/annurev.immunol.23.021704.115526. [DOI] [PubMed] [Google Scholar]
- Lanier LL. Up on the tightrope: natural killer cell activation and inhibition. Nat Immunol. 2008;9:495–502. doi: 10.1038/ni1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanier LL, Le AM, Civin CI, Loken MR, Phillips JH. The relationship of CD16 (Leu-11) and Leu-19 (NKH-1) antigen expression on human peripheral blood NK cells and cytotoxic T lymphocytes. J Immunol. 1986a;136:4480–6. [PubMed] [Google Scholar]
- Lanier LL, Phillips JH, Hackett J, Jr, Tutt M, Kumar V. Natural killer cells: definition of a cell type rather than a function. J Immunol. 1986b;137:2735–9. [PubMed] [Google Scholar]
- Lanier LL, Le AM, Phillips JH, Warner NL, Babcock GF. Subpopulations of human natural killer cells defined by expression of the Leu-7 (HNK-1) and Leu-11 (NK-15) antigens. J Immunol. 1983;131:1789–96. [PubMed] [Google Scholar]
- Leavenworth JW, Schellack C, Kim HJ, Lu L, Spee P, Cantor H. Analysis of the cellular mechanism underlying inhibition of EAE after treatment with anti-NKG2A F(ab’)2. Proc Natl Acad Sci USA. 2010;107:2562–7. doi: 10.1073/pnas.0914732107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N, Llano M, Carretero M, Ishitani A, Navarro F, Lopez-Botet M, et al. HLA-E is a major ligand for the natural killer inhibitory receptor CD94/NKG2A. Proc Natl Acad Sci USA. 1998;95:5199–204. doi: 10.1073/pnas.95.9.5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Pascal V, Martin MP, Carrington M, Anderson SK. Genetic control of variegated KIR gene expression: polymorphisms of the bi-directional KIR3DL1 promoter are associated with distinct frequencies of gene expression. PLoS Genet. 2008;4:e1000254. doi: 10.1371/journal.pgen.1000254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Morris DL, Willcox BE, Steinle A, Spies T, Strong RK. Complex structure of the activating immunoreceptor NKG2D and its MHC class I-like ligand MICA. Nat Immunol. 2001;2:443–51. doi: 10.1038/87757. [DOI] [PubMed] [Google Scholar]
- Li Y, Wang Q, Mariuzza RA. Structure of the human activating natural cytotoxicity receptor NKp30 bound to its tumor cell ligand B7-H6. J Exp Med. 2011;208:703–14. doi: 10.1084/jem.20102548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao NS, Bix M, Zijlstra M, Jaenisch R, Raulet D. MHC class I deficiency: susceptibility to natural killer (NK) cells and impaired NK activity. Science. 1991;253:199–202. doi: 10.1126/science.1853205. [DOI] [PubMed] [Google Scholar]
- Lieberman J. Anatomy of a murder: how cytotoxic T cells and NK cells are activated, develop, and eliminate their targets. Immunol Rev. 2010;235:5–9. doi: 10.1111/j.0105-2896.2010.00914.x. [DOI] [PubMed] [Google Scholar]
- Link J, Lorentzen AR, Kockum I, Duvefelt K, Lie BA, Celius EG, et al. Two HLA class I genes independently associated with multiple sclerosis. J Neuroimmunol. 2010;226:172–6. doi: 10.1016/j.jneuroim.2010.07.006. [DOI] [PubMed] [Google Scholar]
- Ljunggren HG, Karre K. In search of the ‘missing self’: MHC molecules and NK cell recognition. Immunol Today. 1990;11:237–44. doi: 10.1016/0167-5699(90)90097-s. [DOI] [PubMed] [Google Scholar]
- Lorentzen AR, Karlsen TH, Olsson M, Smestad C, Mero IL, Woldseth B, et al. Killer immunoglobulin-like receptor ligand HLA-Bw4 protects against multiple sclerosis. Ann Neurol. 2009;65:658–66. doi: 10.1002/ana.21695. [DOI] [PubMed] [Google Scholar]
- Lu L, Ikizawa K, Hu D, Werneck MB, Wucherpfennig KW, Cantor H. Regulation of activated CD4+ T cells by NK cells via the Qa-1-NKG2A inhibitory pathway. Immunity. 2007;26:593–604. doi: 10.1016/j.immuni.2007.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L, Kim HJ, Werneck MB, Cantor H. Regulation of CD8+ regulatory T cells: Interruption of the NKG2A-Qa-1 interaction allows robust suppressive activity and resolution of autoimmune disease. Proc Natl Acad Sci USA. 2008;105:19420–5. doi: 10.1073/pnas.0810383105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luci C, Reynders A, Ivanov II, Cognet C, Chiche L, Chasson L, et al. Influence of the transcription factor RORgammat on the development of NKp46+ cell populations in gut and skin. Nat Immunol. 2009;10:75–82. doi: 10.1038/ni.1681. [DOI] [PubMed] [Google Scholar]
- Lunemann A, Lunemann JD, Roberts S, Messmer B, Barreira da Silva R, Raine CS, et al. Human NK cells kill resting but not activated microglia via NKG2D- and NKp46-mediated recognition. J Immunol. 2008;181:6170–7. doi: 10.4049/jimmunol.181.9.6170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunemann A, Tackenberg B, DeAngelis T, da Silva RB, Messmer B, Vanoaica LD, et al. Impaired IFN-gamma production and proliferation of NK cells in multiple sclerosis. Int Immunol. 2011;23:139–48. doi: 10.1093/intimm/dxq463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen LS, Andersson EC, Jansson L, krogsgaard M, Andersen CB, Engberg J, et al. A humanized model for multiple sclerosis using HLA-DR2 and a human T-cell receptor. Nat Genet. 1999;23:343–7. doi: 10.1038/15525. [DOI] [PubMed] [Google Scholar]
- Maenaka K, Juji T, Stuart DI, Jones EY. Crystal structure of the human p58 killer cell inhibitory receptor (KIR2DL3) specific for HLA-Cw3-related MHC class I. Structure. 1999;7:391–8. doi: 10.1016/s0969-2126(99)80052-5. [DOI] [PubMed] [Google Scholar]
- Malek TR, Yu A, Vincek V, Scibelli P, Kong L. CD4 regulatory T cells prevent lethal autoimmunity in IL-2Rbeta-deficient mice. Implications for the nonredundant function of IL-2. Immunity. 2002;17:167–78. doi: 10.1016/s1074-7613(02)00367-9. [DOI] [PubMed] [Google Scholar]
- Mandelboim O, Lieberman N, Lev M, Paul L, Arnon TI, Bushkin Y, et al. Recognition of haemagglutinins on virus-infected cells by NKp46 activates lysis by human NK cells. Nature. 2001;409:1055–60. doi: 10.1038/35059110. [DOI] [PubMed] [Google Scholar]
- Marquardt N, Wilk E, Pokoyski C, Schmidt RE, Jacobs R. Murine CXCR3+CD27bright NK cells resemble the human CD56bright NK-cell population. Eur J Immunol. 2010;40:1428–39. doi: 10.1002/eji.200940056. [DOI] [PubMed] [Google Scholar]
- Mars LT, Araujo L, Kerschen P, Diem S, Bourgeois E, Van LP, et al. Invariant NKT cells inhibit development of the Th17 lineage. Proc Natl Acad Sci USA. 2009;106:6238–43. doi: 10.1073/pnas.0809317106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh SG, Parham P, Dupont B, Geraghty DE, Trowsdale J, Middleton D, et al. Killer-cell immunoglobulin-like receptor (KIR) nomenclature report, 2002. Immunogenetics. 2003;55:220–6. doi: 10.1007/s00251-003-0571-z. [DOI] [PubMed] [Google Scholar]
- Martin R. Anti-CD25 (daclizumab) monoclonal antibody therapy in relapsing-remitting multiple sclerosis. Clin Immunol. 2012;142:9–14. doi: 10.1016/j.clim.2011.10.008. [DOI] [PubMed] [Google Scholar]
- Martinez-Rodriguez JE, Lopez-Botet M, Munteis E, Rio J, Roquer J, Montalban X, et al. Natural killer cell phenotype and clinical response to interferon-beta therapy in multiple sclerosis. Clin Immunol. 2011;141:348–56. doi: 10.1016/j.clim.2011.09.006. [DOI] [PubMed] [Google Scholar]
- Martinez-Rodriguez JE, Saez-Borderias A, Munteis E, Romo N, Roquer J, Lopez-Botet M. Natural killer receptors distribution in multiple sclerosis: Relation to clinical course and interferon-beta therapy. Clin Immunol. 2010;137:41–50. doi: 10.1016/j.clim.2010.06.002. [DOI] [PubMed] [Google Scholar]
- Matsumoto Y, Kohyama K, Aikawa Y, Shin T, Kawazoe Y, Suzuki Y, et al. Role of natural killer cells and TCR gamma delta T cells in acute autoimmune encephalomyelitis. Eur J Immunol. 1998;28:1681–8. doi: 10.1002/(SICI)1521-4141(199805)28:05<1681::AID-IMMU1681>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- McGarry RC, Helfand SL, Quarles RH, Roder JC. Recognition of myelin-associated glycoprotein by the monoclonal antibody HNK-1. Nature. 1983;306:376–8. doi: 10.1038/306376a0. [DOI] [PubMed] [Google Scholar]
- McMahon EJ, Bailey SL, Castenada CV, Waldner H, Miller SD. Epitope spreading initiates in the CNS in two mouse models of multiple sclerosis. Nat Med. 2005;11:335–9. doi: 10.1038/nm1202. [DOI] [PubMed] [Google Scholar]
- Middleton D, Gonzalez A, Gilmore PM. Studies on the expression of the deleted KIR2DS4*003 gene product and distribution of KIR2DS4 deleted and nondeleted versions in different populations. Hum Immunol. 2007;68:128–34. doi: 10.1016/j.humimm.2006.12.007. [DOI] [PubMed] [Google Scholar]
- Miyamoto K, Miyake S, Yamamura T. A synthetic glycolipid prevents autoimmune encephalomyelitis by inducing TH2 bias of natural killer T cells. Nature. 2001;413:531–4. doi: 10.1038/35097097. [DOI] [PubMed] [Google Scholar]
- Moesta AK, Norman PJ, Yawata M, Yawata N, Gleimer M, Parham P. Synergistic polymorphism at two positions distal to the ligand-binding site makes KIR2DL2 a stronger receptor for HLA-C than KIR2DL3. J Immunol. 2008;180:3969–79. doi: 10.4049/jimmunol.180.6.3969. [DOI] [PubMed] [Google Scholar]
- Molinero LL, Domaica CI, Fuertes MB, Girart MV, Rossi LE, Zwirner NW. Intracellular expression of MICA in activated CD4 T lymphocytes and protection from NK cell-mediated MICA-dependent cytotoxicity. Hum Immunol. 2006;67:170–82. doi: 10.1016/j.humimm.2006.02.010. [DOI] [PubMed] [Google Scholar]
- Moretta A, Bottino C, Vitale M, Pende D, Cantoni C, Mingari MC, et al. Activating receptors and coreceptors involved in human natural killer cell-mediated cytolysis. Annu Rev Immunol. 2001;19:197–223. doi: 10.1146/annurev.immunol.19.1.197. [DOI] [PubMed] [Google Scholar]
- Moretta A, Marcenaro E, Parolini S, Ferlazzo G, Moretta L. NK cells at the interface between innate and adaptive immunity. Cell Death Differ. 2008;15:226–33. doi: 10.1038/sj.cdd.4402170. [DOI] [PubMed] [Google Scholar]
- Moretta A, Sivori S, Vitale M, Pende D, Morelli L, Augugliaro R, et al. Existence of both inhibitory (p58) and activatory (p50) receptors for HLA-C molecules in human natural killer cells. J Exp Med. 1995;182:875–84. doi: 10.1084/jem.182.3.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretta A, Vitale M, Bottino C, Orengo AM, Morelli L, Augugliaro R, et al. P58 molecules as putative receptors for major histocompatibility complex (MHC) class I molecules in human natural killer (NK) cells. Anti-p58 antibodies reconstitute lysis of MHC class I-protected cells in NK clones displaying different specificities. J Exp Med. 1993;178:597–604. doi: 10.1084/jem.178.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretta L, Ferlazzo G, Bottino C, Vitale M, Pende D, Mingari MC, et al. Effector and regulatory events during natural killer-dendritic cell interactions. Immunol Rev. 2006;214:219–28. doi: 10.1111/j.1600-065X.2006.00450.x. [DOI] [PubMed] [Google Scholar]
- Muller CA, Engler-Blum G, Gekeler V, Steiert I, Weiss E, Schmidt H. Genetic and serological heterogeneity of the supertypic HLA-B locus specificities Bw4 and Bw6. Immunogenetics. 1989;30:200–7. doi: 10.1007/BF02421207. [DOI] [PubMed] [Google Scholar]
- Munschauer FE, Hartrich LA, Stewart CC, Jacobs L. Circulating natural killer cells but not cytotoxic T lymphocytes are reduced in patients with active relapsing multiple sclerosis and little clinical disability as compared to controls. J Neuroimmunol. 1995;62:177–81. doi: 10.1016/0165-5728(95)00115-9. [DOI] [PubMed] [Google Scholar]
- Narni-Mancinelli E, Jaeger BN, Bernat C, Fenis A, Kung S, De Gassart A, et al. Tuning of natural killer cell reactivity by NKp46 and Helios calibrates T cell responses. Science. 2012;335:344–8. doi: 10.1126/science.1215621. [DOI] [PubMed] [Google Scholar]
- Neumann H, Cavalie A, Jenne DE, Wekerle H. Induction of MHC class I genes in neurons. Science. 1995;269:549–52. doi: 10.1126/science.7624779. [DOI] [PubMed] [Google Scholar]
- Neumann H, Schmidt H, Cavalie A, Jenne D, Wekerle H. Major histocompatibility complex (MHC) class I gene expression in single neurons of the central nervous system: differential regulation by interferon (IFN)-gamma and tumor necrosis factor (TNF)-alpha. J Exp Med. 1997;185:305–16. doi: 10.1084/jem.185.2.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman PJ, Abi-Rached L, Gendzekhadze K, Korbel D, Gleimer M, Rowley D, et al. Unusual selection on the KIR3DL1/S1 natural killer cell receptor in Africans. Nat Genet. 2007;39:1092–9. doi: 10.1038/ng2111. [DOI] [PubMed] [Google Scholar]
- Norman PJ, Carey BS, Stephens HA, Vaughan RW. DNA sequence variation and molecular genotyping of natural killer leukocyte immunoglobulin-like receptor, LILRA3. Immunogenetics. 2003;55:165–71. doi: 10.1007/s00251-003-0561-1. [DOI] [PubMed] [Google Scholar]
- Novak J, Lehuen A. Mechanism of regulation of autoimmunity by iNKT cells. Cytokine. 2011;53:263–70. doi: 10.1016/j.cyto.2010.11.001. [DOI] [PubMed] [Google Scholar]
- O’Connor GM, Guinan KJ, Cunningham RT, Middleton D, Parham P, Gardiner CM. Functional polymorphism of the KIR3DL1/S1 receptor on human NK cells. J Immunol. 2007;178:235–41. doi: 10.4049/jimmunol.178.1.235. [DOI] [PubMed] [Google Scholar]
- Oger J, Kastrukoff LF, Li DK, Paty DW. Multiple sclerosis: in relapsing patients, immune functions vary with disease activity as assessed by MRI. Neurology. 1988;38:1739–44. doi: 10.1212/wnl.38.11.1739. [DOI] [PubMed] [Google Scholar]
- Oh U, Blevins G, Griffith C, Richert N, Maric D, Lee CR, et al. Regulatory T cells are reduced during anti-CD25 antibody treatment of multiple sclerosis. Arch Neurol. 2009;66:471–9. doi: 10.1001/archneurol.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Leary JG, Goodarzi M, Drayton DL, von Andrian UH. T cell- and B cell-independent adaptive immunity mediated by natural killer cells. Nat Immunol. 2006;7:507–16. doi: 10.1038/ni1332. [DOI] [PubMed] [Google Scholar]
- Oppenheim DE, Roberts SJ, Clarke SL, Filler R, Lewis JM, Tigelaar RE, et al. Sustained localized expression of ligand for the activating NKG2D receptor impairs natural cytotoxicity in vivo and reduces tumor immunosurveillance. Nat Immunol. 2005;6:928–37. doi: 10.1038/ni1239. [DOI] [PubMed] [Google Scholar]
- Ordonez D, Sanchez AJ, Martinez-Rodriguez JE, Cisneros E, Ramil E, Romo N, et al. Multiple sclerosis associates with LILRA3 deletion in Spanish patients. Genes Immun. 2009;10:579–85. doi: 10.1038/gene.2009.34. [DOI] [PubMed] [Google Scholar]
- Orr MT, Lanier LL. Natural killer cell education and tolerance. Cell. 2010;142:847–56. doi: 10.1016/j.cell.2010.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal E, Tabira T, Kawano T, Taniguchi M, Miyake S, Yamamura T. Costimulation-dependent modulation of experimental autoimmune encephalomyelitis by ligand stimulation of V alpha 14 NK T cells. J Immunol. 2001;166:662–8. doi: 10.4049/jimmunol.166.1.662. [DOI] [PubMed] [Google Scholar]
- Parham P. Immunogenetics of killer-cell immunoglobulin-like receptors. Tissue Antigens. 2003;62:194–200. doi: 10.1034/j.1399-0039.2003.00126.x. [DOI] [PubMed] [Google Scholar]
- Parham P. Killer cell immunoglobulin-like receptor diversity: balancing signals in the natural killer cell response. Immunol Lett. 2004;92:11–3. doi: 10.1016/j.imlet.2003.11.016. [DOI] [PubMed] [Google Scholar]
- Parham P. MHC class I molecules and KIRs in human history, health and survival. Nat Rev Immunol. 2005;5:201–14. doi: 10.1038/nri1570. [DOI] [PubMed] [Google Scholar]
- Park YW, Kee SJ, Cho YN, Lee EH, Lee HY, Kim EM, et al. Impaired differentiation and cytotoxicity of natural killer cells in systemic lupus erythematosus. Arthritis Rheum. 2009;60:1753–63. doi: 10.1002/art.24556. [DOI] [PubMed] [Google Scholar]
- Pegram HJ, Andrews DM, Smyth MJ, Darcy PK, Kershaw MH. Activating and inhibitory receptors of natural killer cells. Immunol Cell Biol. 2011;89:216–24. doi: 10.1038/icb.2010.78. [DOI] [PubMed] [Google Scholar]
- Perini P, Wadhwa M, Buttarello M, Meager A, Facchinetti A, Thorpe R, et al. Effect of IFNbeta and anti-IFNbeta antibodies on NK cells in multiple sclerosis patients. J Neuroimmunol. 2000;105:91–5. doi: 10.1016/s0165-5728(00)00196-x. [DOI] [PubMed] [Google Scholar]
- Perricone R, Perricone C, De Carolis C, Shoenfeld Y. NK cells in autoimmunity: a two-edg’d weapon of the immune system. Autoimmun Rev. 2008;7:384–90. doi: 10.1016/j.autrev.2008.03.002. [DOI] [PubMed] [Google Scholar]
- Petrie EJ, Clements CS, Lin J, Sullivan LC, Johnson D, Huyton T, et al. CD94-NKG2A recognition of human leukocyte antigen (HLA)-E bound to an HLA class I leader sequence. J Exp Med. 2008;205:725–35. doi: 10.1084/jem.20072525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogge von Strandmann E, Simhadri VR, von Tresckow B, Sasse S, Reiners KS, Hansen HP, et al. Human leukocyte antigen-B-associated transcript 3 is released from tumor cells and engages the NKp30 receptor on natural killer cells. Immunity. 2007;27:965–74. doi: 10.1016/j.immuni.2007.10.010. [DOI] [PubMed] [Google Scholar]
- Poli A, Michel T, Theresine M, Andres E, Hentges F, Zimmer J. CD56bright natural killer (NK) cells: an important NK cell subset. Immunology. 2009;126:458–65. doi: 10.1111/j.1365-2567.2008.03027.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polman CH, O’Connor PW, Havrdova E, Hutchinson M, Kappos L, Miller DH, et al. A randomized, placebo-controlled trial of natalizumab for relapsing multiple sclerosis. N Engl J Med. 2006;354:899–910. doi: 10.1056/NEJMoa044397. [DOI] [PubMed] [Google Scholar]
- Purdy AK, Campbell KS. Natural killer cells and cancer: regulation by the killer cell Ig-like receptors (KIR) Cancer Biol Ther. 2009;8:2211–20. doi: 10.4161/cbt.8.23.10455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinovich BA, Li J, Shannon J, Hurren R, Chalupny J, Cosman D, et al. Activated, but not resting, T cells can be recognized and killed by syngeneic NK cells. J Immunol. 2003;170:3572–6. doi: 10.4049/jimmunol.170.7.3572. [DOI] [PubMed] [Google Scholar]
- Radue EW, Stuart WH, Calabresi PA, Confavreux C, Galetta SL, Rudick RA, et al. Natalizumab plus interferon beta-1a reduces lesion formation in relapsing multiple sclerosis. J Neurol Sci. 2010;292:28–35. doi: 10.1016/j.jns.2010.02.012. [DOI] [PubMed] [Google Scholar]
- Rajagopalan S, Long EO. A human histocompatibility leukocyte antigen (HLA)-G-specific receptor expressed on all natural killer cells. J Exp Med. 1999;189:1093–100. doi: 10.1084/jem.189.7.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan S, Long EO. Understanding how combinations of HLA and KIR genes influence disease. J Exp Med. 2005;201:1025–9. doi: 10.1084/jem.20050499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raulet DH. Roles of the NKG2D immunoreceptor and its ligands. Nat Rev Immunol. 2003;3:781–90. doi: 10.1038/nri1199. [DOI] [PubMed] [Google Scholar]
- Raulet DH, Vance RE, McMahon CW. Regulation of the natural killer cell receptor repertoire. Annu Rev Immunol. 2001;19:291–330. doi: 10.1146/annurev.immunol.19.1.291. [DOI] [PubMed] [Google Scholar]
- Reynders A, Yessaad N, Vu Manh TP, Dalod M, Fenis A, Aubry C, et al. Identity, regulation and in vivo function of gut NKp46+RORgammat+ and NKp46+RORgammat- lymphoid cells. EMBO J. 2011;30:2934–47. doi: 10.1038/emboj.2011.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rioux JD, Goyette P, Vyse TJ, Hammarstrom L, Fernando MM, Green T, et al. Mapping of multiple susceptibility variants within the MHC region for 7 immune-mediated diseases. Proc Natl Acad Sci USA. 2009;106:18680–5. doi: 10.1073/pnas.0909307106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodacki M, Svoren B, Butty V, Besse W, Laffel L, Benoist C, et al. Altered natural killer cells in type 1 diabetic patients. Diabetes. 2007;56:177–85. doi: 10.2337/db06-0493. [DOI] [PubMed] [Google Scholar]
- Rose JW, Burns JB, Bjorklund J, Klein J, Watt HE, Carlson NG. Daclizumab phase II trial in relapsing and remitting multiple sclerosis: MRI and clinical results. Neurology. 2007;69:785–9. doi: 10.1212/01.wnl.0000267662.41734.1f. [DOI] [PubMed] [Google Scholar]
- Rose JW, Watt HE, White AT, Carlson NG. Treatment of multiple sclerosis with an anti-interleukin-2 receptor monoclonal antibody. Ann Neurol. 2004;56:864–7. doi: 10.1002/ana.20287. [DOI] [PubMed] [Google Scholar]
- Rosental B, Brusilovsky M, Hadad U, Oz D, Appel MY, Afergan F, et al. Proliferating cell nuclear antigen is a novel inhibitory ligand for the natural cytotoxicity receptor NKp44. J Immunol. 2011;187:5693–702. doi: 10.4049/jimmunol.1102267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu M, Chen Y, Qi J, Liu J, Fan Z, Nam G, et al. LILRA3 binds both classical and non-classical HLA class I molecules but with reduced affinities compared to LILRB1/LILRB2: structural evidence. PLoS One. 2011;6:e19245. doi: 10.1371/journal.pone.0019245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saikali P, Antel JP, Newcombe J, Chen Z, Freedman M, Blain M, et al. NKG2D-mediated cytotoxicity toward oligodendrocytes suggests a mechanism for tissue injury in multiple sclerosis. J Neurosci. 2007;27:1220–8. doi: 10.1523/JNEUROSCI.4402-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuishi K, Miyake S, Yamamura T. Role of NK cells and invariant NKT cells in multiple sclerosis. Results Probl Cell Differ. 2010;51:127–47. doi: 10.1007/400_2009_11. [DOI] [PubMed] [Google Scholar]
- Sambrook JG, Bashirova A, Andersen H, Piatak M, Vernikos GS, Coggill P, et al. Identification of the ancestral killer immunoglobulin-like receptor gene in primates. BMC Genomics. 2006;7:209. doi: 10.1186/1471-2164-7-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sand KL, Knudsen E, Rolin J, Al-Falahi Y, Maghazachi AA. Modulation of natural killer cell cytotoxicity and cytokine release by the drug glatiramer acetate. Cell Mol Life Sci. 2009;66:1446–56. doi: 10.1007/s00018-009-8726-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraste M, Irjala H, Airas L. Expansion of CD56Bright natural killer cells in the peripheral blood of multiple sclerosis patients treated with interferon-beta. Neurol Sci. 2007a;28:121–6. doi: 10.1007/s10072-007-0803-3. [DOI] [PubMed] [Google Scholar]
- Saraste M, Vaisanen S, Alanen A, Airas L. Clinical and immunologic evaluation of women with multiple sclerosis during and after pregnancy. Gend Med. 2007b;4:45–55. doi: 10.1016/s1550-8579(07)80008-8. [DOI] [PubMed] [Google Scholar]
- Satoh-Takayama N, Vosshenrich CA, Lesjean-Pottier S, Sawa S, Lochner M, Rattis F, et al. Microbial flora drives interleukin 22 production in intestinal NKp46+ cells that provide innate mucosal immune defense. Immunity. 2008;29:958–70. doi: 10.1016/j.immuni.2008.11.001. [DOI] [PubMed] [Google Scholar]
- Saulquin X, Gastinel LN, Vivier E. Crystal structure of the human natural killer cell activating receptor KIR2DS2 (CD158j) J Exp Med. 2003;197:933–8. doi: 10.1084/jem.20021624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawcer S, Hellenthal G, Pirinen M, Spencer CCA, Patsopoulos NA, Moutsianas L, et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature. 2011;476:214–9. doi: 10.1038/nature10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleinitz N, March ME, Long EO. Recruitment of activation receptors at inhibitory NK cell immune synapses. PLoS One. 2008;3 doi: 10.1371/journal.pone.0003278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenker N, Flanagan JM. Intragenic DNA methylation: implications of this epigenetic mechanism for cancer research. Br J Cancer. 2012;106:248–53. doi: 10.1038/bjc.2011.550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi FD, Ljunggren HG, La Cava A, van Kaer L. Organ-specific features of natural killer cells. Nat Rev Immunol. 2011;11:658–71. doi: 10.1038/nri3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi FD, Takeda K, Akira S, Sarvetnick N, Ljunggren HG. IL-18 directs autoreactive T cells and promotes autodestruction in the central nervous system via induction of IFN-gamma by NK cells. J Immunol. 2000;165:3099–104. doi: 10.4049/jimmunol.165.6.3099. [DOI] [PubMed] [Google Scholar]
- Shiroishi M, Kajikawa M, Kuroki K, Ose T, Kohda D, Maenaka K. Crystal structure of the human monocyte-activating receptor, “Group 2” leukocyte Ig-like receptor A5 (LILRA5/LIR9/ILT11) J Biol Chem. 2006a;281:19536–44. doi: 10.1074/jbc.M603076200. [DOI] [PubMed] [Google Scholar]
- Shiroishi M, Kuroki K, Rasubala L, Tsumoto K, Kumagai I, Kurimoto E, et al. Structural basis for recognition of the nonclassical MHC molecule HLA-G by the leukocyte Ig-like receptor B2 (LILRB2/LIR2/ILT4/CD85d) Proc Natl Acad Sci USA. 2006b;103:16412–7. doi: 10.1073/pnas.0605228103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiroishi M, Tsumoto K, Amano K, Shirakihara Y, Colonna M, Braud VM, et al. Human inhibitory receptors Ig-like transcript 2 (ILT2) and ILT4 compete with CD8 for MHC class I binding and bind preferentially to HLA-G. Proc Natl Acad Sci USA. 2003;100:8856–61. doi: 10.1073/pnas.1431057100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh AK, Wilson MT, Hong S, Olivares-Villagomez D, Du C, Stanic AK, et al. Natural killer T cell activation protects mice against experimental autoimmune encephalomyelitis. J Exp Med. 2001;194:1801–11. doi: 10.1084/jem.194.12.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Single RM, Martin MP, Gao X, Meyer D, Yeager M, Kidd JR, et al. Global diversity and evidence for coevolution of KIR and HLA. Nat Genet. 2007;39:1114–9. doi: 10.1038/ng2077. [DOI] [PubMed] [Google Scholar]
- Sivori S, Falco M, Carlomagno S, Romeo E, Soldani C, Bensussan A, et al. A novel KIR-associated function: evidence that CpG DNA uptake and shuttling to early endosomes is mediated by KIR3DL2. Blood. 2010;116:1637–47. doi: 10.1182/blood-2009-12-256586. [DOI] [PubMed] [Google Scholar]
- Sivori S, Pende D, Bottino C, Marcenaro E, Pessino A, Biassoni R, et al. NKp46 is the major triggering receptor involved in the natural cytotoxicity of fresh or cultured human NK cells. Correlation between surface density of NKp46 and natural cytotoxicity against autologous, allogeneic or xenogeneic target cells. Eur J Immunol. 1999;29:1656–66. doi: 10.1002/(SICI)1521-4141(199905)29:05<1656::AID-IMMU1656>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Smeltz RB, Wolf NA, Swanborg RH. Inhibition of autoimmune T cell responses in the DA rat by bone marrow-derived NK cells in vitro: implications for autoimmunity. J Immunol. 1999;163:1390–7. [PubMed] [Google Scholar]
- Smyth MJ, Godfrey DI, Trapani JA. A fresh look at tumor immunosurveillance and immunotherapy. Nat Immunol. 2001;2:293–9. doi: 10.1038/86297. [DOI] [PubMed] [Google Scholar]
- Snyder GA, Brooks AG, Sun PD. Crystal structure of the HLA-Cw3 allotype-specific killer cell inhibitory receptor KIR2DL2. Proc Natl Acad Sci USA. 1999;96:3864–9. doi: 10.1073/pnas.96.7.3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sospedra M, Martin R. Immunology of multiple sclerosis. Annu Rev Immunol. 2005;23:683–747. doi: 10.1146/annurev.immunol.23.021704.115707. [DOI] [PubMed] [Google Scholar]
- Steinman L. Assessment of animal models for MS and demyelinating disease in the design of rational therapy. Neuron. 1999;24:511–4. doi: 10.1016/s0896-6273(00)81107-1. [DOI] [PubMed] [Google Scholar]
- Stern M, Ruggeri L, Capanni M, Mancusi A, Velardi A. Human leukocyte antigens A23, A24, and A32 but not A25 are ligands for KIR3DL1. Blood. 2008;112:708–10. doi: 10.1182/blood-2008-02-137521. [DOI] [PubMed] [Google Scholar]
- Stewart CA, Laugier-Anfossi F, Vely F, Saulquin X, Riedmuller J, Tisserant A, et al. Recognition of peptide-MHC class I complexes by activating killer immunoglobulin-like receptors. Proc Natl Acad Sci USA. 2005;102:13224–9. doi: 10.1073/pnas.0503594102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart CA, Walzer T, Robbins SH, Malissen B, Vivier E, Prinz I. Germ-line and rearranged Tcrd transcription distinguish bona fide NK cells and NK-like gammadelta T cells. Eur J Immunol. 2007;37:1442–52. doi: 10.1002/eji.200737354. [DOI] [PubMed] [Google Scholar]
- Sullivan LC, Clements CS, Beddoe T, Johnson D, Hoare HL, Lin J, et al. The heterodimeric assembly of the CD94-NKG2 receptor family and implications for human leukocyte antigen-E recognition. Immunity. 2007;27:900–11. doi: 10.1016/j.immuni.2007.10.013. [DOI] [PubMed] [Google Scholar]
- Sun JC, Beilke JN, Lanier LL. Adaptive immune features of natural killer cells. Nature. 2009;457:557–61. doi: 10.1038/nature07665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun JC, Beilke JN, Lanier LL. Immune memory redefined: characterizing the longevity of natural killer cells. Immunol Rev. 2010;236:83–94. doi: 10.1111/j.1600-065X.2010.00900.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun JC, Lanier LL. Tolerance of NK cells encountering their viral ligand during development. J Exp Med. 2008;205:1819–28. doi: 10.1084/jem.20072448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun JC, Lanier LL. NK cell development, homeostasis and function: parallels with CD8 T cells. Nat Rev Immunol. 2011;11:645–57. doi: 10.1038/nri3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Aranami T, Endoh M, Miyake S, Yamamura T. The regulatory role of natural killer cells in multiple sclerosis. Brain. 2004;127 (Pt 9):1917–27. doi: 10.1093/brain/awh219. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Miyake S, Kondo T, Terao K, Hatakenaka M, Hashimoto S, et al. Natural killer type 2 bias in remission of multiple sclerosis. J Clin Invest. 2001;107:R23–9. doi: 10.1172/JCI11819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thananchai H, Gillespie G, Martin MP, Bashirova A, Yawata N, Yawata M, et al. Cutting Edge: Allele-specific and peptide-dependent interactions between KIR3DL1 and HLA-A and HLA-B. J Immunol. 2007;178:33–7. doi: 10.4049/jimmunol.178.1.33. [DOI] [PubMed] [Google Scholar]
- Thomas R, Matthias T, Witte T. Leukocyte immunoglobulin-like receptors as new players in autoimmunity. Clin Rev Allergy Immunol. 2010;38:159–62. doi: 10.1007/s12016-009-8148-8. [DOI] [PubMed] [Google Scholar]
- Torkar M, Haude A, Milne S, Beck S, Trowsdale J, Wilson MJ. Arrangement of the ILT gene cluster: a common null allele of the ILT6 gene results from a 6.7-kbp deletion. Eur J Immunol. 2000;30:3655–62. doi: 10.1002/1521-4141(200012)30:12<3655::AID-IMMU3655>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Townsend MJ, Monroe JG, Chan AC. B-cell targeted therapies in human autoimmune diseases: an updated perspective. Immunol Rev. 2010;237:264–83. doi: 10.1111/j.1600-065X.2010.00945.x. [DOI] [PubMed] [Google Scholar]
- Traherne JA, Martin M, Ward R, Ohashi M, Pellett F, Gladman D, et al. Mechanisms of copy number variation and hybrid gene formation in the KIR immune gene complex. Hum Mol Genet. 2010;19:737–51. doi: 10.1093/hmg/ddp538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traugott U, Raine CS. Further lymphocyte characterization in the central nervous system in multiple sclerosis. Ann NY Acad Sci. 1984;436:163–80. doi: 10.1111/j.1749-6632.1984.tb14788.x. [DOI] [PubMed] [Google Scholar]
- Tripathy SK, Keyel PA, Yang L, Pingel JT, Cheng TP, Schneeberger A, et al. Continuous engagement of a self-specific activation receptor induces NK cell tolerance. J Exp Med. 2008;205:1829–41. doi: 10.1084/jem.20072446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trowsdale J. Genetic and functional relationships between MHC and NK receptor genes. Immunity. 2001;15:363–74. doi: 10.1016/s1074-7613(01)00197-2. [DOI] [PubMed] [Google Scholar]
- Uchida A, Maida EM, Lenzhofer R, Micksche M. Natural killer cell activity in patients with multiple sclerosis: interferon and plasmapheresis. Immunobiology. 1982;160:392–402. doi: 10.1016/S0171-2985(82)80003-X. [DOI] [PubMed] [Google Scholar]
- Uhrberg M. The KIR gene family: life in the fast lane of evolution. Eur J Immunol. 2005;35:10–5. doi: 10.1002/eji.200425743. [DOI] [PubMed] [Google Scholar]
- Uhrberg M, Parham P, Wernet P. Definition of gene content for nine common group B haplotypes of the Caucasoid population: KIR haplotypes contain between seven and eleven KIR genes. Immunogenetics. 2002;54:221–9. doi: 10.1007/s00251-002-0463-7. [DOI] [PubMed] [Google Scholar]
- Uhrberg M, Valiante NM, Shum BP, Shilling HG, Lienert-Weidenbach K, Corliss B, et al. Human diversity in killer cell inhibitory receptor genes. Immunity. 1997;7:753–63. doi: 10.1016/s1074-7613(00)80394-5. [DOI] [PubMed] [Google Scholar]
- Vales-Gomez M, Reyburn HT, Erskine RA, Lopez-Botet M, Strominger JL. Kinetics and peptide dependency of the binding of the inhibitory NK receptor CD94/NKG2-A and the activating receptor CD94/NKG2-C to HLA-E. EMBO J. 1999;18:4250–60. doi: 10.1093/emboj/18.15.4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vliet HJ, von Blomberg BM, Nishi N, Reijm M, Voskuyl AE, van Bodegraven AA, et al. Circulating V(alpha24+) Vbeta11+ NKT cell numbers are decreased in a wide variety of diseases that are characterized by autoreactive tissue damage. Clin Immunol. 2001;100:144–8. doi: 10.1006/clim.2001.5060. [DOI] [PubMed] [Google Scholar]
- van Kaer L. NKT cells: T lymphocytes with innate effector functions. Curr Opin Immunol. 2007;19:354–64. doi: 10.1016/j.coi.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Vance RE, Jamieson AM, Raulet DH. Recognition of the class Ib molecule Qa-1(b) by putative activating receptors CD94/NKG2C and CD94/NKG2E on mouse natural killer cells. J Exp Med. 1999;190:1801–12. doi: 10.1084/jem.190.12.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance RE, Kraft JR, Altman JD, Jensen PE, Raulet DH. Mouse CD94/NKG2A is a natural killer cell receptor for the nonclassical major histocompatibility complex (MHC) class I molecule Qa-1(b) J Exp Med. 1998;188:1841–8. doi: 10.1084/jem.188.10.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbark AA, Huan J, Agotsch M, La Tocha D, Goelz S, Offner H, et al. Interferon-beta-1a treatment increases CD56bright natural killer cells and CD4+CD25+ Foxp3 expression in subjects with multiple sclerosis. J Neuroimmunol. 2009;215:125–8. doi: 10.1016/j.jneuroim.2009.08.007. [DOI] [PubMed] [Google Scholar]
- Vilches C, Pando MJ, Parham P. Genes encoding human killer-cell Ig-like receptors with D1 and D2 extracellular domains all contain untranslated pseudoexons encoding a third Ig-like domain. Immunogenetics. 2000a;51:639–46. doi: 10.1007/s002510000184. [DOI] [PubMed] [Google Scholar]
- Vilches C, Rajalingam R, Uhrberg M, Gardiner CM, Young NT, Parham P. KIR2DL5, a novel killer-cell receptor with a D0-D2 configuration of Ig-like domains. J Immunol. 2000b;164:5797–804. doi: 10.4049/jimmunol.164.11.5797. [DOI] [PubMed] [Google Scholar]
- Vilches C, Parham P. KIR: diverse, rapidly evolving receptors of innate and adaptive immunity. Annu Rev Immunol. 2002;20:217–51. doi: 10.1146/annurev.immunol.20.092501.134942. [DOI] [PubMed] [Google Scholar]
- Vitale M, Castriconi R, Parolini S, Pende D, Hsu ML, Moretta L, et al. The leukocyte Ig-like receptor (LIR)-1 for the cytomegalovirus UL18 protein displays a broad specificity for different HLA class I alleles: analysis of LIR-1 + NK cell clones. Int Immunol. 1999;11:29–35. doi: 10.1093/intimm/11.1.29. [DOI] [PubMed] [Google Scholar]
- Vitale M, Zimmer J, Castriconi R, Hanau D, Donato L, Bottino C, et al. Analysis of natural killer cells in TAP2-deficient patients: expression of functional triggering receptors and evidence for the existence of inhibitory receptor(s) that prevent lysis of normal autologous cells. Blood. 2002;99:1723–9. doi: 10.1182/blood.v99.5.1723. [DOI] [PubMed] [Google Scholar]
- Vivian JP, Duncan RC, Berry R, O’Connor GM, Reid HH, Beddoe T, et al. Killer cell immunoglobulin-like receptor 3DL1-mediated recognition of human leukocyte antigen B. Nature. 2011;479:401–5. doi: 10.1038/nature10517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivier E, Anfossi N. Inhibitory NK-cell receptors on T cells: witness of the past, actors of the future. Nat Rev Immunol. 2004;4:190–8. doi: 10.1038/nri1306. [DOI] [PubMed] [Google Scholar]
- Vivier E, Tomasello E, Baratin M, Walzer T, Ugolini S. Functions of natural killer cells. Nat Immunol. 2008;9:503–10. doi: 10.1038/ni1582. [DOI] [PubMed] [Google Scholar]
- Vollmer TL, Liu R, Price M, Rhodes S, La Cava A, Shi FD. Differential effects of IL-21 during initiation and progression of autoimmunity against neuroantigen. J Immunol. 2005;174:2696–701. doi: 10.4049/jimmunol.174.5.2696. [DOI] [PubMed] [Google Scholar]
- Volz A, Wende H, Laun K, Ziegler A. Genesis of the ILT/LIR/MIR clusters within the human leukocyte receptor complex. Immunol Rev. 2001;181:39–51. doi: 10.1034/j.1600-065x.2001.1810103.x. [DOI] [PubMed] [Google Scholar]
- Vosshenrich CA, Garcia-Ojeda ME, Samson-Villeger SI, Pasqualetto V, Enault L, Richard-Le Goff O, et al. A thymic pathway of mouse natural killer cell development characterized by expression of GATA-3 and CD127. Nat Immunol. 2006;7:1217–24. doi: 10.1038/ni1395. [DOI] [PubMed] [Google Scholar]
- Vranes Z, Poljakovic Z, Marusic M. Natural killer cell number and activity in multiple sclerosis. J Neurol Sci. 1989;94:115–23. doi: 10.1016/0022-510x(89)90222-0. [DOI] [PubMed] [Google Scholar]
- Wagtmann N, Rajagopalan S, Winter CC, Peruzzi M, Long EO. Killer cell inhibitory receptors specific for HLA-C and HLA-B identified by direct binding and by functional transfer. Immunity. 1995;3:801–9. doi: 10.1016/1074-7613(95)90069-1. [DOI] [PubMed] [Google Scholar]
- Walzer T, Blery M, Chaix J, Fuseri N, Chasson L, Robbins SH, et al. Identification, activation, and selective in vivo ablation of mouse NK cells via NKp46. Proc Natl Acad Sci USA. 2007;104:3384–9. doi: 10.1073/pnas.0609692104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber MS, Hemmer B. Cooperation of B cells and T cells in the pathogenesis of multiple sclerosis. Results Probl Cell Differ. 2010;51:115–26. doi: 10.1007/400_2009_21. [DOI] [PubMed] [Google Scholar]
- Wiendl H, Feger U, Mittelbronn M, Jack C, Schreiner B, Stadelmann C, et al. Expression of the immune-tolerogenic major histocompatibility molecule HLA-G in multiple sclerosis: implications for CNS immunity. Brain. 2005;128(Pt 11):2689–704. doi: 10.1093/brain/awh609. [DOI] [PubMed] [Google Scholar]
- Willcox BE, Thomas LM, Bjorkman PJ. Crystal structure of HLA-A2 bound to LIR-1, a host and viral major histocompatibility complex receptor. Nat Immunol. 2003;4:913–9. doi: 10.1038/ni961. [DOI] [PubMed] [Google Scholar]
- Willcox BE, Thomas LM, Chapman TL, Heikema AP, West AP, Jr, Bjorkman PJ. Crystal structure of LIR-2 (ILT4) at 1.8 A: differences from LIR-1 (ILT2) in regions implicated in the binding of the Human Cytomegalovirus class I MHC homolog UL18. BMC Struct Biol. 2002;2:6. doi: 10.1186/1472-6807-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams AP, Bateman AR, Khakoo SI. Hanging in the balance. KIR and their role in disease. Mol Interv. 2005;5:226–40. doi: 10.1124/mi.5.4.6. [DOI] [PubMed] [Google Scholar]
- Wilson MJ, Torkar M, Haude A, Milne S, Jones T, Sheer D, et al. Plasticity in the organization and sequences of human KIR/ILT gene families. Proc Natl Acad Sci USA. 2000;97:4778–83. doi: 10.1073/pnas.080588597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler-Pickett R, Young HA, Cherry JM, Diehl J, Wine J, Back T, et al. In vivo regulation of experimental autoimmune encephalomyelitis by NK cells: alteration of primary adaptive responses. J Immunol. 2008;180:4495–506. doi: 10.4049/jimmunol.180.7.4495. [DOI] [PubMed] [Google Scholar]
- Winter CC, Gumperz JE, Parham P, Long EO, Wagtmann N. Direct binding and functional transfer of NK cell inhibitory receptors reveal novel patterns of HLA-C allotype recognition. J Immunol. 1998;161:571–7. [PubMed] [Google Scholar]
- Wolan DW, Teyton L, Rudolph MG, Villmow B, Bauer S, Busch DH, et al. Crystal structure of the murine NK cell-activating receptor NKG2D at 1.95 A. Nat Immunol. 2001;2:248–54. doi: 10.1038/85311. [DOI] [PubMed] [Google Scholar]
- Wu GF, Laufer TM. The role of dendritic cells in multiple sclerosis. Curr Neurol Neurosci Rep. 2007;7:245–52. doi: 10.1007/s11910-007-0037-z. [DOI] [PubMed] [Google Scholar]
- Wynn D, Kaufman M, Montalban X, Vollmer T, Simon J, Elkins J, et al. Daclizumab in active relapsing multiple sclerosis (CHOICE study): a phase 2, randomised, double-blind, placebo-controlled, add-on trial with interferon beta. Lancet Neurol. 2010;9:381–90. doi: 10.1016/S1474-4422(10)70033-8. [DOI] [PubMed] [Google Scholar]
- Xu W, Fazekas G, Hara H, Tabira T. Mechanism of natural killer (NK) cell regulatory role in experimental autoimmune encephalomyelitis. J Neuroimmunol. 2005;163:24–30. doi: 10.1016/j.jneuroim.2005.02.011. [DOI] [PubMed] [Google Scholar]
- Yeo TW, De Jager PL, Gregory SG, Barcellos LF, Walton A, Goris A, et al. A second major histocompatibility complex susceptibility locus for multiple sclerosis. Ann Neurol. 2007;61:228–36. doi: 10.1002/ana.21063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama WM, Kim S. Licensing of natural killer cells by self-major histocompatibility complex class I. Immunol Rev. 2006;214:143–54. doi: 10.1111/j.1600-065X.2006.00458.x. [DOI] [PubMed] [Google Scholar]
- Young NT, Uhrberg M. KIR expression shapes cytotoxic repertoires: a developmental program of survival. Trends Immunol. 2002;23:71–5. doi: 10.1016/s1471-4906(01)02113-5. [DOI] [PubMed] [Google Scholar]
- Young NT, Waller EC, Patel R, Roghanian A, Austyn JM, Trowsdale J. The inhibitory receptor LILRB1 modulates the differentiation and regulatory potential of human dendritic cells. Blood. 2008;111:3090–6. doi: 10.1182/blood-2007-05-089771. [DOI] [PubMed] [Google Scholar]
- Yu YY, Kumar V, Bennett M. Murine natural killer cells and marrow graft rejection. Annu Rev Immunol. 1992;10:189–213. doi: 10.1146/annurev.iy.10.040192.001201. [DOI] [PubMed] [Google Scholar]
- Zafirova B, Wensveen FM, Gulin M, Polic B. Regulation of immune cell function and differentiation by the NKG2D receptor. Cell Mol Life Sci. 2011;68:3519–29. doi: 10.1007/s00018-011-0797-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Yamamura T, Kondo T, Fujiwara M, Tabira T. Regulation of experimental autoimmune encephalomyelitis by natural killer (NK) cells. J Exp Med. 1997;186:1677–87. doi: 10.1084/jem.186.10.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer J, Donato L, Hanau D, Cazenave JP, Tongio MM, Moretta A, et al. Activity and phenotype of natural killer cells in peptide transporter (TAP)-deficient patients (Type I bare lymphocyte syndrome) J Exp Med. 1998;187:117–22. doi: 10.1084/jem.187.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]